Abstract
The nucleotide sequence of the Thermus sp. strain T2 DNA coding for a thermostable α-galactosidase was determined. The deduced amino acid sequence of the enzyme predicts a polypeptide of 474 amino acids (Mr, 53,514). The observed homology between the deduced amino acid sequences of the enzyme and α-galactosidase from Thermus brockianus was over 70%. Thermus sp. strain T2 α-galactosidase was expressed in its active form in Escherichia coli and purified. Native polyacrylamide gel electrophoresis and gel filtration chromatography data suggest that the enzyme is octameric. The enzyme was most active at 75°C for p-nitrophenyl-α-d-galactopyranoside hydrolysis, and it retained 50% of its initial activity after 1 h of incubation at 70°C. The enzyme was extremely stable over a broad range of pH (pH 6 to 13) after treatment at 40°C for 1 h. The enzyme acted on the terminal α-galactosyl residue, not on the side chain residue, of the galactomanno-oligosaccharides as well as those of yeasts and Mortierella vinacea α-galactosidase I. The enzyme has only one Cys residue in the molecule. para-Chloromercuribenzoic acid completely inhibited the enzyme but did not affect the mutant enzyme which contained Ala instead of Cys, indicating that this Cys residue is not responsible for its catalytic function.
α-Galactosidases (α-Gals) are known to occur widely in microorganisms, plants, and animals, and some of them have been purified and characterized (5). α-Gals catalyze the hydrolysis of 1,6-linked α-galactose residues from oligosaccharides and polymeric galactomannans (19, 27, 28). In the sugar beet industry, α-Gals have been used to increase the sucrose yield by eliminating raffinose, which prevents the crystallization of beet sugar (31). Raffinose and stachyose in beans are known to cause flatulence. α-Gal has the potential to alleviate these symptoms, for instance, in the treatment of soybean milk (6).
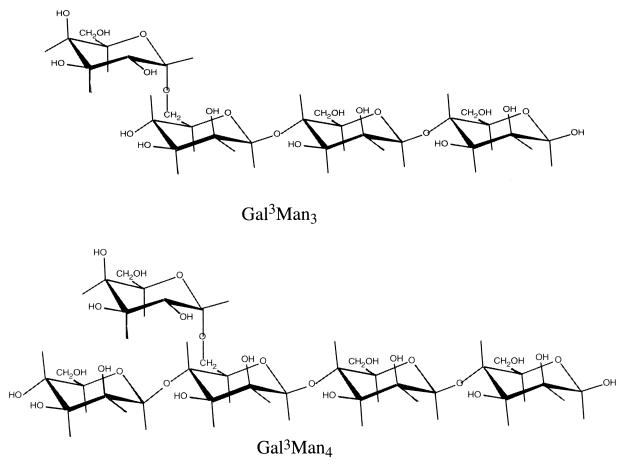
We have studied the substrate specificity of α-Gals from eukaryotes by using galactomanno-oligosaccharides, such as 63-mono-α-d-galactopyranosyl-β-1,4-mannotriose (Gal3Man3) and 63-mono-α-d-galactopyranosyl-β-1,4-mannotetraose (Gal3Man4). The structures of these galactomanno-oligosaccharides are shown in Fig. 1. Mortierella vinacea α-Gal I (11) and yeast α-Gals (32) are specific for Gal3Man3, having an α-galactosyl residue (designated the terminal α-galactosyl residue) attached to the O-to-6 position of the nonreducing end mannose of β-1,4-mannotriose. On the other hand, Aspergillus niger 5-16 α-Gal (12) and Penicillium purpurogenum α-Gal (27) show a preference for Gal3Man4, having an α-galactosyl residue (designated as the side chain α-galactosyl residue) attached to the O-to-6 position of the third mannose from the reducing end of β-1,4-mannotetraose. The M. vinacea α-Gal II (28) acts on both substrates to almost equal extents. These facts indicate that eukaryotic α-Gals were classified into three groups based on the substrate specificity of these galactomanno-oligosaccharides.
FIG. 1.
Structures of galactomanno-oligosaccharides.
