ABSTRACT
Despite the rapid deployment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines, the emergence of SARS-CoV-2 variants and reports of their immune evasion characteristics have led to an urgent need for novel vaccines that confer potent cross-protective immunity. In this study, we constructed three different SARS-CoV-2 spike S1-conjugated nanoparticle vaccine candidates that exhibited high structural homogeneity and stability. Notably, these vaccines elicited up to 50-times-higher neutralizing antibody titers than the S1 monomer in mice. Crucially, it was found that the S1-conjugated nanoparticle vaccine could elicit comparable levels of neutralizing antibodies against wild-type or emerging variant SARS-CoV-2, with cross-reactivity to SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), the effect of which could be further enhanced using our designed nanoparticles. Our results indicate that the S1-conjugated nanoparticles are promising vaccine candidates with the potential to elicit potent and cross-reactive immunity against not only wild-type SARS-CoV-2, but also its variants of concern, variants of interest, and even other pathogenic betacoronaviruses.
IMPORTANCE The emergence of SARS-CoV-2 variants led to an urgent demand for a broadly effective vaccine against the threat of variant infection. The spike protein S1-based nanoparticle designed in our study could elicit a comprehensive humoral response toward different SARS-CoV-2 variants of concern and variants of interest and will be helpful to combat COVID-19 globally.
KEYWORDS: SARS-CoV-2, variants of concern, Omicron, nanoparticle vaccine, S1
INTRODUCTION
The pandemic of COVID-19 continues to strain the global public health system and has caused millions of deaths (1). Despite strict restrictions of entry-and-exit policies implemented by all countries, global transmission of COVID-19 is not well contained, and broad vaccination is touted as the only effective method for the control of the pandemic (2).
The causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a member of the betacoronavirus family, which includes other highly pathogenic human coronaviruses, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (3, 4). It is a single-stranded RNA virus containing four major structural proteins, whereby the spike (S) protein on the surface of viral particles plays a critical role in virus-host cell membrane attachment and fusion (5, 6). The spike protein comprises two subunits, S1 and S2, of which S1 contains the broadly known receptor-binding domain (RBD) recognizing angiotensin-converting enzyme 2 (ACE2) as the receptor, and an N-terminal domain (NTD) recently found to be involved in viral infection by interacting with another host tyrosine-protein kinase receptor, UFO (AXL), as well as a C-terminal domain (CTD) required for membrane fusion (7–13). Accordingly, most neutralizing antibodies were found to target S1, which suggests that could be an ideal vaccine antigen (14, 15).
Since the outbreak of COVID-19, there have been intensive efforts to develop a vaccine, mostly focusing on the whole virus, spike protein, or the RBD as antigen (16, 17). However, when the antigen is designed to be more immune focused, such as the RBD, the pathogen specificity increases, leading to a decrease of cross-reactivity toward multiple variants or sibling species (13, 18). Conversely, overexpansion of antigenic epitopes could undermine immune efficacy due to the induction of low-quality antibodies (19). Therefore, a balanced strategy of antigen selection is critical for successful vaccine design. As antibodies against the NTD and the NTD-recognizing receptor were found, the importance of the NTD in eliciting additional neutralizing antibodies beyond RBD-specific antibodies suggests that S1 may provide better protection than RBD, without introducing relatively immune-redundant domains by using the whole spike protein as an immunogen (20–24). Accordingly, S1 could be an ideal antigen to counter the current threat of the wide transmission of SARS-CoV-2 variants.
SARS-CoV-2 variants have become a major threat to global disease control, and the variants of concern (VOCs) identified by the World Health Organization are characterized by different degrees of resistance to available antibodies and vaccines, as well as a higher propensity to cause severe disease, necessitating the development of vaccines that confer robust cross-reactive immunity against rapidly emerging SARS-CoV-2 variants (25–31). Thus, whether S1 could be a vaccine candidate to meet this urgent need to counter VOCs deserves further investigation. However, the isolation of S1 from the trimeric spike protein, similar to the RBD, results in an alternation of its state into a monomer and loss of orientation found in the original protein structure, which could impair its immunogenicity due to a lack of multivalency and decrease in antigen size (32–37). Nanoparticles (NPs) are gradually becoming a mature platform for antigen delivery with multivalency and increased size, and recombinant expression of antigen-bearing nanoparticles has become a new challenge for vaccine development due to increased structural complexity during nanoparticle assembly (38–40). Since covalent protein conjugation has been widely used in protein modification, we adopted a similar strategy to construct S1 nanoparticles (41–43).
Here, we produced S1 nanoparticles with different degrees of multivalency. All nanoparticles were thermostable and homogenous under typical conditions, allowing easy transport and storage. Immunization of mice showed that S1 nanoparticles could elicit more robust humoral immunity than the S1 monomer, leading to higher neutralizing antibody titers against both ancestral SARS-CoV-2 and VOCs in a dose-dependent manner. Notably, the S1 nanoparticle-elicited sera exhibited cross-reactivity to SARS-CoV and MERS-CoV. Thus, this platform may be more competitive for vaccine development in the fight against future SARS-CoV-2 variants or other betacoronaviruses.
RESULTS
Design of SARS-CoV-2 S1-based nanoparticle immunogens.
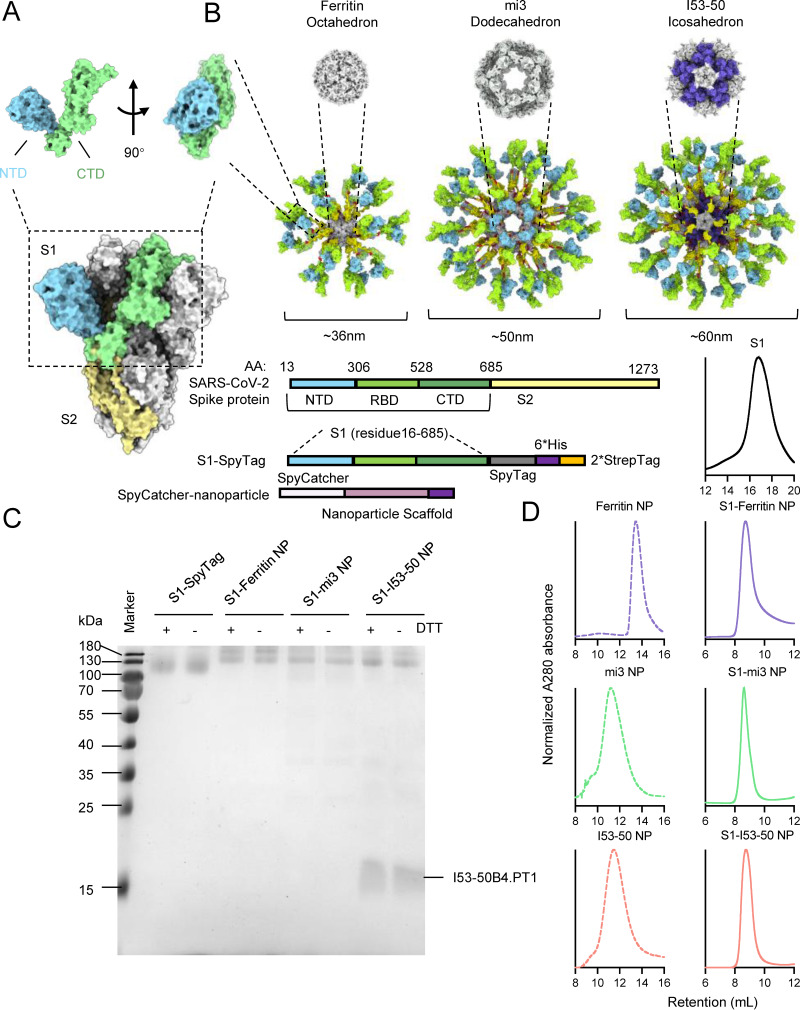
SARS-CoV-2 spike protein has been exploited as a major antigen for protein- or nucleic acid-based vaccines, which have been widely used. The spike protein can be proteolytically cleaved into a membrane-distal S1 subunit and a membrane-proximal S2 subunit at the furin cleavage site (Fig. 1A) (10). The S1 subunit consists of a receptor-binding domain (RBD) and an N-terminal domain (NTD), which binds to the host receptor, human angiotensin-converting enzyme 2 (ACE2), to initiate the process of viral attachment, and a C-terminal domain (CTD) (Fig. 1B). To date, most potent neutralizing antibodies against SARS-CoV-2 target the RBD and NTD, and thus we adopted the S1 subunit as the antigen for vaccine design (15, 44). To improve immunogenicity and adjust the orientation of the S1 monomer, we presented S1 antigen on the surface of the protein nanoparticles through the covalent amide bond formed by the SpyTag-SpyCatcher system. The SpyCatcher-SpyTag system consists of the 138-residue SpyCatcher and the 15-residue SpyTag, which autocatalytically form a covalent bond, making this system ideal for designing complex macromolecular assemblies with different compositions (45).
FIG 1.
Design and construction of S1-conjugated nanoparticles. Shown is a structural representation of SARS-CoV-2 spike protein. (A) The SARS-CoV-2 spike consists of two segments, S1 and S2 (light yellow), and the S1 segment includes NTD (light blue), RBD (dark green), and CTD (light green). (B) Structural representation and construction of S1-conjugated nanoparticle components. The S1 domain was conjugated to the nanoparticle ferritin, mi3, or I53-50 by the formation of a covalent bond between SpyTag and SpyCatcher. (C) SDS-PAGE of the S1 monomer and three S1-conjugated nanoparticles. The conjugated nanoparticles were resisted to the DTT reduction. (D) Size exclusion chromatography (SEC) of the S1 monomer, unconjugated nanoparticles, and S1-conjugated nanoparticles. After conjugation, the molecular weight of nanoparticles significantly increased. The normalized A280 absorbance curve is presented.
