Abstract
Conventional drug development strategies typically use pocket in protein structures as drug-target sites. They overlook the plausible effects of protein evolvability and resistant mutations on protein structure which in turn may impair protein-drug interaction. In this study, we used an integrated evolution and structure guided strategy to develop potential evolutionary-escape resistant therapeutics using receptor binding domain (RBD) of SARS-CoV-2 spike-protein/S-protein as a model. Deploying an ensemble of sequence space exploratory tools including co-evolutionary analysis and deep mutational scans we provide a quantitative insight into the evolutionarily constrained subspace of the RBD sequence-space. Guided by molecular simulation and structure network analysis we highlight regions inside the RBD, which are critical for providing structural integrity and conformational flexibility. Using fuzzy C-means clustering we combined evolutionary and structural features of RBD and identified a critical region. Subsequently, we used computational drug screening using a library of 1615 small molecules and identified one lead molecule, which is expected to target the identified region, critical for evolvability and structural stability of RBD. This integrated evolution-structure guided strategy to develop evolutionary-escape resistant lead molecules have potential general applications beyond SARS-CoV-2.
Keywords: Covid-19, SARS-CoV-2, Receptor binding domain, Sequence space analysis, Co-evolution, Deep mutation scan, Molecular dynamic simulation, Structure network analysis, Fuzzy C-means clustering, Druggability, Machine learning
1. Introduction
Covid-19 has emerged as the worst pandemic [1] of recent times with catastrophic impacts on human lives and global economy [2]. Many countries have already experienced unprecedented escalation of Covid-19 infections, and the emergence of different variants of concerns has been a continuing problem [3], [4], [5], [6], [7]. Although global vaccination efforts have been ramped up, the immune-escape variants of SARS-CoV-2 with increased infectivity can be a bottleneck towards developing therapeutic and preventive strategies [8], [9], [10], [11], [12], [13]. In the recent past different variants of concerns (e.g. alpha, beta, delta) had resulted in successive episodes of rapid infection in major parts of the world [14]. More recently emerged SARS-CoV-2 Omicron variant with higher infectivity rates is a global concern [15], [16]. The evolutionary process leading to the emergence of new variants of a disease related protein would allow it to potentially evade the action of drugs as well as vaccine induced antibodies. As a result, better understanding of the evolutionary trends of escape-variant emergence and accordingly restructuring current therapeutic protocol could be a promising way to have a solution of many diseases, including Covid-19 [17].
For Covid-19 majority of the therapeutic strategies are aimed at blocking the virus entry into the host cells [18], which is primarily driven by the SARS-CoV-2 spike (S) protein through its interaction with the angiotensin-converting enzyme 2 (ACE2) [18], [19], [20], [21], [22], [23], [24]. ACE2 receptors are located at the outer membranes of cells in the lungs, arteries, heart, kidney, and intestines. The coronavirus entry into the host cell is a complex process, which involves multiple processing stages of the homo-trimeric S-Protein [21], [22] (comprised of S1 and S2 subunit in each monomer).
The S-Protein undergoes structural reorganization to allow the fusion of the viral membrane with the host cell [25], [26], [27], [28] using its receptor binding domain (RBD). Receptor binding destabilizes the pre-fusion trimer leading to the dissociation of the S1 subunit with ACE2, and stimulating the transition of S2 subunit from a metastable pre-fusion state to a more-stable post-fusion state [21], [29], [30]. Previous report suggested that the RBD of S1 subunit, as in other coronaviruses [30], [31], follows a hinge-like movement that triggers the transition between receptor inaccessible “down” conformation and receptor accessible “up” conformation [21], [27], [32]. The loop dominant RBD region of the S-protein has been reported to be the key in the process of receptor binding [33].
A number of potential FDA-approved drugs of different classes have been under trial or used for therapeutic interventions against Covid-19 [34], [35], [36], [37]. Although mutational episodes in proteins is critical for viral evolution (appreciated well in the S-Protein of SARS-CoV-2), traditional drug development efforts typically ignore the complications associated with rapid mutations [34]. However, there are available previous studies, which have focused on the evolutionary trends of the S-Protein and its similarity with other members of coronavirus family [33], [38], [39], [40]. Recent reports have also predicted the mutability of the S-Protein as well as its inter-human transmissibility and immune-escape ability [41], [42]. In addition, one of the recent structural studies have generated model structures of S-protein to predict the mutations of higher stability [43]. In other studies, the interactions of RBD region with human ACE2 have been investigated to obtain insights into virulence strengths of some of the variants [44], [45], [46]. There are previous reports of designing peptides as well as small molecules to target the RBD region, which would potentially inhibit virus entry [47], [48], [49], [50]. In this context the strategy we present in our story stands unique as we aimed to identify RBD sub-structure which is critical for its evolvability and structural stability. We further went on to assess its druggability. Thus, targeting this identified region/RBD-sub-structure could potentially disrupt viral evolvability.
We devised a strategy to develop small molecule therapeutics, which can potentially inhibit the emergence of SARS CoV-2 evolutionary escape variants using RBD as a possible model (Fig. 1 ). This proposed strategy involves a two-tier approach (as shown in the Schematic in Fig. 2 ) encompassing evolutionary and structural analysis. The evolutionary analyses described here provides an insight into the evolvability scope of the RBD, while the molecular dynamic simulation investigated the flexibility of different region in the RBD. Also, the structure network analysis presented the residue distribution and their pair-wise interaction pattern by grouping the highly interacting residues together to form the clusters/structure blocks (SBs). We then integrated these two approaches by applying fuzzy logic principles to the structure blocks (SBs) with higher quantified evolutionary and structural features. Using this strategy, we have identified a structure block (SB10), which presented the highest evolutionary and structural impact on spike-RBD. Any disruption of this SB would not only impact the RBD structure but would also perturb its evolvability. This potentially makes SB10 an interesting pocket for drug targeting, which was further validated using a number of physical and biochemical parameters. We then screened a library of small molecules and ranked them by using a machine-learning model with defined molecular descriptors. We further studied the interactions between the molecules and SB10 and identified a potential lead as a plausible therapeutic lead disrupting viral evolvability.
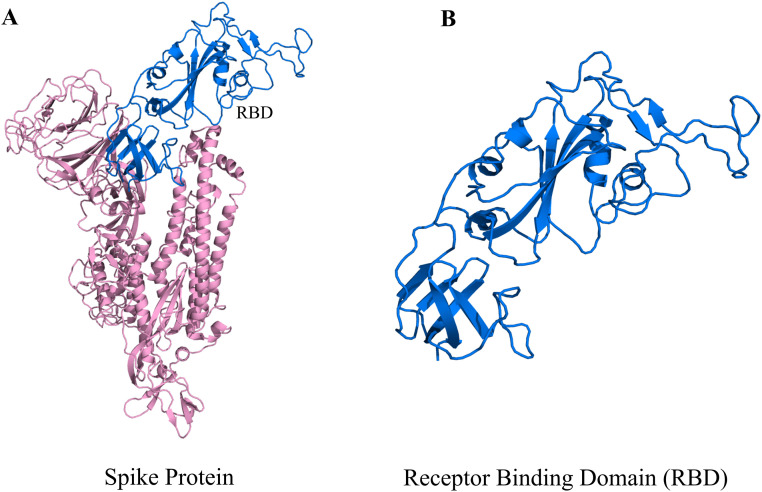
Fig. 1.
A representation of SARS-CoV-2 Spike protein monomer and the selection logistics of the protein segment. (A) In the monomeric Spike protein. Here RBD has been highlighted as blue whereas rest part of the Spike monomer is indicated by pink colour. (B) The RBD, which is a 273 amino acid, segment stretching from the residue 319 to 591 in the S-protein.
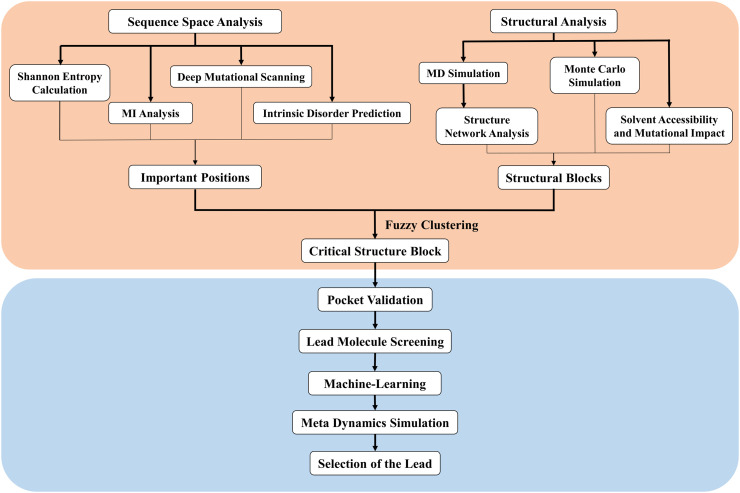
Fig. 2.