Genes encoding α-Gals have been cloned from various sources, including humans (3), plants (20, 33), yeasts (12), filamentous fungi (4, 19, 26, 28), and bacteria (1, 2, 13, 17, 18). α-Gals from eukaryotes show a significant degree of similarity and are grouped into family 27. On the other hand, bacterial α-Gals have been placed in family 36 (10), even though these enzymes display a low-level amino acid sequence similarity among them.
The genes encoding the thermostable α- and β-galactosidases from a thermophilic bacterium, Thermus sp. strain T2, have been cloned in Escherichia coli (14). The α-Gal gene was located just downstream from the β-galactosidase gene, and the α-Gal gene was expressed in E. coli by using the expression vector pQE30. Here we describe the sequencing of the α-Gal gene of the Thermus sp. strain T2, its expression in E. coli, and the purification and characterization of the recombinant enzyme.
MATERIALS AND METHODS
Materials.
Melibiose, raffinose, stachyose, p-nitrophenyl-α-d-galactopyranoside (pNP-α-Gal), other p-nitrophenyl glycosides, and other chemicals were purchased from the Sigma Chemical Co. Restriction endonucleases and other enzymes were purchased from the Takara Shuzo Co. and used in accordance with the manufacturer's instructions.
Bacterial strains, plasmids, growth conditions, and sequencing procedures.
E. coli JM109 and M15 (30), pQE30, and plasmid pOS105 carrying the Thermus sp. strain T2 α-Gal gene were used for cloning and gene expression. E. coli cells were cultured in Luria-Bertani (LB) broth at 30°C with ampicillin (100 μg/ml). Recombinant DNA techniques were performed by conventional protocols (21). DNA sequencing was performed by the dideoxy chain termination method (22) with a dRhodamine Terminator Cycle Sequencing Reaction kit and ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif.).
Thermus sp. strain T2 chromosomal DNA was prepared as described previously (14), and the sequence of the 16S rRNA gene was determined by the method described by Saul et al. (23).
Construction of expression system.
PCR amplification of the gene was performed with 2.5 U of Taq DNA polymerase (Takara Shuzo Co.), 10 ng of plasmid pOS105, a 0.2 μM concentration of each synthetic primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 2 mM MgCl2 in the buffer recommended by the manufacturer. Amplification was achieved with 30 cycles of 0.5 min of denaturation at 95°C, 0.5 min of annealing at 50°C, and 2.5 min of polymerase extension at 72°C, plus an additional extension at 72°C for 7 min using a Perkin-Elmer thermal cycler (GeneAmp PCR System 2400). The synthetic oligonucleotide primers used for the PCR amplification were P1 (5′-GGGGGATCCATGAGGCTTGTACTGG-3′) and P2 (5′-GGGAAGCTTATGGAAAGGGGGCATA-3′) (the BamHI and HindIII restriction sites are underlined). The obtained PCR product cloned in pCRII was digested with BamHI and HindIII and was ligated with pQE30 between the BamHI and HindIII sites. The plasmid was transferred into competent E. coli M15 cells.
Site-directed mutagenesis.
Site-directed mutagenesis replacing Cys159 by Ala was performed by the improved megaprimer PCR mutagenesis strategy that was originally described by Seraphin and Kandels-Lewis (25).
Enzymatic assay and measurement of protein concentration.
α-Gal standard assays were performed with pNP-α-Gal at 70°C in 50 mM sodium phosphate, pH 6.0. After 10 min, an equal volume of 0.2 M Na2CO3 was added to stop the reaction and absorbance at 408 nm was measured. pNP-α-Gal was dissolved in 0.1 M sodium phosphate buffer (pH 6.0) and used at a final concentration of 10 mM. One unit of purified α-Gal activity was defined as 1 μmol of p-nitrophenol released per min under the conditions described above.