Three kinds of nanoparticles were designed, including octahedral 24-meric ferritin, dodecahedral 60-meric mi3, and icosahedral 120-meric I53-50 (Fig. 1B). The nanoparticles were fused with a SpyCatcher on the N terminus, while S1 was fused with a SpyTag on the C terminus. The nanoparticle based on ferritin from Helicobacter pylori was self-assembled from 24 identical subunits into a spherical particle octahedral symmetry (46). The mi3 NP protein, optimized from the computationally designed I3-01 to increase uniformity and stability, could self-assemble into a dodecahedral particle (47). Finally, the I53-50 NP was coassembled from 20 trimeric I53-50A and 12 pentameric I53-50B subunits to form an icosahedral nanoparticle (48, 49). These three nanoparticles were found to be safe, stable, and easy to produce in previous studies (32, 50–52).
Preparation and characterization of S1-based nanoparticle vaccines.
S1 genetically fused with a C-terminal SpyTag was cloned into a mammalian expression vector for transient transfection of Expi293F cells. The supernatant of cell cultures was collected for immobilized-metal affinity chromatography (IMAC) and size exclusion chromatography (SEC) to purify the recombinant S1-SpyTag monomer. Subunits of the nanoparticles, including SpyCatcher-ferritin, SpyCatcher-mi3, and SpyCatcher-I53-50A and -I53-50B proteins, were expressed in Escherichia coli and purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography followed by SEC (Fig. 1C and D).
The ferritin and mi3 subunits could spontaneously self-assemble into a nanoparticle (NP) upon expression in host cells, while the two components of I53-50 NP, I53-50A and I53-50B, were incubated together to assemble into complete particles in vitro. In order to conjugate S1-SpyTag to the SpyCatcher-NP, the SpyCatcher-NPs were incubated with S1-SpyTag for covalent conjugation, and SEC was then applied to further purify the S1-conjugated nanoparticles and remove the remnants. The SDS-PAGE results showed that S1 was successfully conjugated to the nanoparticles, and the resulting covalent bond was resistant to the denaturant dithiothreitol (DTT) (Fig. 1C). Following conjugation of S1, the overall size and total weight of the nanoparticles significantly increased in comparison with those of the S1 monomer or unconjugated NPs, as determined by SEC (Fig. 1D).
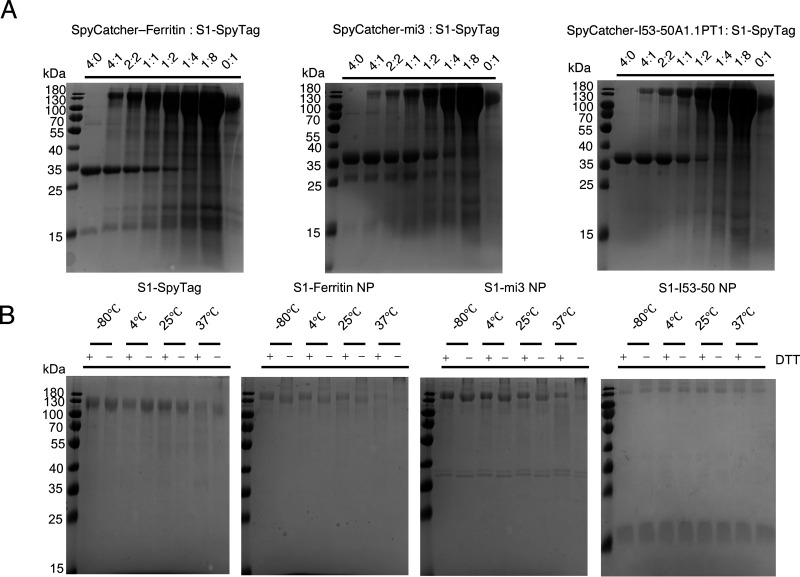
To further investigate the conjugation efficiency and protein stability, we tested different incubation ratios of S1-SpyTag and SpyCatcher-NP, as well as different storage conditions. It was first observed that unconjugated SpyCatcher-NPs were gradually depleted with the increased amount of S1-SpyTag, and at a SpyCatcher-NP ratio of 1:8, nearly complete conjugation to S1-SpyTag was achieved (Fig. 2A). This result demonstrated that the 1:8 molar concentration ratio of SpyCatcher-NPs and S1-SpyTag was appropriate for in vitro conjugation, since the complete conjugation was crucial for ensuring the uniformity of the produced S1 nanoparticles. Then, we evaluated the protein stability of S1 nanoparticles at different storage temperatures. We stored the S1-SpyTag monomer and three S1-conjugated nanoparticles at –80°C, 4°C, 25°C, and 37°C for 2 weeks to determine the occurrence of any degradation, deconjugation, or disassembly. The results showed that all nanoparticles remained stable after 2 weeks of storage at the tested temperatures (Fig. 2B).
FIG 2.
Assembly efficiency and the storage stability of S1-conjugated nanoparticles. (A) SDS-PAGE assembling assays of S1-SpyTag and SpyCatcher-nanoparticle scaffolds at different mixing ratios. As the ratio of SpyCatcher-ferritin/mi3/I53-50A1.PT1 and S1 reached 1:8, the remnant unconjugated nanoparticle scaffolds were almost depleted, suggesting that the nanoparticles were fully conjugated with S1 at high superficial abundance. (B) SDS-PAGE assembling assays of S1-SpyTag and SpyCatcher-nanoparticle at different temperatures. No significant degradation or aggregation was observed among S1 monomer or S1 nanoparticles.
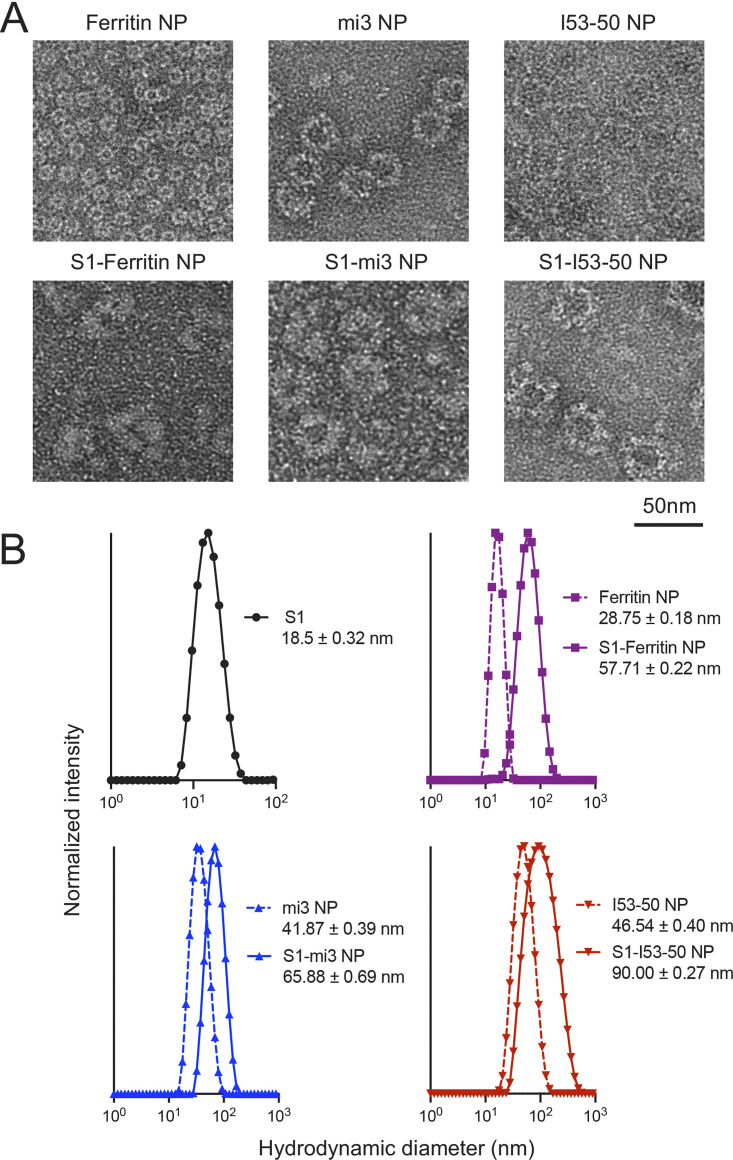
In order to directly observe the structure of S1 nanoparticles, we used negative-staining electron microscopy (EM) (Fig. 3A). The images showed that all the nanoparticles were monodispersed particles, and S1-conjugated nanoparticles displayed a rougher surface than the unconjugated nanoparticles. Dynamic light scattering (DLS) was then used to measure the diameters of the nanoparticles (Fig. 3B). After conjugation, the hydrodynamic diameter of S1-conjugated nanoparticles was increased, while the homogeneity of each particle type remained unaffected, which was in accordance to the predicted structural diagram and structures deduced from EM images.
FIG 3.
Structural characterization of S1 nanoparticles shows increased size of nanoparticles and preserved high uniformity. (A) Negative-staining electronic microscopy (EM) of unconjugated nanoparticles and S1-conjugated nanoparticles. After conjugation, the outer surface of nanoparticles was thickened, and the diameters of nanoparticles were increased. (B) Dynamic light scattering (DLS) of unconjugated nanoparticles and S1-conjugated nanoparticles. The hydrodynamic diameter of S1 antigen, unconjugated nanoparticles, and S1-conjugated nanoparticles is displayed. The mean normalized intensity from 3 replicate experiments is presented.
To summarize, we successfully produced three S1 nanoparticles of different sizes and S1 valences and determined their structural characteristics and protein stability. It was shown that the production of S1 nanoparticles through the covalent conjugation strategy could achieve both high efficiency and great structural stability, while the increased valency and overall size could benefit antigen presentation and recognition, facilitating the immune response, which was further investigated in our study.
