Abstract
SARS-CoV-2 has caused a global pandemic that has infected more than 250 million people worldwide. Although several vaccine candidates have received emergency use authorization, there is still limited knowledge on how vaccine dosing affects immune responses. We performed mechanistic studies in mice to understand how the priming dose of an adenovirus-based SARS-CoV-2 vaccine affects long-term immunity to SARS-CoV-2. We first primed C57BL/6 mice with an adenovirus serotype 5 vaccine encoding the SARS-CoV-2 spike protein, similar to that used in the CanSino and Sputnik V vaccines. The vaccine prime was administered at either a standard dose or 1000-fold lower dose, followed by a boost with the standard dose 4 weeks later. Initially, the low dose prime induced lower immune responses relative to the standard dose prime. However, the low dose prime elicited immune responses that were qualitatively superior and, upon boosting, exhibited substantially more potent recall and functional capacity. We also report similar effects with a simian immunodeficiency virus (SIV) vaccine. These findings show an unexpected advantage of fractionating vaccine prime doses, warranting a reevaluation of vaccine trial protocols for SARS-CoV-2 and other pathogens.
INTRODUCTION
Adenoviruses are widely used in vaccine research due to their high insert capacity and robust immunogenicity (1). In particular, adenovirus serotype 5 (Ad5) is among the most immunogenic vaccine vectors available, but over the past decade, its clinical use has been hampered by its high seroprevalence in the human population (2–4). Recently, the coronavirus disease 2019 (COVID-19) pandemic has reinvigorated the clinical use of this widely characterized vector as a vaccine platform for preventing COVID-19.
Vaccines based on Ad5, such as the CanSino vaccine and the Sputnik V vaccine, as well as vaccines based on adenovirus serotype 26 (Ad26) and chimpanzee adenovirus (ChAdOx1), have been used in millions of people worldwide and have shown potent protection against severe COVID-19 disease (5–7). However, there are current attempts to enhance the overall magnitude and durability of immune responses, for example, by altering vaccine dosing and boosting regimens. Optimization of prime-boost regimens may obviate the need for “third boosters,” which are now recommended for people over 65 years old, as well as patients at high risk of severe illness, including immunosuppressed individuals (8, 9). The waning of immunity several months after vaccination is associated with increase in the number of breakthrough infections (10), leading experts to consider additional boosters for the general population. Before licensure, the clinical testing of vaccine candidates consists of three phases aimed at evaluating safety, immunogenicity, and efficacy. In particular, phase 1 typically involves dose-escalation studies comparing a range of vaccine doses in people who receive the same dose of the vaccine during the prime and the boost. However, vaccine trials do not typically evaluate “intragroup dose escalation,” in which individuals would first receive a prime with a low dose (LD) and then a boost with a higher dose. Here, we performed studies in mice to determine the immunological effects of intragroup dose escalation. We first primed mice with an LD or a standard dose (SD) of an Ad5-based COVID-19 vaccine, followed by homologous boosting with an SD to measure anamnestic immune responses. Our data show that limiting the priming dose of Ad5-vectored vaccines improves the boosting capacity of CD8+ T cell responses and antibody responses. This beneficial effect of limiting the priming dose was associated with reduced generation of vector-specific immune responses and cell-intrinsic differences in memory T cell differentiation.
RESULTS
Low-dose Ad5-based SARS-CoV-2 vaccine prime elicits T cells with high anamnestic capacity
We first primed C57BL/6 mice intramuscularly with an Ad5 adenovirus vector expressing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein [Ad5-SARS-CoV-2 spike (11)] at either an LD [106 plaque-forming units (PFU)] or an SD (109 PFU). After 4 weeks, mice were boosted with the SD and CD8+ T cell responses were analyzed by major histocompatibility complex (MHC) tetramer binding assays (Fig. 1A). We tracked a CD8+ T cell response against an epitope (VNFNFNGL) that is highly conserved in bat and SARS-like coronaviruses, including SARS-CoV-1, SARS-CoV-2, RatG13, HKU3, WIV1, WIV16, RsSHC014, Rs3367, Shaanxi2011, Rm1/2004, YN2018B, SC2018, HuB2013, GX2013, and BM48–31/BGR/2008/Yunnan2011, among other coronaviruses. We will refer to this conserved CD8+ T cell response as SARS-CoV-2–specific response, or Kb VL8 for simplicity, where V represents the first amino acid, L represents the last amino acid, and 8 represents the number of amino acids in the peptide sequence.
Fig. 1. A low-dose Ad5-SARS-CoV-2 spike vaccine prime elicits more SARS-CoV-2 CD8+ T cells after boosting compared with an SD prime.
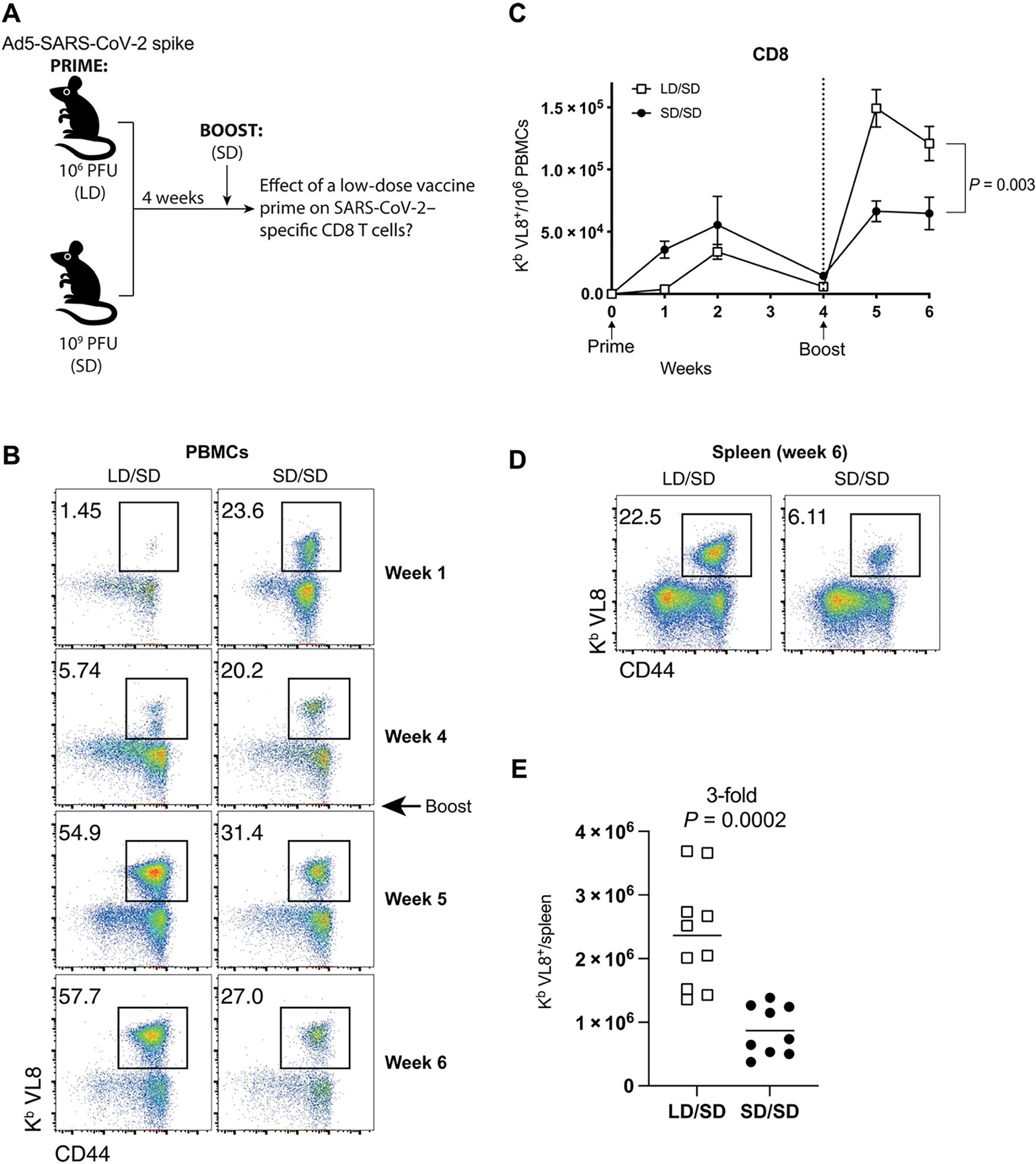
(A) Experimental approach for evaluating how the priming dose, either 106 PFU (LD) or 109 PFU (SD), of an Ad5-SARS-CoV-2 spike vaccine affects CD8+ T cell responses after an SD boost dose 4 weeks later in C57BL/6 mice. (B) Representative FACS plots showing the frequencies of SARS-CoV-2–specific CD8+ T cells (Kb VL8+) in PBMCs. (C) Summary of SARS-CoV-2–specific CD8+ T cell responses in PBMCs. (D) Representative FACS plots showing the frequencies of SARS-CoV-2–specific CD8+ T cells (Kb VL8+) in spleen. (E) Summary of SARS-CoV-2–specific CD8+ T cell responses in spleen at day 14 after boost. Data are from two experiments, one with n = 5 mice per group and another with n = 4 to 5 mice per group. All data are shown. Indicated P value was determined by two-way ANOVA in (C) and by unpaired t test in (E). Error bars represent SEM.