A flowchart of the integrated evolution and structure guided workflow as discussed in this manuscript. The proposed workflow has two components: first, the identification of a critical region of RBD based on a combination of evolutionary and structural studies (pink); and second, a druggability assessment of small molecule leads targeting the above critical region (blue). In the first section (pink), we performed the mentioned analyses to identify the region of RBD that is evolutionarily very much significant. Also, we performed Structural analysis to find out important Structure Block (SB) from structural perspective. Then we combined these two information by applying Fuzzy Clustering analysis and identified the most important SB in the RBD which can be used as the druggable pocket. Then in the second section (Blue region), we validated the residues of the identified important SB as potential drug binding sites, considered drug molecules from database, selected top 100 molecules and performed Docking study and Metadynamic Simulation to select potential drug candidate against Covid-19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Interestingly a validation of the presented strategy comes from the recently emerged variants (e.g. Alpha variant, Beta variant, Delta variant, and Omicron variant), where majority of the mutations are either in or around the identified SB10. We further observe that most of these positions are evolutionarily highly interdependent. As a result, we argue that any therapeutic lead targeting SB10 would potentially be useful against the newly emerged strains. While this evolution and structure guided framework to identify critical regions in a protein is developed using SARS-CoV-2 as a potential test case example, we believe that this strategy can be explored in other disease systems involving mutating proteins.
2. Methods
For this study, we used the Cryo-EM structure (PDB ID: 6VSB) of SARS-CoV-2 spike protein. From there, we extracted the amino acid sequence information of the stretch, which constitutes the ACE2 receptor binding domain (RBD) as our query sequence. We used BlastP and selected 1000 non-redundant protein sequences [51]. We further used this dataset in Clustal Omega to obtain multiple sequence alignment (MSA) [52], [53] and performed sequence space analysis.
2.1. Sequence space analysis
We analyzed the MSA of homologous proteins to explore two evolutionary features; 1) amino acid sequence conservation and 2) co-varying/interdependent positions throughout the course of evolution [54]. We calculated Shannon's entropy (Eq. (1)) and performed mutual information (MI, Eq. (2)) study to predict positional correlations in MSA to determine the conserved as well as interdependent positions respectively [54], [55].
| (1) |
In Eq. (1), ‘i’ represented the sequence position, P(a i) designated the probability of amino acid ‘a’ to be present at the ‘i'th column of MSA. S(i) represented the Shannon's entropy score, its lower values correspond to the fully conserved amino acid residues at ith position [55]. Whereas increase in Shannon's entropy score indicated the probability of that particular position to be less conserved, i.e. more random. Gaps in each column were treated as uniformly distributed amino acids [55].
| (2) |
where P(ai, bj) described the probability of finding amino acids of type “a” and “b” at the respective sequence positions i and j simultaneously [56]. MI(i,j) indicated the coevolution propensity of position i and j. The gaps were treated as the 21st amino acid type. This MI(i,j) value spans in the range from 0 to MImax where 0 corresponds to the fully uncorrelated residues and the highest value indicate the most interdependent pairs of residues. In order to capture higher co-evolutionary signals in RBD, which in turn could reliably indicate the sequence space constraints, we considered top 30 % of the co-varying pairs, and hence the cut off value for MI was selected as 0.75. We used this positional information to generate co-evolutionary matrix and represented as a heatmap.
We then used EVcouplings [57] to perform deep mutational scan to assess the quantitative effects of mutations in RBD [58] and generated the mutational landscape. Co-evolutionary network was generated using the program Gephi by using Thomas Fruchterman & Edward Reingold graph layout [59]. It simulates the graph as a system of particles in a force directed layout. In the presented layout, the nodes that the drawing algorithm assumes as particles are the residue positions and the edges refer to the position-position MI values. Edge width is defined by the MI values associated with position-position pairs. We constructed the Phylogenetic tree using neighbor-joining (NJ), algorithm based on the aligned sequence as implemented in the Rate4Site program [60].
2.2. Intrinsic disorder analysis
We evaluated the predisposition for intrinsic disorder for each of the residues of the 15 most evolutionary similar Spike RBD sequences by PONDR® VSL2 [61]. Access to this disorder predictor is provided by the PONDR platform (http://www.pondr.com/). This predictor combines two predictors optimized for long (>30 residues) and short (≤30 residues) disordered regions, respectively, using weights generated by a third meta-predictor. All three component predictors are logistic regression models built on balanced training sets. Attribute selection and window length optimization were performed independently for the three component predictors to maximize prediction accuracy. PONDR® VSL2 is one of the most accurate stand-alone disorder predictors, which is statistically better for proteins containing both structured and disordered regions.
2.3. Structure-based analysis
2.3.1. Model preparation and MD simulations
We retrieved the S-Protein sequence from the recently solved Cryo-EM S-protein structure (6VSB) and selected the structural-stretch with residues, which constitute the receptor binding domain (RBD), i.e. from residue 319 to residue 591. We manually refined the structure by incorporating the missing residues at their positions (according to the amino acid sequence information) using an in-built builder tool of PyMOL (Version 2.4.1) [62], [63]. We performed an in vacuo energy minimization with this above refined RBD structure in order to minimize unfavorable interactions and steric clashes using previous protocol [64]. We used this in vacuo energy-minimized structure for final system preparation.
When the study was carried out, we did not find out any structure of the isolated RBD in the literature. Most of the available structures were ACE2 receptor bound and in many of them large number of residues were missing. Hence we considered the above mentioned Cryo-EM S-protein structure.
We solvated the system with SPC/E water model and neutralized it with an appropriate number of counter ions using 4 chlorine ions. We equilibrated the system twice under NVT and NPT conditions at 27 °C and 1 bar pressure for 100 ps each with position restraints. Thereafter, we performed the MD simulation at 27 °C and 1 bar pressure with Gromos54a7 force field using the GROMACS 2019 software package. We performed this simulation for up to 1.5 μs and saved the conformations at 10 ps interval, yielding 150,000 conformations. Here we used V-rescale algorithm [65] and Parrinello and Rahman algorithm [66] to maintain constant temp and pressure respectively. We calculated the long-range electrostatic interactions by means of particle mesh Ewald (PME) method [67] and treated the short-range electrostatic as well as van der Waal interactions with a 10 Å of cut-off. In this simulation method, we used LINCS algorithm to constrain all bond lengths [68]. We deployed the leap-frog algorithm in order to integrate the equations of motion in every 2 fs.
Here we used the tools integrated with GROMACS package to analyze the data and PyMOL to visualize the structure. We applied a 2 Å cut off in order to cluster the conformations using gmx cluster tool and selected the centre conformation of the largest cluster for further structure analysis. By deploying the gmx rmsf tool for Cα atoms of each residue, we calculated root-mean-square fluctuation (RMSF).
2.3.2. Structure network analysis
The structure network illustration of a protein is a depiction of topological analysis of 3D structure irrespective of its secondary structure and folding type [69]. The internal motions as well as structural dynamics of proteins are directly associated with their function and activity; hence we used the normal mode analysis (NMA) for the prediction of functional motions in the protein segment [70]. Followed by NMA, we performed a correlation analysis to generate cross-correlation matrix. Further, by using correlation network analysis, we generated the all-residue network using the energy minimized structure of RBD of the spike protein. We further split this all residue network into a highly correlated coarse grained community cluster network by using Girvan-Newman clustering method where the highly interacting residues were grouped together in the clusters, each of which we referred in our studies as structure blocks (SBs).
2.3.3. Monte-Carlo simulation
To understand the dynamics of the RBD, we resorted to Monte Carlo simulation technique to simulate the RBD dynamics deploying CABS (C-alpha, beta, and side chain) coarse grained protein model [71]. The simulation parameters were modified at the number of cycle (Ncycle) and number of models skipped keeping the seed for random number generator at 3864. The ‘Number of cycles’ (Ncycle) field was set at 100 resulting in 20 × 100 = 2000 models in the trajectory. The ‘Cycles between trajectory frames’ (Nskipped) which refers to the number of models skipped on saving models was kept at 100. Total numbers of generated models were thus 20 × 100 × 100 = 200,000. We used a T = 1.2 which is close to the native state temperature.
We further deployed Tanford-Kirkwood model (TK) in which protein molecule is treated by a spherical cavity with dielectric constant ϵp and radius b surrounded by an electrolyte solution modeled by the Debye-Hückel theory. We resorted to the modification of the model, which included solvent static accessibility rectification for each of the residues which are ionisable and which takes into account the irregular protein-solvent interface. The model is referred as the Tanford-Kirkwood model with solvent accessibility and we in our study would refer to it as TKSA.
2.4. Combination of sequence and structure index
To integrate evolutionary and structural features of selected significant structure blocks (SBs) (1, 6, 9, 10 and 12) of the receptor binding domain/RBD, we used two subsets of properties that decipher structural characteristics and two subsets decoding the sequence space information of the SBs. Here our focus was only on the above-mentioned SBs as they have significantly higher number of constituent residues and influence the internal dynamics of RBD. We used Mean Hydrophobicity (Mean HP), and Order Index (extent of structural integrity) to quantify structural traits. Similarly, we also calculated conservation index for conserved positions and coevolution index for the highly interdependent positional patches in the sequence.
