Abstract
mRNA is like Hermes, delivering the genetic code to cellular construction sites, so it has long been of interest, but only to a small group of scientists, and only demonstrating its remarkable efficacy in coronavirus disease 2019 (COVID-19) vaccines allowed it to go out into the open. Therefore, now is the right timing to delve into the stepping stones that underpin this success and pay tribute to the underlying scientists. From this perspective, advances in mRNA engineering have proven crucial to the rapidly growing role of this molecule in healthcare. Development of consecutive generations of cap analogs, including anti-reverse cap analogs (ARCAs), has significantly boosted translation efficacy and maintained an enthusiasm for mRNA research. Nucleotide modification to protect mRNA molecules from the host’s immune system, followed by finding appropriate purification and packaging methods, were other links in the chain enabling medical breakthroughs. Currently, vaccines are the central area of mRNA research, but it will reach far beyond COVID-19. Supplementation of missing or abnormal proteins is another large field of mRNA research. Ex vivo cell engineering and genome editing have been expanding recently. Thus, it is time to recognize mRNA pioneers while building upon their legacy.
Keywords: MT: Oligonucleotides: Therapies and Applications, mRNA, SARS-CoV-2, COVID-19, DNA, oligonucleotide, nucleotide, stem cells, vaccine, protein supplementation, nucleoside
Graphical abstract
mRNA science culminated in widespread application of COVID-19 vaccines. At the core of this success is more than half a century of research. Therefore, it is of utmost importance to pay tribute to the pioneers who made early engineering efforts by awarding them the Nobel Prize.
Introduction to nucleic acid research
DNA discovery
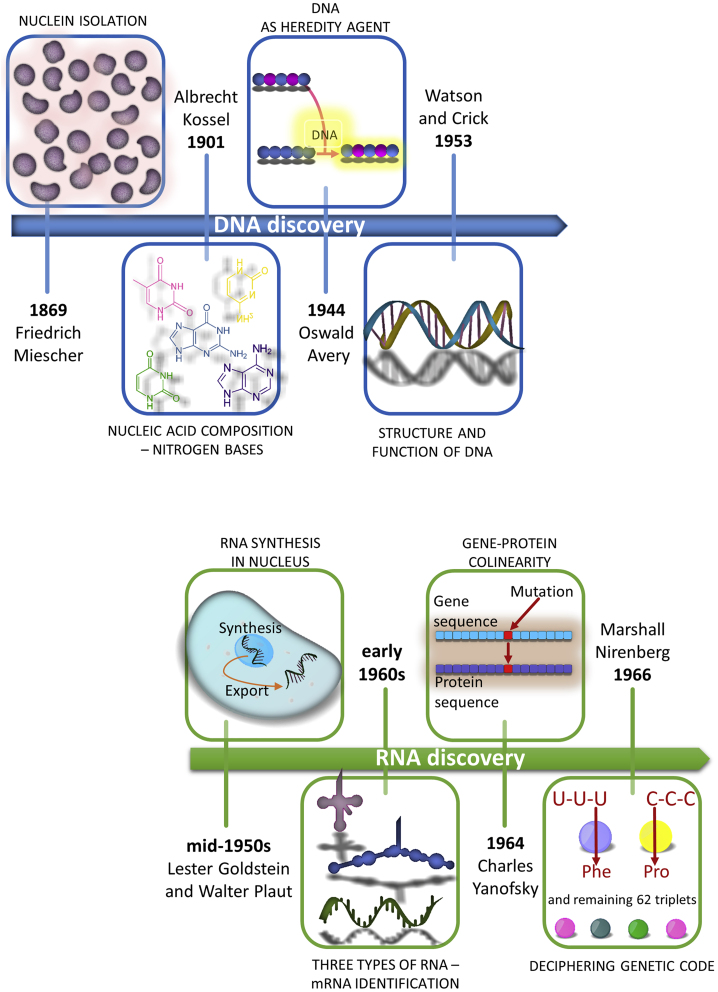
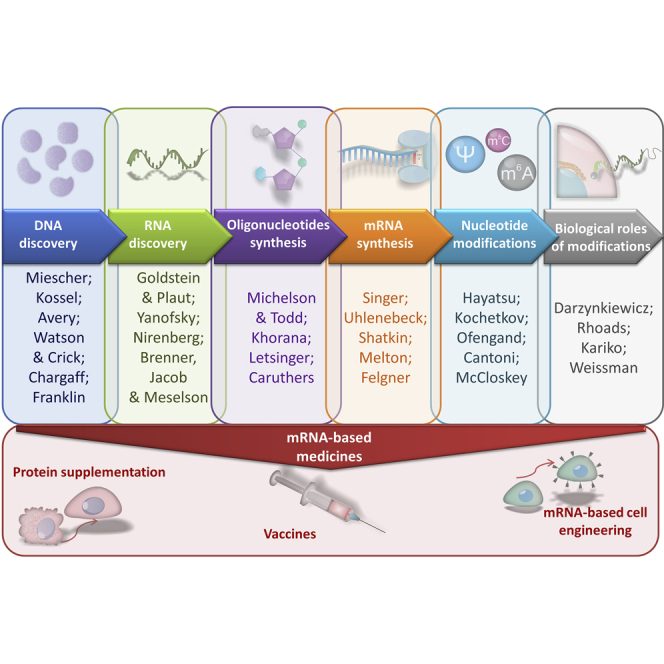
The previous two centuries were marked by elucidation of the structure and function of nucleic acids (Figure 1).1 Friedrich Miescher, a Swiss physician and biologist, isolated nuclein in 1869. The name nucleic acid was coined, and at the end of the 19th century, Albrecht Kossel, a German physician and biochemist, divided this into five bases: adenine, cytosine, guanine, thymine, and uracil. Another half century passed before Oswald Avery, an American bacteriologist, discovered that deoxyribonucleic acid (DNA) is responsible for heredity, thus laying the foundation for molecular biology genetics.2 Erwin Chargaff’s biochemistry discovery of the fixed ratio of certain bases, along with the crystallography studies of Rosalind Franklin, led to discovery of the structure and function of DNA in 1953 by Watson and Crick.3 However, the role of RNA and its link to DNA and proteins remained unclear.
Figure 1.
Key events in the history of the discovery of nucleic acids
mRNA discovery
Nuclear transfer in amebas by Goldstein and Plaut4 proved the location of RNA synthesis in the mid-1950s, followed by identification of ribosomal5 and transfer6 RNA. However, it was not until the early 1960s that an avalanche of work demonstrated a third type of RNA: soluble and short-lived messenger RNA (mRNA). Two publications distinguished mRNA from other kinds of cellular RNA.7,8 Others demonstrated similar-sized RNA molecules attached to ribosomes at that time.9 Steps toward solving the puzzle of code transfer from DNA to proteins was further facilitated by an article demonstrating the complementarity of DNA and transitory RNA.10 In 1964, the collinearity of gene structure and protein structure was reported by Charles Yanofsky, who later received the Lasker Award for his work.11 Ultimately, Nirenberg cracked the genetic code in 1966,12, 13, 14 and he was awarded the Nobel Prize 2 years later. In this way, the era of mRNA discovery unfolded.
