Abstract
Gastric cancer (GC) is an aggressive malignancy with a high mortality rate and poor prognosis, primarily caused by metastatic lesions. Improved understanding of GC metastasis at the molecular level yields meaningful insights into potential biomarkers and therapeutic targets. Covalently closed circular RNAs (circRNAs) have emerged as crucial regulators in diverse human cancers including GC. Furthermore, accumulating evidence has demonstrated that circRNAs exhibit the dysregulated patterns in GC and have emerged as crucial regulators in GC invasion and metastasis. However, systematic knowledge regarding the involvement of circRNAs in metastatic GC remains obscure. In this review, we outline the functional circRNAs related to GC metastasis and drug resistance and discuss their underlying mechanisms, providing a comprehensive delineation of circRNA functions on metastatic GC and shedding new light on future therapeutic interventions for GC metastases.
Keywords: circRNA, Gastric cancer, Metastasis, miRNA sponge, RNA binding protein, Drug resistance
Background
Gastric cancer (GC) is an aggressive and heterogeneous malignancy [1, 2]. With a median overall survival (OS) of 16 months among all patients, GC remains the fourth leading cause of cancer-related mortality worldwide [1–3]. Metastasis is a crucial process characterized by increased invasion and the ability of cancer to spread from its site of origin to other regions of the body, accounting for 90% of cancer-related deaths [4, 5]. Most GC patients are diagnosed at advanced stages and are frequently accompanied by invasion and metastasis, such as lymph node and peritoneum metastases [6, 7]. In metastatic (late) GC patients, the clinical outcomes are extremely poor, while the 5-year overall survival rate of early GC patients can reach over 90% [4, 7]. In addition, metastatic GC has long been considered less effective for surgical treatment and more resistant to drug therapy [8, 9]. Up to date, no effective methods or approaches are applied to treat metastatic GC [8, 9]. Recently, significant advances have been made in clarifying GC metastasis [5, 10]; however, the overall delineation of the molecular mechanisms is limited and ambiguous. Therefore, an in-depth understanding of GC metastasis at the molecular and cellular levels is imperative to identify potential biomarkers for diagnosis and therapeutic targets for intervention.
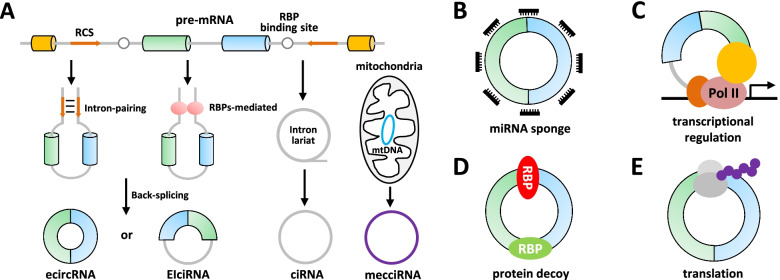
Covalently closed circular RNAs (circRNAs) are single-stranded endogenous RNA molecules with loop structures and are resistant to exonuclease activity [11–13]. The biogenesis of circRNAs is widely acknowledged via a back-splicing event from precursor RNA (pre-RNA), which is facilitated by the flanking reverse complementary sequences, such as Alu elements, and is regulated by some RNA binding proteins (RBPs), including QKI, DHX9, FUS, Sam68, hnRNP L, hnRNPM and ADARs (Fig. 1A) [14–23].
Fig. 1.
Biogenesis and function of circRNAs. A. The flanking introns of circularized exons contain reverse complementary sequences, which form a circular structure through direct base pairing, or RBP binding sites, which generate a circular structure via RBPs dimerization. The introns are removed or retained to form exonic circRNA (ecircRNA) or exon-intron circRNA (EIciRNA). Intronic circRNA (ciRNA) is derived from an intron by preventing intron debranching after splicing. Mitochondria encoded circRNAs (mecciRNAs) probably circularize via a splicing-independent mechanism. B. CircRNAs serving as miRNA sponges. C. CircRNAs promoting the transcription of targets through interacting with proteins or complexes such as U1 snRNP. D. CircRNAs behaving as protein decoys. E. CircRNAs working via generating polypeptides
Thousands of circRNAs across species have been identified and characterized through high-throughput sequencing combined with bioinformatic analyses in the past decade [24, 25]. Most circRNAs are chiefly derived from known protein-coding genes, consist of a single or multiple exon(s) (exonic circRNAs, ecircRNAs), and generally localize to the cytoplasm [13]. The most prominent function of cytoplasmic ecircRNAs is to serve as competing endogenous RNAs (ceRNAs) or miRNA sponges to lift the inhibitory effects of miRNAs on their downstream targets (Fig. 1B) [24, 26–28]. Interestingly, the intronic sequences between the circularized exons may be retained, forming exon-intron circRNAs (EIciRNAs) [29]. EIciRNAs are proved to enhance their parental gene expressions in cis via binding to the U1 small nuclear ribonucleoprotein (snRNP) complex in the nucleus (Fig. 1C) [29]. Intronic lariat precursors escaping from debranching produce intronic circRNAs (ciRNAs), which could regulate RNA polymerase II (Pol II)-mediated transcription in the nucleus [30, 31]. Besides, circRNAs directly interact with RBPs to regulate key targets as protein scaffolds or antagonists in various biological processes as well (Fig. 1D) [32–34]. In addition, a small fraction of ecircRNAs undergoes cap-independent translation to encode small peptides through the internal ribosome entry site (IRES)-driven mechanisms, although the vast majority of circRNAs are thought to be non-coding RNAs (Fig. 1E) [35–37]. Recently, a novel class of circRNAs encoded by mitochondria (mecciRNAs) has been reported to facilitate the mitochondrial entry of nuclear-encoded proteins by serving as molecular chaperones [38].
Accumulating evidence has pointed out the aberrant expression patterns of circRNAs and their regulatory roles in cancer progression and metastasis [39–44]. Systematic and comprehensive knowledge regarding circRNAs related to GC metastasis expands our understanding of the underlying mechanisms of metastatic GC. In the present review, we overview the current research status of circRNAs in GC metastasis, including modulating epithelial-mesenchymal transition (EMT), regulating angiogenesis, exosomal circRNAs, and drug resistance, and discuss the potential clinical application value of circRNAs in GC. We hope to provide insights into circRNAs-mediated GC metastasis and their potential as putative biomarkers or therapeutic targets of GC in the future.
CircRNAs participate in EMT
EMT, a highly complex and dynamic process, is recognized as a vital step driving the early phase of cancer metastasis [45, 46]. Recently, several circRNAs have been reported to participate in EMT by modulating various signaling pathways, such as TGF-β/SMAD, Wnt/β-catenin, and PI3K/AKT pathways [47]; thereby, we summarized up-to-date information on circRNAs engaged in these signaling pathways in GC metastasis (Table 1).
Table 1.