To identify the important SB of the RBD in terms of the above-mentioned sequence and structural properties we performed Principal component analysis (PCA) followed by Fuzzy clustering analysis.
PCA is a technique of dimensionality reduction by means of data projection, where multidimensional data is reduced to a few orthogonal, uncorrelated principal components, while preserving the information (variance) as much as possible [72]. We applied PCA to reduce the sequence-based as well as structural indices separately into principal sequence component and principal structure component respectively. Then we projected the sequence and structural indices data on to the sequence-structure 2-D subspace comprising of the principal components.
To identify structure blocks (SBs) in RBD which are evolutionarily constrained and structurally important, we used Fuzzy clustering on the reduced 2-D data based on fuzzy logic principle. FCM or fuzzy C-means clustering technique was utilized to partition a finite collection of “n” elements X = {x1, …xn} into a collection of c fuzzy clusters [73]. With a finite set of data, the algorithm returns a list of c cluster centers as C = {c1……cn) along with a partition matrix. Any point x has a set of coefficients, and this indicate the degree of being in the kth cluster wk(x). With fuzzy c-means, the mean of all points is the centroid of a cluster and is weighted by their degree of belonging to the cluster. Mathematically it can be represented as:
Here, m is the hyper- parameter that decides how fuzzy the cluster will be. The higher it is, the fuzzier the cluster will be in the end.
2.5. Pocket validation
To validate the potential druggability of the identified pockets using specific physical and biochemical properties we used two different computational methods. In one method, we considered solvent-accessible volume as a distinguishing parameter [74]. In other approach, we considered some properties, like polarity score, hydrophobic density, number of alpha spheres, density of cavities etc. and also Voronoi tessellation [75].
2.6. Screening of drug molecules
In order to identify the suitable FDA-approved drug molecules, we resorted to a dataset of drugs from the database ZINC [76] accessed on April 2021. From this dataset, we applied the “Fda” filter and selected the dataset of 1615 FDA-approved drugs. Then we sorted the selected drugs in the decreasing order of their Quantitative Estimation of Drug-likeness (QED) values [77]. We also applied a machine learning model (see Supplementary Data, Screening of Drug molecule) that had already been trained using Delaney Solubility Dataset based on an extra trees regressor model (Supplementary Fig. 3) in order to select top 100 drug molecules (Supplementary Table 1) according to their aqueous solubility. We used these 100 molecules for docking study with the predicted important SB in the RBD and to identify top 10 drug molecules that interact with the SB10 with the highest binding affinity. The details of their QED and solubility values have been shown in Supplementary Table 2.
2.7. Molecular docking
Using the AutoDock Vina 1.5.6 program, we carried out the molecular docking study [78]. Our docking study focused on the SB10 of RBD. We resorted the 3D structures of the selected ligand molecules (drugs) from Pubchem [79] (in sdf format) and converted them into PDB format using OpenBabel program [80]. We then selected the rotatable bonds in the ligand molecules and prepared pdbqt files of the drug molecules using the AutoDock Tools [81] to construct a library of previously selected 100 molecules. Next, we prepared the protein (spike RBD) with Autodock Tool 1.5.6 [81], added polar hydrogens, and calculated Kollman charges. We then selected the residues of the identified potential SB (SB10) as drug target sides. After the construction of ligand library, preparation of receptor molecules and the protein, we virtually docked all 100 compounds in the library at a time into the target binding site using Perl code in AutoDock Vina 1.5.678 program. We conducted the docking with exhaustiveness of 28, number modes of 10 and energy range of 4 by considering the spike RBD as rigid, whereas ligands were flexible in nature.
In addition to AutoDock Vina, we also used the MedusaDock server [82], [83], [84] to perform docking study using SB10 of RBD as the ligand binding region in order to verify the consistency.
2.8. Metadynamic simulation
In order to establish that the docked configuration indeed corresponds to a stable, minimum energy configuration, we performed meta-dynamics simulations, an enhanced sampling technique that is used to construct a free energy profile along a collective variable. Using a history-dependent biasing potential, the technique ensures efficient sampling of all meta-states (ligand bound and unbound configurations). In order to ensure that the system is not trapped in local minima, an energy Gaussian is deposited corresponding to every state that is visited. Therefore, each time the simulation visits a configuration, the deposited energy ensures that other states are sampled at simulation timescales. The deposited potential at the end of the simulation (when all states along the free energy landscape have been sampled), therefore corresponds to the free energy of the states.
In order to ascertain the relative free energies of the ligand-bound and unbound states, we run a metadynamics simulation using the distance between the center of mass of the ligand and the binding pocket (ascertained from the docked configuration) as the collective variable. The binding pocket comprises of residue numbers 402–405, 408, 416–417, 418–421, 452–455, 480–483, 489. The height of the Gaussian hills deposited was set to 0.2 kcal/mol while the width of 2 Å was used to construct this landscape. Gaussian hills were deposited to the metadynamics potential every 1000 simulation steps. A 250-ns metadynamics trajectory was used to compute the free energy landscape along the aforementioned collective variable. The system sampled all states along the CV within the simulation timescale and the free energy profiles showed convergence. The metadynamics trajectory was simulated in NAMD and the CHARMM27 force field was employed. An electrostatic cutoff of 12 Å units and a van der Waals cutoff of 10 Å units along with periodic boundary conditions was used for pairwise calculations. The systems were first energy minimized and scaled heating to 310 K was performed. A 20-ns equilibration run was performed (at NPT) before we ran the metadynamics simulation for free energy calculations.
2.9. Drug selection
We used Discovery Studio Visualizer and PyMOL [62], [63] to visualize and analyze the interactions. Finally, we selected the drug molecules that had the lowest binding free energy (more negative) and had interactions with the key target sites of the spike RBD to analyze their conformations.
3. Results
Fig. 2 represents the research workflow, which is described in the manuscript. We discussed the methodical details of individual components of Fig. 2 in the materials and methods section.
3.1. Sequence space analysis has provided critical information on the evolutionarily conserved and co-varying positions/regions, sequence variability and mutational tolerance
For the sequence analysis, we need to refer the RBD residue stretch from 319 to 591 as positions 1 to 273, which was required to simplify the script. Hence, to avoid confusion, in the result portion of the sequence analyses, we would mention both the changed numbers for RBD (as required by the script) and the original residue number for the S-protein. As a part of Sequence Space Analyses, we performed Shannon's Entropy calculation (Fig. 3A) to identify the sequence patches, which are not expected to change or may change slowly during the course of evolution. Using this calculation, we found that the evolutionarily highly conserved residues (which would have zero Shannon's entropy) are scattered throughout the RBD sequence space. As Shannon's entropy increased, the extent of conservation decreased; which means that these residues would be more interdependent. In a broad sequence patch between 122 and 232 in RBD (i.e., residues 440 to 550 in the S-protein) the entropy profile displayed some spikes (high values). These spikes indicate less conservation (Fig. 3A).
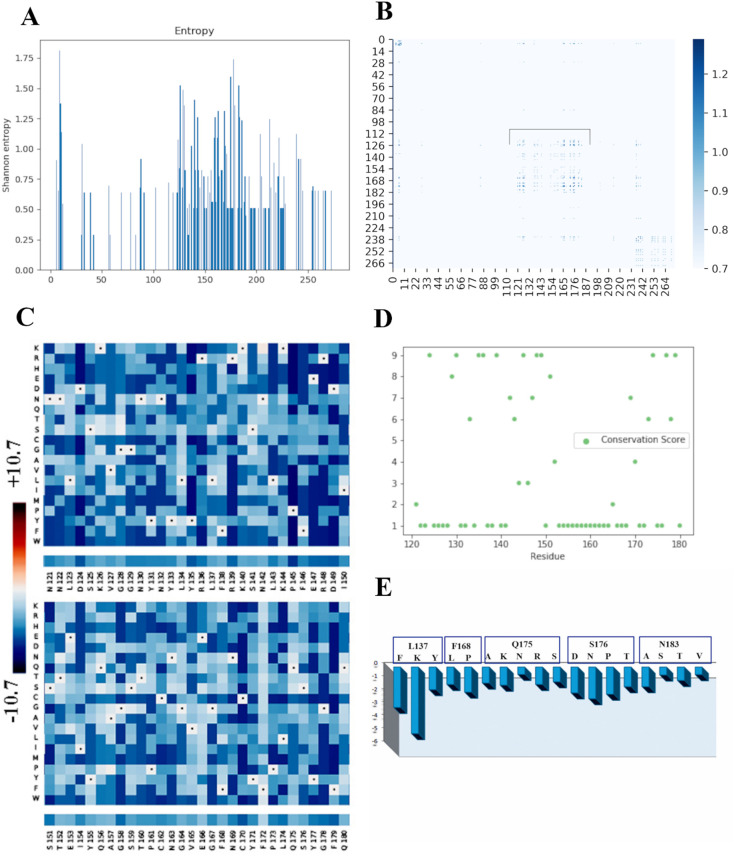
Fig. 3.
