Summary
Background
Pre-treatment FDG-PET/CT scans were analyzed with machine learning to predict progression of lung malignancies and overall survival (OS).
Methods
A retrospective review across three institutions identified patients with a pre-procedure FDG-PET/CT and an associated malignancy diagnosis. Lesions were manually and automatically segmented, and convolutional neural networks (CNNs) were trained using FDG-PET/CT inputs to predict malignancy progression. Performance was evaluated using area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, and specificity. Image features were extracted from CNNs and by radiomics feature extraction, and random survival forests (RSF) were constructed to predict OS. Concordance index (C-index) and integrated brier score (IBS) were used to evaluate OS prediction.
Findings
1168 nodules (n=965 patients) were identified. 792 nodules had progression and 376 were progression-free. The most common malignancies were adenocarcinoma (n=740) and squamous cell carcinoma (n=179). For progression risk, the PET+CT ensemble model with manual segmentation (accuracy=0.790, AUC=0.876) performed similarly to the CT only (accuracy=0.723, AUC=0.888) and better compared to the PET only (accuracy=0.664, AUC=0.669) models. For OS prediction with deep learning features, the PET+CT+clinical RSF ensemble model (C-index=0.737) performed similarly to the CT only (C-index=0.730) and better than the PET only (C-index=0.595), and clinical only (C-index=0.595) models. RSF models constructed with radiomics features had comparable performance to those with CNN features.
Interpretation
CNNs trained using pre-treatment FDG-PET/CT and extracted performed well in predicting lung malignancy progression and OS. OS prediction performance with CNN features was comparable to a radiomics approach. The prognostic models could inform treatment options and improve patient care.
Funding
NIH NHLBI training grant (5T35HL094308-12, John Sollee).
Keywords: Machine learning, Artificial intelligence, Deep learning, Lung cancer, FDG-PET/CT, Prognosis
Research in context.
Evidence before this study
We searched PubMed for articles published from database inception to June 1, 2021, with the search terms (“lung nodule” OR “lung malignancy”) AND (“artificial intelligence” OR “machine learning”) AND “FDG-PET/CT,” with no language restrictions, and found 89 publications. Nine focused entirely on methodological considerations of machine learning, and four did not mention machine learning. Of the clinically relevant articles, forty-six focused on a type of cancer not involving the lungs or an unrelated disease. Twenty-six used machine learning and FDG-PET/CT scans to detect, classify, and diagnose lung malignancies. Four used machine learning and radiology to make a treatment decision or a treatment outcome prediction. This search indicated that, despite an abundance of studies using machine learning with radiology to detect and diagnose lung cancer, there is a scarcity of studies related to its use as a tool for prognostication. Moreover, the studies aimed at prognostication with machine learning have mostly relied on handcrafted features rather than deep learning. Additionally, they have considered either PET or CT individually without investigating their additive values in ensemble models.
Added value of this study
We used machine learning based on pre-treatment FDG-PET/CT scans to predict progression of lung malignancies and overall survival in a multicenter and multi-national cohort. A deep learning model was used to automatically identify and extract important prognostic features. Models were generated based on PET features alone, CT features alone, and clinical features alone, and ensemble models were created to evaluate the additive prognostic value of each input. Radiomics features were also extracted and used to construct models for comparison. The study demonstrated that deep learning models trained using pre-treatment FDG-PET/CT data perform well in predicting lung malignancy progression. Features extracted from the deep learning models predict overall survival well and is comparable to performance features derived from a radiomics approach. Furthermore, overall survival models based on CT perform better than those based on PET, and the addition of PET to CT only insignificantly improves performance. As such, there is unclear added value of PET for prognosis of lung cancer with machine learning based on CT. After prospective validation, this model could help inform treatment options and improve patient care.
Implications of all the available evidence
Efficient and accurate prognostication of patients with lung malignancies is important for informing treatment options. With rapid advances in computing power and algorithm development, there is an opportunity to use machine learning to assist physician decision making. While future work is needed, the available evidence suggests that machine learning models based on FDG-PET/CT can predict lung malignancy progression and overall survival with high accuracy. It remains unclear whether PET provides additional prognostic information beyond CT in lung cancer.
Alt-text: Unlabelled box
Introduction
Despite advances in prevention and treatment in the past decade, lung cancer remains the leading cause of cancer death worldwide.1, 2, 3 In the United States, overall survival (OS) remains low even with improvements in understanding of disease etiology and treatment options.1 There is only 15% five-year survivorship following diagnoses,4 likely given that approximately 62-70% of individuals with lung cancer are diagnosed at an advanced stage.2,3 However, the majority of early stage lung cancers are treatable. For instance, in non-small cell lung carcinomas (NSCLC), the five-year survival rate at diagnosis is up to 92% for stage IA, compared to a survival rate of 10% for stage IV.5 Treatment options are typically based on cancer subtype and prognosis, the latter of which is highly variable and challenging to predict. Prognosis may depend on a variety of factors, such stage of disease, molecular composition, histopathological characteristics, patient age, sex, and baseline functional status, and the existence of comorbidities.1
Computed tomography (CT) and fluorodeoxyglucose positron emission tomography (FDG-PET) are two imaging modalities that are commonly used for lung cancer diagnosis and staging. Their combined use (FDG-PET/CT) provides both anatomical and metabolic information that is important for determining an accurate prognosis and informing treatment options.6 Patients that are identified as having a low progression or mortality risk based on FDG-PET/CT may be candidates for invasive procedures with curative intent, while those that are higher risk may be amendable to chemotherapy, radiation, targeted therapy, or immunotherapy with palliative intent. Advanced thoracic surgery techniques, such as video-assisted thoracoscopic surgery, are highly effective, with five-year survivorship for early stage patients up to 80%.7 Those with early stage cancer but with lower risk of progression may alternatively be treated with radiotherapy or microwave ablation, especially if the existence of comorbidities would increase the likelihood of post-surgical complications.2 Finally, high risk patients may be started with palliative drug therapy and early palliative care, which can significantly extend survival time.8
Although FDG-PET/CT is important for prognosis in lung malignancy, manual interpretation of imaging is imperfect and inefficient, with low inter-reader agreement.9 Recent developments in artificial intelligence (AI) and machine learning (ML) have demonstrated potential for improved accuracy in the characterization of lung malignancies with FDG-PET/CT.10, 11, 12 The majority of existing AI studies have focused on diagnosis and staging, and those aimed at prognostication have mostly relied on handcrafted features rather than deep learning.10,11 Most studies have also considered the prognostic utilities of CT and PET individually without investigating their additive value. The purpose of this study was to use deep learning with pre-treatment FDG-PET/CT to predict progression of lung malignancies and OS.
Methods
Patient cohort
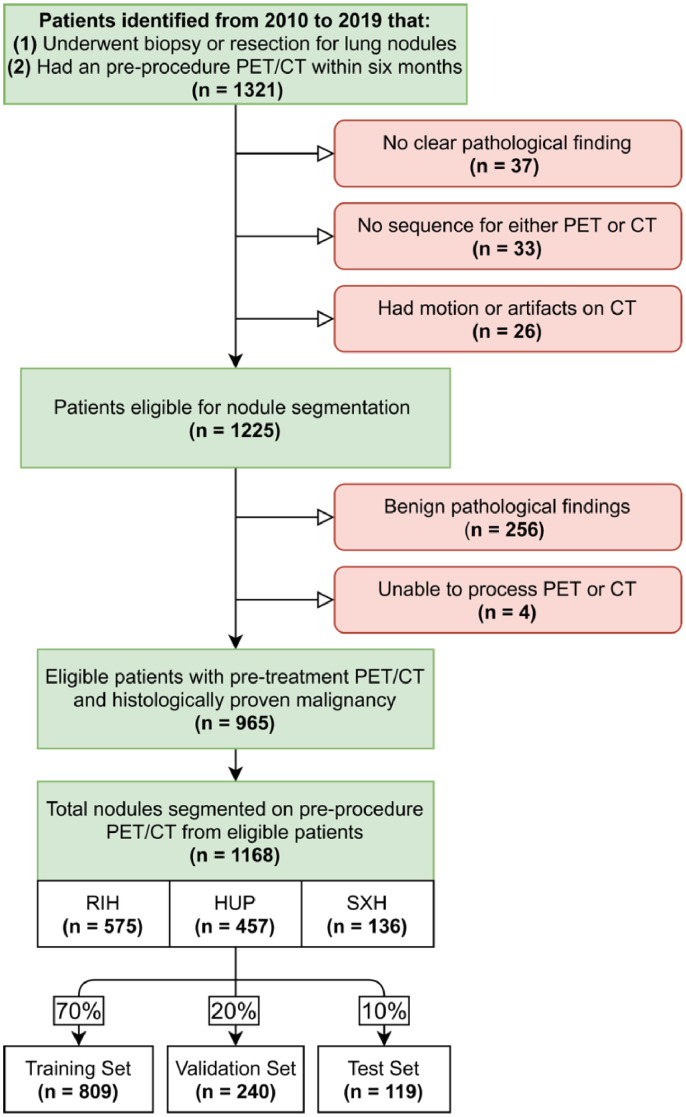
A retrospective review between 2010 and 2019 was performed across two major institutions in the United States (Rhode Island Hospital [RIH] and the Hospital of the University of Pennsylvania [HUP]) and one in the People's Republic of China (Xiangya Second Hospital [XSH]). Patients who had a histologically diagnosed lung malignancy by biopsy or surgery and had undergone an FDG-PET/CT scan up to six months prior to biopsy or resection were identified. Demographic and clinical information such as age, sex, race, and cancer type were recorded. Overall survival time and the occurrence of progression were also determined. Progression was defined as local-regional recurrence or metastasis following treatment. A detailed flow diagram of the patient selection process is shown in Figure 1. The dataset collected for this study is not publicly available due to patient privacy concerns but can be available from the corresponding authors if there is a reasonable request and approval from affiliated ethics board.