A list of circRNAs related to GC metastasis
| CircRNA | CircBase ID | Expression | Property in metastasis | Molecular mechanism | Refs |
|---|---|---|---|---|---|
| circTHBS1 | hsa_circ_0034536 | Up | Enhancer | Modulate the miR-204-5p/INHBA axis and interact with the RBP, HuR | [48] |
| circCCDC66 | hsa_circ_0001313 | Up | Enhancer | Activate c-Myc/TGF-β signaling pathway | [49] |
| circ_0001829 | hsa_circ_0001829 | Up | Enhancer | Sponge miR-155-5p to upregulate SMAD | [50] |
| circOXCT1 | hsa_circ_0004873 | Down | Repressor | Sponge miR-136 to upregulate SMAD4 | [51] |
| circAXIN1 | hsa_circ_0005838 | Up | Enhancer | Encode a novel protein, AXIN1-295aa | [52] |
| circFGD4 | hsa_circ_0000390 | Down | Repressor | Sponge miR-532-3p to upregulate APC | [53] |
| circREPS2 | hsa_circ_0139996 | Down | Repressor | Sponge miR-558 to upregulate RUNX3 | [54] |
| circAKT3 | hsa_circ_0000199 | Up | Enhancer | Sponge miR-198 to upregulate PIK3R1 | [55] |
| circ_0023409 | hsa_circ_0023409 | Up | Enhancer | Sponge miR-542-3p to upregulate IRS4 | [56] |
| ciRS-7 | hsa_circ_0001946 | Up | Enhancer | Sponge miR-7 to upregulate PTEN | [57] |
| circTNPO3 | hsa_circ_0001741 | Down | Repressor | Interact with the RBP, IGF2BP3 | [58] |
| circFNDC3B | hsa_circ_0006156 | Up | Enhancer | Interact with the RBP, IGF2BP3 | [59] |
| circ_100876 | hsa_circ_0023404 | Up | Enhancer | Sponge miR-665 to upregulate YAP1 | [60] |
| circPRRX1 | hsa_circ_0004370 | Up | Enhancer | Sponge miR-665 to upregulate YWHAZ | [61] |
| circRanGAP1 | hsa_circ_0063526 | Up | Enhancer | Regulate the miR-877-3p/VEGFA axis | [62] |
| circ_0044366 | hsa_circ_0044366 | Up | Enhancer | Sponge miR-29a to upregulate VEGF | [63] |
| circURI1 | hsa_circ_0000921 | Up | Repressor | Interact with the splicing factor hnRNPM | [64] |
| ebv-circLMP2A | - | Up | Enhancer | Form a positive feedback loop with HIF1α | [65] |
| circNRIP1 | hsa_circ_0004771 | Up | Enhancer | Sponge miR-149-5p to upregulate AKT1 | [66] |
| circNEK9 | hsa_circ_0032683 | Up | Enhancer | Sponge miR-409-3p to upregulate MAP7 | [67] |
| circRELL1 | hsa_circ_0001400 | Down | Repressor | Sponge miR-637 to upregulate EPHB3 | [68] |
| circSHKBP1 | hsa_circ_0000936 | Up | Enhancer | Modulate the miR-582-3p/HuR/VEGF axis and interact with HSP90 | [69] |
| circMRPS35 | hsa_circ_0000384 | Down | Repressor | Recruit the histone modifier, KAT7 | [70] |
| circMAPK1 | hsa_circ_0004872 | Down | Repressor | Encode a MAPK1-109aa protein | [71] |
| circRPL15 | hsa_circ_0064574 | Up | Enhancer | Sponge miR-502-3p to upregulate OLFM4 | [72] |
| circUBE2Q2 | hsa_circ_0005151 | Up | Enhancer | Modulate the miR-370-3p/STAT3 axis | [73] |
| circAGO2 | hsa_circ_0135889 | Up | Enhancer | Interact with the RBP, HuR | [74] |
| circHuR | hsa_circ_0049027 | Down | Repressor | Transcriptionally repression in cis | [75] |
TGF-β/SMAD signaling pathway
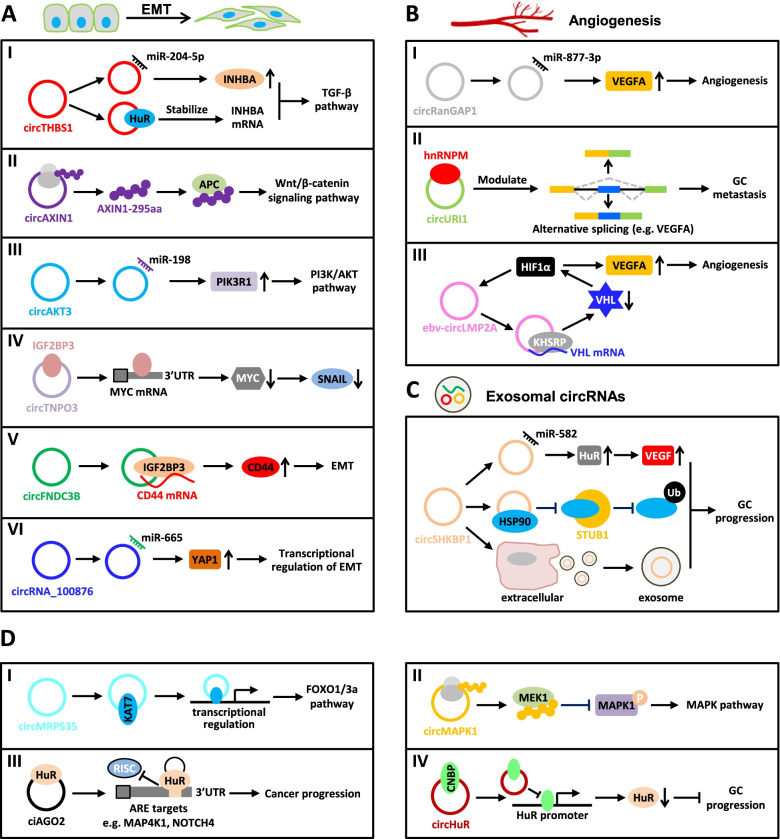
The TGF-β/SMAD signaling is a classic pathway in cancer metastasis [47]. The circRNA circTHBS1, which is highly expressed in GC and associated with poor prognosis, is reported to promote the malignant behaviors and EMT of GC cells by triggering the INHBA/TGF-β pathway [48]. Mechanistically, circTHBS1 behaves as a miR-204-5p sponge to enhance the INHBA expression, and it also stabilizes the INHBA mRNA mediated by HuR, consequently activating the TGF-β pathway (Fig. 2AI) [48]. The circCCDC66 expression is elevated in GC and related to tumor stage and lymphatic metastasis [49]. Gain- and loss-of-function studies have revealed that circCCDC66 promotes GC metastasis by activating c-Myc and the TGF-β signaling pathways [49]. In another case, hsa_circ_0001829 promotes GC cell migration and invasion in vitro and GC metastasis in vivo via modulating the miR-155-5p/SMAD axis [50]. A similar ceRNA mechanism also applies to circOXCT1, which interacts with miR-136 to relieve the repressive effect on its target SMAD4, inhibiting GC EMT and metastasis [51].
Fig. 2.
