Abstract
Objective
The increasing translation of artificial intelligence (AI)/machine learning (ML) models into clinical practice brings an increased risk of direct harm from modeling bias; however, bias remains incompletely measured in many medical AI applications. This article aims to provide a framework for objective evaluation of medical AI from multiple aspects, focusing on binary classification models.
Materials and Methods
Using data from over 56 000 Mass General Brigham (MGB) patients with confirmed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), we evaluate unrecognized bias in 4 AI models developed during the early months of the pandemic in Boston, Massachusetts that predict risks of hospital admission, ICU admission, mechanical ventilation, and death after a SARS-CoV-2 infection purely based on their pre-infection longitudinal medical records. Models were evaluated both retrospectively and prospectively using model-level metrics of discrimination, accuracy, and reliability, and a novel individual-level metric for error.
Results
We found inconsistent instances of model-level bias in the prediction models. From an individual-level aspect, however, we found most all models performing with slightly higher error rates for older patients.
Discussion
While a model can be biased against certain protected groups (ie, perform worse) in certain tasks, it can be at the same time biased towards another protected group (ie, perform better). As such, current bias evaluation studies may lack a full depiction of the variable effects of a model on its subpopulations.
Conclusion
Only a holistic evaluation, a diligent search for unrecognized bias, can provide enough information for an unbiased judgment of AI bias that can invigorate follow-up investigations on identifying the underlying roots of bias and ultimately make a change.
Keywords: medical AI, bias, COVID-19, predictive model, electronic health records
INTRODUCTION
The healthcare research and industry have been increasingly progressive on the translation and implementation of artificial intelligence (AI)/machine learning (ML) to improve outcomes and lower costs. Diligently identifying and addressing biases in AI/ML algorithms (hereafter, referred to as “algorithms”) have garnered widespread public attention as pressing ethical and technical challenges.1–5 For instance, there is growing concern that algorithms may import and/or exacerbate ethno-racial and gender disparities/inequities through the data used to train them, due to their math, or the people who develop them.6,7
The costs of deploying algorithms in healthcare carelessly could exacerbate the very health inequalities society is working to address.8,9 The Algorithmic Accountability Act of 201910 requires businesses to evaluate risks associated with algorithm fairness and bias.11 Nevertheless, regulating algorithm biases in healthcare remains a difficult task. Eminent cases of algorithm bias have been documented, for example, in facial recognition and natural language processing (NLP) algorithms. Facial recognition systems, for instance, that are being increasingly utilized in law enforcement often perform poorly in recognizing faces of women and Black individuals.12–14 In NLP, language representations have been shown to capture human-like gender and other biases.15–17
These issues are relevant in the healthcare domain, with different caveats at the bench and at the bedside. The demographics (eg, ethnic, racial) of the patients used to train algorithms are often unknown for external evaluation.8 As a result, algorithms have been observed to produce inferior performance in detecting melanoma and health risk estimation in disadvantaged poorer African-American populations.7,18,19 Such biases in healthcare may be caused by missing data (eg, higher rates of missingness in minority populations due to decreased access to healthcare or lower healthcare utilization), observational error, misapplication, and overfitting due to small sample sizes or limited population and practice heterogeneity.5,20–22
In general, bias in AI/ML can be categorized under statistical and social. Statistical bias, which is common in predictive algorithms, refers to algorithmic inaccuracies in producing estimates that significantly differ from the underlying truth. Social bias embodies systemic inequities in care delivery leading to suboptimal health outcomes for certain populations.23 Social bias can underly statistical bias. In healthcare, we could have a third category of “latent” biases, which encompasses increases in social or statistical biases over time due to the complexities of the healthcare processes.5
Despite the eminent work in other fields, bias often remains unmeasured or partially measured in healthcare domains. Most published research articles only provide information about very few performance metrics—mostly through measures of algorithm’s discrimination power, such as the area under the receiving operating characteristics curve (AUROC). The few studies that officially aim at addressing bias, usually utilize single measures (eg, model calibration7) that do not portray a full picture of the story on bias. Addressing bias in medical AI requires a framework for a holistic search for bias, which can invigorate follow-up investigations to identify the underlying roots of bias.
The COVID-19 pandemic resulted in the generation of new data and data infrastructures related to both pandemic illness and healthcare more broadly. In this article, we evaluate unrecognized statistical and latent biases from multiple perspectives using a set of AI prediction models developed and validated retrospectively during the first 6 months of the COVID-19 pandemic. These models predict risks of mortality, hospitalization, ICU admission, and ventilation due to COVID-19 infection.24 We characterize the evaluation of bias into model-level metrics and propose a new approach for evaluating bias from an individual level. Here, we provide a framework for a holistic search for bias in medical AI that the user/model developer can utilize to ensure her search of bias (1) includes multiple aspects of technical and practical considerations and (2) can invigorate follow-up investigations for identifying the underlying roots of bias, rather than providing a partial perspective that may not lead to constructive improvement.