Oligonucleotide synthesis
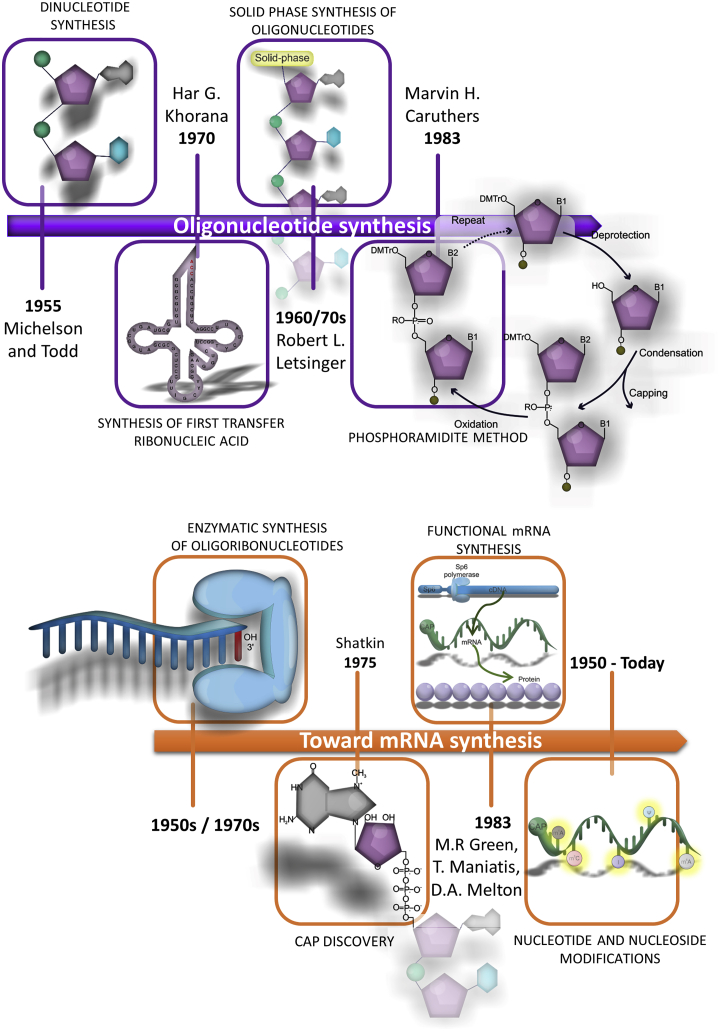
The discovery that nucleic acids are the source of genetic information rapidly fueled scientific investigation to produce synthetic code, starting with Michelson and Todd15 in 1955 with chemical synthesis of dinucleotide (Figure 2). Synthesis of longer chains of oligonucleotides was made feasible by a new phosphodiester method introduced by Rammler and Khorana et al.16 in the early 1960s, including protection of 2′- and 5′-hydroxyl groups in ribonucleosides and subsequent condensation. The focus was on oligodeoxyribonucleotides because of their stability and known role in storage of genetic information. Synthetic oligonucleotides were critical for cracking the genetic code and resulted in the Nobel Prize for Khorana and Nirenberg (see above). This method also proved robust enough to produce synthetic oligodeoxyribonucleotides, which could then be joined to double-stranded DNA to serve as a template for the first biologically meaningful molecule of RNA, alanine transfer ribonucleic acid, in 1970.17 However, the lack of phosphate protection contributed to the branching of oligonucleotide chains, which required a laborious purification task at each step, proving impractical for longer oligonucleotides. The drawbacks of Khorana’s approach have been addressed by the phosphotriester, solid-state approach of Letsinger introduced in the 1960s and 1970s.18, 19, 20 This method was sufficiently simple to quickly reproduce and served as a basis for the first oligonucleotide synthesizers. However, stepwise efficiency and a long coupling time made extending a chain of nucleotides beyond 20 bases complex. Fine tuning via replacement of the trouble-making polymer with inorganic support and a chloride group by amine was necessary to facilitate large-scale nucleic acid because phosphoramidites could be prepared in advance and then stored and easily activated before use.20,21
Figure 2.
Stages of scientific and technological development leading to the synthesis of functional mRNA
Toward mRNA production
Enzymatic synthesis of oligoribonucleotides using polynucleotide phosphorylase and RNAse A was achieved as early as the 1950s22 and further developed in the 1960s.23 Chemical synthesis of oligoribonucleotide from nucleoside 2′-O-benzyl ethers offered another option.24 Application of ribonuclease T1 in 1969 allowed enzymatic synthesis of oligoribonucleotides of a defined base sequence.25 In the same year, a method of DNA-dependent RNA polymerase isolation was described.26 In vitro production of short ribonucleotides based on a DNA template was demonstrated in 1973.27 Stepwise enzymatic oligoribonucleotide synthesis, including modified nucleotides, was described 2 years later.28 However, in this era before critical in vitro protein synthesis discoveries, none of these ribonucleotides were capable of protein production. In parallel, studies on mRNA structure were undertaken, which, together with the synthetic developments described above, enabled mRNA production. Single-stranded, adenine-rich RNAs were discovered during studies of reoviruses to correspond to the poly(A) tail of mRNA.29 Then it was determined that a 5′-terminal 7-methylguanosine cap is necessary for translation of eukaryotic mRNA,30 and the translation initiation region of eukaryotic mRNA was characterized.31 Finally, all of the components needed to produce functional mRNA were available, culminating in 1983 with the first in vitro production of functional mRNA using SP6 bacteriophage promoter fused to the human gene as a template and SP6 polymerase.32 Nearly a decade later, direct mRNA-based in vivo gene transfer to mouse muscle was reported.33 However, the developments have been hampered by the short duration of protein production and elicitation of the immune response.34, 35, 36, 37
Nucleotide and nucleoside modifications
Initial studies on nucleoside modifications were undertaken to search for therapeutic targets of alkylating agents.38,39 Subsequent research in 1964 revealed selective modification of the cytidine residue in RNA by semicarbazide.40 An avalanche of similar studies followed.41, 42, 43, 44, 45 However, the majority of these research efforts were directed toward rRNA.46 Thus, the artificial modifications ran in parallel with the investigation of naturally occurring RNA modifications, which were most frequently detected in tRNA (79), rRNA (28), mRNA (12), small nuclear RNA (snRNA; 11), and other small RNAs (3), until 1994.47 tRNA nucleotide modifications contribute to the speed of codon-dependent nucleotide polymerization, which is another source of biological variability and a mechanism for regulating cell function, providing an opportunity for mRNA codon optimization to accelerate protein production.48 Therefore, knowledge of artificial and naturally occurring nucleotide and nucleoside modifications accumulated very quickly.49
Transformative biological roles of nucleotide modifications
Nucleotide and nucleoside modifications were a driving force in overcoming challenges preventing the widespread utility of mRNA as a therapeutic agent. They also constitute the most important events in the history of mRNA development. The two most important aspects to be addressed were translational efficacy and immunogenicity. Pioneering work in these fields contributed the most to mRNA’s tremendous success, including highly effective coronavirus disease 2019 (COVID-19) vaccines. However, none of the discoveries in the field of mRNA have so far been recognized with the Nobel Prize.
The efficacy of mRNA translation is essential for a number of reasons. Higher efficacy translates to a lower load of mRNA and, thus, decreased immunogenicity. The flanking mRNA regions are the most critical regulators of translational efficacy. As early as 1981, Darzynkiewicz, who worked with Shatkin, showed that methyl esterification of m7G5′p reversibly blocks its activity as an analog of eukaryotic mRNA 5′-caps.50 Then Darzynkiewicz contributed a series of cap analogs over the next two decades.51, 52, 53, 54, 55 However, in 1995, it was demonstrated that up to half of the traditional m7GpppG cap is incorporated into mRNA in reverse orientation during in vitro transcription, which leads to loss of functionality.56 In 2001, Darzynkiewicz, in collaboration with Rhoads, published an antidote in the form of the anti-reverse cap analog (ARCA), a new analog that overcomes this limitation and is always incorporated in the proper orientation.57 ARCA was patented and licensed; it has been available through multiple manufacturers and used widely for the last two decades, showing outstanding results. Initially, it was thought to just double translation efficiency, but in a study of lipofection of dendritic cells, it was shown that ARCA increases translation efficiency 20-fold and acts synergistically with elongation of a poly(A) tail from 64–100 adenosines, which further increases 35-fold, so altogether, the production of reporter genes jumped 700-fold.58 ARCA has also been astoundingly more effective for stem cell modifications.59 ARCA served as an inspiration and basis for the next series of cap analogs designed over the next two decades.60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 Phosphorothioate cap analogs based on the ARCA concept were patented and exclusively acquired by BioNTech, the designer of the anti-COVID-19 vaccine, and used in their research.72 Recently, Cap 1 mRNA has been synthesized by TriLink with the co-transcriptional CleanCap analog with a higher efficiency of reaction than enzymatic reaction, which is particularly compelling when a great deal of mRNA needs to be produced in a very short time.73 Therefore, four decades of research on cap analogs led to mRNA molecules’ outstanding efficiency and stability, enabling their vast therapeutic use. It was also a source of motivation dearly needed to advance science. Considering the essential role of the cap in mRNA function, it seems highly justifiable to award a Nobel Prize for cap analog discoveries, and Darzynkiewicz, through his continuous contributions to cap analog design, including ARCA, over the last 40 years, is undoubtedly worthy of such consideration.
In addition to problems with translation efficacy, mRNA immunogenicity quickly dampened enthusiasm for swift translation of mRNA-based medicines to clinical practice. However, the subsequent extraordinary diligence of Karikó and Weissman provided essential insight into the fact that the majority of natural mRNA molecules include modified nucleotides, which may shield them from the cellular innate immune system.74 Indeed, Karikó et al.68 demonstrated that incorporation of pseudouridine into mRNA prevented an immediate immune response in the form of a systemic interferon-alpha (IFN-alpha) spike after intravenous administration. Pseudouridine also enhances translation by less pronounced activation of RNA-dependent protein kinase (PKR), which then phosphorylates translation initiation factor 2-alpha (eIF-2α) and inhibits translation of uridine-containing transcripts.75 Nucleoside modifications in RNA also limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L, another cellular sensing system.76
Further lowering of the cellular immune response has been achieved by improving mRNA purification using high-performance liquid chromatography (HPLC).77 Such prepared mRNA proved highly effective with submicrogram quantities in vivo for erythropoietin supplementation in mice and monkeys.78 In this way, mRNA proved to be fulfilling the promise of gene therapy.79 Therefore, Karikó also appears to be a certain candidate for the Nobel Prize.