Molecular mechanisms of circRNAs related to GC metastasis. A. Roles of circRNAs in signaling pathways associated with EMT. I. The circRNA circTHBS1 increases the INHBA level via adsorbing miR-204-5p in a sponge form, and stabilizes the INHBA mRNA via sequestering HuR protein, leading to the activation of the TGF-β pathway. II. CircAXIN1-encoded a novel protein, AXIN1-295aa interacts with APC to activate the canonical Wnt/β-catenin signaling pathway. III. The circRNA circAKT3 activates the PI3K/AKT signaling by serving as a ceRNA against miR-198 to upregulate PIK3R1. IV. CircTNPO3 competitively binds to IGF2BP3, leading to the destabilization of the MYC mRNA, consequently repressing the expressions of MYC and its target SNAIL. V. The circFNDC3B level is significantly increased in GC and circFNDC3B interacts with IGF2BP3 protein and CD44 mRNA to form a ternary complex, resulting in the upregulation of CD44, which facilitates EMT in GC. VI. The circRNA circ_100876 interacts with miR-665 to relieve the repressive effect on its target YAP1, which is involved in the transcriptional activation of EMT-related genes. B. Roles of circRNAs engaged in angiogenesis. I. CircRanGAP1 is validated to stimulate angiogenesis via modulating the miR-877-3/VEGFA axis. II. CircURI1, a highly expressed circRNA in GC, sequesters the splicing factor, hnRNPM protein in a sequence-dependent manner to modulate alternative splicing of a subset of migration-related genes, such as VEGFA, consequently inhibiting GC metastasis. III. Ebv-circLMP2A promotes angiogenesis through forming a positive feedback loop with HIF1α to improve the VEGFA expression. Under hypoxia, HIF1α up-regulates ebv-circLMP2A, and ebv-circLMP2A interacts with KHSRP to destabilize the VHL mRNA, resulting in VHL down-regulation and HIF1α accumulation. C. Exosomal circRNA in GC. The circRNA circSHKBP1 promotes GC progression via the miR-582-3p/HuR/VEGF axis, and sequestering HSP90 to suppress STUB1-mediated HSP90 ubiquitination. Additionally, increased exosomal circSHKBP1 could facilitate co-cultured cell growth. D. Other pivotal pathways or targets involved in GC metastasis. I. The circRNA circMRPS35 inhibits GC tumorigenesis through the recruitment of histone acetyltransferase KAT7 to the promoters of FOXO1/3a genes, activating the FOXO1/3a transcription, consequently triggering the FOXO1/3a pathway. II. The circRNA circMAPK1 exerted an anti-tumor effect on GC invasion via generating a 109aa protein forming as a molecular sponge for MEK1, thus inhibiting the phosphorylation of MAPK1 and eventually leading to the inactivation of the MAPK pathway. III. The ciRNA circAGO2 interacts with HuR protein to promote its activation and enrichment on the 3’ UTR of HuR targets, resulting in repressing the AGO2/miRNA-mediated gene silencing involved in cancer progression. IV. The circRNA circHuR sequesters CNBP from the HuR’s promoter, leading to the repressions of HuR and GC progression
Wnt/β-catenin signaling pathway
The Wnt/β-catenin signaling pathway is indispensable among the pathways regulated by circRNAs in EMT [47, 52–54]. The circAXIN1 expression is significantly up-regulated in GC compared to the corresponding non-tumor gastric tissues [52]. Silencing of circAXIN1 suppresses GC cell proliferation, migration, and invasion, whereas the ectopic expression of circAXIN1 promotes GC malignancy in vitro and in vivo [52]. Mechanistically, a novel protein AXIN1-295aa encoded by circAXIN1 competes with parental AXIN1 protein to bind APC and release β-catenin, consequently activating the canonical Wnt/β-catenin signaling pathway to facilitate GC progression (Fig. 2AII) [52]. Additionally, Dai et al. have proposed that the circFGD4 expression is markedly attenuated in GC tissues and negatively correlated with lymphatic metastasis and the short prognosis of GC patients [53]. Furthermore, circFGD4 shows its anti-tumor effect on GC tumorigenesis and metastasis by modulating the miR-532-3p/APC/β-catenin axis [53]. Similarly, circREPS2 exhibits a decreased level in GC and inhibits GC migration and invasion via repression of the RUNX3/β-catenin pathway by sequestering miR-558 [54].
PI3K/AKT signaling pathway
The PI3K/AKT signaling pathway is frequently activated in EMT during metastasis and a series of dysregulated circRNAs have been found to interfere with this pathway [47, 55–57]. For example, GC-specific circAKT3 activates the PI3K/AKT signaling by repressing miR-198-mediated inhibition of PIK3R1, a regulatory subunit of PI3K (Fig. 2AIII) [55]. The circRNA hsa_circ_0023409 is highly expressed in GC tissues and markedly correlated with tumor size, histological grade, and TNM staging, nominating it as a potential prognostic marker for GC [56]. Functionally, hsa_circ_0023409 exerts the oncogenic effects on GC progression and metastasis by competitively sponging miR-542-3p to enhance the expression of IRS4, which contributes to activating the PI3K/AKT pathway [56]. A well-characterized circRNA, CDR1as (ciRS-7), is markedly up-regulated in GC and linked to poor survival in an independent validation cohort, and promotes GC cell migration and metastasis via antagonizing the miR-7-mediated expression of PTEN, which is broadly regarded as a negative regulator of the PI3K/AKT signaling pathway [57, 76].
Other pathways
Several additional circRNAs have been gradually characterized to engage in other EMT signaling pathways [58–61]. For example, circTNPO3 is significantly downregulated in GC compared with matched noncancerous tissues and plasma circTNPO3 owns the ability to serve as a potential diagnostic biomarker [58]. In vitro and in vivo observations reveal that circTNPO3 suppresses GC proliferation and metastasis [58]. Mechanistically, circTNPO3 competitively interacts with IGF2BP3 and subsequently destabilizes the MYC mRNA, ultimately inhibiting MYC and its target SNAIL, a primary and key inducer of EMT (Fig. 2AIV) [58]. The circRNA circFNDC3B appears to be increased in GC significantly and facilitates cell migration, invasion and EMT of GC cells by forming a ternary complex of circFNDC3B-IGF2BP3-CD44 mRNA (Fig. 2AV) [59]. In addition, circ_100876, a significantly up-regulated circRNA in GC, contributes to GC migration and invasion by serving as a molecular sponge for miR-665 to regulate the expression of YAP1, which activates a transcriptional program involved in EMT (Fig. 2AVI) [60].
Collectively, these findings strongly indicate that circRNAs can modify several critical biological pathways relevant to GC metastasis.
CircRNAs regulate angiogenesis
Angiogenesis, defined as the formation of new blood vessels sprouting from preexisting vessels, is well-regarded as an important initial step in cancer metastasis [77–79]. Several signaling pathways, including VEGFA and HIF1α signaling, can continuously induce angiogenesis, aggravating cancer progression [80, 81]. Recently, several circRNAs have been reported to participate in GC metastasis by regulating VEGFA- or HIF1α-mediated angiogenesis [62–65].
The circRNA circRanGAP1 is validated to sponge miR-877-3p to increase the VEGFA expression, stimulate angiogenesis and promote GC metastasis (Fig. 2BI) [62]. A similar ceRNA mechanism also applies to circ_0044366, which binds to miR-29a to derepress the VEGF expression and thus facilitates angiogenesis and migration in GC [63]. The circRNA circURI1 back-spliced from exons 3–4 of URI1 has been identified from circRNA profiling of 5 paired GC and adjacent non-cancerous (paraGC) specimens [64]. CircURI1 exhibits a remarkably higher expression in GC than paraGC tissues and is negatively associated with metastasis in GC patients [64]. Functional studies perform that circURI1 inhibits GC metastasis in vitro and in vivo. Mechanistically, circURI1 behaved as a decoy of hnRNPM in a sequence-dependent manner to modulate alternative splicing of a subset of genes related to cell migration, thus suppressing GC metastasis (Fig. 2BII) [64]. VEGFA is a direct and functional target of circURI1, and circURI1 can promote exon 7 inclusion of VEGFA (VEGFAe7IN) [64]. CircURI1-induced VEGFAe7IN possesses a greater ability to prevent the circURI1-silencing-mediated promoting effect on GC cell invasion than exon 7 exclusion of VEGFA [64, 82]. This study firstly reported the engagement of circRNA-modulated alternative splicing in cancer metastasis [64].
Additionally, virus-encoded circRNA has also been found to engage in angiogenesis in GC [65, 83]. Epstein-Barr virus (EBV)-derived circRNA LMP2A (ebv-circLMP2A) is correlated with distant metastasis and poor prognosis in EBV-associated GC (EBVaGC) [65]. Furthermore, the ebv-circLMP2A expression is positively correlated with the expressions of HIF1α and VEGF in clinical samples of EBVaGC and a mouse model [65]. Ectopic expression of ebv-circLMP2A promotes angiogenesis and GC cell migration under hypoxia, while ebv-circLMP2A knockdown reverses these effects [65]. Mechanistic studies reveal that HIF1α and ebv-circLMP2A form a positive feedback loop, which promotes angiogenesis in EBVaGC [65]. Briefly, under hypoxia, HIF1α induces the ebv-circLMP2A expression, and ebv-circLMP2A interacts with KHSRP to enhance the VHL mRNA decoy mediated by KHSRP, resulting in HIF1α accumulation (Fig. 2BIII) [65].