Figure 1.
Patient Selection Flow Diagram.
Red boxes represent patients that were excluded for the stated reason. RIH: Rhode Island Hospital; HUP: Hospital of the University of Pennsylvania; SXH: Second Xiangya Hospital.
Manual and automatic tumor segmentation
PET and CT scans were exported in Digital Imaging and Communications in Medicine (DICOM) format in their original resolutions. The FDG-PET/CT scan protocols are detailed in Appendix A. Nodules were manually segmented on both CT and PET modalities by an experienced radiologist (Y.L.) with seven years of experience.
For automatic tumor segmentation of PET and CT scans, lesions were randomly split 7:2:1 among training, validation and test sets. All lesions from the same patient were grouped together to prevent information leak between datasets. Patients represented in the training set were distributed 47.5% RIH, 41.6% HUP, and 10.9% from XSH, while patients in the validation set were distributed 47.7% RIH, 41.8% HUP, and 10.4% XSH. Patients represented in the test set consisted of 42.6% RIH, 42.6% HUP, and 14.7% XSH. Automatic segmentation of CT and PET modalities were then performed using the out-of-the-box nnU-Net segmentation tool with the default nnU-Net parameters. Automated lesion segmentation performance on the training, validation, and test sets were evaluated on AUC, accuracy, sensitivity, and specificity. A repeat manual segmentation of the test set was also performed by a second experienced radiologist (J.W.) with five years of experience.
Image pre-processing
The window width for CT images was 1500 Hounsfield units; a window level of -400 Hounsfield units was used to disregard non-pulmonary regions. No windowing was applied to the PET images. Lesion volumes were then computed for PET and CT images independently. For each lesion on PET, maximum standardized uptake value (maxSUV) was also extracted. For both CT and PET lesions, image slices were cropped to include a fixed range of background surrounding each lesion and scaled to 224 square pixels. Pixel values were then normalized from a range of 0-255 to 0-1.
Deep learning model architecture
A convolutional neural network (CNN) with the pre-trained EfficientNetB4 architecture was applied as a backbone network. A series of dense (fully connected) layers with 256 and 16 neurons with a rectified linear unit (ReLU) activation function and batch normalization constituted the top layers of the neural network. The final classification layer was composed of a single neuron with a sigmoid activation function. The image inputs for the three channels of the EfficientNet consisted of a single slice each from the sagittal, coronal, and axial dimensions of the lesion, selected by maximal cross sectional lesion area in each axis. Ensemble models were created by taking an average of the final predicted progression probabilities from the strongest individual PET and CT models by test set area under the receiver operating characteristic curve (AUC).
Deep learning model training and testing
Deep learning models using manual and automated masks were trained and tested using the same 7:2:1 training, validation, and test split as the automated lesion segmentation. Each model was trained for a maximum of 150 epochs with a patience of 50, early stopping, and a fixed batch size of 32. The training set was augmented by using vertical flips, horizontal flips, shears, and zooms on the original images. Validation accuracy was measured at each epoch, and the point with the highest validation accuracy was selected as the final model. Models were trained with stochastic gradient descent with Nesterov momentum using a learning rate of 0.001 and a dropout rate of 0.5. Five models were trained for each modality using the Keras library with Tensorflow backend on two Nvidia Quadro GV100 GPUs. Performance of the progression risk model was evaluated using AUC, accuracy, sensitivity, and specificity. Performance was additionally re-evaluated with the second manual segmentation. Heatmaps generated using the Grad-CAM algorithm with absolute gradients from the CT and PET CNN models with manual or automatically segmented masks were generated with and overlaid onto axial slices of the corresponding CT or PET lesions to visualize salient lesion regions identified by the models.
Deep learning and radiomics feature extraction
For deep learning features, 16 image features for the training, validation, and test sets were extracted as the output of the penultimate dense 16 neuron layer of the strongest performing CNNs of PET and CT modalities for both manually and automatically segmented masks. Radiomics feature extraction was performed by using the PyRadiomics feature extractor13 on the preprocessed CT and PET data. First-order features, 2D and 3D shape features, and textural features were obtained from the extraction process. Missing feature values were estimated through mean-value imputation. Additional deep learning and feature sets were also constructed by applying the ComBAT data harmonization algorithm to examine inter-site variability.
Random survival forest model
Training and validation sets for deep learning and radiomics features were merged to form new training sets for the random survival forest (RSF) model. GridsearchCV was performed to select optimal hyperparameters for the RSF model, including number of estimators, maximum tree depth, minimum samples to split a node, minimum samples per leaf node, and maximum features used at each split. CT features, PET features, and clinical features were each utilized to train three RSF models. The clinical feature model included age, sex, race, maxSUV, and lesion volume (CT and PET). Each model was fitted on the training set to predict OS. Two ensemble RSF models (CT and PET together and CT and PET along with clinical features) were also created by taking the average risk prediction of the individual RSFs. Model performance was evaluated on the test set by concordance index (C-index) and integrated brier score (IBS). C-index calculations on the test set were also performed using direct CNN model prediction outputs, maxSUV alone, average of CT and PET tumor volume alone, or AJCC (American Joint Committee on Cancer) stage as risk scores for comparison.
Test set nodules were also separated into two groups based on whether their predicted risk score was above or below the mean predicted risk score from the CT+PET+clinical feature ensemble RSF models. Kaplan Meier survival curves were generated for the high and low risk nodules. Deep learning features were ordered by RSF feature importance and absolute value correlation heatmaps with radiomics features were generated.14
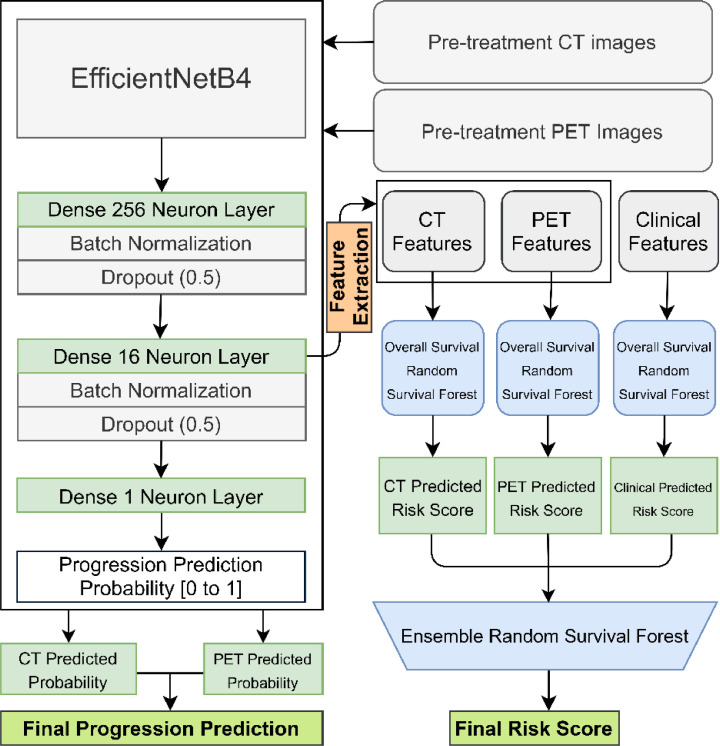
A full flow diagram for the machine learning models is displayed in Figure 2. The codebase used in this study is available online (https://github.com/BrianHeHuang/Lung-PET-CT-Survival). The full dataset used to train and evaluate the models in this study is not available for public access because of patient privacy concerns but is available from the corresponding authors if there is a reasonable request and approval from the institutional review boards of the affiliated institutions.