METHODS
We study unrecognized bias in 4 validated prediction models of COVID-19 outcomes to investigate whether (1) the models were biased when developed (we refer to this as a retrospective evaluation) and (2) the bias changed over time when applying the models on new COVID-19 patients who were infected after the models were trained (we refer to this as a prospective evaluation).
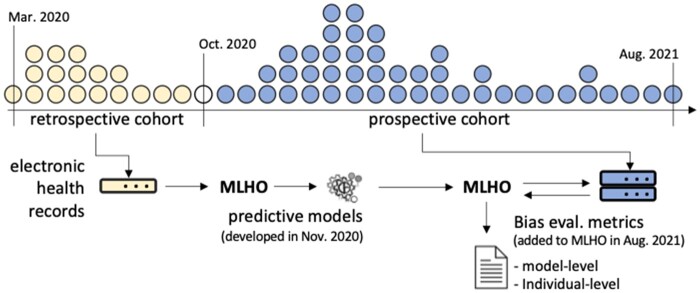
We recently developed the thinkin’ Machine Learning pipeline for modeling Health Outcomes (MLHO), for predicting risks of hospital admission, ICU admission, invasive ventilation, and death in patients who were infected with COVID-19, only using the data from prior to the COVID-19 infection.24,25 MLHO models were developed on data from the first 6 months of the pandemic in Boston—that is, between March and October 2020. MLHO produces and evaluates several models using different classification algorithms and train-test sampling iterations—for more details see ref.24 For each outcome, MLHO developed several models using different classification algorithms and/or train-test sampling. To evaluate possible unrecognized bias across patient sub-groups and time while developing the models retrospectively, we first select the top 10 models for each outcome based on their retrospective AUROC—that is, the discrimination metric obtained on the test set when the models were tested retrospectively. Then we apply the models to data from the retrospective cohort (who were infected with COVID-19 after the models were trained) to evaluate retrospective bias as a baseline. In addition to retrospective evaluations, we also perform prospective bias evaluations by applying these models to patient data from the subsequent 10 months to evaluate temporal changes in discrimination, accuracy, and reliability metrics (Figure 1). We evaluate bias by race, ethnicity, gender (biological sex), and across time, by comparing the multiple bias metrics against the overall models, which were trained on all patients.
Figure 1.
Generating bias metrics from MLHO models using EHR data from retrospective and prospective COVID-19 cohorts. *The dot plot is a schematic of the COVID-19 patient population over time. MLHO was applied to EHR data from the retrospective cohort to develop predictive models and produce bias metrics. Prospective bias metrics were generated by applying the retrospective predictive models to prospective cohorts.
DATA
Data from 56 590 Mass General Brigham (MGB) patients, with a positive reverse transcription-polymerase chain reaction (RT-PCR) test for SARS-CoV-2 between March 2020 and September 2021 were analyzed (Supplementary Table S1). Features utilized in MLHO models included transitive sequential patterns,26,27 where we mined sequences of EHR diagnoses, procedures, and medications extracted from these patients’ electronic health records between 2016 and 14 days before their positive reverse transcription-polymerase chain reaction (RT-PCR) test.
Measuring bias
There are multiple metrics for measuring bias/fairness in the broader AI community. To measure bias in binary clinical predictive models, we adapt the definitions in refs28–30 where an unbiased algorithm reflects the same likelihood of the outcome, irrespective of the individual’s group membership, . This definition is referred to as “test fairness” in Mehrabi et al.28 Thus, in an unbiased predictive model, for any predicted probability score , people in all groups have equal probability of correctly belonging to the positive class—for example, .
MLHO’s performance metrics
MLHO is equipped with functionality to provide a comprehensive evaluation of model performance from different standpoints, including both model-level and individual-level bias.
