Abstract
Fumonisins are a group of mycotoxins produced in corn kernels by the plant-pathogenic fungus Fusarium verticillioides. A mutant of the fungus, FT536, carrying a disrupted gene named FCC1 (for Fusarium cyclin C1) resulting in altered fumonisin B1 biosynthesis was generated. FCC1 contains an open reading frame of 1,018 bp, with one intron, and encodes a putative 319-amino-acid polypeptide. This protein is similar to UME3 (also called SRB11 or SSN8), a cyclin C of Saccharomyces cerevisiae, and contains three conserved motifs: a cyclin box, a PEST-rich region, and a destruction box. Also similar to the case for C-type cyclins, FCC1 was constitutively expressed during growth. When strain FT536 was grown on corn kernels or on defined minimal medium at pH 6, conidiation was reduced and FUM5, the polyketide synthase gene involved in fumonisin B1 biosynthesis, was not expressed. However, when the mutant was grown on a defined minimal medium at pH 3, conidiation was restored, and the blocks in expression of FUM5 and fumonisin B1 production were suppressed. Our data suggest that FCC1 plays an important role in signal transduction regulating secondary metabolism (fumonisin biosynthesis) and fungal development (conidiation) in F. verticillioides.
Fumonisins are a group of mycotoxins produced by Fusarium verticillioides (Sacc.) Nirenberg (synonym Fusarium moniliforme Sheldon, teleomorph Gibberella moniliformis Wineland, synonym Gibberella fujikuroi mating population A) that contaminate maize and maize-based products (3, 22). Since the discovery of fumonisin B1 in 1988, more than 10 fumonisins have been isolated and characterized. Of these, fumonisin B1 (FB1), FB2, and FB3 are the major fumonisins found under field conditions. Fumonisins have been linked to various animal and human mycotoxicoses, such as leukoencephalomalacia in horses, pulmonary edema in pigs, and cancer in rats and humans (11, 36). The onset and progression of fumonisin-associated diseases are closely correlated with the disruption of sphingolipid metabolism. FB1 inhibits ceramide synthase (sphinganine N-acyltransferase), the enzyme responsible for the acylation of sphinganine in the de novo biosynthetic pathway for sphingolipids (32). In cells exposed to FB1, sphinganine accumulates rapidly and ceramides decrease, concomitant with increased DNA fragmentation, decreased viability, loss of regulation of differentiation, and apoptotic morphology (20, 28). Over the past few years, the U.S. Food and Drug Administration has been evaluating the carcinogenic nature of fumonisins, and guidelines have been set for fumonisins in food (http://vm.cfsan.fda.gov/∼dms/fumongui.html).
Structurally, fumonisins have a linear 19- or 20-carbon backbone with hydroxyl, methyl, and tricarballylic acid moieties at various positions along the backbone. Radiolabeling experiments suggest that the backbone is produced by the polyketide pathway of secondary metabolism (4). The genes involved in fumonisin biosynthesis appear to be clustered (23), and five gene sequences, those of FUM5, FUM6, FUM7, FUM8, and FUM9, have been deposited in GenBank (accession number AF155773). FUM5, which encodes the polyketide synthase of F. verticillioides, has been cloned and characterized (23). Also, four additional loci involved in fumonisin production were identified by genetic analyses of field isolates (8). Knowledge of the regulation of the fumonisin biosynthetic pathway is limited to evidence indicating that fumonisins are synthesized under conditions of nitrogen stress and acidic pH (14, 27).
Here we describe a mutant of F. verticillioides that is blocked in FB1 biosynthesis when grown on cracked corn. The mutant locus, FCC1 (for Fusarium cyclin C1), encodes a putative polypeptide with similarities to C-type cyclins of the yeast Saccharomyces cerevisiae. We hypothesize that FCC1 is not essential for vegetative growth but that it plays an important role in signal transduction regulating secondary metabolism (e.g., fumonisin biosynthesis) and fungal development (e.g., conidiation).
MATERIALS AND METHODS
Fungal strains.
F. verticillioides strains 7600 and 7598 (Fungal Genetics Stock Center, Kansas City, Kans.) were stored in 20% glycerin at −80°C. Conidia were produced for inoculum by growing the fungus on potato dextrose agar (Difco, Detroit, Mich.) at 28°C.
Fungal transformation.
Conidia (108) of F. verticillioides strain 7600 (wild type) were inoculated into 100 ml of yeast-peptone-dextrose (YPD) (Difco) broth and incubated for 18 h at 28°C on a rotary shaker (150 rpm). Protoplasts were prepared as described by Upchurch et al. (31) except that the mycelium (1 g [wet weight]) was resuspended in 20 ml of an enzyme solution containing β-glucuronidase (5,200 U per ml) (Sigma, St. Louis, Mo.), mureinase (2 mg per ml) (Amersham, Arlington Heights, Ill.), 10 mM NaH2PO4 (pH 5.8), 20 mM CaCl2, and 1.2 M KCl.
