Abstract
Ovarian cancer is the most fatal tumor characterized by an abundance of tumor-associated macrophage (TAM) infiltrations in women. Functional TAMs, which mainly present M2-like phenotypes and perform key functions on tumor progress, have been considered an attractive target for ovarian cancer therapy. Cardamonin showed an excellent antitumor activity in multiple tumor cells. This study aimed to investigate the role of cardamonin on TAMs. With the conditioned medium of ovarian cancer cells, macrophages were induced to TAMs and, accordingly, promoted the proliferation, migration, and invasion of ovarian cancer cells. Cardamonin suppressed alternatively activated (M2) polarization of TAMs and downregulated TAM-secreted tumorigenic factors, thereby hindering the pro-tumor function of TAMs on ovarian cancer cells. Moreover, cardamonin inhibited tumor growth in xenograft nude mice and lowered the expression of CD163 and CD206. Mechanistically, cardamonin inhibited the phosphorylation of mammalian target of rapamycin (mTOR) and signal transducer and activator of transcription 3 (STAT3), resulting in the suppression of M2 polarization. Furthermore, STAT3 is tightly related with mTOR activity. Altogether, these findings implied that cardamonin suppresses the pro-tumor function of TAMs by decreasing M2 polarization via mTOR inhibition, and cardamonin may be a potential therapeutic agent for ovarian cancer.
Keywords: cardamonin, TAMs, mTOR, STAT3, ovarian cancer
Graphical abstract
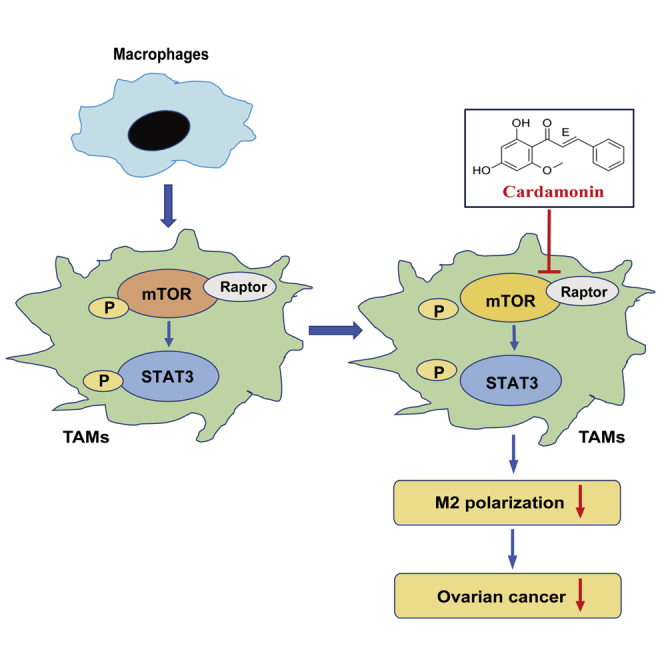
This study reports that cardamonin, a natural chalcone compound, reduces tumor-associated macrophage M2 polarization and impedes their pro-tumor function via mTOR inhibition, thereby suppressing the proliferation and migration of ovarian cancer cells, which implies that cardamonin may be a potential therapeutic agent for ovarian cancer.
Introduction
Increased infiltration of tumor-associated macrophages (TAMs) was observed in most human cancers, especially in ovarian cancer. Evidence shows that the high density of TAMs is closely related to reduced relapse-free survival and poor prognosis,1 suggesting that TAMs play a crucial role in tumor progress. TAMs are heterogeneous and plastic cells, which can be extremely induced to either classically activated (M1) subtype with antitumor effect or alternatively activated (M2) phenotype with function of tumor promotion.2 Previous studies have shown that macrophages in tumor microenvironment (TME) are gradually educated by cancer cells and transformed into TAMs, which exhibit an M2-like subtype.3,4 Subsequently, functional TAMs secret a variety of cytokines, chemokines, and enzymes, including interleukin-6 (IL-6), IL-10, vascular endothelial growth factor α (VEGFα), matrix metalloproteinase2 (MMP2), and MMP9, which accelerate tumor progression in tumor growth, angiogenesis, metastasis, and chemoresistance.5,6 Therefore, targeting TAMs by polarization regulation and function modulation may be a potential strategy for ovarian cancer therapy.
Convincing facts indicate that the phenotypes and functions of macrophages are orchestrated by the mammalian target of rapamycin (mTOR) signaling pathway.7, 8, 9 mTOR is a conserved serine/threonine protein kinase that plays a central role in cell biological processes.10 By sensing and integrating various environmental cues including growth factors, amino acids, inflammatory factors, and stress status, mTOR controls cell growth, metabolism, and differentiation in immune cells.11,12 Extensive studies have demonstrated that mTOR is very important in terms of macrophage function in disease progression and tissue homeostasis.8,13,14 mTOR governs the cancer-promoting macrophage responses in the TME and elicits TAMs to a M2-like polarization.15,16 Coincidentally, TAMs are usually accompanied by high mTOR activity.17,18 In addition, previous studies revealed that mTOR modulated macrophage polarization and functional transformation by crosstalk with other pathways, such as signal transducer and activator of transcription 3 (STAT3).14,18 Therefore, mTOR is closely related to macrophages’ functional modulation and is a potential option for human M2 macrophage-related disease treatment.
Cardamonin (CAR), a natural chalcone compound, showed excellent antitumor activity in many kinds of tumors cells including ovarian cancer. By inducing apoptosis, cell-cycle arrest, and autophagy, cardamonin suppresses tumor growth and metastasis in vivo and in vitro.19,20 Moreover, our previous studies and other research have confirmed that the antitumor effect of cardamonin was related to mTOR inhibition.20, 21, 22 Recent studies have verified that cardamonin induced immune responses and enhanced survival rate in WEHI-3 cell-generated mouse leukemia,23 suggesting a potential role of cardamonin in immune regulation. However, it is not clear whether the antitumor effect of cardamonin is related to TAM regulation. This study aimed to explore the role of cardamonin in TAM regulation by focusing on the polarization and functional change of macrophages and to confirm whether mTOR is involved in the regulation of TAMs by cardamonin.
Results
Macrophages were induced into functional TAMs by conditioned medium from ovarian cancer cells
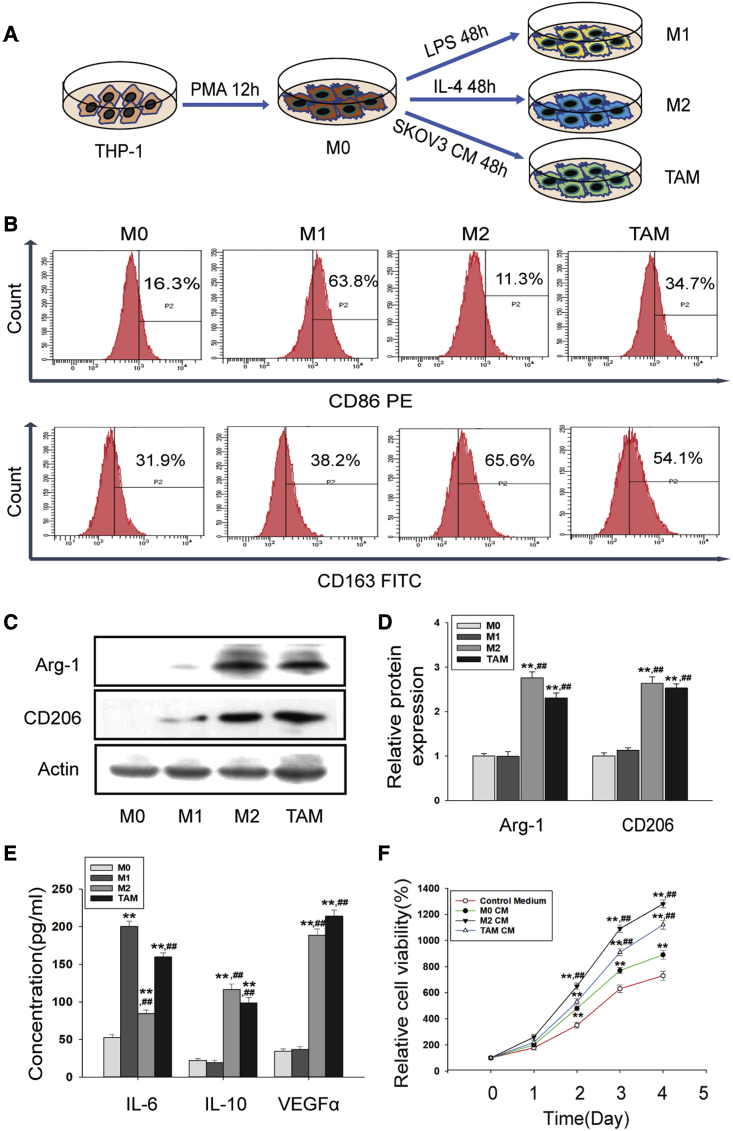
Macrophages are plastic and can be induced to different phenotypes and functions according to different stimulations. As shown in Figure 1A, THP-1 monocytes were induced to M0 (resting macrophages), M1 and M2 macrophages, and TAMs. After stimulation, the suspended cells attached to the dishes, accompanied by significant changes in cell morphology with obvious pseudopodia (Figure S1A). Then, we examined the expression of specific markers in THP-1-induced macrophages. Our results showed that CD86 was upregulated in M1 macrophages, while CD163 was highly expressed in M2 macrophages and TAMs (Figure 1B). Protein levels of Arginase-1 (Arg-1) and CD206 were upregulated in both M2 macrophages and TAMs (Figures 1C and 1D). Compared with M0, the expression of IL-6 was more highly expressed in M1 macrophages and TAMs, while IL-10 and VEGFα were more highly expressed in M2 macrophages and TAMs (Figures 1E and S1B).
