Abstract
Motivation
An important step in the transcriptomic analysis of individual cells involves manually determining the cellular identities. To ease this labor-intensive annotation of cell-types, there has been a growing interest in automated cell annotation, which can be achieved by training classification algorithms on previously annotated datasets. Existing pipelines employ dataset integration methods to remove potential batch effects between source (annotated) and target (unannotated) datasets. However, the integration and classification steps are usually independent of each other and performed by different tools. We propose JIND (joint integration and discrimination for automated single-cell annotation), a neural-network-based framework for automated cell-type identification that performs integration in a space suitably chosen to facilitate cell classification. To account for batch effects, JIND performs a novel asymmetric alignment in which unseen cells are mapped onto the previously learned latent space, avoiding the need of retraining the classification model for new datasets. JIND also learns cell-type-specific confidence thresholds to identify cells that cannot be reliably classified.
Results
We show on several batched datasets that the joint approach to integration and classification of JIND outperforms in accuracy existing pipelines, and a smaller fraction of cells is rejected as unlabeled as a result of the cell-specific confidence thresholds. Moreover, we investigate cells misclassified by JIND and provide evidence suggesting that they could be due to outliers in the annotated datasets or errors in the original approach used for annotation of the target batch.
Availability and implementation
Implementation for JIND is available at https://github.com/mohit1997/JIND and the data underlying this article can be accessed at https://doi.org/10.5281/zenodo.6246322.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Recent developments in single-cell RNA sequencing (scRNA-seq) technologies have made it possible to profile the transcriptome of thousands of single cells in parallel. Massive amounts of single-cell RNA-seq data can now be generated enabling data-driven studies of gene expression at the single-cell resolution. An important step in single-cell genomic data analysis is the characterization of cell-types in a large mixture of cells. A typical pipeline involves a clustering algorithm to group cells with similar transcriptomic profiles, followed by manual labeling of the clusters based on appropriate biological markers identified in prior studies. However, the variability in clustering methods and the lack of standardized ontologies of cell labels creates a bottleneck in scalability of such methods (Abdelaal et al., 2019; Diaz-Mejia et al., 2019).
With increasing popularity of single-cell RNA sequencing and creation of large reference datasets (Regev et al., 2017; Schaum et al., 2018), supervised classification presents a natural framework for automating the cumbersome cell annotation process. Based on this idea, several methods have been proposed to transfer labels from an annotated scRNA-seq dataset (source batch) to an unannotated dataset (target batch) (Abdelaal et al., 2019; Alavi et al., 2018; Alquicira-Hernandez et al., 2019; Diaz-Mejia et al., 2019; Lopez et al., 2018; Ma and Pellegrini, 2019; Stuart et al., 2019).
One challenge associated with transferring cell annotations between datasets is the potential existence of batch effects that arise due to technological variability in the data collection process and sample preparation. In the presence of batch effects, batch integration techniques are utilized with the goal of removing any distributional differences (Johnson et al., 2007; Stuart et al., 2019) by transforming the source and target batches. This is followed by a supervised classification technique that allows cell-type transfer in the transformed space (Alquicira-Hernandez et al., 2019; Ma and Pellegrini, 2019; Stuart et al., 2019). There also exist methods which use neural networks to perform generative modeling of the single-cell gene expression data while accounting for batch effects and limited sensitivity (Lopez et al., 2018).
Our main insight is that cell-type information is not typically used in the batch integration step, while it could potentially inform the transformation function improving label-transfer accuracy. To this end, we propose a joint approach to batch integration and cell classification called JIND (joint integration and discrimination for automated single-cell annotation). The key idea behind JIND is to perform integration in a manner that is optimized for cell classification. For alignment, JIND utilizes ideas from the literature on Generative Adversarial Networks (GANs) (Goodfellow et al., 2014) and asymmetrically aligns the target batch to the source batch which remains fixed, in contrast to other symmetric approaches (Stuart et al., 2019). This allows faster inference on new target batches since the classifier training does not need to be performed again (refer to Methods).
In addition to the asymmetric batch integration, JIND estimates cell-type-specific confidence thresholds during training, which are then used to filter out low confidence prediction to reduce the misclassification rate. Finally, JIND allows the refinement of the parameters of the prediction model via self-training (Lee, 2013), by treating the high confidence predictions on the target batch as new labeled data. We refer to this extension as JIND+.
We show that JIND outperforms state-of-the-art methods on most datasets, achieving approximately 97% classification accuracy on average. We also show that the proposed thresholding scheme is robust to datasets of varying difficulties, rejecting only about 4% of cells, while state-of-the-art methods reject considerably higher proportions of cells on average. The misclassification rate can be further reduced with JIND+.
We point out that the benchmarking of JIND and other methods is performed on previously annotated batched datasets which may themselves contain incorrectly labeled cells. These original annotations are typically obtained using clustering-based approaches which are highly sensitive to noise and outliers. Since label transfer methods are trained and evaluated on these noisy annotations, it is expected that misclassification rates would be non-zero even for an optimal classifier. To investigate this point, we perform a careful analysis of the misclassifications made by JIND on the considered datasets. We observe that, for many of the misclassified cells, key marker genes of their ground-truth label are under-expressed, suggesting that these cells may not be unambiguously assigned to a specific cell-type, or that these are incorrectly annotated as the manual annotation process is prone to errors, something that JIND aims at solving.