In vitro antigenicity characterization of S1 nanoparticles.
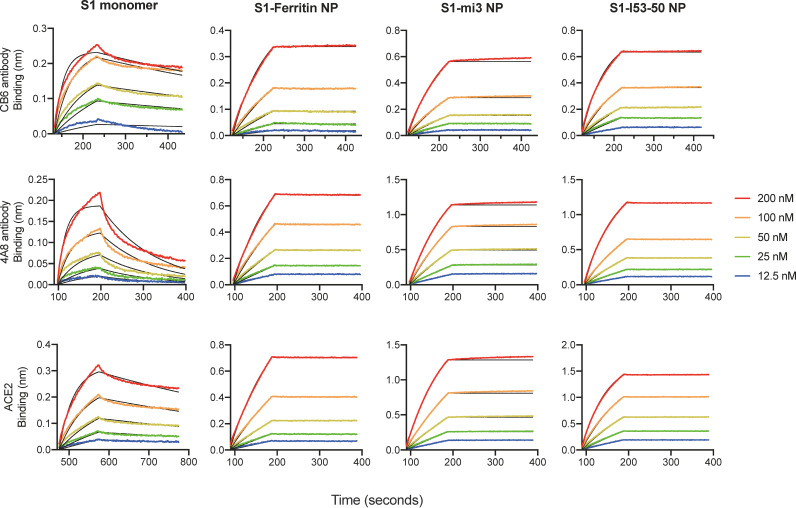
To study the in vitro antigenicity of S1 nanoparticles, we purified human ACE2 and two neutralizing antibodies, CB6 against RBD and 4A8 against NTD. We further measured the binding affinity of monomeric S1 and three nanoparticles for the purified receptor and antibodies by biolayer interferometry (BLI), to assess any changes of S1 antigenicity after conjugation to the nanoparticles (Fig. 4 and Table 1). As the receptor of SARS-CoV-2, ACE2 binds to the RBD and mediates viral entrance into host cells, and its binding affinity reflects the integrity of the RBD as part of the S1 antigen. In contrast, CB6 is a neutralizing monoclonal antibody (MAb) isolated from a patient convalescing from COVID-19, which was reported to bind the RBD and introduce steric hindrance as well as direct competition to the binding of ACE2 (53). Similarly, 4A8, is another potent neutralizing monoclonal antibody that binds to the NTD and is speculated to restrain the conformational change of the S protein (22). Determination of the binding affinity to these antibodies can be used as a proxy for the preservation of the neutralizing epitopes on the RBD and NTD of S1, reflecting antigen integrity as well as the exposure of neutralizing epitopes in our designed vaccine candidates (54).
FIG 4.
Antigenicity characterization of S1-SpyTag and S1-conjugated nanoparticles. The S1 monomer and S1-based nanoparticles are associated with the CB6 and 4A8 captured on the ProA biosensors or with ACE2 captured on SA by biolayer interferometry to determine the binding capabilities. The detailed kinetic parameters (KD, Kon, and Koff) are presented in Table 1.
TABLE 1.
Kinetic analysis of reactions of the S1 monomer and S1-conjugated nanoparticle vaccines to SARS-CoV-2 MAbs and ACE2 receptor
| Kinetic assay pair | Result fora: |
|||||
|---|---|---|---|---|---|---|
|
KD
|
K
on
|
K
off
|
||||
| M | SE | 1/M · s | SE | 1/s | SE | |
| CB6 antibody | ||||||
| S1 | 5.48E−09 | 1.52E−10 | 2.43E+05 | 4.26E+03 | 1.33E−03 | 2.86E−05 |
| S1-ferritin NP | <1.0E−12 | 1.78E−10 | 6.07E+04 | 7.90E+02 | <1.0E−07 | |
| S1-mi3 NP | <1.0E−12 | 2.69E−10 | 5.71E+04 | 1.11E+03 | <1.0E−07 | |
| S1-I53-50 NP | <1.0E−12 | 1.01E−10 | 8.64E+04 | 7.07E+02 | <1.0E−07 | |
| 4A8 antibody | ||||||
| S1 | 3.41E−08 | 1.03E−09 | 2.34E+05 | 6.63E+03 | 7.97E−03 | 7.83E−05 |
| S1-ferritin NP | 1.18E−09 | 1.53E−10 | 5.14E+04 | 5.96E+02 | 6.07E−05 | 7.83E−06 |
| S1-mi3 NP | <1.0E−12 | 1.89E−10 | 5.49E+04 | 8.23E+02 | <1.0E−07 | |
| S1-I53-50 NP | 7.36E−11 | 5.86E−11 | 7.39E+04 | 3.36E+02 | 5.44E−06 | 4.33E−06 |
| ACE2 receptor | ||||||
| S1 | 1.13E−08 | 2.30E−10 | 1.34E+05 | 1.94E+03 | 1.51E−03 | 2.15E−05 |
| S1-ferritin NP | 5.47E−10 | 1.50E−10 | 4.35E+04 | 4.68E+02 | 2.38E−05 | 6.52E−06 |
| S1-mi3 NP | <1.0E−12 | 1.95E−10 | 5.59E+04 | 8.29E+02 | <1.0E−07 | |
| S1-I53-50 NP | 1.04E−10 | 5.77E−11 | 8.90E+04 | 4.45E+02 | 9.23E−06 | 5.13E−06 |
Shown are the binding kinetics of S1-based immunogens and SARS-CoV-2 MAbs (CB6 and 4A8) or ACE2 receptor determined by biolayer interferometry. The kinetic parameter is summarized in the table. KD, affinity constant; Kon, kinetic constant of associations; Koff, kinetic constant of dissociations. The KD value is calculated from Koff/Kon.
Notably, all three S1-conjugated nanoparticles displayed higher binding affinity to ACE2 and the two antibodies recognizing different domains of S1 (Table 1), and the dissociation of the nanoparticles from both receptor and antibodies was notably delayed. Especially, after conjugation to nanoparticles, the binding affinity for the NTD-specific antibody 4A8 was significantly increased over 30-fold. These results indicated that the S1-conjugated nanoparticles had a superior binding affinity to the specific antibodies or B cell receptor (BCR) compared to the S1 monomer.
Immunization with S1-based nanoparticles induced potent neutralizing antibodies against SARS-CoV-2.
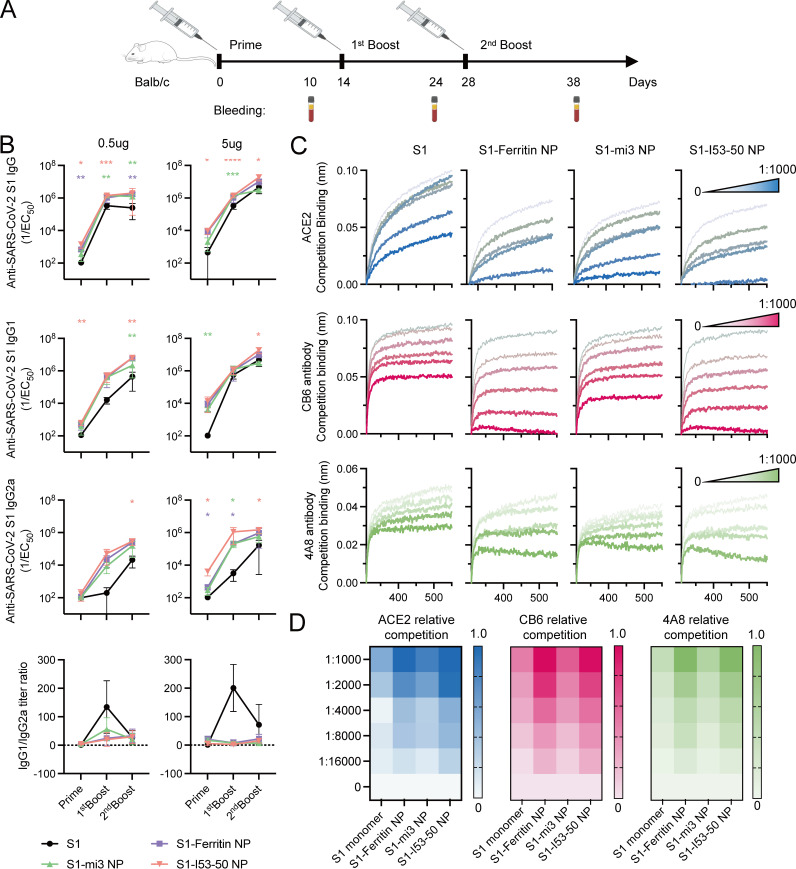
Since S1-conjugated nanoparticles exhibited high stability and improved antigenicity, we examined their efficacy in eliciting an immune response in vivo. BALB/c mice were immunized with 0.5 or 5 μg S1 monomer or an equimolar amount of the monomer displayed on S1-mi3 NP, S1-ferritin NP, or S1-I53-50 NP at 0, 2 and 4 weeks (Fig. 5A). The injected immunogens were all formulated with 50% (vol/vol) MF59 adjuvant, and mice injected with phosphate-buffered saline (PBS) incorporating 50% adjuvant were included as a negative-control group. Serum samples were collected 10 days after each immunization.
FIG 5.