Mutations were generated by a modification of restriction enzyme-mediated integration (REMI) (17, 19, 26). A transformation vector, pUCH1 (5 μg) (30), which contains the hygromycin B phosphotransferase (HYG) gene and ampicillin resistance (AMP) gene as selectable markers, was linearized with HindIII. After digestion, the entire 20-μl reaction mixture was added to the protoplasts (100 μl), and the transformation was performed as previously described (19). Protoplasts were plated in regeneration agar medium (1 M sucrose, 0.02% yeast extract, and 1% agar) containing hygromycin B (150 μg/ml) (Roche, Indianapolis, Ind.) and incubated at 28°C. Hygromycin B-resistant transformants appeared after 5 to 7 days. For further studies, single-spore isolates were obtained by dilution plating and microscopically selecting and transferring germinated conidia.
Screening for transformants affected in fumonisin biosynthesis.
Transformants were grown on cracked-corn medium (27). FB1 was extracted and analyzed by thin-layer chromatography (TLC) as previously described (24). Transformants that did not produce FB1 based on the TLC analysis were analyzed by high-pressure liquid chromatography (HPLC), as previously described (27). This subset of transformants also was grown in defined liquid (DL) medium (27) and analyzed by HPLC for FB1.
Sexual crosses and random ascospore analysis.
Sexual crosses were performed as described by Klittich and Leslie (15). Transformant FT536 served as the male strain in crosses to F. verticillioides strain 7598. After 21 days of incubation at 28°C with a 12-h light-dark cycle, ascospores were collected from 25 perithecia and pooled. Hygromycin B-resistant (Hygr) and hygromycin B-sensitive (Hygs) isolates were tested for FB1 production on cracked-corn medium.
Nucleic acid manipulation.
Bacterial plasmids were isolated with the Wizard miniprep DNA purification system (Promega, Madison, Wis.). Fungal genomic DNA was isolated from mycelium grown in potato dextrose broth (Difco) as described previously (34). Total RNA was isolated with Trizol reagent (Gibco BRL, Grand Island, N.Y.) by following the manufacturer's suggested protocol and by a phenol-LiCl method (33). Southern analysis and Northern analysis were performed as previously described (34). DNA probes were 32P labeled with a Prime-It II random primer labeling kit (Stratagene, La Jolla, Calif.). A genomic DNA cosmid library made from F. verticillioides strain 7600 was obtained from Robert Proctor (National Center for Agricultural Utilization Research, USDA Agricultural Research Service, Peoria, Ill.).
cDNA subtraction library construction.
Poly(A)+ RNAs from total RNAs isolated from the wild-type and mutant strains grown on cracked-corn medium were purified with Oligotex mRNA spin columns (Qiagen, Valencia, Calif.). The PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, Calif.) was used to construct a wild-type subtraction library (genes expressed only in the wild type) and an FT536 subtraction library (genes expressed only in FT536). Amplified PCR products were cloned in the pGEM-T Easy cloning vector (Promega), and Library Efficiency DH5α competent cells (Gibco BRL) were used for library construction. DNA sequencing and analysis were performed at the Agricultural Genome Center, Purdue University.
Targeted gene disruption of FCC1.
Disruption vector pKB7XM was generated from pKB7 (Fig. 1A) by removing the XbaI site with mung bean nuclease. Wild-type protoplasts were transformed with pKB7XM linearized by HindIII. Transformants that had FCC1 replaced with pKB7XM by homologous recombination were identified by Southern analysis.
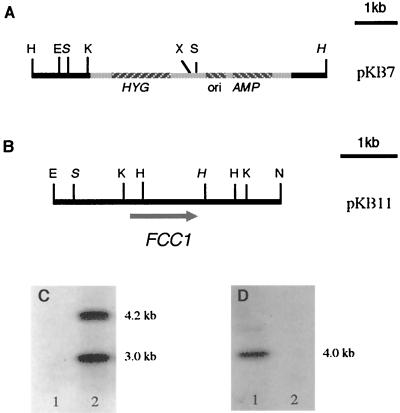
FIG. 1.
Disruption of the FCC1 locus in F. verticillioides by REMI mutagenesis. (A and B) Restriction maps of pKB7 from FT536 (A) and pKB11 from the wild type (B) containing the FCC1 locus. Restriction enzyme sites: H, HindIII; S, SalI; X, XbaI; E, EcoRI; K, KpnI; N, NotI. (C and D) Genomic DNAs (3 μg) from the wild type and FT536 were digested with SalI, electrophoresed in a 1.0% agarose gel, transferred to a nylon membrane, and probed with 32P-labeled vector (pUCH1) DNA (C) and FCC1 DNA fragment (KB500) (D). Lanes 1, wild type; lanes 2, FT536.