Initially, priming with an LD resulted in lower SARS-CoV-2–specific CD8+ T cell responses, relative to priming with an SD (Fig. 1B). This result is consistent with the well-reported observation that immunogenicity after adenovirus vaccination is dose dependent (12, 13). However, an unexpected effect was observed after the booster immunization 4 weeks later. Mice that were initially primed with an LD exhibited significantly greater CD8+ T cell expansion upon boosting relative to mice that were initially primed with the higher SD (Fig. 1, B to E).
SARS-CoV-2–specific CD8+ T cells induced by the LD/SD regimen exhibited enhanced interferon γ (IFNγ) expression based on in vitro stimulation of splenocytes with overlapping SARS-CoV-2 peptide pools at day 14 after boost (Fig. 2, A and B). Moreover, there was a pattern of improved CD4+ T cell responses in the LD/SD regimen relative to the SD/SD regimen (Fig. 2C). We did not observe differences in CD4+ T cell subset differentiation (fig. S1). SARS-CoV-2–specific CD8+ T cells induced by the LD/SD regimen showed more robust granzyme B and Ki67 expression relative to the SD/SD regimen (Fig. 2, D to F), suggesting enhanced cytotoxicity and proliferation upon boosting. Collectively, these results demonstrate that limiting the priming dose of Ad5-SARS-CoV-2 spike elicits SARS-CoV-2–specific T cell responses with superior anamnestic and functional capacity.
Fig. 2. An LD/SD vaccine regimen elicits more functional CD8+ T cell responses compared with an SD/SD vaccine regimen.
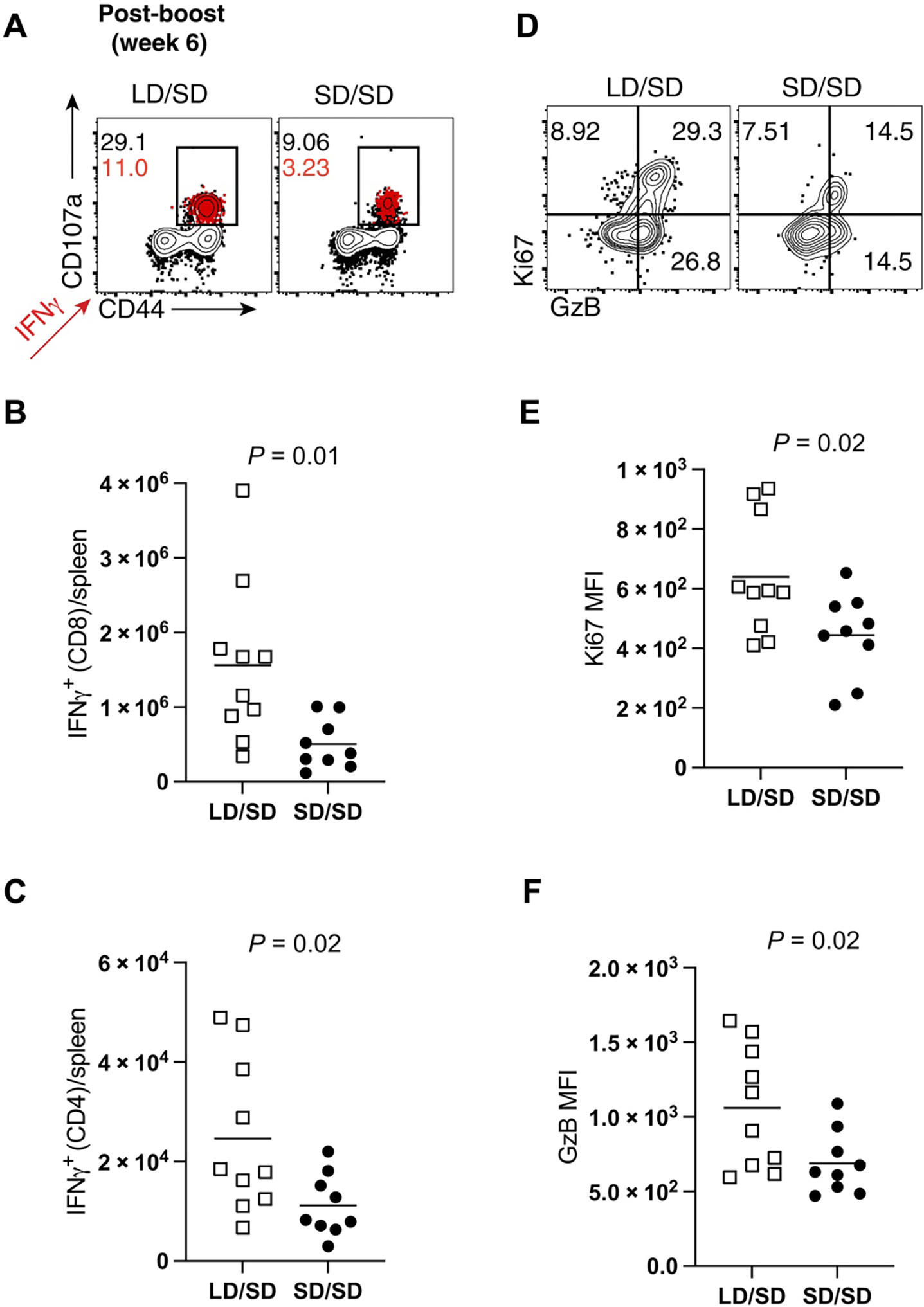
(A to C) Splenocytes were incubated with overlapping SARS-CoV-2 peptide pools for 5 hours at 37°C in the presence of GolgiStop and GolgiPlug to detect SARS-CoV-2–specific CD8+ and CD4+ T cell responses at day 14 after boost. (A) Representative FACS plots showing the frequencies of SARS-CoV-2–specific CD8+ T cells expressing the degranulation marker CD107a. IFNγ+ cells are overlaid in red, and their frequencies are indicated in red. (B) Summary of SARS-CoV-2–specific CD8+ T cells that express IFNγ. (C) Summary of SARS-CoV-2–specific CD4+ T cells that express IFNγ. (D) Representative FACS plots showing the frequencies of granzyme B– and Ki67-expressing CD8+ T cells in spleen (gated from Kb VL8+ cells). (E) Summary of Ki67 expression. (F) Summary of granzyme B expression. Data from (E) and (F) are indicated as mean fluorescence intensity (MFI). Data are from two experiments, one with n = 5 per group and another with n = 4 to 5 per group. All data are shown (day 14 after boost). Indicated P values were determined by parametric test (unpaired t test).
Effects of vaccine dose on T cell phenotype
The data above suggested qualitative differences in T cell responses following a single injection with either an LD or an SD of vaccine based on the functional differences observed after boosting with an SD. It is known that after an initial antigen encounter, T cells differentiate into distinct subsets, including effector cells (Teff), effector memory cells (Tem), and central memory cells (Tcm). Teff and Tem subsets exhibit rapid cytotoxicity but tend to be more short-lived than Tcm. Conversely, the Tcm subset exhibits superior recall capacity (14–16). To evaluate whether the priming dose affected the differentiation of these T cells subsets, we immunophenotyped SARS-CoV-2–specific CD8+ T cells at week 4 after prime. Primary CD8+ T cell responses generated by an LD prime exhibited more pronounced central memory markers, such as CD62L and CD127 (Fig. 3, A to D). In addition, CD8+ T cells induced by an LD prime showed higher levels of the CD44 activation marker (Fig. 3E) and lower levels of the inhibitory PD-1 receptor (Fig. 3F) and Tim-3 receptor (Fig. 3G), relative to CD8+ T cells induced after an SD prime. We also fluorescence-activated cell sorting (FACS)–sorted SARS-CoV-2–specific CD8+ T cells at week 4 after vaccination (prime only) and performed single-cell RNA sequencing (scRNA-seq). SARS-CoV-2–specific CD8+ T cells elicited by an LD and an SD clustered separately, suggesting distinct transcriptional signatures (fig. S2A). CD8+ T cells elicited by an LD prime showed lower transcription of genes associated with effector and terminal differentiation, and higher transcription of genes associated with long-lived memory T cells (fig. S2, B and C, and data files S1 and S2). We also interrogated whether these effects on T cell differentiation could be due to distinct TCR usage by performing single-cell T cell receptor (TCR) sequencing on SARS-CoV-2–specific CD8+ T cells at week 4 after prime. In both LD and SD, TCR usage was biased for Vβ11 (encoded by TRBV16 gene) (fig. S2, D and E), suggesting that the prime dose did not substantially affect TCR usage. Together, our functional data, phenotypic data, and transcriptomics data show that the strength of an initial antigen encounter has profound long-term effects on T cell differentiation after SARS-CoV-2 vaccination in mice.
Fig. 3. Phenotype of CD8+ T cell responses after a single prime with the Ad5-SARS-CoV-2 spike vaccine.