Figure 1.
THP-1 cells were induced to functional TAMs
(A) Schematic diagram of macrophages induction. (B) The expression of CD86 and CD163 protein was measured by flow cytometry after macrophage induction. (C and D) The protein expression of Arg-1 and CD206 was detected by western blotting. The relative density ratio of each band was normalized to actin. (E) The concentration of IL-6, IL-10, and VEGFα protein was measured by ELISA assay. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus M0. ##p < 0.01 versus M1. (F) Cell viability of SKOV3 cells was measured by CCK-8 assay. SKOV3 cells were incubated with the specified conditioned medium (CM) for 5 days. Quantitative is relative to each group of 0 day, set to 100%. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus control medium. ##p < 0.01 versus M0 CM.
It has been demonstrated that functional TAMs play an essential role in tumor growth and metastasis. Conditioned medium (CM) of THP-1-induced macrophages was collected for ovarian cancer cell incubation (Figure S2A). Indeed, a 5-Ethynyl-2′-deoxyuridine (EdU) test showed that both M2 CM (CM from M2 macrophages) and TAM CM (CM from TAMs) could significantly promote cell proliferation of SKOV3 and A2780 cells (Figures S2B and S2C). Consistently, a Cell Counting Kit-8 (CCK-8) assay displayed similar results (Figures 1F and S2D). In addition, scratch experiment and Transwell assay showed that the migrated cells were obviously increased in both kinds of cells with TAM CM incubation (Figures S3A–3E). These results indicated that SKOV3 CM inclined macrophages to M2 polarization with pro-tumor functions.
Cardamonin impeded pro-tumor functions of TAMs on ovarian cancer cells
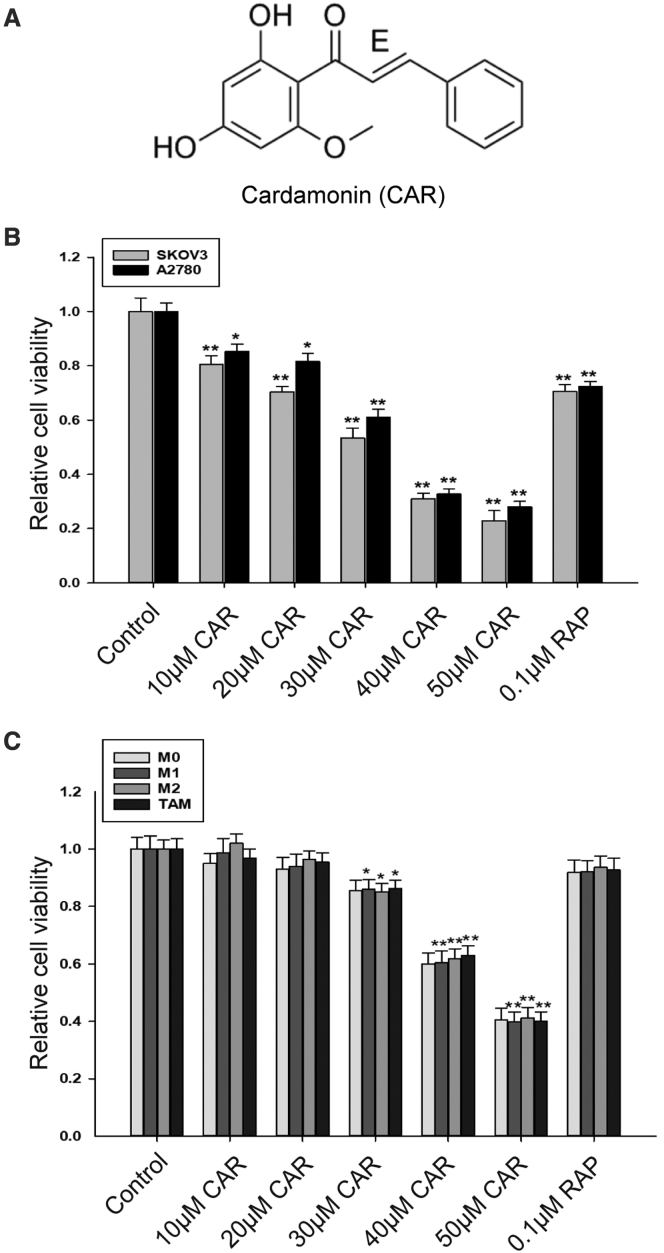
Cardamonin (chemical structure shown in Figure 2A) shows excellent antitumor activity; our results indicated that cardamonin inhibited the cell viability of SKOV3 and A2780 cells in a concentration-dependent manner (Figure 2B), while the effect of cardamonin on TAMs was still unclear. In the CCK-8 assay, cell viability of TAMs had no significant changes by cardamonin at a concentration less than 20 μM; however, it declined observably by cardamonin at a concentration more than 30 μM (Figure 2C). Similar results were observed in M0, M1, and M2 macrophages. In the subsequent experiments, we selected the concentration of cardamonin at 10 and 20 μM (respectively) to avoid the interference effect from drug inhibition.
Figure 2.
Cytotoxicity of cardamonin on ovarian cancer cells and macrophages
(A) Chemical structure of cardamonin. (B and C) Cell viability was conducted with CCK-8 assay. SKOV3 cells and A2780 cells (B) and THP-1-induced macrophages (C) were treated with different concentrations of cardamonin (0, 10, 20, 30, 40, and 50 μM) or rapamycin (0.1 μM) for 24 h, respectively. Data are presented as the mean ± SD (n = 5). ∗p < 0.05, ∗∗p < 0.01 versus control.
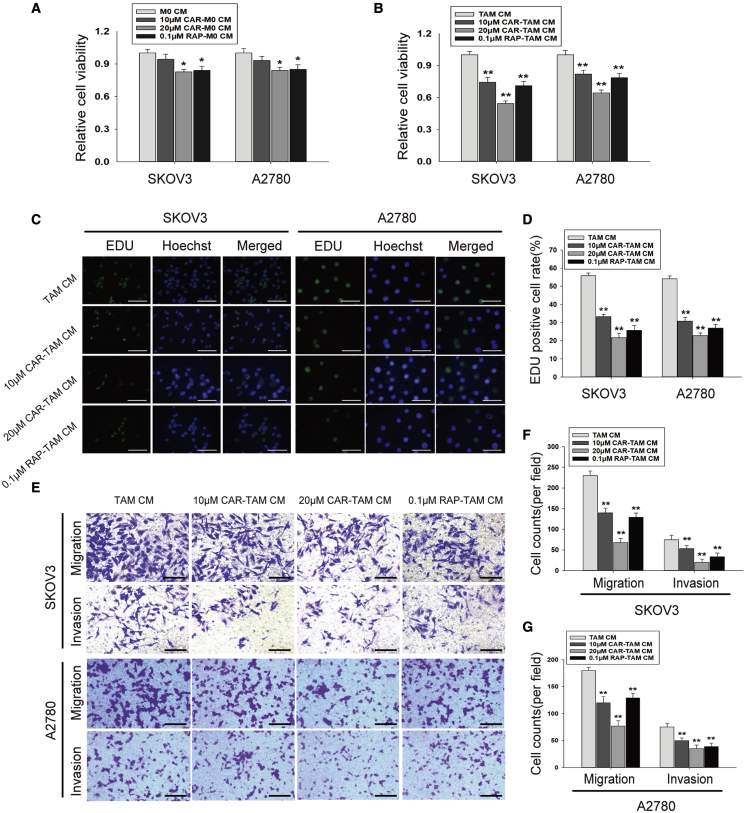
To clarify the effect of cardamonin on TAM functions, CM of cardamonin-pretreated TAMs was collected and then incubated with SKOV3 and A2780 cells (Figure S4A). We found that proliferation of both kind cells induced by TAM CM was impeded by cardamonin pretreatment (Figures 3A–3D). A slower migration was observed in ovarian cancer cells incubated with CM from cardamonin-treated TAMs by scratch test (Figures S4B and S4C). Consistently, Transwell assays revealed that cardamonin decreased the migration and invasion of both cells induced by TAM CM (Figures 3E–3G), suggesting that cardamonin treatment inhibits the pro-tumor functions of TAMs.