PCR and RT-PCR.
PCRs and reverse transcription-PCRs (RT-PCRs) were performed in an Omn-E thermocycler (Hybaid, Teddington, United Kingdom). The primers for the FCC1 PCR were FCC1F1 (5′-CGGTCCGACAAATGACTGG-3′) and FCC1R1 (5′-GGACACGTGCAGACATCATCC-3′). DNA amplification was performed in a 25-μl mixture with Taq DNA polymerase (Promega). The reactions were carried out for 30 cycles of 40 s of denaturation at 94°C, 1 min of annealing at 56°C, and 1 min of extension at 72°C. The primers for RT-PCRs were as follows: (i) for FCC1, FCC1RTF (5′-CACTTCGTCGTCCACCAACG-3′) and FCC1RTR (5′-CGACACAATGTCGCTTCTGG-3′); (ii) for FUM5, FUM5F21 (5′-CATACGTGATGGAGGCATGG-3′) and FUM5R1 (5′-TCAGAACCAGAGCAGACTGG-3′); (iii) for AREA (29), AREF1 (5′-GCTGCTATTCACAACGCTCC-3′) and ARER1 (5′-GAGTAGCTTGGTGAGCTG-3′); and (iv) for TUB2 (35), TUBF1 (5′-GAGCCGTCCTCGTCGACC-3′) and TUBR31 (5′-GAGCTCCTGGATAGAAGTGG-3′). GenBank accession numbers are AF155773 (FUM5), Y11006 (AREA), and U27303 (TUB2). RT-PCR was performed with an Access RT-PCR kit (Promega). The reactions were carried out according to the manufacturer's suggested protocol except that the annealing temperature was 56°C. When necessary, PCR and RT-PCR products were cloned with a TA Cloning kit (Invitrogen, Carlsbad, Calif.). Sequencing was performed on both DNA strands at the DNA Sequencing Facility, Purdue University. DNA sequences were analyzed and amino acid sequences were deduced with the MacDNAsis program (Hitachi Software Engineering Co., San Bruno, Calif.) (10, 25). Similarity searches were done via the BLAST algorithm, version 3.6 (1).
Effect of pH on FB1 production.
The wild-type and FT536 strains were grown on BSAL medium (modified DL medium with bovine serum albumin [BSA] [0.1 g/100 ml] replacing ammonium phosphate) with the pH adjusted to 3, 6, or 9 with 14.7 N phosphoric acid or 10 N sodium hydroxide and on carnation leaf agar (CLA), pH 5.2 (21). BSAL medium (100 ml) in a 250-ml flask was inoculated with 108 conidia and incubated at 28°C for 7 days on a rotary shaker at 150 rpm. Dry mass, sporulation, FB1 production, and final pH were measured after 7 days of incubation. Conidia were produced on CLA as previously described (21) and quantified with a hemacytometer.
Nucleotide sequence accession number.
The nucleotide sequence and the predicted polypeptide sequence of FCC1 have been submitted to GenBank (accession no. AF294431).
RESULTS
Isolation of FT536.
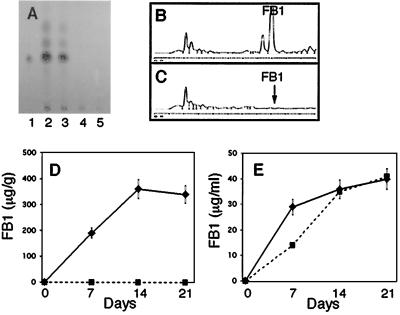
A total of 760 transformants were screened for vector integration events that affected FB1 production. One transformant, FT536, produced no FB1 when grown on cracked-corn medium for 7 days, while the wild type produced over 200 μg of FB1/g of corn (Fig. 2A, B, and C). Radial growth of FT536 was similar to that of the wild type when grown on potato dextrose agar, YPD agar, or Czapek-Dox (Difco) agar (data not shown). FB1 production by the wild type and FT536, grown on cracked-corn medium and DL medium, was measured over a 21-day incubation period (Fig. 2D and E). Growth on the cracked-corn medium by FT536 and the wild type appeared to be similar, and mycelial dry weights of the two strains grown in DL medium were similar (data not shown). However, FT536 produced a dark purple metabolite when grown on cracked-corn medium. The wild type produced over 300 μg of FB1/g of cracked-corn medium after 21 days, whereas FT536 did not produce detectable FB1 (Fig. 2D). In contrast, the two strains produced similar amounts of FB1 when grown in DL medium (Fig. 2E).
FIG. 2.