Model-level metrics
The model-level performance metrics in MLHO provide an overall description of the model’s performance, including standard metrics for discrimination, accuracy, and reliability (a.k.a., calibration). For discrimination, in this study, we use the widely used AUROC. Several model-level metrics are also available to evaluate the model’s accuracy such as the Brier score,31 which is the mean squared error between the observed outcome and the estimated probabilities for the outcome, including components of both discrimination and calibration.32 As a demonstration, we break down the AUC and Brier metrics retrospectively in aggregate, and prospectively by month. To statistically compare model-level metrics across patient sub-groups, we apply the Wilcoxon rank-sum test with Benjamini, Hochberg, and Yekutieli P-value correction.33
Reliability is a key factor in AI/ML models’ utility in clinical care, which is also known as calibration. Reliability refers to the extent to which the observed value of an outcome matches the risk score produced by a predictive model.7,29 Several measures have been recommended for measuring model calibration in binary classifiers. For a review of the available techniques, see Huang et al.34 However, many medical AI/ML models developed in healthcare settings ignore reliability and only report discrimination power although the AUROC, also known as the concordance statistic or c-statistic.35 MLHO’s performance report provides the ability to assess the models’ reliability for clinical interpretation using diagnostic reliability diagrams. The diagnostic reliability diagrams are produced from the raw predicted probabilities computed by each algorithm (X-axis) against the true probabilities of patients falling under probability bins (Y-axis). In a reliable model, the reliability diagrams appear along the main diagonal—the closer to the line, the more reliable. To evaluate diagnostic reliability diagrams, we compare the retrospective performance with aggregated prospective performance—that is, we do not break down this measure by month prospectively.
Individual-level metric
In contrast to model-level metrics that provide an overall description of the model’s performance, MLHO also provides the capability for evaluating model performance at an individual level, when the variable of interest is numeric (vs categorical). This is important when assessing whether a model is biased against an individual, for example, an older patient or a sicker patient (ie, having more medical encounters). To do that, MLHO computes and records the mean absolute error (MAE) for each patient that can be visualized to illustrate changes across numeric indices of interest. MAE is the absolute distance between the computed probability of the outcome to the actual outcome.
Where is the number of models (10 models here), is the predicted probability for patient and is the observed outcome for patient .
To visualize the MAE patterns, we plot the numeric variables (in this study, age) on the X-axis and the MAE on the Y-axis and fit a generalized additive model (GAM) with integrated smoothness36 from R package.37
RESULTS
Data from 56 590 patients with a positive COVID test were analyzed. Over 15 000 of these patients constituted the retrospective cohort—that is, whose data were used to train and test the retrospective models. More than 41 000 of the patients were infected between November 2020 and August 2021, who composed our prospective cohort. Supplementary Figure S1 and Table S1 provide a demographic breakdown of the patient population over time.
Model-level evaluations
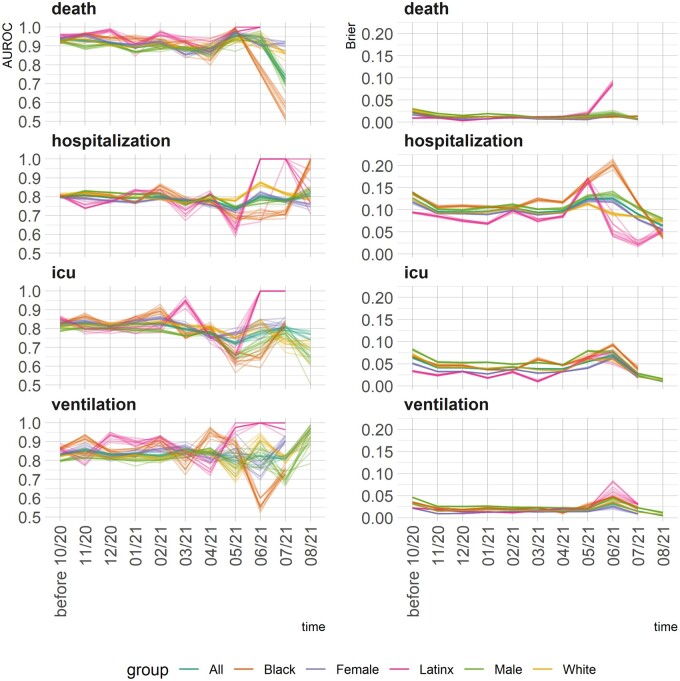
Figures 2 and 3 illustrate temporal changes in the AUROC and Brier scores across the models. The 2 figures demonstrate complimentary information. Where Figure 2 shows change over time and across groups, Figure 3 visualizes a non-parametric test of the equality of means in 2 independent samples. Unlike AUROC, A higher Brier score means lower accuracy. The models’ performance metrics remained stable until June 2021. That is, the models that were developed with data from March to September 2020 were still able to perform similarly up until May–June 2021. Starting June 2021, both AUROC and Brier scores exhibit variabilities, in general providing better discrimination power for Latinx and female COVID-19 patients compared with male patients. In other words, the models did not demonstrate temporal bias until June 2021, when applied prospectively.
Figure 2.
Changes in the 2 model-level metrics for discrimination (AUROC—left panels) and error (Brier score—right panel) by group and over time. *The top-10 models for each outcome are broken down by race, ethnicity, gender, and over time.