Immunization of S1 nanoparticle vaccine in mice elicited a higher antibody titer with stronger competition to ACE2 and neutralizing antibodies. (A) Diagram of the 3-dose mouse immunization schedule. All mice received 3-dose vaccinations on days 0, 14, and 28, and their blood samples were harvested 10 days after each immunization for the experiment. (B) Serum total IgG, IgG1, and IgG2a antibody titers of mice immunized with the S1 monomer or S1 nanoparticles at an equal molar ratio of 0.5 μg or 5 μg S1. The mean titer from 5 mice from each group with the standard deviation (SD) of each group is presented. Two-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test was performed to calculate the statistical significance between S1 and each S1 nanoparticle at different immunization points. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Serum competitive assay with ACE2 and neutralizing antibodies CB6 and 4A8. The competitive binding signal of ACE2, CB6, and 4A8 in the presence of serially diluted serum from the 5-μg dose groups at day 38 is presented. A lower binding signal indicated a stronger competition between certain dilution level of serum and ACE2, CB6, or 4A8. (D) Heat maps of the relative competition level of serum to ACE2 and neutralizing antibodies CB6 and 4A8. The relative competition at each dilution level was based on ratio of the final binding signal of a certain diluted serum curve (Bindc) and the non-serum curve (Bind0) and was calculated by (1 − Bindc)/Bind0. A darker color in each heat map indicates a stronger competition.
We first measured the serum binding titer (total anti-S1 IgG) as well as the IgG1 and IgG2a titers of each group (Fig. 5B). The S1 nanoparticles elicited binding antibody titers equivalent or superior to those elicited by the S1 monomer, regardless of antibody subtypes. Furthermore, mice immunized with vaccines containing a higher valency of the S1 component were able to generate higher peak serum binding titers. Additionally, it was observed that the three S1 nanoparticles could elicit higher titers of the IgG2a subtype than monomeric S1, indicating the transition to a preferentially Th1-type immune response, which may benefit the clearance of pathogens (55–57). However, the S1-mi3 nanoparticle elicited an inferior response at a dose of 5 μg, which was similar to the effect of the same dose of S1 monomer in terms of total binding titer. To further investigate the effect of the elicited sera in blocking viral infection, we used a competitive binding assay to determine the binding epitopes of serum antibodies, which was directly associated with the neutralization efficiency. By saturation of SARS-CoV-2 S1 captured on the biosensors with different concentrations of mouse sera from groups immunized with NP amounts equimolar to 5 μg S1 at day 38, the competition of ACE2 or antibodies CB6 and 4A8 by serum antibody in binding with S1 could be observed (Fig. 5C and D). It was observed that the overall competition was in accordance with the binding titer. The S1-ferritin NP and S1-I53-50 NP groups displayed approximately 4- to 5-times-stronger competition with ACE2 and the two antibodies than the S1 monomer, while S1-mi3 NP was similar to the S1 monomer. Nevertheless, the sera from all groups immunized with S1 components included antibodies targeting the RBD and NTD regions, as suggested by the strong competition with CB6 binding to the RBD and 4A8 binding to the NTD.
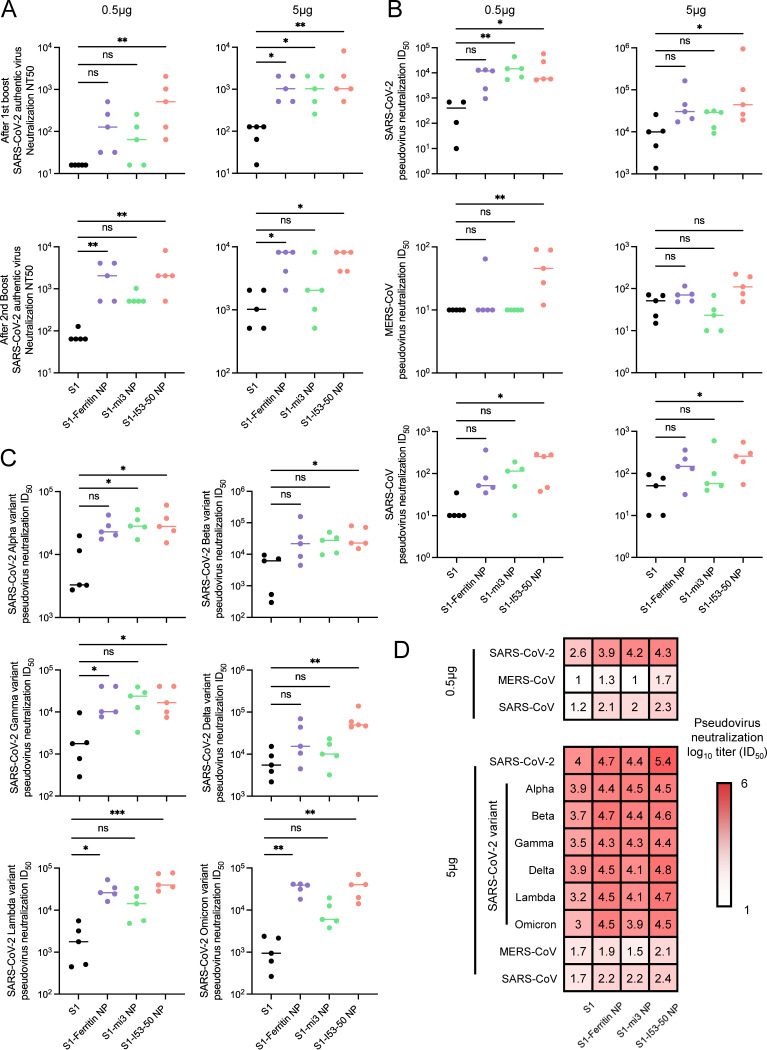
To confirm whether the generated antibodies could neutralize viral infection, we performed authentic virus and pseudovirus neutralization assays to determine the efficacy of the elicited mouse sera. With the increase of vaccination dose and with revaccination, the 50% neutralizing titer (NT50) of mouse sera against authentic wild-type SARS-CoV-2 increased. Moreover, S1 nanoparticles displayed a significant advantage over the S1 monomer (Fig. 6A). After a 3-dose immunization with a dose of NPs equimolar to 5 μg of S1, S1-ferritin NP and S1-I53-50 NP could elicit over 5-times-higher neutralizing antibody titers than S1 monomer, and even the titer elicited by S1-mi3 NP was nearly 3 times higher. Moreover, when the dose of each immunization was limited to 0.5 μg, the superiority of S1 nanoparticles was even more dramatic, and the average titer ratios of S1-ferritin NP, S1-mi3 NP, and S1-I53-50 NP to S1 monomer were 30, 8, and 39, respectively. Although S1-mi3 NP was inferior to the other two nanoparticles in terms of the elicited neutralizing antibody titer, which exhibited a similar trend to the serum binding titer, the S1-conjugated nanoparticles showed a clear advantage in inducing neutralizing antibodies against SARS-CoV-2 infection, especially when the antigen dose was relatively lower.
FIG 6.
Authentic and pseudovirus neutralization assays displayed robust and cross-reactivity of serum antibody against SARS-CoV-2 variants and other betacoronaviruses elicited by S1 nanoparticle vaccines. (A) Authentic SARS-CoV-2 virus neutralization assay of mice sera from 0.5-μg and 5-μg dose groups after 1st boost and 2nd boost immunization based on the cytopathic effect (CPE) method. The neutralization titer is represented by NT50, indicating the serum titer inhibiting 50% of CPE. (B) Pseudovirus SARS-CoV-2, MERS-CoV, and SARS-CoV neutralization assay of mice sera from the 0.5-μg and 5-μg dose groups after 2nd boost immunization. (C) Variant neutralization assay of pseudovirus SARS-CoV-2 variants of concern (VOCs) Alpha, Beta, Gamma, Delta, Lambda, and Omicron of mice sera from the 5-μg dose group after the 2nd boost immunization. The neutralization titer of groups in panels B and C is represented by ID50, indicating the serum titer inhibiting 50% of the luciferase signal. (D) Pseudovirus neutralization titer ID50 heat map. Darker red represents a higher neutralization titer. The individual and mean titer from all 5 mice in each group are presented in panels B to D. The Kruskal-Wallis test was performed to calculate the statistical significance between S1 and each S1 nanoparticle. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
S1 nanoparticles elicited higher titers of cross-reactive neutralizing antibodies against SARS-CoV-2 variants and other coronaviruses.
After confirming the protective effect of the S1 nanoparticle vaccination against wild-type SARS-CoV-2, we explored whether there was still a neutralizing effect of the elicited antibodies against common VOCs, including the Alpha, Beta, Gamma, Delta, Lambda, and Omicron variants. Corresponding pseudoviruses were produced by introducing genes encoding the spike proteins from different variants into a lentiviral vector, which produced a luciferase signal after infection. The results confirmed that S1 nanoparticles elicited neutralizing antibody titers against wild-type SARS-CoV-2 pseudovirus equivalent or superior to those of the S1 monomer, regardless of the 0.5- or 5-μg immunization dose (Fig. 6B). Moreover, similar results were observed with the tested SARS-CoV-2 variants (Fig. 6C). In detail, the ratio of the advantage of the neutralizing antibody titer elicited by S1 nanoparticles over the titer elicited by S1 monomer varied from 4 to 10 across the assays, and the overall titers against different strains were quite close, suggesting that neutralizing antibodies induced by wild-type S1 antigen still effectively blocked the cell entry of SARS-CoV-2 variant pseudoviruses (Fig. 6D).
As SARS-CoV-2 belongs to the betacoronavirus family, which includes other human-pathogenic viruses, the homology of spike protein sequences may allow S1-based immunogens to elicit cross-reactive antibodies against sibling species (5, 58–61). Therefore, we also produced SARS-CoV and MERS-CoV pseudoviruses to investigate the cross-neutralization effect. As expected, the neutralizing antibody titers against SARS-CoV and MERS-CoV of sera from SARS-CoV-2 S1-immunized mice were significantly lower than those against SARS-CoV-2 pseudovirus (Fig. 6B). However, the S1 nanoparticles still elicited neutralizing titers equivalent or superior to those of the S1 monomer, in the same manner as in the SARS-CoV-2 neutralization assays.