FB1 production by the wild type and FT536. (A) FB1 was assayed by TLC after 7 days of growth on cracked-corn medium. Lane 1, FB1 standard (1 μg); lanes 2 and 3, wild type; lanes 4 and 5, FT536. (B and C) FB1 from the wild type (B) and FT536 (C) was analyzed by HPLC after 7 days of growth on cracked-corn medium. (D and E) FB1 production by the wild type (♦) and FT536 (▪) was measured by HPLC after 7, 14, and 21 days of growth on cracked-corn medium (D) and DL medium (E). Results are means and standard errors from three replicates.
Random ascospore analysis.
We obtained 500 ascospores from a cross between FT536 and F. verticillioides strain 7598. Of these, 213 were Hygr and 287 were Hygs. Twenty ascospores were randomly selected from each Hygr and Hygs group and tested for FB1 production on cracked-corn medium. None of the Hygr isolates produced FB1, whereas all 20 Hygs isolates produced over 200 μg of FB1/g. If the Hygr and fumonisin nonproduction phenotypes are not due to the same mutation, then the two mutations responsible for these phenotypic changes are 95% certain to be within a 7.2-centimorgan interval.
Characterization of the disrupted locus
A Southern blot of FT536 genomic DNA digested with SalI was probed with labeled vector. Two bands of hybridization were observed, indicating that a single copy of the transformation vector was inserted into the wild-type genome (Fig. 1C). A 7-kb HindIII DNA fragment, pKB7, containing the entire transformation vector and the DNA flanking both sides of the insertion point was recovered from FT536 (Fig. 1A). In addition, a cosmid clone containing DNA from the disrupted locus was isolated from a wild-type genomic library from which a 4-kb NotI-EcoRI DNA fragment was subcloned (pKB11 [Fig. 1B]). Comparison of the DNA sequences of pKB7 and pKB11 indicated that during the REMI mutagenesis 1,069 bp was deleted (Fig. 3A). PCR primers FCC1RTF and FCC1RTR were used to amplify a 500-bp fragment (KB500) from the wild-type genomic DNA. A Southern blot of genomic DNAs from the wild type and FT536 digested with SalI was probed with labeled KB500 DNA. A single band of hybridization was detected in the wild type, and no band was observed in FT536, confirming the deletion in FT536 (Fig. 1D).
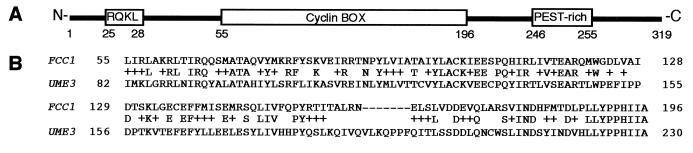
FIG. 3.
Sequence analysis of the FCC1 locus in F. verticillioides. (A) Diagram of the deduced 319-amino-acid polypeptide of FCC1 (GenBank accession number AF294431), showing conserved motifs, i.e., the destruction box-like motif (RQKL), the cyclin box motif, and the PEST-rich motif. Numbers indicate the locations of the amino acids in the polypeptide. (B) Amino acid sequence alignment of the cyclin box motifs from FCC1 and UME3, a C-type cyclin of S. cerevisiae. +, similar amino acids. The sequence alignment was performed with BLAST2. The GenBank accession number of UME3 is S59373.
The deleted region contained a fragment of FCC1, a gene with an open reading frame of 1,018 bp including one intron (62 bp), which encodes a putative 319-amino-acid polypeptide. The deletion in FT536 occurred between nucleotides at positions 417 and 1487 of the FCC1 DNA sequence submitted to GenBank. FCC1 is similar to UME3 (also known as SRB11 [accession number U20221] and SSN8 [accession number U20635]), a cyclin C of S. cerevisiae (7, 16, 18). FCC1 and UME3 share 36% overall identity. Furthermore, these proteins share 43% identity and 71% similarity within the cyclin box domain (Fig. 3B). FCC1 also shows high similarity with cyclin C proteins from human (57%), fruit fly (59%), and mouse (57%). In addition to the cyclin box domain, FCC1 contains a destruction box-like motif (RQKL) and a PEST-rich region (7).
Targeted gene disruption of FCC1.
Complementation in FT536 with FCC1 would have provided additional proof that the phenotype of FT536 resulted from the deletion of FCC1. However, in our hands F. verticillioides was not sensitive to other selectable inhibitors such as bialophos and bleomycin, and thus we were not able to complement the mutation. As an alternative approach, FCC1 was disrupted in the wild-type strain by homologous recombination with the vector pKB7XM (pKB7 lacking the XbaI restriction site [Fig. 1A]). Southern analysis of transformant TK19 showed that FCC1 was replaced by pKB7XM (data not shown). TK19 also failed to produce FB1 when grown on cracked-corn medium (data not shown). These data provided independent evidence that the deletion of FCC1 leads to the observed phenotype in FT536, and they support the genetic data.
Gene expression in FT536.
