Abstract
During organogenesis, cell proliferation is followed by the differentiation of specific cell types to form an organ. Any aberration in differentiation can result in developmental defects, which can result in a near-complete loss of an organ. We employ the Drosophila eye model to understand the genetic and molecular mechanisms involved in the process of differentiation. In a forward genetic screen, we identified, cullin-4 (cul-4), which encodes an E3 ubiquitin ligase, to play an important role in retinal differentiation. During development, cul-4 is known to be involved in protein degradation, regulation of genomic stability, and regulation of cell cycle. Previously, we have reported that cul-4 regulates cell death during eye development by downregulating Wingless (Wg)/ Wnt signaling pathway. We found that loss-of-function of cul-4 results in a reduced eye phenotype, which can be due to onset of cell death. However, we found that loss-of-function of cul-4 also affects retinal development by downregulating RD gene expression. Early markers of retinal differentiation are dysregulated in cul-4 loss of function conditions, indicating that cul-4 is necessary for differentiation. Furthermore, loss-of-function of cul-4 ectopically induces expression of negative regulators of eye development like Wg and Homothorax (Hth). During eye development, Wg is known to block the progression of a synchronous wave of differentiation referred to as Morphogenetic furrow (MF). In cul-4 loss-of-function background, expression of dpp-lacZ, a MF marker is significantly downregulated. Our data suggest a new role of cul-4 in retinal differentiation. These studies may have significant bearings on our understanding of early eye development.
Keywords: Drosophila melanogaster, Eye development, Retinal differentiation, Retinal determination, Cullin-4
Introduction
In all multicellular organisms, organogenesis involves various fundamental processes like cell proliferation, cell death, cell fate specification and differentiation to generate the final shape and size of an organ. A fine balance between these cell biological processes is crucial for organogenesis and to maintain the shape of an organ (Gogia, Puli, Raj, and, & Singh, 2020; Kango-Singh & Singh, 2009; Mehta & Singh, 2019; A Singh, 2012; M. Tare, Puli Roy, O., Singh, A., 2013). Drosophila melanogaster a.k.a. fruit fly, a holometabolous insect, has served as an excellent model for understanding the genetic and molecular mechanisms involved in these processes. Since the genetic machinery is conserved between Drosophila and humans, the information generated in Drosophila model can be extrapolated to humans (Bier, 2005; A. Singh & Irvine, 2012).
In Drosophila, the progenitors for all adult appendages are housed inside the larva as a monolayer epithelium called as imaginal discs (Poulson, 1950). Imaginal discs, which are derived from the embryonic ectoderm, are a favored model system to understand how fields of cells can autonomously regulate patterning, growth and differentiation (Atkins & Mardon, 2009; Cohen, 1993; Dominguez & Casares, 2005; Held, 2002; A. Singh & Choi, 2003; Verghese, Bedi, & Kango-Singh, 2012). Among various appendages, the Drosophila eye has been extensively used to study patterning, growth and differentiation (A Singh, 2012; A. Singh & Irvine, 2012). Drosophila eye begins from approximately 20 eye-antennal primordial cells that are set aside during embryogenesis (Garcia-Bellido & Merriam, 1969; Held, 2002; Poulson, 1950). These eye-antennal primordial cells undergo proliferation during first- and second- instar stages of larval development, followed by differentiation during late second or early third instar of larval development, which give rise to the adult compound eye, antenna and head cuticle (Atkins & Mardon, 2009; Gogia, Puli, et al., 2020; Kumar, 2011, 2020b; Mishra & Sprecher, 2020; Ready, Hanson, & Benzer, 1976; M. Tare, Puli Roy, O., Singh, A., 2013).
During Drosophila eye development, a core cascade of genes, referred to as the retinal determination and differentiation (RD) gene network, which comprises of PAX-6 homolog eyeless (ey), twin of eyeless (toy), eye gone (eyg), twin of eyegone (toe), eyes absent (eya), sine oculis (so), and dachshund (dac) are required to form an adult eye (Gogia, Puli, et al., 2020; Jang et al., 2003; Kango-Singh, Singh, & Sun, 2003; Mishra & Sprecher, 2020; A. Singh, Lim, & Choi, 2005; M. Tare, Puli Roy, O., Singh, A., 2013). Most of the core members of RD gene network except Eya, a tyrosine phosphatase, are transcription factors. These RD genes are necessary and sufficient for the normal development of the eye, and have capability to induce ectopic eyes in other imaginal discs like wing and leg imaginal discs (Halder et al., 1998; Kango-Singh et al., 2003; Kumar, 2011; Mishra & Sprecher, 2020). In first instar eye primordial cells, ey is expressed in the entire eye disc and its expression is activated by Toy (Czerny et al., 1999). Ey expression is downregulated at the MF as well as in the cells posterior to the MF (Czerny et al., 1999), which in turn activates the expression of downstream genes eya, so, and dac to promote retinal differentiation (Bui, Zimmerman, Liu, & Bonini, 2000; Halder et al., 1998; Kango-Singh et al., 2003; Mishra & Sprecher, 2020; Niimi, Seimiya, Kloter, Flister, & Gehring, 1999; Serikaku & O’Tousa, 1994).
During late second instar stage, the retinal or photoreceptor neurons begin to differentiate from their precursor cells. The process of retinal differentiation, marked by a significant change in the shape of the cells, begins at the posterior end of the eye imaginal disc, and moves toward the anterior end as a synchronous wave. This wave of differentiation originates at the posterior margin of larval eye imaginal disc and is referred to as the morphogenetic furrow (MF) (Kumar, 2020a; Ready et al., 1976; Wolff & Ready, 1991). As the MF progresses, it leaves behind rows of gradually assembling ommatidial clusters. Consequently, the older clusters are positioned farthest from the MF. The cells posterior to the MF are differentiated photoreceptor neurons, which make the compound eye whereas the cells anterior to MF remain in proliferating states until the end of the third instar larval stage (Kumar, 2020a).
The differentiation along the MF is also guided by signaling molecules, such as Hedgehog (Hh), Decapentaplegic (Dpp) and other transcription factors (Dominguez & Hafen, 1997; Kumar, 2020a). In the developing eye imaginal disc, dpp expression initiates at the posterior margin and moves dynamically with the MF. Therefore, dpp serves as an excellent marker for MF progression (Kumar, 2020a; Sarkar, Gogia, Farley, Payton, & Singh, 2018). Dpp promotes MF progression by downregulating wingless (wg), a negative regulator of eye development, and is known to block MF progression (Ma & Moses, 1995; Treisman & Rubin, 1995). Loss-of-function of wg in the developing eye results in MF enlargement along the DV margins. Wg is expressed along the antero-lateral margins of the third instar developing eye imaginal disc (Royet & Finkelstein, 1997; Won et al., 2015a, 2015b). In the developing eye, Wg, a ligand for evolutionarily conserved Wg/ WNT signaling pathway, is involved in various diverse functions of cell proliferation, differentiation, and cell death. In the developing eye, Wg is also known to regulate expression of homothorax (hth), to suppress eye fate and thereby define the boundary of the developing eye. Hth encodes a TALE (Three Amino-Acid Loop Extension) type homeodomain protein (Pai et al., 1998; Pichaud & Casares, 2000; Rieckhof, Casares, Ryoo, Abu-Shaar, & Mann, 1997; A. Singh et al., 2011). Hth expresses uniformly in the first instar larval eye-antennal disc, but it retracts from the posterior region of the second instar larval eye-antennal imaginal disc. In the third instar larval eye-antennal imaginal disc, Hth expression is in the anterior region of the eye disc (Bessa, Gebelein, Pichaud, Casares, & Mann, 2002; Moran, Tare, Kango-Singh, & Singh, 2013; Pai et al., 1998; A. Singh, Kango-Singh, Choi, & Sun, 2004; A. Singh, Kango-Singh, & Sun, 2002; A. Singh et al., 2011).