Figure 2.
Machine Learning Architecture and Flow Diagram.
The EfficientNetB4 architecture was used for the convolutional neural network with three dense layers with batch normalization and dropout. Ensemble models were generated by taking an average of the final predicted progression probabilities from the strongest individual PET and CT models. Features were extracted from the penultimate dense layer for the random survival forest model for overall survival risk. CT: computed tomography; PET: positron emission tomography.
Ethics
This study met eligibility for review exemption and HIPAA waiver by the institutional review boards of the University of Pennsylvania (Protocol # 8499999) and was approved by the institutional review board of Rhode Island Hospital (Protocol # 1666262-2). This study was also approved by the institutional review board of Xiangya Second Hospital (Protocol # 2020144). A HIPAA authorization waiver was granted based on minimal risk to the privacy of individuals, so informed consent was not obtained for participants.
Statistics
The significance level used throughout this study was 0.05. Two-sample T-tests were used to evaluate differences in means between patients with progressive and non-progressive nodules for continuous variables, and Chi-squared tests were used to evaluate differences in proportions between the groups for categorical variables.
For progression CNN model evaluation metrics, 95% confidence intervals were computed using the adjusted Wald interval and comparisons between model performance metrics were performed using a McNemar test for paired proportions.
Percentile 95% confidence intervals were generated RSF models by performing a bootstrap on the test set.15 Two-thirds of the set were sampled without replacement for 100 total iterations. Comparisons between RSF model results were performed by computing percentile bootstrap p-values on the differences in performance across the 100 iterations. For the Kaplan Meier survival curves, a log rank test for survival was conducted between the high risk and low risk survival groups.
Role of Funders
The funding source had no role in the study design, data collection, data analysis, interpretation, or writing of the report. All the authors have full access to the data and take full responsibility for the contents of this report and the decision to submit it for publication.
Results
Patient cohort and clinical features
1168 nodules from 965 patients were identified. Of these nodules, 792 had progression and 376 were progression-free. The most common malignancies were adenocarcinoma (n=740 nodules) and squamous cell carcinoma (n=179 nodules). Progression-free nodules occurred in older patients (p<0.001 [t-test]) and had increased maxSUV uptake (p=0.020 [t-test]), but did not have significantly increased volume. Progressive nodules were also significantly more likely to have received chemotherapy, radiation therapy, and immunotherapy at some point during treatment. A detailed summary of demographic and clinical information is shown in Table 1.
Table 1.
Demographic and Clinical Features. Data for progressive (n=792) and non-progressive (n=376) nodules are displayed as mean (standard deviation) for continuous variables and percent (count) for categorical variables. Age is represented in years, and lesion volumes are measured in voxels. P-values were computed for difference in means (two-sample T-test) or proportions (Chi-squared test) between the progression and non-progression groups for continuous and categorical variables, respectively. Statistically significant p-values are highlighted in bold. MaxSUV: maximum standardized uptake value, SD: standard deviation; SCLC: small cell lung carcinoma; NSCLC: non-small cell lung carcinoma.
| Clinical Feature Summary | |||
|---|---|---|---|
| Variable | Progression | No Progression | P-value |
| Total Nodule Count | 792 | 376 | |
| Age (SD) | 66.54 (13.93) | 69.66 (10.69) | <0.001 |
| MaxSUV Uptake (SD) | 8.64 (7.40) | 9.72 (7.34) | 0.020 |
| CT Lesion Volume (SD) | 26,097 (66,850) | 26,012 (72,518) | 0.984 |
| PET Lesion Volume (SD) | 27,575 (68,507) | 27,626 (73,039) | 0.991 |
| Sex (Nodule Count) | 0.953 | ||
| Male | 48.5% (384) | 48.7% (183) | |
| Female | 51.5% (408) | 51.3% (193) | |
| Diagnosis Method (Nodule Count) | 0.235 | ||
| Biopsy | 41.8% (331) | 45.5% (171) | |
| Surgery | 58.2% (461) | 54.5% (205) | |
| Race (Nodule Count) | <0.001 | ||
| White | 68.6% (543) | 75.3% (283) | |
| African American | 15.9% (126) | 12.0% (45) | |
| Asian/Pacific Islander | 12.5% (99) | 12.8% (48) | |
| Other | 1.1% (9) | 0% (0) | |
| Unknown | 1.9% (15) | 0% (0) | |
| Treatments (Nodule Count) | |||
| Chemotherapy | 27.0% (215) | 3.0% (11) | <0.001 |
| Radiation therapy | 26.5% (211) | 16.5% (53) | <0.001 |
| Immunotherapy | 8.1% (64) | 3.3% (12) | 0.002 |
| Histologically Confirmed Diagnosis Summary | |||
|---|---|---|---|
| Progression (Nodule Count) | No Progression (Nodule Count) | ||
| Adenocarcinoma | 62.4% (494) | Adenocarcinoma | 65.4% (246) |
| Squamous Cell Carcinoma | 14.9% (118) | Squamous Cell Carcinoma | 16.2% (61) |
| Lymphoma | 4.2% (33) | Lymphoma | 1.6% (6) |
| Metastasis | 7.2% (57) | Metastasis | 2.7% (10) |
| Unspecified SCLC | 4.7% (37) | Unspecified SCLC | 7.2% (27) |
| Unspecified NSCLC | 2.9% (23) | Unspecified NSCLC | 4.8% (18) |
| Carcinoid | 1.5% (12) | Carcinoid | 1.3% (5) |
| Neuroendocrine | 1.3% (10) | Neuroendocrine | 0% (0) |
| Other | 1.0% (8) | Other | 0.8% (3) |
nnU-Net automatic segmentation performance
The nnU-Net AUC on the test set was 0.828 for CT and 0.755 for PET, while sensitivity was 0.657 for CT and 0.510 for PET, defined as proportion of nodule correctly annotated. Accuracy and specificity were greater than 0.998 across both CT and PET segmentations, defined as percentage of overall area and healthy area annotated correctly respectively. The full results of the nnU-Net segmentation, including training and test set performance, are displayed in Table 2.
Table 2.
nnU-Net Automatic Segmentation Performance. Accuracy represents percentage of overall percentage of correctly predicted areas in the overall image. Sensitivity corresponds to the percentage of nodules that were correctly predicted and annotated. Specificity refers to the percentage of healthy area that was correctly predicted and annotated.
| CT Automatic Segmentation Performance | ||||
|---|---|---|---|---|
| AUC | Accuracy | Sensitivity | Specificity | |
| Training Set | 0.865 | 0.999 | 0.730 | 0.999 |
| Validation Set | 0.848 | 0.999 | 0.696 | 0.999 |
| Test Set | 0.828 | 0.999 | 0.657 | 0.999 |
| PET Automatic Segmentation Performance | ||||
|---|---|---|---|---|
| AUC | Accuracy | Sensitivity | Specificity | |
| Training Set | 0.747 | 0.998 | 0.495 | 0.999 |
| Validation Set | 0.741 | 0.999 | 0.489 | 0.999 |
| Test Set | 0.755 | 0.999 | 0.510 | 0.999 |
Progression model performance
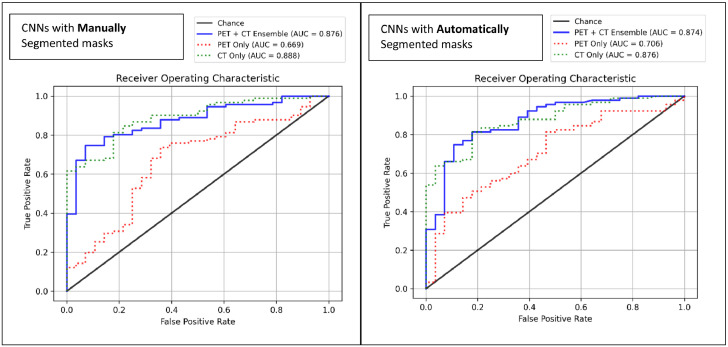
The CT only CNN model with manually segmented masks predicted progression with an accuracy of 0.723, AUC of 0.888, sensitivity of 0.681, and specificity of 0.857. The corresponding PET only progression model achieved an accuracy of 0.664, AUC of 0.669, sensitivity of 0.659, and specificity of 0.679. The PET+CT ensemble model achieved an accuracy of 0.790, AUC of 0.876, sensitivity of 0.769, and specificity of 0.857. The ensemble had significantly stronger accuracy and sensitivity than the PET model (p = 0.029, p =0.031 [McNemar test]) but had no significant differences compared to the CT only model.