Figure 3.
Cardamonin impeded pro-tumor functions of TAMs on SKOV3 cells and A2780 cells
The specified CM was collected and incubated with SKOV3 cells and A2780 cells, respectively. (A and B) Cell viability was tested with CCK-8 assays. (A) ∗p < 0.05 versus M0 CM. (B) ∗∗p < 0.01 versus TAM CM. (C and D) Cell proliferation was measured by EDU test (scale bar, 100 μm). (E–G) Cell migration and invasion were evaluated by Transwell assays (scale bar, 100 μm). Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus TAM CM.
Cardamonin decreased M2 polarization and partially inhibited pro-tumorigenic factors in TAMs
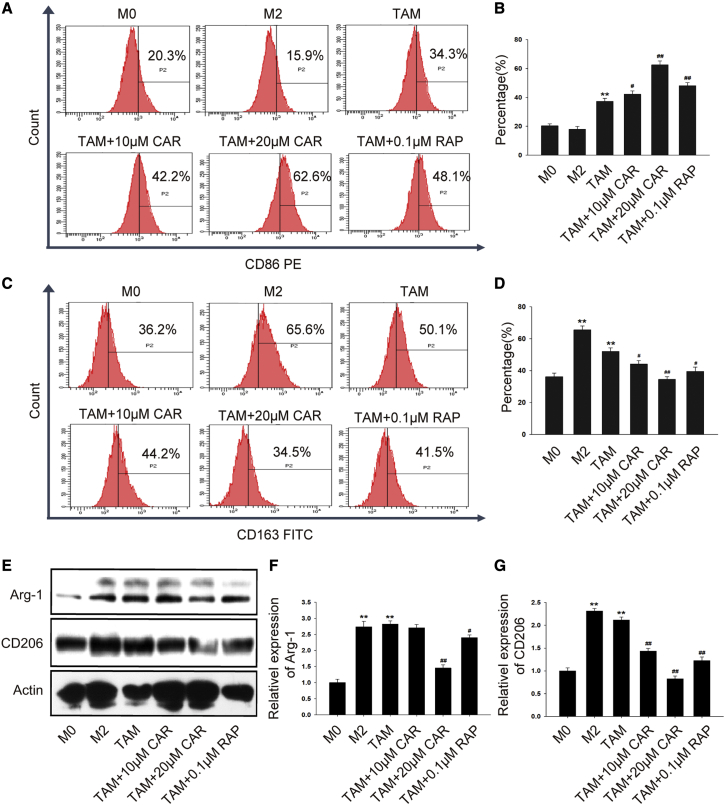
The functions of macrophages are closely related to their phenotype. Next, we evaluated the effect of cardamonin on TAM polarization. Treated with cardamonin for 24 h, expression of CD86 was promoted (Figures 4A and 4B), while the expression of CD163 was obviously inhibited (Figures 4C and 4D). Interestingly, Arg-1 showed no significant change in a lower concentration but was inhibited by a higher concentration of cardamonin (Figures 4E and 4F), while the protein level of CD206 was downregulated by both concentrations of cardamonin (Figure 4G).
Figure 4.
Cardamonin decreased M2 polarization in TAMs
THP-1 cells were induced and treated with or without cardamonin (10 or 20 μM) and rapamycin (0.1 μM) for 24 h. (A–D) Surface markers CD86 and CD163 were analyzed by flow cytometry. The percentage of dyed cells for each group is shown as the bar graph. (E–G) Expression of the Arg-1 and CD206 protein was detected by western blotting. Relative protein expression was normalized to actin. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus M0. #p < 0.05, ##p < 0.01 versus TAM.
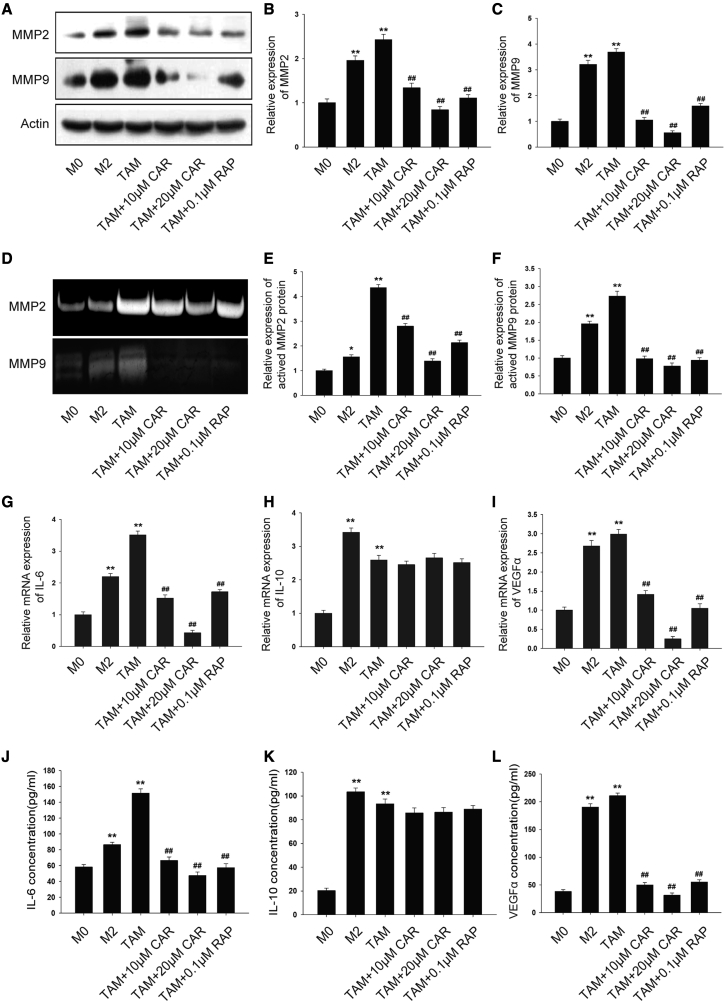
TAMs establish a pro-tumor microenvironment by secreting a variety of tumorigenic factors. We hypothesized that cardamonin inhibits tumor-promoting effects of TAMs by reducing the secretion of pro-tumor factors. As expected, protein levels of MMP2 and MMP9 were upregulated in TAMs compared with M0 macrophages (Figure 5A). Notably, a dramatic suppression was observed on protein levels of MMP2 and MMP9 by cardamonin treatment (Figures 5B and 5C). Similar results were observed in a gelatin zymography assay, which tested the enzymatic activity of MMP2 and MMP9 (Figures 5D–5F). Additionally, the mRNA expression of IL-6 and VEGFα was remarkably reduced by cardamonin. However, no significant effect of cardamonin on IL-10 expression was observed (Figures 5G–5I). Consistently, ELISA assays showed a similar result (Figures 5J–5L). These results revealed that cardamonin decreased M2 polarization and inhibited pro-tumorigenic factors secretion in TAMs.
Figure 5.
Cardamonin inhibited pro-tumorigenic factors in TAMs
THP-1-induced macrophages were treated with or without cardamonin (10 or 20 μM) and rapamycin (0.1 μM) for 24 h. (A–C) The expression of MMP2 and MMP9 protein was detected by western blotting. Relative density ratios of MMP2 and MMP9 were normalized to actin. (D) Expression of activated MMP2 and MMP9 was analyzed using gelatin zymography. (E and F) The relative expression of activated MMP2 and MMP9 is shown as the bar graph, with the M0 group set to 1. (G–I) mRNA expression of IL-6, IL-10, and VEGFα was measured by real-time PCR. Relative mRNA expression was normalized to actin. (J–L) The concentration of IL-6, IL-10, and VEGFα protein was measured by ELISA assay. Data are presented as the mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01 versus M0. ##p < 0.01 versus TAM.