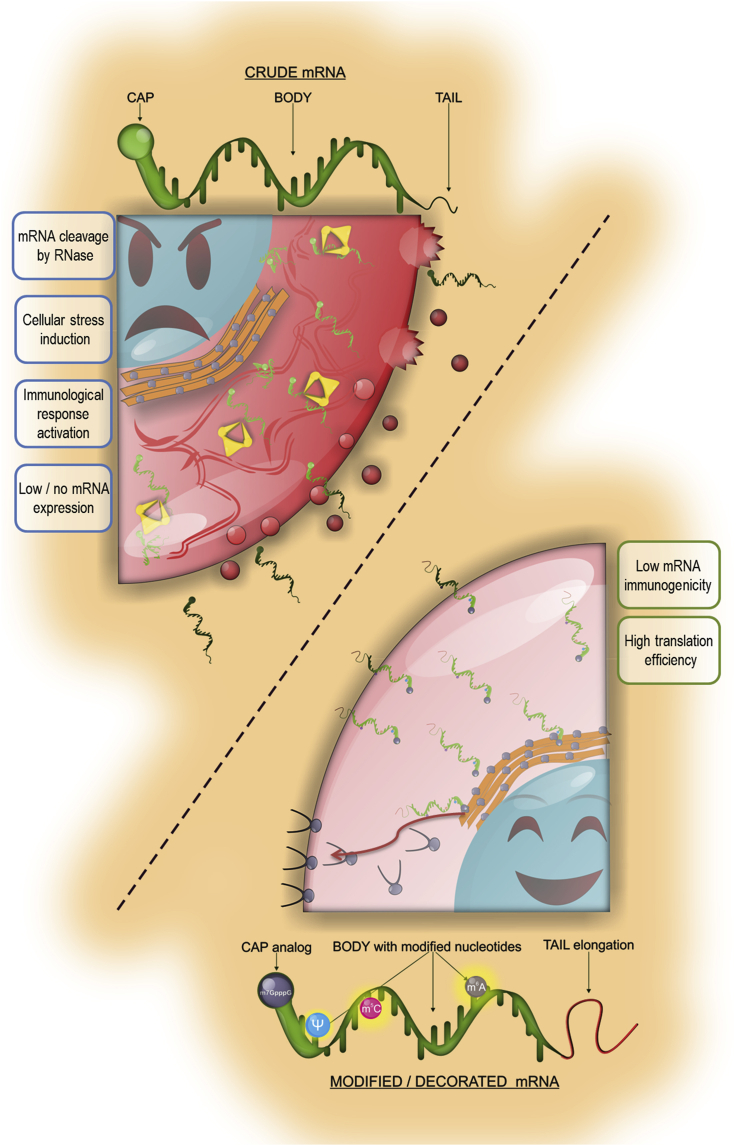
Efforts to improve mRNA quality and biocompatibility continue. Modifications in the bioprocess of pseudouridine production decreased the immune-activating profile of therapeutic mRNA through further elimination of impurities.80 Incorporation of pseudouridine and ARCA significantly increases the yield per reaction, improves modRNA (chemically modified mRNA) translation, and reduces its immunogenicity in vitro. It seems to be an ideal solution to address clinical challenges.81 Therefore, it appears that decorating crude mRNA with modified nucleotides ultimately made it highly welcome by living organisms (Figure 3). We may have reached a plateau in nucleotide modifications to make mRNA a very attractive medicine applicable to several fields, including vaccines, protein supplementation, and stem cell engineering. All RNA modification pathways are stored in a MODOMICS database, which is an excellent source for advancing research on this topic.82
Figure 3.
Differences in cell response to transfection with crude and modified mRNA
Biological roles of mRNA-based medicines
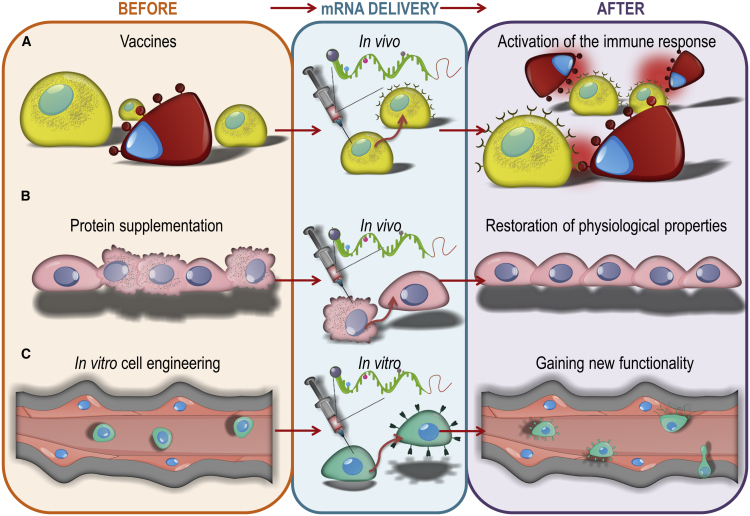
Vaccines
Vaccines are an indispensable tool for public health,83 preventing loss of many lives and occupying a prominent place in medicine. However, their efficacy and safety requirements are rigorous because typically they are applied to vast populations of healthy people. The early approaches with attenuated or inactivated microbes were quite effective, but the issue of safety was debatable, and production, purification, and quality control were cumbersome and expensive.84 Protein-based vaccines have fewer drawbacks but require immune adjuvants, which may involve awakening dormant autoimmune processes.85 In contrast, DNA-based vaccines are plagued by ineffective cellular and nuclear entry and the need for additional devices to support this process.86 This challenge has been addressed recently by packing DNA into a harmless virus, but it can still evoke bad associations.87 Regardless, nearly all of these approaches require living or structurally intact organisms to produce, making development slower and costlier. Against this backdrop, the concept of using mRNA seems to be the holy grail in vaccine development (Figure 4A). mRNA vaccines can be designed within days by simply mimicking the nucleotide sequence of receptor binding domains (RBDs) after sequencing microbe genetic material and then synthetically producing them in robotized factories. Therefore, the timespan from microbe discovery to clinical-grade vaccine candidate is extremely short. The simplicity and speed of the entire procedure also allow fast reaction and facile tweaking of the primary vaccine to respond to the genetic drift of microbes and formation of variants. There is also no risk of contamination with xenogeneic material, which is reassuring to the public. It seems that these features favor mRNA as an ideal vaccine solution.88
Figure 4.
Recent advances in medicine and science because of the introduction of mRNA-based technologies
(A) vaccines, (B) protein supplementation and (C) cell engineering.
When the COVID-19 pandemic emerged, mRNA vaccines already had been tried against previous threats. The first mRNA vaccines took shape in the field of oncology.89, 90, 91 A few years later, they began to be used for treatment of infectious diseases, especially influenza.92,93 Then the preclinical efficacy of the mRNA vaccine was demonstrated against rabies.94 Subsequently, it has been shown that a single dose of a mRNA vaccine is sufficient to induce durable protection of mice and monkeys against the Zika virus.95 However, this virus has never gone global and become a pandemic, so further effort toward vaccine development did not materialize. It has also been shown to be effective in the guinea pig model of another potential global threat: the Ebola virus.96 mRNA vaccines were proposed to combat outbreaks even before the coronavirus pandemic.97 Other preclinical targets for mRNA vaccines include cytomegalovirus,98 tick-transmitted flavivirus,99 human immunodeficiency virus (HIV),100 dengue virus,101 Crimean-Congo hemorrhagic fever virus (CCHFV),102 herpes simplex virus 2 (HSV-2),102 Nipah virus,103 and respiratory syncytial virus (RSV).104 The outstanding safety of mRNA vaccines even prompted the scientific community to propose its use to treat allergies.105 Therefore, it was not surprising that mRNA vaccines were also immediately designed against the novel virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In mice, a single immunization with a nucleoside-modified mRNA vaccine elicited strong cellular and humoral immune responses against SARS-CoV-2.106 Because of the advantages mentioned above, mRNA vaccines have been found to be not only effective, but they were developed the fastest and received emergency use authorization first. Arriving in huge quantities, they were true stars, and they had a huge impact on taming the COVID-19 pandemic in many geographical regions.107
Protein supplementation
Because proteins are critical molecules of intracellular and extracellular compartments of all organisms, their loss or abnormality usually leads to pathological consequences. Therefore, protein supplementation is a valid therapeutic approach. Although there are various approaches to delivering proteins, such as virus-based gene replacement or direct protein delivery, there is also a growing interest in using mRNA to achieve this goal (Figure 4B). There is a specific advantage to this approach. As opposed to DNA, mRNA does not integrate with a genome, obviating many safety concerns. Physicochemically, all mRNA molecules are relatively similar, so the same formulation may serve many genes. However, considerable differences in protein properties cause the formulation and delivery of every protein to be separately developed. The therapeutic effects of mRNA-based protein delivery in an animal model have been demonstrated as far back as nearly three decades ago.108 In comparison with vaccines, preparation of mRNA for protein supplementation is more demanding. Although induction of the host immune response is welcome in vaccine applications, it is of utmost importance to prevent immunogenicity of mRNA delivered for protein supplementation, especially because protein supplementation may be given throughout the entire lifespan, and immunogenic material may cause chronic inflammation.