DISCUSSION
The pandemic of SARS-CoV-2 has caused over 250 million confirmed cases globally, including over 5 million deaths (1). Since the outbreak, intensive efforts have been focused on vaccine development, and several vaccines entered clinical trials at unprecedented speed and have been widely applied in the population (62, 63). However, the rapid emergence of SARS-CoV-2 variants tempered the initial optimism of containing the pandemic through vaccination. Among the currently documented VOCs, the Beta, Gamma, Delta, and Omicron variants were reported to exhibit a significant capacity to escape neutralization by wild-type-elicited antibodies, and clinical investigations indicated that Beta, Omicron, and Delta variants possibly reduced the protective effect of vaccination against symptomatic disease (64, 65). The present epidemiological situation required new vaccination strategies that elicit potent and cross-protective immune response. We therefore also investigated whether our designed vaccines could effectively elicit protection against spreading SARS-CoV-2 variants, in addition to measuring neutralizing antibody titers against wild-type SARS-CoV-2.
Most vaccines against SARS-CoV-2 currently target the S protein, which consists of a membrane-distal S1 subunit and a membrane-proximal S2 subunit. The S1 subunit includes an N-terminal domain (NTD), a receptor-binding domain (RBD) that binds to the cellular ACE2 receptor, and a C-terminal domain (CTD) (12). Numerous studies found that neutralizing antibodies target the RBD, further confirming its promising potential as an immunogen for vaccine design (14, 66, 67). However, the newly emerging VOCs harbor a variety of mutations in the RBD region that significantly alter its antigenicity, decreasing the binding affinity of neutralizing antibodies and diminishing the sensitivity to antibodies and vaccine-elicited sera (68–70). In contrast to the RBD, the NTD received far less attention due to its uncertain role in viral infection. Recently, the neutralizing antibody 4A8 targeting NTD was found to have strong neutralizing activity against SARS-CoV-2, and AXL was found to be a host receptor for SARS-CoV-2 infection binding to the NTD region of spike protein. These results suggested that NTD may be critical for viral infection as well, and its value in vaccine development deserved further investigation.
Since the emergence of SARS-CoV-2 VOCs, the efficacy and breadth of licensed vaccine-elicited sera to neutralization activity have diminished. However, all SARS-CoV-2 vaccines approved and authorized for emergency use were developed and produced based on the sequence of the wild-type strain, so the neutralization capacity of vaccine-induced sera in mice or nonhuman primates to circulating VOCs remains poorly understood, especially for the emerging Omicron variants (71). In this study, we observed that three S1-conjugated nanoparticle vaccines induced a robust and broad antibody response to SARS-CoV-2 VOCs and also elicited a certain level of neutralizing antibodies against the other betacoronaviruses. Dose-sparing vaccination also induced comparable neutralizing antibody titers, which is essential for “good manufacturing practice” (GMP) production and for popularization and use in low-income countries. Due to the induction of a potent and durable antibody response by the vaccine, the protein-based nanoparticle platforms for next-generation vaccines, including genetic fusion, chemical conjugation, and protein coupling methods to attach antigens on nanoparticles, have been widely developed to combat global emerging infectious diseases in recent years, such as influenza and AIDS. Our and other research groups developed the protein-based presentation platforms (such as 24-meric ferritin, 60-meric LS, I3-01 [mi3], and E2P, and 120-meric I53-50) using genetic fusion or coupling strategies to design and produce self-assembled RBD or prefusion spike-based nanoparticle vaccine (72–75). Although these nanoparticle vaccines induced high antibody titers to neutralize SARS-CoV-2 and provided protection against SARS-CoV-2 infection, discrepancies may arise due to differences in the size, flexibility, antigen valency, and spacing of nanoparticle immunogens and adjuvant. In addition, how these critical factors affect the humoral immune responses induced by nanoparticle vaccine needs to be further explored. Prior studies have clarified that multivalent HIV antigens with more valency (8-mer and 60-mer) preferentially activated the cognate B cell response and induced higher titers than antigens with less valency (1-mer and 4-mer) (33). Here, we observed that vaccination with 24-mer S1-ferritin NP and 120-mer S1-I53-50 induced higher neutralizing antibodies in mice than vaccination with 60-mer S1-mi3. We speculated the possible reason is that the orientations and angles of S1 antigen displayed on the surface of mi3 nanoparticles are different, which affects the conjugated efficacy and results in less exposure of accessible epitopes.
In this study, the SpyCatcher-SpyTag system was used to covalently link S1 to protein-based self-assembling nanoparticles. This design allowed the independent production and purification of antigen and nanoparticles, accelerating the general vaccine development process. Homogeneity and stability were then tested to validate successful production and characterize the nanoparticle vaccine candidates. All three designs exhibited considerable homogeneity, especially S1-ferritin NP and S1-I53-50 NP, which is vital to avoid unnecessary risks of vaccination, as well as stability at different storage temperatures, which is critical to lower the costs of storage and transportation and make vaccines more accessible. We then compared the efficacy of our designed S1-conjugated nanoparticles with that of the S1 monomer in eliciting protection against SARS-CoV-2, its variants, and other pathogenic betacoronaviruses. We found that presenting S1 on the surface of our designed nanoparticles greatly enhanced its antigenicity and induced much higher neutralizing activities against pseudoviruses. Furthermore, these S1-based antigens elicited comparable levels of neutralizing antibodies against wild-type SARS-CoV-2 and its variants. Moreover, S1-I53-50 NP also induced considerable protection against cell entry by SARS-CoV and MERS-CoV pseudoviruses. As SARS-CoV-2 VOCs are spreading widely and new variants are expected to emerge, our study provides important tools for the development of more advanced vaccines in the near future.
MATERIALS AND METHODS
Cells and viruses.
HEK293T cells stably expressing human ACE2 (hACE2-293T) were generated by transducing HEK293T cells with recombinant lentivirus expressing hACE2. The positive hACE2-293T cells were selected based on puromycin resistance. Vero E6, Vero, and Huh-7 cells were purchased from ATCC. All adherent cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37°C with 5% CO2. Expi293F cells were cultured in 293 cell culture medium (Union-Biotech [Shanghai] Co., Ltd.; catalog no. UP1000) in conical shake flasks at 120 rpm, 37°C, and 5% CO2. All cultured cell lines were free of mycoplasma. The SARS-CoV-2 virus strain (Guangdong/2020XN4276-P0037/2020) was isolated from a female COVID-19 patient in Guangzhou (GISAID no. EPI_ISL_413859), passaged six times in Vero E6 cells, and frozen at –80°C in a biosafety level 3 (BSL3) laboratory.
Gene synthesis and plasmid construction.
The gene fragments encoding SARS-CoV-2 S1 (residues 16 to 685, GenBank no. MN908947) and human ACE2 (residues 19 to 615, GenBank no. NM_021804) were codon optimized for human cells and synthesized by GenScript, and then they were cloned into mammalian expression vector VRC8400 using a single restriction site (BamHI). All plasmids constructed for eukaryotic expression contained a consensus Kozak sequence and signal peptide sequence at the N terminus of the protein of interest for protein secretion, as well as an octahistidine tag at the C terminus for purification. A 13-residue SpyTag was added to the C terminus of S1 for the construction of S1-SpyTag. SpyCatcher-ferritin, SpyCatcher-mi3, SpyCatcher-I53-50A1.1PT1, and I53-50B.4PT1 fused with a hexahistidine tag were codon optimized and synthesized by GenScript and then cloned into a modified pET28a+ expression vector as described previously. To generate SARS-CoV-2-specific monoclonal antibodies (MAbs), the IgG heavy- and light-chain variable region gene sequences of CB6 MAb (GenBank no. MT470196 and MT470197) and 4A8 MAb (GenBank no. MT622682.1 and MT622683.1) were codon optimized for human cells and synthesized by GenScript, and then they were cloned into expression vectors containing the IgG1 constant region.
Protein expression and purification in HEK293F cells.
Expi293F cells at a concentration of 1.5 × 106/mL were transiently transfected with plasmid expressing soluble SARS-CoV-2 S1 with or without a 13-residue SpyTag or soluble hACE2 using polyethylenimine (PEI) (Polysciences, Inc.; catalog no. 24675-1) to generate soluble S1-SpyTag and hACE2 proteins. Five days after transfection, culture supernatants containing the recombinant proteins were collected and centrifuged to discard cell debris. The supernatants were filtered through a 0.22-μm-pore-size vacuum-driven filter (Guangzhou Jet Bio-Filtration Co., Ltd.; catalog no. FPE204000) and applied to a gravity column loaded with Ni Sepharose Excel nickel resin (Cytiva, Inc.; catalog no. 17371201) and washed with a mixture of 50 mM Tris-HCl (pH 7.0), 300 mM NaCl, and 30 mM imidazole, followed by elution of the protein of interest with a mixture of 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 300 mM imidazole. After elution, the purified proteins were concentrated and additionally purified by size exclusion chromatography (SEC) using a Superose 200 Increase 10/300 GL column equilibrated with 50 mM HEPES (pH 8.0) and 300 mM NaCl. SARS-CoV-2 S-specific monoclonal antibodies CB6 (53) and 4A8 (22) were produced and purified similarly. The filtered supernatants were applied to gravity column loaded with protein A resin (GenScript; catalog no. L00210-50), washed with PBS, and eluted with 0.2 M glycine (pH 3.0).
Protein expression and purification in E. coli.