The determination of the enzyme activity versus pH or temperature profiles was done in the buffers (pH 1.5 to 3, 50 mM Gly-HCl; pH 3 to 5, 100 mM sodium acetate; pH 5 to 7, 100 mM sodium phosphate; pH 7 to 9, 100 mM Tris-HCl; pH 9 to 13, 50 mM Gly-NaOH) at 70°C and in sodium phosphate buffer (pH 6.0) from 40 to 90°C, using pNP-α-Gal as the substrate. The heat stability was investigated by preincubating the purified α-Gal at a concentration of 0.05 mg/ml in 10 mM sodium phosphate, pH 7.0, at various temperatures. After various periods of time, aliquots were withdrawn and the residual activity was measured under the standard assay conditions. The influence of the pH on enzyme stability was studied by incubating the enzyme at 0.05 mg/ml for 1 h at 40°C in the buffers adjusted to various pH values between 1.5 and 13.
The protein contents of the enzyme preparations were measured with a Bio-Rad DC Protein Assay Kit with bovine serum albumin as the standard.
Enzyme purification.
E. coli M15 cells were grown in 500 ml of LB broth supplemented with ampicillin (100 μg/ml) at 37°C for 16 h with shaking. After the A600 reached 0.6, 2.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture and the culture continued to grow at 37°C for 5 h. The cells were harvested; suspended in 5 ml of 50 mM sodium phosphate, pH 7.8, containing 300 mM NaCl; and sonicated on ice. The majority of the heat-labile proteins were precipitated by the heat treatment at 70°C for 10 min and removed by centrifugation. The supernatant was applied to the Chelating Sepharose FF column (0.6 by 4.5 cm; Amersham, Pharmacia Biotech, Little Chalfont, Buckinghamshire, England), which was equilibrated with 10 ml of 20 mM sodium phosphate, pH 7.4, containing 10 mM imidazole and 0.5 M NaCl. After a washing with the buffer containing 150 mM imidazole, the enzyme was eluted with 4 ml of the buffer containing 300 mM imidazole. The enzyme solution was desalted by dialysis against 20 mM sodium phosphate, pH 7.0, and stored at 4°C.
Electrophoretic analysis.
Sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously by Laemmli (16). Native PAGE was carried out using an acrylamide gradient gel (4 to 15% [wt/vol]) that was electrophoresed in 192 mM glycine buffer, pH 8.4. After electrophoresis, the protein band was stained with CBB R-250.
Amino acid sequencing of recombinant α-Gal.
After the protein in the SDS-polyacrylamide gel was blotted on a polyvinylidene difluoride membrane, the membrane was stained with CBB R-250 to detect the protein. The protein band was cut out and put on a protein sequencer (G1005A; Hewlett-Packard Co.).
Preparation of galactomanno-oligosaccharides.
The galactomanno-oligosaccharide having an α-1,6-galactosyl side chain on β-1,4-mannotetraose, Gal3Man4, was prepared from a hydrolyzate of copra galactomannan using Streptomyces β-mannanase (11). In addition, galactomanno-oligosaccharide with a terminal galactose at the nonreducing end of β-1,4-mannotriose, Gal3Man3, was prepared from Gal3Man4 by cutting off the nonreducing end mannosyl residue of the saccharide with A. niger β-mannosidase (15). The structures of Gal3Man3 and Gal3Man4 are shown in Fig. 1.
Substrate specificity.
Hydrolysis of the galacto-oligosaccharides (such as melibiose, raffinose, and stachyose) and of the galactomanno-oligosaccharides (such as Gal3Man3 and Gal3Man4) by the purified α-Gal was done at pH 6.0 (0.1 M sodium phosphate buffer) and 70°C. The sugar sample obtained after the enzyme reaction was analyzed by thin-layer chromatography (TLC) (Silica gel 60; Merck) for the characterization of the hydrolysis products. The reaction products were developed with 1-propanol-nitromethane-water (5:2:3, vol/vol). The sugars on the plate were detected by heating at 140°C for 5 min after spraying with sulfuric acid.
Determination of kinetic properties.
The Km and Vmax values were graphically determined from the Lineweaver-Burk plots of the initial rate of the hydrolyzing reactions. The enzyme reactions were performed in 0.1 M sodium phosphate buffer, pH 6.0, at 70°C for pNP-α-Gal. pNP-α-Gal was used in the range of 0.1 to 10 mM.