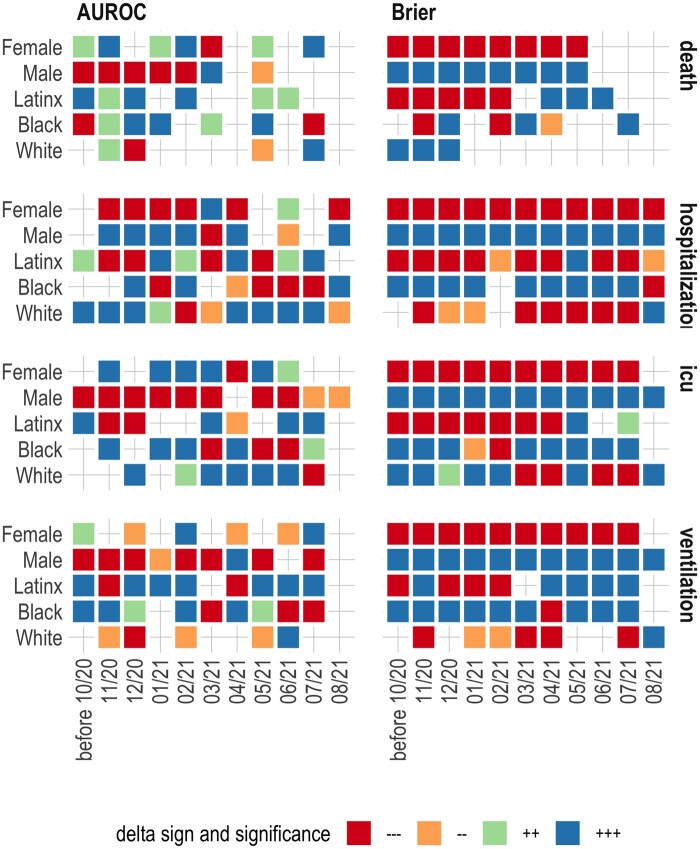
Figure 3.
Comparing model-level performance metrics using the Wilcoxon rank-sum test. *A color-coded cell means some type of bias compared to the overall model. −−− and −− represent significantly smaller than the overall model (at P < .001 and P < .01, respectively). +++ and ++ represent significantly larger than the overall model (at P < .001 and P < .01, respectively). **Discrimination power and error are opposing measures—better discrimination means smaller error.
The models that were developed with data from March to September 2020, provided relatively stable predictive performance prospectively up until May–June 2021. Despite the increased variability, the prospective modeling performance remained high for predicting hospitalization and the need for mechanical ventilators.
To facilitate understanding Figure 2, we provide Figure 3 in which we compare model-level performance metrics using the Wilcoxon rank-sum test. The figure combines an illustration of statistical significance and sign for comparing a given metric for a demographic group to the overall model at a point in time. For example, +++ under AUROC for the female patients in November 2020 means that the AUROC was higher for females compared with the overall model and the difference was statistically significant at P < .001.
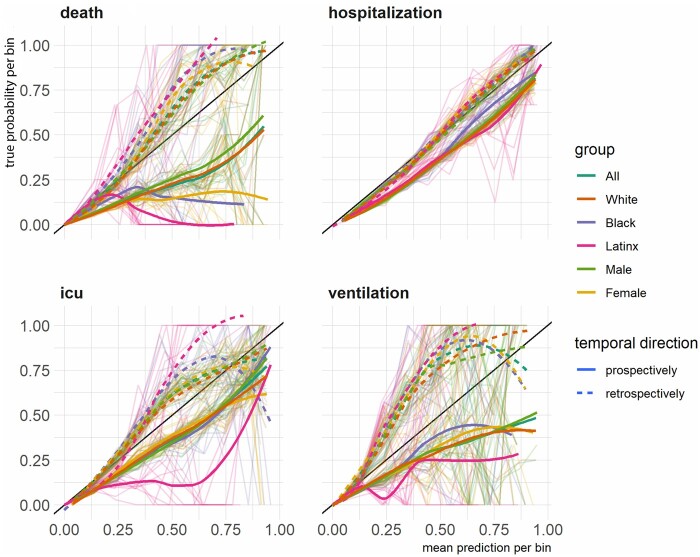
Figure 4 presents diagnostic reliability diagrams, broken down by demographic group and temporal direction of the evaluation (retrospective vs prospective). Any divergence from the diagonal line in the diagnostic reliability diagrams means lower reliability. The diagrams show that, retrospectively, models’ predicted probabilities were similar across groups for predicting mortality, hospitalization, and ventilation. Prospectively, between-group variability in models fared similarly, although the uncalibrated predicted probabilities were less reliable for mortality prediction, specifically, among Latinx, Black, and Female patients.