Pulmonary diseases are particularly compelling applications because of the potential for mRNA to be administered easily and locally through inhalation.109 Along this line, it has been demonstrated in a mouse model of asthma that inhalation of mRNA for Foxp3 reduces pulmonary inflammation by increasing the presence of T regulatory (Treg) cells in the lungs.110 Local delivery of mRNA for Foxp3 may avoid systemic activation of Treg cells, which, in turn, could facilitate tumor formation111 and the spread of infection.112 Non-fenestrated endothelial cells in pulmonary capillaries are an additional obstacle when systemically delivering mRNA to the lungs.113 mRNA-mediated gene supplementation of Toll-like receptors is another strategy investigated in the mouse model of asthma.114 Inhalation of mRNA for the CFTR gene proved effective in a small animal model of monogenetic lung diseases such as cystic fibrosis.115 Translate Bio (acquired recently by Sanofi) is currently running a clinical trial to cure cystic fibrosis with a drug composed of mRNA holding the CFTR gene. Positive effects of mRNA-based therapeutic agents in an animal model of another monogenetic disorder, a1-antitrypsin deficiency, has also been demonstrated recently.116 Metabolic diseases are another group of illnesses potentially curable by mRNA encoding missing or abnormal enzymes.117 Methylmalonic aciduria is addressed by mRNA encoding methylmalonyl-coenzyme A (CoA) mutase.118 mRNA also has been tried as a drug in the context of regenerative medicine to improve revascularization of the heart119 and skin120 as well as in cancer immunotherapy.121
mRNA-based cell engineering
Stem and immune cell therapies are becoming an important field of medical research.122, 123, 124 Cell engineering is frequently used to increase therapeutic potential. Several cellular functions can be achieved through administration of mRNA. This technology is essential for transient cellular modifications (Figure 4C).
Cellular trafficking and migration can be particularly well addressed by mRNA-based engineering. Indeed, it has been shown that adhesion molecule integrin alpha 4 (ITGA4) can be expressed on the surface of mesenchymal stem cells (MSCs) after administration of encoding mRNA,59 acquiring their function in vitro,125 whereas the process of in vivo diapedesis was independent.126 MSC migration has also been increased in vitro by administration of mRNA encoding CXCR4.127 The same strategy is also capable of enhancing the homing of natural killer cells.128
The expression of active immune molecules is another large field of mRNA-based cell engineering. Administration of mRNA for chimeric antigen receptor (CAR) or T cell receptor (TCR) to lymphocytes produces effective immune cells. The transient nature may be overcome with multiple infusions, whereas short-term expression can prevent severe complications because of immune cell auto-aggression.129 mRNA-based programming of dendritic cells to provide relevant instructions to lymphocytes is another attractive therapeutic strategy to reach clinical trials.130
mRNA-based technology is also perfectly suitable for genome editing. The mRNA encoding genome-integrating enzyme transposase can be co-injected with the transposon to deliver a DNA payload.131 Recent addition of site specificity to transposase through CRISPR technology has finally allowed editing of the genome precisely and efficiently with large DNA payloads.132 The Cas9 enzyme can also be delivered to cells in the form of mRNA along with relevant guide RNA (gRNA) sequences.133 mRNA is also a viable strategy for induction of pluripotency in somatic cells.134
mRNA packaging and delivery
mRNA is a fragile and negatively charged molecule; therefore, appropriate packaging and delivery systems are critical to fully exploit its potential. There are many approaches to accomplish this task. PEGylated lipid nanoparticles (LNPs) are currently a mainstay of mRNA enveloping, mainly because of their success in delivering COVID-19 vaccines. However, besides LNPs, there are many other ways to deliver mRNA, such as polymers, inorganic structures, proteins, hydrogels, etc.
Lipid-based carriers have a very long history. Liposomes have been used to deliver a variety of therapeutic compounds. Thus, they were the first obvious choice for nucleic acid delivery. However, they ultimately failed in in vivo small interfering RNA (siRNA) delivery and became a motivation to further develop carriers, which resulted in LNPs.135 Therefore, the rising star of mRNA therapeutic agents immediately enjoyed a fertile ground of powerful carriers capable of effective in vivo payload delivery.
Inorganic components such as apatite have been shown to increase the efficacy of mRNA.136,137 Silica nanoparticles are another option to facilitate nucleic acid delivery138,139 and are considered an emerging vaccine delivery system for COVID-19.140 Silica is a relatively low-cost and easy-to-scale solution for safe mRNA delivery and enjoys an extensive functionalization portfolio with a very encouraging storage profile.141 Recently, room-temperature synthesis of dendritic mesoporous silica nanoparticles with small sizes and large pores enhanced mRNA delivery performance.142
Polymers are an extensive group of natural and synthetic compounds. They are characterized by their high versatility in terms of nucleic acid binding and release as well as relative stability. Cationic polymers such as polyethyleneimine-stearic acid (PSA) can self-assemble and encapsulate nucleic acids.143 Ionizable amino-polyesters synthesized via ring-opening polymerization of tertiary amino alcohols is another group of exciting polymers for mRNA administration.144 They are devoid of a strong positive charge and, thus, are more neutral for cells. Amphiphilic polyhydrazones are another type of polymer candidate for mRNA delivery.145 A microneedle patch (RNApatch) composed of low-molecular-weight polyvinylpyrrolidone (PVP) can address the essential aspect of very low mRNA stability by preserving mRNA functionality for up to 2 weeks under ambient conditions.146 Lipid polymer hybrids are another approach, capitalizing on the best features of lipids and polymers.147
Hydrogels are somewhat related to polymers, which are the main components, although with hydrogels, different properties are used as carriers. Hydrogels are typically characterized by a highly tunable structure capable of meeting various requirements. Hydrogels’ structure and injectability allow localized deployment of mRNA to body cavities or other relevant locations. However, hydrogel particles can also be used as any other nucleic acid carrier. The tunable size of such particles could perfectly suit the inhalation route. Therefore, hydrogels are a highly versatile carrier. Typically, mRNA can be entrapped within hydrogels and then slowly released over a relatively long period. Different release rates characterize various hydrogel systems, and blending multiple hydrogels presents an opportunity to discover optimal release conditions. One group revealed that chitosan-alginate hybrids encourage local and sustained expression of exogenous proteins in cells.148 Hydrogels from graphene oxide and polyethylenimine (PEI) can deliver mRNA to tissues, and such mRNA remains functional for up to a month.149 DNA hydrogels are another concept for small interfering mRNA production, but they could potentially be adapted to mRNA production as well.149 Because hydrogels can be labeled and imaged relatively quickly, they offer seamless precision for procedures.150
RNA viruses capitalize on the ability of nucleoproteins to package mRNA. It has been shown that, under some circumstances, transmembrane proteins can interact and package other mRNA-bearing RNA-packaging signals in the form of virus-like particles.151 There are also other endogenous proteins, which selectively bind to their mRNA and protect it. One of these proteins (polietylenoglikol 10, PEG10) has been engineered to selectively wrap and package other mRNAs. It is accomplished by flanking genes of interest with PEG10 untranslated regions.152 Such mRNA-PEG10 complexes can be shuttled between cells as virus-like particles (VLPs). The high modularity of this platform to fit a variety of mRNAs and the endogenous character of proteins to avoid immune activation are advantages that may lead to the rapid adoption of this approach to achieve highly effective and non-toxic mRNA delivery.
Summary and future outlook
The history of nucleic acid discoveries makes for one of the most fabulous stories ever. The chemical findings and physical discoveries applied to a biological context contributed to the revelation of the greatest secret of life. It, in turn, launched an era of miraculous medical discoveries. For decades, stable and easy-to-handle DNA was the focus of research, whereas in recent years, mRNA came into the spotlight. As always, the devil is in the details, and the tireless efforts of scientists allowed us to uncover the intricacies of mRNA molecules. Engineering of cap analogs and nucleotide modification was a recipe for potent mRNA, which became the basis for a new generation of powerful medicines. The entire mRNA orchestra became harmonious when it was most needed to tackle the worst pandemic in 100 years. Therefore, there is a strong need to pay tribute to the early pioneers of mRNA and those still alive, such as Darzynkiewicz, Karikó, and others, and recognize them with a Nobel Prize.
We expect such remarkable mRNAs to prompt further breakthroughs in medicine in the near future. We anticipate other advancements in mRNA technology and continuous contributions to healthcare. Purely chemical, DNA template-free synthesis of mRNA may be another step toward widespread use of mRNA.153 The alternative genetic code may inflate the potency of mRNA therapeutic agents.154 Unnatural base pairs are another biotechnological frontier.155 Artificial intelligence adds yet another dimension to RNA research.156 Thus, our supply of mRNA-related ingredients is rapidly expanding to meet the needs of successive healthcare developments. the discoveries made in genetics could move outside the box and be applied in other fields, such as computing, information storage, sensors, robotics, and more.