Cardamonin inhibited tumor growth and decreased M2 macrophages in vivo
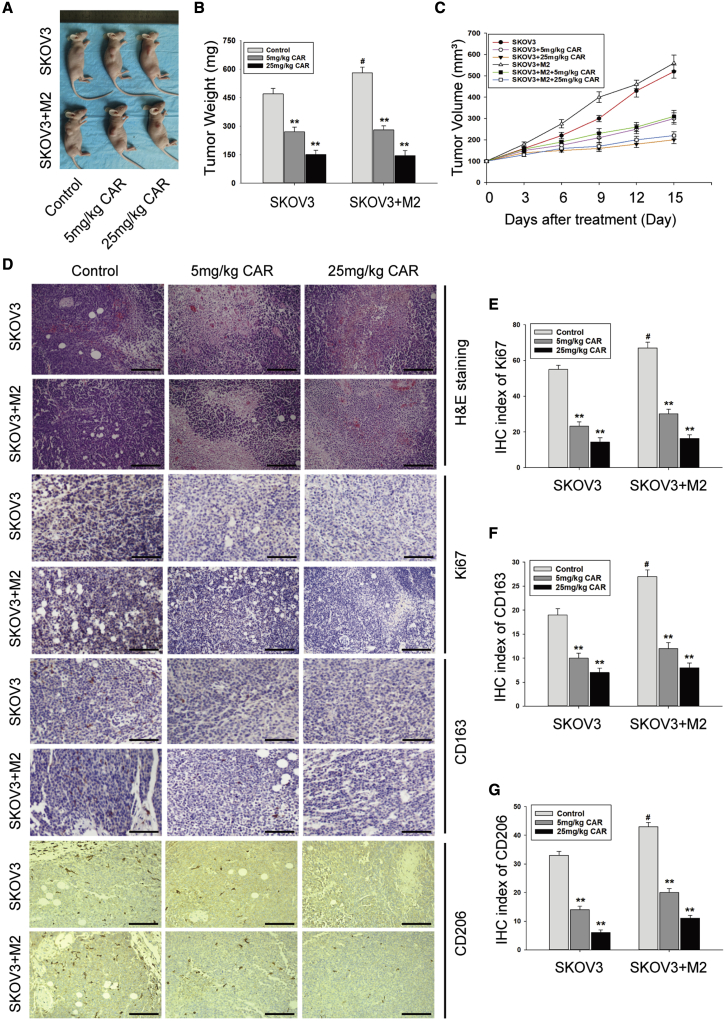
Then, we assessed the anticancer effect of cardamonin in vivo. SKOV3 cells alone or mixed with M2 macrophages were inoculated into mice. No mice died during cardamonin administration. Tumor growth displayed faster in the co-injected group than the SKOV3 group. Conversely, tumor growth was significantly inhibited in both groups by low- and high-doses of cardamonin (Figures 6A–6C). Additionally, a histological examination showed that, compared with the control group with larger solid tumor tissues and bigger cell nuclei, mice treated with cardamonin displayed a small island of tumor tissues with larger areas of tumor necrosis, which was characterized by cell shrinkage, fragmentation, and chromatin disappearance (Figure 6D). Moreover, the expression of Ki67 was inhibited by cardamonin (Figure 6E). Consistent with the experiments in vitro, cardamonin decreased the expression of CD163 and CD206 in both the SKOV3 and co-injected groups (Figures 6F and 6G).
Figure 6.
Cardamonin inhibited tumors growth and decreased M2 macrophages in vivo
BALB/c nude mice were injected subcutaneously with SKOV3 cells or a mixture of SKOV3 cells and M2 macrophages. Then mice were treated with or without indicated drugs for 15 days (n = 6 per group). (A) Representative images of sacrificed mice. (B) Tumor weight. (C) Change of tumor volume during drug administration. Tumor size was calculated according to the formula: [length × (width)2]/2. (D) Representative images of hematoxylin and eosin (H&E) staining and immunohistochemistry for tumor histology analysis (scale bar, 200 μm). (E–G) The immunohistochemistry (IHC) index of Ki67(E), CD163 (F), and CD206 (G) in tumor tissue. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus control group. #p < 0.05 versus control group of SKOV3 injection.
Cardamonin downregulated mTOR and STAT3 expression in TAMs
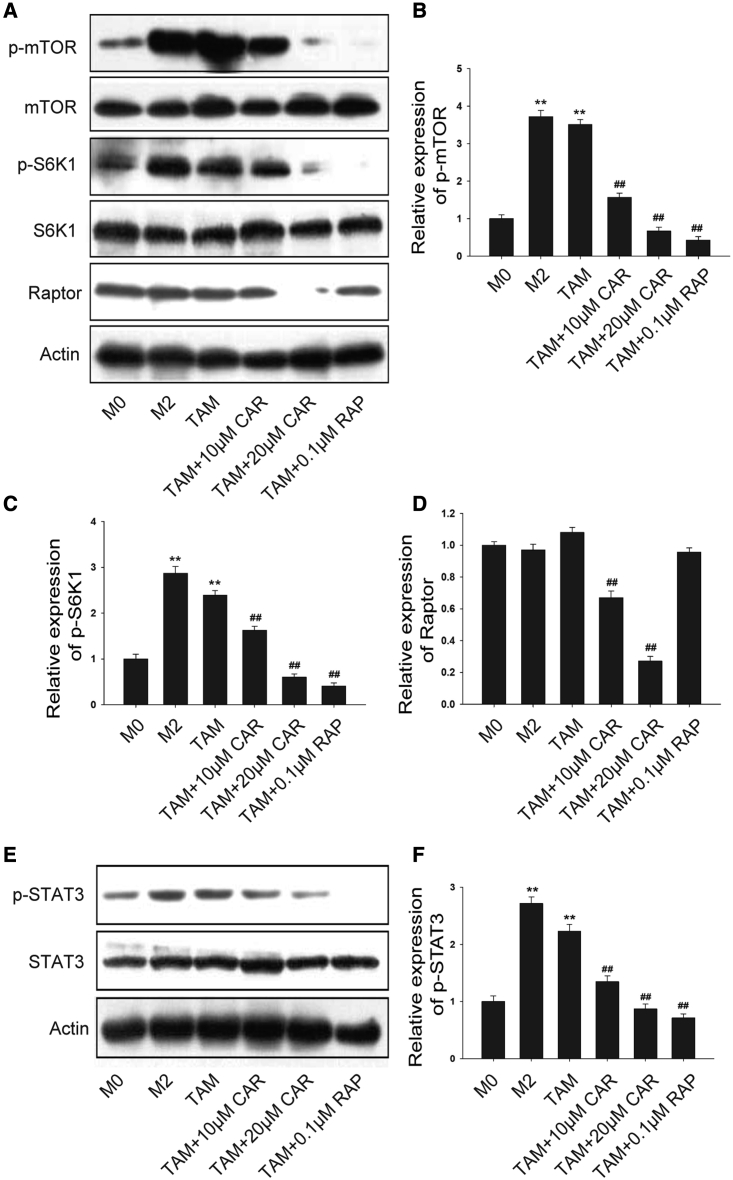
It has been described that mTOR is very important in terms of macrophage function. Here, we first examined the protein expression of mTOR in TAMs. Compared with the M0 macrophage, a higher level of p-mTOR and p-S6K1 (phosphorylation of p70 S6 kinase) was expressed in M2 and TAMs (Figure 7A), suggesting that mTOR was activated in TAMs. Cardamonin and rapamycin downregulated the protein level of phosphorylation but did not affect the total expression of mTOR and S6K1 (Figures 7B and 7C). Interestingly, the expression level of regulatory-associated protein of mTOR (Raptor), a specific protein of mTOR complex 1 (mTORC1), was significantly inhibited by cardamonin but not by rapamycin (Figure 7D). In addition, we detected a significant increased expression of STAT3 in TAMs (Figure 7E). Similarly, cardamonin decreased the protein level of phosphorylation of STAT3, while it had no influence on the total expression of STAT3 (Figure 7F), indicating that the inhibition effects of cardamonin on functional TAMs may be associated with STAT3 and mTOR activation.
Figure 7.