Figure 4.
The diagnostic reliability (calibration) diagrams for each outcome broken by group and temporal direction. *The background lines represent reliability curves from each of the top models selected for prospective evaluation.
Compared to the overall population, retrospectively and prospectively across time, the models marginally performed worse for male patients and better for Latinx and female patients, as measured by AUROC and higher Brier scores. We use the term “marginal” as the range of delta between performance metrics within demographics groups and the overall model was relatively small. For the rest of the demographic groups, the performances were more mixed. From the diagnostic reliability diagrams, the divergence from the diagonal line is present in 3 of the 4 prediction tasks, but there are variabilities across groups in both with no consistent pattern. The only exception in this regard was diminished reliability in prospectively predicted probabilities of COVID-19 mortality among Latinx, female, and Black patients.
Individual-level evaluation
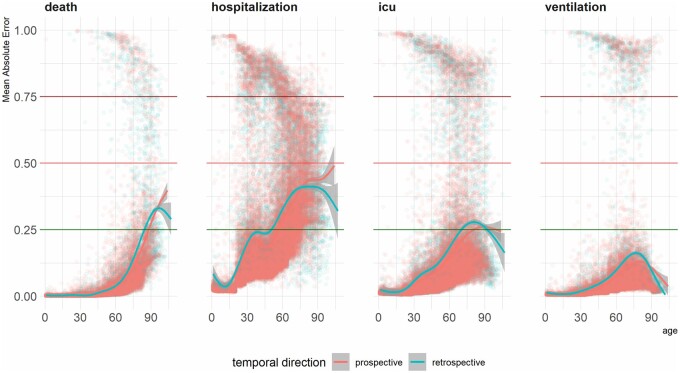
For the individual-level evaluation of the bias, we looked at the mean absolute error across age (Figure 5). We evaluated whether the models’ average error rates (ie, the absolute difference between the actual outcome and the predicted probabilities) change as patients’ age increases. Conventionally assuming 0.5 as the operating point for assigning patients to positive/negative groups, an MAE smaller than 0.5 would indicate that the model predicted probability was not far off from the actual outcome. For example, the patient could have the outcome and the computed probability would be above 50% and therefore the MAE would be smaller than 0.5. To visualize the trends, we fit a smoothed trendline using generalized additive models. For all outcomes, modeling error seemed to increase as the patients became older, and the patterns were almost identical retrospectively and prospectively. None of the trend lines passed the 0.5 threshold, which means despite the higher error rates for the older patients, the models provide acceptable errors for most of the patients. The lowest error rates were observed in predicting ventilation. In the case of predicting mortality and hospitalization, the error rates increasingly escalated by age, whereas in predicting ICU admission and need for mechanical ventilator, error rates peaked at around 75 years and then diminished for older patients.
Figure 5.
Mean absolute error of predictions across patient age.
DISCUSSION
From a model-level perspective, we did not find consistent biased behaviors in predictive models against all underrepresented groups. From the individual-level, we found consistent bias in increasing error rates for older patients. It is known that a predictive model’s reliability (calibration) and discrimination cannot both be maximized simultaneously.35 That is, for example, improving reliability may not meaningfully improve discrimination.38 Yet, there are ad hoc calibration methodologies to scale predicted probabilities for better clinical interpretation. We argue that proper evaluation of bias in medical AI requires a holistic framework (which we provide here) that can invigorate follow-up investigations for identifying the underlying roots of bias, rather than providing a partial perspective that may not lead to constructive improvements.
Compared to the overall population, retrospectively and prospectively across time, the models marginally performed worse for male patients and better for Latinx and female patients, as measured by AUROC and Brier scores. The range of delta between these performance metrics within demographics groups and the overall model was relatively small. For the rest of the demographic groups, the performances were more mixed. The models’ performance metrics remained stable until June 2021. That is, the models that were developed with data from March to September 2020, provided relatively stable predictive performance prospectively up until May–June 2021. Despite the increased variability, the prospective modeling performance remained high for predicting hospitalization and the need for mechanical ventilators.
COVID-19 vaccinations became widely available in the spring of 2021. It is possible that the widespread use of vaccinations throughout Massachusetts, along with the incorporation of other proven therapies including dexamethasone39 and Remdesivir,40 changed outcomes for patients. Also, the case rate in Massachusetts was very low in July,41 which may have resulted in increased capacity compared to the outset of the pandemic when the healthcare system was stressed. Additionally, the Delta variant was expected to be the dominant strain of Coronavirus in Massachusetts.42 The mutations to the virus itself could potentially change outcomes for patients. While we do not know exactly what led to the decreased performance of the model in July, future studies should consider characterizing whether the model overestimates or underestimates an outcome in certain populations, which could give further insight into how these changes are cumulatively having a favorable or adverse impact on patient care.