Related work: Scanorama (Hie et al., 2019) is a single-cell data annotation tool that, to allow batch alignment through a simple subtraction operation, makes stringent assumptions on the nature of batch effects, such as orthogonality between technical variability and biological variability in the data. It has been shown that such stringent assumptions do not generalize well in the many cases where they are invalid (Tran et al., 2020). In contrast, JIND does not require such assumptions to perform the asymmetric alignment. ItClust is a cell-assignment tool that is inspired from the batch effect removal tool DESC (Li et al., 2020), and does not rely on prior integration of the source and target batches. ItClust uses a deep embedding network learned from the cell-type annotations of the source batch to perform iterative clustering on the target batch. While ItClust enables cell-type label transfer from the source to the target batch, it uses, unlike JIND, the target batch statistics for selecting genes prior to training, and is therefore not purely asymmetric in nature. Moreover, clustering techniques typically require large number of data points and hence this approach does not work well on small datasets. Finally, there are methods that, unlike JIND, utilize multiple annotated datasets (instead of one), such as scQuery (Alavi et al., 2018), which learns discriminative embeddings using contrastive learning, and MARS (Brbić et al., 2020), which learns a batch invariant classifier. However, in this work, we focus on the problem of transferring cell-type annotations from a single annotated source batch to a target batch where the cell-types need to be determined.
2 Materials and methods
2.1 JIND
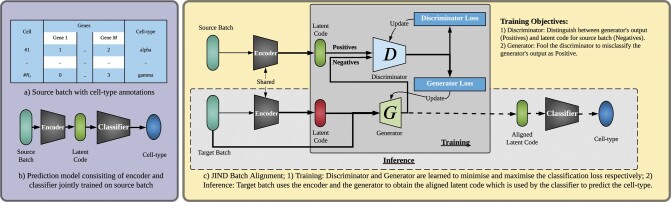
JIND tackles the problem of supervised transfer of cell-type annotations across single-cell RNA sequencing datasets. We denote the source batch (see Fig. 1a) used for training the prediction model by (), and the corresponding cell annotations by . We assume the cell annotations are represented as one-hot encoded K-dimensional vectors, where K is the number of cell-types in the source batch, such that . At the prediction step, we denote the gene expression matrix for the target batch by ().
Fig. 1.
Overview of JIND. (a) We assume access to a source batch containing the gene expression matrix accompanied with the corresponding cell-types. (b) A Neural Network-based prediction model, consisting of an encoder and a classifier, is trained on the source batch. The low-dimensional representation output by the encoder subnetwork is denoted as the latent code. Note that this prediction model should not be directly used to annotate the target batch due to batch effects. (c) JIND uses adversarial training via a generator and discriminator pair to align the source and target latent codes. The discriminator is trained to classify an input latent code either as a latent code produced by the generator (negative label) or as the source latent code produced by the encoder (positive label). In contrast, the generator is trained to fool the discriminator into misclassifying the generator’s output as source latent code. Finally, the output of the trained generator (the aligned latent code) is used by the classifier subnetwork to infer the cell-types of the target batch
2.1.1 Training stage
Prediction model: The prediction model used in JIND is based on Neural Networks (NNs) and consists of two subnetworks (Fig. 1b): (i) an encoder, which contains one hidden layer and (ii) a classifier consisting of one hidden layer followed by a softmax layer which outputs K probabilities (with K being the number of distinct cell-types in the source batch). The output of the encoder network is denoted as the latent code, and the output of the prediction model is a K-dimensional vector representing the probabilities of the cell belonging to each of the cell-types.
We denote the expression data for one cell by the vector x containing the expression for M genes (5000 by default); and the corresponding cell-type annotation, encoded as a one-hot encoded K-dimensional vector, by y. The network parameters are trained by minimizing the weighted categorical cross-entropy loss given by
| (1) |
where yk and denote the kth entry of the vectors y and , respectively, λ is a hyperparameter set to 2 by default, wk is a constant scalar determined as a function of the proportion of cells annotated as the kth type and ρk denotes the fraction of the kth cell-type in the dataset. A weighted loss is used to account for a potential class imbalance in the dataset and during computation ϵ (0.01 by default) is added to the denominator to avoid assignment of an exceedingly high weight to scarce cell-types. Note that we only train on 80% of the source batch and allocate the rest for validation (necessary for filtering).
Filtering: To minimize the misclassification rate, JIND uses K confidence thresholds, one per cell-type, denoted by τk, with . At inference time, before mapping a cell to the kth cell-type corresponding to the highest probability, we cross check whether the probability is greater than the corresponding threshold τk, resulting in an ‘unassigned’ label upon failure. The thresholds are determined as a part of the training process. We use the validation dataset to select a threshold for each cell-type based on an outlier fraction θ (set to 0.05 by default). Specifically, the threshold τk is the highest predicted probability of the bottom θ-quantile of the cells assigned to kth cell-type. These thresholds can be conveniently estimated at the inference time for any θ.
2.1.2 Inference stage
Below we describe how the trained prediction model is used to infer the cell-types for the target batch with a GAN-based alignment approach.
Asymmetric integration: JIND aligns the target batch onto the source batch in the latent space learned by the encoder subnetwork after training the NN-based prediction model (Fig. 1c). This alignment is attained by transforming the latent code obtained from the encoder subnetwork to the target batch so that it is indistinguishable from the latent code obtained from the source batch. Let the function learned by the encoder subnetwork be represented by F and the one learned by the classifier subnetwork by P. Thus, the predictions are produced as . We denote the corresponding hidden representations obtained from F (the latent code) by . We learn a function G(x, h) such that where (source batch), is indistinguishable from where (target batch). With such a G, we expect to produce accurate cell-type predictions.