The compound eye of Drosophila consists of 750–800 unit eyes, each termed as an ommatidium. Each ommatidium is a hexagonal arrangement of eight precisely arranged photoreceptors R1-R8 (Ready et al., 1976). These photoreceptors differentiate along with the movement of MF in a precise order. The differentiation begins with R8 photoreceptor(s) developing first (Jarman, Grell, Ackerman, Jan, & Jan, 1994; Tomlinson & Ready, 1987) followed by pairwise differentiation of R2/R5, R3/R4 and R1/R6 and at the end, R7 is formed (Ready et al., 1976; Tomlinson & Ready, 1987). These events for conversion of a monolayer epithelium into a three dimensional neuro-crystalline lattice are kept under tight spatio-temporal regulation by different gene products and signaling pathways (Kumar, 2020a). As the differentiation begins at the end of second instar stage, the RD genes are expressed and are necessary in the cells posterior to the MF (Kumar, 2020a; Silver & Rebay, 2005). At the MF, a crucial interaction between Eya, Dac and So is required for the activation of atonal (ato), which is required for the initiation of R8 differentiation (Jarman et al., 1994; Lim, Lee, Hsu, Singh, & Choi, 2007; Tanaka-Matakatsu & Du, 2008; Zhang, Ranade, Cai, Clouser, & Pignoni, 2006). While photoreceptors differentiate, a uniform space between the differentiated clusters is maintained by a fibrinogen-related secreted protein Scabrous (Sca) (Baker, Mlodzik, & Rubin, 1990).
In a genetic screen to look for genes involved in eye development, we identified cullin-4 (cul-4) as a genetic modifier of reduced eye phenotype seen in axial patterning gene mutant(s) (A. Singh, Chan, Chern, & Choi, 2005). The cul-4, a member of evolutionarily conserved Cullin family, encodes an E3 ubiquitin ligase, and is ubiquitously expressed during development (Jackson & Xiong, 2009). Previously it has been shown that Cul-4 is involved in maintenance of genomic integrity by promoting degradation of cell cycle progression factors and cell survival (Braun et al., 2011; M. Tare, Sarkar, Bedi, Kango-Singh, & Singh, 2016; Zielke et al., 2011). Earlier, we have shown that cul-4 regulates Wg and c-Jun-N-terminal kinas (JNK) signaling during early development to limit cell death (Tare et al, 2016). Here we present a new role of cul-4 in retinal development and differentiation. Here, we show that cul-4 is required for differentiation of the eye via regulating the core members of the RD gene network.
Materials and Methods
Fly stocks
The fly stocks used in this study are described in Flybase (http://flybase.bio.indiana.edu). The fly stocks used in this study are, Canton-S (Wild-type), cul-4JJ11/ twi>GFP, CyO, which carries a nonsense mutation at Trp199 position (Lin, Wu, Tan, & Chien, 2009) and eyFLP; FRT42D, cl-w+/CyO-GFP. We used an enhancer trap line, dpp-lacZ (Blackman, Sanicola, Raftery, Gillevet, & Gelbart, 1991; A Singh, 1995) (BL5528) to study dpp expression, which was obtained from Bloomington Drosophila Stock Center (BDSC).
Genetic Crosses
We employed the FLP/FRT mediated genetic mosaic approach to generate loss-of-function clones of cul-4 in the eye (Blair, 2003). We employed the cell lethal (cl) approach to characterize loss-of-function phenotypes of cul-4 in the developing eye. Using cl approach, nearly 80% of the cell population of the disc was made mutant for cul-4, since cl mutation leads to elimination of wild-type cells (Newsome, Asling, & Dickson, 2000).The virgins of eyFLP; FRT42D, cl-w+/CyO-GFP were crossed to cul-4JJ11/twi>GFP, CyO (Lin et al., 2009; M. Tare et al., 2016).
Immunohistochemistry
Eye-antennal imaginal discs were dissected in 1X phosphate buffered saline (PBS) from the wandering third-instar larvae and fixed in 4% paraformaldehyde in PBS (fixative) for 20 minutes and washed in PBST (three times)(A. Singh et al., 2002; M. Tare, Puli, Moran, Kango-Singh, & Singh, 2013). The tissues were stained with combination(s) of antibodies following the standard protocol. Antibodies used were rat anti-Elav (1:100, Developmental Studies Hybridoma Bank, DSHB), mouse anti-Wg (1:50, DSHB), mouse anti-Sca (1:200, DSHB), goat anti-Ato (1:100, Santa Cruz Biotechnology), guinea pig anti-Homothorax (1:200, SantaCruz Biotechnology), mouse anti-β galactosidase (1:100, Promega) and rabbit anti-Eyeless (a kind gift from Uwe Walldorf and Patrick Callaerts). The discs were washed in PBST thrice for 10 minutes. Secondary antibodies used were donkey anti-rat IgG conjugated to Cy5 (1:250), donkey anti-rabbit IgG conjugated to Cy3 (1:300) or goat anti-mouse IgG conjugated to FITC (1:200) (Jackson Laboratories). The discs were mounted in Vectashield and photo-documented on a Fluoview 3000 Laser Scanning Confocal Microscope. The image analysis and preparation of the final figures was carried out using Adobe Photoshop CS6 software.
EdU Staining
EdU staining is a technique to detect cells in S –phase of cell cycle by incorporation of 5-ethynyl-2’ deoxyuridine (EdU), which is a thymidine analogue. EdU staining was performed following manufacturer’s recommendations (Invitrogen™ Clitck-iT® EdU kit, Cat# 10339) with some modifications (Dichtel-Danjoy et al., 2013). All the steps were performed at room temperature, incubations were carried out using a rocker/shaker. Third instar eye-antennal imaginal discs were dissected in 1X PBS and incubated for 60 minutes in 20mM EdU, diluted with Schneider’s medium. The samples were then fixed with 4% para-formaldehyde (PFA) solution for 30 minutes and washed twice with 3% Bovine Serum albumin (BSA) in 1XPBS. Following this, eye imaginal discs were permeabilized by adding PBST (PBS with 1% Triton X-100) for 30 minutes at room temperature and washed again with 3% BSA in 1XPBS. Tissues were permeabilized with 1XPBST and washed thrice with 3% BSA (in 1XPBS). Tissues were incubated for 30 minutes in room temperature with 0.5 mL of reaction cocktail (prepared as per kit instructions). The discs were then washed with 3% BSA (in 1X PBS) and mounted on a slide using Vectashield (Sarkar, Gogia, Glenn, et al., 2018). Images were captured using Fluoview 3000 Laser Scanning Confocal Microscope (A Singh & Gopinathan, 1998) and Adobe Photoshop CS6 software was used for analysis and final image preparation.
Adult eye Imaging
The adult eye images were taken after freezing flies at −20°C for ∼4 hours. The frozen flies were mounted on a needle after removing the legs and wings of the flies. The flies were positioned on a glass slide using mounting putty. Images were taken on a MrC5 color camera mounted on an Axioimager.Z1 Zeiss Apotome using a Z-sectioning function of Axiovision software 4.6.3 (Gogia, Sarkar, et al., 2020; Irwin et al., 2020). The final images were prepared using Adobe Photoshop CS6 software.
Statistical Analysis
Statistical analysis was performed using Microsoft excel software. The P-values were calculated using student’s t-test and the error bars represent Standard deviation from Mean. Statistical significance is shown by P-value: *** P<0.001; ** P<0.01; * P<0.05 (Cutler et al., 2015; Gogia, Sarkar, et al., 2020; Steffensmeier et al., 2013; M. Tare et al., 2011).