The CT only CNN model with automatically segmented masks achieved an accuracy of 0.798, AUC of 0.876, sensitivity of 0.791, and specificity of 0.821. The corresponding PET only model had an accuracy of 0.571, AUC of 0.706, sensitivity of 0.495, and specificity of 0.821. The PET+CT ensemble had an accuracy of 0.815, AUC of 0.874, specificity of 0.813, and sensitivity of 0.821. Both the CT and ensemble models outperformed the PET model in accuracy (p < 0.001, <0.001 [McNemar test]), and sensitivity (p < 0.001, <0.001 [McNemar]), but there were no significant differences between CT and CT+PET ensemble performance.
The CT CNN model with automatically segmented masks had significantly higher accuracy and sensitivity than the corresponding manual mask model (p = 0.049, p = 0.006 [McNemar test]), while the PET CNN model with manually segmented masks had significantly higher sensitivity compared to corresponding automatic mask model (p = 0.004, [McNemar test]). There were no significant differences in performance of the CT+PET ensemble models between manually and automatically segmented masks. Model performance and comparisons are summarized in Table 3 and AUC curves are displayed in Figure 3. Testing with the second independent manual segmentation yielded comparable results, apart from a significantly lower CT+PET specificity for the model trained with automated masks (p = 0.039, [McNemar test]), which are shown in Appendix B.
Table 3.
Model Performance. Results for performance metrics on the test set are displayed for the machine learning models using manually and automatically segmented masks. Accuracy, sensitivity, and specificity for the CNN models are shown with 95% confidence intervals in parentheses. Confidence intervals were calculated using the adjusted Wald method. Comparisons between performance metrics between models was performed with a McNemar test for paired proportions. Statistically significant p-values are highlighted in bold (p<0.05); CNN: convolutional neural network; RSF: random survival forest; AUC: area under the receiver operating characteristic curve.
| Modality | AUC | Accuracy (Acc) | Sensitivity (Sens) | Specificity (Spec) |
|---|---|---|---|---|
| CNN Models – Progression (Manually segmented masks) | ||||
| CT | 0.888 | 0.723 (0.643, 0.803) | 0.681 (0.587, 0.775) | 0.857 (0.722, 0.992) |
| PET | 0.669 | 0.664 (0.580, 0.748) | 0.659 (0.563, 0.755) | 0.679 (0.514, 0.843) |
| PET+CT Ensemble | 0.876 | 0.790 (0.717, 0.863) | 0.769 (0.683, 0.855) | 0.857 (0.722, 0.992) |
| CNN Models – Progression (Automatically segmented masks) | ||||
| CT | 0.876 | 0.798 (0.726, 0.870) | 0.791 (0.708, 0.875) | 0.821 (0.678, 0.965) |
| PET | 0.706 | 0.571 (0.484, 0.659) | 0.495 (0.394, 0.595) | 0.821 (0.678, 0.965) |
| PET+CT Ensemble | 0.874 | 0.815 (0.745, 0.885) | 0.813 (0.733, 0.894) | 0.821 (0.678, 0.965) |
|
P-values [McNemar], comparisons between CNNs (Manual Masks) |
P-values [McNemar], comparisons between CNNs (Automated Masks) |
P-values [McNemar], comparisons between manual (M) and automated (A) CNNs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Comp. | Acc. | Sens. | Spec. | Comp. | Acc. | Sens. | Spec. | Comp. | Acc. | Sens. | Spec. |
| CT vs. PET | 0.371 | 0.864 | 0.227 | CT vs. PET | <0.001 | <0.001 | 1.00 | CT (M vs. A) | 0.049 | 0.006 | 1.00 |
| CT vs. PET+CT | 0.115 | 0.077 | 1.00 | CT vs. PET+CT | 0.754 | 0.727 | 1.00 | PET (M vs. A) | 0.090 | 0.004 | 0.344 |
| PET vs. PET+CT | 0.004 | 0.031 | 0.125 | PET vs. PET+CT | <0.001 | <0.001 | 1.00 | PET + CT (M vs. A) | 0.629 | 0.388 | 1.00 |
Figure 3.
Receiver Operating Characteristic Curves for Progression Risk.
AUC: area under the receiver operating characteristic curve.
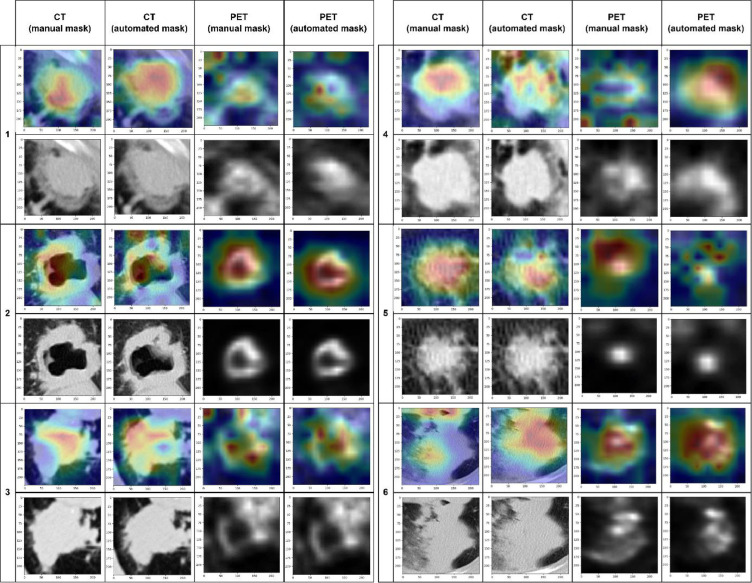
The GradCAM algorithm results showed that CT models tended to highlight intra-tumoral regions with frequent extension and crossing over an edge of the lesion. PET models frequently annotated a more diffuse region around the center of the lesion. Images of overlaid Grad-CAM heatmaps of six representative lesions are displayed in Figure 4.
Figure 4.
Grad-CAM Results.
Heatmaps generated using the Grad-CAM algorithm with absolute gradients from the CT and PET CNN models with manual/automatically segmented masks were generated with and overlaid onto axial slices of corresponding lesions. Images of six representative lesions across the four CNNs are displayed.
Overall survival model performance
For deep learning features derived with manually segmented masks, the CT feature only RSF had a C-index of 7.30, while the PET only RSF had a C-index of 0.595 and the clinical RSF had a C-index of 0.595. The combined PET+CT and PET+CT+clinical ensemble models achieved C-indices of 0.741 and 0.737 respectively. For deep learning features from automatically segmented masks, the CT, PET, and clinical only C-indices were 0.739, 0.567, and 0.587 respectively, while the CT+PET ensemble had a C-index of 0.740 and the full CT+PET+clinical ensemble had a C-index of 0.732.
For radiomics features derived with manually segmented masks, the CT only RSF had a C-index of 0.735, the PET only had a C-index of 0.572, and the clinical only model had a C-index of 0.597. The PET+CT ensemble C-index was 0.725 and the PET+CT+clinical ensemble C-index was 0.717. The radiomic feature RSF models with automated masks had a CT only C-index of 0.718, PET only C-index of 0.526, and clinical only C-index of 0.584. PET+CT ensemble had a C-index of 0.703, while the PET+CT+clinical ensemble achieved a C-index of 0.700.
For all sets of models, the CT only, CT+PET, and CT+PET+clinical models had a significantly higher C-index than the PET only and clinical feature only models (p <0.001, [bootstrap]). There were no significant differences when comparing C-indices between any of the deep learning feature RSF models (single modality and ensemble) and the corresponding radiomics feature RSF model. There were also no significant differences comparing models with manually segmented masks and automatically segmented masks. Table 4 contains the full summary of RSF results and Appendix C contains a chart of p-values for all comparisons performed.
Table 4.