The SpyCatcher-ferritin, SpyCatcher-mi3, SpyCatcher-I53-50A1.1PT1 and I53-50B.4PT1 proteins were expressed and purified as described previously, with minor modifications as follows. Rosetta(DE3) cells carrying the indicated expression plasmid were cultured in Terrific Broth (TB) medium supplemented with 50 μg/mL kanamycin and 33 μg/mL chloramphenicol at 37°C with shaking at 200 rpm. After incubation for 16 h, 3-L baffled shaker flasks containing 1 L of TB medium with 50 μg/mL kanamycin and 33 μg/mL chloramphenicol were inoculated with the 10-mL culture, incubated for an additional 3 h under the same conditions, and induced by the addition of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, followed by protein expression for 20 h at 18°C. Cells were collected by centrifugation and resuspended in lysis buffer consisting of 50 mM HEPES (pH 8.0), 300 mM NaCl, 30 mM imidazole, 0.75% 3-[(3-cholamidepropyl)dimethylammonio]-1-propanylsulfonate (CHAPS), 1 mM dithiothreitol (DTT), and EDTA-free protease inhibitor cocktail, followed by lysis in a high-pressure cell homogenizer (Union-Biotech). The cleared lysates were loaded onto Ni-NTA resin (Roche) on a WET FRED gravity flow column (IBA), washed with lysis buffer, and then eluted with a mixture of 50 mM HEPES (pH 8.0), 300 mM NaCl, 300 mM imidazole, 0.75% CHAPS, 1 mM DTT, and EDTA-free protease inhibitor cocktail. The eluted fractions were further purified by SEC on a Superose 6 Increase 10/300 GL column. Then, endotoxin was removed using ToxinEraser endotoxin removal kit (GenScript) according to the manufacturer’s protocol. The ToxinSensor chromogenic LAL endotoxin assay kit (GenScript) was used to qualify the content of remnant bacterial endotoxin in protein purified from E. coli, and protein samples assaying at less than 0.1 EU/mL were considered endotoxin-free.
Preparation of S1-conjugated nanoparticles.
To generate the SARS-CoV-2 S1-conjugated nanoparticle vaccine, a subunit concentration of 50 to 100 μM purified SpyCatcher-ferritin NP and SpyCatcher-mi3 NP was mixed with an 8-fold molar excess of S1-SpyTag in a mixture of 50 mM HEPES (pH 8.0) and 300 mM NaCl and incubated at 4°C overnight to allow autocatalytic covalent conjugation in vitro. The mixture was separated to remove unconjugated S1-SpyTag by SEC on a Superose 6 Increase 10/300 GL column preequilibrated with PBS. For S1-I53-50 NP assembly, 50 μM purified SpyCatcher-I53-50A1.1PT1 trimer was mixed with an 8-fold molar excess of S1-SpyTag and incubated at 4°C overnight in conjugation buffer (50 mM HEPES [pH 8.0] and 300 mM NaCl). After SEC on a Superose 200 10/300 Increase GL column, target proteins were immediately collected and pooled and then incubated with an equimolar ratio of purified I53-50B.4PT1 at room temperature for 2 h, followed by further purification by SEC using a Superose 6 Increase 10/300 GL column preequilibrated with PBS. The concentrations of all purified proteins were determined using a bicinchoninic acid (BCA) assay kit according to the manufacturer ‘s instructions.
Dynamic light scattering.
Dynamic light scattering (DLS) was applied to observe the particle diameter and size distribution of purified S1-conjugated nanoparticles. Briefly, S1-SpyTag monomer and S1-conjugated nanoparticles (S1-ferritin NP, S1-mi3 NP, and S1-I53-50 NP) were diluted to a concentration of 0.5 mg/mL in PBS, after which 40 μL of target protein was added to a solvent-resistant disposable microcuvette and measured on a Zetasizer Ultra instrument (Malvern Panalytical). The polydispersity index and hydrodynamic diameter of purified proteins were measured in triplicate and analyzed according to the manufacturer’s protocol (Malvern Panalytical).
Negative-staining electron microscopy.
Samples comprising 5 μL of purified S1-ferritin NP, S1-mi3 NP, or S1-I53-50 NP at a concentration of 0.5 mg/mL in PBS were dripped onto the surface of parafilm and adsorbed onto glow-discharged 400-mesh carbon-coated copper grids (Ted Pella) for 1 min. Excess protein solution was gently wicked away using absorbing paper. The grids were negatively stained with 2% (wt/vol) uranyl acetate for 1 min. Excess staining solution was wicked away in the same manner. Samples were imaged at 120 kV using a Talos L120C transmission electron microscope (FEI) equipped with a CETA 16M CMOS camera.
SDS-PAGE analysis.
Purified proteins were mixed with 2× sample buffer containing bromophenol blue, oiled at 100°C for 5 min, loaded onto the 10.5% Tris-glycine gel, and subjected to electrophoresis for 30 min at 300 V in running buffer. The gel was stained with 0.125% (wt/vol) Coomassie brilliant blue R-250, destained in buffer containing ethanol and acetic acid, and photographed using a ChemiDoc system (Bio-Rad).
Biolayer interferometry.
Biolayer interferometry (BLI) was performed on an Octet R8 instrument (Sartorius) to determine the antigenicity of S1 and S1 nanoparticles (kinetic assay) and serum competition with ACE2 and the antibodies CB6 and 4A8 (serum competition assay).
For the kinetic assay, ACE2 was labeled with biotin using the EZ-Link NHS-LC-LC-biotin labeling kit (ThermoFisher), and biotinylated ACE2 or the CB6 and 4A8 antibodies were captured on the SA or ProA biosensors. Then, different concentrations of S1 or S1 nanoparticles were associated with the biosensors, followed by a dissociation process using PBS with 0.5% Tween 20. The raw curves were recorded and processed using the Octet Analysis Studio software, and a 1:1 binding model was used to fit the curves and calculate the kinetic parameters.
To perform the serum competition assay, S1 was biotinylated and captured on the SA biosensors. Mouse serum samples were taken at 10 weeks postimmunization with 5 μg of S1 or the equimolar dose of S1 nanoparticles, and sera from the same group were mixed as representative samples. Then, the S1 on the biosensors was saturated with serially diluted serum samples for 300 s to simulate the blocking of spike protein on the SARS-CoV-2 virion. Afterwards, 500 nM ACE2 or 200 nM CB6 or 4A8 was further associated with the biosensors to observe the competitive binding. When more antibody bound to the corresponding epitope of ACE2 or antibodies in sera at a certain dilution level, less ACE2 or antibodies could bind to S1, leading to a low binding signal during the association with the biosensor. The ratio of the peak binding signal from each curve representing different dilution levels of sera to the peak from the curve without serum was recorded as the relative competition level.
SARS-CoV-2 pseudovirus production.
Pseudoviruses corresponding to the SARS-CoV-2 prototype and variants (including Alpha, Beta, Gamma, Delta, Lambda, and Omicron), as well as two additional human-pathogenic betacoronaviruses (SARS-CoV and MERS-CoV), were produced using the classical HIV-based method as previously described (76). Briefly, HEK293T cells were cotransfected with the reporter plasmid pNL4-3-Luc-R(−)E(−), expressing a firefly luciferase gene, and eukaryotic expression vector pCMV14, harboring the indicated spike proteins, using PEI. After incubation for 48 to ~72 h, pseudovirus-containing supernatants were harvested, filtered through a 0.45-μm-pore-size membrane, aliquoted, and frozen at –80°C. Viral stocks were 2-fold serially diluted, and viral titers were determined based on the luciferase activity of infected cells expressed in relative light units (RLU).
BALB/c mouse vaccination and sample collection.
All immunization studies were conducted according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Sun Yat-sen University Cancer Center. The project was approved by the ethics committee of the Sun Yat-sen University Cancer Center (approval no. L102042020000A). Six-week-old female BALB/c mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd., divided into 9 groups (n = 5 mice each), and housed in a quarantine room at the experimental animal facility of Sun Yat-sen University Cancer Center. After quarantine for 1 week, the mice were move into ventilated cages in an environmental barrier room. The mice were subcutaneously immunized with 5 μg S1-SpyTag or an equimolar amount of S1-ferritin NP (6.46 μg), S1-mi3 NP (6.46 μg), or S1-I53-50 NP (7.21 μg) or 0.5 μg S1-SpyTag or an equimolar amount of S1-ferritin NP (0.646 μg), S1-mi3 NP (0.646 μg), or S1-I53-50 NP (0.721 μg) formulated with MF59-like adjuvant (0.5% [vol/vol] Span 85, 0.5% [vol/vol] Tween 80, and 4.3% [vol/vol] squalene buffered with 10 mM sodium citrate) (77) at weeks 0, 2, and 4. MF59-like adjuvant alone was used a negative control. After 10 days following each immunization, mice were bled, and serum samples were harvested, heat inactivated at 56°C for 30 min, and then stored at –20°C for further use.
Serum antibody ELISA.
Polystyrene high-bind flat-bottom 96-well microplates (Corning; product no. 42592) were precoated with 100 μL of 1 μg/mL purified S1-SpyTag in PBS and incubated overnight at 4°C. The plates were washed three times with PBST and then blocked with 1× blocking buffer for 12 h at 4°C, followed by washed three times with PBST. Mouse sera were serially 5-fold diluted with blocking buffer, added to the plates, incubated for 1 h at 37°C, and washed five times. Horseradish peroxidase (HRP)-conjugated goat anti-mouse total IgG, IgG1, or IgG2a secondary antibody (Abcam) was diluted 5,000-fold in blocking buffer and added for 45 min at 37°C, followed by five washes. The reaction was developed using EL-TMB chromogenic substrate for 15 min at 37°C and stopped according to the manufacturer’s protocol (Sangon Biotech [Shanghai] Co., Ltd.). The absorbance at 450 and 630 nm was measured using a SpectraMax Plus plate reader (Molecular Devices, USA). The half-maximal effective concentration (EC50) values were calculated from the recorded curves using four-parameter nonlinear fitting in GraphPad Prism 9.0.
Pseudovirus neutralization assay.