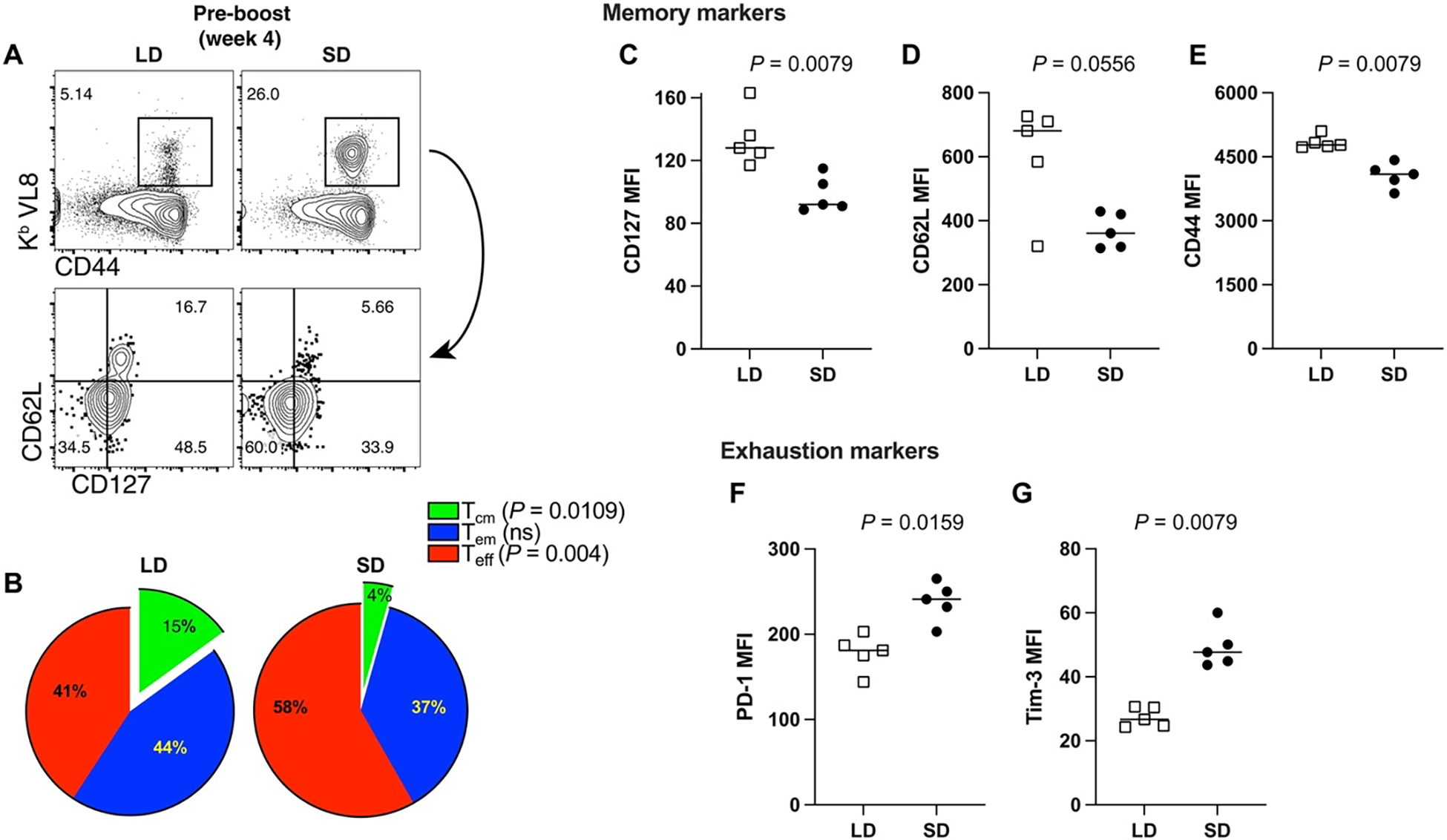
(A) Representative FACS plots showing the frequencies of peripheral blood SARS-CoV-2–specific CD8+ T cells (Kb VL8) that differentiate into Tem and Tcm cell subsets. (B) Summary of Teff, Tem, and Tcm cell subsets. (C) CD127 expression. (D) CD62L expression. (E) CD44 expression. (F) PD-1 expression. (G) Tim-3 expression. Data are gated from SARS-CoV-2–specific CD8+ T cells (Kb VL8) from PBMCs. All data are from day 28 after prime. Data are from one experiment with n = 5 per group. Experiment was repeated with similar results (see data file S3). Indicated P values were determined by parametric test (unpaired t test).
Preexisting immunity to adenoviral vectors can negatively affect vaccine-elicited immunity. We reasoned that because mice primed with an SD initially display a stronger virus-specific immune responses, this may result in more stringent competition for antigen during a subsequent booster immunization, impinging upon SARS-CoV-2–specific recall responses. To rule out the potential effects of preexisting T cell–mediated immunity, we evaluated recall CD8+ T cells in the absence of other preexisting responses, by performing adoptive transfers of low numbers of purified SARS-CoV-2–specific CD8+ T cells into naïve hosts. Four weeks after priming mice with an LD or an SD, we FACS-purified splenic SARS-CoV-2–specific CD8+ T cells (Kb VL8+) and transferred these at low numbers (500 cells) into congenically distinct naïve mice. One day after adoptive transfer, all recipient mice were vaccinated with the same SD (109 PFU) of the vaccine, and secondary CD8+ T cell expansion was assessed by flow cytometry (Fig. 4A). We show that donor CD8+ T cells from mice that were primed with an LD exhibited more robust secondary expansion compared with those primed with an SD (Fig. 4, B to D). These data show that a “gentle” antigenic prime elicits CD8+ T cells that are intrinsically superior on a per-cell basis and better able to mount a secondary response upon a booster immunization. Our adoptive transfer experiment with normalized numbers of CD8+ T cells was also consistent with our earlier phenotypic analyses showing that an LD prime preferentially elicits highly anamnestic Tcm rather than Tem cells.
Fig. 4. An LD prime elicits CD8+ T cell responses with intrinsically superior anamnestic capacity.
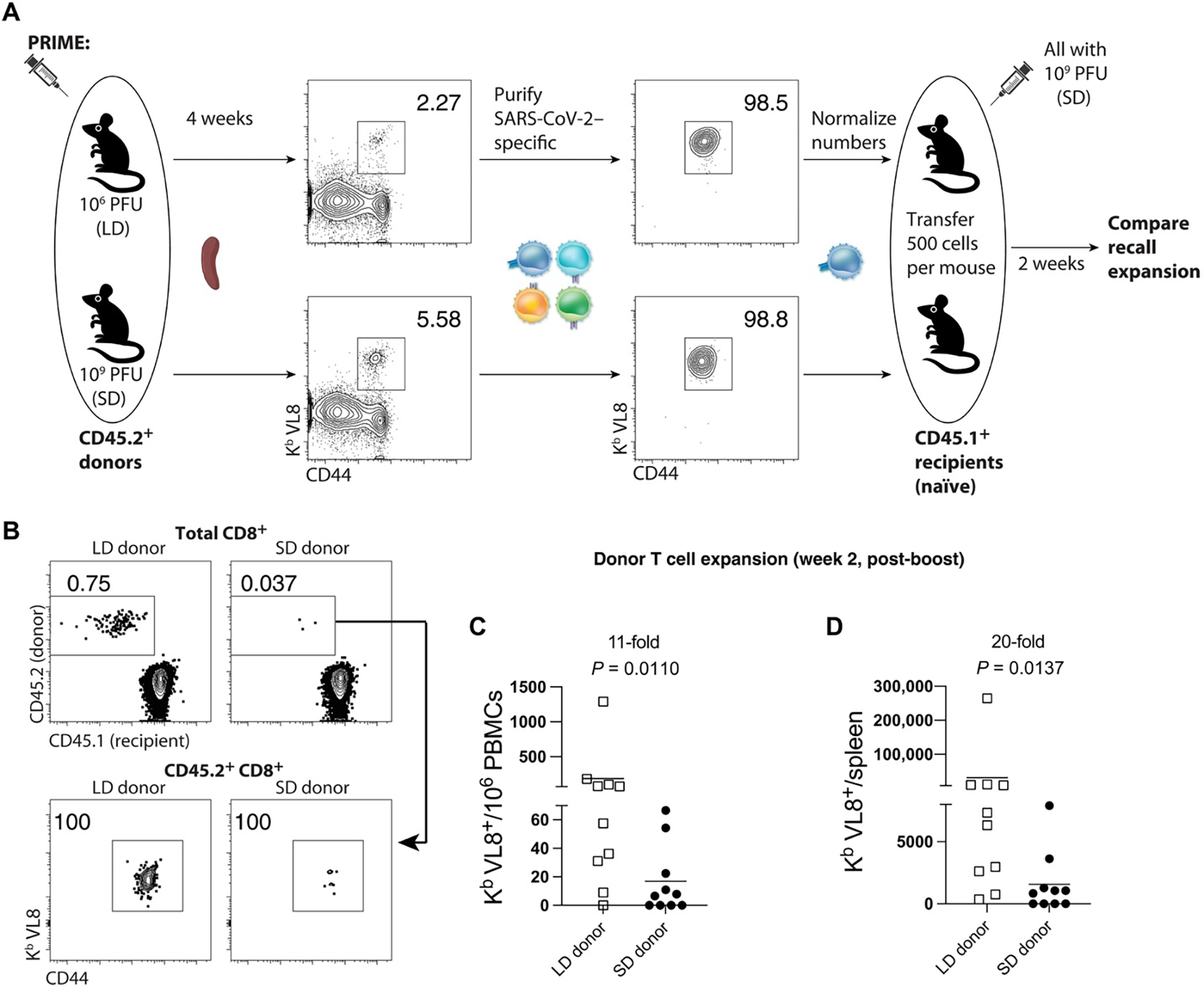
CD45.2+ mice were immunized intramuscularly with 106 or 109 PFU of Ad5-SARS-CoV-2 spike, and at day 28, splenic CD8+ T cells were MACS-sorted. Subsequently, live Kb VL8+ CD8+ T cells were FACS-sorted to ~99% purity, and numbers were normalized for adoptive transfer into CD45.1+ recipient mice. (A) Experimental design for evaluating secondary expansion of donor CD8+ T cells. (B) Representative FACS plots showing the frequencies of donor CD8+ T cells in PBMCs after boosting (gated from live CD8+ T cells). (C) Summary of donor-derived CD8+ T cells in PBMCs. (D) Summary of donor-derived CD8+ T cells in spleen. Data from (B) to (D) are from day 14 after boost. Data are from two experiments, each with n = 5 per group. All data are shown. Indicated P value was determined by nonparametric (Mann-Whitney U) test.