RSF Model Performance. Mean results for performance metrics with associated bootstrap percentile 95% confidence intervals are displayed for deep learning and radiomics features with manually or automatically segmented masks. C-index: concordance index; IBS: integrated brier score.
| Modality | C-Index | IBS | Modality | C-Index | IBS | ||
|---|---|---|---|---|---|---|---|
| RSF model with deep learning features (Manual masks) | RSF model with deep learning features (Automated masks) | ||||||
| CT | 0.730 (0.687, 0.774) | 0.171 (0.139, 0.208) | CT | 0.739 (0.703, 0.793) | 0.159 (0.13, 0.193) | ||
| PET | 0.595 (0.543, 0.663) | 0.177 (0.15, 0.204) | PET | 0.568 (0.512, 0.620) | 0.185 (0.165, 0.202) | ||
| Clinical | 0.595 (0.529, 0.659) | 0.177 (0.15, 0.204) | Clinical | 0.587 (0.536, 0.658) | 0.185 (0.165, 0.202) | ||
| PET + CT Ensemble | 0.741 (0.695, 0.802) | 0.168 (0.141, 0.197) | PET + CT Ensemble | 0.740 (0.695, 0.792) | 0.165 (0.142, 0.189) | ||
| PET + CT + Clinical Ensemble | 0.737 (0.680, 0.804) | 0.169 (0.14, 0.199) | PET + CT + Clinical Ensemble | 0.732 (0.682, 0.783) | 0.166 (0.141, 0.193) | ||
| RSF model with radiomics features (Manual masks) | RSF model with radiomics features (Automated masks) | ||||||
| CT | 0.735 (0.687, 0.787) | 0.159 (0.135, 0.181) | CT | 0.718 (0.665, 0.771) | 0.160 (0.139, 0.187) | ||
| PET | 0.572 (0.520, 0.630) | 0.191 (0.174, 0.207) | PET | 0.526 (0.502, 0.565) | 0.211 (0.197, 0.242) | ||
| Clinical | 0.597 (0.542, 0.655) | 0.191 (0.174, 0.207) | Clinical | 0.584 (0.52, 0.631) | 0.211 (0.197, 0.242) | ||
| PET + CT Ensemble | 0.725 (0.689, 0.769) | 0.169 (0.153, 0.187) | PET + CT Ensemble | 0.703 (0.654, 0.764) | 0.179 (0.163, 0.204) | ||
| PET + CT + Clinical Ensemble | 0.717 (0.674, 0.757) | 0.168 (0.149, 0.189) | PET + CT + Clinical Ensemble | 0.700 (0.645, 0.754) | 0.176 (0.158, 0.206) | ||
In comparison, the direct CT CNN risk score model had a C-index of 0.544, the corresponding PET model had a C-index of 0.536, and the CT+PET ensemble had a C-index of 0.543. With automatic masks, the direct CT, PET, and CT+PET models had C-indices of 0.537, 0.540, and 0.539 respectively. The corresponding RSF models had significantly stronger performance on CT and CT+PET models for both manual and automated masks (all p < 0.001, [bootstrap]). The direct maxSUV model had a C-index of 0.537, the average CT and PET volume model had a C-index of 0.541, and the AJCC stage model had a C-index of 0.538. All three of the models had comparable performance to a corresponding RSF clinical model. Full results and comparisons are displayed in Appendix D.
In the Kaplan Meier survival analysis, 62 nodules had predicted risk scores higher than the mean score from the CT+PET+clinical feature DL manual mask RSF model, while 57 nodules had scores lower than the mean. For the corresponding ensemble model with automatic masks, 66 nodules had higher than mean risk scores, while 53 had lower than mean risk. In the radiomics model CT+PET+clinical RSF model with manual masks, 59 nodules had above average risk, while 60 nodules have below average. The corresponding automatic mask model had 69 lesions with above average risk and 50 lesions with below average risk. Nodules deemed high risk had a significantly lower survival distribution than nodules deemed low risk for all four categories (p<0.001, [log rank test]). Full curves are displayed in Appendix E.
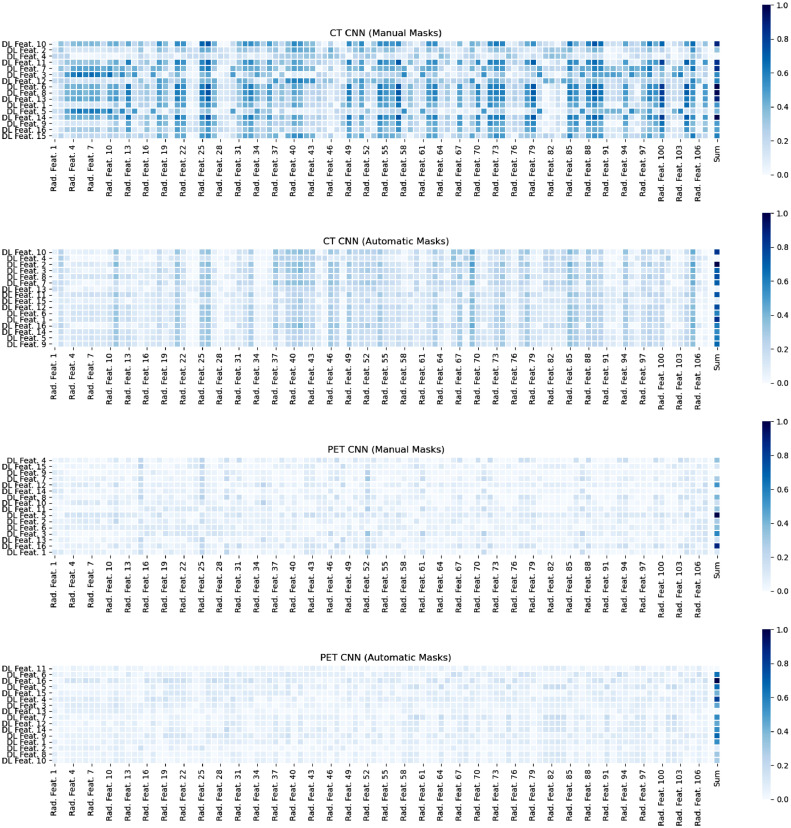
Correlation heatmaps sorted by RSF feature importance demonstrated that the manual CT deep learning features were most strongly correlated with corresponding radiomics features, followed by the automatic CT deep learning features. PET deep learning features for both manual and automatic masks showed weak correlation with radiomics features. There were no noticeable patterns between feature importance and correlation strength. The generated heatmaps are shown in Figure 5.
Figure 5.
Correlation Heatmaps between Deep Learning Features and Radiomics Features.
Absolute value of correlations are displayed. Deep learning features are ordered from top to bottom by relative importance within the random survival forest model. The sum of all correlations for each deep learning feature, normalized to 0 to 1, is shown in the final column.
ComBAT Data harmonization results
For manual mask RSF models trained with deep learning features following ComBAT harmonization, the CT, PET, and clinical modalities had C-indices of 0.649, 0.716, and 0.591 respectively. The PET+CT ensemble and PET+CT+clinical ensemble models had C-indices of 0.711 and 0.709 respectively. For the corresponding CT, PET, and clinical models with automatic masks, C-indices were 0.748, 0.717, and 0.590, while the PET+CT and PET+CT+clinical models had C-indices of 0.729 and 0.727.
For manual mask RSF models with radiomics features following harmonization, the CT, PET, and clinical modalities had C-indices of 0.738, 0.629, and 0.600 respectively. PET+CT and PET+CT+clinical ensemble models had C-indices of 0.680 and 0.676. For corresponding radiomics models with automatic masks, CT, PET, and clinical models had C-indices of 0.680, 0.653, and 0.589, while the PET+CT and PET+CT+clinical models had C-indices of 0.664 and 0.659.
Compared to the corresponding models trained with non-harmonized features, both deep learning PET models and the radiomics PET model with automatic masks had significantly higher C-index (all p < 0.001, [bootstrap]). The manual deep learning CT model had a significantly lower C-index (p < 0.001, [bootstrap]), but all other CT models and both ensemble models had no significant differences compared to their non-harmonized equivalents. The full RSF model results are displayed in Table 5 and a table of p-values for comparisons is displayed in Appendix F.
Table 5.
Harmonized Feature RSF Model Performance. Mean results for performance metrics with associated bootstrap percentile 95% confidence intervals are displayed for deep learning and radiomics features with manually or automatically segmented masks. C-index: concordance index; IBS: integrated brier score.
| Modality | C-Index | IBS | Modality | C-Index | IBS | ||
|---|---|---|---|---|---|---|---|
| RSF model with deep learning features (Manual masks) | RSF model with deep learning features (Automatic masks) | ||||||
| CT | 0.649 (0.596, 0.708) | 0.207 (0.189, 0.238) | CT | 0.748 (0.700, 0.801) | 0.168 (0.139, 0.200) | ||
| PET | 0.716 (0.657, 0.776) | 0.175 (0.138, 0.203) | PET | 0.717 (0.671, 0.768) | 0.176 (0.150, 0.210) | ||
| Clinical | 0.591 (0.525, 0.647) | 0.175 (0.138, 0.203) | Clinical | 0.590 (0.538, 0.663) | 0.176 (0.150, 0.21) | ||
| PET + CT Ensemble | 0.711 (0.661, 0.767) | 0.179 (0.154, 0.207) | PET + CT Ensemble | 0.729 (0.682, 0.774) | 0.169 (0.141, 0.20) | ||
| PET + CT + Clinical Ensemble | 0.709 (0.655, 0.774) | 0.176 (0.148, 0.207) | PET + CT + Clinical Ensemble | 0.727 (0.679, 0.774) | 0.169 (0.142, 0.201) | ||
| RSF model with radiomics features (Manual masks) | RSF model with radiomics features (Automatic masks) | ||||||
| CT | 0.738 (0.677, 0.796) | 0.183 (0.162, 0.217) | CT | 0.680 (0.638, 0.722) | 0.196 (0.173, 0.223) | ||
| PET | 0.629 (0.581, 0.682) | 0.225 (0.194, 0.273) | PET | 0.653 (0.602, 0.704) | 0.204 (0.173, 0.235) | ||
| Clinical | 0.600 (0.541, 0.673) | 0.225 (0.194, 0.273) | Clinical | 0.589 (0.534, 0.647) | 0.204 (0.173, 0.235) | ||
| PET + CT Ensemble | 0.680 (0.619, 0.743) | 0.197 (0.174, 0.235) | PET + CT Ensemble | 0.664 (0.616, 0.705) | 0.195 (0.167, 0.223) | ||
| PET + CT + Clinical Ensemble | 0.676 (0.611, 0.733) | 0.187 (0.162, 0.228) | PET + CT + Clinical Ensemble | 0.659 (0.613, 0.712) | 0.186 (0.159, 0.214) | ||
Discussion
The study demonstrates that CNNs trained using pre-treatment FDG-PET/CT data performed well in predicting lung malignancy progression and OS. Ensemble progression models had significantly improved performance compared to PET only but comparable results to CT, and performed similarly with manual and automated segmentations. All CT models and ensemble models for overall survival were significantly stronger than PET only and clinical only models, but had comparable performance to each other. Deep learning models had generally higher C-indices than corresponding radiomics models, but these differences were not significant. Data harmonization significantly increased the performance of multiple PET RSF models, but did not change ensemble performance.