Cardamonin suppressed mTOR and STAT3 expression in TAMs
THP-1 monocytes were induced and then treated with or without cardamonin (10 or 20 μM) and rapamycin (0.1 μM) for 24 h. (A–D) Protein level of mTOR, p-mTOR, S6K1, p-S6K1, and Raptor was determined by western blotting. Relative density ratios of p-mTOR, p-S6K1, and Raptor were normalized to actin. (E and F) Protein level of p-STAT3 and STAT3 was determined. Relative density ratios of p-STAT3 protein were normalized to actin. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus M0. ##p < 0.01 versus TAM.
mTOR mediated the inhibition effect of cardamonin on TAMs
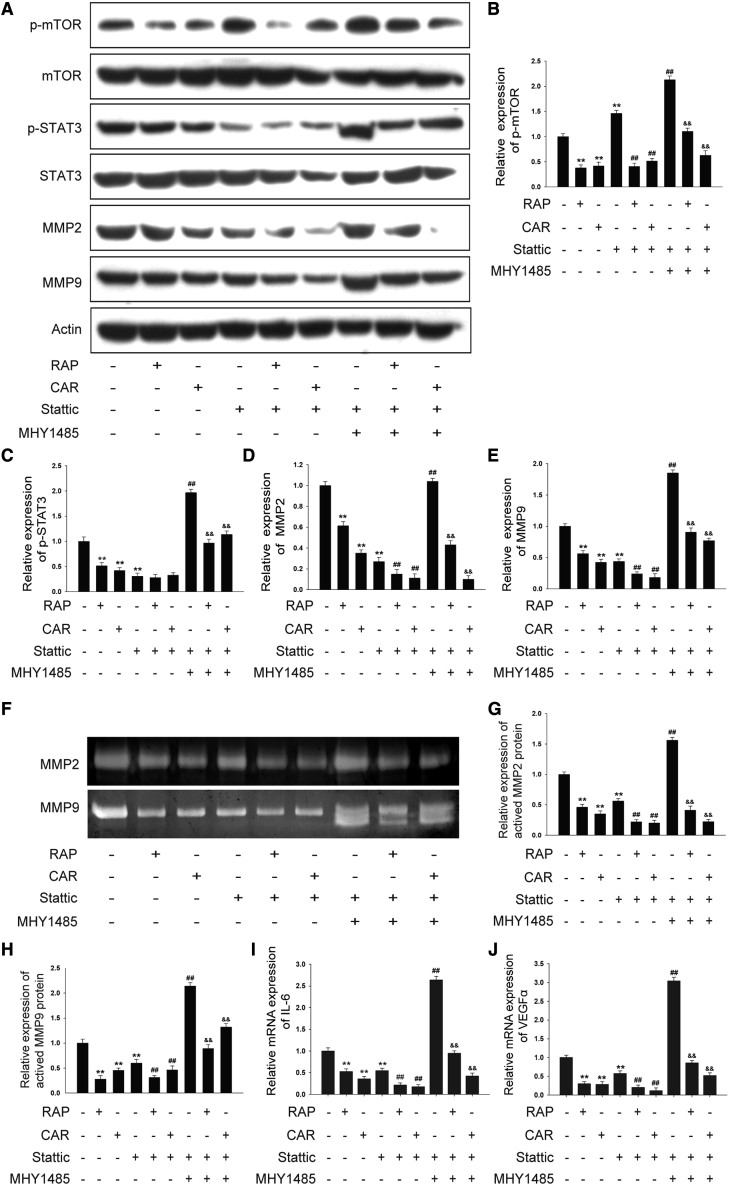
To investigate the mechanisms responsible for cardamonin-mediated TAMs’ functional inhibition, we treated TAMs with cardamonin in the presence of Stattic, a specific inhibitor of STAT3 (Figure 8A). As expected, cardamonin significantly inhibited both mTOR and STAT3 expression, while Stattic suppressed the phosphorylation of STAT3 and did not lower p-mTOR (Figures 8B and 8C). Both cardamonin and Stattic reduced protein levels of MMP2 and MMP9. Interestingly, levels of MMP2 and MMP9 were furtherly reduced by cardamonin in cells pretreated with Stattic, accompanied by a decreased expression of p-mTOR but not of p-STAT3. To further verify the relationship between mTOR and STAT3, we added MHY1485, a specific activator of mTOR, to cells pretreated with Stattic. Notably, MHY1485 not only upregulated the activity of mTOR but also increased the expression of p-STAT3, MMP2, and MMP9. Moreover, the promotion effects of MHY1485 were reversed by the subsequent treatment with cardamonin (Figures 8D and 8E). Similar results were observed in a gelatin zymography assay (Figures 8F–8H). Consistent with MMP2 and MMP9, the expression of IL-6 and VEGFα was inhibited by Stattic, and it was furtherly reduced following cardamonin treatment. Additionally, the expression of IL-6 and VEGFα inhibited by Stattic was increased by MHY1485, while it was downregulated by following with cardamonin administration (Figures 8I and 8J). These findings indicate that mTOR regulates STAT3 activity and mediates the effect of cardamonin on TAMs’ functional inhibition.
Figure 8.
mTOR regulated STAT3 expression and mediated the effect of cardamonin on TAMs
THP-1-induced TAMs were cultured with or without cardamonin (20 μM), rapamycin (0.1 μM), and Stattic (20 μM). In the combined group, cells were induced to TAMs and pretreated with Stattic (20 μM) for 2 h in the presence of MHY1485 (10 μM) or not, and then cells were cultured with cardamonin (20 μM) or rapamycin (0.1 μM) for 24 h. (A) Protein level of mTOR, p-mTOR, p-STAT3, STAT3, MMP2, and MMP9 was determined by western Blotting. (B–E) Relative density ratios of p-mTOR, p-STAT3, MMP2, and MMP9 were normalized to actin. (F) Enzyme activity of MMP2 and MMP9 was determined by gelatin zymography assay. (G and H) The relative enzyme activity of MMP2 and MMP9 is shown as the bar graph, with the TAM group set to 1. (I and J) mRNA expressions of IL-6 and VEGFα were measured by real-time PCR. Relative mRNA expression was normalized to actin. Data are presented as the mean ± SD (n = 3). ∗∗p < 0.01 versus Control. ##p < 0.01 versus Stattic. &&p < 0.01 versus Stattic + MHY1485.
Discussion
Numerous studies have confirmed that TAMs are mostly biased toward M2 polarization and that they facilitate tumor progression.3,5 Some herbs and chemicals play a pivotal role in TAM polarization and contribute to tumor inhibition.24, 25, 26 Cardamonin shows excellent antitumor effects in several cancer cells.19,27,28 Recent studies have demonstrated a potential role of cardamonin in tumor immunity.23 However, whether the antitumor effect of cardamonin associated with TAM regulation is not of concern. In the present study, our results showed that M2 polarization of TAMs was decreased by cardamonin via mTOR and STAT3 inhibition, which, in turn, suppressed the proliferation and migration of ovarian cancer cells (Figure S5).
It is reported that functions of TAMs are closely related to their phenotypes.4, 5, 6 In the present study, SKOV3-CM-induced macrophages expressed high levels of M2 markers and promoted the proliferation and invasion of ovarian cancer cells, which indicated that SKOV3 CM successfully induced THP-1 cells to functional TAMs. Moreover, cardamonin inhibited TAMs’ M2 polarization for the decreased M2 markers. Moreover, cardamonin dramatically suppressed TAM-derived pro-tumorigenic factors (including IL-6, VEGFα, MMP2, and MMP9). It is suggested that functional TAMs secrete numbers of cytokines that promote tumor growth and metastasis.5,6 These results indicate that cardamonin plays an important role in the polarization and functional regulation of TAMs.
As a super central regulator, mTOR has become an attractive target for tumor therapy.29 Recent studies have shown that mTOR is involved in macrophage activation and polarization.7,30 Overactivation of mTOR was observed on TAMs, and it skewed macrophages toward M2 polarization and promoted the aggressive progression of cancer cells.18 Based on the important role of mTOR in macrophage regulation, we hypothesized that cardamonin regulates TAMs via mTOR inhibition. As expected, cardamonin suppressed the phosphorylation of mTOR in TAMs. The expression level of Raptor, a specific protein of mTORC1, was significantly inhibited by cardamonin. Our previous studies have confirmed that cardamonin decreased mTOR activity and inhibited the expression of Raptor, indicating that cardamonin may be a specific mTORC1 inhibitor.31 Coincidently, previous studies have confirmed that mTOR inhibition impinged on macrophage polarization. For example, mTOR inhibition increased M1 polarization.32,33 Conversely, other studies supported that mTOR polarized macrophages to the M1 subtype during infection and inflammation.34 Interestingly, it has been shown that another mTOR complex, mTORC2, controls the immune suppressive phenotype of TAMs.35 Therefore, the role of mTOR remains controversial in macrophage regulation, especially in different tissues and different tissue states. Nevertheless, our finding indicates that mTOR plays an important role in macrophage polarization and that it mediated the inhibition effects of cardamonin on TAMs.