LD/SD elicits superior SARS-CoV-2–specific antibody responses relative to SD/SD
Our analyses, so far, have been focused on T cell responses, but we also report profound differences in antibody responses. After a single prime immunization, the LD elicited antibody responses that were expectedly lower compared with the SD. However, antibody responses elicited by the LD expanded more robustly upon boosting, resulting in a sixfold increase in SARS-CoV-2 spike–specific immunoglobulin G (IgG) by 6 weeks after the initial prime dose (Fig. 5A). The LD/SD regimen resulted in a twofold increase in germinal center (GC) B cells upon boosting (week 6) relative to the SD/SD regimen (Fig. 5, B and C). Generation of GC B cells after SARS-CoV-2 vaccination is associated with induction of a SARS-CoV-2–neutralizing antibody response (17). Therefore, we performed neutralization assays using SARS-CoV-2 pseudovirus to measure the ability of vaccine-elicited antibodies to block initial viral entry. Consistent with the increase in GC B cells, the LD/SD regimen elicited a 72-fold improvement in neutralizing antibodies compared with SD/SD (Fig. 5D).
Fig. 5. An LD/SD vaccine regimen elicits antibody responses with superior recall potential compared with an SD/SD vaccine regimen.
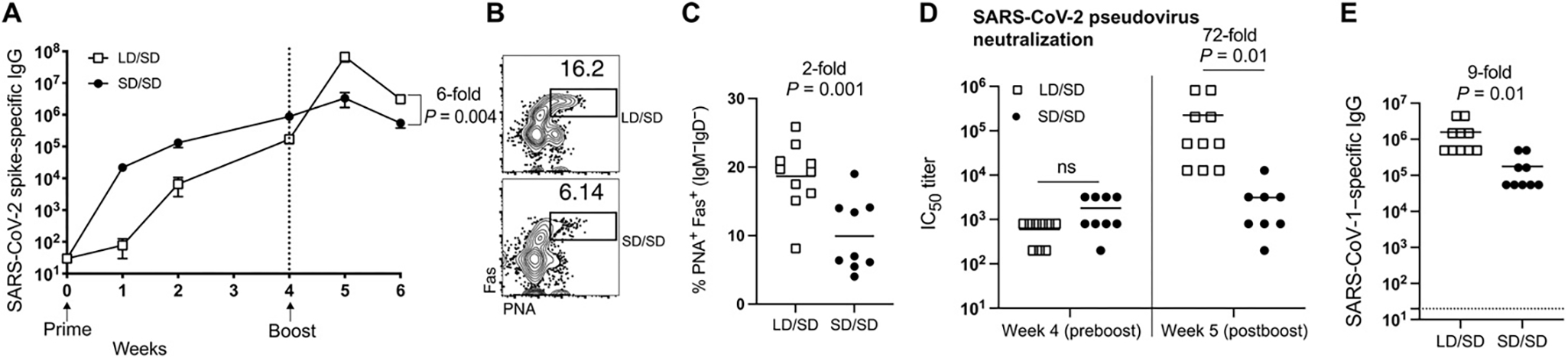
Vaccination regimen was the same as Fig. 1A. (A) Summary of SARS-CoV-2–specific antibody responses in sera of mice treated with either an LD or SD prime dose and an SD boost dose 4 weeks later. (B) Representative FACS plots showing the frequencies of GC B cells in spleen. Cells were gated from CD3− B220+ IgM− IgD−, as this gating strategy can be used to identify GC B cells (55). (C) Summary of GC B cells in spleen. (B) and (C) are from week 6. (D) Summary of SARS-CoV-2–neutralizing antibody responses in sera. IC50, median inhibitory concentration. (E) Summary of SARS-CoV-1–binding antibody responses in sera at week 5. Data are from two experiments, one with n = 5 per group and another with n = 4 to 5 per group. All data are shown. In (D) (week 5), insufficient sera were obtained from one SD/SD mouse after boost, so the total number of replicates in this group is n = 8. Indicated P value was determined by two-way ANOVA in (A), by parametric test (unpaired t test) in (C) and (E), and by two-way ANOVA (Dunnett’s multiple comparisons test) in (D). Dashed line indicates limit of detection. Error bars represent SEM.
A current concern is that SARS-CoV-1 may reenter the human population in the future, and it is currently unknown whether SARS-CoV-2 vaccines could confer cross-protection against SARS-CoV-1. We interrogated whether changing the priming dose of our SARS-CoV-2 vaccine could affect immune cross-reactivity to SARS-CoV-1. To answer this question, we measured antibody responses that bound to the original SARS-CoV-1 that was first identified in 2003. The LD/SD regimen resulted in a ninefold increase in SARS-CoV-1–binding antibody responses relative to the SD/SD regimen (Fig. 5E). Together, these findings demonstrate that cross-reactive antibody responses to SARS-CoV-1 are also significantly enhanced when the initial priming dose is reduced.
Effects of extending the prime-boost interval and narrowing the priming dose
A previous clinical trial with an adenovirus-based vaccine (AstraZeneca ChAdoX1) showed that a lower prime dose was associated with enhanced protection against SARS-CoV-2 (18). However, it was unclear whether the prime-boost interval could have also played a role. Our studies above did not rule out potential effects by the prime-boost interval, so we performed additional experiments to examine the effect of a protracted prime-boost interval. We observed improved antibody responses with more protracted prime-boost intervals (fig. S3), suggesting that the prime-boost interval could also influence vaccine-elicited immunity.
Our studies have compared immune responses elicited by two distinct priming doses that were 1000-fold apart (106 versus 109 PFU). We then interrogated whether a similar pattern could be observed with a narrower range of priming doses that were only 10-fold apart (106, 107, 108 PFU, with all mice receiving the same boosting dose of 108 PFU). These dose-escalation studies also showed a pattern of enhanced secondary responses in mice that received lower priming doses, but the differences were only statistically significant for antibodies when the doses were at least 100-fold apart (fig. S4). These data suggest that recall immune responses are inversely proportional to the priming dose, within the dose range tested.
Effects of priming dose on vector-specific responses
Because the studies above were focused on transgene-specific responses, we next evaluated vector-specific responses. The LD prime elicited substantially lowered vector (Ad5 hexon)–specific antibody responses relative to the SD prime (fig. S5). Even after boost, mice that were initially primed with an LD elicited lower Ad5 hexon–specific antibody responses. This result shows that the improvement of humoral immune responses in the LD/SD regimen is specific to transgene-specific responses.
Preexisting Ad5-specific humoral immunity accelerates transgene clearance
Previous studies, in particular, the HIV-1 STEP trial, have suggested that preexisting immunity to viral vectors may be detrimental for vector-based vaccines (3). However, the exact mechanism by which preexisting anti-vector immunity impairs vaccine efficacy is not fully understood. A prevailing wisdom is that preexisting antibodies neutralize Ad5, precluding entry into host cells (19–22). However, it is not clear whether Ad5-specific antibodies (which we observed were more highly induced by an SD prime) can play a more critical role via antibody effector mechanisms, such as antibody-dependent cellular cytotoxicity (ADCC). We interrogated these two possibilities by transferring naïve or Ad5-immune sera into naïve mice (via intraperitoneal injection), followed by immunization with a luciferase-expressing Ad5 vector 24 hours later. Luciferase expression was evaluated longitudinally by injecting mice intraperitoneally with luciferin, followed by in vivo luciferase quantification using in vivo imaging system (IVIS), as performed previously (23). This model allowed visualization of both initial transduction of the vector (breakthrough virus) and long-term transgene expression by in vivo luminescence (Fig. 6A). Ad5-specific humoral responses did not appear to abrogate the initial entry of Ad5 into muscle cells; transgene expression was comparable in the two groups during the hyperacute phase (≤6 hours after vaccination) (Fig. 6, B and C). However, Ad5-specific humoral responses quickly mitigated transgene expression 24 hours after vaccination. These data suggest that Ad5-specific antibodies may not fully prevent the entry of Ad5 into cells. Instead, Ad5-specific antibodies seem to play a more critical role in accelerating the clearance of Ad5-transduced cells at the site of vaccination, subsequently limiting transgene expression likely due to antibody effector mechanisms.
Fig. 6. Ad5-specific humoral responses limit the duration of transgene expression after Ad5 immunization.