Adversarial training: To learn the generator G, we use adversarial training where a discriminator function D parameterized by ΘD is trained to distinguish between , and (Supplementary Fig. S2). D is a NN-based classifier which estimates the probability of the input latent code coming from the source batch. Therefore, an ideal discriminator would produce and . Simultaneously, G is optimized to fool the discriminator into misinterpreting as h. The two models G and D are trained to learn parameters , by iteratively minimizing the corresponding generator and discriminator losses and given by,
| (2) |
| (3) |
While minimizing and , ΘD and ΘG are kept fixed, respectively. Note that, the generator’s objective function only depends on , whereas the discriminator’s objective function depends on both and h (Supplementary Fig. S2).
Final prediction model: To infer cell-types for target batch, the encoder subnetwork takes as input the cell’s gene expression vector x and produces the latent code h. Both x and h are then input to the generator, which produces the corrected latent code . Finally, the corrected latent code goes through the classifier subnetwork, which produces the final probabilities of the cell belonging to each of the considered K cell-types (Fig. 1c).
Finally, an extension to the JIND framework based on self-training (Lee, 2013), termed JIND+, is proposed. In JIND+, additionally, the confident predictions made on the target batch by JIND are used to further fine-tune the parameters of the encoder subnetwork. For a detailed description of the neural architectures used by JIND/JIND+ and relevant training details, refer to the Supplementary Section S1.1.
2.2 Datasets
To assess the performance of the proposed method JIND, we consider eight scRNA-seq datasets (Supplementary Table S1). Specifically, we consider the following datasets used for creating batched experiments: (i) PBMC (Peripheral Blood Mononuclear Cells) dataset (Park et al., 2018) ‘10x_v3’ and PBMC ‘10x_v5’ which differ in the type of assay used during library preparation; and (ii) a collection of three different human pancreas datasets, namely Pancreas ‘Bar16’ (Baron et al., 2016), Pancreas ‘Mur16’ (Muraro et al., 2016) and Pancreas ‘Seg16’ (Segerstolpe et al., 2016). We create three pairs of datasets for our experiments on transferring labels across different batches, (i) PBMC 10x_v3-10x_v5, (ii) Pancreas Bar16-Mur16 and (iii) Pancreas Bar16-Seg16, in each of which the source (10x_v3, Bar16, Bar16) and target (10x_v5, Mur16, Seg16) batches are collected under different settings or are obtained from separate studies. These three pancreas datasets were collected using different protocols and hence exhibit significant technical variability. As such, they are widely used to benchmark batch correction methods (Abdelaal et al., 2019). We also consider three datasets for experiments on transferring labels across datasets without batch effects: (i) Human Hematopoiesis dataset (Granja et al., 2019) (referred to as Human-Hemato) (GEO Accession ID GSE139369) with 26 annotated cell-types collected from Human blood; (ii) Mouse Cortex (Zeisel et al., 2015) (GEO Accession ID GSE60361) with 7 annotated cell-types collected from the mouse brain cortex and (iii) the Mouse Cell Atlas (Schaum et al., 2018) dataset with 13 annotated cell-types and more than 250 thousand cells.
2.3 Evaluation metrics
Cell identification methods are commonly benchmarked based on the classification accuracy on the target batch (Boufea et al., 2020; Kiselev et al., 2018; Ma and Pellegrini, 2019). However, most current methods incorporate an option to reject cells with low-confidence predictions. Thus, it becomes essential to also quantify the proportion of rejected cells, together with the accuracy on the remaining ones. In this context, we use ‘raw accuracy’ (raw) to refer to the accuracy of the classifier on the target batch prior to any rejection; ‘rejection rate’ (rej) to refer to the percentage of cells rejected by the classification method; and ‘effective accuracy’ (eff) to refer to the accuracy of the classifier on the cells that were not rejected by the method. We also provide three more metrics to better compare different methods: ‘weighted F1 score’ (wf1), which refers to F1 scores averaged across cell-types weighted by their respective proportions, ‘mean F1 score’ (mf1), which is the average F1 score across cell-types, and ‘median F1 score’ (medf1), which is the median F1 score among cell-types.
2.4 Experimental setup
We compare JIND and JIND+ with SVM (Abdelaal et al., 2019), scPred (Alquicira-Hernandez et al., 2019), Seurat-LT (Seurat Label Transfer) (Stuart et al., 2019), ItClust (Hu et al., 2020) and ACTINN (Ma and Pellegrini, 2019). In a recent study (Abdelaal et al., 2019), SVMRej was shown to perform better than most existing automated cell identification methods, including methods incorporating prior knowledge in the form of marker genes. In addition, ACTINN and scPred were among the best performing methods. Seurat-LT was chosen as it is widely used in practice. We also compare with ItClust, since it is the most recently proposed method to the best of our knowledge. In addition, both Seurat-LT and ItClust capture batch effects and do not rely on external tools to align or integrate source and target batches, unlike the other considered methods.