STAT3 is another important modulator in the alternative activation of macrophages.36 Our results demonstrated that STAT3 was upregulated in TAMs and was inhibited by cardamonin, indicating that the inhibition effects of cardamonin on TAMs may be associated with both STAT3 and mTOR. However, the relationship between mTOR and STAT3 in TAMs is unclear. In the present study, we found that mTOR regulates STAT3 activation. The suppressed expression of p-STAT3 by Stattic was reversed by mTOR activator MHY1485. Coincidently, previous studies have shown that mTOR acts as a major regulator in the STAT3 pathway.37,38 mTOR regulates STAT3 activation via phosphorylation of STAT3 in macrophages.39,40 Interestingly, the inhibitory effect of cardamonin on p-STAT3 was blocked in cells pretreated with Stattic, while the expressions of TAM-secreted factors were furtherly reduced in the same condition. Discordant suppression on STAT3 and TAM-secreted factors by cardamonin may be explained by how not only STAT3, but also another mTOR-dependent pathway, such as nuclear factor κB (NF-κB), regulates the secretion of pro-tumor factors in TAMs.33,41
Previous studies have shown that the antitumor effect of cardamonin was related to mTOR inhibition by structure-activity relationship (SAR) analysis,42 while its anti-inflammatory effect was related to NF-κB.43 However, the specific mechanism of the cardamonin effect also needs to be further verified by SAR analysis.
In summary, our results indicate that cardamonin inhibited M2 polarization and suppressed pro-tumorigenic factor secretion in TAMs, which, accordingly, impeded the pro-tumor function of TAMs on ovarian cancer cells. Furthermore, the underlying mechanism of cardamonin on TAMs may be closely related to inhibition of mTOR and STAT3. These findings reveal that cardamonin is a potential therapeutic agent for ovarian cancer.
Materials and methods
Chemicals and reagents
Cardamonin (no. 110763, purity ≥98%) was purchased from China Food and Drug Research Institute (Beijing, China). Rapamycin (RAP), phorbol 12-myristate 13-acetate (PMA), lipopolysaccharide (LPS), IL-4, EdU, and CCK-8 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Stattic and MHY1485 were purchased from MedChemExpress (Princeton, NJ, USA).
Cell culture and induction
Human THP-1 monocytes, SKOV3 cells, and A2780 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). THP-1 cells and A2780 cells were maintained in RPMI-1640, and SKOV3 cells were cultured with McCoy’s 5A medium. These media are all from Hyclone (Logan, UT, USA). The cell lines were cultured in medium supplemented with 10% FBS (Gibco, Rockville, MD, USA), 100 μg/mL streptomycin, and 100 U/mL penicillin (Gibco, Rockville, MD, USA) at 37°C in a humidified atmosphere with 5% CO2. THP-1 cells were seeded at 1 × 105 cells per well in 6-well plates and incubated with PMA at 50 ng/mL for 12 h to induce M0 macrophages. Then, the M0 macrophages were induced to TAMs with 50% (v/v) SKOV3 CM for another 48 h. In parallel, M0 macrophages were directed to the M1 phenotype with LPS at a final concentration of 20 ng/mL or were polarized to M2 with 20 ng/mL IL-4.
CM collection
SKOV3 cells grow to 80%, then the cells were incubated with FBS-free medium for 24 h, and finally, the medium was collected for TAM induction. Similarly, THP-1 cells were induced to different phenotypes of macrophages as described above, followed by incubating with or without cardamonin (10 or 20 μM) or RAP (0.1 μM) for 24 h, respectively, then cells were washed with PBS twice and cultured in FBS-free medium for 24 h. Finally, the medium was gathered and stored at −20°C, centrifuging for 10 min before used.
Cell viability assay
Cell viability assays were conducted with the CCK-8 reagent in accordance with the manufacturer’s protocol. Cells were seeded in 96-well plates with 5,000 cells per well. After cell induction or drug treatment, 10 μL CCK-8 solution was added to each well and incubated with the cells at 37°C for 2 h. The optical density (OD) values were assessed by scanning with a microplate reader (Model 1680, Bio-Rad, Hercules, CA, USA) at 450 nm. Additionally, cell proliferation was observed by EdU test. EdU detection reagent was added to each well, and images for EdU-positive cells were acquired with a fluorescence inverted microscope (Olympus, Tokyo, Japan). Ten randomly chosen fields per sample were counted to determine the percentage of labeled nuclei. The ratio of EdU-positive cells in each field was calculated by a formula: green fluorescence cells (positive cells)/blue fluorescence cells (total cells) × 100%.
Scratch test
SKOV3 cells and A2780 cells were seeded in 6-well plates and grow to a confluence of 90%. Then, the cell layer was scratched by a sterile 200-μL pipette tip. Following twice washing with PBS, the cells were incubated with the indicated CM. Images were captured under an inverted microscope (Olympus, Tokyo, Japan) at 0 and 24 h. Wound area was measured to assess cell migration. Quantitative analysis of the wound area was measured using the free image-processing software ImageJ v.1.47.
Transwell assay
Transwell cell culture chambers containing Matrigel or not were used for invasive and migration evaluation, respectively. SKOV3 cells or A2780 cells (1 × 105 cells/well) were seeded in upper chambers with serum-free medium, and the indicated CM was loaded to the lower chambers. After 24 h co-incubation, the migrated cells were fixed with methanol at room temperature for 30 min and stained with 0.1% crystal violet for 20 min. The remaining cells were gently wiped with a cotton swab. Finally, cells in the lower layer of the membrane were counted in at least three randomized fields using a microscope (Olympus, Tokyo, Japan).
Flow cytometry
After induction and drug treatment, cells were scraped and collected for flow cytometry. M0 cells were used as normal control. A total of 1 × 106 cells were counted in each flow tube, then washed twice with PBS. Fluorescein isothiocyanate (FITC) anti-Human CD163 (BD Biosciences, #563697), PE anti-Human CD86 (BD Biosciences, #305405), and isotype control antibodies (BD Biosciences) were then added and incubated for 30 min in the dark at 4°C. Upon washing twice, cells were resuspended in 500 μL PBS and finally examined with a FacsCantoII flow cytometer (BD Biosciences). The results were analyzed using BD FACSDiva 6.1 software (BD Biosciences).
Real-time PCR
Total RNA was extracted by using Trizol (Takara). One μg RNA was converted into cDNA by use of Primer Buffer (5×), Primase, Oligonucleotide DT Primer, and Reverse Transcriptase (Takara). Real-time PCR was performed using a SYBR GREEN PCR mixture, and gene expression was standardized for actin. According to the standard curve drawn by the gradient dilution cDNA, the relative content of the target gene mRNA expression was calculated. The primers were designed and synthesized by Shanghai Shenggong BioengineeringZ (Shanghai, China). The primer sequences were as follows: human IL-6 forward, 5′-ACTCACCTCTTCAGAACGAATTG-3′ and reverse, 5′-CCATCTTTGGAAGGTTCAGGTTG-3′; human IL-10 forward, 5′-TACGGCGCTGTCATCGATTT-3′ and reverse, 5′-TAGAGTCGCCACCCTGATGT-3′; human VEGFα forward, 5′-GAAGTGGTGAAGTTCATGGATGTC-3′ and reverse, 5′-CGATCGTTCTGTATCAGTCTTTCC-3′; human actin forward, 5′-ACTACCTCATGAAGATC-3′ and reverse, 5′-GATCCACATCTGCTGGAA-3′.
ELISA assay
After induction and drug treatment, cell supernatants were collected and clarified by centrifugation. Concentrations of IL-6, IL-10, and VEGFα were measured by a commercial ELISA kit from R&D Systems following the manufacturer’s instructions. Results were normalized to cell counts.
Gel zymography
Expression of activated MMP2 and MMP9 protein was analyzed using gelatin zymography. Culture medium was collected and concentrated by ultrafiltration. Then, samples were separated by 10% SDS-PAGE in the presence of gelatin (1 mg/mL, Sigma-Aldrich). After renaturing in a 2.5% Triton X-100 solution for 1 h, gels were immersed in a buffer that contained 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM CaCl2, and 5 μM ZnCl2 for 48 h. Then, gels were stained with 0.5% Coomassie brilliant blue R-250 for 2 h and followed by destaining with 50% methanol and 5% acetic acid. Finally, proteolytic bands were visualized for densitometry analysis.
Western blotting
After drug treatment, cells were collected and lysed by radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) containing 1 mM phenylmethylsulfonyl fluoride (PMSF; Beyotime Biotechnology, Shanghai, China) for 30 min. After centrifugation at 12,000 × g for 20 min at 4°C, the supernatant was collected, and the total protein concentration was determined by the bicinchoninic acid protein assay kit (Applygen Technologies). Protein samples (30 μg/lane) were separated by 8%–10% SDS-PAGE and transferred to PVDF membranes (Thermo Fisher Scientific). After blocking with 5% bovine serum albumin at room temperature for 1 h, the membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were purchased from Cell Signaling and used at a 1:1,000 dilution: phospho-mTOR (Ser244, #5536P); mTOR (#2983); phospho-S6K1 (Thr389, #9205S); S6K1 (#9202S); Raptor (24C12, #2280S); phospho-STAT3 (Tyr705, #9131); STAT3 (#12640); MMP2 (#87809); MMP9 (#13667); Arg-1 (#93668); mannose receptor antibody (CD206, #24595); and actin (#4970). Subsequently, the membranes were probed with secondary antibodies (anti-rabbit immunoglobulin G, horseradish peroxidase [HRP]-linked antibody, CST, #7074S) at a 1:2,500 dilution for 1 h at room temperature. Finally, the bound antibodies were detected by the HRP-enhanced chemiluminescence (ECL) reagents (Thermo Fisher Scientific) and developed by autoradiography (Kodak Film, Tokyo, Japan) and quantified by BioImaging Systems.