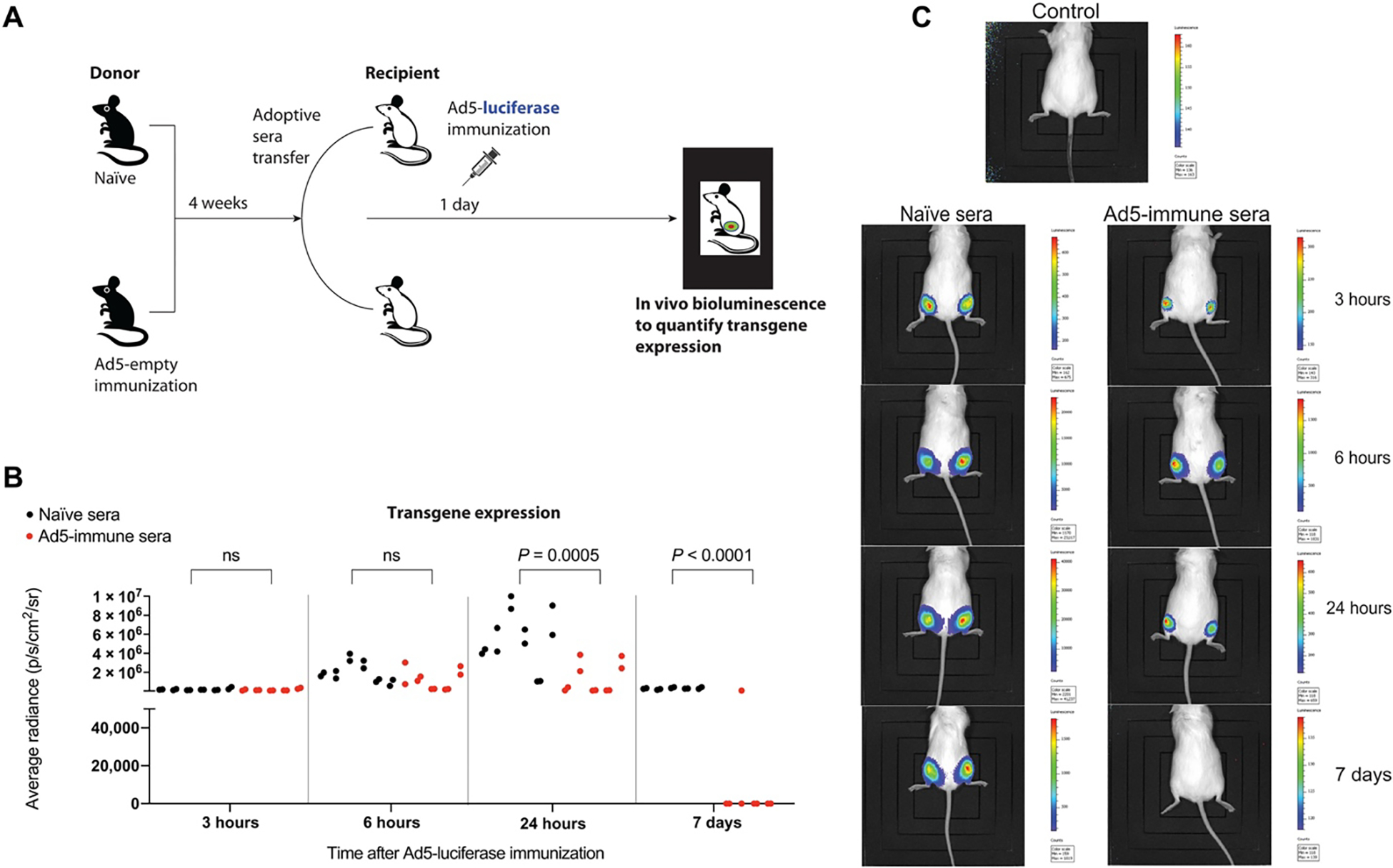
(A) Experimental approach for evaluating the effect of Ad5-specific humoral responses on transgene expression. Sera donors were Ad5-immune (week 4 after immunization) or naïve C57BL/6 mice, and recipients were naïve A/J mice (with white coat) to allow for improved visualization of luciferase by in vivo bioluminescence. One day after passive immunization (1000 μl of sera per mouse intraperitoneally), recipient A/J mice were immunized intramuscularly with 109 PFU of Ad5-luciferase. (B) Summary of transgene expression by in vivo bioluminescence imaging. Each adjacent dot represents the luciferase signal from the left and right quadriceps (adjacent dots are from the same mouse). Each quadriceps represents an independent data point because it is a separate site of immunization. (C) Representative bioluminescence images. Data are from two experiments, one with n = 4 to 5 per group and another with n = 1 per group. All data are shown. Indicated P value was determined by parametric test (unpaired t test).
During a booster immunization, both secondary and primary (de novo) T cell responses contribute to the effector response (24, 25). Because preexisting immunity to Ad5 downmodulates transgene expression after Ad5 immunization, we hypothesized that an LD prime would facilitate the future reutilization of Ad5, by promoting antigen presentation and subsequent priming of immune responses. To test this, we primed mice with an LD or an SD of Ad5-SARS-CoV-2 spike. After 3 weeks, we boosted these mice with an Ad5 vector expressing a mismatched antigen [Ad5–ovalbumin (OVA)] and then measured primary (OVA-specific) CD8+ T cells (fig. S6A). Mice that were initially primed with an LD of Ad5-SARS-CoV-2 spike generated more primary (OVA-specific) CD8+ T cells in draining lymph nodes, after the Ad5-OVA boost (fig. S6, B and D). Together, our results indicate that an LD prime not only elicits memory T cells with enhanced recall capacity but also facilitates de novo priming of T cells, following subsequent reutilization of Ad5 vector expressing an unrelated antigen.
The studies above using Ad5-SARS-CoV-2 spike and Ad5-OVA vaccines have shown that transgene-specific responses are enhanced when the priming dose is reduced. We interrogated whether similar effects could be observed with another antigen. We first primed mice with an Ad5 vector expressing simian immunodeficiency virus (SIV) Gag protein (Ad5-SIV) and then boosted mice after 4 weeks (fig. S7A). Consistent with our previous results, we observed a significant improvement of SIV-specific CD8+ T cell responses with the LD/SD regimen, relative to the SD/SD regimen (fig. S7, B and C), showing that our observations generalize to other antigen systems. In conclusion, we show that limiting the priming dose of Ad5-based vaccines results in substantial improvement of adaptive immune responses after boosting in mice.
DISCUSSION
Adenovirus-based SARS-CoV-2 vaccines are currently being deployed in humans to prevent COVID-19, including the Johnson & Johnson’s Janssen vaccine, AstraZeneca/Oxford vaccine, CanSino vaccine, and the Sputnik V vaccine. These vaccines have already been administered to millions of people and have shown potent immunogenicity, safety, and efficacy against severe SARS-CoV-2 infection. The CanSino and Sputnik V vaccines specifically use Ad5, which is the same vector platform used in our studies. Ad5 is among the most well-studied vaccine vectors due, in part, to its extraordinary immunogenicity (2, 26). Although SARS-CoV-2 vaccines prevent severe disease and death, they do not always confer durable sterilizing immunity, warranting the further optimization of current vaccine regimens.
Here, we show that fractionating the priming dose of an adenovirus-based SARS-CoV-2 vaccine confers an unexpected immunological benefit after boosting. A possible trade-off of fractionating the priming dose is that it may initially result in lower antigen-specific immune responses, which may render the host transiently more susceptible to infection, although this requires investigation. While it remains unclear what the minimum level of immune responses required to protect against severe COVID-19 is, a recent study demonstrated that even an LD single shot of an Ad26-based SARS-CoV-2 vaccine is sufficient to confer protection against severe disease in primates (27). Similarly, a single dose of a DNA-based SARS-CoV-2 vaccine, which is ~1000-fold less immunogenic than an LD single shot of the Ad5-based vaccine, was able to protect macaques from a SARS-CoV-2 challenge (28). These previous papers suggest that low levels of immune responses are sufficient to protect against severe COVID-19 (at least during the short time frame when the vaccines were tested), which can explain the extraordinary success of multiple SARS-CoV-2 vaccines in the last year.
There are historical examples of vaccine dose fractionation, also known as dose sparing, as a way to enable more people to get vaccinated. For instance, previous studies have shown that administering a fifth of the recommended dose of the yellow fever virus vaccine results in comparable immune responses relative to the recommended dose, without selection of escape variants (29). Similar results have been reported for influenza, measles, polio, and typhoid vaccines (30–32), which has led to discussions of whether SARS-CoV-2 vaccine doses should be fractionated to enable more people to get vaccinated. However, a counterargument against vaccine fractionation is that it may result in suboptimal immune protection (relative to SD) before the boost is administered.
What is the possible mechanistic basis for the improved recall response observed in the LD/SD regimen? First, we observed that an LD prime induced lower expression of inhibitory receptors and higher expression of central memory markers on CD8+ T cells relative to an SD prime. Second, anti-vector immunity was substantially lower after an LD prime relative to an SD prime. Lower anti-vector immunity may facilitate transgene expression and antigen presentation, as well as more de novo priming after a subsequent boost. Ad5-specific antibodies did not prevent Ad5 from entering muscle cells but resulted in more rapid elimination of Ad5 after 24 hours, likely via effector mechanisms. Whether blocking effector functions (such as ADCC) can allow the reutilization of Ad5 vectors, especially in the context of high preexisting immunity, requires further investigation. Our data with an SIV vaccine and an OVA vaccine suggest that our findings may also be useful to improve other Ad5-based vaccines. Nevertheless, it is unclear why only the transgene-specific response (but not the vector-specific response) was increased in the LD/SD regimen. The hexon is the most abundant adenovirus capsid protein and is considered to be the main target for adenovirus-neutralizing antibodies (33, 34). Future studies should determine whether the improvement of recall responses in the LD-primed mice is dependent on the level of antigen expression and whether similar effects can be observed with other vector platforms different than Ad5.