From the reliability/calibration perspective, except in the case of prospective evaluation of mortality predictions among Latinx, female, and Black patients, the diagnostic reliability diagrams did not show consistent bias towards or against a certain group.
In terms of the mean absolute error between the actual outcome and the estimated probabilities, we did see error rates increase over age, but the error rates were not critical in that one could still assign the patients to the correct group based on the produced probabilities. Although the age-based analysis is reported as a summarization of a group-level trend, the individual-level metric enables further investigation of modeling error in user-defined sub-groups of the patient population. More numeric metrics need to be evaluated at the patient level for a comprehensive view of changes in AI bias against or towards certain patients.
To an AI algorithm, bias can happen due to the signal strength (or lack thereof) in one or more of the features (ie, variables, covariates, predictors). That is, the model which has been trained on a certain predictor may not predict well for a certain protected group because the important predictors are not available or are noisy in that population. This, in turn, could have multiple underlying causes, such as healthcare disparities that can influence access to care, systematic inequalities, data quality issues, biological factors, and/or socio-economic and environmental determinants. Some of this bias can be addressed by post-processing techniques, depending on which aspect of bias one aims to address. We concluded that medical AI bias is multi-faceted and requires multiple perspectives to be practically addressed. Nevertheless, the first step for addressing the bias in medical AI is to identify bias in a way that can be traced back to its root.
We evaluated raw predicted scores. There is a large body of work on calibrating prediction scores for improving the reliability of prediction models in clinical settings.43–46 Calibration methods are useful ad hoc solutions for increasing the reliability of the prediction models. We show in Supplementary Figure S3 that isotonic calibration,47 for instance, can provide more reliable predictive scores and may reduce bias. However, unless calibration methods are embedded into a predictive modeling pipeline, their impact on improving or aggravating bias in medical AI needs to be fully evaluated as a post-processing step.
Given that we face systemic bias in our country’s core institutions, we need technologies that will reduce these disparities and not exacerbate them.48 There are efforts from the larger AI community, such as AI Fairness 36049 and Fairlearn,50 to develop open-source software systems for measuring and mitigating bias. These programs are often ad hoc or work as standalone post-processing solutions. We plan to compare these model independent methods and add relevant functionalities to our domain-specific approach.
The premise for evaluating these predictive models was to create a framework for discovering and quantifying the various types of biases towards different sub-groups that were encoded unintentionally. The population sub-groups selected by the model evaluator can include minority ethno-racial groups or intersectional sub-groups, such as Black women. We have incorporated the presented bias measurement framework within the MLHO pipeline,24 which is specifically designed for modeling clinical data. We hope that providing means to evaluate such unrecognized biases within a data-centric modeling pipeline will enable the generation of medical AI that considers various biases while in development and addresses them in production.
AUTHOR CONTRIBUTIONS
Conceptualization, HE; Methodology, HE; Formal Analysis, HE; Investigation, HE and ZHS. Writing—Original Draft, HE and ZHS; Writing—Review & Editing, HE, ZHS, SR, JGK, THM, KBW, and SNM.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTERESTS STATEMENT
None declared.
DATA AVAILABILITY
Protected Health Information restrictions apply to the availability of the clinical data here, which were used under IRB approval for use only in the current study. As a result, this dataset is not publicly available. Qualified researchers affiliated with the Mass General Brigham (MGB) may apply for access to these data through the MGB Institutional Review Board.
CODE AVAILABILITY STATEMENT
The R code to perform this analysis is available at https://hestiri.github.io/mlho.
Supplementary Material
Contributor Information
Hossein Estiri, Laboratory of Computer Science, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA.
Zachary H Strasser, Laboratory of Computer Science, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA.
Sina Rashidian, Verily Life Sciences, Boston, Massachusetts, USA; Massachusetts General Hospital, Boston, MA 02114, USA.
Jeffrey G Klann, Laboratory of Computer Science, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA; Research Information Science and Computing, Mass General Brigham, Somerville, Massachusetts, USA.
Kavishwar B Wagholikar, Laboratory of Computer Science, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA.
Thomas H McCoy, Jr, Center for Quantitative Health, Massachusetts General Hospital, Boston, Massachusetts, USA.