SVMRej uses a linear Support Vector Machine (SVM) classifier followed by probability calibration using Platt’s method, and sets a confidence threshold of 0.7 to reject cells. scPred also uses a linear SVM classifier but uses only the first few PCA components as input features. scPred uses a confidence threshold of 0.9 for rejecting low confidence predictions. Seurat-LT projects the PCA structure of the source batch onto the target batch so as to transfer labels (Stuart et al., 2019). ItClust, on the other hand, learns on the source batch a NN-based encoder by means of a clustering objective. This encoder is then optimized to improve clustering on the target batch, thereby transferring the cell-type labels. We note that ItClust filters out cells based on their gene expression profile, which are then not considered for classification. For a fair comparison with the other methods, in the conducted experiments we skip the filtering of cells in ItClust. Interestingly, skipping this step yields the same or an improved accuracy on all considered datasets (Supplementary Table S5). Also, while ItClust is not designed to be run on preprocessed datasets, due to unavailability of raw data for Mouse Atlas and PBMC datasets, we modify the preprocessing step of ItClust for these two datasets (see Supplementary Section S3). Since Seurat-LT and ItClust do not use any rejection scheme, rej and eff metrics are not reported for these two methods. Finally, ACTINN uses a three hidden layer NN as the base classifier. As done with scPred, we use the confidence threshold of 0.9 for rejecting ACTINN predictions.
For datasets lacking batch effects, we randomly split the dataset into a 7:3 ratio to generate source and target batches. For the datasets containing batches, we specify the source and the target batch. For the Pancreas-based benchmark experiments, while generating the source and target batches we only retained cells whose cell-type is present in both batches as done in Abdelaal et al. (2019).
3 Results
3.1 JIND can accurately annotate scRNA-seq datasets with batch effects
To assess the classification performance of JIND on transferring labels across datasets with batch effects, we consider: PBMC 10x_v3-10x_v5, Pancreas Bar16-Mur16 and Pancreas Bar16-Seg16, where the task is to transfer labels from the source batch to the target batch. On these paired datasets, we compare JIND, JIND+, SVMRej, Seurat-LT, ItClust, scPred and ACTINN and report respective raw accuracy, rejection rate, effective accuracy after rejection and weighted F1 score.
First, we observe that JIND+ consistently achieves better or similar performance than JIND on all datasets (Table 1a). Moreover, JIND+ reduces the rejection rates of JIND by a factor of 2 in most cases while maintaining the effective accuracy. We also observe that JIND and JIND+ consistently outperform previously proposed methods in both raw accuracy and weighted F1 score on all datasets, except for the PBMC datasets in which JIND and JIND+ perform competitively to the best performing methods. Nonetheless, on Pancreas datasets, JIND+ outperforms Seurat-LT by 9% on average. In comparison to ItClust, JIND and JIND+ achieve similar accuracy on PBMC dataset; while on the Pancreas datasets, JIND and JIND+ outperform ItClust by around 2% in raw accuracy. For comparison with SVMRej, scPred and ACTINN, which do not internally perform any batch alignment, we report their performance after aligning source and target batches using two integration techniques, Seurat integration (Stuart et al., 2019) and Harmony (Korsunsky et al., 2019) integration (refer to Table 1a). On the raw Pancreas datasets, we observe that Harmony integration works better than Seurat integration for SVMRej and ACTINN, in contrast to scPred, which always performs better with Seurat integration. On all three datasets, we observe that JIND+ performs on average 1–2% better than ACTINN, which outperforms both SVMRej and scPred. For PBMC dataset, we observe that Seurat integration works better than Harmony integration, since for all three classifiers SVMRej, scPred and ACTINN, better classification accuracy was obtained. This behavior can also be attributed to the fact that PBMC dataset is already preprocessed and Harmony expects raw counts.
Table 1.
Benchmarking: tabular comparison for (a) batched datasets and (b) non-batched datasets based on four metrics: raw the initial accuracy of the classifier, rej the percentage of cells rejected by the classifier (if supported by the method), eff is the effective accuracy after rejecting unconfident predictions and wf1 the weighted F1 score based on the predicted probabilities
|
(a) Batched datasets | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Datasets | Metrics | JIND | JIND+ | Seurat-LT | ItClust | SVMRej |
scPred |
ACTINN |
|||
| HInt | SInt | HInt | SInt | HInt | SInt | ||||||
| Pancreas Bar16-Mur16 | raw | 0.963 | 0.963 | 0.870 | 0.945 | 0.959 | 0.932 | 0.856 | 0.914 | 0.955 | 0.932 |
| rej | 0.05 | 0.02 | — | — | 0.07 | 0.02 | 0.39 | 0.08 | 0.05 | 0.05 | |
| eff | 0.979 | 0.971 | — | — | 0.986 | 0.944 | 0.945 | 0.942 | 0.976 | 0.954 | |
| wf1 | 0.964 | 0.965 | 0.853 | 0.944 | 0.960 | 0.933 | 0.834 | 0.914 | 0.955 | 0.933 | |
| Pancreas Bar16-Seg16 | raw | 0.995 | 0.997 | 0.929 | 0.978 | 0.970 | 0.964 | 0.726 | 0.945 | 0.976 | 0.963 |
| rej | 0.05 | 0.02 | — | — | 0.10 | 0.02 | 0.43 | 0.11 | 0.06 | 0.04 | |
| eff | 1.000 | 1.000 | — | — | 0.981 | 0.973 | 0.906 | 0.971 | 0.992 | 0.982 | |
| wf1 | 0.995 | 0.997 | 0.915 | 0.977 | 0.968 | 0.962 | 0.661 | 0.942 | 0.975 | 0.960 | |
| PBMC 10x_v3-10x_v5 | raw | 0.966 | 0.973 | 0.981 | 0.969a | 0.939 | 0.962 | 0.341 | 0.946 | 0.943 | 0.965 |
| rej | 0.08 | 0.06 | — | — | 0.99 | 0.05 | 0.49 | 0.10 | 0.49 | 0.05 | |