Conceptually, our findings are counterintuitive to conventional wisdom because we show that when it comes to the immune system, sometimes “less is more.” Antigen is the first signal required for the activation of the adaptive immune system, but here, we show an example where a lower amount of priming antigen results in superior recall response later during boosting. We also show that reducing the priming dose of a vaccine favors Tcm differentiation, which is characterized by high expression of the interleukin-7 (IL-7) receptor α chain (CD127) that promotes long-term T cell survival. Tcm CD8+ T cells also exhibit enhanced recall capacity (14), which may explain the improved secondary expansion that was observed in our LD/SD regimen.
The LD/SD regimen also induced superior antibody responses compared with the SD/SD regimen. Strikingly, the LD/SD regimen elicited a 72-fold improvement in the neutralizing antibody response, compared with the SD/SD regimen. In vitro antibody neutralization is directly correlated with in vivo protection against SARS-CoV-2 challenges (28), but we did not perform in vivo SARS-CoV-2 challenges to compare immune protection, because in our pilot studies in k18-ACE2 mice, even a single shot of the Ad5-SARS-CoV-2 spike vaccine provided robust protection. Similar efficacy has been described after a single dose with other adenovirus-based SARS-CoV-2 vaccines (35–37). It is plausible that an LD/SD regimen may confer a protective advantage over an SD/SD regimen months or years after vaccination when responses wane, which might reduce the need for a third boost. The LD/SD regimen may also be useful in individuals who develop suboptimal immune responses to vaccines or those who may benefit from having a higher level of immune responses. Moreover, reducing vaccine dose may result in fewer side effects or adverse events, because reactogenicity is directly proportional to vaccine dose (38–40). There are concerns that SARS-CoV-1 may reenter the human population, and an important question is whether the current SARS-CoV-2 vaccines could cross-protect against SARS-CoV-1. We analyzed cross-reactive antibody (binding to SARS-CoV-1 spike), and we show that the LD/SD regimen resulted in higher cross-reactive antibody levels compared with the SD/SD regimen. Notably, the CD8+ T cell response that we measured in these studies was specific for a highly conserved epitope (VNFNFNGL) that is present across multiple coronaviruses, including SARS-CoV-2, SARS-CoV-1, RatG13, HKU3, and WIV1, among others. Together, the LD/SD regimen may be particularly effective in the context of universal coronavirus vaccines, because this regimen induces potent levels of cross-reactive antibody and CD8+ T cell responses.
The extent to which our results generalize to humans has not been determined, but a recent clinical trial with an adenovirus-based SARS-CoV-2 vaccine (ChAdOx1, nCoV-19), in which the priming dose was unintentionally reduced to half, reported superior efficacy relative to SD (90% efficacy for LD/SD versus 62% efficacy for SD/SD) (18). However, in those studies, it was unclear whether the improvement in the LD/SD group was due to prolonging the prime-boost interval or due to the priming dose itself. Our results bring more clarity to this issue, as we show that the priming dose alone can have a substantial qualitative effect on immune responses in mice. Historically, phase 1 vaccine trials are designed to compare a range of vaccine doses among different groups of individuals. However, they do not typically analyze the effect of dose escalation within the same individual. The data presented support exploring intragroup vaccine dose escalation in future clinical trials and may be useful to understand the immunological effects of vaccine fractionation.
MATERIALS AND METHODS
Study design
The goal of this study was to understand how an Ad5 prime dose affects T cell and antibody responses in mice. Mice were first immunized intramuscularly with an LD versus an SD of an Ad5-based SARS-CoV-2 vaccine, and after several weeks, they were all boosted with an SD. Blood was harvested at different time points to measure SARS-CoV-2–specific T cell responses in peripheral blood mononuclear cells (PBMCs), and SARS-CoV-2– and vector-specific antibody responses in sera. Neutralizing antibody responses were also assessed before and after vaccination using luciferase-based SARS-CoV-2 pseudovirus assays.
Ad5-SARS-CoV-2 spike vaccine and other vaccines
The nonreplicating Ad5 vector is E1/E3-deleted and expresses the SARS-CoV-2 spike protein (strain 2019-nCoV-WIV04) within the putative E1 site (11, 41). This vector contains a cytomegalovirus promoter driving the expression of SARS-CoV-2 spike protein. The Ad5-SIV, Ad5-OVA, and Ad5-luciferase vectors have a similar genetic makeup but express their respective transgene in the putative E1 site, instead of spike. The Ad5-SARS-CoV-2 spike, Ad5-OVA, and Ad5-luciferase vectors were constructed and propagated at the Iowa Vector Core on trans-complementing human embryonic kidney (HEK)–293 cells (American Type Culture Collection), purified by cesium chloride density gradient centrifugation, titrated, and then frozen at −80°C. The Ad5-SIV (expressing Gag from SIVmac239) was obtained from J. Mascola [National Institutes of Health (NIH)] and propagated in our laboratory as indicated above. The Ad5-OVA vector was originally developed by T. Griffith (University of Minnesota).
Mice and vaccinations
Six- to 8-week-old C57BL/6 mice were used. Mice were purchased from the Jackson Laboratory (approximately half males and half females). Mice were immunized intramuscularly (50 μl per quadriceps) with an Ad5 vector–expressing SARS-CoV-2 spike protein (Ad5-SARS-CoV-2 spike) or the indicated Ad5 vector diluted in sterile phosphate-buffered saline (PBS). Mice were housed at the Northwestern University Center for Comparative Medicine in downtown Chicago. All mouse experiments were performed with approval of the Northwestern University Institutional Animal Care and Use Committee.
Reagents, flow cytometry, and equipment
Single-cell suspensions were obtained from PBMCs and various tissues as described previously (42). Dead cells were gated out using Live/Dead fixable dead cell stain (Invitrogen). SARS-CoV-2 spike peptide pools were used for intracellular cytokine staining and were obtained from BEI Resources. Biotinylated MHC class I monomers (Kb VL8, sequence VNFNFNGL), as well as the other tetramers used in this study, were obtained from the NIH tetramer facility at Emory University. The VNFNFNGL epitope is highly conserved among multiple coronaviruses, representing a cross-reactive CD8+ T cell response. It is located in position 539–546 of the SARS-CoV-2 spike protein or position 525–532 of the SARS-CoV-1 spike protein. Cells were stained with fluorescently labeled antibodies against CD8α [53–6.7 on peridinin chlorophyll protein (PerCP)–Cy5.5], CD44 (IM7 on Pacific Blue), IFNγ [XMG1.2 on allophycocyanin (APC)], peanut agglutinin (PNA) (conjugated to fluorescein), Fas [Jo2 on phycoerythrin (PE)], IgD (11–26 on Pacific Blue), IgM (RMM-1 on PE-Cy7), B220 (RA3–6B2 on PerCP-Cy5.5), CXCR5 (SPRCL5 conjugated to biotin and then to streptavidin-PE), Ly6C (HK1.4 on Pacific Blue), granzyme B (GB11 on PE), Ki67 [B56 on fluorescein isothiocyanate (FITC)], CD107a (1D4B on FITC), CD45.1 (A20 on PE-Cy7), CD45.2 (104 on FITC), CD25 (PC61 on PerCP-Cy5.5), FoxP3 (FJK.16S on APC), IL-4 (BVD6–24G2 on PE-Cy7), CD4 (RM4–5 on FITC), CD3 (145–2c11 on FITC), Tim-3 (RMT3–23 on PE), PD-1 (RMPI-30 on APC), CD62L (MEL-14 on PE-Cy7), and CD127 (A7R34 on FITC). CD4+ T cell subsets [e.g., T helper 1 (TH1), T follicular helper (Tfh), and T regulatory (Treg)] were identified following established phenotypes from previous papers (43–46). Fluorescently labeled antibodies were purchased from BD Pharmingen, except for anti-CD44 (which was from BioLegend). Flow cytometry samples were acquired with Becton Dickinson Canto II or LSRII and analyzed using FlowJo (Treestar).
Ad5 hexon, homologous (SARS-CoV-2 spike), and heterologous (SARS-CoV-1 spike) virus-specific ELISA
Binding antibody titers were measured using enzyme-linked immunosorbent assay (ELISA) as described previously (47–49) but using spike protein instead of viral lysates. Briefly, 96-well flat bottom plates (MaxiSorp, Thermo Fisher Scientific) were coated with 0.1 μg per well of the respective spike protein for 48 hours at 4°C. Plates were washed with PBS + 0.05% Tween 20. Blocking was performed for 4 hours at room temperature with 200 μl of PBS + 0.05% Tween 20 + bovine serum albumin. Six microliters of sera was added to 144 μl of blocking solution in the first column of the plate, 1:3 serial dilutions were performed until row 12 for each sample, and plates were incubated for 60 min at room temperature. Plates were washed three times followed by addition of goat anti-mouse IgG horseradish peroxidase–conjugated (Southern Biotech) diluted in blocking solution 1:1000, at 100 μl per well, and incubated for 60 min at room temperature. Plates were washed three times, and 100 μl per well of SureBlue substrate (SeraCare) was added for approximately 8 min. Reaction was stopped using 100 μl per well of KPL TMB stop solution (SeraCare). Absorbance was measured at 450 nm using SpectraMax Plus 384 (Molecular Devices). Ad5 hexon protein was purchased from Abcam (ab123995). SARS-CoV-2 spike protein was made at the Northwestern Recombinant Protein Production Core by S. Pshenychnyi using a plasmid that was produced under HHSN272201400008C and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), NIH: Vector pCAGGS containing the SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike Glycoprotein Gene (soluble, stabilized), NR-52394. SARS-CoV-1 spike protein was obtained through BEI Resources, NIAID, NIH: SARS-CoV Spike (S) Protein deltaTM, Recombinant from Baculovirus, NR-722.