The majority of existing studies focusing on PET and CT analysis for lung cancer outcome prediction have used radiomics,16 which have had varied success. For instance, 3D CT features were used to predict survival time with decision trees in patients with adenocarcinoma, achieving an accuracy of 0.775 and an AUC of 0.712.17 Similarly, CT radiomics features were used to predict OS with ML in NSCLC, achieving a C-index of 0.68, less than our CT feature only and combined ensemble RSF models across feature and mask types.18 RSFs have also been used with PET radiomics to predict three-year disease recurrence with a strong accuracy of 0.901 and an AUC of 0.956.19 Our results highlight that DL feature based RSF models have at least comparably strong performance to a corresponding radiomics feature model and offers some preliminary evidence of potential improvement in performance with the DL models. The GradCAM visualization demonstrates that these features are likely to correlate with meaningful regions and edges of the lesions. The heatmap results provide further clinical validation for these features, showing that CT DL features are well and more strongly correlated with known engineered radiomics features compared with PET DL features, which parallels the improved CT performance with survival prediction.
While most existing work has used radiomics, several studies have also proposed deep learning models like those in our study. For instance, a 3D CNN was used to predict two-year OS based on CT in NSCLC, achieving AUCs ranging from 0.70 to 0.73.20 A U-net CNN trained on the same CT dataset achieved an AUC of 0.88 for two-year OS, and the CNN image features were better correlated with OS than traditional radiomics features.21 In the analysis of PET with deep learning, Tau et al. used CNNs to predict lymph node and distant metastasis in patients with NSCLC.22 While the CNNs performed well in predicting N staging category (accuracy=0.80), they performed poorly in predicting distant metastasis at 6-month follow up (accuracy=0.63). While these results are strong, they have focused on prediction of a timepoint-specific binary outcome. This study extends prior work done with this methodology to prediction of survival as a continuous outcome through incorporation of the RSF model, which outperformed corresponding direct CNN CT and ensemble models.
This study additionally compared the survival model performance between manual and automated lesion masks. The nnU-NET segmentation had a very high accuracy and specificity due to the high proportion of healthy background tissue; while sensitivity performance was weaker, AUC remained high for both modalities. The non-significant differences in ensemble model performance between manual and automatic segmentation suggest that an end-to-end generalizable model could offer comparably strong results, which would be beneficial for reduction of inter-reader variability.
Another strength of this study is evaluation individual CT and PET modalities alongside ensemble models, showing that CT models generally outperformed PET and that addition of PET to CT caused non-significant changes in model performance. Whether PET provides additional prognostic information beyond CT in lung cancer prognosis remains controversial. In the literature, the PET derived value maxSUV is most often included in time-to-event models. Some studies have indicated that while maxSUV may be helpful for predicting OS,23 local control,23 and recurrence free survival24 when combined with other metrics, it is a poor independent predictor,25 which is supported by the weaker performance of our maxSUV only OS prediction model. A recent multivariate analysis in 1500 patients found SUV to be predictive of prognosis in stage I to III disease but not in stage IV disease.26 Interestingly, maxSUV was greater in non-progressing nodules than in progressing nodules, despite studies indicating that increased maxSUV may be a biomarker for nodule aggressiveness.27 Inconsistent findings regarding the prognostic value of maxSUV may be explained by high degrees of variability based on factors such as blood glucose level, time of imaging following injection, lesion size, age, sex, and body mass index.27, 28, 29 The independent predictive value of AJCC stage or total tumor volume, both of which performed similar to maxSUV in this study, have also had significantly variability in reported results. One study found that AJCC stage alone achieved a strong c-index of 0.833 on non-metastatic NSCLC,30 while another large study examining general NSCLC found that stage only achieved a c-index of 0.624.31 In a study examining subsets within Stage I NSCLC, stage had a c-index of 0.56,32 and in another analysis with SCLC, stage achieved a c-index of 0.65.33 For total tumor volume, one study on Stage IA NSCLC found volume to predict recurrence with a modest AUC-ROC of 0.58,34 but another study with Stage III NSCLC predicted PFS following chemoradiation with a c-index of 0.68.35
Radiomics and deep learning studies with both PET and CT features also offer mixed results on the prognostic utility of PET. In one recent radiomics study for OS prediction in NSCLC based on longitudinal FDG-PET/CT, a support vector machine classifier with PET features outperformed one with CT features, and the combination of PET and CT features slightly increased performance compared to PET alone.10 Another study in NSCLC found PET features to be more predictive of local recurrence than CT features, and the combination of PET and CT features increased performance.11 However, features from neither modality were predictive of OS. A radiomics-based study examining disease-free survival prediction with PET/CT using Cox models found that a CT only model had an AUC of 0.75, while a PET only and PET+CT models both had AUCs of 0.68.36 Another recent deep learning study employed CT and PET CNNs to derive single risk scores for OS prediction in lung cancer instead of using extracted features.14 The CT risk score was found to be a better predictor of OS than PET, and combining the CT risk, PET risk, and age in a Cox model predicted OS with a C-index of 0.68, although they did not specifically examine single modality results.
Prognostication based on PET may be inherently more difficult than that based on CT imaging alone, since patients sent for FDG-PET/CT often have indeterminate lesions that physicians have deemed difficult to evaluate with sequential CT.37 Furthermore, the decision to pursue an FDG-PET/CT scan is often dependent on clinical judgement and institutional practices, which may lead to high variability in patient datasets and associated studies. The ComBAT data harmonization algorithm has been shown to remove site bias in imaging and radiomics features and improve prediction results in PET, CT, and MRI imaging for neuroimaging, NSCLC, cervical cancer, among others.38, 39, 40 In this study, ComBAT harmonization did significantly improve multiple PET model performances, suggesting that the lower non-harmonized PET performance could be attributable to inter-site variation. However, ensemble performance did not have a corresponding increase, even when CT model performance was comparable. This indicates that site variability may be a cause of decreased PET model performance, but that harmonization may not necessarily improve the prognostic utility of adding PET information to CT.
This study has several limitations. Firstly, the manual segmentation of the nodules was done predominantly by one radiologist, which could have artificially introduced variability into the dataset and influenced the manual mask results. As discussed, most patients that undergo FDG-PET/CT scans have specifically indeterminate nodules. Consequently, the study may be limited by selection bias and have less generalizability to a broader lung cancer population. A further limitation is that while this study examined a set of well-studied radiomics features, multiple other radiomics approaches exist and could provide differing results.14,18 Although the GradCAM results and heatmaps lend some insight into the deep learning features in this study, they still lack some clinical interpretability as we were not able to define a direct relationship between these features and known clinical markers or outcomes. Finally, the data harmonization work done within this study was following feature extraction; additional analysis with harmonization techniques prior to image processing could provide additional performance benefits not represented within our results.
There are several potential avenues for future work. A similar analysis of longitudinal FDG-PET/CT imaging before and after treatment incorporating additional information on clinical course with recurrent neural networks could improve progression and OS risk prediction. More analysis could also be done examining if there are more optimal or robust strategies for extracting features from these CNNs than using the penultimate layer of features. While our data was derived from three independent institutions, generalizability can still be improved if data from additional institutions is added to the training set. A possible approach is federated learning, which facilitates distribution of model training between multi-institutional datasets without requiring data sharing.41 Proof of concept for this approach has recently been demonstrated in lung cancer.42 Finally, any prognostic ML model must be evaluated prospectively in a variety of institutions and patient populations for real clinical implementation. Computational efficiency should also be prioritized if real-time use is to be achieved.