Results
Loss of function of cul-4 results in reduced eye phenotype
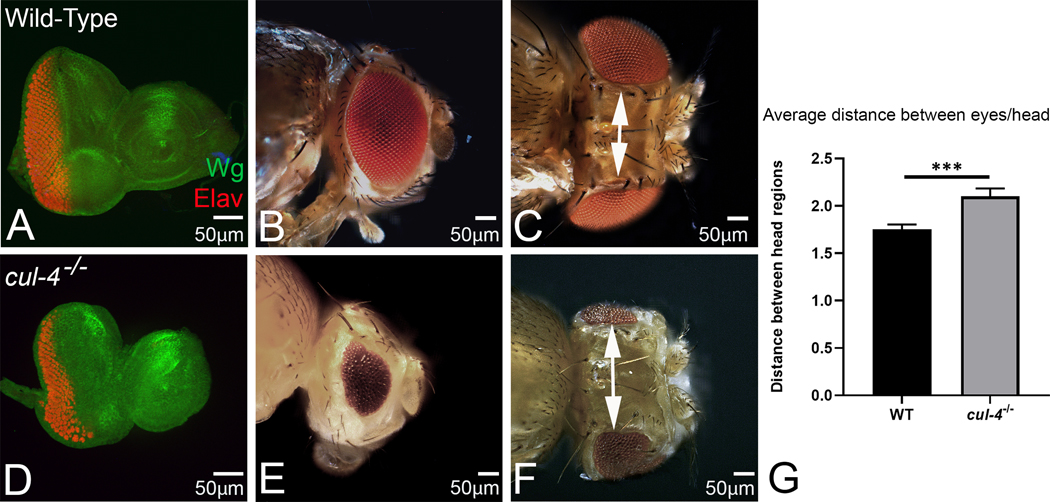
The Drosophila adult eye, a compound eye comprising of 600–800 ommatidia, develops from the larval eye imaginal disc (Fig. 1A, B). The eye-antennal imaginal discs were stained with neuronal marker Elav (red) that marks the nuclei of photoreceptor neuron and a morphogen Wg (green). In wild-type eye imaginal disc, Wg is expressed in anterolateral margin of the developing eye imaginal disc (Fig. 1A). Loss-of-function clones of cul-4, generated by conventional genetic mosaic approach, fail to grow. Therefore, we employed the cell lethal (cl) approach to characterize loss-of-function phenotypes of cul-4 in the eye. Using cl approach, nearly 80% of the cell population of the eye disc comprise of cul-4 mutant, since cl mutation leads to elimination of wild-type cells (Newsome et al., 2000). In comparison to the wild-type eye imaginal disc, which results in an adult compound eye (Fig.1A, B), cul-4 loss-of-function exhibit reduced eye phenotype as seen in the eye imaginal disc and the adult eye (Fig. 1D, E). Furthermore, there is a robust induction of Wg expression in cul-4 loss-of-function background (Fig. 1D). We observed that all the cul-4 mutant flies have much reduced eyes (n=212/212, 100%, Fig. 1E). Despite the smaller eyes, the size of antenna remains unaffected in the cul-4 mutant background (Fig. 1 B, E). Interestingly, the placement of the eyes on the adult head capsule is affected in the cul-4 loss-of-function (Fig. 1 C, F). We observed significant increase in the distance between the two reduced eyes in adult head of cul-4 mutant background (Fig. 1 G). These results suggest that head capsule is enlarged in cul-4−/− flies, whereas the eye field is reduced.
Fig.1. Loss of cul-4 functions exhibits a reduced eye phenotype.
(A) Wild-type eye-antennal imaginal disc showing expression of Wg (green); and a pan-neuronal marker, Elav (red), (B) and the adult eye. In comparison to the wild type eye imaginal disc, loss of function of cul-4, using cl-w+ approach results in a reduced eye phenotype as seen in (D) a third instar eye imaginal disc. Note that Elav staining (red), which marks the photoreceptor neurons of the eye, is significantly reduced. (E) The adult compound eye of cul-4 mutant’s exhibit reduced eye phenotype with fewer ommatidia as compared to (B) wild-type. (F) Additionally, cul-4 mutant’s dorsal head capsule is increased in size as compared to the (C) wild-type. (G) The length of the dorsal head cuticle was quantified for determining average distance between the two eyes in wild-type and cul-4 mutants by using Image J software (NIH).The length of dorsal head cuticle between the two eyes is significantly longer (p<0.001; ***) in cul-4 mutant head as compared to the wild-type. The p values for the length (μm) were calculated in a set of five (n=5) using Student’s t-test in MS Excel Software. A total of 5 heads were used for quantification. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. The magnification of all eye-antennal imaginal disc is 20X and the adult eye is 10X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Loss of function of cul-4 results into more proliferation in the head capsule
The enlarged head cuticle between the two compound eyes on the head of cul-4−/− adult fly (Fig. 1) led us to test if there are more proliferating cells in the head capsule. We used EdU staining that marks newly synthesized DNA of the proliferating cells (Dichtel-Danjoy et al., 2013). In the wild-type eye disc, these cells can be seen as a band of cells at the MF and in a broad domain, anterior to the MF, in the head capsule region (Fig 2A, A’). We found that in the cul-4 mutant background the proliferating cells population is increased in the undifferentiated region (anterior to the MF) as well as at the MF (Fig 2 B, B’). Interestingly, we did not see a significantly higher number of EdU positive cells in the posterior differentiated region of the eye discs. Together these results suggest that head capsules are enlarged due to increased cells proliferation in the anterior region of the eye antennal imaginal disc. This can explain the increase in the distance between the two reduced eyes on adult head of cul-4 mutant due to enlarged head cuticle.
Fig. 2. Loss-of-function of cul-4 exhibit proliferation defects in the anterior eye.
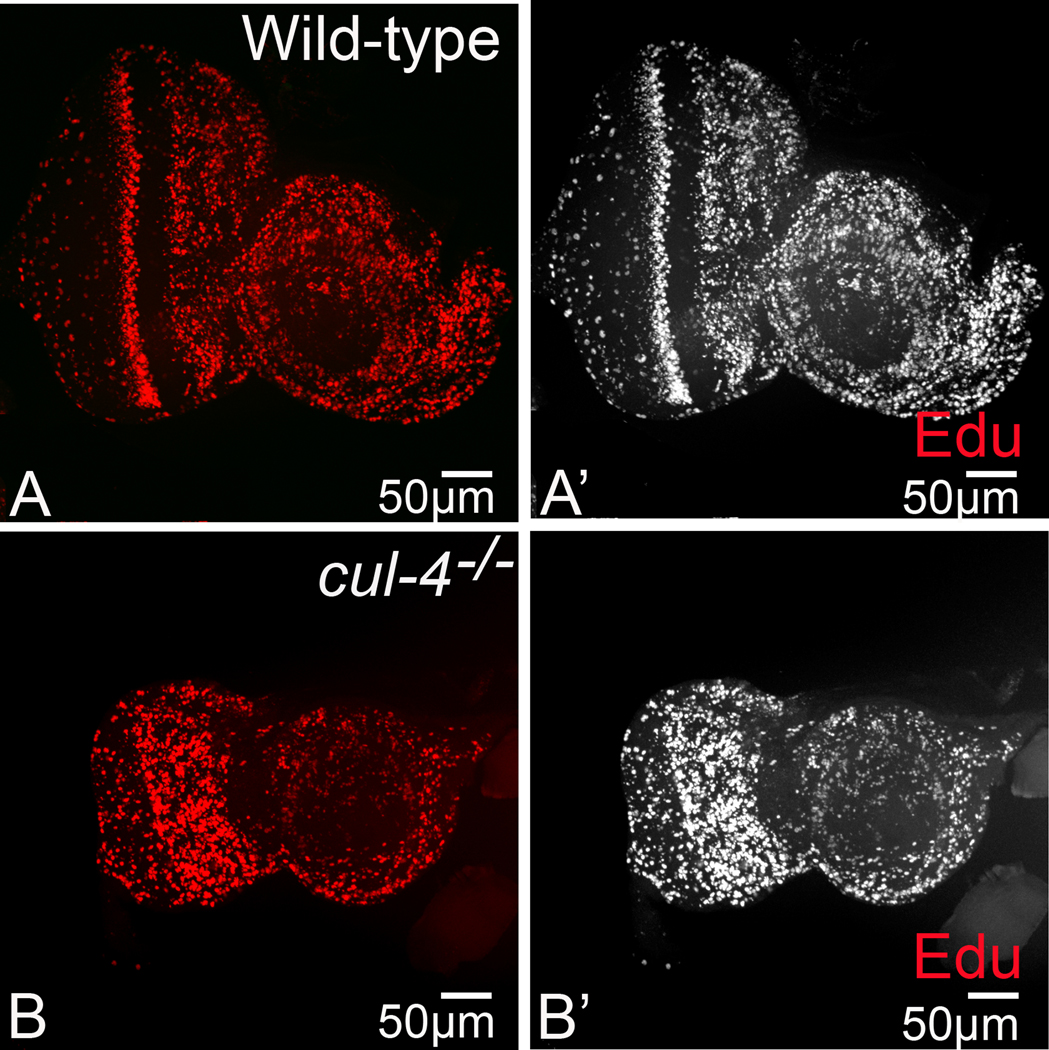
(A, A’). Wild-type eye disc are marked by EdU representing a band of proliferating cells posterior to the MF, (B, B’) cul-4 mutant cells exhibit a disruption of this pattern in the region anterior to MF. The magnification of all eye-antennal imaginal disc is 20X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Loss of cul-4 affects the retinal determination gene expression
To understand the genetic mechanism behind the reduced eye phenotype, we analyzed the expression of retinal determination (RD) genes in the developing eye (Fig. 3A) and compared their expression in cul-4 mutant eye imaginal disc generated by cell lethal approach(M. Tare et al., 2016). The master regulator Ey is expressed in the undifferentiated early primordial cells, and, as photoreceptor differentiation occurs; Ey expression retracts from the differentiating eye region and continues to express in the region, anterior to MF in the third instar eye imaginal discs (Fig. 3B, B’) (Bessa et al., 2002; Halder et al., 1998; Quiring, Walldorf, Kloter, & Gehring, 1994; A. Singh et al., 2002). We observed elevated levels of Ey posterior to the MF in cul-4 mutant third instar larval eye imaginal discs (Fig. 3 C, C’). Interestingly, retraction of Ey expression is required for the initiation of the downstream RD genes like eya, and dac (Bessa et al., 2002; A Singh, 2012; M. Tare, Puli Roy, O., Singh, A., 2013). In a third instar eye antennal imaginal disc, a tyrosine phosphatase, Eya is predominantly expressed both in the differentiated photoreceptor neurons posterior to the MF and undifferentiated retinal precursor cells anterior to the MF (Bonini, Leiserson, & Benzer, 1993) (Fig. 3D, D’). In cul-4 mutant background, Eya levels were significantly reduced (Fig. 3E, E’). Interestingly, this reduction was much enhanced at the midline of the eye imaginal disc. Another RD genes hierarchy member, Dac is expressed in two different domains one anterior and other posterior to the MF. Furthermore, Dac expression spans until R1, R6 and R7 photoreceptors to a few rows down to MF (Mardon, Solomon, & Rubin, 1994) (Fig. 3F, F’). In cul-4 mutant eye imaginal disc, Dac levels are significantly downregulated in the posterior as well as anterior to the MF (Fig. 3G, G’). Furthermore, we observed a significant downregulation in the Dac expression at the midline of the eye disc. Taken together, these observations suggest that cul-4 loss of function downregulates the retinal determination genes, which are required to form the eye field.