Shawn N Murphy, Laboratory of Computer Science, Massachusetts General Hospital, Boston, Massachusetts, USA; Research Information Science and Computing, Mass General Brigham, Somerville, Massachusetts, USA; Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA; Department of Neurology, Massachusetts General Hospital, Boston, Massachusetts, USA.
REFERENCES
- 1. Vayena E, Blasimme A, Cohen IG. Machine learning in medicine: addressing ethical challenges. PLoS Med 2018; 15 (11): e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Char DS, Shah NH, Magnus D. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med 2018; 378 (11): 981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moratinos GL, Lazcoz Moratinos G, de Miguel Beriain I. Big data analysis and machine learning in intensive care medicine: identifying new ethical and legal challenges. Med Intensiva (Engl Ed) 2020; 44 (5): 319–20. [DOI] [PubMed] [Google Scholar]
- 4. Hajjo R. The ethical challenges of applying machine learning and artificial intelligence in cancer care. In: 2018 1st International Conference on Cancer Care Informatics (CCI). 2018. doi: 10.1109/cancercare.2018.8618186. [DOI]
- 5. DeCamp M, Lindvall C. Latent bias and the implementation of artificial intelligence in medicine. J Am Med Inform Assoc 2020; 27 (12): 2020–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chouldechova A, Roth A. A snapshot of the frontiers of fairness in machine learning. Commun ACM 2020; 63 (5): 82–9. [Google Scholar]
- 7. Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019; 366 (6464): 447–53. [DOI] [PubMed] [Google Scholar]
- 8. Noor P. Can we trust AI not to further embed racial bias and prejudice? BMJ 2020; 368: m363. [DOI] [PubMed] [Google Scholar]
- 9. Adamson AS, Smith A. Machine learning and health care disparities in dermatology. JAMA Dermatol 2018; 154 (11): 1247–8. [DOI] [PubMed] [Google Scholar]
- 10. Clarke YD. Algorithmic Accountability Act of 2019. 2019. https://www.congress.gov/bill/116th-congress/house-bill/2231. Accessed July 20, 2021.
- 11. Floridi L, Cowls J, Beltrametti M, et al. AI4People—an ethical framework for a good AI society: opportunities, risks, principles, and recommendations. Minds Mach (Dordr) 2018; 28 (4): 689–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klare BF, Burge MJ, Klontz JC, Vorder Bruegge RW, Jain AK. Face recognition performance: role of demographic information. IEEE Trans Inf Forensic Secur 2012; 7 (6): 1789–801. [Google Scholar]
- 13. O’Toole AJ, Phillips PJ, An X, Dunlop J. Demographic effects on estimates of automatic face recognition performance. Image Vis Comput 2012; 30 (3): 169–76.
- 14. Hupont I, Fernandez C. DemogPairs: quantifying the impact of demographic imbalance in deep face recognition. In: 2019 14th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2019). 2019. doi: 10.1109/fg.2019.8756625. [DOI]
- 15. Caliskan A, Bryson JJ, Narayanan A. Semantics derived automatically from language corpora contain human-like biases. Science 2017; 356 (6334): 183–6. [DOI] [PubMed] [Google Scholar]
- 16. Aran XF, Such JM, Criado N. Attesting Biases and Discrimination using Language Semantics. arXiv [cs.AI] 2019. https://arxiv.org/abs/1909.04386.
- 17. Rice JJ, Norel R. Faculty Opinions recommendation of Semantics derived automatically from language corpora contain human-like biases. Faculty Opinions—Post-Publication Peer Review of the Biomedical Literature. 2017; doi:10.3410/f.727506427.793532942.