In conclusion, CNNs trained using pre-treatment FDG-PET/CT perform well in predicting lung malignancy progression. Features extracted with the CNN had strong performance in OS prediction with a RSF model that was comparable to a radiomics feature extraction approach. Models based on CT performed better than those based on PET, and the addition of PET to CT in an ensemble model only provides non-significant improvements in performance over CT alone. By identifying lung malignancies with high progression and mortality risk, ML based on FDG-PET/CT can improve prognostication and planning of care.
Contributors
Brian Huang: Methodology, Software, Formal analysis, Investigation, Data curation, Validation, Writing – review and editing, Visualization. John Sollee: Writing – original draft, Writing – review and editing, Data curation, Visualization, Project administration. Yong-Heng Luo: Conceptualization, Project Administration, Data curation, Methodology, Investigation, Verification of underlying data, Supervision. Ashwin Reddy: Methodology, Software, Investigation, Formal Analysis, Writing-review and editing. Zhusi Zhong: Methodology, Software, Investigation, Formal Analysis, Writing-review and editing. Jing Wu: Conceptualization, Data curation, Supervision, Project administration. Joseph Mammarappallil: Resources, Supervision, Data Curation. Terrance Healey: Resources, Supervision, Data Curation. Gang Cheng: Resources, Supervision, Data Curation. Christopher Azzoli: Resources, Supervision, Data Curation. Dana Korogodsky: Writing – original draft preparation, Writing – review and editing. Paul Zhang: Resources, Supervision, Data Curation. Xue Feng: Resources, Supervision, Software. Jie Li: Methodology, Software, Supervision. Li Yang: Conceptualization, Resources, Data Curation, Writing – review and editing, Supervision, Project administration, funding acquisition. Zhicheng Jiao: Conceptualization, Supervision, Methodology, Writing – review and editing, Project administration. Harrison X. Bai: Conceptualization, Supervision, Writing – review and editing, Project administration, Verification of underlying data, Resources.
All contributors have read and approved this manuscript.
Data sharing statement
The data are not available for public access because of patient privacy concerns but are available from the corresponding authors if there is a reasonable request and approval from the institutional review boards of the affiliated institutions. The codebase used in this study is available online (https://github.com/BrianHeHuang/Lung-PET-CT-Survival). All implementation details are described thoroughly in the Methods and Appendix sections.
Declaration of interests
Dr. Feng reports personal fees from Carina Medical LLC, outside the submitted work. The remaining authors declare that they have no conflicts of interest and nothing to disclose.
Acknowledgements
We would like to thank Drs. Man-Jun Xiao, Juan Chen, Ting Huang, and Shan-Shan Chen from the Second Xiangya Hospital for their assistance with data evaluation.
Research reported in this publication was partially supported by a training grant from the National Institute of Health (NIH), National Heart, Lung, and Blood Institute (NHLBI) (5T35HL094308-12, John Sollee) and from the RSNA Medical Student Grant (Brian Huang). Jing Wu is supported by the Huxiang High-level Talent Gathering Project-Innovation Talent(No. 2021RC5003) and Natural Science Foundation of Hunan Province(No. 2021JJ40876); Yongheng Luo is supported by the Natural Science Foundation of Hunan Province(No. 2021JJ30945). This research did not receive any other specific grant from funding agencies in the public, commercial, or not-for-profit sectors. All authors confirm that they have full access to all the data in the study and accept responsibility to submit the report for publication.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104127.
Contributor Information
Li Yang, Email: Yangli762@csu.edu.cn.
Zhicheng Jiao, Email: Zhicheng_Jiao@Brown.edu.
Harrison Xiao Bai, Email: Hbai7@jhu.edu.
Appendix. Supplementary materials
References
- 1.Bade BC, Dela Cruz CS. Clinics in Chest Medicine. W.B. Saunders; 2020. Lung cancer 2020: epidemiology, etiology, and prevention [Internet] pp. 1–24.https://pubmed.ncbi.nlm.nih.gov/32008623/ Vol. 41. [cited 2021 May 27]. Available from: [DOI] [PubMed] [Google Scholar]
- 2.Jones GS, Baldwin DR. Recent Advances in the Management of Lung Cancer [Internet] Clin Med (Lond) 2018;18:s41–s46. doi: 10.7861/clinmedicine.18-2s-s41. [cited 2021 May 27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med [Internet] 2020;382(6):503–513. doi: 10.1056/NEJMoa1911793. https://www.nejm.org/doi/full/10.1056/NEJMoa1911793 [cited 2021 May 27]. Available from: [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin [Internet] 2018;68(1):7–30. doi: 10.3322/caac.21442. https://pubmed.ncbi.nlm.nih.gov/29313949/ [cited 2021 May 27]. Available from: [DOI] [PubMed] [Google Scholar]
- 5.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. Chest. 2017. The eighth edition lung cancer stage classification [Internet]http://journal.chestnet.org/article/S0012369216607808/fulltext 151:193–203. [cited 2021 May 27]. Available from: [DOI] [PubMed] [Google Scholar]
- 6.Kandathil A, Kay FU, Butt YM, Wachsmann JW, Subramaniam RM. Role of FDG PET/CT in the eighth edition of TNM staging of non– small cell lung cancer. Radiographics [Internet] 2018;38(7):2134–2149. doi: 10.1148/rg.2018180060. https://pubmed.ncbi.nlm.nih.gov/30422775/ [cited 2021 May 27]. Available from: [DOI] [PubMed] [Google Scholar]
- 7.Falcoz PE, Puyraveau M, Thomas PA, et al. Video-assisted thoracoscopic surgery versus open lobectomy for primary non-small-cell lung cancer: a propensity-matched analysis of outcome from the European society of thoracic surgeon database. Eur J Cardio-thoracic Surg [Internet] 2016;49(2):602–609. doi: 10.1093/ejcts/ezv154. https://pubmed.ncbi.nlm.nih.gov/25913824/ [cited 2021 Jun 16]. Available from: [DOI] [PubMed] [Google Scholar]
- 8.Temel JS, Greer JA, Muzikansky A, et al. Early palliative care for patients with metastatic non–small-cell lung cancer. N Engl J Med [Internet] 2010;363(8):733–742. doi: 10.1056/NEJMoa1000678. https://www.nejm.org/doi/full/10.1056/NEJMoa1000678 [cited 2021 Jun 16]. Available from: [DOI] [PubMed] [Google Scholar]
- 9.Rubin GD. J Thorac Imag. Lippincott Williams and Wilkins Ltd.; 2015. Lung nodule and cancer detection in computed tomography screening. 30(2):130–138. [Internet] [cited 2021 May 27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astaraki M, Wang C, Buizza G, Toma-Dasu I, Lazzeroni M, Smedby Ö. Early survival prediction in non-small cell lung cancer from PET/CT images using an intra-tumor partitioning method. Phys Medica [Internet] 2019;60:58–65. doi: 10.1016/j.ejmp.2019.03.024. https://pubmed.ncbi.nlm.nih.gov/31000087/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 11.Dissaux G, Visvikis D, Da-Ano R, et al. Pretreatment 18F-FDG PET/CT radiomics predict local recurrence in patients treated with stereotactic body radiotherapy for early-stage non-small cell lung cancer: a multicentric study. J Nucl Med [Internet] 2020;61(6):814–820. doi: 10.2967/jnumed.119.228106. https://pubmed.ncbi.nlm.nih.gov/31732678/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 12.Amini M, Hajianfar G, Haddadi Avval A, et al. Multiple machine learning algorithms for overall survival modeling of NSCLC patients using PET-, CT-, and fusion-based radiomics. J Nucl Med. 2021;62(suppl 1):1192. [Google Scholar]
- 13.Van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res [Internet] 2017;77(21):e104. doi: 10.1158/0008-5472.CAN-17-0339. [cited 2022 Mar 7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afshar P, Mohammadi A, Tyrrell PN, et al. DRTOP: deep learning-based radiomics for the time-to-event outcome prediction in lung cancer. Sci Rep [Internet] 2020;10(1):1–5. doi: 10.1038/s41598-020-69106-8. https://pubmed.ncbi.nlm.nih.gov/32703973/ [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K, Lee J, Gupta V, Cho G. Comparison of bootstrap confidence interval methods for GSCA using a monte carlo simulation. Front Psychol. 2019;10:2215. doi: 10.3389/fpsyg.2019.02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manafi-Farid R, Karamzade-Ziarati N, Vali R, Mottaghy FM, Beheshti M. Methods. Academic Press Inc.; 2021. 2-[18F]FDG PET/CT radiomics in lung cancer: an overview of the technical aspect and its emerging role in management of the disease [Internet]https://pubmed.ncbi.nlm.nih.gov/32497604/ Vol. 188. 188:84–97. [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 17.Hawkins SH, Korecki JN, Balagurunathan Y, et al. Predicting outcomes of nonsmall cell lung cancer using CT image features. IEEE Access. 2014;2:1418–1426. [Google Scholar]