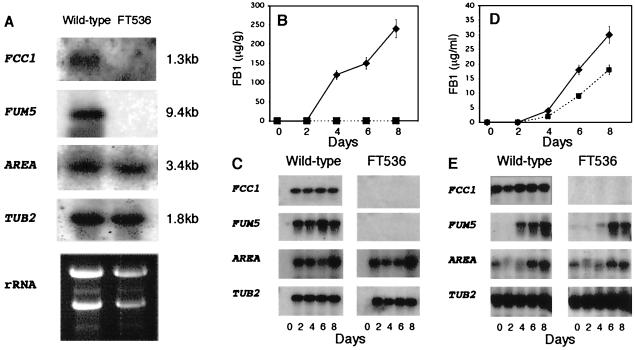
Northern blots of total RNAs obtained from the wild type and FT536 grown on cracked corn for 8 days were probed with labeled FCC1. A 1.3-kb band of hybridization was detected in the wild type, while no band was detected in FT536 (Fig. 4A). Similarly, when the blots were probed with labeled FUM5, a 9.4-kb band was detected only in the wild type. Hybridization bands of 3.4 and 1.8 kb were obtained in both the wild type and FT536 when the blots were probed with AREA, the nitrogen metabolism regulatory gene, and TUB2, which encodes β-tubulin, respectively (Fig. 4A). Gene expression and FB1 in the wild type and FT536 were analyzed over 8 days after inoculation to cracked-corn medium and DL medium. When the wild type was grown on cracked corn, FCC1 and FUM5 were expressed after 2 days, and the expression remained constant throughout the study (Fig. 4C). FB1 was not detected in the wild-type culture after 2 days of incubation; however, by 4 days the concentration was over 100 μg of FB1/g of corn (Fig. 4B). In contrast, FT536 did not express FUM5 during growth on cracked corn, and no FB1 was detected (Fig. 4B and C). After 2 days of incubation, expression of both AREA and TUB2 was detected in both the wild type and FT536 (Fig. 4C).
FIG. 4.
Gene expression and FB1 biosynthesis in the wild type and FT536. (A) Northern blot analysis of the wild type and FT536 grown on cracked-corn medium for 8 days. Total RNA (15 μg) was electrophoresed in 1.2% agarose-formaldehyde gels, transferred to a nylon membrane, and hybridized with radiolabeled FCC1, FUM5, AREA, and TUB2. The molecular sizes of the transcripts are indicated on the right. rRNA, photograph of the ethidium bromide-stained gel. (B and D) FB1 production by the wild type (♦) and FT536 (▪) grown on cracked-corn (B) and DL (D) media. Values are means and standard errors from three replicates. For points lacking error bars, the standard error was less than 10% of the value. (C and E) RT-PCR analysis of FCC1, FUM5, AREA, and TUB2 in the wild type and FT536 grown on cracked-corn (C) and DL (E) media. Day zero samples were from uninoculated corn (C) and wild type and FT536 grown for 4 days on YPD medium (E). The total RNA used as template for each RT-PCR was standardized before the reactions (2 μg/reaction). RT-PCR products were electrophoresed in a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with radiolabeled FCC1, FUM5, AREA, and TUB2.
Mycelia collected from wild-type and FT536 cultures grown on YPD medium, which does not support fumonisin production, were used to inoculate DL medium. Fumonisin biosynthesis in the DL medium began 4 days after inoculation (Fig. 4D). Expression of AREA was detected at the 6-day time point in both the wild type and FT536 (Fig. 4E). FUM5 expression was first detected after 4 days and was independent of FCC1. No differences in TUB2 expression were observed. FCC1 was expressed constitutively throughout the study in the wild type and was never detected in FT536. These data are consistent with the hypothesis that FCC1 regulates expression of genes involved in FB1 biosynthesis when grown on cracked corn.
Effect of pH on conidiation.
Both the wild type and FT536, when grown on DL medium, caused the pH of the medium to drop from an initial value of 5.9 to 2.4 after 7 days of growth (Table 1). The dry weights and the numbers of conidia produced by the strains were also similar. When ammonium was replaced with BSA as the sole nitrogen source in DL medium, conidiation by FT536 was influenced by pH. In BSAL medium at pH 6 or 9, the wild type produced 100-fold more conidia than FT536 (Table 1). In contrast, the wild type grown in BSAL medium at pH 3 produced one-third of the conidia seen in media with a higher pH. In the same medium, FT536 conidiation increased nearly 50-fold, although it was still less than the number produced by the wild type (Table 1). On cracked-corn medium, the wild type produced over 1,000-fold more conidia than FT536 (data not shown). When grown on CLA medium (pH 5.2), the wild type produced abundant microconidial chains, characteristic of F. verticillioides, whereas the mycelium of FT536 rarely produced microconidial chains.