Exosomal circRNAs and GC metastasis
Exosomes are small extracellular vesicles with an average diameter of ~100 nanometers, containing an abundant cargo of proteins and different RNA species, including circRNAs, which can enhance substance exchange between cells and improve signal transduction [84, 85]. Accumulating evidence has demonstrated that exosomes play emerging roles in regulating cancer metastasis and treatment through the transfer and exchange of molecules during cell-cell communications [86, 87]. Recently, circRNAs have been shown to be abundant in exosomes and exosomal circRNAs might be regarded as circulating biomarkers for metastatic disease in GC patients [88, 89].
Multiple exosomal circRNAs from the plasmas of GC patients are involved in GC invasion and metastasis [66–69]. CircNRIP1 possesses a significantly higher expression level in exosomes from GC plasma than in normal tissues and engages in exosomal crosstalk between GC cells [66]. GC cells co-cultured with exosomes derived from circNRIP1-overexpressed cells exhibit higher metastatic potential than control cells via the tail vein metastasis model [66]. Simultaneously, exosomal circNRIP1 promotes GC metastasis in vivo and regulates EMT by activating the AKT1/mTOR signaling pathway via sponging miR-149-5p [66]. Similarly, circNEK9, an up-regulated circRNA in GC tissues, accelerates GC proliferation by serving as a ceRNA against miR-409-3p to target MAP7 [67]. Additionally, the exosome-mediated transfer of circNEK9 performs promotive effects on GC cell migration and invasion [67]. Sang et al. have uncovered that exosomal circRELL1 is down-regulated in GC, and its delivery mediated by GC cells-derived exosomes stimulate autophagy by modulating the miR-637/EPHB3 axis in GC progression [68]. In another case, circSHKBP1 is remarkably upregulated in both GC tissues and serum and is significantly associated with advanced TNM stage and poor survival [69]. Mechanistically, exosomal circSHKBP1 promotes GC cell migration and invasion via modulating the miR-582-3p/HuR/VEGF axis, and inhibiting HSP90 ubiquitination through sequestering HSP90 to obstruct its interaction with STUB1 (Fig. 2C) [69]. These promising results provide novel insights into therapy and the predictions of GC prognosis.
Other metastasis-related pivotal pathways or targets
FOXO1/3a pathway
The FOXO1/3a pathway stimulates the expressions of the downstream targets, including p21, p27, Twist1, and E-cadherin [70, 90]. The circRNA circMRPS35 is identified from circRNA profiles of three paired GC and the corresponding non-tumor tissues, whose level is associated with clinicopathological characteristics and prognosis in GC patients [70]. Biologically, in vivo observations and in vitro experiments reveal that circMRPS35 inhibits GC cell proliferation and invasion [70]. Furthermore, mechanistic studies reveal that circMRPS35 combats GC tumorigenesis by recruiting histone acetyltransferase KAT7 to transcriptionally activate the FOXO1/3a genes, consequently triggering the FOXO1/3a pathway (Fig. 2DI) [70].
MEK-MAPK pathway
The MEK-MAPK signaling pathway is mainly involved in GC proliferation and metastasis [71, 91]. The circRNA circMAPK1 exhibits a decreased level in GC compared to the corresponding adjacent non-tumor tissues and is inversely correlated with GC tumor size, lymphatic invasion, TNM stage, and poor OS [71]. Functional investigations implicate that circMAPK1 suppresses GC proliferation and invasion in vitro and in vivo [71]. Mechanistically, circMAPK1 exerts the anti-tumor effect through encoding a MAPK1-109aa protein as a molecular sponge for MEK1, thus suppressing the phosphorylation of MAPK1 and eventually resulting in the inactivation of the MAPK pathway (Fig. 2DII) [71].
STAT3 pathway
Signal transducer and activator of transcription 3 (STAT3) is a widely-characterized oncogene in diverse human cancers [92, 93]. The circRNA circRPL15, up-regulated in GC tissues and correlated with short survival, enhances GC cell migration and invasion, and inhibits apoptosis by sequestering miR-502-3p from the OLFM4 mRNA to activate the STAT3 pathway [72]. A similar ceRNA mechanism also applies to circUBE2Q2, which interacts with miR-370-3p to relieve the inhibitory effect on its target STAT3 in GC, promoting proliferation, glycolysis, and metastasis [73].
Human antigen R
Human antigen R (HuR), a classic RBP, is frequently up-regulated in multiple human cancers including GC and plays a vital role in cancer progression and metastasis [94]. An intronic circRNA circAGO2 generated from the first intron of AGO2 is increased in GC and boosts GC metastasis in vitro and in vivo [74]. Mechanistic studies reveal that circAGO2 physically interacts with HuR protein to facilitate its activation and enrichment on the 3’ UTR of HuR targets, inhibiting AGO2/miRNA-mediated gene silencing associated with cancer progression (Fig. 2DIII) [74]. In another case, circHuR, predominantly localized in the nucleus, is downregulated in GC tissues and suppresses GC cell growth, invasion, and metastasis [75]. Mechanistically, circHuR interacts with CNBP and subsequently restrains its binding to the promoter of HuR, leading to the repressions of HuR and GC progression (Fig. 2DIV) [75].
Interplay between circRNAs and drug resistance in GC
Although chemo- and radio-therapy are recognized as the most effective and extensive treatment methods for GC patients after surgery during the past few decades, the clinical applications are still limited owing to the intrinsic and acquired resistance, resulting in the occurrence of distant metastasis in GC patients [1, 3, 95]. Additionally, targeted therapy and immunotherapy with immune checkpoint inhibitors for GC have emerged [96]. Convincing evidence has confirmed that diverse circRNAs influence drug resistance in GC therapeutic responses (Table 2) [55, 112].
Table 2.