Vaccine-elicited mouse sera were 4-fold serially diluted with complete DMEM at a starting ratio of 10, mixed with an equal volume of pseudoviruses, and incubated at 37°C for 2 h. The mixtures were added to 96-well Nunclon Delta-treated microporous plates (Thermo Fisher Scientific) preseeded with 1.75 × 104 HEK293T-ACE2 or Huh-7 cells per well the night before, incubated for 12 h, and replaced with fresh complete medium. Corresponding pseudoviruses and medium were included as positive and negative controls in each plate, respectively. After 48 h postinfection, the overlying medium was removed, and cells were lysed with 100 μL per well of culture medium containing Steady-Glo buffer and substrate (Promega) for 10 min at room temperature. The firefly luciferase activity of infected and uninfected cells was measured using a GloMax 96 microplate luminometer (Promega) in accordance to manufacturer’s protocol. The inhibition rate was calculated as ×100%. Neutralization curves were recorded, and half-maximal inhibitory doses (ID50s) of sera against pseudoviruses were calculated using four-parameter logistic regression in GraphPad Prism 8.0.1 software.
Authentic SARS-CoV-2 virus neutralization assay.
The authentic SARS-CoV-2 neutralization assay was performed in the BSL3 facility, which strictly complied with operating rules and regulations. The night before the infection, 1.5 × 105 Vero E6 cells/mL were seeded into 96-well microplates (100 μL/well). Serum samples were serially 4-fold diluted from 16 to 16,384 in minimal essential medium (MEM) supplemented with 2% FBS and 1% penicillin-streptomycin and incubated with an equal volume of 100 tissue culture half-infective doses (TCID50s) of live SARS-CoV-2 virus 2020XN4276 at 37°C. After incubation for 2 h, the mixtures were added to the 96-well plates. The cytopathic effect (CPE) induced by SARS-CoV-2 was observed daily for 4 days. Cells, virus, and minimum dilution of serum samples were included in each plate as controls. All serum samples were measured in duplicate. Authentic SARS-CoV-2 viruses were also serially 10-fold diluted and back-titrated in each assay. The 50% neutralization titers (NT50s) of serum samples were defined as the highest dilution that results in a 50% reduction of infection by authentic SARS-CoV-2 virus in Vero E6 cells by 4 days postinfection.
Statistical analysis.
Statistical analysis and visualization of the experimental data were performed using GraphPad Prism 9.0 software. The method used for determining statistical significance between groups is presented in the figure legends.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (no. 2017YFA0505600) and by the National Natural Science Foundation of China (no. 82030046).
Mu-Sheng Zeng and Yin-Feng Kang conceived and designed the study and revised the manuscript. Cong Sun performed kinetic and competitive assays. Cong Sun, Yin-Feng Kang, and Chu Xie drafted the manuscript. Yin-Feng Kang constructed plasmids and performed animal experiments. Run-Yu Yuan and Jiu-Feng Sun performed the authentic virus neutralization assay. Xin-Yan Fang performed the negative-staining electronic microscopy assay. Yi-Sha Hu performed visualization of the simulated protein structure. Xiao-Hui Yu and Zheng Liu provided constructive suggestions.
We declare no conflict of interest.
Contributor Information
Mu-Sheng Zeng, Email: zengmsh@sysucc.org.cn.
Yin-Feng Kang, Email: kangyf@sysucc.org.cn.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.WHO. 2021. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 24 November 2021.
- 2.Shang W, Yang Y, Rao Y, Rao X. 2020. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines 5:18. 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J, Coronaviridae Study Group. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. 2020. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect 26:729–734. 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Rescue Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, Wu P, Xie S, Bian W, Zhang C, Sun Z, Liu K, Shan C, Lin A, Jiang S, Xie Y, Zhou Q, Lu L, Huang J, Li X. 2021. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31:126–140. 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Xiao T, Cai Y, Chen B. 2021. Structure of SARS-CoV-2 spike protein. Curr Opin Virol 50:173–182. 10.1016/j.coviro.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai L, Gao GF. 2021. Viral targets for vaccines against COVID-19. Nat Rev Immunol 21:73–82. 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haslwanter D, Dieterle ME, Wec AZ, O'Brien CM, Sakharkar M, Florez C, Tong K, Rappazzo CG, Lasso G, Vergnolle O, Wirchnianski AS, Bortz RH, III, Laudermilch E, Fels JM, Mengotto A, Malonis RJ, Georgiev GI, Quiroz JA, Wrapp D, Wang N, Dye KE, Barnhill J, Dye JM, McLellan JS, Daily JP, Lai JR, Herbert AS, Walker LM, Chandran K, Jangra RK. 2021. A combination of receptor-binding domain and N-terminal domain neutralizing antibodies limits the generation of SARS-CoV-2 spike neutralization-escape mutants. mBio 12:e0247321. 10.1128/mBio.02473-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krammer F. 2020. SARS-CoV-2 vaccines in development. Nature 586:516–527. 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 17.WHO. 2021. COVID-19 vaccine tracker and landscape. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Accessed 24 November 2021.
- 18.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 19.Ravichandran S, Coyle EM, Klenow L, Tang JJ, Grubbs G, Liu SF, Wang T, Golding H, Khurana S. 2020. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci Transl Med 12:eabc3539. 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, Nargi RS, Sutton RE, Winkler ES, Chen EC, Fouch ME, Davidson E, Doranz BJ, Chen RE, Shi P-Y, Carnahan RH, Thackray LB, Diamond MS, Crowe JE, Jr.. 2021. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 184:2316–2331.e15. 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCallum M, De Marco A, Lempp F, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:2332–2347.e16. 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. 2020. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369:650–655. 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang G-Y, Ho DD, Sheng Z, Kwong PD, Shapiro L. 2021. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29:819–833.e7. 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Cao L, Gao X-S, Zheng B-Y, Deng Y-Q, Li J-X, Feng R, Bian Q, Guo X-L, Wang N, Qiu H-Y, Wang L, Cui Z, Ye Q, Chen G, Lu K-K, Chen Y, Chen Y-T, Pan H-X, Yu J, Yao W, Zhu B-L, Chen J, Liu Y, Qin C-F, Wang X, Zhu F-C. 2021. A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2. Natl Sci Rev 8:nwab053. 10.1093/nsr/nwab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grint DJ, Wing K, Houlihan C, Gibbs HP, Evans SJW, Williamson E, McDonald HI, Bhaskaran K, Evans D, Walker AJ, Hickman G, Nightingale E, Schultze A, Rentsch CT, Bates C, Cockburn J, Curtis HJ, Morton CE, Bacon S, Davy S, Wong AYS, Mehrkar A, Tomlinson L, Douglas IJ, Mathur R, MacKenna B, Ingelsby P, Croker R, Parry J, Hester F, Harper S, DeVito NJ, Hulme W, Tazare J, Smeeth L, Goldacre B, Eggo RM. 2021. Severity of severe acute respiratory system coronavirus 2 (SARS-CoV-2) Alpha variant (B.1.1.7) in England. Clin Infect Dis 2021:ciab754. 10.1093/cid/ciab754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2021. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184:2372–2383.e9. 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O'Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE, Corti D, Virgin HW, Ellebedy AH, Shi P-Y, Diamond MS. 2021. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27:717–726. 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdool Karim SS, de Oliveira T. 2021. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N Engl J Med 384:1866–1868. 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes MI, Lunn SM, Famulare M, Frisbie LA, Painter I, Burstein R, Roychoudhury P, Xie H, Mohamed Bakhash SA, Perez R, Lukes M, Ellis S, Sathees S, Mathias PC, Greninger A, Starita LM, Frazar CD, Ryke E, Zhong W, Gamboa L, Threlkeld M, Lee J, McDermot E, Truong M, Nickerson DA, Bates DL, Hartman ME, Haugen E, Nguyen TN, Richards JD, Rodriguez JL, Stamatoyannopoulos JA, Thorland E, Melly G, Dykema PE, MacKellar DC, Gray HK, Singh A, Peterson JM, Russell D, Marcela Torres L, Lindquist S, Bedford T, Allen KJ, Oltean HN. 2022. Associations between SARS-CoV-2 variants and risk of COVID-19 hospitalization among confirmed cases in Washington State: a retrospective cohort study. Clin Infect Dis. 10.1093/cid/ciac279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freitas ARR, Beckedorff OA, Cavalcanti LPG, Siqueira AM, Castro DB, Costa CFD, Lemos DRQ, Barros ENC. 2021. The emergence of novel SARS-CoV-2 variant P.1 in Amazonas (Brazil) was temporally associated with a change in the age and sex profile of COVID-19 mortality: a population based ecological study. Lancet Reg Health Am 1:100021. 10.1016/j.lana.2021.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang Y-F, Sun C, Zhuang Z, Yuan R-Y, Zheng Q, Li J-P, Zhou P-P, Chen X-C, Liu Z, Zhang X, Yu X-H, Kong X-W, Zhu Q-Y, Zhong Q, Xu M, Zhong N-S, Zeng Y-X, Feng G-K, Ke C, Zhao J-C, Zeng M-S. 2021. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano 15:2738–2752. 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 33.Kato Y, Abbott RK, Freeman BL, Haupt S, Groschel B, Silva M, Menis S, Irvine DJ, Schief WR, Crotty S. 2020. Multifaceted effects of antigen valency on B cell response composition and differentiation in vivo. Immunity 53:548–563.e8. 10.1016/j.immuni.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshyar N, Gray S, Han H, Bao G. 2016. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine (Lond) 11:673–692. 10.2217/nnm.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foged C, Brodin B, Frokjaer S, Sundblad A. 2005. Particle size and surface charge affect particle uptake by human dendritic cells in an in vitro model. Int J Pharm 298:315–322. 10.1016/j.ijpharm.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y-N, Lazarovits J, Poon W, Ouyang B, Nguyen LNM, Kingston BR, Chan WCW. 2019. Nanoparticle size influences antigen retention and presentation in lymph node follicles for humoral immunity. Nano Lett 19:7226–7235. 10.1021/acs.nanolett.9b02834. [DOI] [PubMed] [Google Scholar]