Fig.3. Loss of function of cul-4 affects the retinal determination (RD) gene network.
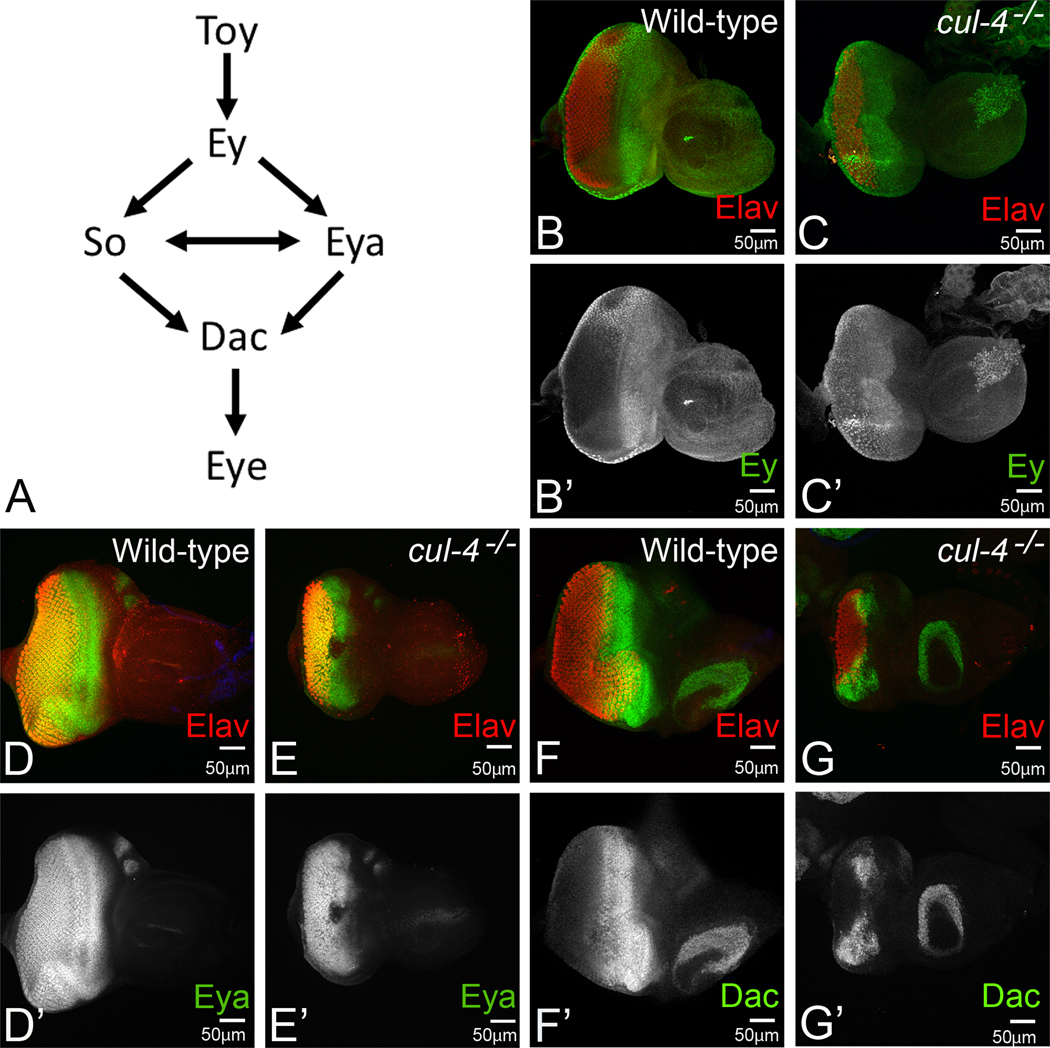
(A)The RD gene network in developing Drosophila eye includes a hierarchy of interaction(s) between Twin of Eyeless (Toy), Eyelsss (Ey), Eyes absent, (Eya), Sine oculis (So) and Dacshund (Dac). (B-G) Third instar eye-antennal imaginal disc stained for retinal determination markers and pan-neural marker Elav (red), which marks the photoreceptors. Note that the wild-type expression of (B) Ey (green, B’ split channel), (D) Eya (green, D’ split channel), and (F) Dac (green, F’ split channel) is significantly affected in (C, E, G) cul-4 mutant. Note that levels of RD genes remain unaffected in the antenna. The magnification of all eye-antennal imaginal disc is 20X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Loss-of-function of cul-4 triggers defective retinal differentiation
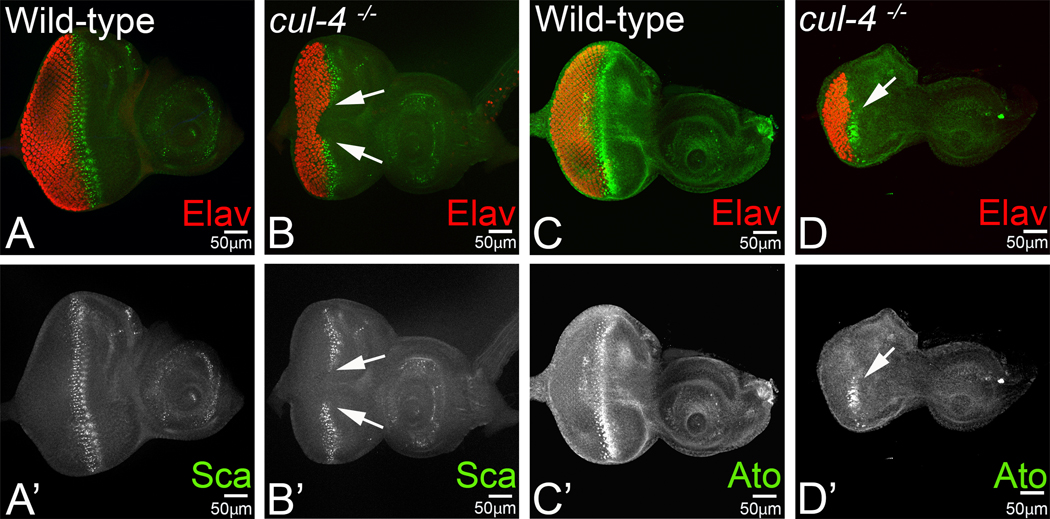
Since cul-4 loss-of-function results in the reduced eye phenotype, it can be argued that cul-4 is required for maintenance of retinal fate in the developing eye field. In order to test its role in retinal development and differentiation, we analyzed the expression of retinal differentiation fate markers in the developing eye (Wittkorn, Sarkar, Garcia, Kango-Singh, & Singh, 2015). We analyzed the spaces between the newly formed R8 cells by staining eye discs with antibody against fibrinogen like secreted protein Scabrous (Sca) (Baker et al., 1990; Mlodzik, Baker, & Rubin, 1990). Sca is expressed in several cells of each precluster and in a few rows posterior to the MF in wild type imaginal discs (Fig. 4A, A’) (Baker et al., 1990; Frankfort & Mardon, 2002; Mlodzik et al., 1990). We found that Sca expression is disrupted in cul-4 mutant eye imaginal discs near the middle of the eye imaginal disc (Fig. 4B, B’, arrows), indicating that spacing between R8 cells is disrupted. Furthermore, the Sca expression is completely missing in these cul-4 mutant eye disc near the equator (Fig. 4B, B’, arrow). Another R8 specification marker, the proneural protein Atonal (Ato) marks R8 fate of the retinal cell types, which is the first photoreceptor neuron that differentiates, hence marks 3–4 rows posterior to MF (Fig. 4C, C’). Mutant cul-4 eye imaginal discs stained for the proneural protein Atonal exhibits the defective pattern for R8 positive cells, compared to the wild type eye discs (Fig. 4C, C’, D, D’). Our data suggests that cul-4 function is also required for retinal differentiation.