CircRNAs involved in drug resistance in GC
| CircRNA | CircBase ID | Drug | Expression | Drug resistance | Targets | Refs |
|---|---|---|---|---|---|---|
| circVAPA | hsa_circ_0006990 | Cisplatin | Up | Enhance | miR-125b-3p, STAT3 | [97] |
| circAKT3 | hsa_circ_0000199 | Cisplatin | Up | Enhance | miR-198, PIK3R1 | [55] |
| circARVCF | hsa_circ_0092330 | Cisplatin | Up | Enhance | miR-1205, FGFR1 | [98] |
| circCCDC6 | hsa_circ_0001313 | Cisplatin | Up | Enhance | miR-618, BCL-2 | [99] |
| circFN1 | hsa_circ_0058147 | Cisplatin | Up | Enhance | miR-182-5p | [100] |
| circPVT1 | - | Cisplatin | Up | Enhance | miR-30a-5p, YAP1 | [101] |
| circ_0000260 | hsa_circ_0000260 | Cisplatin | Up | Enhance | miR-129-5p, MMP11 | [102] |
| circ_0032821 | hsa_circ_0032821 | Oxaliplatin | Up | Enhance | miR-515-5p, SOX9 | [103] |
| circPVT1 | - | Paclitaxel | Up | Enhance | miR-124-3p, ZEB1 | [104] |
| circNRIP1 | hsa_circ_0004771 | 5-fluorouracil | Up | Enhance | miR-138-5p, HIF-1α | [105] |
| circDLG1 | hsa_circ_0008583 | anti-PD-1 | Up | Enhance | miR-141-3p, CXCL12 | [106] |
| circCUL2 | hsa_circ_0000234 | Cisplatin | Down | Suppress | miR-142-3p, ROCK2 | [107] |
| circMCTP2 | hsa_circ_0000657 | Cisplatin | Down | Suppress | miR-99a-5p, MTMR3 | [108] |
| circ_0000144 | hsa_circ_0000144 | Oxaliplatin | Down | Suppress | miR-502-5p, ADAM9 | [109] |
| circ_0000376 | hsa_circ_0000376 | Bupivacaine | Down | Suppress | miR-145-5p | [110] |
| circ_0000520 | hsa_circ_0000520 | Herceptin | Down | Suppress | PI3K-AKT pathway | [111] |
Cisplatin (CDDP) is one of the most effective chemotherapeutic agents for patients with GC, especially those in advanced stages [113, 114]. The circVAPA expression is elevated in CDDP-resistant GC cells, and circVAPA facilitates GC cell migration, invasion, and CDDP resistance [97]. Further mechanistic investigations indicate that circVAPA exerts its oncogenic activity through sponging with miR-125b-5p to increase the STAT3 expression [97]. Similarly, several other circRNAs such as circAKT3, circPVT1, circFN1, and circ_0000260, also enhance CDDP resistance and malignant progression in GC [55, 98–102]. Oxaliplatin (OXA) is a widely used anti-cancer medicine [115]. The circRNA circ_0032821 is significantly increased in OXA-resistant GC cells and their derived exosomes, and contributes to OXA resistance, GC cell migration and invasion through derepressing SOX9 via sequestering miR-515-5p [103]. Paclitaxel (PTX) is an effective first-line chemotherapy drug in GC treatment, and circPVT1 contributes to PTX resistance and GC cell invasion via serving as a ceRNA against miR-124-3p to target ZEB1, a crucial transcriptional inhibitor of E-cadherin [104]. 5-fluorouracil (5-FU) is currently a first-line agent for the clinical treatment of GC, and circNRIP1 promotes hypoxia-induced 5-FU resistance via modulating the miR-138-5p/HIF-1α axis in GC [105]. Anti-programmed cell death protein 1 (PD-1) monoclonal antibody is a commonly used immune-checkpoint blockade agent for GC immunotherapy [116]. The circRNA circDLG1 facilitates GC progression and anti-PD-1 resistance via miR-141-3p-mediated the regulation of CXCL12 [106].
On the other hand, various circRNAs reverse drug resistance in GC treatment [107–109]. Peng et al. have unveiled that circCUL2 displays a decreased level in GC tissues and possesses a repressively regulatory function in CDDP resistance, GC cell migration, and invasion via miR-142-3p/ROCK2-mediated autophagy activation [107]. Another circRNA circMCTP2 is reported to inhibit CDDP resistance of GC cells via the miR-99a-5p/MTMR3 axis [108]. The circRNA hsa_circ_0000144 exerts inhibitory effects on OXA resistance, GC cell proliferation, and metastasis through up-regulating ADAM9 mediated by miR-502-5p [109]. Bupivacaine, a local anesthetic commonly used in the resection operation of GC patients, reduces the circ_0000376 level in GC cells, and circ_0000376 partially reverses bupivacaine-mediated repressive effects on GC cell viability and metastasis via sponging miR-145-5p [110]. Herceptin, a targeted therapy drug, is a humanized monoclonal antibody specifically binding to HER2 and acts as an antitumor role in GC [117]. The circRNA hsa_circ_0000520 is significantly reduced in GC and reverses the Herceptin resistance of GC cells by inhibiting the PI3K/AKT signaling pathway [111].
Taken together, these studies provide the possibility that a combination of circRNAs-based therapy with chemotherapy, targeted therapy or immunotherapy may be a valuable approach to overcome drug resistance and prevent metastasis in GC in the future.
Clinical significance of circRNAs in GC
CircRNAs have multiple remarkable characteristics which provide tremendous potential for serving as biomarkers and therapeutic targets owing to the covalently closed-loop structure, disease-specific and dynamic expression pattern and high conservation across species [118–122]. For example, according to a study by Liang and colleagues, hsa_circ_0110389 has been identified as a diagnostic/prognostic biomarker and therapeutic target for GC [123]. Similarly, circOSBPL10 might serve as a novel proliferation factor and prognostic marker of the OS and disease-free survival (DFS) of GC patients [124]. In another case, Chen et al. have displayed that the circPVT1 level is an independent prognostic biomarker for OS and DFS in GC patients [125].
Since exosomes can be detected in various body fluids, including plasma, saliva, urine, and cerebrospinal fluid, exosomal circRNAs might be ideal noninvasive biomarkers for the diagnosis and/or prognosis of gastric cancer [88, 126]. For instance, the circSHKBP1 expression is significantly increased in GC serum and positively correlated with advanced TNM stage and poor survival [69]. Furthermore, GC cell exosomes enhance co-cultured cell growth by delivering circSHKBP1 [69]. These findings indicate that circSHKBP1 is a promising circulating biomarker for GC diagnosis and prognosis [69]. Additionally, the circRNA circRanGAP1 exhibits a significantly higher expression in plasma exosomes derived from GC patients than the healthy controls. It promotes GC cell migration and invasion, indicating that plasma exosomal circRanGAP1 might serve as a promising biomarker for GC patients [62]. The circRNAs that show potential as biomarkers in GC are summarized in Table 3.
Table 3.
Clinical significance of circRNAs in GC (Cases more than 50)
| CircRNA | CircBase ID | Sample | Expression | Clinicopathologic Features | Prognosis | Refs |
|---|---|---|---|---|---|---|
| circTHBS1 | hsa_circ_0034536 | Tissue | Up | Size, stage, grade, LNM | OS | [48] |
| circCCDC66 | hsa_circ_0001313 | Tissue | Up | Stage, LNM | - | [49] |
| circOXCT1 | hsa_circ_0004873 | Tissue | Down | Stage, LNM | OS | [51] |
| circAXIN1 | hsa_circ_0005838 | Tissue | Up | Stage, grade, LNM | - | [52] |
| circFGD4 | hsa_circ_0000390 | Tissue | Down | Grade, LNM | OS | [53] |
| circREPS2 | hsa_circ_0139996 | Tissue | Down | Size, stage, grade | - | [54] |
| circAKT3 | hsa_circ_0000199 | Tissue | Up | Size, stage, grade, chemoresistance | OS | [55] |
| circ_0023409 | hsa_circ_0023409 | Tissue | Up | Size, stage, grade | OS | [56] |
| ciRS-7 | hsa_circ_0001946 | Tissue | Up | Stage, LNM | OS | [57] |
| circTNPO3 | hsa_circ_0001741 | Tissue, plasma | Down | Differentiation | - | [58] |
| circ_100876 | hsa_circ_0023404 | Tissue | Up | Stage, LNM, BVI, LVI | DFS | [60] |
| circRanGAP1 | hsa_circ_0063526 | Tissue, plasma | Up | Size, stage, LNM | OS | [62] |
| circURI1 | hsa_circ_0000921 | Tissue | Up | Stage, tumor metastasis | - | [64] |
| ebv-circLMP2A | - | Tissue | Up | Stage, LNM, tumor metastasis | OS, DFS | [65] |
| circNRIP1 | hsa_circ_0004771 | Tissue | Up | Size, LNM | OS, DFS | [66] |
| circRELL1 | hsa_circ_0001400 | Tissue, plasma | Down | Stage, LNM, differentiation | OS, DFS | [68] |
| circSHKBP1 | hsa_circ_0000936 | Tissue | Up | Size, stage, vascular invasion | OS | [69] |
| circMRPS35 | hsa_circ_0000384 | Tissue | Down | Size, stage, LNM | OS | [70] |
| circMAPK1 | hsa_circ_0004872 | Tissue | Down | Size, stage, LNM | OS | [71] |
| circUBE2Q2 | hsa_circ_0005151 | Tissue, plasma | Up | Size, lymphatic invasion | - | [73] |
| circAGO2 | hsa_circ_0135889 | Tissue | Up | - | OS | [74] |
| circHuR | hsa_circ_0049027 | Tissue | Down | Stage, tumor metastasis | OS | [75] |
| circVAPA | hsa_circ_0006990 | Tissue | Up | - | - | [97] |
| circFN1 | hsa_circ_0058147 | Tissue | Up | Stage, grade, chemoresistance | - | [100] |
| circCUL2 | hsa_circ_0000234 | Tissue | Down | Stage, LNM, differentiation | OS | [107] |
| circMCTP2 | hsa_circ_0000657 | Tissue | Down | Size, stage, grade, chemoresistance | OS, DFS | [108] |
| circ_0110389 | hsa_circ_0110389 | Tissue | Up | Stage, differentiation | OS, DFS | [123] |
| circOSBPL10 | hsa_circ_0008549 | Tissue | Up | Stage, grade | OS, DFS | [124] |
| circPVT1 | - | Tissue | Up | Stage, nervous invasion | OS, DFS | [125] |
Conclusions and future perspectives
Current active research in circRNAs has brought us a range of exciting findings implying that circRNAs are of great importance in various diseases [11, 118, 127–129]. A tremendous amount of evidence has demonstrated the abnormal expression pattern of circRNAs in GC and the involvement of circRNAs in GC metastasis and drug resistance [11, 64, 126]. We have systematically described a series of dysregulated circRNAs in GC and elucidated their underlying molecular mechanisms in GC metastasis and drug resistance (Tables 1 and 2).