Inhibition study.
The purified enzyme was incubated with 1 mM chemicals, including pCMB and HgCl2, at 30°C for 30 min. The remaining activity was then determined as described above.
Nucleotide sequence accession number.
The α-Gal DNA sequence and the 16S rRNA gene sequence are available in the DDBJ, EMBL, and GenBank databases under accession no. AB018548 and AB054646, respectively.
RESULTS AND DISCUSSION
Sequencing analysis of 16S rRNA gene of Thermus sp. strain T2.
The sequence of the 16S rRNA gene of Thermus sp. strain T2 was determined. Compared with other Thermus species, the sequence of Thermus sp. strain T2 exhibited the highest similarity (99.9%) to the sequence of Thermus oshimai (EMBL database accession no. Y18416). There are only two nucleotide differences between the 16S rRNA genes of Thermus sp. strain T2 and T. oshimai, suggesting that these strains are closely related to each other.
Sequencing analysis of the DNA encoding Thermus sp. strain T2 α-Gal.
The plasmid pOS105, containing one open reading frame (ORF) of 1,425 bp, was sequenced. The gene encodes a polypeptide of 474 amino acids with a calculated molecular mass of 53,514 Da. The deduced amino acid sequence of the α-Gal gene was compared with α-Gal sequences available from DDBJ. The sequence identities of Thermus T2 α-Gal with the enzymes from Thermus brockianus (8), Thermotoga neapolitana (7), and Thermotoga maritima (17) were 74.7, 25.7, and 25.7%, respectively (Fig. 2). The G+C content of the ORF is 62.3%, and no similarity with the eukaryotic α-Gals of family 27 was observed. There was only one cysteine residue in the molecule of Thermus sp. strain T2 α-Gal. This cysteine residue (residue 189 in Fig. 2) is conserved among all the enzymes.
FIG. 2.
Sequence homology of α-Gals from different sources. The amino acid sequences of Thermus sp. strain T2, T. brockianus (GenBank/EMBL accession no. AF135398), Thermotoga neapolitana (accession no. AF011400), and Thermotoga maritima (AJ001776) were aligned for optimal sequence similarity using the program CLUSTAL W, which is available in the DDBJ site (www.ddbj.nig.ac.jp). Hyphens indicate gaps. Identical amino acid residues, three of four or more at the same position, are shaded.
Purification of recombinant α-Gal and its molecular mass.
Purification was carried out using a Chelating Sepharose FF column because it has a histidine tag (His tag) at its N-terminal end. More than a 2,000-fold purification was obtained, with 53% recovery of the activity from the crude enzyme solution (Table 1). SDS-PAGE of the fraction corresponding to the peak of activity revealed a single protein band with a molecular mass of 55 kDa, which agrees with the sum of the molecular mass of α-Gal (53.5 kDa) calculated from the nucleotide sequence and the additional His tag sequence (1.4 kDa).
TABLE 1.
Purification of Thermus sp. strain T2 α-Gal
| Substance used | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude enzyme | 9.28 | 84.0 | 0.11 | 100 | 1 |
| Chelating Sepharose FF | 4.94 | 0.02 | 247 | 53.2 | 2,245 |
Bacterial α-Gals can be classified into two groups depending on their molecular sizes. α-Gals from Streptococcus mutans (1), Bacillus stearothermophilus (9), Pediococcus pentosaceus (9), and E. coli Raf A (2) belong to the first group, which had molecular sizes of more than 80 kDa, while α-Gals from Thermus sp. strain T2, T. brockianus, Thermotoga maritima, and Thermotoga neapolitana belong to the second group, which had smaller sizes ranging from 53 to 65 kDa.
The molecular mass of the Thermus sp. strain T2 enzyme was estimated to be more than 400 kDa by use of a calibrated Superose 12-gel filtration column and native PAGE (data not shown). These results indicate an octameric form of the native enzyme in solution. α-Gals from E. coli (24), S. mutans (1), B. stearothermophilus (9), and T. brockianus (8) existed in the tetrameric structure; on the other hand, the hyperthermophilic enzymes from Thermotoga existed in the monomeric or dimeric structure (17). The enzyme from Thermus sp. strain T2 is very unique because it probably existed as an octameric structure in solution.