Xenograft tumor models
Pathogen-free female BALB/c nude mice (3–5 weeks, 10–15 g body weight) were purchased from National Rodent Laboratory Animal Resource (Shanghai, China) and kept in the Experimental Animal Center of Fujian Medical University under a specific pathogen-free (SPF) condition. Mice were allowed free access to water and food with a 12 h light-dark cycle. All procedures involving animals were conducted in accordance with national and institutional guidelines. Animal experiments were approved by the institutional Ethics Committee of Fujian Maternity and Child Health Hospital (2021KD017). Animal experiments were divided into two groups including SKOV3 cells (1 × 107/mouse) subcutaneous injection and SKOV3 cells (7.5 × 106/mouse) combined with THP-1-induced M2 macrophages (2.5×106/mouse) subcutaneous injection. Mice in each set were randomly divided into three groups (n = 6 per group) and intraperitoneally treated with physiological saline (control group), 5 mg/kg cardamonin (5 mg/kg cardamonin group), or 25 mg/kg cardamonin (25 mg/kg cardamonin group), respectively, once a day. Tumor size was calculated according to the formula: [length × (width)2]/2. After 15 days of drug administration, all mice were sacrificed, and tumor tissues were isolated for further analysis.
Immunohistochemistry and histology analysis
The tumor tissues were formalin fixed and paraffin embedded and then cut into 3-μm sections. After deparaffinating and rehydrating, the sections were immersed with 10 mM sodium citrate buffer in a microwave for antigen retrieval. Then, the samples were treated with 0.3% H2O2 at room temperature for 15 min and washed with PBS 3 times, followed by incubating with primary antibodies including Ki67 (1:100, Affinity Biosciences, #AF0198), CD163 (1:200, Abcam, #ab182422), and CD206 (1:200, CST, #24595) for 15 min. Subsequently, the samples were incubated with second antibody and then with diaminobenzidine (DAB)-substrate chromogen. The brown staining represented positive antigens, and five randomly selected fields were evaluated. The area and OD of Ki67 and CD163 in each field were assessed by a computer-aided automatic image analysis system (Qiu Wei, Shanghai, China). The immunohistochemistry (IHC) index was calculated as the average integral OD: [(positive area × OD)/total area]. Sections were also stained with hematoxylin and eosin (H&E) and examined under a microscope (Olympus, Tokyo, Japan) for histological analysis.
Statistical analysis
Statistical analyses were conducted by Prism software 6 (GraphPad Software). All data were expressed as mean ± SD. Differences between groups were evaluated by the one-way analysis of variance (ANOVA) and Dunnett’s test. p <0.05 was considered statistically significant.
Data availability
All data included in this study are available upon request by contact with the corresponding author.
Acknowledgments
This research was supported by the Natural Science Foundation of Fujian Province, China (grant nos. 2021J01416 and 2020J01620) and the Fujian Provincial Maternity and Children Hospital Science and Technology Innovation Fund (CN) (grant no. YCXZ 18-16). We would like to thank Xiao-dan Mao (Laboratory of Gynecologic Oncology of Fujian Maternity and Child Health Hospital), Li-Ping Zhu (Medical Research Center of Fujian Maternity and Child Health Hospital), and Jian-Fang Zhu (Department of pathology of Fujian Maternity and Child Health Hospital) for their contributions and technical work in this study.
Author contributions
D.S. and H.C. conceived and designed the research; H.C. and S.H. performed the main experiments; P.N., Y.Z., and J.Z. assisted with the experiments; L.J. and D.L. assisted in analyzing the data; H.C. and S.H. wrote the manuscript; D.S. and H.C. reviewed and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.06.009.
Supplemental information
References
- 1.Yuan X., Zhang J., Li D., Mao Y., Mo F., Du W., Ma X. Prognostic significance of tumor-associated macrophages in ovarian cancer: a meta-analysis. Gynecol. Oncol. 2017;147:181–187. doi: 10.1016/j.ygyno.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Yunna C., Mengru H., Lei W., Weidong C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020;877:173090. doi: 10.1016/j.ejphar.2020.173090. [DOI] [PubMed] [Google Scholar]
- 3.Yin M., Shen J., Yu S., Fei J., Zhu X., Zhao J., Zhai L., Sadhukhan A., Zhou J. Tumor-associated macrophages (TAMs): a critical activator in ovarian cancer metastasis. Onco. Targets Ther. 2019;12:8687–8699. doi: 10.2147/ott.s216355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagemann T., Wilson J., Burke F., Kulbe H., Li N.F., Plüddemann A., Charles K., Gordon S., Balkwill F.R. Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J. Immunol. 2006;176:5023–5032. doi: 10.4049/jimmunol.176.8.5023. [DOI] [PubMed] [Google Scholar]
- 5.Yousefzadeh Y., Hallaj S., Baghi Moornani M., Asghary A., Azizi G., Hojjat-Farsangi M., Ghalamfarsa G., Jadidi-Niaragh F. Tumor associated macrophages in the molecular pathogenesis of ovarian cancer. Int. Immunopharmacol. 2020;84:106471. doi: 10.1016/j.intimp.2020.106471. [DOI] [PubMed] [Google Scholar]
- 6.Ke X., Zhang S., Wu M., Lou J., Zhang J., Xu T., Huang L., Huang P., Wang F., Pan S. Tumor-associated macrophages promote invasion via Toll-like receptors signaling in patients with ovarian cancer. Int. Immunopharmacol. 2016;40:184–195. doi: 10.1016/j.intimp.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Byles V., Covarrubias A.J., Ben-Sahra I., Lamming D.W., Sabatini D.M., Manning B.D., Horng T. The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed I., Ismail N. M1 and M2 macrophages polarization via mTORC1 influences innate immunity and outcome of ehrlichia infection. J. Cell Immunol. 2020;2:108–115. doi: 10.33696/immunology.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H., Han Y., Rodriguez Sillke Y., Deng H., Siddiqui S., Treese C., Schmidt F., Friedrich M., Keye J., Wan J., et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019;11:e10698. doi: 10.15252/emmm.201910698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Festuccia W.T. Regulation of adipocyte and macrophage functions by mTORC1 and 2 in metabolic diseases. Mol. Nutr. Food Res. 2020:e1900768. doi: 10.1002/mnfr.201900768. [DOI] [PubMed] [Google Scholar]
- 12.Yan J., Wang R., Horng T. mTOR is key to T cell transdifferentiation. Cell Metab. 2019;29:241–242. doi: 10.1016/j.cmet.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocher C., Singla D.K. SMAD-PI3K-Akt-mTOR pathway mediates BMP-7 polarization of monocytes into M2 macrophages. PLoS One. 2013;8:e84009. doi: 10.1371/journal.pone.0084009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A., Das S., Mandal A., Verma S., Abhishek K., Kumar A., Kumar V., Ghosh A.K., Das P. Leishmania infection activates host mTOR for its survival by M2 macrophage polarization. Parasite Immunol. 2018;40:e12586. doi: 10.1111/pim.12586. [DOI] [PubMed] [Google Scholar]
- 15.Tan X., Zhang Z., Yao H., Shen L. Tim-4 promotes the growth of colorectal cancer by activating angiogenesis and recruiting tumor-associated macrophages via the PI3K/AKT/mTOR signaling pathway. Cancer Lett. 2018;436:119–128. doi: 10.1016/j.canlet.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Liu Y., Guo J., He H., Mi X., Chen C., Xie J., Wang S., Wu P., Cao F., et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis. 2018;7:97. doi: 10.1038/s41389-018-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisi L., Ciotti G.M.P., Chiavari M., Pizzoferrato M., Mangiola A., Kalinin S., Feinstein D.L., Navarra P. Phospho-mTOR expression in human glioblastoma microglia-macrophage cells. Neurochem. Int. 2019;129:104485. doi: 10.1016/j.neuint.2019.104485. [DOI] [PubMed] [Google Scholar]