To date, various circRNA candidates have been validated and engaged in GC metastasis based on a series of molecular and cellular experiments [64, 66–69, 124, 125]. However, a global and comprehensive understanding of circRNAs related to GC metastasis is still scarce. To gain better and deeper insight into the aberrant expression pattern of circRNAs involved in GC metastasis, genome-wide circRNA profiling with high throughput sequencing from metastatic and non-metastatic GC tissues is a powerful approach to address this issue.
Four subclasses of circRNAs have been identified, including ecircRNAs, EIciRNAs, ciRNAs and mitochondria-encoded circRNAs (mecciRNAs) [11, 38, 130]. Current literature about circRNAs in GC metastasis generally includes ecircRNAs and ciRNAs, their functions and the molecular mechanisms [72–75, 94, 126]. Nevertheless, two other kinds of circRNAs and their functions have not been evaluated, which presents an exciting field to explore further.
The well-characterized mechanism of circRNAs is to sequester miRNAs to regulate the expressions of targeted genes [11–13]. A single circRNA could function as a scaffold for several different miRNAs [123]. Conversely, a miRNA can target multiple circRNAs as well [60, 61]. Identification and construction of the circRNA-miRNA regulatory network will help to systematically decipher the roles of circRNAs in GC metastasis in the future. In addition to the ceRNA mechanism, circRNAs have various molecular modes of action, including participating in epigenetic regulations, modulating alternative splicing, and generating protein [64, 71, 75]. We expect a burst of circRNA studies to elucidate some novel mechanisms of action in GC metastasis in the upcoming years.
Considering that circRNAs possess unique features such as tissue- and developmental stage-specific patterns, structural resistance to exonucleases and longer half-lives, and specific circRNAs play essential roles in GC metastasis and drug resistance, manipulating circRNA abundance appears to be a promising therapeutic strategy for the advanced GC treatment [126–128, 131, 132]. Furthermore, combining circRNAs-based therapeutic interventions with traditional chemotherapy or targeted therapy offers a unique opportunity to conquer drug resistance in advanced GC patients [97–111, 113–117]. However, choosing crucial target circRNAs of interest is still a problem. Furthermore, precisely and effectively delivering circRNAs into targeted cells for tumor treatment is also a significant issue that needs to be solved.
Conclusions
In summary, the advances in circRNAs research will be essential to unravel their potential significance in GC. Furthermore, a better understanding of the association between circRNAs and GC would make circRNAs promising candidates as valuable biomarkers or potential targets in GC treatment.
Acknowledgments
A portion of this work was supported by the High Magnetic Field Laboratory of Anhui Province.
Abbreviations
- 5-FU
5-fluorouracil
- BVI
Blood vessel infiltration
- CDDP
Cisplatin
- ceRNA
Competing endogenous RNA
- circRNA
Circular RNA
- ciRNA
Intronic circRNA
- DFS
Disease-free survival
- ecircRNA
exonic circRNA
- EIciRNA
exon-intron circRNA
- EMT
Epithelial-mesenchymal transition
- EBV
Epstein-Barr virus
- EBVaGC
EBV-associated GC
- GC
Gastric cancer
- HuR
Human antigen R
- IRES
Internal ribosome entry site
- LNM
Lymph node metastasis
- LVI
Lymphatic vessel infiltration
- mecciRNA
mitochondria-encoded circRNA
- OS
Overall survival
- OXA
Oxaliplatin
- paraGC
adjacent non-cancerous GC
- PD-1
Programmed cell death protein 1
- Pol II
Polymerase II
- pre-RNA
precursor RNA
- PTX
Paclitaxel
- RBP
RNA binding protein
- snRNP
small nuclear ribonucleoprotein
- STAT3
Signal transducer and activator of transcription 3
- VEGFAe7IN
Exon 7 inclusion of VEGFA
Authors’ contributions
X.W. was responsible for the table and figure generation. X.W., G.S. and W.L. wrote this manuscript. X.W., J.Z., G.C., J.H. and W.L. discussed and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2019YFA0802600 and 2018YFC1004500), National Natural Science Foundation of China (81972191 and 81672647), and Science and Technology Major Project of Anhui Province (18030801140).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree to the content of the paper and are listed as co-authors of the paper.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ge Shan, Email: shange@ustc.edu.cn.
Wenchu Lin, Email: wenchu@hmfl.ac.cn.