| eff | 0.968 | 0.984 | — | — | 1.000 | 0.975 | 0.545 | 0.971 | 0.990 | 0.980 | |
| wf1 | 0.966 | 0.972 | 0.980 | 0.968 | 0.940 | 0.962 | 0.223 | 0.942 | 0.943 | 0.964 | |
|
(b) Unbatched datasets | ||||||||
|---|---|---|---|---|---|---|---|---|
| Datasets | Metrics | JIND | JIND+ | Seurat-LT | ItClust | SVMRej | scPred | ACTINN |
| Human-Hemato | raw | 0.929 | 0.928 | 0.897 | 0.905 | 0.904 | 0.796 | 0.922 |
| rej | 0.06 | 0.03 | — | — | 0.24 | 0.54 | 0.18 | |
| eff | 0.948 | 0.941 | — | — | 0.966 | 0.952 | 0.974 | |
| wf1 | 0.929 | 0.928 | 0.89 | 0.905 | 0.905 | 0.809 | 0.921 | |
| Mouse Cortex | raw | 0.973 | 0.977 | 0.957 | 0.910 | 0.976 | 0.969 | 0.969 |
| rej | 0.07 | 0.04 | — | — | 0.05 | 0.12 | 0.04 | |
| eff | 0.989 | 0.988 | — | — | 0.997 | 0.996 | 0.986 | |
| wf1 | 0.973 | 0.977 | 0.957 | 0.911 | 0.969 | 0.969 | 0.969 | |
| Mouse Atlas | raw | 0.983 | 0.981 | 0.958 | 0.984 a | 0.977 | — | 0.984 |
| rej | 0.04 | 0.04 | — | — | 0.05 | — | 0.07 | |
| eff | 0.992 | 0.988 | — | — | 0.994 | — | 0.998 | |
| wf1 | 0.983 | 0.981 | 0.957 | 0.984 | 0.977 | — | 0.984 | |
Note: On batched datasets, for SVMRej, scPred and ACTINN, we report results with batch alignment prior to classification using using Seurat (SInt) and Harmony (HInt). Best raw accuracy rates are bold faced and rejection rates above 0.1 are colored red.
Since ItClust is designed to run on raw datasets and the Mouse Atlas and PBMC datasets are already processed, we modified the preprocessing step in ItClust to annotate these datasets.
To further compare the classification performance of JIND+ against Seurat-LT and ItClust, we also provide confusion matrices for these methods on the conduced experiments (Supplementary Figs S3–S5). We observe that for the Pancreas experiments, which exhibit prominent batch effects between source and target batch, JIND+ without rejection provides better classification performance than Seurat-LT and ItClust, while JIND+ with rejection improves upon JIND, Seurat-LT and ItClust on all datasets. Moreover, upon visualizing the cells filtered by JIND+, we clearly observe that filtered cells were either outliers or originally misclassified by JIND+ (see Supplementary Section S2.3). We also demonstrate robustness of JIND and JIND+ by running them with five different intializations, and showing that both achieve consistent classification performance (Supplementary Table S2). We note that JIND and JIND+ use the same neural network architecture for all datasets unlike ItClust, which selects from a predefined set of different neural network architectures depending on the number of cells in the dataset. For completeness, we also provide mean and median F1 scores for all the considered methods (Supplementary Table S3).
Furthermore, we also run JIND and JIND+ after Seurat and Harmony integration and observe that the classification performance slightly degrades in most cases (Supplementary Table S4). These results demonstrate the benefit of JIND, which does not perform alignment and classification as two independent steps.
3.2 JIND achieves low rejection rates with high accuracy on non-batched datasets
While the main benefit of JIND is in the presence of batch effects between the source and target batches, we also perform an assessment on non-batched data. For the analysis, we consider the three datasets Human-Hemato, Mouse Cortex and Mouse Atlas. On these datasets which do not exhibit discerning batch effects, JIND and JIND+ achieve a rejection rate of less than 8% and 4% for all tested datasets, respectively, while maintaining an effective accuracy ranging from 0.94 to 0.99 (Table 1b). In addition, we consistently obtain competitive performance on all three datasets while previously proposed methods reject a varying proportion of cells. Specifically, on Human-Hemato dataset, SVMRej rejects 24% of cells, scPred 54% and ACTINN 18% (Table 1b). In comparison, on the other two datasets, Mouse Cortex and Mouse Atlas, these methods reject a much lower fraction of cells (between 5% and 12%). As we discuss below (see next section), this is likely due to Human-Hemato containing a significantly larger number of distinct cell-types. We note that we were unable to run scPred on the Mouse Atlas dataset (due to large dataset size).
While there is a natural trade-off between rejection rate and effective accuracy on the filtered cells, JIND provides an efficient way of controlling the rejection rate. Specifically, the outlier fraction specified during inference to estimate the cell-type specific thresholds (set to 5% by default, see Supplementary Section S1.1) coincides approximately with the percentage of rejected cells from the target batch, which is on average 6% in all tested datasets (this also applies to batched data). This percentage is further reduced to 3.3% with JIND+.
Lastly, JIND+ achieves approximately an average raw accuracy and weighted F1 score that is roughly similar to ACTINN and SVMRej. In comparison to scPred, the performance of JIND+ is 13% higher on Human-Hemato dataset and 1% higher on Mouse Cortex. In comparison to Seurat-LT, JIND+ achieves a raw accuracy which is on average 3–4% higher on all datasets. When compared with ItClust, JIND+ achieves 2% and 6% percent higher raw accuracy on the Human-Hemato and Mouse Cortex datasets, respectively; while we observe similar performance on the Mouse Atlas dataset. We note that on the Mouse Cortex dataset, ItClust’s performance drops as clustering the Mouse Cortex dataset is harder since it contains fewer number of cells (see Methods). We also provide the confusion matrices obtained in each case (Supplementary Figs S6–S8). Similarly, we also provide mean and median F1 scores for all the considered methods (Supplementary Table S3).