N-terminal amino acid sequencing of purified α-Gal.
The purified α-Gal was subjected to SDS-PAGE and blotted on a polyvinylidene difluoride membrane. The N-terminal amino acid sequence was determined as M-R-G-S-H-H-H-H-H-H-G-S-M-R-L-V-L-G-G-L-E-V-P-L-K-A. It corresponds to the His tag sequence followed by the N-terminal deduced amino acid sequence of the α-Gal ORF, which is underlined.
Enzymatic properties.
The purified Thermus sp. strain T2 α-Gal was most active at 75°C for pNP-α-Gal hydrolysis and was stable up to 60°C at pH 7.0 for 1 h. The maximum activity of the enzyme was observed at pH 6.0, and the enzyme was stable between pH 6.0 and 13.0 at 40°C for a 1-h incubation. This temperature dependence of the activity of the enzyme is the same as that of B. stearothermophilus (75°C) and is not as high as those of the T. brockianus and Thermotoga enzymes (90 to 95°C).
Substrate specificity.
The α-Gal was specific for α-galactopyranosidic compounds. In contrast to pNP-α-Gal, it did not hydrolyze pNP-α-fucopyranoside, pNP-β-fucopyranoside, pNP-α-arabinofuranoside, pNP-β-arabinopyranoside, pNP-α-glucopyranoside, pNP-β-glucopyranoside, pNP-β-galactopyranoside, pNP-α-xylopyranoside, pNP-β-xylopyranoside, pNP-α-rhamnopyranoside, pNP-α-mannopyranoside, or pNP-β-mannopyranoside. Thermus sp. strain T2 α-Gal exhibited a Km for pNP-α-Gal of 4.7 mM, which is similar to the one obtained for α-Gal from T. brockianus (2.5 mM).
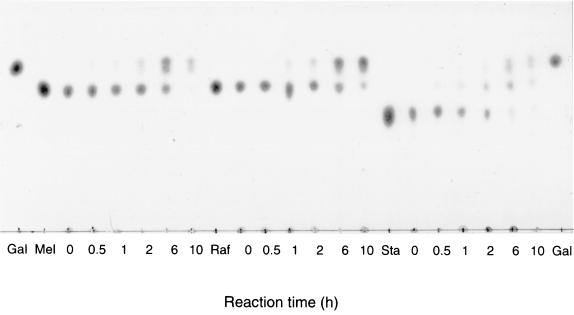
The enzymatic properties of the recombinant α-Gal were studied by using substrates with galacto-oligosaccharides, such as melibiose, raffinose, and stachyose. The enzyme hydrolyzed these substrates in the order of stachyose>melibiose>raffinose, as shown in Fig. 3. α-Gals usually degrade raffinose quickly and stachyose slowly (24), but the Thermus sp. strain T2 enzyme showed a different specificity against the galacto-oligosaccharides.
FIG. 3.
Action of α-Gal on galacto-oligosaccharides. The reaction mixture was composed of 80 μl of 1% (wt/vol) substrate, 80 μl of 0.2 M sodium phosphate buffer (pH 6.0), and 40 μl (0.4 U) of enzyme solution. The reaction was done at 70°C, and 20 μl of the reaction mixture was withdrawn at each indicated time. Three microliters of the mixture was used for the TLC. Gal, authentic galactose; Mel, authentic melibiose; Raf, authentic raffinose; Sta, authentic stachyose.