TABLE 1.
Effect of pH on conidiation and fumonisin productiona
| Mediumb | Strain | Final pHc | Conidia (105)d | Dry wt (g) | FB1 concn (μg/ml)e |
|---|---|---|---|---|---|
| DL | Wild type | 2.4 ± 0.10 | 16 ± 1.2 | 1.0 ± 0.05 | 15 ± 0.71 |
| FT536 | 2.4 ± 0.11 | 14 ± 1.4 | 0.9 ± 0.11 | 5.7 ± 0.54 | |
| BSAL3 | Wild type | 2.6 ± 0.10 | 82 ± 13 | 1.4 ± 0.02 | 14 ± 0.60 |
| FT536 | 2.7 ± 0.06 | 57 ± 16 | 1.1 ± 0.05 | 8.7 ± 0.98 | |
| BSAL6 | Wild type | 5.6 ± 0.15 | 230 ± 14 | 1.8 ± 0.05 | 3.9 ± 0.91 |
| FT536 | 5.6 ± 0.16 | 1.8 ± 0.22 | 1.8 ± 0.05 | NDf | |
| BSAL9 | Wild type | 6.8 ± 0.05 | 240 ± 9.8 | 1.9 ± 0.05 | ND |
| FT536 | 6.5 ± 0.05 | 1.9 ± 0.11 | 1.5 ± 0.05 | ND |
All data are means and standard deviations from three replicates.
DL, defined liquid medium (pH 5.9); BSAL, defined liquid medium with BSA as the sole nitrogen source (numbers indicate the initial pH of the medium).
Final pH of the fungal cultures after 7 days of incubation.
Determined with a hemacytometer.
Analyzed by HPLC.
ND, none detected.
Wild-type and FT536 cDNA subtraction libraries.
Subtraction libraries were made with RNAs isolated from the wild type and FT536 grown on cracked corn. Approximately 800 clones from each library were sequenced and analyzed. Sequences with high similarity (P <10−5) to genes with known function were categorized into eight groups: carbohydrate metabolism, protein metabolism, fatty acid metabolism, secondary metabolism, cell differentiation, pH responsiveness, stress responsiveness, and signal transduction. The two libraries differ particularly in the stress responsiveness, pH responsiveness, fatty acid metabolism, and carbohydrate metabolism categories (data not shown). Five sequences from the fumonisin biosynthetic gene cluster (GenBank accession number AF155773) were identified in the wild-type subtraction library but not in the FT536 subtraction library.
DISCUSSION
We identified a mutant of F. verticillioides that lacks the FCC1 open reading frame, which encodes a putative 319-amino-acid polypeptide. The FCC1 translation product is closely related to UME3, the cyclin C of S. cerevisiae (7, 16, 18). Cyclins are essential activating subunits of cyclin-dependent kinases (CDKs) and comprise a large family of proteins conserved in function in many organisms from yeast to human (2). Although originally identified as cell cycle-regulatory proteins, C-type cyclins also are involved in transcriptional activation or repression of genes associated with stress responses and development (7, 16, 18). In S. cerevisiae, UME3 forms a complex with the CDK-like protein UME5 (also known as SRB10 and SSN3) to regulate transcription by phosphorylation of the RNA polymerase II carboxy-terminal domain (18). The mRNA and protein levels of C-type cyclins, unlike those of other cyclins, do not fluctuate during mitotic cell division (7). C-type cyclins also contain three conserved regions that are important for function: the PEST-rich, cyclin box, and destruction box-like motifs (7, 16). The cyclin box is required for interaction and activation of the CDK partners. The PEST-rich and destruction box-like motifs are required for the degradation of C-type cyclins in response to external signals, such as heat shock (7). Prior to this study, C-type cyclins from filamentous fungi have not been described. Our data suggest that FCC1 is a C-type cyclin similar to UME3. FCC1 contains the cyclin box-like region, a destruction box-like motif, and a PEST-rich region, is constitutively expressed, and appears to regulate genes involved in fumonisin biosynthesis.
Prior to this study, the role of cyclin C as a regulator of genes involved in conidiation and secondary metabolism linked to pH-dependent gene expression has not been tested in filamentous fungi. However, PHOA, a cyclin-dependent kinase in Aspergillus nidulans, is required for conidial development under phosphate stress and alkaline pH (5). The cyclin component for PHOA has not been identified. The phenotypes of FT536 and the phoA mutant of A. nidulans share some interesting similarities. Both mutants accumulate pigments when grown in alkaline pH, and in both mutants conidiation is affected by the pH of the growth medium. Neither mutant produces conidia at pH 6, and at lower pH, both mutants sporulate similarly to wild-type strains. These characteristics suggest that FCC1 may be a homolog of the cyclin that complexes with PHOA in A. nidulans. One of the cDNA clones in the wild-type subtraction library has high sequence similarity to PHOA.