Fig.4. Loss of function of cul-4 affects the retinal differentiation markers.
Eye-antennal imaginal disc of third instar larva stained for pan-neural marker Elav (red), which marks the photoreceptors and retinal differentiation markers (A, B) Sca, (A’, B’) Sca in split channel (C, D) Ato and (C’, D’) Ato in split channel. Note that wild-type expression of (A) Sca (green) and (C) Ato (green), is reduced and disrupted in (B, D) cul-4 mutants (arrow). The magnification of all eye-antennal imaginal disc is 20X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Negative regulators of retinal differentiation are up-regulated in the cul-4 mutants
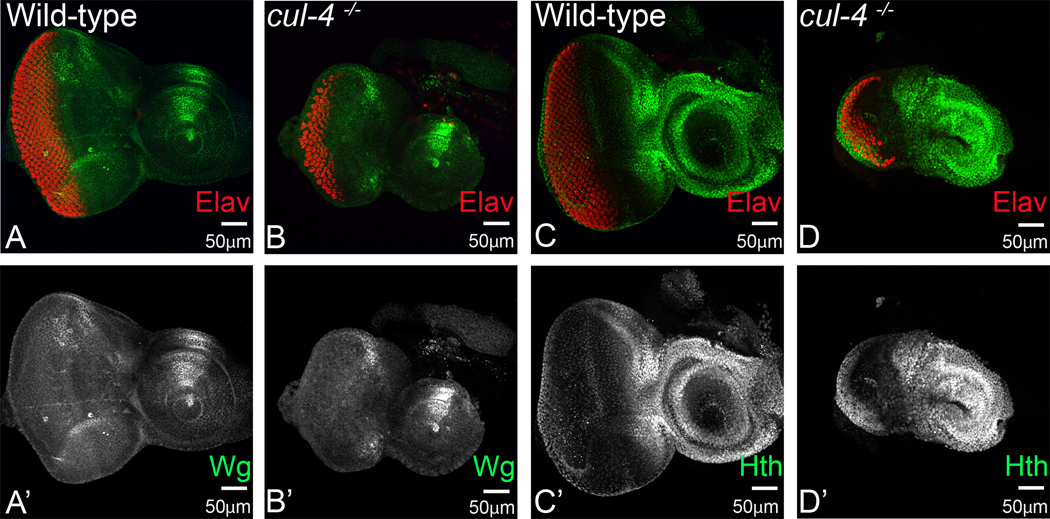
Defective photoreceptor differentiation phenotypes were congruent with defective Ey, Eya and Dac expression as well. We argued that negative regulators of eye development, which prevent retinal differentiation in the anterior region, by maintaining undifferentiated fate for cells, should also be affected in cul-4 mutant eye discs. Previously, we have reported aberrant higher levels of Wg in the cul-4 mutant eye disc, leading to cell death in early stages of eye development (Tare et al, 2016). In context to differentiation defects, we tested levels of Wg, as a negative regulator of retinal determination (Oros, Tare, Kango-Singh, & Singh, 2010; Pichaud & Casares, 2000; Silver & Rebay, 2005; M. Tare, Puli Roy, O., Singh, A., 2013; M. Tare et al., 2016). We found that in cul-4 mutant eye discs, higher levels of Wg prevents retinal fate by blocking retinal differentiation (Fig. 5 A, A’; B, B’). It has been reported that a combinatorial control by Ey, Hth and Tsh is required to prevent differentiation in the anterior region of the eye disc (Bessa et al, 2002). Meis class protein Hth is also required for maintaining the progenitor population in the anterior region of the eye discs. Hth is expressed in the region, anterior to MF in the undifferentiated proliferating cells (Pai et al., 1998; A. Singh, Gogia, Chang, & Sun, 2019; A. Singh et al., 2002; A. Singh et al., 2011). We found aberrantly high levels of Hth in the cul-4 mutant eye discs in the posterior as well as anterior region, along with loss of photoreceptors as indicated by loss of Elav positive cells (Fig. 5C, C’; D, D’). These observations are consistent with enlarged head capsules, and higher number of proliferating cells in the anterior region of the eye discs.
Fig. 5. Loss of function of cul-4 promotes ectopic expression of negative regulators of retinal development.
Secreted morphogen Wingless (Wg) is required for antenna and head development but is a negative regulator of MF progression as well as retinal development. Eye antennal imaginal discs are stained for pan-neural marker Elav (red). (A, A’) Wg (green) is expressed in dorso-lateral margins of the developing eye-antennal imaginal disc. (B, B’) Loss of function of cul-4 results in increase in the levels of Wg along these margins. (C, C’) Meis class protein Homothorax (Hth, green) is a negative regulator of the eye development and is expressed only in the undifferentiated head cuticle region and in the peripodial epithelial cells. (D, D’) Loss of function of cul-4 results in extension of Hth levels all the way into differentiated photoreceptor neuron region. The magnification of all eye-antennal imaginal disc is 20X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Loss-of-function of cul-4 blocks Morphogenetic Furrow (MF) progression
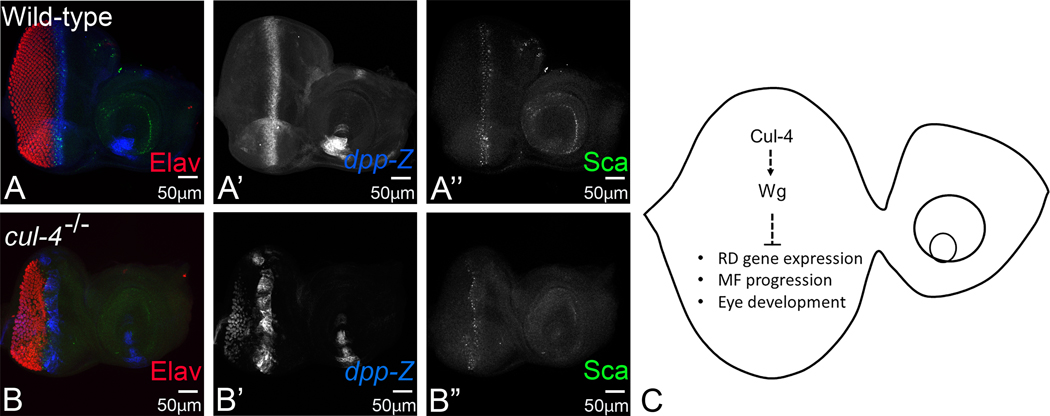
Our data suggests that the reduced eye phenotypes of cul-4 mutant eye imaginal disc is due to defective differentiation. This led us to test the MF progression in cul-4 mutant background. This dynamic process bears the guides for progression of differentiation in the developing eye imaginal disc, which is initiated and maintained by signaling cues of Hh and Dpp. High levels of Dpp signaling in the posterior margins is required to repress Wg, allowing movement of MF, permitting differentiation (Hazelett et al, 1998). However, in the third instar eye imaginal discs, Dpp expression is restricted to the MF (Silver and Rebay, 2005). We tested expression of dpp, using a dpp-lacZ reporter, along with R8 marker Sca (Fig. 6A, A’, A”). We used lac-Z transgene of dpp, which indicates expression pattern of dpp (Fig. 6A, A’) (Blackman et al., 1991; Sarkar, Gogia, Farley, et al., 2018; Won et al., 2015a). As expected, we did see disrupted pattern of Sca and dpp at MF, indicating that progression of MF is defective in cul-4 mutants (Fig. 6B, B’, B”). We detected aberrant dpp expression in the posterior most regions of the disc, spanning within a few of the photoreceptor neurons as well.