References
- 1.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Ducreux M. Colorectal and gastric cancer in 2015: The development of new agents and molecular classifications. Nat Rev Clin Oncol. 2016;13:69–70. doi: 10.1038/nrclinonc.2015.217. [DOI] [PubMed] [Google Scholar]
- 4.Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat Rev Cancer. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21:446–460. doi: 10.1038/s41568-021-00353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 8.Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E, et al. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537–548. doi: 10.1007/s10555-019-09803-7. [DOI] [PubMed] [Google Scholar]
- 9.Li GZ, Doherty GM, Wang J. Surgical Management of Gastric Cancer: A Review. JAMA Surg. 2022;157:446–454. doi: 10.1001/jamasurg.2022.0182. [DOI] [PubMed] [Google Scholar]
- 10.Yeoh KG, Tan P. Mapping the genomic diaspora of gastric cancer. Nat Rev Cancer. 2022;22:71–84. doi: 10.1038/s41568-021-00412-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Huang C, Shan G. Circular RNAs in physiology and non-immunological diseases. Trends Biochem Sci. 2022;47:250–264. doi: 10.1016/j.tibs.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 13.Kristensen LS, Andersen MS, Stagsted LV, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Huang C, Wang X, Shan G. Circular RNAs in eukaryotic cells. Curr Genomics. 2015;16:312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 19.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1612235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagliarini V, Jolly A, Bielli P, Di Rosa V, De la Grange P, Sette C. Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization. Nucleic Acids Res. 2020;48:633–645. doi: 10.1093/nar/gkz1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho JS, Di Tullio F, Schwarz M, Low D, Incarnato D, Gay F, et al. HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness. Elife. 2021;10:e59654. doi: 10.7554/eLife.59654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H, An O, Ren X, Song Y, Tang SJ, Ke XY, et al. ADARs act as potent regulators of circular transcriptome in cancer. Nat Commun. 2022;13:1508. doi: 10.1038/s41467-022-29138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 25.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen X, et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc Natl Acad Sci U S A. 2019;116:7455–7464. doi: 10.1073/pnas.1814874116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y, et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol. 2021;39:836–845. doi: 10.1038/s41587-021-00842-6. [DOI] [PubMed] [Google Scholar]
- 32.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, et al. A novel circular RNA, circFAT1 (e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 34.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-Encoded Peptides or Proteins and Cancer. Mol Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Wang X, Li J, Hu S, Deng Y, Yin H, et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429–1449. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer. 2020;6:319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Hua J, Wang X, Ma L, Li J, Cao G, Zhang S, et al. CircVAPA promotes small cell lung cancer progression by modulating the miR-377-3p and miR-494-3p/IGF1R/AKT axis. Mol Cancer. 2022;21:123. doi: 10.1186/s12943-022-01595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu G, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14:404–418. doi: 10.1080/15548627.2017.1414755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 45.Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol. 2020;30:764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu S, Li B, Xia Y, Xuan Z, Li Z, Xie L, et al. CircTHBS1 drives gastric cancer progression by increasing INHBA mRNA expression and stability in a ceRNA- and RBP-dependent manner. Cell Death Dis. 2022;13:266. doi: 10.1038/s41419-022-04720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu G, Chen Y, Fu M, Zang X, Cang M, Niu Y, et al. Circular RNA CCDC66 promotes gastric cancer progression by regulating c-Myc and TGF-β signaling pathways. J Cancer. 2020;11:2759–2768. doi: 10.7150/jca.37718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niu Q, Dong Z, Liang M, Luo Y, Lin H, Lin M, et al. Circular RNA hsa_circ_0001829 promotes gastric cancer progression through miR-155-5p/SMAD2 axis. J Exp Clin Cancer Res. 2020;39:280. doi: 10.1186/s13046-020-01790-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Liu J, Dai X, Guo X, Cheng A, Mac SM, Wang Z. Circ-OXCT1 suppresses gastric cancer EMT and metastasis by attenuating TGF-beta pathway through the circ-OXCT1/miR-136/SMAD4 axis. Onco Targets Ther. 2020;13:3987–3998. doi: 10.2147/OTT.S239789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo X, et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol Cancer. 2021;20:158. doi: 10.1186/s12943-021-01457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai X, Liu J, Guo X, Cheng A, Deng X, Guo L, et al. Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/beta-catenin signalling pathway. Clin Sci (Lond). 2020;134:1821–1839. doi: 10.1042/CS20191043. [DOI] [PubMed] [Google Scholar]
- 54.Guo X, Dai X, Liu J, Cheng A, Qin C, Wang Z. Circular RNA circREPS2 acts as a sponge of miR-558 to suppress gastric cancer progression by regulating RUNX3/ beta-catenin signaling. Mol Ther Nucleic Acids. 2020;21:577–591. doi: 10.1016/j.omtn.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Yang Y, Xu D, Cao L. hsa_circ_0023409 accelerates gastric cancer cell growth and metastasis through regulating the IRS4/PI3K/AKT pathway. Cell Transplant. 2021;30:963689720975390. doi: 10.1177/0963689720975390. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 58.Yu T, Ran L, Zhao H, Yin P, Li W, Lin J, et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649–664. doi: 10.1016/j.omtn.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong Y, Qin H, Li Y, Zhang Y, Zhuang X, Liu L, et al. FNDC3B circular RNA promotes the migration and invasion of gastric cancer cells via the regulation of E-cadherin and CD44 expression. J Cell Physiol. 2019;234:19895–19910. doi: 10.1002/jcp.28588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin X, Huang C, Chen Z, Wang H, Zeng Y. CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells' growth, migration and invasion via miR-665/YAP1 signaling. Front Genet. 2020;11:546275. doi: 10.3389/fgene.2020.546275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W, Hu W, Hou K, Zhu S. Circular RNA paired-related homeobox 1 promotes gastric carcinoma cell progression via regulating microRNA-665/YWHAZ axis. Dig Dis Sci. 2021;66:3842–3853. doi: 10.1007/s10620-020-06705-5. [DOI] [PubMed] [Google Scholar]
- 62.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Li J, Zhang H, Zhang Y, Wang X, Yang H, et al. Gastric cancer derived exosomes mediate the delivery of circRNA to promote angiogenesis by targeting miR-29a/VEGF axis in endothelial cells. Biochem Biophys Res Commun. 2021;560:37–44. doi: 10.1016/j.bbrc.2021.04.099. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci U S A. 2021;118:e2012881118. doi: 10.1073/pnas.2012881118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du Y, Zhang JY, Gong LP, Feng ZY, Wang D, Pan YH, et al. Hypoxia-induced ebv-circLMP2A promotes angiogenesis in EBV-associated gastric carcinoma through the KHSRP/VHL/HIF1α/VEGFA pathway. Cancer Lett. 2022;526:259–272. doi: 10.1016/j.canlet.2021.11.031. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu L, Xie J, Liu X, Yu Y, Wang S. Plasma exosomal circNEK9 accelerates the progression of gastric cancer via miR-409-3p/MAP7 axis. Dig Dis Sci. 2021;66:4274–4289. doi: 10.1007/s10620-020-06816-z. [DOI] [PubMed] [Google Scholar]
- 68.Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, et al. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis. 2022;13:56. doi: 10.1038/s41419-021-04364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19:56. doi: 10.1186/s12943-020-01160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20:66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Gong Y, Ma J, Gong X. Overexpressed circ-RPL15 predicts poor survival and promotes the progression of gastric cancer via regulating miR-502-3p/OLFM4/STAT3 pathway. Biomed Pharmacother. 2020;127:110219. doi: 10.1016/j.biopha.2020.110219. [DOI] [PubMed] [Google Scholar]
- 73.Yang J, Zhang X, Cao J, Xu P, Chen Z, Wang S, et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis. 2021;12:910. doi: 10.1038/s41419-021-04216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158. doi: 10.1186/s12943-019-1094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM, et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6:7769. doi: 10.1038/ncomms8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 78.Roviello G, Petrioli R, Marano L, Polom K, Marrelli D, Perrella A, et al. Angiogenesis inhibitors in gastric and gastroesophageal junction cancer. Gastric Cancer. 2016;19:31–41. doi: 10.1007/s10120-015-0537-5. [DOI] [PubMed] [Google Scholar]