A striking difference between the phenotypes of FT536 and the phoA mutant of A. nidulans is that the phoA phenotype is apparent at low phosphate concentrations (0.1 mM) and reverts to the wild-type phenotype at 11 mM phosphate. In contrast, the phenotype of FT536 was not affected by phosphate concentration, and the mutant phenotype was observed in BSAL medium containing 22 mM phosphate (5). However, we cannot completely rule out the involvement of phosphate regulation, because genes similar to the yeast genes PHO80 and PHO85 (12, 13), coding for cyclin and CDK, respectively, were expressed in the FT536 subtraction library. This cyclin-CDK complex is involved in activation of alkaline phosphatase under extracellular phosphate stress. The fact that genes similar to PHO80 and PHO85 were identified in the FT536 subtraction library suggests that FCC1 may regulate gene expression by affecting downstream cyclins.
FCC1 is clearly involved in the regulation of conidiation and fumonisin biosynthesis, perhaps in signal transduction and perhaps at the level of gene expression. FCC1 may form a cyclin-CDK complex that acts as a putative receptor or directly links to receptors that sense the environment. Alternatively, the cyclin-CDK complex could directly regulate transcription in response to extracellular stress, similar to UME3-UME5 (12). The question of why FT536 produces FB1 in the DL medium at lower pH remains unanswered. We hypothesize that another cyclin forms a complex with the CDK in FT536 or that the regulatory circuit through FCC1 is bypassed under low-pH growth conditions.
Sequences identified in the wild-type and FT536 subtraction libraries will provide significant information toward understanding the role of FCC1 in fumonisin biosynthesis and fungal development. Finding fumonisin genes, pH-responsive genes, and conidial development genes in our wild-type subtraction library provides new avenues for further investigation. A number of fatty acid metabolism-related genes were also observed in the wild-type subtraction library. Among the cDNAs is one similar to that of linoleate diol synthase, which catalyzes dioxygenation of polyunsaturated fatty acids. Calvo et al. (6) reported that polyunsaturated fatty acids, such as linoleic acid and its lipoxygenase-derived derivatives, stimulate conidiation in A. nidulans. Furthermore, linolenic acid has been shown to affect perithecial development in Nectria haematococca (Fusarium sobni) (9). It remains to be determined if FCC1 regulates fatty acid metabolism and if these fatty acids act as factors in regulating fungal differentiation. Further study of the genes identified in the subtraction libraries will help elucidate how C-type cyclins regulate signal transduction pathways of secondary metabolism and development in F. verticillioides as well as other filamentous fungi.
ACKNOWLEDGMENTS
We thank Ray Bressan, Larry Dunkle, and Jin-Rong Xu for their helpful discussion and review of this work. We also thank Robert Proctor for providing us with the F. verticillioides genomic DNA library and sharing sequence data.
Financial support was provided by Pioneer Hi-Bred International, Inc., and USDA NRI Competitive Grants Program award no. 99–35201-8124.
Footnotes
Journal publication 16348 of the Purdue University Agricultural Research Program.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 3.Bezuidenhout S C, Gelderblom W C A, Gorstallman C P, Horak R M, Marasas W F O, Spiteller G, Vleggaar R. Structure elucidation of the fumonisins, mycotoxins from Fusarium moniliforme. J Chem Soc Chem Commun. 1988;11:743–745. [Google Scholar]
- 4.Blackwell B A, Miller J D, Savard M E. Production of carbon 14-labeled fumonisin in liquid culture. J AOAC Int. 1994;77:506–511. [Google Scholar]
- 5.Bussink H J, Osmani S A. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 1998;17:3990–4003. doi: 10.1093/emboj/17.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo A M, Hinze L L, Gardner H W, Keller N P. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl Environ Microbiol. 1999;65:3668–3673. doi: 10.1128/aem.65.8.3668-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper K F, Mallory M J, Smith J B, Strich R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjardins A E, Plattner R D, Proctor R H. Linkage among genes responsible for fumonisin biosynthesis in Gibberella fujikuroi mating population A. Appl Environ Microbiol. 1996;62:2571–2576. doi: 10.1128/aem.62.7.2571-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer P S, Ingram D S, Johnstone R. Evidence for the involvement of linolenic acid and other endogenous lipid factors in perithecial development of Nectria haematococca mating population VI. Mycol Res. 1993;97:485–498. [Google Scholar]