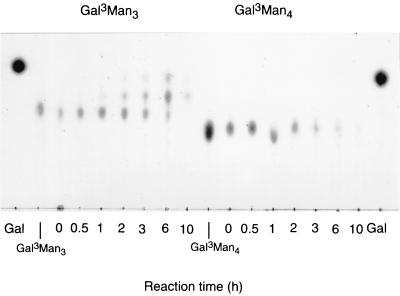
In order to investigate the substrate specificity of Thermus sp. strain T2 α-Gal (prokaryotic enzyme) on the galactomanno-oligosaccharides, the purified enzyme was incubated with galactomanno-oligosaccharides and the degradation products were analyzed by TLC. As shown in Fig. 4, the enzyme acted only on Gal3Man3 as well as on M. vinacea α-Gal I and the yeast enzymes. This is the first paper to describe the substrate specificity of bacterial α-Gal toward galactomanno-oligosaccharides. As previously described, eukaryotic α-Gals are classified into three groups depending on the specificity on the galactomanno-oligosaccharides; i.e., the first group contains enzymes, such as M. vinacea α-Gal I and yeast α-Gals, which act on the terminal α-galactosyl residue of Gal3Man3; the second group contains enzymes, such as P. purpurogenum α-Gal and A. niger 5-16 α-Gal, which act only on side chain α-galactosyl residue of Gal3Man4; and the last group contains enzymes, such as M. vinacea α-Gal II, which can act on both substrates. Thermus sp. strain T2 α-Gal could liberate the galactose residue from Gal3Man3 but could not act on Gal3Man4, indicating that the enzyme can act only on the terminal α-Gal residues of the substrate as well as on M. vinacea α-Gal I and the yeast enzymes. Consequently, Thermus sp. strain T2 α-Gal was classified into the group which can act only on the terminal α-galactosyl residue of the substrate, indicating that this is the first bacterial enzyme which can act only on the terminal α-galactosyl residue.
FIG. 4.
Action of α-Gal on galactomanno-oligosaccharides. The enzymatic reaction was done as described for Fig. 3. Gal, authentic galactose. The structures of Gal3Man3 and Gal3Man4 are shown in Fig. 1.
Inhibition study.
Some α-Gals are reported to be inhibited by SH reagents, such as pCMB. Thermus sp. strain T2 α-Gal was also completely inactivated (less than 1% of the control) after the treatment with 1 mM pCMB at 30°C for 30 min. One millimolar metal ions, including Co2+, Ca2+, Mg2+, Ni2+, Cu2+, Zn2+, and Mn2+, did not affect the enzymatic activity, but Hg2+ and Ag+ significantly inactivated the enzyme (less than 5 and 20% of the control, respectively). Fridjonsson et al. (8) reported that α-Gal from T. brockianus contained three Cys residues and was almost completely inhibited by HgCl2 and pCMB. They determined the presence of a thiol group at or near the catalytic site of the enzyme. Among the two conserved Cys residues (161 and 336), Cys residue 336, according to T. brockianus α-Gal numbering, could be the conserved Cys residue found in their alignment with T. maritima and T. neapolitana. However, Cys residue 189, not residue 371 (according to our numbering in Fig. 2), corresponding to residue 161 of T. brockianus, could be considered the Cys residue which is modified by pCMB, because the Thermus sp. strain T2 enzyme has only 1 Cys residue in the molecule.
Purification and characterization of mutant enzyme Cys159Ala.
The replacement of Cys by Ala of the enzyme was carried out to analyze the role of the enzyme's Cys residue. The expression and purification of the mutant enzyme were carried out as described in Materials and Methods. The purified mutant enzyme showed a single protein band in SDS-PAGE (data not shown). The mutant enzyme also showed the same specific activity against pNP-α-Gal with the native enzyme, and p-chloromercuribenzoic acid did not affect the enzymatic activity of the mutant enzyme, indicating that the Cys residue is not responsible for the catalytic function. This modification of the Cys residue of the native enzyme by pCMB probably introduced a conformational change in the enzyme by adding the large hydrophobicity and also the negative charge of the compound into the protein.
ACKNOWLEDGMENTS
This study was supported in part by a grant of Rice Genome Project PR-2206, MAFF, Japan. The work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences.