Acknowledgments
This work was supported by Mossakowski Medical Research Institute Statutory Grant No. 6 funded by the Ministry of Science and Higher Education (Poland) and start-up funds from the University of Maryland School of Medicine (Baltimore, USA).
Author contributions
M.J. reviewed literature, drafted the manuscript, and approved the final version. A.A. provided illustrations and drafted the manuscript.
Declaration of interests
M.J. is co-founder and co-owner of Ti-com, LLC and IntraART, LLC, but there is no direct relation of the manuscript’s content to the activity of either company.
References
- 1.Cobb M. Who discovered messenger RNA? Curr. Biol. 2015;25:R526–R532. doi: 10.1016/j.cub.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 2.Avery O.T., Macleod C.M., McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types : induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type iii. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson J.D., Crick F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein L., Plaut W. Direct evidence for nuclear synthesis of cytoplasmic ribose nucleic acid. Proc. Natl. Acad. Sci. USA. 1955;41:874–880. doi: 10.1073/pnas.41.11.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson J.D. Involvement of RNA in the synthesis of proteins. Science. 1963;140:17–26. doi: 10.1126/science.140.3562.17. [DOI] [PubMed] [Google Scholar]
- 6.Hoagland M.B., Stephenson M.L., Scott J.F., Hecht L.I., Zamecnik P.C. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958;231:241–257. [PubMed] [Google Scholar]
- 7.Brenner S., Jacob F., Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190:576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 8.Gros F., Hiatt H., Gilbert W., Kurland C.G., Risebrough R.W., Watson J.D. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature. 1961;190:581–585. doi: 10.1038/190581a0. [DOI] [PubMed] [Google Scholar]
- 9.Aronson A.I., McCARTHY B.J. Studies of E. coli ribosomal RNA and its degradation products. Biophys. J. 1961;1:215–226. doi: 10.1016/s0006-3495(61)86885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall B.D., Spiegelman S. Sequence complementarity of T2-DNA and T2-specific RNA. Proc. Natl. Acad. Sci. USA. 1961;47:137–163. doi: 10.1073/pnas.47.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanofsky C., Carlton B.C., Guest J.R., Helinski D.R., Henning U. On the colinearity of gene structure and protein structure. Proc. Natl. Acad. Sci. USA. 1964;51:266–272. doi: 10.1073/pnas.51.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernfield M.R., Nirenberg M.W. Rna codewords and protein synthesis. The nucleotide sequences of multiple codewords for phenylalanine, serine, leucine, and proline. Science. 1965;147:479–484. doi: 10.1126/science.147.3657.479. [DOI] [PubMed] [Google Scholar]
- 13.Trupin J.S., Rottman F.M., Brimacombe R.L., Leder P., Bernfield M.R., Nirenberg M.W. Rna codewords and protein synthesis, vi. On the nucleotide sequences of degenerate codeword sets for isoleucine, tyrosine, asparagine, and lysine. Proc. Natl. Acad. Sci. USA. 1965;53:807–811. doi: 10.1073/pnas.53.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellogg D.A., Doctor B.P., Loebel J.E., Nirenberg M.W. RNA codons and protein synthesis. IX. Synonym codon recognition by multiple species of valine-alanine-and methionine-sRNA. Proc. Natl. Acad. Sci. USA. 1966;55:912–919. doi: 10.1073/pnas.55.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michelson A.M., Todd A.R. Nucleotides part XXXII. Synthesis of a dithymidine dinucleotide containing a 3′: 5′-internucleotidic linkage. J. Chem. Soc. 1955;0:2632–2638. doi: 10.1039/JR9550002632. [DOI] [Google Scholar]
- 16.Rammler D.H., Khorana H.G. A new approach to the specific synthesis of the C3'-C5' inter-ribonucleotide linkage. Biochem. Biophys. Res. Commun. 1962;7:147–150. doi: 10.1016/0006-291x(62)90164-x. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal K.L., Büchi H., Caruthers M.H., Gupta N., Khorana H.G., Kleppe K., Kumar A., Ohtsuka E., Rajbhandary U.L., Van de Sande J.H., et al. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature. 1970;227:27–34. doi: 10.1038/227027a0. [DOI] [PubMed] [Google Scholar]
- 18.Letsinger R.L., Mahadevan V. Oligonucleotide synthesis on a polymer support. J. Am. Chem. Soc. 1965;87:3526–3527. doi: 10.1021/ja01093a058. [DOI] [PubMed] [Google Scholar]
- 19.Letsinger R.L., Caruthers M.H., Jerina D.M. Reactions of nucleosides on polymer supports. Synthesis of thymidylylthymidylylthymidine. Biochemistry. 1967;6:1379–1388. doi: 10.1021/bi00857a021. [DOI] [PubMed] [Google Scholar]
- 20.Letsinger R.L., Lunsford W.B. Synthesis of thymidine oligonucleotides by phosphite triester intermediates. J. Am. Chem. Soc. 1976;98:3655–3661. doi: 10.1021/ja00428a045. [DOI] [PubMed] [Google Scholar]
- 21.Caruthers M.H., Beaucage S.L., Becker C., Efcavitch J.W., Fisher E.F., Galluppi G., Goldman R., deHaseth P., Matteucci M., McBride L., et al. Deoxyoligonucleotide synthesis via the phosphoramidite method. Gene Amplif. Anal. 1983;3:1–26. [PubMed] [Google Scholar]
- 22.Singer M.F. Phosphorolysis of oligoribonucleotides by polynucleotide phosphorylase. J. Biol. Chem. 1958;232:211–228. [PubMed] [Google Scholar]
- 23.Bernfield M.R. Ribonuclease and oligoribonucleotide synthesis. I. Synthetic activity of bovine pancreatic ribonuclease derivatives. J. Biol. Chem. 1965;240:4753–4762. [PubMed] [Google Scholar]
- 24.Griffin B.E., Reese C.B., Stephenson G.F., Trentham D.R. Oligoribonucleotide synthesis from nucleoside 2'-O-benzyl ethers. Tetrahedron Lett. 1966;36:4349–4354. doi: 10.1016/s0040-4039(00)76063-1. [DOI] [PubMed] [Google Scholar]
- 25.Mohr S.C., Thach R.E. Application of ribonuclease T1 to the synthesis of oligoribonucleotides of defined base sequence. J. Biol. Chem. 1969;244:6566–6576. [PubMed] [Google Scholar]
- 26.Stevens A. Studies of the ribonucleic acid polymerase from Escherichia coli. V. Studies of its complexes with polyribonucleotides. J. Biol. Chem. 1969;244:425–429. [PubMed] [Google Scholar]
- 27.Niyogi S.K. Annealing properties of oligoribonucleotides derived from T5 DNA-directed RNA compared to those from T2 and T7 DNA-directed RNAs. J. Biol. Chem. 1973;248:2323–2327. [PubMed] [Google Scholar]
- 28.Walker G.C., Uhlenbeck O.C. Stepwise enzymatic oligoribonucleotide synthesis including modified nucleotides. Biochemistry. 1975;14:817–824. doi: 10.1021/bi00675a027. [DOI] [PubMed] [Google Scholar]
- 29.Shatkin A.J., Sipe J.D. Single-stranded, adenine-rich RNA from purified reoviruses. Proc. Natl. Acad. Sci. USA. 1968;59:246–253. doi: 10.1073/pnas.59.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muthukrishnan S., Both G.W., Furuichi Y., Shatkin A.J. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M., Shatkin A.J. Characterization of translational initiation regions from eukaryotic messenger RNAs. Methods Enzymol. 1979;60:360–375. doi: 10.1016/s0076-6879(79)60034-4. [DOI] [PubMed] [Google Scholar]
- 32.Green M.R., Maniatis T., Melton D.A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32:681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- 33.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 34.Karikó K., Keller J.M., Harris V.A., Langer D.J., Welsh F.A. In vivo protein expression from mRNA delivered into adult rat brain. J. Neurosci. Methods. 2001;105:77–86. doi: 10.1016/s0165-0270(00)00355-1. [DOI] [PubMed] [Google Scholar]
- 35.Ni H., Capodici J., Cannon G., Communi D., Boeynaems J.M., Karikó K., Weissman D. Extracellular mRNA induces dendritic cell activation by stimulating tumor necrosis factor-alpha secretion and signaling through a nucleotide receptor. J. Biol. Chem. 2002;277:12689–12696. doi: 10.1074/jbc.M110729200. [DOI] [PubMed] [Google Scholar]
- 36.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 37.Koski G.K., Karikó K., Xu S., Weissman D., Cohen P.A., Czerniecki B.J. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J. Immunol. 2004;172:3989–3993. doi: 10.4049/jimmunol.172.7.3989. [DOI] [PubMed] [Google Scholar]
- 38.Stacey K.A., Cobb M., Cousens S.F., Alexander P. The reactions of the radiomimetic alkylating agents with macromolecules in vitro. Ann. N. Y. Acad. Sci. 1958;68:682–701. doi: 10.1111/j.1749-6632.1958.tb42634.x. [DOI] [PubMed] [Google Scholar]