- 10.Fickett J W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelderblom W C A, Jaskiewicz K, Marasas W F O, Thiel P G, Horak R M, Vleggaar R, Kriek N P J. Fumonisins: novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl Environ Microbiol. 1988;54:1806–1811. doi: 10.1128/aem.54.7.1806-1811.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaffman A, Herskowitz I, Tjian R, Oshea E K. Phosphorylation of the transcription factor-PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 14.Keller S E, Sullivan T M, Chirtel S. Factors affecting the growth of Fusarium proliferatum and the production of fumonisin B1: oxygen and pH. J Indust Microbiol Biotechnol. 1997;19:305–309. doi: 10.1038/sj.jim.2900466. [DOI] [PubMed] [Google Scholar]
- 15.Klittich C J R, Leslie J F. Chlorate-resistant, nitrate-utilizing (crn) mutants of Fusarium moniliforme (Gibberella fujikuroi) J Gen Microbiol. 1989;135:721–727. doi: 10.1093/genetics/118.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein-kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuspa A, Loomis W F. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao S M, Zhang J H, Jeffrey D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, Vanvuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 19.Lu S W, Lyngholm L, Yang G, Bronson C, Yoder O C, Turgeon B G. Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc Natl Acad Sci USA. 1994;91:12649–12653. doi: 10.1073/pnas.91.26.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrill A H, Liotta D C, Riley R T. Fumonisins: fungal toxins that shed light on sphingolipid function. Trends Cell Biol. 1996;6:218–223. doi: 10.1016/0962-8924(96)10021-0. [DOI] [PubMed] [Google Scholar]
- 21.Nelson P E, Marasas W F O, Toussoun T A. Fusarium species; an illustrated manual for identification. University Park: Pennsylvania State University Press; 1983. [Google Scholar]
- 22.Nelson P E, Desjardins A E, Plattner R D. Fumonisins, mycotoxins produced by Fusarium species—biology, chemistry, and significance. Annu Rev Phytopathol. 1993;31:233–252. doi: 10.1146/annurev.py.31.090193.001313. [DOI] [PubMed] [Google Scholar]
- 23.Proctor R H, Desjardins A E, Plattner R D, Hohn T M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet Biol. 1999;27:100–112. doi: 10.1006/fgbi.1999.1141. [DOI] [PubMed] [Google Scholar]
- 24.Ross P F, Nelson P E, Richard J L, Osweiler G D, Rice L G, Plattner R D, Wilson T M. Production of fumonisins by Fusarium moniliforme and Fusarium proliferatum isolates associated with equine leukoencephalomalacia and a pulmonary edema syndrome in swine. Appl Environ Microbiol. 1990;56:3225–3226. doi: 10.1128/aem.56.10.3225-3226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rychlik W, Rhoads R E. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim W B, Woloshuk C P. Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi. FEMS Microbiol Lett. 1999;177:109–116. doi: 10.1111/j.1574-6968.1999.tb13720.x. [DOI] [PubMed] [Google Scholar]
- 28.Tolleson W H, Couch L H, Melchior W B, Jenkins G R, Muskhelishvili M, Muskhelishvili L, McGarrity L J, Domon O, Morris S M, Howard P C. Fumonisin B1 induces apoptosis in cultured human keratinocytes through sphinganine accumulation and ceramide depletion. Int J Oncol. 1999;14:833–843. doi: 10.3892/ijo.14.5.833. [DOI] [PubMed] [Google Scholar]
- 29.Tudzynski B, Homann V, Feng B, Marzluf G A. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol Gen Genet. 1999;261:106–114. doi: 10.1007/s004380050947. [DOI] [PubMed] [Google Scholar]
- 30.Turgeon B G, Garber R C, Yoder O C. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987;7:3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upchurch R G, Ehrenshaft M, Walker D C, Sanders L A. Genetic transformation system for the fungal soybean pathogen Cercospora kikuchii. Appl Environ Microbiol. 1991;57:2935–2939. doi: 10.1128/aem.57.10.2935-2939.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang E, Norred W P, Bacon C W, Riley R T, Merrill A H. Inhibition of sphingolipid biosynthesis by fumonisins—implications for diseases associated with Fusarium moniliforme. J Biol Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 33.Woloshuk C P, Payne G A. The alcohol dehydrogenase gene Adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:670–676. doi: 10.1128/aem.60.2.670-676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woloshuk C P, Yousibova G L, Rollins J A, Bhatnagar D, Payne G A. Molecular characterization of the Afl-1 locus in Aspergillus flavus. Appl Environ Microbiol. 1995;61:3019–3023. doi: 10.1128/aem.61.8.3019-3023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan K Y, Dickman M B. Isolation of a β-tubulin gene from Fusarium moniliforme that confers cold-sensitive benomyl resistance. Appl Environ Microbiol. 1996;62:3053–3056. doi: 10.1128/aem.62.8.3053-3056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizawa T, Yamashita A, Luo Y. Fumonisin occurrence in corn from high-risk and low-risk areas for human esophageal cancer in China. Appl Environ Microbiol. 1994;60:1626–1629. doi: 10.1128/aem.60.5.1626-1629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]