- 79.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gonzalez FJ, Xie C, Jiang C. The role of hypoxia-inducible factors in metabolic diseases. Nat Rev Endocrinol. 2018;15:21–32. doi: 10.1038/s41574-018-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, Hua J, Li J, Zhang J, Dzakah EE, Cao G, et al. Mechanisms of non-coding RNA-modulated alternative splicing in cancer. RNA Biol. 2022;19:541–547. doi: 10.1080/15476286.2022.2062846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gong LP, Chen JN, Dong M, Xiao ZD, Feng ZY, Pan YH, et al. Epstein-Barr virus-derived circular RNA LMP2A induces stemness in EBV-associated gastric cancer. EMBO Rep. 2020;21:e49689. doi: 10.15252/embr.201949689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 86.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 87.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin S, Buel GR, Nagiec MJ, Han MJ, Roux PP, Blenis J, et al. ERK2 regulates epithelial-to-mesenchymal plasticity through DOCK10-dependent Rac1/FoxO1 activation. Proc Natl Acad Sci U S A. 2019;116:2967–2976. doi: 10.1073/pnas.1811923116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 92.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Timofeeva OA, Tarasova NI, Zhang X, Chasovskikh S, Cheema AK, Wang H, et al. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci U S A. 2013;110:1267–1272. doi: 10.1073/pnas.1211805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR) Wiley Interdiscip Rev RNA. 2020;11:e1581. doi: 10.1002/wrna.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee J, Kim ST, Kim K, Lee H, Kozarewa I, Mortimer PGS, et al. Tumor genomic profiling guides patients with metastatic gastric cancer to targeted treatment: The VIKTORY umbrella trial. Cancer Discov. 2019;9:1388–1405. doi: 10.1158/2159-8290.CD-19-0442. [DOI] [PubMed] [Google Scholar]
- 96.Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264–279. doi: 10.3322/caac.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng P, Sun M, Zhao WY, Hou B, Li K, Zhang T, et al. Circular RNA circVAPA promotes chemotherapy drug resistance in gastric cancer progression by regulating miR-125b-5p/STAT3 axis. World J Gastroenterol. 2021;27:487–500. doi: 10.3748/wjg.v27.i6.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang R, Zhao H, Yuan H, Wu J, Liu H, Sun S, et al. CircARVCF contributes to cisplatin resistance in gastric cancer by altering miR-1205 and FGFR1. Front Genet. 2021;12:767590. doi: 10.3389/fgene.2021.767590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng A, et al. CircRNACCDC66 regulates cisplatin resistance in gastric cancer via the miR-618/BCL2 axis. Biochem Biophys Res Commun. 2020;526:713–720. doi: 10.1016/j.bbrc.2020.03.156. [DOI] [PubMed] [Google Scholar]
- 100.Huang XX, Zhang Q, Hu H, Jin Y, Zeng AL, Xia YB, et al. A novel circular RNA circFN1 enhances cisplatin resistance in gastric cancer via sponging miR-182-5p. J Cell Biochem. 2020;122:1009–1020. doi: 10.1002/jcb.29641. [DOI] [PubMed] [Google Scholar]
- 101.Yao W, Guo P, Mu Q, Wang Y. Exosome-derived circ-PVT1 contributes to Cisplatin resistance by regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 2021;36:347–359. doi: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- 102.Liu S, Wu M, Peng M. Circ_0000260 regulates the development and deterioration of gastric adenocarcinoma with Cisplatin resistance by upregulating MMP11 via targeting miR-129-5p. Cancer Manag Res. 2020;12:10505–10519. doi: 10.2147/CMAR.S272324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA circ_0032821 contributes to oxaliplatin (OXA) resistance of gastric cancer cells by regulating SOX9 via miR-515-5p. Biotechnol Lett. 2021;43:339–351. doi: 10.1007/s10529-020-03036-3. [DOI] [PubMed] [Google Scholar]
- 104.Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep. 2019;39:BSR20193045. doi: 10.1042/BSR20193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu G, Li M, Wu J, Qin C, Tao Y, He H. Circular RNA circNRIP1 sponges microRNA-138-5p to maintain hypoxia-induced resistance to 5-Fluorouracil through HIF-1α-dependent glucose metabolism in gastric carcinoma. Cancer Manag Res. 2020;12:2789–2802. doi: 10.2147/CMAR.S246272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen DL, Sheng H, Zhang DS, Jin Y, Zhao BT, Chen N, et al. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol Cancer. 2021;20:166. doi: 10.1186/s12943-021-01475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. doi: 10.1186/s12943-020-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 2020;39:246. doi: 10.1186/s13046-020-01758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gao H, Xu J, Qiao F, Xue L. Depletion of hsa_circ_0000144 suppresses Oxaliplatin resistance of gastric cancer cells by regulating miR-502-5p/ADAM9 axis. Onco Targets Ther. 2021;14:2773–2787. doi: 10.2147/OTT.S281238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ju C, Zhou J, Miao H, Chen X, Zhang Q. Bupivacaine suppresses the progression of gastric cancer through regulating circ_0000376/miR-145-5p axis. BMC Anesthesiol. 2020;20:275. doi: 10.1186/s12871-020-01179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lv X, Li P, Wang J, Gao H, Hei Y, Zhang J, et al. hsa_circ_0000520 influences herceptin resistance in gastric cancer cells through PI3K-Akt signaling pathway. J Clin Lab Anal. 2020;34:e23449. doi: 10.1002/jcla.23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu W, Zhen T, Yu J, Yang Q. Circular RNAs as new regulators in gastric cancer: diagnosis and cancer therapy. Front Oncol. 2020;10:1526. doi: 10.3389/fonc.2020.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol. 2008;9:215–221. doi: 10.1016/S1470-2045(08)70035-4. [DOI] [PubMed] [Google Scholar]
- 114.Mo D, Fang H, Niu K, Liu J, Wu M, Li S, et al. Human helicase RECQL4 drives Cisplatin resistance in gastric cancer by activating an AKT-YB1-MDR1 signaling pathway. Cancer Res. 2016;76:3057–3066. doi: 10.1158/0008-5472.CAN-15-2361. [DOI] [PubMed] [Google Scholar]
- 115.Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–4153. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- 116.Rizvi NA, Mazières J, Planchard D, Stinchcombe TE, Dy GK, Antonia SJ, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsurutani J, Iwata H, Krop I, Jänne PA, Doi T, Takahashi S, et al. Targeting HER2 with trastuzumab deruxtecan: A dose-expansion, phase I study in multiple advanced solid tumors. Cancer Discov. 2020;10:688–701. doi: 10.1158/2159-8290.CD-19-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Q, Li F, He AT, Yang BB. Circular RNAs: Expression, localization, and therapeutic potentials. Mol Ther. 2021;29:1683–1702. doi: 10.1016/j.ymthe.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2015;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 120.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 121.Gao L, Chang S, Xia W, Wang X, Zhang C, Cheng L, et al. Circular RNAs from BOULE play conserved roles in protection against stress-induced fertility decline. Sci Adv. 2020;6:eabb7426. doi: 10.1126/sciadv.abb7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suenkel C, Cavalli D, Massalini S, Calegari F, Rajewsky N. A highly conserved circular RNA is required to keep neural cells in a progenitor state in the mammalian brain. Cell Rep. 2020;30:2170–2179. doi: 10.1016/j.celrep.2020.01.083. [DOI] [PubMed] [Google Scholar]
- 123.Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z, et al. Circular RNA hsa_circ_0110389 promotes gastric cancer progression through upregulating SORT1 via sponging miR-127-5p and miR-136-5p. Cell Death Dis. 2021;12:639. doi: 10.1038/s41419-021-03903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang S, Zhang X, Li Z, Wang W, Li B, Huang X, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene. 2019;38:6985–7001. doi: 10.1038/s41388-019-0933-0. [DOI] [PubMed] [Google Scholar]
- 125.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 126.Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu L, Wang J, Khanabdali R, Kalionis B, Tai X, Xia S. Circular RNAs: Isolation, characterization and their potential role in diseases. RNA Biol. 2017;14:1715–1721. doi: 10.1080/15476286.2017.1367886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57. doi: 10.1016/j.canlet.2021.02.004. [DOI] [PubMed] [Google Scholar]
- 129.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao Q, Liu J, Deng H, Ma R, Liao JY, Liang H, et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 131.Li F, Yang Q, He AT, Yang BB. Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin Cancer Biol. 2021;75:49–61. doi: 10.1016/j.semcancer.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 132.Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. doi: 10.1186/s12943-020-01185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.