- 18.Shan M., Qin J., Jin F., Han X., Guan H., Li X., Zhang J., Zhang H., Wang Y. Autophagy suppresses isoprenaline-induced M2 macrophage polarization via the ROS/ERK and mTOR signaling pathway. Free Radic. Biol. Med. 2017;110:432–443. doi: 10.1016/j.freeradbiomed.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Nawaz J., Rasul A., Shah M.A., Hussain G., Riaz A., Sarfraz I., Zafar S., Adnan M., Khan A.H., Selamoglu Z. Cardamonin: a new player to fight cancer via multiple cancer signaling pathways. Life Sci. 2020;250:117591. doi: 10.1016/j.lfs.2020.117591. [DOI] [PubMed] [Google Scholar]
- 20.Jin J., Qiu S., Wang P., Liang X., Huang F., Wu H., Zhang B., Zhang W., Tian X., Xu R., et al. Cardamonin inhibits breast cancer growth by repressing HIF-1α-dependent metabolic reprogramming. J. Exp. Clin. Cancer Res. 2019;38:377. doi: 10.1186/s13046-019-1351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Shi D., Niu P., Zhu Y., Zhou J. Anti-inflammatory effects of cardamonin in ovarian cancer cells are mediated via mTOR suppression. Planta Med. 2018;84:1183–1190. doi: 10.1055/a-0626-7426. [DOI] [PubMed] [Google Scholar]
- 22.Niu P.G., Zhang Y.X., Shi D.H., Liu Y., Chen Y.Y., Deng J. Cardamonin inhibits metastasis of lewis lung carcinoma cells by decreasing mTOR activity. PLoS One. 2015;10:e0127778. doi: 10.1371/journal.pone.0127778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao N.C., Shih Y.L., Ho M.T., Lu T.J., Lee C.H., Peng S.F., Leu S.J., Chung J.G. Cardamonin induces immune responses and enhances survival rate in WEHI-3 cell-generated mouse leukemia in vivo. Environ. Toxicol. 2020;35:457–467. doi: 10.1002/tox.22881. [DOI] [PubMed] [Google Scholar]
- 24.Tsuboki J., Fujiwara Y., Horlad H., Shiraishi D., Nohara T., Tayama S., Motohara T., Saito Y., Ikeda T., Takaishi K., et al. Onionin A inhibits ovarian cancer progression by suppressing cancer cell proliferation and the protumour function of macrophages. Sci. Rep. 2016;6:29588. doi: 10.1038/srep29588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q., Li Y., Miao C., Wang Y., Xu Y., Dong R., Zhang Z., Griffin B.B., Yuan C., Yan S., et al. Anti-angiogenesis effect of Neferine via regulating autophagy and polarization of tumor-associated macrophages in high-grade serous ovarian carcinoma. Cancer Lett. 2018;432:144–155. doi: 10.1016/j.canlet.2018.05.049. [DOI] [PubMed] [Google Scholar]
- 26.Park H.J., Chi G.Y., Choi Y.H., Park S.H. The root bark of <i>Morus alba</i> L. regulates tumor-associated macrophages by blocking recruitment and M2 polarization of macrophages. Phytother. Res. 2020;34:3333–3344. doi: 10.1002/ptr.6783. [DOI] [PubMed] [Google Scholar]
- 27.Wu N., Liu J., Zhao X., Yan Z., Jiang B., Wang L., Cao S., Shi D., Lin X. Cardamonin induces apoptosis by suppressing STAT3 signaling pathway in glioblastoma stem cells. Tumour Biol. 2015;36:9667–9676. doi: 10.1007/s13277-015-3673-y. [DOI] [PubMed] [Google Scholar]
- 28.Liao N.C., Shih Y.L., Chou J.S., Chen K.W., Chen Y.L., Lee M.H., Peng S.F., Leu S.J., Chung J.G. Cardamonin induces cell cycle arrest, apoptosis and alters apoptosis associated gene expression in WEHI-3 mouse leukemia cells. Am. J. Chin. Med. 2019;47:635–656. doi: 10.1142/s0192415x19500332. [DOI] [PubMed] [Google Scholar]
- 29.Yang D., Chen T., Zhan M., Xu S., Yin X., Liu Q., Chen W., Zhang Y., Liu D., Yan J., et al. Modulation of mTOR and epigenetic pathways as therapeutics in gallbladder cancer. Mol. Ther. Oncolytics. 2021;20:59–70. doi: 10.1016/j.omto.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Covarrubias A.J., Aksoylar H.I., Horng T. Control of macrophage metabolism and activation by mTOR and Akt signaling. Semin. Immunol. 2015;27:286–296. doi: 10.1016/j.smim.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi D., Zhu Y., Niu P., Zhou J., Chen H. Raptor mediates the antiproliferation of cardamonin by mTORC1 inhibition in SKOV3 cells. Onco. Targets Ther. 2018;11:757–767. doi: 10.2147/ott.s155065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercalli A., Calavita I., Dugnani E., Citro A., Cantarelli E., Nano R., Melzi R., Maffi P., Secchi A., Sordi V., Piemonti L. Rapamycin unbalances the polarization of human macrophages to M1. Immunology. 2013;140:179–190. doi: 10.1111/imm.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paschoal V.A., Amano M.T., Belchior T., Magdalon J., Chimin P., Andrade M.L., Ortiz-Silva M., Castro É., Yamashita A.S., Rosa Neto J.C., et al. mTORC1 inhibition with rapamycin exacerbates adipose tissue inflammation in obese mice and dissociates macrophage phenotype from function. Immunobiology. 2017;222:261–271. doi: 10.1016/j.imbio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Lin L.R., Gao Z.X., Lin Y., Zhu X.Z., Liu W., Liu D., Gao K., Tong M.L., Zhang H.L., Liu L.L., et al. Akt, mTOR and NF-κB pathway activation in Treponema pallidum stimulates M1 macrophages. Int. Immunopharmacol. 2018;59:181–186. doi: 10.1016/j.intimp.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 35.Shrivastava R., Asif M., Singh V., Dubey P., Ahmad Malik S., Lone M.U.D., Tewari B.N., Baghel K.S., Pal S., Nagar G.K., et al. M2 polarization of macrophages by Oncostatin M in hypoxic tumor microenvironment is mediated by mTORC2 and promotes tumor growth and metastasis. Cytokine. 2019;118:130–143. doi: 10.1016/j.cyto.2018.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Mu X., Shi W., Xu Y., Xu C., Zhao T., Geng B., Yang J., Pan J., Hu S., Zhang C., et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle. 2018;17:428–438. doi: 10.1080/15384101.2018.1444305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J.H., Lee N.K., Lim H.J., Ji S.T., Kim Y.J., Jang W.B., Kim D.Y., Kang S., Yun J., Ha J.S., et al. Pharmacological inhibition of mTOR attenuates replicative cell senescence and improves cellular function via regulating the STAT3-PIM1 axis in human cardiac progenitor cells. Exp. Mol. Med. 2020;52:615–628. doi: 10.1038/s12276-020-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue J., Ge X., Zhao W., Xue L., Dai C., Lin F., Peng W. PIPKIγ regulates CCL2 expression in colorectal cancer by activating AKT-STAT3 signaling. J. Immunol. Res. 2019;2019 doi: 10.1155/2019/3690561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ai D., Jiang H., Westerterp M., Murphy A.J., Wang M., Ganda A., Abramowicz S., Welch C., Almazan F., Zhu Y., et al. Disruption of mammalian target of rapamycin complex 1 in macrophages decreases chemokine gene expression and atherosclerosis. Circ. Res. 2014;114:1576–1584. doi: 10.1161/circresaha.114.302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao W., Wang Y., Fu Y., Jia X., Li J., Vangan N., Bao L., Hao H., Wang Z. mTORC1 regulates flagellin-induced inflammatory response in macrophages. PLoS One. 2015;10:e0125910. doi: 10.1371/journal.pone.0125910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumas A.A., Pomella N., Rosser G., Guglielmi L., Vinel C., Millner T.O., Rees J., Aley N., Sheer D., andWei J., et al. Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment. EMBO J. 2020;39:e103790. doi: 10.15252/embj.2019103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Break M.K.B., Hossan M.S., Khoo Y., Qazzaz M.E., Al-Hayali M.Z.K., Chow S.C., Wiart C., Bradshaw T.D., Collins H., Khoo T.J. Discovery of a highly active anticancer analogue of cardamonin that acts as an inducer of caspase-dependent apoptosis and modulator of the mTOR pathway. Fitoterapia. 2018;125:161–173. doi: 10.1016/j.fitote.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Ur Rashid H., Xu Y., Ahmad N., Muhammad Y., Wang L. Promising anti-inflammatory effects of chalcones via inhibition of cyclooxygenase, prostaglandin E2, inducible NO synthase and nuclear factor κb activities. Bioorg. Chem. 2019;87:335–365. doi: 10.1016/j.bioorg.2019.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.