Fig. 6. Loss of function of cul-4 downregulates decapentaplegic (dpp), a marker for Morphogenetic Furrow (MF) progression.
Dpp- lacZ reporter marks the progression of the MF during eye development (blue; A, A’), which also colocalize with proneural marker Scabrous (Sca) (green; A, A”) in the wild-type eye imaginal disc. The expression of dpp-lacZ in cul-4 loss of function of eye-imaginal disc is lost in patches of cells along the MF (blue; B, B’). Note that proneural marker Sca expression is also dysregulated, instead of two rows of photoreceptors at furrow, only one of the rows of photoreceptors is seen positive for Scabrous (green; B, B”). (C) A schematic presentation to demonstrate the role of cul-4 in eye development. The magnification of all eye-antennal imaginal disc is 20X. A total of five eye-antennal imaginal discs (n=5) for each genotype were analyzed for respective immunohistochemistry staining.
Discussion
Differentiation during process of organogenesis is an important event to generate a three dimensional organ from a monolayer epithelial sheet of cells. In a forward genetic screen, we identified an E3 ubiquitin ligase encoding cul-4 as a genetic modifier of an eye mutant (A. Singh et al., 2005; M. Tare et al., 2016). Cul-4 is known to be involved in regulation of chromatin function, maintenance of genomic integrity, protein degradation via ubiquitylation, cell cycle and cell survival (Higa & Zhang, 2007; Hu et al., 2008; J. Kim & Kipreos, 2008; Y. Kim, Starostina, & Kipreos, 2008; Shibutani et al., 2008; M. Tare et al., 2016). Earlier we have reported that cul-4 plays an important role in preventing cell death and thereby promoting cell survival in the developing eye, via regulation of Wnt/Wg and c- Jun- amino terminal kinase (JNK) pathway (M. Tare et al., 2016).
It has been shown that several other E3 ligases such as Neuralized, Mindbomb, Slimb, and DIAP-1 are also involved in regulating signaling pathways, to fine tune the developmental processes (Jiang & Struhl, 1998; Jumpertz et al., 2014; Nagaraj & Banerjee, 2007; Yamazaki et al., 2016; Yamazaki et al., 2013). Cul-4 has been reported in regulating surface expression of smoothened via ubiquitylation, for its surface expression, thereby regulating Hedgehog signaling, in the wing discs (Li, Cho, Wang, Li, & Jiang, 2018). Our studies point to a new role of cul-4 in regulating retinal differentiation during eye development.
We have observed that cul-4 loss-of-function results in a reduced eye phenotype (Fig. 1). This reduced eye phenotype can be detected even in the larval eye imaginal disc stage. In cul-4 mutant eye imaginal disc, a reduced number of neuronal marker Elav positive nuclei are seen (Fig.1). Earlier we have shown that loss of cul-4 induces Wg expression, which results in the induction of cell death (A. Singh, Shi, & Choi, 2006; M. Tare, Puli Roy, O., Singh, A., 2013). Furthermore, blocking the cell death in cul-4 mutant background, significantly rescues the reduced eye phenotype. However, there is no complete rescue to wild-type eye (M. Tare et al., 2016). This suggests that loss-of-function of cul-4 is not only affecting cell death but also some other developmental process during eye development (M. Tare et al., 2016). In the developing eye, Wg is involved in various diverse functions of cell proliferation, differentiation, and cell death (Swarup & Verheyen, 2012). Wg is known to be involved in blocking retinal differentiation during eye imaginal disc development (Ma & Moses, 1995; A. Singh et al., 2002; A. Singh et al., 2006; Treisman & Rubin, 1995). In cul-4 loss-of-function background the retinal determination genes expression was downregulated suggesting that cul-4 also plays a role in retinal determination (Fig. 3). Furthermore, we found that R8 specification markers like Ato and ommatidial spacing marker Sca was downregulated in cul-4 loss-of-function background (Fig. 4). Thus, downregulation of differentiation markers from RD pathway is indicative of the defective differentiation (Fig. 4). It has been reported that Notch (N) signaling plays an important role in cell proliferation and differentiation in the developing eye. Furthermore, N acts upstream to Ey and is involved in regulation of eye specification (Kumar & Moses, 2001). We also tested if cul-4 regulates N and thereby play a role in eye development, however, we did not find any change(s) in N levels in cul-4 loss of function discs (data not shown). Instead, we found that cul-4 loss-of-function mediated defects are due to ectopic induction of Ey, Hth and Wg proteins in the eye imaginal (Fig.5). Both Hth and Wg are known negative regulators of retinal development (Pichaud & Casares, 2000; A. Singh & Irvine, 2012; A. Singh et al., 2011). Wg is known to antagonize Dpp signaling in the developing eye. A dpp-lacZ reporter insertion serves as a reporter of MF progression in the developing eye. Loss-of-function of cul-4 clones exhibit downregulation of Dpp levels in the developing eye. Furthermore, dpp-lacZ expression is not continuous along the MF but exhibits some holes in middle of MF (Fig. 6). Taken together, these results point to a new role of cul-4 in promoting retinal differentiation, MF progression and eye development (Fig. 6C). In future, it will be interesting to find out, if these RD pathway members are target substrates of Cul-4; or are regulated via a non- proteasomal degradation pathway.
The reduced eye phenotype of cul-4 mutant clones was also accompanied with changes in the placement of eyes on the head (Fig. 1). Interestingly, we have reported earlier that placement of eyes on the head of the fly can be due to extension of proximo-distal axis as seen in the stalk eyed fly (A. Singh et al., 2019). Furthermore, this extension is due to enhanced Hth expression, which marks the proximal fate. Interestingly, we found that cul-4 loss-of-function results in increased spacing between the two compound eyes on the adult head (Fig.1). Furthermore, it is also accompanied by ectopic induction of Hth (Fig. 5). In addition, the cell proliferation rates of cells anterior to the MF are higher in loss-of-function of cul-4 in the eye imaginal disc (Fig. 2). This data suggests that cul-4 maybe playing a role in axial patterning, a process required for delineation of antero-posterior (AP), dorso-ventral (DV) and proximo-dital (PD) axes (Gogia, Puli, et al., 2020; A. Singh & Irvine, 2012; M. Tare, Puli Roy, O., Singh, A., 2013). Our studies will have significant bearing on understanding various functions of ubiquitin ligases during development.
Acknowledgement:
The authors thank the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank (DSHB) for the antibodies. The authors also thank Dr. Madhuri Kango-Singh, and the members of Singh lab for critical comments on the manuscript. Confocal microscopy was supported by the core facility at the University of Dayton. For this work MT is supported by professional contingency funds from BITS Pilani, Pilani, India. NG, ACV and PD are supported by the University of Dayton graduate program. SN is supported by BITS Pilani graduate program. This work is supported by NIH1R15GM124654-01 from NIH, Schuellein Chair Endowment Fund, and STEM Catalyst Grant from the University of Dayton to AS.