Sequence space study of the spike RBD reveals critical areas, which are under evolutionary constraints. (A) Shannon's Entropy profile of the RBD sequence space. Here the spike height is inversely proportional to the conservation propensity of the residue position. This profile shows that mostly less conserved residues (having high spikes) are positioned between 122 and 232 (i.e., residues 440 to 550 in the S-protein). (B) Heat-map showing co-evolution as quantified using MI values, in the RBD sequence space. Colour bar in the left shows the gradient of MI values in the co-evolving sites. Bracketed (black) segment, i.e. residue patch from 121 to 180 (439 to 498 w.r.t. the S-protein) are highly interdependent as they show the highest co-evolution. (C) Mutational landscape as generated from the Deep Mutation Scan is shown as a heat-map with colour bar in the left showing the gradient of Statistical Energy. The bottom bars below each of the sub maps depict the sequence information for the spike RBD. This profile indicates how much a protein structure can tolerate the effect of mutation at a particular residue position. The intensity of the blue colour is inversely proportional to the extent of stability of a particular position w.r.t. an amino acid substitution. (D) Conservation profile of the spike RDB as calculated by Bayesian Method. The high conservation score is proportional to the extent of conservation of a particular position. (E) Bar plots depicting differences in Statistical Energy (dE) profiles upon substitution mutations with residues derived from variations observed among the most likely closest relatives at positions known to be critical in RBD-ACE2 interaction. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although Shannon's Entropy Calculation captures the rate of evolution, it does not provide any information on the residue-residue pairwise interaction in evolutionary timescale, i.e., the coupling propensity of amino acid positions. This was achieved by performing the Co-variation Analysis (Mutual Information or MI calculation). Mutations in a non-conserved position would result in a compensatory substitution at another position to preserve overall structure of the protein. This compensatory mutation would hence determine the position-position interdependencies. We performed the MI analysis by using a large dataset of 1000 similar sequences to understand the positional interdependencies. As discussed in detail in the Methods section, these 1000 similar sequences were obtained from the Multiple Sequence Alignment (MSA). A high MI value corresponds to higher positional inter-dependency and hence higher co-evolutionary signal from the sequence space. The densely dotted area in the MI heat-map (position 121–180, indicated by the bracket) (Fig. 3B) reflects the sub-region in the sequence space of RBD with maximal positional inter-dependent residues. We then used Deep Mutational Scan Analyses on the above-mentioned positions of 121 to 180 (439 to 498 w.r.t. the entire protein) to predict possible effects of the substitution mutations on the overall stability of this segment. Since viral genomes are prone for mutation events, which, in turn, shape their evolutionary fitness, this Deep Mutational Scan would provide a map of positional tolerance and its potential effect on biophysical fitness on the structure. Mutational landscape (Fig. 3C) shows a relatively high position-wide residue tolerance, which in our calculation is a function of ∆E (statistical energy) with particularly higher tolerance at positions 134, 137, 138, 140, 141, 142, 152, 153, 156, 157, 166, 168, 172, 175, 176 (positions 452, 455, 456, 458, 459, 460, 470, 471, 474, 475, 484, 486, 490, 493 and 494 with respect to the entire S-protein), where specific residue-based substitution is likely to confer higher stability. Intensity of the blue colour in individual cell (Fig. 3C) is inversely proportional to the extent of stability of a particular position w.r.t. an amino acid substitution.
We then took the position stretch spanning between 121 and 180 (from position 439 to 498 in case of the S-Protein), which represents maximal co-evolution signal, and calculated the conservation score (Fig. 3D). Fig. 3D shows that higher the conservation score of a position, higher is its conservation propensity and vice versa. It is interesting to note that positions spanning from 150 to 164; i.e., 468 to 482 w.r.t. the S-Protein (save 150 and 158) do show very low conservation, and, when compared to the mutational landscape, these regions also exhibit high mutation tolerance. On the other hand, the positions with higher conservation (spanning from 143 to 149; i.e., 461 to 467 of the S-protein) have relatively more restricted bias towards substitution mutations and show lesser tolerance (Fig. 3C).
We then generated the Statistical Energy profiles of the most commonly encountered substitution mutations in the sequence space for those residues, which participate in ACE2 receptor recognition. We retrieved the residue substitution information from those evolutionary branches, which were most likely the closest and were used for generating the phylogenetic tree (Supplementary Fig. 1A). Statistical Energy profile (Fig. 3E) gives a quantitative idea as to which mutations would vastly stabilize the protein. Save the lysine substitution at position 137 (which designates residue 455 in the S-protein) others mostly show comparable extent of stability and hence explain why they could occur in the other close relatives of RBD yet retaining the same structural integrity.
We then used graph theoretical approach on the co-evolutionary signal to generate a weighted matrix to map the highest co-evolving residues. Using the Fruchterman-Reingold graph layout (Supplementary Fig. 1B) we observed that positions 123,125, 126, 127, 137, 148, and 165 (441, 443, 444, 445, 455, 466 and 483 respectively w.r.t. S-protein) represent the nodes with maximum edges. This indicates that these nodes (positions) are highly interdependent and hence significant in terms of preserving the structure of the RBD. It is worth noting that these positions (except 148; i.e., 466 w.r.t. S-protein) also exhibited higher tolerance in the mutational landscape.
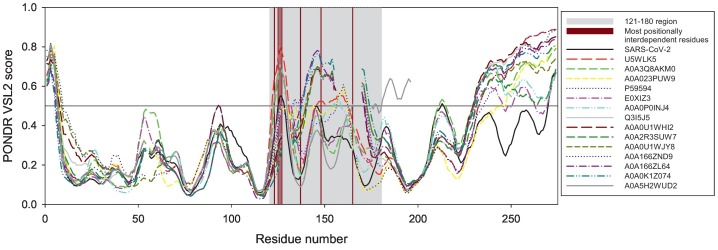
Since RBD contains substantial unstructured segments [85], we then analyzed the preservation of the intrinsic disorder predisposition within the amino acid sequences of the evolutionarily closest Spike RBDs. The phylogenetic tree analysis as described above showed 15 most likely closest proteins retrieved from the aligned sequences (Supplementary Fig. 1A) and further revealed that Spike RBD of SARS-CoV-2 has high degrees of sequence identity with other members of the spike glycoprotein family of Bat coronavirus origin [24], [86], [87]. We found that most likely the closest relative of the Spike RBD is the Spike protein (Fragment) n = 1 Tax = Bat coronavirus TaxID = 1,508,220 (UniRef90: A0A5H2WUD2). Fig. 4 summarizes the results of the sequence-based intrinsic disorder analysis, and shows that the overall shapes of the disorder profiles of these domains are characterized by the close similarity. One can clearly observe comparable patterns, where more flexible or even intrinsically disordered regions (i.e., regions with the disorder scores between 0.2 and 0.5 and above 0.5, respectively) are interspersed among more ordered segments. Many of these flexible/disordered regions correspond to the RBD loops. Fig. 4 also demonstrates that the largest variability in the per-residue disorder predisposition is observed for the C-tail of RBDs and their centrally located, 60-residue-long region (residues 121–180). We then compared the intrinsic disorder profiles with the mutational landscape as obtained from the deep mutation scanning. We found that many of the residues with high mutational tolerance were located within the intrinsically disordered or flexible regions (highlighted by dark red colour in Fig. 4).
Fig. 4.
Per-residue intrinsic disorder predisposition of the 15 most evolutionary similar Spike RBDs evaluated by PONDR® VSL2 (33). Breaks in the disorder profiles correspond to the breaks in sequence alignments. In this analysis, disorder scores between 0.2 and 0.5 indicate flexible regions, whereas regions with the disorder scores ≥0.5 are expected to be intrinsically disordered. Position of the 121–180 region corresponding to the area with the highest co-evolutionary signal and thus with the maximal positionally interdependent residues is shown by light grey shading. Dark red bars show position of residues, which are most positionally interdependent (123, 125, 126, 127, 137, 148, and 165). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To summarize, this sequence space analysis provides crucial information about the evolutionarily conserved and co-varying positions, sequence variability and mutational tolerance. It further identifies specific positions spanning between 121 and 180 (439 to 498 w.r.t. S-protein), which are extremely critical in defining the constraints in the evolutionary sequence space of RBD.
3.2. Structure analysis has provided important information on the regions of RBD which play critical role in ACE2 receptor recognition and associated conformational dynamics
By means of sequence space analysis, we have identified a sequence patch in RBD, which is evolutionarily interdependent. Now we performed an independent assessment using structural analysis to investigate important region in RBD that would influence the internal dynamics as well as the receptor recognition.