Since JIND+ either outperforms JIND or achieves similar performance, henceforth, we focus mainly on the performance of JIND+.
3.3 JIND+ misclassified cells differentially express biomarker genes associated to their originally annotated cell-type
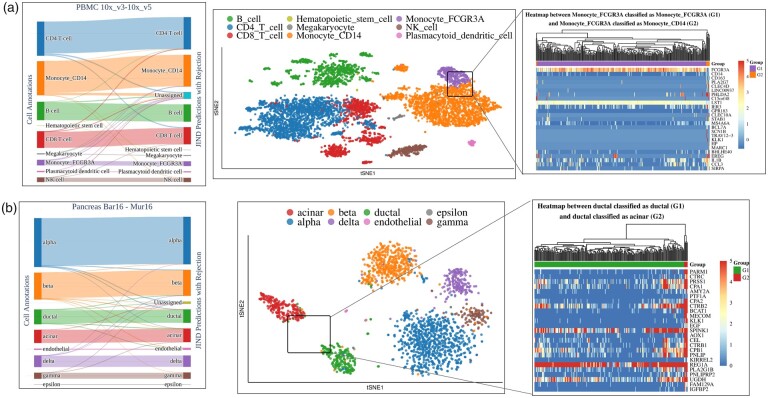
To better understand the misclassifications made by JIND+, we further analyze the results obtained on PBMC 10x_v3-10x_v5 and Pancreas Bar16-Mur16 experiments. On the PBMC 10x_v3-10x_v5 experiment, we observe that JIND+ misclassifies approximately 1.6% of cells after rejection (Fig. 2a-left). To identify which cell-types can result in misclassifications due to cluster overlaps, we visualize the target batch (PBMC 10x_v5) using tSNE dimensionality reduction (Fig. 2a-middle). We observe that two subpopulations of Monocytes, namely, Monocyte FCGR3A and Monocyte CD14, lie close to each other with a noticeable overlap in the tSNE-reduced space. Since some of the Monocyte FCGR3A cells are misclassified by JIND+ as Monocyte CD14, we conduct a differential expression (DE) analysis using Limma (Law et al., 2014) between the misclassified Monocyte FCGR3A cells (Monocyte FCGR3A predicted as Monocyte CD14) and the correctly classified Monocyte FCGR3A cells (Monocyte FCGR3A predicted as Monocyte FCGR3A).
Fig. 2.
Performance evaluation and differential expression analysis on two datasets. The alluvial plots (left) reflect the performance of JIND+ on (a) PBMC 10x_v3-10x_v5 and (b) Pancreas Bar16-Mur16 datasets. The tSNE plots (middle) illustrate the cell-type clusters of the target batch, and highlight the two cell-types with the highest misclassification rates: (a) Monocyte_FCGR3A and Monocyte_CD14 and (b) Acinar and Ductal. The heatmaps (right) show the top 25 differentially expressed genes between (a) Monocyte_FCGR3A cells classified as Monocyte_FCGR3A (G1) and Monocyte_FCGR3A classified as Monocyte_CD14 (G2), and between (b) Ductal cells classified as Ductal (G1) and Ductal cells classified as Acinar (G2). The shown hierarchical clustering is performed using all the differentially expressed genes
We identify 90 significantly differentially expressed genes (FDR , where FDR refers to P-values with false discovery rate correction) between correctly predicted Monocyte FCGR3A cells and Monocyte FCGR3A cells predicted as Monocyte CD14 (Fig. 2a-right, Supplementary Fig. S9 and Supplementary Excel File S1). We observe that the positive biomarker gene FCGR3A for cell-type Monocyte FCGR3A (Sampath et al., 2018) is clearly overexpressed in the group of cells classified by JIND+ as Monocyte FCGR3A but underexpressed on the misclassified cells. Interestingly, the CD14 gene (a positive biomarker gene for Monocyte CD14 (Sampath et al., 2018)), is differentially expressed between the two groups, although we do not observe an overexpression of this gene on the cells classified as Monocyte CD14. This suggests that the cells misclassified by JIND+ are actually outliers (or possibly mislabeled in the original dataset), thus being intrinsically difficult to classify. Therefore, these misclassifications do not correspond to arbitrary mistakes made by the prediction model and rather suggest potential mislabeling in the cell-type annotations. We also conduct a similar DE analysis on the PBMC 10x_v3 dataset (source batch) between the cells annotated as Monocyte FCGR3A cells and as Monocyte CD14 cells. We observe that the biomarker gene FCGR3A is almost never underexpressed on any of the cells annotated as Monocyte FCGR3A (Supplementary Fig. S10 and Supplementary Excel File S2). However, most of the Monocyte FCGR3A cells misclassified as Monocyte CD14 by JIND+ in the target batch underexpress this gene (Fig. 2a-right). Regarding gene CD14, we observe that on cells annotated as Monocyte CD14 it is almost always overexpressed. Nevertheless, we observe a group of Monocyte CD14 cells for which both FCGR3A and CD14 genes are underexpressed, similarly to the Monocyte FCGR3A cells misclassified by JIND+ as Monocyte CD14 on the target batch.