Pseudovirus neutralization assays
A SARS-CoV-2 spike pseudotyped lentivirus kit was obtained through BEI Resources, NIAID, NIH (SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike-Pseudotyped Lentiviral Kit V2, NR-53816). We used HEK-293T–expressing human angiotensin-converting enzyme 2, also known as HEK-293T–hACE2, which are susceptible to SARS-CoV-2. This cell line was obtained through BEI Resources, NIAID, NIH, NR-52511. Serial dilutions of sera were incubated with the SARS-CoV-2 spike pseudotyped lentivirus, following a protocol by Balazs and Bloom (50). Cells were lysed using luciferase cell culture lysis buffer (Promega). Luciferase reaction was performed using 30 μl of cell lysis (Promega). The reaction was added to 96-well black optiplates (PerkinElmer). Luminescence was measured using a PerkinElmer Victor3 luminometer.
scRNA-seq data acquisition and analysis
C57BL/6 mice were immunized with an LD (106 PFU) or an SD (109 PFU) of Ad5-SARS-CoV-2 spike, and at day 28, splenic CD8+ T cells were MACS-sorted with a MACS-negative selection kit (STEMCELL Technologies). Purified CD8+ T cells were stained with Kb VL8 tetramer, live dead stain, and flow cytometry antibodies for CD8+ and CD44 to gate on activated CD8+ T cells. Live, CD8+, CD44+, Kb VL8+ cells were FACS-sorted to ~99% purity on a FACSAria cytometer (BD Biosciences) and delivered to the Northwestern University NUSeq core for scRNA-seq using a Chromium Next GEM 5′ v2 kit (10X Genomics). After the library was sequenced, the output file in BCL format was converted to fastq files and aligned to mouse genome to generate a matrix file using the Cell Ranger pipeline (10X Genomics). These upstream QC steps were performed by C. M. Wai and M. Schipma at the Northwestern University NUSeq core. Further analyses were performed in R using the Seurat package v4.0, as previously described (51). Terminal effector gene signatures were derived using the edgeR package (52), comparing Tem with terminal effector CD8+ T cells (53). Clusters representing less than 4% of each population were excluded from downstream analyses. TCR analyses were performed using the scRepertoire package (54). Only cells expressing both TCRa and TCRb chains were selected. For cells with more than two TCR chains, only the top 2 expressed chains were used.
Adoptive CD8+ T cell transfers
CD45.2+ C57BL/6 mice were immunized intramuscularly with an LD (106 PFU) or an SD (109 PFU) of Ad5-SARS-CoV-2 spike, and at day 28, splenic CD8+ T cells were enriched using a MACS-negative selection kit (STEMCELL Technologies). Purified CD8+ T cells were stained with Kb VL8 tetramer and live dead stain and FACS-sorted to ~99% purity on a FACSAria cytometer (BD Biosciences). Live Kb VL8 tetramer+ cells were transferred intravenously into naïve CD45.1 C57BL/6 mice (~500 cells per mouse). Recipient mice were vaccinated intramuscularly with an SD (109 PFU) of Ad5-SARS-CoV-2 spike 24 hours later. PBMCs and spleens were harvested ~2 weeks after boost for flow cytometric analyses.
In vivo bioluminescence
Sera from C57BL/6 mice immunized with 109 PFU of Ad5-empty (week 4 after immunization) or naïve C57BL/6 mice were collected. Immune (1000 μl) or naïve sera were then transferred intraperitoneally into white (A/J) recipient mice. Recipient A/J mice were immunized intramuscularly with Ad5-luciferase (109 PFU) 24 hours after sera transfer. Mice were imaged at various times after immunization. To quantify luciferase expression, luciferin (GoldBio, catalog no. LUCK-100; weight of 10 μl/g) was administered intraperitoneally 15 min before imaging, and mice were imaged for 2 min using the Kino IVIS Imager (Spectral Instruments Imaging). Region of interest (ROI) bioluminescence was used to quantify response.
Statistical analysis
Statistical tests used are indicated on each figure legend. Dashed lines in data figures represent limit of detection. Data were analyzed using Prism version 9 (GraphPad).
Supplementary Material
Acknowledgments
We thank S. Weiss, S. Perlman, and T. Gallagher for discussions.
Funding:
This work was possible with a grant from the Emerging and Re-Emerging Pathogens Program (EREPP) at Northwestern University and a grant from the National Institute on Drug Abuse (NIDA, DP2DA051912) to P.P.-M.
Footnotes
Competing interests: P.P.-M. reports being Task Force Advisor to the Illinois Department of Public Health (IDPH) on SARS-CoV-2 vaccine approval and implementation in the state of Illinois. P.P.-M. is also a member of the Northwestern COVID-19 Vaccine Communication and Evaluation Network (CoVAXCEN) at Northwestern University’s Institute for Global Health.
Data and materials availability: RNA and TCR sequencing data are available in the Gene Expression Omnibus under accession number GSE173567. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
REFERENCES AND NOTES
- 1.Tatsis N, Ertl HCJ, Adenoviruses as vaccine vectors. Mol. Ther. 10, 616–629 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alayo QA, Provine NM, Penaloza-MacMaster P, Novel concepts for HIV vaccine vector design. mSphere 2, e00415–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN; Step Study Protocol Team, Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372, 1881–1893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, Kik SV, Weverling GJ, Dilan R, King SL, Maxfield LF, Clark S, Ng’ang’a D, Brandariz KL, Abbink P, Sinangil F, de Bruyn G, Gray GE, Roux S, Bekker LG, Dilraj A, Kibuuka H, Robb ML, Michael NL, Anzala O, Amornkul PN, Gilmour, Hural J, Buchbinder SP, Seaman MS, Dolin R, Baden LR, Carville A, Mansfield G, Pau MG, Goudsmit J, International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine 29, 5203–5209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyriakidis NC, Lopez-Cortes A, Gonzalez EV, Grimaldos AB, Prado EO, SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 6, 28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creech CB, Walker SC, Samuels RJ, SARS-CoV-2 vaccines. JAMA 325, 1318–1320 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Subbarao K, The success of SARS-CoV-2 vaccines and challenges ahead. Cell Host Microbe 29, 1111–1123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall VG, Ferreira VH, Ku T, Ierullo M, Majchrzak-Kita B, Chaparro C, Selzner N, Schiff J, McDonald M, Tomlinson G, Kulasingam V, Kumar D, Humar A, Randomized trial of a third dose of mRNA-1273 vaccine in transplant recipients. N. Engl. J. Med. 385, 1244–1246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall VG, Ferreira VH, Ierullo M, Ku T, Marinelli T, Majchrzak-Kita B, Yousuf A, Kulasingam V, Humar A, Kumar D, Humoral and cellular immune response and safety of two-dose SARS-CoV-2 mRNA-1273 vaccine in solid organ transplant recipients. Am. J. Transplant. 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergwerk M, Gonen T, Lustig Y, Amit S, Lipsitch M, Cohen C, Mandelboim M, Gal Levin E, Rubin C, Indenbaum V, Tal I, Zavitan M, Zuckerman N, Bar-Chaim A, Kreiss Y, Regev-Yochay G, Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 385, 1474–1484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dangi T, Class J, Palacio N, Richner JM, Penaloza MacMaster P, Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 36, 109664 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinn KM, Zak DE, Costa A, Yamamoto A, Kastenmuller K, Hill BJ, Lynn GM, Darrah PA, Lindsay RW, Wang L, Cheng C, Nicosia A, Folgori A, Colloca S, Cortese R, Gostick E, Price DA, Gall JG, Roederer M, Aderem A, Seder RA, Antigen expression determines adenoviral vaccine potency independent of IFN and STING signaling. J. Clin. Invest. 125, 1129–1146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weaver EA, Dose effects of recombinant adenovirus immunization in rodents. Vaccines (Basel) 7, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R, Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4, 225–234 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Sallusto F, Geginat J, Lanzavecchia A, Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Geginat J, Lanzavecchia A, Sallusto F, Proliferation and differentiation potential of human CD8+ memory T-cell subsets in response to antigen or homeostatic cytokines. Blood 101, 4260–4266 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Lederer K, Castano D, Gómez Atria D, Oguin III TH, Wang S, Manzoni TB, Muramatsu H, Hogan MJ, Amanat F, Cherubin P, Lundgreen KA, Tam YK, Fan SHY, Eisenlohr LC, Maillard I, Weissman D, Bates P, Krammer F, Sempowski GD, Pardi N, Locci M, SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53, 1281–1295.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoll MD, Wonodi C, Oxford-AstraZeneca COVID-19 vaccine efficacy. Lancet 397, 72–74 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gahéry-Ségard H, Farace F, Godfrin D, Gaston J, Lengagne R, Tursz T, Boulanger P, Guillet JG, Immune response to recombinant capsid proteins of adenovirus in humans: Antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72, 2388–2397 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kass-Eisler A, Leinwand L, Gall J, Bloom B, Falck-Pedersen E, Circumventing the immune response to adenovirus-mediated gene therapy. Gene Ther. 3, 154–162 (1996). [PubMed] [Google Scholar]