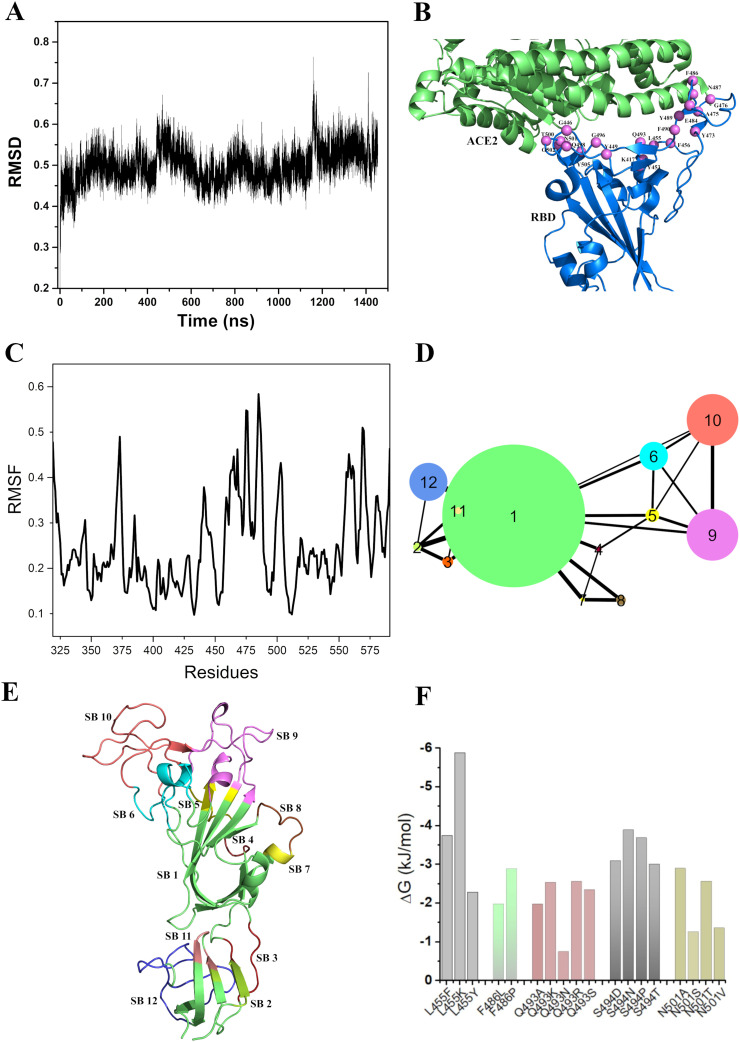
Since no experimental structure of unbound RBD in open conformation was available at the time of this study, we used a Cryo-EM structure of the S-protein by computationally building the missing residues (for details see Methods). As an initial stage of the structure analyses, we subjected this structure an all-atom MD simulation for 1.5 μs. We found that during the simulation the RBD structure was fairly stable (Fig. 5A). Now to obtain the equilibrated structure of RBD, we performed the cluster analysis and selected the centre conformation of the largest cluster (Fig. 5B). To investigate the backbone flexibility of RBD, we computed root-mean-square fluctuations (RMSF) for the Cα atoms of each residue from the simulation trajectory, in which we observed the presence of three highly dynamic regions spanning residue 360–375, 457–488 and 550–570 (Fig. 5C). Interestingly, we also observed that residue positions from 457 to 488 are housed into the evolutionary highly interdependent region (Fig. 3B). Furthermore, some positions from this region showed high tolerance towards residue substitution (Fig. 3C).
Fig. 5.
Structural analysis of RBD to identify the region, which plays critical role in ACE2 receptor recognition and associated conformational dynamics. (A) Root mean square deviation (RMSD) (in Ǻ) of protein Cα atoms with respect to the initial structure of RBD. It shows that the RBD structure was fairly stable during the simulation of 1.5 μs. (B) ACE2 receptor bound RBD structure. Violet spheres represent the ACE2 binding residues. We have labelled the spheres using single letter amino acid code. Blue and green colours represent RBD and ACE2 receptor respectively. (C) The RMSF profile (in Ǻ) represents the backbone flexibility of the equilibrated structure of RBD. It identifies three highly dynamic regions; 360–375, 457–488 and 550–570. (D) Structure Network plot of spike RBD where the size of the clusters/structure blocks (nodes) refers to the number of constituent residues. The width of the edges represents the magnitude of the interaction between the SBs. The RBD structure has been found to be comprised of 12 SBs. (E) The equilibrated RBD structure where different SBs has been designated by their corresponding colours and labelled alongside. (F) Plot showing the effects of destabilizing mutation (ΔG, kJ/mol) as indicated by the negative y-axis values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to unravel the internal arrangement and inter-dependency of the residues in terms of their pairwise interaction, we carried out structure network analysis [54]. We found that the residues of the RBD were split into twelve SBs or clusters (Fig. 5A) (Table 1 ). Among these, five SBs (namely, 1, 6, 9, 10 and 12) contained large numbers of residues (Fig. 5D) with dense connections with other SBs. We chose these five SBs (1, 6, 9, 10 and 12) for further investigation to understand how the residues in these clusters are significant in terms of evolutionary features, receptor binding, as well as local and global motions.
Table 1.
RBD Structure Block/Cluster Members.
| Structure block/cluster ID | Residue members |
|---|---|
| 1 | 319:322, 330:338, 355:366, 377:399, 411:414, 425:435, 510:527, 534:540, 544, 548:554, 572:573, 585:591 |
| 2 | 323:327, 541 |
| 3 | 328:329, 528:533 |
| 4 | 339:342 |
| 5 | 343:349, 400:401, 509 |
| 6 | 350:354, 415:424, 461:466 |
| 7 | 367:369 |
| 8 | 370:376 |
| 9 | 402:410, 436:450, 495:508 |
| 10 | 451:460, 467:494 |
| 11 | 542:543, 545:547 |
| 12 | 555:571, 574:584 |
We complemented the MD simulation using a course-grained Monte Carlo simulation (MC Simulation) and sampled 2 × 105 conformations. RMSF profiles from this MC simulation showed very high fluctuations (>6 Å, Fig. 5C) for the stretches 477 to 484 and 441 to 445. Incidentally, three residues (477,478 and 484) from this stretch were reported to be mutated in the recently emerged omicron variant [16]. This region is also highly disordered (Fig. 4), and hence would be important for the allostery and conformational flexibility guiding effective receptor binding.
The TKSA-MC (Tanford-Kirkwood model with solvent accessibility method) is a tool we used to determine residues with a destabilizing contribution to the protein native state. The algorithm calculates protein electrostatic energy, taking into account the contribution of each residue with charged side chain. The bar plots (Fig. 5F) show charge-charge energy contribution of residues, which are ionisable with respect to the protein native state stability. The bars (with negative y-axis) refer to the residues to be mutated to decrease the protein thermal stability. As per Ibarra-Molero model, residues with unfavorable energy values show ΔGqq ≥ 0 and are exposed to solvent with SASA ≥50% [88]. We found that a small number of potentially destabilizing mutations got picked up in the evolutionarily constrained segment of RBD further indicating low mutational tolerance of these positions.
The structure analyses indicate that residues 441 to 488 are important having significant influence on the allostery and conformational flexibility that lead to the receptor binding. It further identifies five highly interacting SBs (1, 6, 9, 10 and 12) which are comprised of large number of residues, which in turn can also influence the receptor binding.
3.3. Combined sequence and structure index analysis using fuzzy clustering identifies SB10 to be a critical region
Based on the comprehensive evolutionary and structural analysis of RBD we went on to identify a segment in the RBD structure, which is evolutionarily constrained and is structurally critical.
We calculated the evolutionary traits i.e. conservation and coevolution index of the selected SBs that represent the conservation and interdependency of the amino acid positions in the sequence respectively. We defined two structure indices viz. Mean HP (hydrophobicity) and Order Index (extent of structural integrity) that represent the structural features of individual SBs (structure blocks) of the RBD.