REFERENCES
- 1.Aduse-Opoku J, Tao L, Ferretti J J, Russell R R B. Biochemical and genetic analysis of Streptococcus mutans α-galactosidase. J Gen Microbiol. 1991;137:757–764. doi: 10.1099/00221287-137-4-757. [DOI] [PubMed] [Google Scholar]
- 2.Aslanidis C, Schmid K, Schmitt R. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J Bacteriol. 1989;171:6753–6763. doi: 10.1128/jb.171.12.6753-6763.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop F F, Calhoun D H, Bernstein H S, Hantsopoulos P, Quinn M, Desnick R J. Human α-galactosidase A: nucleotide sequence of a cDNA clone encoding the mature enzyme. Proc Natl Acad Sci USA. 1986;83:4859–4863. doi: 10.1073/pnas.83.13.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Herder I F, Rosell A M, van Zuilen C M, Punt P J, van den Hondel C A. Cloning and expression of a member of the Aspergillus niger gene family encoding α-galactosidase. Mol Gen Genet. 1992;233:404–411. doi: 10.1007/BF00265437. [DOI] [PubMed] [Google Scholar]
- 5.Dey P M, Del Campillo E. Biochemistry of the multiple forms of glycosidases in plants. Adv Enzymol Relat Areas Mol Biol. 1984;56:141–249. doi: 10.1002/9780470123027.ch3. [DOI] [PubMed] [Google Scholar]
- 6.Dey P M, Patel S, Brownleader M D. Induction of α-galactosidase in Penicillium ochrochloron by guar (Cyamopsis tetragonoloba) gum. Biotechnol Appl Biochem. 1993;17:361–371. [PubMed] [Google Scholar]
- 7.Duffaud G D, McCutchen C M, Leduc P, Parker K N, Kelly R M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridjonsson O, Watzlawick H, Gehweiler A, Rohrfisch T, Mattes R. Cloning of the gene encoding a novel thermostable α-galactosidase from Thermus brockianus ITI360. Appl Environ Microbiol. 1999;65:3955–3963. doi: 10.1128/aem.65.9.3955-3963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridjonsson O, Watzlawick H, Gehweiler A, Mattes R. Thermostable α-galactosidase from Bacillus stearothermophilus NUB3621: cloning, sequencing and characterization. FEMS Microbiol Lett. 1999;176:147–153. doi: 10.1111/j.1574-6968.1999.tb13655.x. [DOI] [PubMed] [Google Scholar]
- 10.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko R, Kusakabe I, Sakai Y, Murakami K. Substrate specificity of α-galactosidase from Mortierella vinacea. Agric Biol Chem. 1990;54:237–238. [Google Scholar]
- 12.Kaneko R, Kusakabe I, Ida E, Murakami K. Substrate specificity of α-galactosidase from Aspergillus niger 5-16. Agric Biol Chem. 1991;55:109–115. [PubMed] [Google Scholar]
- 13.King M R, Yernool D A, Eveleigh D E, Chassy B M. Thermostable α-galactosidase from Thermotoga neapolitana: cloning, sequencing and expression. FEMS Microbiol Lett. 1998;163:37–42. doi: 10.1111/j.1574-6968.1998.tb13023.x. [DOI] [PubMed] [Google Scholar]
- 14.Koyama Y, Okamoto S, Furukawa K. Cloning of α- and β-galactosidase genes from an extreme thermophile, Thermus strain T2, and their expression in Thermus thermophilus HB27. Appl Environ Microbiol. 1990;56:2251–2254. doi: 10.1128/aem.56.7.2251-2254.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusakabe I, Kaneko R, Tanaka N, Zamora A F, Fernandez W L, Murakami K. A simple method for elucidating structures of galactomanno-oligosaccharides by sequential actions of β-mannosidase and α-galactosidase. Agric Biol Chem. 1990;54:1081–1083. [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Liebl W, Wagner B, Schellhase J. Properties of an α-galactosidase, and structure of its gene GalA, within an α- and β-galactosidase utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst Appl Microbiol. 1998;21:1–11. doi: 10.1016/s0723-2020(98)80002-7. [DOI] [PubMed] [Google Scholar]