- 21.Toogood CI, Crompton J, Hay RT, Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J. Gen. Virol. 73 (Pt 6), 1429–1435 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Wohlfart C, Neutralization of adenoviruses: Kinetics, stoichiometry, and mechanisms. J. Virol. 62, 2321–2328 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larocca RA, Provine NM, Aid M, Iampietro MJ, Borducchi EN, Badamchi-Zadeh A, Abbink P, Ng’ang’a D, Bricault CA, Blass E, Penaloza-MacMaster P, Stephenson KE, Barouch DH, Adenovirus serotype 5 vaccine vectors trigger IL-27-dependent inhibitory CD4+T cell responses that impair CD8+T cell function. Sci. Immunol 1, eaaf7643 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan WG, Jin HT, West EE, Penaloza-MacMaster P, Wieland A, Zilliox MJ, McElrath MJ, Barouch DH, Ahmed R, Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J. Virol. 87, 1359–1372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penaloza-MacMaster P, Teigler JE, Obeng RC, Kang ZH, Provine NM, Parenteau L, Blackmore S, Ra J, Borducchi EN, Barouch DH, Augmented replicative capacity of the boosting antigen improves the protective efficacy of heterologous prime-boost vaccine regimens. J. Virol. 88, 6243–6254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penaloza-MacMaster P, Provine NM, Ra J, Borducchi EN, McNally A, Simmons NL, Iampietro MJ, Barouch DH, Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J. Virol. 87, 1373–1384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X, Chandrashekar A, Zahn R, Wegmann F, Yu J, Mercado NB, McMahan K, Martinot AJ, Piedra-Mora C, Beecy S, Ducat S, Chamanza R, Huber SR, van der Fits L, Borducchi EN, Lifton M, Liu J, Nampanya F, Patel S, Peter L, Tostanoski LH, Pessaint, Van Ry A, Finneyfrock B, Velasco J, Teow E, Brown R, Cook A, Andersen H, Lewis G, Schuitemaker H, Barouch DH, Low-dose Ad26.COV2.S protection against SARS-CoV-2 challenge in rhesus macaques. Cell 184, 3467–3473.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JY, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolol JP, Liu JY, Li ZF, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen YZ, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin ZJ, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan HH, Cai YF, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH, DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JT, Peak CM, Leung GM, Lipsitch M, Fractional dosing of yellow fever vaccine to extend supply: A modelling study. Lancet 388, 2904–2911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley S, Wu JT, Leung GM, Optimizing the dose of pre-pandemic influenza vaccines to reduce the infection attack rate. PLOS Med. 4, e218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnyder JL, De Pijper CA, Garcia Garrido HM, Daams JG, Goorhuis A, Stijnis C, Schaumburg F, Grobusch MP, Fractional dose of intradermal compared to intramuscular and subcutaneous vaccination - A systematic review and meta-analysis. Travel Med. Infect. Dis. 37, 101868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okayasu H, Sein C, Chang Blanc D, Gonzalez AR, Zehrung D, Jarrahian C, Macklin G, Sutter RW, Intradermal administration of fractional doses of inactivated poliovirus vaccine: A dose-sparing option for polio immunization. J Infect Dis 216, S161–S167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, Dmitriev I, Kashentseva E, Seki T, Wang M, Curiel DT, Construction and characterization of adenovirus serotype 5 packaged by serotype 3 hexon. J. Virol. 76, 12775–12782 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner R, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH, Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441, 239–243 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, Kashentseva E, McCune BT, Bailey AL, Zhao H, VanBlargan LA, Dai YN, Ma M, Adams LJ, Shrihari S, Danis JE, Gralinski LE, Hou YJ, Schafer A, Kim AS, Keeler SP, Weiskopf D, Baric RS, Holtzman MJ, Fremont DH, Curiel DT, Diamond MS, A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell 183, 169–184.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Huber KR, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna M, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Van Ry A, Blade K, Strasbaugh A, Cabus M, Brown R, Cook A, Zouantchangadou S, Teow E, Andersen H, Lewis MG, Cai Y, Chen B, Schmidt AG, Reeves RK, Baric RS, Lauffenburger DA, Alter G, Stoffels P, Mammen M, Van Hoof J, Schuitemaker H, Barouch DH, Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S, Zhong G, Zhang J, Shuai L, Zhang Z, Wen Z, Wang B, Zhao Z, Song X, Chen Y, Liu R, Fu L, Zhang J, Guo Q, Wang C, Yang Y, Fang T, Lv P, Wang J, Xu J, Li J, Yu C, Hou L, Bu Z, Chen W, A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 11, 4081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, Stoop J, Tete S, Van Damme W, Leroux-Roels I, Berghmans PJ, Kimmel M, Van Damme P, de Hoon J, Smith W, Stephenson KE, De Rosa SC, Cohen KW, McElrath MJ, Cormier E, Scheper G, Barouch DH, Hendriks J, Struyf F, Douoguih M, Van Hoof J, Schuitemaker H, Interim results of a phase 1–2a trial of Ad26.COV2.S Covid-19 vaccine. N. Engl. J. Med. 384, 1824–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, Voysey M, Aley PK, Angus B, Babbage G, Belij-Rammerstorfer S, Berry L, Bibi S, Bittaye M, Cathie K, Chappell H, Charlton S, Cicconi P, Clutterbuck EA, Colin-Jones R, Dold C, Emary KRW, Fedosyuk S, Fuskova M, Gbesemete D, Green C, Hallis B, Hou MM, Jenkin D, Joe CCD, Kelly EJ, Kerridge S, Lawrie AM, Lelliott A, Lwin MN, Makinson R, Marchevsky NG, Mujadidi Y, Munro APS, Pacurar M, Plested E, Rand J, Rawlinson T, Rhead S, Robinson H, Ritchie AJ, Ross-Russell AL, Saich S, Singh N, Smith CC, Snape MD, Song R, Tarrant R, Themistocleous Y, Thomas KM, Villafana TL, Warren SC, Watson MEE, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Faust SN, Pollard AJ; Oxford COVID Vaccine Trial Group, Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 396, 1979–1993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, Wu SP, Wang BS, Wang Z, Wang L, Jia SY, Jiang HD, Wang L, Jiang T, Hu Y, Gou JB, Xu SB, Xu JJ, Wang XW, Wang W, Chen W, Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395, 1845–1854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng L, Wang Q, Shan C, Yang C, Feng Y, Wu J, Liu X, Zhou Y, Jiang R, Hu P, Liu X, Zhang F, Li P, Niu X, Liu Y, Zheng X, Luo J, Sun J, Gu Y, Liu B, Xu Y, Li C, Pan W, Zhao J, Ke C, Chen X, Xu T, Zhong N, Guan S, Yuan Z, Chen L, An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat. Commun. 11, 4207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masopust D, Vezys V, Marzo AL, Lefrancois L, Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharyya M, Madden P, Henning N, Gregory S, Aid M, Martinot AJ, Barouch DH, Penaloza-MacMaster P, Regulation of CD4 T cells and their effects on immunopathological inflammation following viral infection. Immunology 152, 328–343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R, Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38, 805–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, O’Mara L, Yang S, Konieczny BT, Sharpe AH, Freeman GJ, Rudensky AY, Ahmed R, Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J. Exp. Med. 211, 1905–1918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iyer SS, Latner DR, Zilliox MJ, McCausland M, Akondy RS, Penaloza-Macmaster P, Hale JS, Ye L, Mohammed AU, Yamaguchi T, Sakaguchi S, Amara RR, Ahmed R, Identification of novel markers for mouse CD4+ T follicular helper cells. Eur. J. Immunol. 43, 3219–3232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dangi T, Chung YR, Palacio N, Penaloza-MacMaster P, Interrogating Adaptive Immunity Using LCMV. Curr. Protoc. Immunol. 130, e99 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palacio N, Dangi T, Chung YR, Wang Y, Loredo-Varela JL, Zhang Z, Penaloza-MacMaster P, Early type I IFN blockade improves the efficacy of viral vaccines. J. Exp. Med. 217, e20191220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Chung YR, Eitzinger S, Palacio N, Gregory S, Bhattacharyya M, Penaloza-MacMaster P, TLR4 signaling improves PD-1 blockade therapy during chronic viral infection. PLOS Pathog. 15, e1007583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD, Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, McGavern DB, Bosselut R, The emergence and functional fitness of memory CD4+ T cells require the transcription factor Thpok. Immunity 50, 91–105.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milner JJ, Nguyen H, Omilusik K, Reina-Campos M, Tsai M, Toma C, Delpoux A, Boland BS, Hedrick SM, Chang JT, Goldrath AW, Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl. Acad. Sci. U.S.A. 117, 25667–25678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borcherding N, Bormann NL, Kraus G, scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res. 9, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penaloza-MacMaster P, Barber DL, Wherry EJ, Provine NM, Teigler JE, Parenteau L, Blackmore S, Borducchi EN, Larocca RA, Yates KB, Shen H, Haining WN, Sommerstein R, Pinschewer DD, Ahmed R, Barouch DH, Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science 347, 278–282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