- 39.Ukita T., Okuyama H., Hayatsu H. Modifications of nucleosides and nucleotides. I. Reaction of ethylene oxide with uridine and uridylic acid. Chem. Pharm. Bull. 1963;11:1399–1404. doi: 10.1248/cpb.11.1399. [DOI] [PubMed] [Google Scholar]
- 40.Hayatsu H., Ukita T. Selective modification of cytidine residue in ribonucleic acid by semicarbazide. Biochem. Biophys. Res. Commun. 1964;14:198–203. doi: 10.1016/0006-291x(64)90255-4. [DOI] [PubMed] [Google Scholar]
- 41.Azegami M., Iwai K. Specific modification of nucleic acids and their constituents with trinitrophenyl group. J. Biochem. 1964;55:346–348. doi: 10.1093/oxfordjournals.jbchem.a127892. [DOI] [PubMed] [Google Scholar]
- 42.Kochetkov N.K., Budowsky E.I., Shibaeva R.P. Selective modification of uridine and guanosine. Biochim. Biophys. Acta. 1964;87:515–518. doi: 10.1016/0926-6550(64)90128-8. [DOI] [PubMed] [Google Scholar]
- 43.Ofengand J. A chemical method for the selective modification of pseudouridine in the presence of other nucleosides. Biochem. Biophys. Res. Commun. 1965;18:192–201. doi: 10.1016/0006-291x(65)90739-4. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida M., Ukita T. Selective modifications of inosine and psi-uridine with acrylonitrile out of the other ribonucleosides. J. Biochem. 1965;57:818–821. doi: 10.1093/oxfordjournals.jbchem.a128151. [DOI] [PubMed] [Google Scholar]
- 45.Kochetkov N.K., Budowsky E.I., Demushkin V.P., Turchinsky M.F., Simukova N.A., Sverdlov E.D. The chemical modification of nucleic acids. I. The preparation of deuridylic RNA's. Biochim. Biophys. Acta. 1967;142:35–46. doi: 10.1016/0005-2787(67)90513-8. [DOI] [PubMed] [Google Scholar]
- 46.Molinaro M., Sheiner L.B., Neelon F.A., Cantoni G.L. Effect of chemical modification of dihydrouridine in yeast transfer ribonucleic acid on amino acid acceptor activity and ribosomal binding. J. Biol. Chem. 1968;243:1277–1282. [PubMed] [Google Scholar]
- 47.Limbach P.A., Crain P.F., McCloskey J.A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duechler M., Leszczyńska G., Sochacka E., Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci. 2016;73:3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agris P.F. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 50.Darzynkiewicz E., Antosiewicz J., Ekiel I., Morgan M.A., Tahara S.M., Shatkin A.J. Methyl esterification of m7G5'p reversibly blocks its activity as an analog of eukaryotic mRNA 5'-caps. J. Mol. Biol. 1981;153:451–458. doi: 10.1016/0022-2836(81)90289-8. [DOI] [PubMed] [Google Scholar]
- 51.Cai A., Jankowska-Anyszka M., Centers A., Chlebicka L., Stepinski J., Stolarski R., Darzynkiewicz E., Rhoads R.E. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 52.Darzynkiewicz E., Ekiel I., Lassota P., Tahara S.M. Inhibition of eukaryotic translation by analogues of messenger RNA 5'-cap: chemical and biological consequences of 5'-phosphate modifications of 7-methylguanosine 5'-monophosphate. Biochemistry. 1987;26:4372–4380. doi: 10.1021/bi00388a028. [DOI] [PubMed] [Google Scholar]
- 53.Darzynkiewicz E., Stepinski J., Ekiel I., Jin Y., Haber D., Sijuwade T., Tahara S.M. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darzynkiewicz E., Stepinski J., Ekiel I., Goyer C., Sonenberg N., Temeriusz A., Jin Y., Sijuwade T., Haber D., Tahara S.M. Inhibition of eukaryotic translation by nucleoside 5'-monophosphate analogues of mRNA 5'-cap: changes in N7 substituent affect analogue activity. Biochemistry. 1989;28:4771–4778. doi: 10.1021/bi00437a038. [DOI] [PubMed] [Google Scholar]
- 55.Darzynkiewicz E., Lönnberg H. Base stacking of simple mRNA cap analogues. Association of 7, 9-dimethylguanine, 7-methylguanosine and 7-methylguanosine 5'-monophosphate with indole and purine derivatives in aqueous solution. Biophys. Chem. 1989;33:289–293. doi: 10.1016/0301-4622(89)80030-4. [DOI] [PubMed] [Google Scholar]
- 56.Pasquinelli A.E., Dahlberg J.E., Lund E. Reverse 5' caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 57.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel "anti-reverse" cap analogs 7-methyl(3'-O-methyl)GpppG and 7-methyl (3'-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 58.Mockey M., Gonçalves C., Dupuy F.P., Lemoine F.M., Pichon C., Midoux P. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006;340:1062–1068. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- 59.Nowakowski A., Andrzejewska A., Boltze J., Nitzsche F., Cui L.L., Jolkkonen J., Walczak P., Lukomska B., Janowski M. Translation, but not transfection limits clinically relevant, exogenous mRNA based induction of alpha-4 integrin expression on human mesenchymal stem cells. Sci. Rep. 2017;7:1103. doi: 10.1038/s41598-017-01304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel "anti-reverse" cap analogs with superior translational properties. RNA. 2003;9:1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jemielity J., Stepinski J., Jaremko M., Haber D., Stolarski R., Rhoads R.E., Darzynkiewicz E. Synthesis of novel mRNA 5' cap-analogues: dinucleoside P1, P3-tri-P1, P4-tetra-and P1, P5-pentaphosphates. Nucleosides Nucleotides Nucleic Acids. 2003;22:691–694. doi: 10.1081/NCN-120022611. [DOI] [PubMed] [Google Scholar]
- 62.Grudzien E., Stepinski J., Jankowska-Anyszka M., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA. 2004;10:1479–1487. doi: 10.1261/rna.7380904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kowalska J., Lewdorowicz M., Zuberek J., Bojarska E., Wojcik J., Cohen L.S., Davis R.E., Stepinski J., Stolarski R., Darzynkiewicz E., Jemielity J. Synthesis and properties of mRNA cap analogs containing phosphorothioate moiety in 5', 5'-triphosphate chain. Nucleosides Nucleotides Nucleic Acids. 2005;24:595–600. doi: 10.1081/ncn-200061915. [DOI] [PubMed] [Google Scholar]
- 64.Jemielity J., Heinonen P., Lonnberg H., Darzynkiewicz E. A novel approach to solid phase chemical synthesis of oligonucleotide mRNA cap analogs. Nucleosides Nucleotides Nucleic Acids. 2005;24:601–605. doi: 10.1081/ncn-200061922. [DOI] [PubMed] [Google Scholar]
- 65.Kalek M., Jemielity J., Grudzien E., Zuberek J., Bojarska E., Cohen L.S., Stepinski J., Stolarski R., Davis R.E., Rhoads R.E., Darzynkiewicz E. Synthesis and biochemical properties of novel mRNA 5' cap analogs resistant to enzymatic hydrolysis. Nucleosides Nucleotides Nucleic Acids. 2005;24:615–621. doi: 10.1081/ncn-200060091. [DOI] [PubMed] [Google Scholar]
- 66.Kowalska J., Zuberek J., Darzynkiewicz Z.M., Lukaszewicz M., Darzynkiewicz E., Jemielity J. The first examples of mRNA cap analogs bearing boranophosphate modification. Nucleic Acids Symp. Ser. 2008;52:289–290. doi: 10.1093/nass/nrn146. [DOI] [PubMed] [Google Scholar]
- 67.Kocmik I., Piecyk K., Rudzinska M., Niedzwiecka A., Darzynkiewicz E., Grzela R., Jankowska-Anyszka M. Modified ARCA analogs providing enhanced translational properties of capped mRNAs. Cell Cycle. 2018;17:1624–1636. doi: 10.1080/15384101.2018.1486164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rydzik A.M., Lukaszewicz M., Zuberek J., Kowalska J., Darzynkiewicz Z.M., Darzynkiewicz E., Jemielity J. Synthetic dinucleotide mRNA cap analogs with tetraphosphate 5', 5' bridge containing methylenebis(phosphonate) modification. Org. Biomol. Chem. 2009;7:4763–4776. doi: 10.1039/b911347a. [DOI] [PubMed] [Google Scholar]
- 70.Strenkowska M., Kowalska J., Lukaszewicz M., Zuberek J., Su W., Rhoads R.E., Darzynkiewicz E., Jemielity J. Towards mRNA with superior translational activity: synthesis and properties of ARCA tetraphosphates with single phosphorothioate modifications. New J. Chem. 2010;34:993–1007. doi: 10.1039/b9nj00644c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warminski M., Kowalska J., Nowak E., Kubacka D., Tibble R., Kasprzyk R., Sikorski P.J., Gross J.D., Nowotny M., Jemielity J. Structural insights into the interaction of clinically relevant phosphorothioate mRNA cap analogs with translation initiation factor 4E reveal stabilization via electrostatic thio-effect. ACS Chem. Biol. 2021;16:334–343. doi: 10.1021/acschembio.0c00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 73.Henderson J.M., Ujita A., Hill E., Yousif-Rosales S., Smith C., Ko N., McReynolds T., Cabral C.R., Escamilla-Powers J.R., Houston M.E. Cap 1 messenger RNA synthesis with Co-transcriptional CleanCap((R)) analog by in vitro transcription. Curr. Protoc. 2021;1:e39. doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- 74.Karikó K., Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 75.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weissman D., Karikó K. mRNA: fulfilling the promise of gene therapy. Mol. Ther. 2015;23:1416–1417. doi: 10.1038/mt.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hadas Y., Sultana N., Youssef E., Sharkar M.T.K., Kaur K., Chepurko E., Zangi L. Optimizing modified mRNA in vitro synthesis protocol for heart gene therapy. Mol. Ther. Methods Clin. Dev. 2019;14:300–305. doi: 10.1016/j.omtm.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A., et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Excler J.L., Privor-Dumm L., Kim J.H. Supply and delivery of vaccines for global health. Curr. Opin. Immunol. 2021;71:13–20. doi: 10.1016/j.coi.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gendon Iu Z. [Advantages and disadvantages of inactivated and live influenza vaccine] Vopr. Virusol. 2004;49:4–12. [PubMed] [Google Scholar]