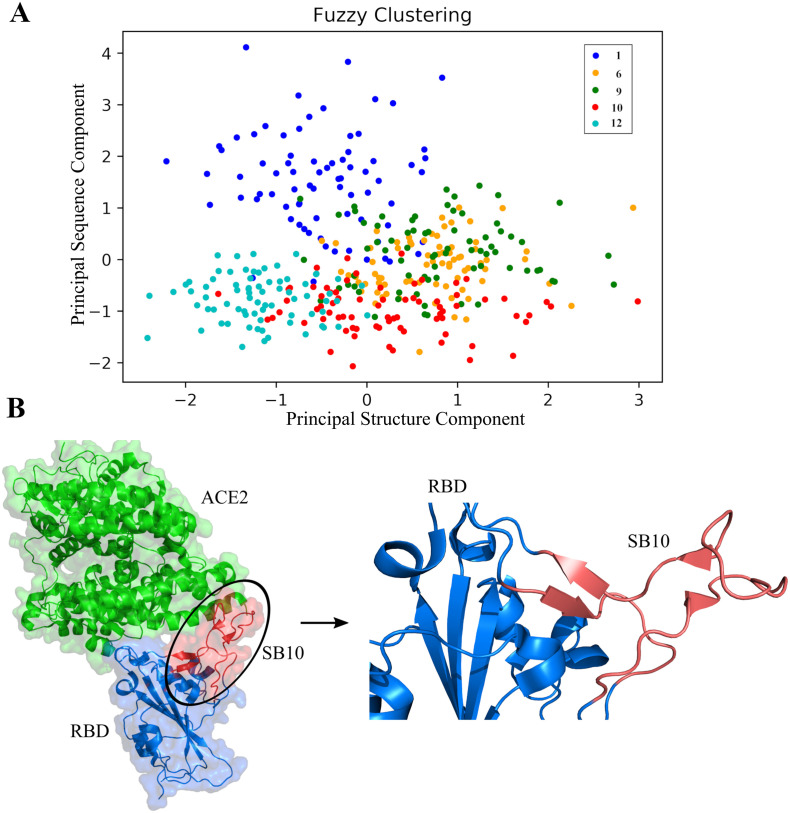
Our fuzzy clustering analysis helped to understand which structure blocks have maximal overlap and which one stands out with unique evolutionary and structural features. We resorted to fuzzy c-means clustering technique as we believed that this approach would essentially capture overlap among the blocks and hence their evolutionary and structural traits. Fuzzy clustering generalizes partition clustering methods like k-means and mediod which in the present study allowed an individual SB to be partially classified into more than one clusters [89]. Being a soft clustering method, it does not force individual SBs to be associated with one specific cluster, hence allowing more information to be parsed and interpreted in the context of overlap of evolutionary and structural features.
By combining the sequence and structure information, we essentially captured the overlaps between the evolutionary and structural traits of the SBs. We also found that SB1 stands out with a distribution pattern having the least overlap, which could be attributed to its difference in structural features and propensity towards conservation compared to other. In addition, we observed significant overlap of SB9 and SB10 with all other SBs in the fuzzy cluster profile. It indicates the importance of these two SBs towards the internal dynamics of the RBD.
3.4. Druggability assessment using computational screening and machine learning provides a potential small molecule lead
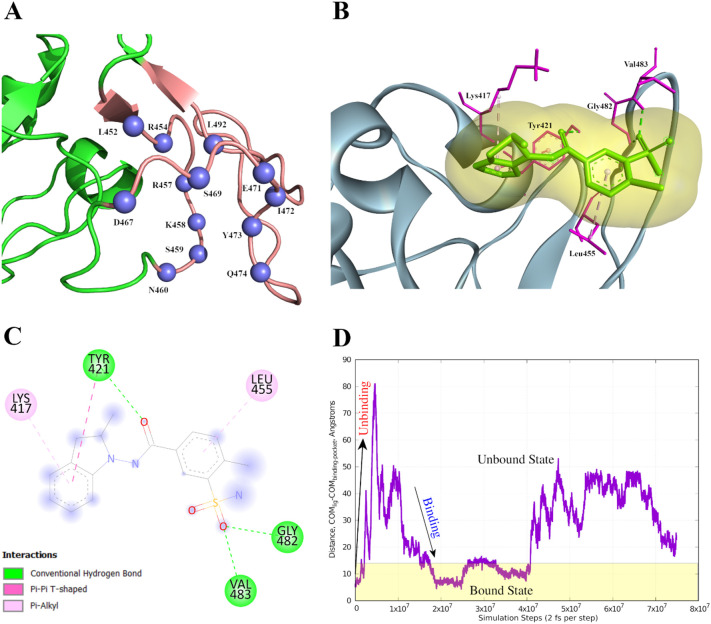
As discussed before, we identified SB10 as a potential pocket towards developing a small molecule/drug. To assess the druggability of SB10 we carried out binding pocket validation study and determined if this stretch contains potential drug binding sites. Considering solvent-accessible volume, which was calculated by combining three computational geometrical components (Voronoi diagram, Delaunay triangulation and alpha spheres), we identified Leu452, Asn460, Asp467, Ser469, Glu471, Ile472, Tyr473, Gln474 and Leu492 in SB10 as the most probable binding sites [74]. We found Arg454, Arg457, Lys458, Ser459 and Asn460 from SB10 to be important constituents of a highest ranked druggable pocket by combining mean polarity of all residues in a binding pocket, hydrophobic density, alpha spheres, cavity density as well as Voronoi tessellation [75]. Integrating the above-mentioned approaches, we identified 13 residues of SB10 as the potential druggable sites (Fig. 7A).
Fig. 7.
Representation of proposed lead molecule binding sites in RBD and its interaction with the therapeutic lead. (A) Blue spheres indicate the residues (of SB10) predicted as binding sites. The tv_red region indicates rest of the residues of SB10. The rest part of the RBD is represented in forestgreen. (B) R-Indapamide bound RBD where R-Indapamide is shown by green sticks with a pale orange surface around it. Its interactions with RBD has been represented as sticks (magenta) while rest of the protein is represented as slate blue. (C) 2D representation of docked complex which indicates the interacting residues. (D) Evolution of reaction coordinate during the metadynamics trajectory. Plot showing unbinding and binding events during the metadynamics simulation. The simulation protocol ensures that the system samples both unbound and bound configurations to compute the relative free energies of both configurations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, using the screening approaches (elaborated in the Method section), we selected top 100 drug molecules for the docking study against the predicted important SB (SB10) in the RBD. Our docking study aimed at predicting the protein-ligand complex structure by exploring the conformational space of the ligand molecules within the selected target sites of the protein. From the molecular docking study, we identified 10 molecules that interacted with the SB10 and have the highest binding affinity. Docking study revealed that R-Indapamide molecule interacted with SB10 with the lowest binding free energy (−7.4 kcal/mol) (Fig. 7B-C). On comparing the ligand conformations obtained after docking with AutoDock Vina and MedusaDock server, we observed nearly similar poses of the ligand while interacting with the SB10 of spike RBD (Supplementary Fig. 4).
In order to establish that the docked configuration indeed corresponds to a stable configuration, we performed metadynamics simulations. As the system revisits the same location along the collective variable space, an energy penalty in the form of a biasing potential ensures that it samples wider region of the phase space. A 250-ns metadynamics trajectory was used to compute the free energy landscape along the aforementioned collective variable. The system sampled all states along the CV within the simulation timescale and the free energy profiles showed convergence. As evident from the above free energy profile, we observed a global free energy minimum for ligand-binding site distance of <10 Angstroms (1 nm). These distances correspond to the R-Indapamide-bound state of RBD and, therefore, represent a stable configuration (Fig. 7D).
Although we do not have any experimental validation, the present study shows that this molecule has the potential to directly interact with the RBD at a region, which is evolutionarily and structurally constrained (SB10).
4. Discussion
The present study integrates an evolutionary and structure-guided approach and provide insights into the hidden traits of RBD, which otherwise remains unexplored by conventional structure-based studies. Sequence space analyses captures the evolutionary trend (of being conserved or co-varying) of the amino acid positions in a protein sequence [55]. Structure based study, in contrast, is an essential aspect to explore the internal orchestration, structural dynamics and functionality of the protein [54]. A combined approach of sequence based evolutionary study and structural exploration unveiled the importance of the evolutionarily significant regions in RBD and their roles in controlling conformation flexibility.
Our evolutionary analyses provided an insight into the constraints of RBD sequence space and reflected the importance of the stretch spanning from 121 to 180 (which is 439 to 498 in the S-protein) in RBD evolvability. Deep mutation studies as well as intrinsic disorder analysis also revealed the importance of the same region. This evolutionarily important region obtained from sequence space analysis was further mapped onto the RBD structure in order to understand how evolutionary traits influence the structural properties. Our structural analysis revealed that evolutionary critical residues, ACE2 receptor binding residues and highly fluctuating regions are mainly positioned into two regions, SB9 and SB10.
The combined strategy presented here explored how the local structural orchestration and the stability factors, i.e. the structural features (discussed in “Comparison between Sequence and Structure Index” in the Method Section) have been associated with the evolutionary context of RBD. We predict that a structure block (SB) with high HP would demonstrate the propensity to become less co-evolving and vice versa. Alternatively, this study implies that majority of interacting residues of RBD would be present in the SBs with less HP. This correlation of structural property with evolutionary trend for the selected SBs uncovers the significance of structural adaptability in sustaining the functional dynamics, along with sequence variations that confer specificity. Our use of fuzzy clustering technique effectively unveils the overlap of SBs based on their cumulative evolutionary and structural traits (Fig. 6A). Interestingly, we observed a significant overlap in the fuzzy cluster profile between SB9 and SB10 based on their evolutionary and structural features. We found from the MI signal in our co-variation analysis that some residue stretches in SB9 and majority of the residues of the SB10 were highly co-varying (interdependent). Being associated with ACE2 interaction and forming the RBD-ACE2 interaction sphere, residues constituting SB9 and SB10 would hence be important for the interaction and the associated conformational changes. Furthermore, the mutation landscape reveals more residues with high tolerance would be present inside SB10 (452, 455, 456, 458, 459, 460, 470, 471, 474, 475, 484, 486, 490, 493 and 494) indicating their implications on structure and evolution.