We perform a similar analysis on the Pancreas Bar16-Mur16 dataset. We observe that JIND+ misclassifies roughly 3% of cells from the Pancreas Mur16 dataset after rejection (Fig. 2b-left). We again visualize the target batch using tSNE dimensionality reduction to identify cells that are hard to classify. Interestingly, JIND+ misclassifies about 3% of the Ductal cells as Acinar, even though the clusters do not overlap in the tSNE space (Fig. 2b-middle). On close observation, we find that some of the Ductal cells actually lie closer to the Acinar cluster centroid than the Ductal centroid. Therefore, we perform a DE analysis between the misclassified Ductal cells (Ductal predicted as Acinar) and correctly classified Ductal cells (Ductal predicted as Ductal). The analysis reveals that 237 genes are differentially expressed (FDR ), among which we also find biomarkers for the two cell-types (Fig. 2b-right, Supplementary Fig. S11 and Supplementary Excel File S3). Specifically, PRSS1, CPA1 PARM1 and CTRC, positive biomarker genes (Baldan et al., 2019; Franzén et al., 2019) for Acinar cell-type, are significantly overexpressed on the misclassified group. These findings suggest that the Ductal cells classified as Acinar are most likely Acinar cells.
We note that the total number of misclassifications in each of the discussed experiments is less than 50. Hence, to assess the statistical significance of our DE findings, we conduct a DE analysis between two randomly selected subsets of Monocyte FCGR3A cells, and two randomly selected subset of Ductal cells. For a fair assessment, we set the sizes of the randomly selected subsets to match those of the previous DE analyses, and sample only cells which were not rejected by JIND+. We observe that any two random subsets of cells typically do not exhibit differentially expressed genes. Specifically, for different random selections, we never observed differentially expressed biomarker genes for both PBMC 10x_v3-10x_v5 and Pancreas Bar16-Mur16 (Supplementary Excel File S4 for PBMC 10x_v3-10x_v5, and Supplementary Excel File S5 for Pancreas Bar16-Mur16). Hence, we conclude that some misclassifications made by JIND+ are explainable based on the differential expression of biomarker genes and hence, are likely due to ambiguities in the cell-type annotations. Finally, we also visualize the location of the misclassified cells in the tSNE reduced space, along with the correctly classified ones, for the considered datasets (Supplementary Figs S12 and S13). This analysis reveals that most misclassified cells are located at the boundary of clusters containing correctly classified cells. Moreover, in some cases, these misclassified cells are closer in the tSNE reduced space to the cluster containing cells of the inferred cell-type, further indicating potential mistakes in the cell-type annotations.
3.4 JIND+ identifies transitioning cells
To further validate the claim on potential mislabeling of cells, we conduct an additional experiment on the Pancreas Bar16-Mur16 dataset. We identify 11 (originally annotated) Alpha cells that are labeled as Beta cells by JIND but then rejected (i.e. they did not satisfy the confident threshold). Upon performing a differential expression analysis between these cells and the Alpha cells correctly classified by JIND+, the gene MAFA is among the top six differentially expressed genes (Supplementary Excel File S6). MAFA is a gene-marker for the Beta cells (Hang and Stein, 2011), indicating that the ‘Unassigned’ group of cells may be Alpha cells transitioning toward Beta cells via the transdifferentiation process (Supplementary Fig. S14).
We repeat the same analysis for scPred and SVMRej, by first integrating the source and target batches with Harmony, and observe that both scPred and SVMRej fail to identify the transitioning cells. The resulting differentially expressed genes for SVMRej are shown in Supplementary Figure S14. For scPred, we did not find any differentially expressed genes between the two considered cell groups.
3.5 Extreme scenarios and runtime
We also experimented with JIND+ on datasets where the source and target batches contained different number of cell-types and we observed that JIND can correctly learn the mapping even when the target batch is missing some cell-types. In cases where the target batch contained a novel cell-type, JIND alignment could still meaningfully learn the right mapping between the datasets (see Supplementary Section S2.4).
In terms of running time, since JIND+ supports CPU and GPU-based parallelization, we observed that JIND is similar in speed to Seurat-LT and 2–6 times faster than ItClust (see Supplementary Section S2.5).
4 Conclusion
In this work, we introduced JIND, an automated cell-type identification tool that utilizes annotated scRNA-seq datasets to reliably annotate unseen sequenced data. Depending on the differences in sequencing protocols or library preparation, there typically exists significant technical variability or batch effects between these datasets, which confounds real biological variability. To transfer cell-type labels while accounting for batch effects, JIND uses supervised learning to learn a latent space suited to classification along with a classifier in this space. Then, JIND utilizes adversarial training to map the unannotated dataset in the learned latent space, which allows inferring cell-types using the classifier trained on the annotated dataset. Since the mapping is learned after training the classifier on the annotated data, any unseen dataset can be annotated directly without the need of retraining the prediction (classification) model. This is in contrast to other cell-type identification methods that rely on symmetric batch integration tools and hence require training the prediction model after performing batch alignment. Moreover, JIND performs alignment in a low-dimensional latent space that explicitly maximizes separation between cell-types. Unlike PCA-based tools, which perform uninformed dimensionality reduction, JIND exploits the cell-type information for projection into the latent space which is suited to the downstream task of classification. JIND also incorporates a robust rejection scheme which filters out low-confidence predictions allowing rejection of cells in ambiguous states or that have highly noisy gene expressions. Finally, we presented JIND+, and extension of JIND that uses the confident predictions made on the unseen data to further fine-tune the parameters of the prediction model.