- 85.Blakney A.K., McKay P.F. Next-generation COVID-19 vaccines: here come the proteins. Lancet. 2021;397:643–645. doi: 10.1016/S0140-6736(21)00258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee J., Arun Kumar S., Jhan Y.Y., Bishop C.J. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018;80:31–47. doi: 10.1016/j.actbio.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen H., Xiang Z.Q., Li Y., Kurupati R.K., Jia B., Bian A., Zhou D.M., Hutnick N., Yuan S., Gray C., et al. Adenovirus-based vaccines: comparison of vectors from three species of adenoviridae. J. Virol. 2010;84:10522–10532. doi: 10.1128/JVI.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandbrink J.B., Shattock R.J. RNA vaccines: a suitable platform for tackling emerging pandemics? Front. Immunol. 2020;11:608460. doi: 10.3389/fimmu.2020.608460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hess P.R., Boczkowski D., Nair S.K., Snyder D., Gilboa E. Vaccination with mRNAs encoding tumor-associated antigens and granulocyte-macrophage colony-stimulating factor efficiently primes CTL responses, but is insufficient to overcome tolerance to a model tumor/self antigen. Cancer Immunol. Immunother. 2006;55:672–683. doi: 10.1007/s00262-005-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T.K., Pawelec G., Hoerr I., Rammensee H.G., Garbe C. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 91.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkić-Zrna S., Probst J., Kallen K.J. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 92.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 93.Dormitzer P.R. Rapid production of synthetic influenza vaccines. Curr. Top. Microbiol. Immunol. 2015;386:237–273. doi: 10.1007/82_2014_399. [DOI] [PubMed] [Google Scholar]
- 94.Schnee M., Vogel A.B., Voss D., Petsch B., Baumhof P., Kramps T., Stitz L. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl. Trop. Dis. 2016;10:e0004746. doi: 10.1371/journal.pntd.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-based vaccines elicit robust immune responses and protect Guinea pigs from Ebola virus disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.John S., Yuzhakov O., Woods A., Deterling J., Hassett K., Shaw C.A., Ciaramella G. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine. 2018;36:1689–1699. doi: 10.1016/j.vaccine.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 99.VanBlargan L.A., Himansu S., Foreman B.M., Ebel G.D., Pierson T.C., Diamond M.S. An mRNA vaccine protects mice against multiple tick-transmitted flavivirus infections. Cell Rep. 2018;25:3382–3392.e3. doi: 10.1016/j.celrep.2018.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pardi N., LaBranche C.C., Ferrari G., Cain D.W., Tombácz I., Parks R.J., Muramatsu H., Mui B.L., Tam Y.K., Karikó K., et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roth C., Cantaert T., Colas C., Prot M., Casadémont I., Levillayer L., Thalmensi J., Langlade-Demoyen P., Gerke C., Bahl K., et al. A modified mRNA vaccine targeting immunodominant NS epitopes protects against dengue virus infection in HLA class I transgenic mice. Front. Immunol. 2019;10:1424. doi: 10.3389/fimmu.2019.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aligholipour Farzani T., Foldes K., Ergunay K., Gurdal H., Bastug A., Ozkul A. Immunological Analysis of a CCHFV mRNA Vaccine Candidate in Mouse Models. Vaccines (Basel) 2019;7 doi: 10.3390/vaccines7030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lo M.K., Spengler J.R., Welch S.R., Harmon J.R., Coleman-McCray J.D., Scholte F.E.M., Shrivastava-Ranjan P., Montgomery J.M., Nichol S.T., Weissman D., Spiropoulou C.F. Evaluation of a single-dose nucleoside-modified messenger RNA vaccine encoding hendra virus-soluble glycoprotein against lethal Nipah virus challenge in Syrian hamsters. J. Infect. Dis. 2020;221:S493–S498. doi: 10.1093/infdis/jiz553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., Donnell G.O., Galli J.D., Swoyer R., Touch S., et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines. 2020;5:16. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weiss R., Scheiblhofer S., Thalhamer J. Generation and evaluation of prophylactic mRNA vaccines against allergy. Methods Mol. Biol. 2017;1499:123–139. doi: 10.1007/978-1-4939-6481-9_7. [DOI] [PubMed] [Google Scholar]
- 106.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walczak P., Janowski M. The COVID-19 menace. Glob. Chall. 2021;5:2100004. doi: 10.1002/gch2.202100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 109.Sahu I., Haque A.K.M.A., Weidensee B., Weinmann P., Kormann M.S.D. Recent developments in mRNA-based protein supplementation therapy to target lung diseases. Mol. Ther. 2019;27:803–823. doi: 10.1016/j.ymthe.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mays L.E., Ammon-Treiber S., Mothes B., Alkhaled M., Rottenberger J., Müller-Hermelink E.S., Grimm M., Mezger M., Beer-Hammer S., von Stebut E., et al. Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J. Clin. Invest. 2013;123:1216–1228. doi: 10.1172/JCI65351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M., et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 112.Belkaid Y., Piccirillo C.A., Mendez S., Shevach E.M., Sacks D.L. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 113.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 114.Zeyer F., Mothes B., Will C., Carevic M., Rottenberger J., Nürnberg B., Hartl D., Handgretinger R., Beer-Hammer S., Kormann M.S.D. mRNA-mediated gene supplementation of toll-like receptors as treatment strategy for asthma in vivo. PLoS One. 2016;11:e0154001. doi: 10.1371/journal.pone.0154001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haque A.K.M.A., Dewerth A., Antony J.S., Riethmüller J., Schweizer G.R., Weinmann P., Latifi N., Yasar H., Pedemonte N., Sondo E., et al. Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis. Sci. Rep. 2018;8:16776. doi: 10.1038/s41598-018-34960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Connolly B., Isaacs C., Cheng L., Asrani K.H., Subramanian R.R. SERPINA1 mRNA as a treatment for alpha-1 antitrypsin deficiency. J. Nucleic Acids. 2018;2018:8247935. doi: 10.1155/2018/8247935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chandler R.J. Messenger RNA therapy as an option for treating metabolic disorders. Proc. Natl. Acad. Sci. USA. 2019;116:20804–20806. doi: 10.1073/pnas.1914673116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S., Theisen M., Hong S.J., Zhou J., Rajendran R., et al. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 2018;24:2520. doi: 10.1016/j.celrep.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 119.Carlsson L., Clarke J.C., Yen C., Gregoire F., Albery T., Billger M., Egnell A.C., Gan L.M., Jennbacken K., Johansson E., et al. Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 Week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 2018;9:330–346. doi: 10.1016/j.omtm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gan L.M., Lagerström-Fermér M., Carlsson L.G., Arfvidsson C., Egnell A.C., Rudvik A., Kjaer M., Collén A., Thompson J.D., Joyal J., et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019;10:871. doi: 10.1038/s41467-019-08852-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Andrzejewska A., Lukomska B., Janowski M. Concise review: mesenchymal stem cells: from roots to boost. Stem Cell. 2019;37:855–864. doi: 10.1002/stem.