Footnotes
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Reference Cited:
- Atkins M, & Mardon G. (2009). Signaling in the third dimension: the peripodial epithelium in eye disc development. Dev Dyn, 238(9), 2139–2148. doi: 10.1002/dvdy.22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Mlodzik M, & Rubin GM. (1990). Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science, 250(4986), 1370–1377. doi: 10.1126/science.2175046 [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, & Mann RS. (2002). Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev, 16(18), 2415–2427. doi: 10.1101/gad.1009002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. (2005). Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet, 6(1), 9–23. doi: 10.1038/nrg1503 [DOI] [PubMed] [Google Scholar]
- Blackman RK, Sanicola M, Raftery LA, Gillevet T, & Gelbart WM. (1991). An extensive 3’ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development, 111(3), 657–666. [DOI] [PubMed] [Google Scholar]
- Blair SS. (2003). Genetic mosaic techniques for studying Drosophila development. Development, 130(21), 5065–5072. doi: 10.1242/dev.00774,130/21/5065 [pii] [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, & Benzer S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell, 72(3), 379–395. doi: 10.1016/0092-8674(93)90115-7 [DOI] [PubMed] [Google Scholar]
- Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, & Madhani HD. (2011). The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell, 144(1), 41–54. doi: 10.1016/j.cell.2010.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui QT, Zimmerman JE, Liu H, & Bonini NM. (2000). Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics, 155(2), 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. (1993). Imaginal Disc development. The development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Cutler T, Sarkar A, Moran M, Steffensmeier A, Puli OR, Mancini G, . . . Singh A. (2015). Drosophila Eye Model to Study Neuroprotective Role of CREB Binding Protein (CBP) in Alzheimer’s Disease. PLoS One, 10(9), e0137691-e0137691. doi: 10.1371/journal.pone.0137691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, & Busslinger M. (1999). twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell, 3(3), 297–307. [DOI] [PubMed] [Google Scholar]
- Dichtel-Danjoy ML, Ma D, Dourlen P, Chatelain G, Napoletano F, Robin M, . . . Mollereau B. (2013). Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ, 20(1), 108–116. doi: 10.1038/cdd.2012.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, & Casares F. (2005). Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev Dyn, 232(3), 673–684. doi: 10.1002/dvdy.20311 [DOI] [PubMed] [Google Scholar]
- Dominguez M, & Hafen E. (1997). Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev, 11(23), 3254–3264. doi: 10.1101/gad.11.23.3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfort BJ, & Mardon G. (2002). R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development, 129(6), 1295–1306. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, & Merriam JR. (1969). Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool, 170(1), 61–75. doi: 10.1002/jez.1401700106 [DOI] [PubMed] [Google Scholar]
- Gogia N, Puli OR, Raj A, and, & Singh A. (2020). Generation of Third Dimension: Axial Patterning in the Developing Drosophila Eye. In Singh A, and, & Kango-Singh M. (Eds.), Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye (Vol. II, pp. 53–95). Springer NewYork Heidelberg Dordrecht London: Springer International Publishing. [Google Scholar]
- Gogia N, Sarkar A, Mehta AS, Ramesh N, Deshpande P, Kango-Singh M, . . . Singh A. (2020). Inactivation of Hippo and cJun-N-terminal Kinase (JNK) signaling mitigate FUS mediated neurodegeneration in vivo. Neurobiol Dis, 140, 104837. doi: 10.1016/j.nbd.2020.104837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, & Gehring WJ. (1998). Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development, 125(12), 2181–2191. [DOI] [PubMed] [Google Scholar]
- Held LIJ. (2002). The eye disc. In Held LI. (Ed.), Imaginal Disc (pp. 197–236): Cambridge University Press. [Google Scholar]
- Higa LA, & Zhang H. (2007). Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div, 2, 5. doi:1747–1028-2–5 [pii] 10.1186/1747-1028-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, & Xiong Y. (2008). WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev, 22(7), 866–871. doi:22/7/866 [pii] 10.1101/gad.1624008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Tare M, Singh A, Puli OR, Gogia N, Riccetti M, . . . Kango-Singh M. (2020). A Positive Feedback Loop of Hippo- and c-Jun-Amino-Terminal Kinase Signaling Pathways Regulates Amyloid-Beta-Mediated Neurodegeneration. Front Cell Dev Biol, 8, 117. doi: 10.3389/fcell.2020.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, & Xiong Y. (2009). Targeting protein ubiquitylation: DDB1 takes its RING off. Nat Cell Biol, 11(4), 379–381. doi:ncb0409–379 [pii] 10.1038/ncb0409-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, . . . Sun YH. (2003). Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development, 130(13), 2939–2951. doi: 10.1242/dev.00522 [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, & Jan YN. (1994). Atonal is the proneural gene for Drosophila photoreceptors. Nature, 369(6479), 398–400. [DOI] [PubMed] [Google Scholar]
- Jiang J, & Struhl G. (1998). Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature, 391(6666), 493–496. doi: 10.1038/35154 [DOI] [PubMed] [Google Scholar]
- Jumpertz S, Hennes T, Asare Y, Vervoorts J, Bernhagen J, & Schutz AK. (2014). The beta-catenin E3 ubiquitin ligase SIAH-1 is regulated by CSN5/JAB1 in CRC cells. Cell Signal, 26(9), 2051–2059. doi: 10.1016/j.cellsig.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, & Singh A. (2009). Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev Dyn, 238(7), 1627–1637. doi: 10.1002/dvdy.21996 [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Singh A, & Sun YH. (2003). Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Developmental Biology, 256(1), 48–60. [DOI] [PubMed] [Google Scholar]
- Kim J, & Kipreos ET. (2008). Control of the Cdc6 replication licensing factor in metazoa: the role of nuclear export and the CUL4 ubiquitin ligase. Cell Cycle, 7(2), 146–150. doi:5282 [pii] [DOI] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, & Kipreos ET. (2008). The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev, 22(18), 2507–2519. doi:22/18/2507 [pii] 10.1101/gad.1703708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. (2011). My what big eyes you have: how the Drosophila retina grows. Dev Neurobiol, 71(12), 1133–1152. doi: 10.1002/dneu.20921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. (2020a). Catching the next wave: patterning of the Drosophila eye by the morphogenetic furrow. In Singh A, and, & Kango-Singh M. (Eds.), Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye (Vol. II, pp. 75–97). Springer NewYork Heidelberg Dordrecht London: Springer International Publishing. [Google Scholar]
- Kumar JP. (2020b). Ghost in the Machine: The Peripodial Epithelium. In Madhuri A. a. K.-S. Singh (Ed.), In Molecular genetics of axial patterning, growth and disease in the Drosophila eye. (Vol. II, pp. 121–141). NewYork: Springer. [Google Scholar]
- Kumar JP, & Moses K. (2001). The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development, 128(14), 2689–2697. [DOI] [PubMed] [Google Scholar]
- Li S, Cho YS, Wang B, Li S, & Jiang J. (2018). Regulation of Smoothened ubiquitylation and cell surface expression through a Cul4-DDB1-Gbeta E3 ubiquitin ligase complex. J Cell Sci, 131(15). doi: 10.1242/jcs.218016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lee OK, Hsu YC, Singh A, & Choi KW. (2007). Drosophila TRAP230/240 are essential coactivators for Atonal in retinal neurogenesis. Dev Biol, 308(2), 322–330. doi:S0012–1606(07)01087–1 [pii] 10.1016/j.ydbio.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HC, Wu JT, Tan BC, & Chien CT. (2009). Cul4 and DDB1 regulate Orc2 localization, BrdU incorporation and Dup stability during gene amplification in Drosophila follicle cells. J Cell Sci, 122(Pt 14), 2393–2401. doi:jcs.042861 [pii] 10.1242/jcs.042861 [DOI] [PubMed] [Google Scholar]
- Ma C, & Moses K. (1995). Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development, 121(8), 2279–2289. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, & Rubin GM. (1994). dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development, 120(12), 3473–3486. [DOI] [PubMed] [Google Scholar]
- Mehta AS, & Singh A. (2019). Insights into regeneration tool box: An animal model approach. Dev Biol, 453(2), 111–129. doi: 10.1016/j.ydbio.2019.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, & Sprecher SG. (2020). Early eye Development: Specification and Determination. In Singh A, and, & Kango-Singh M. (Eds.), Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye (pp. 1–52). Springer NewYork Heidelberg Dordrecht London: Springer. [Google Scholar]
- Mlodzik M, Baker NE, & Rubin GM. (1990). Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev, 4(11), 1848–1861. [DOI] [PubMed] [Google Scholar]
- Moran MT, Tare M, Kango-Singh M, & Singh A. (2013). Homeotic Gene teashirt (tsh) has a neuroprotective function in amyloid-beta 42 mediated neurodegeneration. PLoS One, 8(11), e80829. doi: 10.1371/journal.pone.0080829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, & Banerjee U. (2007). Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development, 134(5), 825–831. doi:dev.02788 [pii] 10.1242/dev.02788 [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, & Dickson BJ. (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development, 127(4), 851–860. [DOI] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, & Gehring WJ. (1999). Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development, 126(10), 2253–2260. [DOI] [PubMed] [Google Scholar]