We demonstrated that both JIND and JIND+ achieve higher accuracy on cell-type identification for datasets containing batch effects as compared to existing state-of-the-art methods. This is accomplished while maintaining a constant rejection rate (about 5%) which can be easily controlled by the user. Upon thorough investigation, we further show that the misclassifications made by JIND+ can be explained by differentially expressed biomarker genes, suggesting potential ambiguity in the cell-type annotations. We also showed that many cells rejected by JIND+ generally correspond to misclassified cells, improving the effective classification accuracy by reducing the misclassification rate. In conclusion, the observed improvements in the performance on cross-batch annotation demonstrate that JIND is highly effective at transferring cell-type labels across datasets, while being computationally fast and scalable to large datasets.
Financial Support: Idoia Ochoa was supported by Ramon y Cajal Grant from the Spanish Government, Gipuzkoa Fellows Grant from the Basque Government. Mikel Hernaez was supported by H2020 Marie S. Curie IF Action, European Commission under Grant Agreement No. 898356. The work of Ilan Shomorony was supported in part by the National Science Foundation under Grant CCF-2046991.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Mohit Goyal, Electrical and Computer Engineering Department, University of Illinois, Urbana, IL 61801, USA.
Guillermo Serrano, Computational Biology Program, Center for Applied Medical Research (CIMA), University of Navarra, Pamplona 31008, Spain.
Josepmaria Argemi, Center for Liver Diseases, Pittsburgh Liver Research Center, Division of Gastroenterology, Hepatology and Nutrition, University of Pittsburgh Medical Center, Pittsburgh, PA 15213, USA; Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas, Madrid 28029, Spain; Liver Unit, Clinica Universitaria de Navarra, Pamplona 31008, Spain; Hepatology Program, Center for Applied Medical Research (CIMA) Universidad de Navarra, Pamplona 31008, Spain.
Ilan Shomorony, Electrical and Computer Engineering Department, University of Illinois, Urbana, IL 61801, USA.
Mikel Hernaez, Computational Biology Program, Center for Applied Medical Research (CIMA), University of Navarra, Pamplona 31008, Spain; Carl R. Woese Institute for Genomic Biology, University of Illinois, Urbana, IL 61801, USA; Artificial Intelligence and Data Science Institute (DATAI), University of Navarra, Pamplona 31008, Spain.
Idoia Ochoa, Electrical and Computer Engineering Department, University of Illinois, Urbana, IL 61801, USA; Artificial Intelligence and Data Science Institute (DATAI), University of Navarra, Pamplona 31008, Spain; Department of Electrical Engineering, Tecnun, University of Navarra, Donostia 20018, Spain.
References
- Abdelaal T. et al. (2019) A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol., 20, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi A. et al. (2018) A web server for comparative analysis of single-cell RNA-seq data. Nat. Commun., 9, 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquicira-Hernandez J. et al. (2019) scpred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol., 20, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldan J. et al. (2019) Adult human pancreatic acinar cells dedifferentiate into an embryonic progenitor-like state in 3D suspension culture. Sci. Rep., 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. et al. (2016) A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst., 3, 346–360.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boufea K. et al. (2020) SCID: identification of equivalent transcriptional cell populations across single cell RNA-seq data using discriminant analysis. IScience, 23(3), 100914. [DOI] [PMC free article] [PubMed]
- Brbić M. et al. (2020) Mars: discovering novel cell types across heterogeneous single-cell experiments. Nat. Methods, 17, 1200–1206. [DOI] [PubMed] [Google Scholar]
- Diaz-Mejia J.J. et al. (2019) Evaluation of methods to assign cell type labels to cell clusters from single-cell RNA-sequencing data. F1000Research, 8, 296–31508207. ISCB Comm J–296. [pmid]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O. et al. (2019) PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I.J. et al. (2014) Generative Adversarial Networks. Adv. Neural Inf. Process. Syst., 27.
- Granja J.M. et al. (2019) Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol., 37, 1458–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang Y., Stein R. (2011) Mafa and mafb activity in pancreatic β cells. Trends Endocrinol. Metab., 22, 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hie B. et al. (2019) Efficient integration of heterogeneous single-cell transcriptomes using scanorama. Nat. Biotechnol., 37, 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. et al. (2020) Iterative transfer learning with neural network for clustering and cell type classification in single-cell RNA-seq analysis. Nat. Mach. Intell., 2, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E. et al. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Kiselev V.Y. et al. (2018) scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods, 15, 359–362. [DOI] [PubMed] [Google Scholar]
- Korsunsky I. et al. (2019) Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods, 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W. et al. (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-H. (2013) Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks. In: ICML 2013 Workshop: Challenges in Representation Learning (WREPL), Atlanta, Vol. 3, 2nd edn., p. 896.
- Li X. et al. (2020) Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun., 11, 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R. et al. (2018) Deep generative modeling for single-cell transcriptomics. Nat. Methods, 15, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Pellegrini M. (2019) Automated identification of cell types in single cell RNA sequencing. bioRxiv. [DOI] [PubMed]
- Muraro M.J. et al. (2016) A single-cell transcriptome atlas of the human pancreas. Cell Syst., 3, 385–394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E. et al. (2018) Fast batch alignment of single cell transcriptomes unifies multiple mouse cell atlases into an integrated landscape. bioRxiv.
- Regev A. et al. (2017) Science forum: the human cell atlas. eLife, 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P. et al. (2018) Monocyte subsets: phenotypes and function in tuberculosis infection. Front. Immunol., 9, 1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N. et al. (2018) Single-cell transcriptomics of 20 mouse organs creates a Tabula muris. Nature, 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe Å. et al. (2016) Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab., 24, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T. et al. (2019) Comprehensive integration of single-cell data. Cell, 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.T.N. et al. (2020) A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol., 21, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A. et al. (2015) Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.