Fig. 6.
Fuzzy clustering analysis integrating evolutionary and structural features of RBD revealed Structure Block (SB) 10 as the most important region of RBD. (A) Fuzzy clustering plot representing overlaps of the Structure Blocks depending on the cumulative contribution of structural and evolutionary features. SB1 is indicated by blue spheres, SB6 is indicated by orange spheres, SB9 is indicated by green spheres, SB10 is indicated by red spheres and SB12 is indicated by cyan spheres. SB9 and SB10 show significant overlap with all other SBs. (B) In the ACE2 receptor (green region) bound RBD structure, the most important SB, SB10 has been highlighted by red colour. Rest part of the RBD has been highlighted by slate blue colour. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The present strategy predicts SB10 to be a potential therapeutic pocket which if targeted would not only destabilize the RBD structure but would also affect the viral evolvability. Alternatively, any alteration at these residues-positions would disrupt the structure, and influence the evolvability. Our structure-guided findings also established that the abovementioned sub-stretch potentially regulates interaction between the RBD and its ACE2 binding interface. In order to validate our prediction, i.e. the potential of SB10 as a druggable pocket, we scrutinized the RBD structure based on some important biochemical and physical properties and observed that >30 % residues of SB10 (13 out of 38) have the ability to directly interact with drug molecules.
Table 2 lists some of the variants of concerns of SARS-CoV-2. Interestingly many mutations associated with different variants of SARS-CoV-2, e.g. Beta, Gamma, Delta and the recently emerged Omicron variant are positioned in SB10. Mutation associated with the alpha variant of SARS-CoV-2 (N501Y) is present at SB9, which is strongly connected with SB10 (Table 2). In case of Beta and Gamma variants, two of the three substituted residue positions (417 and 501) are associated with SBs having strong connection with the predicted important SB10 (Table 2). Another substituted residue 484 itself is housed in the SB10. Interestingly in the Delta variant of SARS-CoV-2, both associated mutations (L452R and T478K) are present in the SB10 (Table 2). More importantly, in recently emerged one of the most infectious Omicron variant, sixteen mutations (out of 30) have been found to be positioned in the RBD. Out of them eight residue positions are highly interdependent (440, 446, 477, 478, 484, 493, 496, 498) and four residues (S477, T478, T484 AND Q493) are housed at the SB10. As a result, targeting SB10 could be a potential “anti-evolution” approach to tackle the problem of SARS-CoV-2, even in the context of emerging strain.
Table 2.
Features of the variants of concerns of SARS-CoV-2 virus.
| Lineage | Variant | Mutations | Presence or absence in the highly co-varying patch | Structure block |
|---|---|---|---|---|
| B.1.1.7 | Alpha | N501Y | No | 9 |
| B.1.351 | Beta | K417N | No | 6 |
| E484K | Yes | 10 | ||
| N501Y | No | 9 | ||
| P.1 | Gamma | K417T | No | 6 |
| E484K | Yes | 10 | ||
| N501Y | No | 9 | ||
| B.1.617.2 | Delta | L452R | Yes | 10 |
| T478K | Yes | 10 | ||
| B.1.1.529 | Omicron | G339D | No | 4 |
| S371L | No | 8 | ||
| S373P | No | 8 | ||
| S375F | No | 8 | ||
| K417N | No | 6 | ||
| N440K | Yes | 9 | ||
| G446S | Yes | 9 | ||
| S477N | Yes | 10 | ||
| T478K | Yes | 10 | ||
| E484A | Yes | 10 | ||
| Q493R | No | 10 | ||
| G496S | No | 9 | ||
| Q498R | No | 9 | ||
| N501Y | No | 9 | ||
| Y505H | No | 9 | ||
| T547K | No | 11 |
Finally, it is important to consider the scopes and caveats of the present study. The most important caveat of this comes from its entirely computational nature and the complete absence of any experimental validation of binding. In addition, we needed to use a fragment of the S protein and modeled few residues-a procedure which is not ideal. However, it is worth noting that therapeutic approaches to block the emergence of evolutionary escape variants presents a promising avenue in ways of tackling diseases and re-orienting the drug development and treatment protocols. Strategies to constraint evolvability like the present one can help to recalibrate the evolutionary arms race in favour of the host. RNA viruses are notoriously known for their evolutionary escape propensities, which help them evade a wide array of selection pressures [90], [91]. Futuristic studies inspired by our approach could be potentially applied to multiple disease models (COVID 19 and/or beyond) where evolution-structure integrated understanding can be of potential benefit in better understanding the disease, identifying drug target and drafting out more perennial drug development protocols.
5. Analysis and representation
Majority of evolutionary and structural analysis were done with Python3. Visual renditions were made using Seaborn library of Python. For statistical analysis and representations Origin Pro 9.0 and Tableau were also used. For Graph theoretical modeling Gephi graphing tool was used. Protein models were represented using PyMOL. Structure network analysis using Bio3D package was carried out on RStudio.
Credit authorship contribution statement
KC and SC planned the overall project outline. DS and SC performed the computational experiments and analysis. AB helped in the MD simulation study and figure preparation. VU critically analyzed the manuscript and performed IDP analysis. DS, SB and VRC performed the druggability analysis and drug screening. DS wrote the first draft of the manuscript, which was then refined by SC and KC. All authors approved the final manuscript.
Declaration of competing interest
Authors do not have any conflict of interest.
Acknowledgment
DS acknowledges the Department of Science and Technology (DST) for doctoral fellowship (DST-INSPIRE). DS and KC acknowledge the Director, IICB. Authors thank Srivastava Ranganathan for his intellectual input in metadynamic simulation study. The project is funded by CSIR-IICB internal funding.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2022.07.022.
Appendix A. Supplementary data
Supplementary material
References
- 1.Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Vol. 91. J.A.B.M.A.P.; 2020. p. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandes N. Economic effects of coronavirus outbreak (COVID-19) on the world economy. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3557504. [DOI] [Google Scholar]
- 3.Kupferschmidt K. New mutations raise specter of ‘immune escape’. Science. 2021;371:329–330. doi: 10.1126/science.371.6527.329. [DOI] [PubMed] [Google Scholar]
- 4.Post L. Vol. 7. 2021. Surveillance of the Second Wave of COVID-19 in Europe: Longitudinal Trend Analyses. e25695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seong H., et al. Comparison of the second and third waves of the COVID-19 pandemic in South Korea: importance of early public health intervention. Int. J. Infect. Dis. 2021;104:742–745. doi: 10.1016/j.ijid.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asrani P., Eapen M.S., Hassan M.I., Sohal S.S. Implications of the second wave of COVID-19 in India. Lancet Respir. Med. 2021;9:e93–e94. doi: 10.1016/S2213-2600(21)00312-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spinello A., Saltalamacchia A., Borišek J., Magistrato A. Allosteric cross-talk among Spike’s receptor-binding domain mutations of the SARS-CoV-2 south african variant triggers an effective hijacking of human cell receptor. J. Phys. Chem. Lett. 2021;12:5987–5993. doi: 10.1021/acs.jpclett.1c01415. [DOI] [PubMed] [Google Scholar]
- 8.Golonka R.M. Harnessing Innate Immunity to Eliminate SARS-CoV-2 and Ameliorate COVID-19 Disease. Vol. 52. 2020. pp. 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F., Lau L.-T., Fok M., Lau J.Y.-N., Zhang K. COVID-19 Delta variants—current status and implications as of august 2021. Precis. Clin. Med. 2021;4:287–292. doi: 10.1093/pcmedi/pbab024. %J Precision Clinical Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Chakraborty M. Bhattacharya A. R. Sharma Present Variants of Concern and Variants of Interest of Severe Acute Respiratory Syndrome Coronavirus 2: Their Significant Mutations in S-glycoprotein, Infectivity, Re-infectivity, Immune Escapeand Vaccines Activity. n/a, e2270, doi:10.1002/rmv.2270.