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Z., Huang X. Cellular immunotherapy for hematological malignancy: recent progress and future perspectives. Cancer Biol. Med. 2021;18:0. doi: 10.20892/j.issn.2095-3941.2020.0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zheng Y., Li Y., Feng J., Li J., Ji J., Wu L., Yu Q., Dai W., Wu J., Zhou Y., Guo C. Cellular based immunotherapy for primary liver cancer. J. Exp. Clin. Cancer Res. 2021;40:250. doi: 10.1186/s13046-021-02030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andrzejewska A., Nowakowski A., Grygorowicz T., Dabrowska S., Orzel J., Walczak P., Lukomska B., Janowski M. Single-cell, high-throughput analysis of cell docking to vessel wall. J. Cereb. Blood Flow Metab. 2019;39:2308–2320. doi: 10.1177/0271678X18805238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Andrzejewska A., Dabrowska S., Nowak B., Walczak P., Lukomska B., Janowski M. Mesenchymal stem cells injected into carotid artery to target focal brain injury home to perivascular space. Theranostics. 2020;10:6615–6628. doi: 10.7150/thno.43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ryser M.F., Ugarte F., Thieme S., Bornhäuser M., Roesen-Wolff A., Brenner S. mRNA transfection of CXCR4-GFP fusion--simply generated by PCR-results in efficient migration of primary human mesenchymal stem cells. Tissue Eng. Part C Methods. 2008;14:179–184. doi: 10.1089/ten.tec.2007.0359. [DOI] [PubMed] [Google Scholar]
- 128.Levy E., Reger R., Segerberg F., Lambert M., Leijonhufvud C., Baumer Y., Carlsten M., Childs R. Enhanced bone marrow homing of natural killer cells following mRNA transfection with gain-of-function variant CXCR4(R334X) Front. Immunol. 2019;10:1262. doi: 10.3389/fimmu.2019.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bertoletti A., Tan A.T. Challenges of CAR- and TCR-T cell-based therapy for chronic infections. J. Exp. Med. 2020;217 doi: 10.1084/jem.20191663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Benteyn D., Heirman C., Bonehill A., Thielemans K., Breckpot K. mRNA-based dendritic cell vaccines. Expert Rev. Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 131.Singh H., Huls H., Kebriaei P., Cooper L.J.N. A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol. Rev. 2014;257:181–190. doi: 10.1111/imr.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vo P.L.H., Acree C., Smith M.L., Sternberg S.H. Unbiased profiling of CRISPR RNA-guided transposition products by long-read sequencing. Mob. DNA. 2021;12:13. doi: 10.1186/s13100-021-00242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qiu M., Glass Z., Chen J., Haas M., Jin X., Zhao X., Rui X., Ye Z., Li Y., Zhang F., Xu Q. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kogut I., McCarthy S.M., Pavlova M., Astling D.P., Chen X., Jakimenko A., Jones K.L., Getahun A., Cambier J.C., Pasmooij A.M.G., et al. High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat. Commun. 2018;9:745. doi: 10.1038/s41467-018-03190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zohra F.T., Chowdhury E.H., Tada S., Hoshiba T., Akaike T. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem. Biophys. Res. Commun. 2007;358:373–378. doi: 10.1016/j.bbrc.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 137.Zohra F.T., Maitani Y., Akaike T. mRNA delivery through fibronectin associated liposome-apatite particles: a new approach for enhanced mRNA transfection to mammalian cell. Biol. Pharm. Bull. 2012;35:111–115. doi: 10.1248/bpb.35.111. [DOI] [PubMed] [Google Scholar]
- 138.Juneja R., Vadarevu H., Halman J., Tarannum M., Rackley L., Dobbs J., Marquez J., Chandler M., Afonin K., Vivero-Escoto J.L. Combination of nucleic acid and mesoporous silica nanoparticles: optimization and therapeutic performance in vitro. ACS Appl. Mater. Interfaces. 2020;12:38873–38886. doi: 10.1021/acsami.0c07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hom C., Lu J., Tamanoi F. Silica nanoparticles as a delivery system for nucleic acid-based reagents. J. Mater. Chem. 2009;19:6308–6316. doi: 10.1039/b904197d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Theobald N. Emerging vaccine delivery systems for COVID-19: functionalised silica nanoparticles offer a potentially safe and effective alternative delivery system for DNA/RNA vaccines and may be useful in the hunt for a COVID-19 vaccine. Drug Discov. Today. 2020;25:1556–1558. doi: 10.1016/j.drudis.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Castillo R.R., Lozano D., Vallet-Regí M. Mesoporous silica nanoparticles as carriers for therapeutic biomolecules. Pharmaceutics. 2020;12:E432. doi: 10.3390/pharmaceutics12050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Y., Song H., Yu M., Xu C., Liu Y., Tang J., Yang Y., Yu C. Room temperature synthesis of dendritic mesoporous silica nanoparticles with small sizes and enhanced mRNA delivery performance. J. Mater. Chem. B. 2018;6:4089–4095. doi: 10.1039/c8tb00544c. [DOI] [PubMed] [Google Scholar]
- 143.Zhao M., Li M., Zhang Z., Gong T., Sun X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016;23:2596–2607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 144.Kowalski P.S., Capasso Palmiero U., Huang Y., Rudra A., Langer R., Anderson D.G. Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv. Mater. 2018;30:e1801151. doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Juanes M., Creese O., Fernández-Trillo P., Montenegro J. Messenger RNA delivery by hydrazone-activated polymers. Medchemcomm. 2019;10:1138–1144. doi: 10.1039/c9md00231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koh K.J., Liu Y., Lim S.H., Loh X.J., Kang L., Lim C.Y., Phua K.K.L. Formulation, characterization and evaluation of mRNA-loaded dissolvable polymeric microneedles (RNApatch) Sci. Rep. 2018;8:11842. doi: 10.1038/s41598-018-30290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao W., Zhang C., Li B., Zhang X., Luo X., Zeng C., Li W., Gao M., Dong Y. Lipid polymer hybrid nanomaterials for mRNA delivery. Cell. Mol. Bioeng. 2018;11:397–406. doi: 10.1007/s12195-018-0536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Steinle H., Ionescu T.M., Schenk S., Golombek S., Kunnakattu S.J., Özbek M.T., Schlensak C., Wendel H.P., Avci-Adali M. Incorporation of synthetic mRNA in injectable chitosan-alginate hybrid hydrogels for local and sustained expression of exogenous proteins in cells. Int. J. Mol. Sci. 2018;19:E1313. doi: 10.3390/ijms19051313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yin Y., Li X., Ma H., Zhang J., Yu D., Zhao R., Yu S., Nie G., Wang H. In situ transforming RNA nanovaccines from polyethylenimine functionalized graphene oxide hydrogel for durable cancer immunotherapy. Nano Lett. 2021;21:2224–2231. doi: 10.1021/acs.nanolett.0c05039. [DOI] [PubMed] [Google Scholar]
- 150.Kalkowski L., Golubczyk D., Kwiatkowska J., Holak P., Milewska K., Janowski M., Oliveira J.M., Walczak P., Malysz-Cymborska I. Two in one: use of divalent manganese ions as both cross-linking and MRI contrast agent for intrathecal injection of hydrogel-embedded stem cells. Pharmaceutics. 2021;13:1076. doi: 10.3390/pharmaceutics13071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/jvi.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Segel M., Lash B., Song J., Ladha A., Liu C.C., Jin X., Mekhedov S.L., Macrae R.K., Koonin E.V., Zhang F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science. 2021;373:882–889. doi: 10.1126/science.abg6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nagata S., Hamasaki T., Uetake K., Masuda H., Takagaki K., Oka N., Wada T., Ohgi T., Yano J. Synthesis and biological activity of artificial mRNA prepared with novel phosphorylating reagents. Nucleic Acids Res. 2010;38:7845–7857. doi: 10.1093/nar/gkq638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hamashima K., Kanai A. Alternative genetic code for amino acids and transfer RNA revisited. Biomol. Concepts. 2013;4:309–318. doi: 10.1515/bmc-2013-0002. [DOI] [PubMed] [Google Scholar]
- 155.Wang Y., Kathiresan V., Chen Y., Hu Y., Jiang W., Bai G., Liu G., Qin P.Z., Fang X. Posttranscriptional site-directed spin labeling of large RNAs with an unnatural base pair system under non-denaturing conditions. Chem. Sci. 2020;11:9655–9664. doi: 10.1039/d0sc01717e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Townshend R.J.L., Eismann S., Watkins A.M., Rangan R., Karelina M., Das R., Dror R.O. Geometric deep learning of RNA structure. Science. 2021;373:1047–1051. doi: 10.1126/science.abe5650. [DOI] [PMC free article] [PubMed] [Google Scholar]