- 18.Sun W, Jiang M, Dang J, Chang P, Yin FF. Effect of machine learning methods on predicting NSCLC overall survival time based on Radiomics analysis. Radiat Oncol [Internet] 2018;13(1):1–8. doi: 10.1186/s13014-018-1140-9. [cited 2021 Jun 4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn HK, Lee H, Kim SG, Hyun SH. Pre-treatment 18F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin Radiol [Internet] 2019;74(6):467–473. doi: 10.1016/j.crad.2019.02.008. https://pubmed.ncbi.nlm.nih.gov/30898382/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 20.Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med [Internet] 2018;15(11) doi: 10.1371/journal.pmed.1002711. https://pubmed.ncbi.nlm.nih.gov/30500819/ :e1002711. [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek S, He Y, Allen BG, et al. Deep segmentation networks predict survival of non-small cell lung cancer. Sci Rep [Internet] 2019;9(1):1–10. doi: 10.1038/s41598-019-53461-2. https://pubmed.ncbi.nlm.nih.gov/31754135/ [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tau N, Stundzia A, Yasufuku K, Hussey D, Metser U. Convolutional neural networks in predicting nodal and distant metastatic potential of newly diagnosed non–small cell lung cancer on FDG PET images. Am J Roentgenol [Internet] 2020;215(1):192–197. doi: 10.2214/AJR.19.22346. https://pubmed.ncbi.nlm.nih.gov/32348182/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 23.Na F, Wang J, Li C, Deng L, Xue J, Lu Y. Primary tumor standardized uptake value measured on F18-fluorodeoxyglucose positron emission tomography is of prediction value for survival and local control in non-small-cell lung cancer receiving radiotherapy: Meta-analysis. J Thorac Oncol [Internet] 2014;9(6):834–842. doi: 10.1097/JTO.0000000000000185. https://pubmed.ncbi.nlm.nih.gov/24787963/ [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon W, Howard BA, Herndon JE, Patz EF. FDG uptake on positron emission tomography correlates with survival and time to recurrence in patients with Stage I non-small-cell lung cancer. J Thorac Oncol [Internet] 2015;10(6):897–902. doi: 10.1097/JTO.0000000000000534. https://pubmed.ncbi.nlm.nih.gov/25811445/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
- 25.Agarwal M, Brahmanday G, Bajaj SK, Ravikrishnan KP, Wong CYO. Revisiting the prognostic value of preoperative 18F-fluoro-2- deoxyglucose (18F-FDG) positron emission tomography (PET) in early-stage (I & II) non-small cell lung cancers (NSCLC) Eur J Nucl Med Mol Imaging [Internet] 2010;37(4):691–698. doi: 10.1007/s00259-009-1291-x. https://pubmed.ncbi.nlm.nih.gov/19915840/ [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paesmans M, Garcia C, Wong CYO, Patz EF, Komaki R, Eschmann S, et al. Primary tumour standardised uptake value is prognostic in nonsmall cell lung cancer: a multivariate pooled analysis of individual data. Eur Respir J [Internet] 2015;46(6):1751–1761. doi: 10.1183/13993003.00099-2015. https://pubmed.ncbi.nlm.nih.gov/26405289/ [cited 2021 Jun 16]. Available from: [DOI] [PubMed] [Google Scholar]
- 27.Duan XY, Wang W, Li M, Li Y, Guo YM. Predictive significance of standardized uptake value parameters of FDG-PET in patients with non-small cell lung carcinoma. Brazilian J Med Biol Res [Internet] 2015;48(3):267–272. doi: 10.1590/1414-431X20144137. [cited 2021 Jun 23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Wang D, Yu L. Prognostic and predictive values of metabolic parameters of 18F-FDG PET/CT in patients with non-small cell lung cancer treated with chemotherapy. Mol Imaging [Internet] 2019;18 doi: 10.1177/1536012119846025. https://pubmed.ncbi.nlm.nih.gov/31144578/ 1536012119846025. [cited 2021 Jun 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groheux D, Quere G, Blanc E, et al. Diagn Interv Imaging. 2016. FDG PET-CT for solitary pulmonary nodule and lung cancer: literature review [Internet]https://pubmed.ncbi.nlm.nih.gov/27567555/ Elsevier Masson SAS. 97:1003–1017. [cited 2021 Jun 8]. Available from: [DOI] [PubMed] [Google Scholar]
- 30.Young KA, Efiong E, Dove JT, et al. External validation of a survival nomogram for non-small cell lung cancer using the national cancer database. Ann Surg Oncol [Internet] 2017;24(6):1459–1464. doi: 10.1245/s10434-017-5795-5. https://link-springer-com.proxy.library.upenn.edu/article/10.1245/s10434-017-5795-5 [cited 2022 May 11]. Available from: [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Wang S, Zhou Y, et al. Evaluation of the 7th and 8th editions of the AJCC/UICC TNM staging systems for lung cancer in a large North American cohort. Oncotarget [Internet] 2017;8(40):66784. doi: 10.18632/oncotarget.18158. [cited 2022 May 11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Y, Mayne N, Yang CFJ, et al. A Nomogram for predicting cancer-specific survival of TNM 8th edition stage i non-small-cell lung cancer. Ann Surg Oncol [Internet] 2019;26(7):2053–2062. doi: 10.1245/s10434-019-07318-7. https://link-springer-com.proxy.library.upenn.edu/article/10.1245/s10434-019-07318-7 [cited 2022 May 11]. Available from: [DOI] [PubMed] [Google Scholar]
- 33.Pan H, Shi X, Xiao D, et al. Nomogram prediction for the survival of the patients with small cell lung cancer. J Thorac Dis [Internet] 2017;9(3):507. doi: 10.21037/jtd.2017.03.121. [cited 2022 May 11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takenaka T, Yamazaki K, Miura N, Mori R, Takeo S. The prognostic impact of tumor volume in patients with clinical stage IA non-small cell lung cancer. J Thorac Oncol [Internet] 2016;11(7):1074–1080. doi: 10.1016/j.jtho.2016.02.005. http://www.jto.org/article/S1556086416004056/fulltext [cited 2022 May 12]. Available from: [DOI] [PubMed] [Google Scholar]
- 35.Zhang N, Liang R, Gensheimer MF, et al. Early response evaluation using primary tumor and nodal imaging features to predict progression-free survival of locally advanced non-small cell lung cancer. Theranostics. 2020;10(25):11707. doi: 10.7150/thno.50565. [Internet] [cited 2022 May 12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirienko M, Cozzi L, Antunovic L, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging [Internet] 2018;45(2):207–217. doi: 10.1007/s00259-017-3837-7. https://pubmed.ncbi.nlm.nih.gov/28944403/ [cited 2022 Mar 13]. Available from: [DOI] [PubMed] [Google Scholar]
- 37.Hadique S, Jain P, Hadi Y, Baig A, Parker JE. Utility of FDG PET/CT for assessment of lung nodules identified during low dose computed tomography screening. BMC Med Imaging [Internet] 2020;20(1) doi: 10.1186/s12880-020-00469-0. https://pubmed.ncbi.nlm.nih.gov/32571221/ [cited 2021 Jun 8]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortin JP, Parker D, Tunç B, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlhac F, Boughdad S, Philippe C, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med [Internet] 2018;59(8):1321–1328. doi: 10.2967/jnumed.117.199935. https://jnm.snmjournals.org/content/59/8/1321 [cited 2022 Mar 6]. Available from: [DOI] [PubMed] [Google Scholar]
- 40.Da-ano R, Masson I, Lucia F, et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci Rep [Internet] 2020;10(1):1–12. doi: 10.1038/s41598-020-66110-w. https://www-nature-com.proxy.library.upenn.edu/articles/s41598-020-66110-w [cited 2022 Mar 6]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheller MJ, Edwards B, Reina GA, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep [Internet] 2020;10(1):1–12. doi: 10.1038/s41598-020-69250-1. https://pubmed.ncbi.nlm.nih.gov/32724046/ [cited 2021 Jul 1]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deist TM, Dankers FJWM, Ojha P, et al. Distributed learning on 20 000+ lung cancer patients – the personal health train. Radiother Oncol [Internet] 2020;144:189–200. doi: 10.1016/j.radonc.2019.11.019. https://pubmed.ncbi.nlm.nih.gov/31911366/ [cited 2021 Jun 4]. Available from: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.