- 11.Zhang X., et al. SARS-CoV-2 omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct. Target. Ther. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs Against SARS-CoV-2/COVID-19. Vol. 9. 2021. p. 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dispinseri S., et al. Seasonal betacoronavirus antibodies’ expansion post-BNT161b2 vaccination associates with reduced SARS-CoV-2 VoC neutralization. J. Clin. Immunol. 2022 doi: 10.1007/s10875-021-01190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontanet A., et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameroni E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 omicron antigenic shift. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem. Biophys. Res. Commun. 2021;538:97–103. doi: 10.1016/j.bbrc.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esler M., Esler D. Can angiotensin receptor-blocking drugs perhaps be harmful in the COVID-19 pandemic? J. Hypertens. 2020;38:781–782. doi: 10.1097/hjh.0000000000002450. [DOI] [PubMed] [Google Scholar]
- 19.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127-00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naqvi A.A.T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 1866;165878:2020. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asrani P., Hasan G.M., Sohal S.S., Hassan M.I. Molecular basis of pathogenesis of Coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS. 2020;24:634–644. doi: 10.1089/omi.2020.0131. [DOI] [PubMed] [Google Scholar]
- 25.Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turoňová B., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8 doi: 10.1038/ncomms15092. 15092-15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gui M., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan M., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asrani P., et al. Clinical features and mechanistic insights into drug repurposing for combating COVID-19. Int. J. Biochem. Cell Biol. 2022;142 doi: 10.1016/j.biocel.2021.106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padhi A.K., Dandapat J., Saudagar P., Uversky V.N., Tripathi T. Interface-based Design of the Favipiravir-binding Site in SARS-CoV-2 RNA-dependent RNA Polymerase Reveals Mutations Conferring Resistance to Chain Termination. Vol. 595. 2021. pp. 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. iScience. 2021:24. doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padhi A.K., Seal A., Khan J.M., Ahamed M., Tripathi T. Unraveling the mechanism of arbidol binding and inhibition of SARS-CoV-2: insights from atomistic simulations. Eur. J. Pharmacol. 2021;894 doi: 10.1016/j.ejphar.2020.173836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Othman H., et al. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghorbani A., et al. Comparative phylogenetic analysis of SARS-CoV-2 spike protein—possibility effect on virus spillover. Brief. Bioinform. 2021;22 doi: 10.1093/bib/bbab144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadam S.B., Sukhramani G.S., Bishnoi P., Pable A.A., Barvkar V.T. SARS-CoV-2, the pandemic coronavirus: molecular and structural insights. J. Basic Microbiol. 2021;61:180–202. doi: 10.1002/jobm.202000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Rivas J., Croce G., Muscat M., Weigt M. Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes. Proc. Natl. Acad. Sci. 2022;119 doi: 10.1073/pnas.2113118119. %J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pucci F., Rooman M. Prediction and Evolution of the Molecular Fitness of SARS-CoV-2 Variants: Introducing SpikePro. Vol. 13. 2021. p. 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2020;22:1239–1253. doi: 10.1093/bib/bbaa233. %J Briefings in Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan A. Higher Infectivity of the SARS-CoV-2 New Variants is Associated With K417N/T, E484K, and N501Y Mutants: An Insight From Structural Data. Vol. 236. 2021. pp. 7045–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan A., et al. Computational modelling of potentially emerging SARS-CoV-2 spike protein RBDs mutations with higher binding affinity towards ACE2: a structural modelling study. Comput. Biol. Med. 2022;141 doi: 10.1016/j.compbiomed.2021.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suleman M., et al. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tallei T.E. Fruit Bromelain-Derived Peptide Potentially Restrains the Attachment of SARS-CoV-2 Variants to hACE2: A Pharmacoinformatics Approach. Vol. 27. 2022. p. 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajpoot S., et al. In-silico design of a novel tridecapeptide targeting spike protein of SARS-CoV-2 variants of concern. Int. J. Pept. Res. Ther. 2021;28:28. doi: 10.1007/s10989-021-10339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav M.K., et al. Predictive modeling and therapeutic repurposing of natural compounds against the receptor-binding domain of SARS-CoV-2. J. Biomol. Struct. Dyn. 2022;1–13 doi: 10.1080/07391102.2021.2021993. [DOI] [PubMed] [Google Scholar]
- 50.Nayak S.K. Inhibition of S-protein RBD and hACE2 interaction for control of SARSCoV- 2 infection (COVID-19) Mini-Rev. Med. Chem. 2021;21:689–703. doi: 10.2174/1389557520666201117111259. [DOI] [PubMed] [Google Scholar]
- 51.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Sievers F., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.75. 539-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F., Higgins D.G. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chowdhury S., et al. Evolutionary analyses of sequence and structure space unravel the structural facets of SOD1. Biomolecules. 2019;9:826. doi: 10.3390/biom9120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Bahar I. Sequence evolution correlates with structural dynamics. Mol. Biol. Evol. 2012;29:2253–2263. doi: 10.1093/molbev/mss097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunn S.D., Wahl L.M., Gloor G.B. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2007;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 57.Hopf T.A., et al. The EVcouplings python framework for coevolutionary sequence analysis. Bioinformatics. 2019;35:1582–1584. doi: 10.1093/bioinformatics/bty862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hopf T.A., et al. The EVcouplings python framework for coevolutionary sequence analysis. Bioinformatics. 2019;35:1582–1584. doi: 10.1093/bioinformatics/bty862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fruchterman T.M.J., Reingold E.M. Graph drawing by force-directed placement. Softw. Pract. Exp. 1991;21:1129–1164. doi: 10.1002/spe.4380211102. [DOI] [Google Scholar]
- 60.Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 61.Uversky V.N. Unreported intrinsic disorder in proteins: disorder emergency room. Intrinsically Disord Proteins. 2015;3 doi: 10.1080/21690707.2015.1010999. e1010999-e1010999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan S., Chan H.C.S., Hu Z. Vol. 7. 2017. Using PyMOL as a Platform for Computational Drug Design. e1298. [DOI] [Google Scholar]
- 63.DeLano W.L. J.H.W.P.O.; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 64.Bej A., Rasquinha J.A., Mukherjee S. Conformational entropy as a determinant of the thermodynamic stability of the p53 Core domain. Biochemistry. 2018;57:6265–6269. doi: 10.1021/acs.biochem.8b00740. [DOI] [PubMed] [Google Scholar]
- 65.Bussi G., Donadio D., Parrinello M. Canonical Sampling Through Velocity Rescaling. Vol. 126. 2007. p. 014101. [DOI] [PubMed] [Google Scholar]
- 66.Parrinello M., Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. Vol. 52. 1981. pp. 7182–7190. [DOI] [Google Scholar]
- 67.Darden T., York D., Pedersen L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. Vol. 98. 1993. pp. 10089–10092. [DOI] [Google Scholar]
- 68.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A Linear constraint Solver for Molecular Simulations. Vol. 18. 1997. pp. 1463–1472. [DOI] [Google Scholar]
- 69.Linding R., et al. Protein disorder prediction. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Alexander M.D. ?True? sporadic ALS associated with a novel SOD-1 mutation. Ann. Neurol. 2002;52:680–683. doi: 10.1002/ana.10369. [DOI] [PubMed] [Google Scholar]
- 71.Kurcinski M., et al. CABS-flex standalone: a simulation environment for fast modeling of protein flexibility. Bioinformatics. 2018;35:694–695. doi: 10.1093/bioinformatics/bty685. %J Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy K.P. Machine Learning: A Probabilistic Perspective. MIT press; 2012. pp. 387–403. [Google Scholar]
- 73.Suganya R., Shanthi R. Fuzzy c-means algorithm-a review. Int. J. Sci. Res. Publ. 2012;2:1. [Google Scholar]
- 74.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky473. W363-W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-168. 168-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim S., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2020;49 doi: 10.1093/nar/gkaa971. %J Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Boyle N.M., et al. Open babel: an open chemical toolbox. J. Cheminform. 2011;3 doi: 10.1186/1758-2946-3-33. 33-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris G.M. AutoDock4 and AutoDockTools4: Automated Docking With Selective Receptor Flexibility. Vol. 30. 2009. pp. 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding F., Yin S., Dokholyan N.V. Rapid flexible docking using a stochastic rotamer library of ligands. J. Chem. Inf. Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin S., Biedermannova L., Vondrasek J., Dokholyan N.V. MedusaScore: an accurate force field-based scoring function for virtual drug screening. J. Chem. Inf. Model. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J., Dokholyan N.V. MedusaDock 2.0: efficient and accurate protein–ligand docking with constraints. J. Chem. Inf. Model. 2019;59:2509–2515. doi: 10.1021/acs.jcim.8b00905. W363-W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anjum F., et al. Identification of intrinsically disorder regions in non-structural proteins of SARS-CoV-2: new insights into drug and vaccine resistance. Mol. Cell. Biochem. 2022;477:1607–1619. doi: 10.1007/s11010-022-04393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu R., et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Vol. 395. The Lancet; 2020. pp. 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ibarra-Molero B., Loladze V.V., Makhatadze G.I., Sanchez-Ruiz J.M. Thermal versus guanidine-induced unfolding of ubiquitin. An analysis in terms of the contributions from charge−charge interactions to protein stability†. Biochemistry. 1999;38:8138–8149. doi: 10.1021/bi9905819. [DOI] [PubMed] [Google Scholar]
- 89.Li J., Lewis H.W. IEEE; 2016. 2016 IEEE International Conference on Smart Cloud (SmartCloud) Fuzzy Clustering Algorithms — Review of the Applications. [Google Scholar]
- 90.Van Blerkom L.M. Role of viruses in human evolution. Am. J. Phys. Anthropol. Suppl. 2003;37:14–46. doi: 10.1002/ajpa.10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharp P.M., Hahn B.H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material