- 18.Liljestrom P L, Liljestrom P. Nucleotide sequence of the melA gene, coding for α-galactosidase in Escherichia coli K-12. Nucleic Acids Res. 1987;15:2213–2220. doi: 10.1093/nar/15.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolles-Clark E, Tenkanen M, Luonteri E, Penttila M. Three α-galactosidase genes of Trichoderma reesei cloned by expression in yeast. Eur J Biochem. 1996;240:104–111. doi: 10.1111/j.1432-1033.1996.0104h.x. [DOI] [PubMed] [Google Scholar]
- 20.Overbeeke N, Fellinger A J, Toonen M Y, van Wassenaar D, Verrips C T. Cloning and nucleotide sequence of the α-galactosidase cDNA from Cyamopsis tetragonoloba (guar) Plant Mol Biol. 1989;13:541–550. doi: 10.1007/BF00027314. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nieklen S, Coulson Q R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saul D J, Rodrigo A G, Reeves R A, Williams L C, Borges K M, Morgan H W, Bergquist P L. Phylogeny of twenty Thermus isolates constructed from 16S rRNA gene sequence data. Int J Syst Bacteriol. 1993;43:754–760. doi: 10.1099/00207713-43-4-754. [DOI] [PubMed] [Google Scholar]
- 24.Schmid K, Schmitt R. Raffinose metabolism in Escherichia coli K12. Purification and properties of a new alpha-galactosidase specified by a transmissible plasmid. Eur J Biochem. 1976;67:95–104. doi: 10.1111/j.1432-1033.1976.tb10637.x. [DOI] [PubMed] [Google Scholar]
- 25.Seraphin B, Kandels-Lewis S. An efficient PCR mutagenesis strategy without gel purification step that is amenable to automation. Nucleic Acids Res. 1996;24:3276–3277. doi: 10.1093/nar/24.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibuya H, Kobayashi H, Kasamo K, Kusakabe I. Nucleotide sequence of α-galactosidase cDNA from Mortierella vinacea. Biosci Biotechnol Biochem. 1995;59:1345–1348. doi: 10.1271/bbb.59.1345. [DOI] [PubMed] [Google Scholar]
- 27.Shibuya H, Kobayashi H, Park G G, Komatsu Y, Sato T, Kaneko R, Nagasaki H, Yoshida S, Kasamo K, Kusakabe I. Purification and some properties of α-galactosidase from Penicillium purpurogenum. Biosci Biotechnol Biochem. 1995;59:2333–2335. doi: 10.1271/bbb.59.2333. [DOI] [PubMed] [Google Scholar]
- 28.Shibuya H, Kobayashi H, Sato T, Kim W-S, Yoshida W, Kaneko S, Kasamo K, Kusakabe I. Purification, characterization and cDNA cloning of a novel α-galactosidase from Mortierella vinacea. Biosci Biotechnol Biochem. 1997;61:592–598. doi: 10.1271/bbb.61.592. [DOI] [PubMed] [Google Scholar]
- 29.Sumner-Smith M, Bozzato R P, Skipper N, Davies R W, Hopper J E. Analysis of the inducible MEL1 gene of Saccharomyces carlsbergensis and its secreted product, α-galactosidase (melibiase) Gene. 1985;36:333–340. doi: 10.1016/0378-1119(85)90188-x. [DOI] [PubMed] [Google Scholar]
- 30.Villarejo M R, Zabin I. β-Galactosidase from termination and deletion mutant strains. J Bacteriol. 1974;120:466–474. doi: 10.1128/jb.120.1.466-474.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamane T. Decomposition of raffinose by α-galactosidase. An enzymatic reaction applied in the factory-process in Japanese beet sugar factories. Sucr Belge/Sugar Ind Abstr. 1971;90:345–348. [Google Scholar]
- 32.Yoshida S, Tan C H, Shimokawa T, Turakainen H, Kusakabe I. Substrate specificity of α-galactosidase from yeasts. Biosci Biotechnol Biochem. 1997;61:359–361. doi: 10.1271/bbb.61.359. [DOI] [PubMed] [Google Scholar]
- 33.Zhu A, Goldstein J. Cloning and functional expression of a cDNA coding coffee bean α-galactosidase. Gene. 1994;140:227–231. doi: 10.1016/0378-1119(94)90548-7. [DOI] [PubMed] [Google Scholar]