- Oros SM, Tare M, Kango-Singh M, & Singh A. (2010). Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev. Biol, 346(2), 258–271. doi:S0012–1606(10)00975–9 [pii] 10.1016/j.ydbio.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, . . . Sun YH. (1998). The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev, 12(3), 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F, & Casares F. (2000). homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev, 96(1), 15–25. doi: 10.1016/s0925-4773(00)00372-5 [DOI] [PubMed] [Google Scholar]
- Poulson DF. (1950). Histogenesis, oogenesis, and differentiation in the embryo of Drosophila melanogaster meigen. In Demerec M. (Ed.), Biology of Drosophila (pp. 168–274). New York: Wiley. [Google Scholar]
- Quiring R, Walldorf U, Kloter U, & Gehring WJ. (1994). Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science, 265(5173), 785–789. doi: 10.1126/science.7914031 [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, & Benzer S. (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol, 53(2), 217–240. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, & Mann RS. (1997). Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell, 91(2), 171–183. doi: 10.1016/s0092-8674(00)80400-6 [DOI] [PubMed] [Google Scholar]
- Royet J, & Finkelstein R. (1997). Establishing primordia in the Drosophila eye-antennal imaginal disc: the roles of decapentaplegic, wingless and hedgehog. Development, 124(23), 4793–4800. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Gogia N, Farley K, Payton L, & Singh A. (2018). Characterization of a morphogenetic furrow specific Gal4 driver in the developing Drosophila eye. PLoS One, 13(4), e0196365. doi: 10.1371/journal.pone.0196365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Gogia N, Glenn N, Singh A, Jones G, Powers N, . . . Singh, A. (2018). A soy protein Lunasin can ameliorate amyloid-beta 42 mediated neurodegeneration in Drosophila eye. Sci Rep, 8(1), 13545. doi: 10.1038/s41598-018-31787-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikaku MA, & O’Tousa JE. (1994). sine oculis is a homeobox gene required for Drosophila visual system development. Genetics, 138(4), 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ 3rd, Reis T, Edgar BA, & Duronio RJ. (2008). Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell, 15(6), 890–900. doi: 10.1016/j.devcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver SJ, & Rebay I. (2005). Signaling circuitries in development: insights from the retinal determination gene network. Development, 132(1), 3–13. doi: 10.1242/dev.01539 [DOI] [PubMed] [Google Scholar]
- Singh A. (1995). Enhancer trap technique- A novel tool for identification and developmental characterization of Drosophila genes. . Current Science, 68(5), 517–525. [Google Scholar]
- Singh A. (2012). Neurodegeneration- A Means to an End. J. Cell Science & Therapy, 3(3), 10000e10107. doi: doi: 10.4172/2157-7013.1000e107 [DOI] [Google Scholar]
- Singh A, Chan J, Chern JJ, & Choi KW. (2005). Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics, 171(1), 169–183. doi:genetics.105.044180 [pii] 10.1534/genetics.105.044180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, & Choi KW. (2003). Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development, 130(25), 6351–6360. doi: 10.1242/dev.00864 [DOI] [PubMed] [Google Scholar]
- Singh A, Gogia N, Chang CY, & Sun YH. (2019). Proximal fate marker homothorax marks the lateral extension of stalk-eyed fly Cyrtodopsis whitei. Genesis, 57(9), e23309. doi: 10.1002/dvg.23309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, & Gopinathan KP. (1998). Confocal microscopy: A powerful technique for biological research. Current Science, 74(10), 841–851. [Google Scholar]
- Singh A, & Irvine KD. (2012). Drosophila as a model for understanding development and disease. Dev Dyn, 241(1), 1–2. doi: 10.1002/dvdy.23712 [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Choi KW, & Sun YH. (2004). Dorso-ventral asymmetric functions of teashirt in Drosophila eye development depend on spatial cues provided by early DV patterning genes. Mech Dev, 121(4), 365–370. doi: 10.1016/j.mod.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, & Sun YH. (2002). Eye suppression, a novel function of teashirt, requires Wingless signaling. Development, 129(18), 4271–4280. [DOI] [PubMed] [Google Scholar]
- Singh A, Lim J, & Choi K-W. (2005). Dorso-ventral boundary is required for organizing growth and planar polarity in the Drosophila eye. In Mlodzik M. (Ed.), “Planar Cell Polarization during Development: Advances in Developmental Biology and Biochemistry” (pp. 59–91): Elsevier Science & Technology Books. . [Google Scholar]
- Singh A, Shi X, & Choi K-W. (2006). Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development, 133(23), 4771. doi: 10.1242/dev.02686 [DOI] [PubMed] [Google Scholar]
- Singh A, Tare M, Kango-Singh M, Son WS, Cho KO, & Choi KW. (2011). Opposing interactions between homothorax and Lobe define the ventral eye margin of Drosophila eye. Dev Biol, 359(2), 199–208. doi: 10.1016/j.ydbio.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensmeier AM, Tare M, Puli OR, Modi R, Nainaparampil J, Kango-Singh M, & Singh A. (2013). Novel Neuroprotective Function of Apical-Basal Polarity Gene Crumbs in Amyloid Beta 42 (Aβ42) Mediated Neurodegeneration. PLoS One, 8(11), e78717. doi: 10.1371/journal.pone.0078717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, & Verheyen EM. (2012). Wnt/Wingless signaling in Drosophila. Cold Spring Harb Perspect Biol, 4(6). doi: 10.1101/cshperspect.a007930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, & Du W. (2008). Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol, 313(2), 787–801. doi: 10.1016/j.ydbio.2007.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M, Modi RM, Nainaparampil J, Puli OR, Bedi S, Fernandez-Funez P, . . . Singh, A. (2011). Activation of JNK Signaling Mediates Amyloid-ß-Dependent Cell Death. PLoS One, 6(9), e24361. doi: 10.1371/journal.pone.0024361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M, Puli OR, Moran MT, Kango-Singh M, & Singh A. (2013). Domain specific genetic mosaic system in the Drosophila eye. Genesis, 51(1), 68–74. doi: 10.1002/dvg.22355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M, Puli Roy O, Singh A. (2013). Molecular Genetic Mechanisms of Axial Patterning: Mechanistic Insights into Generation of Axes in the Developing Eye. In Singh MK-SA. (Ed.), Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye (Vol. I, pp. 37–73): Springer-Verlag New York. [Google Scholar]
- Tare M, Sarkar A, Bedi S, Kango-Singh M, & Singh A. (2016). Cullin-4 regulates Wingless and JNK signaling-mediated cell death in the Drosophila eye. Cell Death Dis, 7(12), e2566. doi: 10.1038/cddis.2016.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, & Ready DF. (1987). Cell fate in the Drosophila ommatidium. Dev Biol, 123(1), 264–275. doi: 10.1016/0012-1606(87)90448-9 [DOI] [PubMed] [Google Scholar]
- Treisman JE, & Rubin GM. (1995). wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development, 121(11), 3519–3527. [DOI] [PubMed] [Google Scholar]
- Verghese S, Bedi S, & Kango-Singh M. (2012). Hippo signalling controls Dronc activity to regulate organ size in Drosophila. Cell Death Differ., 19(10), 1664–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkorn E, Sarkar A, Garcia K, Kango-Singh M, & Singh A. (2015). The Hippo pathway effector Yki downregulates Wg signaling to promote retinal differentiation in the Drosophila eye. Development, 142(11), 2002–2013. doi: 10.1242/dev.117358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, & Ready DF. (1991). The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development, 113(3), 841–850. [DOI] [PubMed] [Google Scholar]
- Won JH, Tsogtbaatar O, Son W, Singh A, Choi KW, & Cho KO. (2015a). Cell type-specific responses to wingless, hedgehog and decapentaplegic are essential for patterning early eye-antenna disc in Drosophila. PLoS One, 10(4), e0121999. doi: 10.1371/journal.pone.0121999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won JH, Tsogtbaatar O, Son W, Singh A, Choi KW, & Cho KO. (2015b). Correction: Cell type-specific responses to wingless, hedgehog and decapentaplegic are essential for patterning early eye-antenna disc in Drosophila. PLoS One, 10(5), e0128169. doi: 10.1371/journal.pone.0128169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Palmer L, Alexandre C, Kakugawa S, Beckett K, Gaugue I, . . . Vincent JP. (2016). Godzilla-dependent transcytosis promotes Wingless signalling in Drosophila wing imaginal discs. Nat Cell Biol, 18(4), 451–457. doi: 10.1038/ncb3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Schonherr C, Varshney GK, Dogru M, Hallberg B, & Palmer RH. (2013). Goliath family E3 ligases regulate the recycling endosome pathway via VAMP3 ubiquitylation. EMBO J, 32(4), 524–537. doi: 10.1038/emboj.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, & Pignoni F. (2006). Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development, 133(24), 4881–4889. doi: 10.1242/dev.02669 [DOI] [PubMed] [Google Scholar]
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S, . . . Edgar BA. (2011). Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature, 480(7375), 123–127. doi: 10.1038/nature10579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.