- 18. Rajkomar A, Hardt M, Howell MD, Corrado G, Chin MH. Ensuring fairness in machine learning to advance health equity. Ann Intern Med 2018; 169 (12): 866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cormier JN, Xing Y, Ding M, et al. Ethnic differences among patients with cutaneous melanoma. Arch Intern Med 2006; 166 (17): 1907–14. [DOI] [PubMed] [Google Scholar]
- 20. Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial intelligence: practical primer for clinical research in cardiovascular disease. J Am Heart Assoc 2019; 8 (17): e012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Jimenez F, Attia Z, Arruda-Olson AM, et al. Artificial intelligence in cardiology: present and future. Mayo Clin Proc 2020; 95 (5): 1015–39. [DOI] [PubMed] [Google Scholar]
- 22. Tat E, Bhatt DL, Rabbat MG. Addressing bias: artificial intelligence in cardiovascular medicine. Lancet Digit Health 2020; 2 (12): e635–36. [DOI] [PubMed] [Google Scholar]
- 23. Parikh RB, Teeple S, Navathe AS. Addressing bias in artificial intelligence in health care. JAMA 2019; 322 (24): 2377–8. [DOI] [PubMed] [Google Scholar]
- 24. Estiri H, Strasser ZH, Murphy SN. Individualized prediction of COVID-19 adverse outcomes with MLHO. Sci Rep 2021; 11 (1): 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Estiri H, Strasser ZH, Klann JG, et al. Predicting COVID-19 mortality with electronic medical records. NPJ Digit Med 2021; 4 (1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Estiri H, Vasey S, Murphy SN. Transitive sequential pattern mining for discrete clinical data. In: Michalowski M, Moskovitch R, eds. Artificial Intelligence in Medicine. Cham: Springer International Publishing; 2020: 414–24. [Google Scholar]
- 27. Estiri H, Strasser ZH, Klann JG, et al. Transitive sequencing medical records for mining predictive and interpretable temporal representations. Patterns (N Y) 2020; 1 (4): 100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. A survey on bias and fairness in machine learning. ACM Comput Surv 2021; 54 (6): 1–35. [Google Scholar]
- 29. Chouldechova A. Fair prediction with disparate impact: a study of bias in recidivism prediction instruments. Big Data 2017; 5 (2): 153–63. [DOI] [PubMed] [Google Scholar]
- 30. Verma S, Rubin J. Fairness definitions explained. In: 2018 IEEE/ACM International Workshop on Software Fairness (FairWare). 2018:1–7.
- 31. Brier GW. Verification of forecasts expressed in terms of probability. Mon Wea Rev 1950; 78 (1): 1–3. [Google Scholar]
- 32. Walsh CG, Sharman K, Hripcsak G. Beyond discrimination: a comparison of calibration methods and clinical usefulness of predictive models of readmission risk. J Biomed Inform 2017; 76: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat 2001; 29: 1165–88. [Google Scholar]
- 34. Huang Y, Li W, Macheret F, Gabriel RA, Ohno-Machado L. A tutorial on calibration measurements and calibration models for clinical prediction models. J Am Med Inform Assoc 2020; 27 (4): 621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Calster B, McLernon DJ, van Smeden M, et al. ; Topic Group ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative. Calibration: the Achilles heel of predictive analytics. BMC Med 2019; 17 (1): 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wood S. Generalized Additive Models: An Introduction with R. 2nd ed. New York: CRC Press; 2017. [Google Scholar]
- 37. Wood S. Package ‘mgcv’. 2021.
- 38. Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem 2008; 54 (1): 17–23. [DOI] [PubMed] [Google Scholar]
- 39.RECOVERY Collaborative Group, Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med 2021; 384: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 2020; 383 (19): 1813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massachusetts Coronavirus Map and Case Count. The New York Times. 2020.
- 42. Markos M. Delta Variant Taking Over as Dominant Strain in Mass., Experts Say. NBC10 Boston. 2021. https://www.nbcboston.com/news/coronavirus/delta-variant-taking-over-as-dominant-strain-in-mass-experts-say/2423599/. Accessed September 24, 2021.
- 43. Benevenuta S, Capriotti E, Fariselli P. Calibrating variant-scoring methods for clinical decision making. Bioinformatics 2021; 36 (24): 5709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alba AC, Agoritsas T, Walsh M, et al. Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA 2017; 318 (14): 1377–84. [DOI] [PubMed] [Google Scholar]
- 45. Van Calster B, Vickers AJ. Calibration of risk prediction models: impact on decision-analytic performance. Med Decis Making 2015; 35 (2): 162–9. [DOI] [PubMed] [Google Scholar]
- 46. Holmberg L, Vickers A. Evaluation of prediction models for decision-making: beyond calibration and discrimination. PLoS Med 2013; 10 (7): e1001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mair P, Hornik K, de Leeuw J. Isotone optimization in R: pool-adjacent-violators algorithm (PAVA) and active set methods. J Stat Softw 2009; 32: 1–24. [Google Scholar]
- 48. Kaushal A, Altman R, Langlotz C. Health care AI systems are biased. Scientific American 2020.
- 49. Bellamy RKE, Mojsilovic A, Nagar S, et al. AI fairness 360: an extensible toolkit for detecting and mitigating algorithmic bias. IBM J Res Dev 2019; 63 (4/5): 4:1–4:15. [Google Scholar]
- 50. Bird S, et al. Fairlearn: a toolkit for assessing and improving fairness in AI. Microsoft, Tech. Rep. MSR-TR-2020-32. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protected Health Information restrictions apply to the availability of the clinical data here, which were used under IRB approval for use only in the current study. As a result, this dataset is not publicly available. Qualified researchers affiliated with the Mass General Brigham (MGB) may apply for access to these data through the MGB Institutional Review Board.