- 37.Routhu NK, Cheedarla N, Bollimpelli VS, Gangadhara S, Edara VV, Lai L, Sahoo A, Shiferaw A, Styles TM, Floyd K, Fischinger S, Atyeo C, Shin SA, Gumber S, Kirejczyk S, Dinnon KH, Shi P-Y, Menachery VD, Tomai M, Fox CB, Alter G, Vanderford TH, Gralinski L, Suthar MS, Amara RR. 2021. SARS-CoV-2 RBD trimer protein adjuvanted with Alum-3M-052 protects from SARS-CoV-2 infection and immune pathology in the lung. Nat Commun 12:3587. 10.1038/s41467-021-23942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaheer T, Pal K, Zaheer I. 2021. Topical review on nano-vaccinology: biochemical promises and key challenges. Process Biochem 100:237–244. 10.1016/j.procbio.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, Seth A, Wibowo N, Zhao CX, Mitter N, Yu CZ, Middelberg APJ. 2014. Nanoparticle vaccines. Vaccine 32:327–337. 10.1016/j.vaccine.2013.11.069. [DOI] [PubMed] [Google Scholar]
- 40.Kheirollahpour M, Mehrabi M, Dounighi NM, Mohammadi M, Masoudi A. 2020. Nanoparticles and vaccine development. Pharm Nanotechnol 8:6–21. 10.2174/2211738507666191024162042. [DOI] [PubMed] [Google Scholar]
- 41.Lu L, Duong VT, Shalash AO, Skwarczynski M, Toth I. 2021. Chemical conjugation strategies for the development of protein-based subunit nanovaccines. Vaccines 9:563. 10.3390/vaccines9060563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee A, Howarth M. 2018. Nanoteamwork: covalent protein assembly beyond duets towards protein ensembles and orchestras. Curr Opin Biotechnol 51:16–23. 10.1016/j.copbio.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Reddington SC, Howarth M. 2015. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr Opin Chem Biol 29:94–99. 10.1016/j.cbpa.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Jiang S, Zhang X, Yang Y, Hotez PJ, Du L. 2020. Neutralizing antibodies for the treatment of COVID-19. Nat Biomed Eng 4:1134–1139. 10.1038/s41551-020-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. 2012. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA 109:E690–E697. 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong W-P, Wang L, Nabel GJ. 2013. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499:102–106. 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruun TU, Andersson A-MC, Draper SJ, Howarth M. 2018. Engineering a rugged nanoscaffold to enhance plug-and-display vaccination. ACS Nano 12:8855–8866. 10.1021/acsnano.8b02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bale JB, Gonen S, Liu Y, Sheffler W, Ellis D, Thomas C, Cascio D, Yeates TO, Gonen T, King NP, Baker D. 2016. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353:389–394. 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcandalli J, Fiala B, Ols S, Perotti M, de van der Schueren W, Snijder J, Hodge E, Benhaim M, Ravichandran R, Carter L, Sheffler W, Brunner L, Lawrenz M, Dubois P, Lanzavecchia A, Sallusto F, Lee KK, Veesler D, Correnti CE, Stewart LJ, Baker D, Loré K, Perez L, King NP. 2019. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176:1420–1431.e17. 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues MQ, Alves PM, Roldão A. 2021. Functionalizing ferritin nanoparticles for vaccine development. Pharmaceutics 13:1621. 10.3390/pharmaceutics13101621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen AA, Yang Z, Gnanapragasam PN, Ou S, Dam K-MA, Wang H, Bjorkman PJ. 2021. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS One 16:e0247963. 10.1371/journal.pone.0247963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, Aye P, Navarro MJ, Lai L, Edara VV, Röltgen K, Rogers K, Shirreff L, Ferrell DE, Wrenn S, Pettie D, Kraft JC, Miranda MC, Kepl E, Sydeman C, Brunette N, Murphy M, Fiala B, Carter L, White AG, Trisal M, Hsieh C-L, Russell-Lodrigue K, Monjure C, Dufour J, Spencer S, Doyle-Meyers L, Bohm RP, Maness NJ, Roy C, Plante JA, Plante KS, Zhu A, Gorman MJ, Shin S, Shen X, Fontenot J, Gupta S, O'Hagan DT, Van Der Most R, Rappuoli R, Coffman RL, Novack D, McLellan JS, Subramaniam S, et al. 2021. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594:253–258. 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 53.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, Gao G, Hu X, Zhang Y, Tong Z, Huang W, Liu WJ, Wu G, Zhang B, Wang L, Qi J, Feng H, Wang F-S, Wang Q, Gao GF, Yuan Z, Yan J. 2020. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584:120–124. 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 54.Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Freund NT, Weiss Y. 2007. Epitope mapping. BioDrugs 21:145–156. 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snapper CM, Paul WE. 1987. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science 236:944–947. 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 56.Stevens TL, Bossie A, Sanders VM, Fernandez-Botran R, Coffman RL, Mosmann TR, Vitetta ES. 1988. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 334:255–258. 10.1038/334255a0. [DOI] [PubMed] [Google Scholar]
- 57.Bretscher P. 2014. On the mechanism determining the TH1/TH2 phenotype of an immune response, and its pertinence to strategies for the prevention, and treatment, of certain infectious diseases. Scand J Immunol 79:361–376. 10.1111/sji.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med 28:174–184. [PubMed] [Google Scholar]
- 60.Lv Z, Deng Y-Q, Ye Q, Cao L, Sun C-Y, Fan C, Huang W, Sun S, Sun Y, Zhu L, Chen Q, Wang N, Nie J, Cui Z, Zhu D, Shaw N, Li X-F, Li Q, Xie L, Wang Y, Rao Z, Qin C-F, Wang X. 2020. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369:1505–1509. 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grobben M, van der Straten K, Brouwer PJ, Brinkkemper M, Maisonnasse P, Dereuddre-Bosquet N, Appelman B, Lavell AA, van Vught LA, Burger JA, Poniman M, Oomen M, Eggink D, Bijl TP, van Willigen HD, Wynberg E, Verkaik BJ, Figaroa OJ, de Vries PJ, Boertien TM, Bomers MK, Sikkens JJ, Le Grand R, de Jong MD, Prins M, Chung AW, de Bree GJ, Sanders RW, van Gils MJ. 2021. Cross-reactive antibodies after SARS-CoV-2 infection and vaccination. eLife 10:e70330. 10.7554/eLife.70330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kashte S, Gulbake A, El-Amin SF, III, Gupta A. 2021. COVID-19 vaccines: rapid development, implications, challenges and future prospects. Human Cell 34:711–733. 10.1007/s13577-021-00512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Tenchov R, Smoot J, Liu C, Watkins S, Zhou Q. 2021. A comprehensive review of the global efforts on COVID-19 vaccine development. ACS Cent Sci 7:512–533. 10.1021/acscentsci.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID 19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:757–773. 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan M, Liu H, Wu NC, Wilson IA. 2021. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem Biophys Res Commun 538:192–203. 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Min L, Sun Q. 2021. Antibodies and vaccines target RBD of SARS-CoV-2. Front Mol Biosci. 10.3389/fmolb.2021.671633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, Pere H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Marti-Carreras J, Cuypers L, Seve A, Hocqueloux L, Prazuck T, Rey FA, Simon-Loriere E, Bruel T, Mouquet H, Andre E, Schwartz O. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 69.Liu L, Iketani S, Guo Y, Chan JF, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KK, Yuen TT, Yoon C, To KK, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen KY, Ho DD. 2022. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602:676–681. 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 70.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, et al. 2022. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602:664–670. 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He X, Hong W, Pan X, Lu G, Wei X. 2021. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm (2020) 2:838–45. 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joyce MG, King HAD, Elakhal-Naouar I, Ahmed A, Peachman KK, Macedo Cincotta C, Subra C, Chen RE, Thomas PV, Chen W-H, Sankhala RS, Hajduczki A, Martinez EJ, Peterson CE, Chang WC, Choe M, Smith C, Lee PJ, Headley JA, Taddese MG, Elyard HA, Cook A, Anderson A, McGuckin Wuertz K, Dong M, Swafford I, Case JB, Currier JR, Lal KG, Molnar S, Nair MS, Dussupt V, Daye SP, Zeng X, Barkei EK, Staples HM, Alfson K, Carrion R, Krebs SJ, Paquin-Proulx D, Karasavva N, Polonis VR, Jagodzinski LL, Amare MF, Vasan S, Scott PT, Huang Y, Ho DD, de Val N, Diamond MS, et al. 2022. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci Transl Med 14:eabi5735. 10.1126/scitranslmed.abi5735. [DOI] [PubMed] [Google Scholar]
- 73.Wuertz KM, Barkei EK, Chen W-H, Martinez EJ, Lakhal-Naouar I, Jagodzinski LL, Paquin-Proulx D, Gromowski GD, Swafford I, Ganesh A, Dong M, Zeng X, Thomas PV, Sankhala RS, Hajduczki A, Peterson CE, Kuklis C, Soman S, Wieczorek L, Zemil M, Anderson A, Darden J, Hernandez H, Grove H, Dussupt V, Hack H, de la Barrera R, Zarling S, Wood JF, Froude JW, Gagne M, Henry AR, Mokhtari EB, Mudvari P, Krebs SJ, Pekosz AS, Currier JR, Kar S, Porto M, Winn A, Radzyminski K, Lewis MG, Vasan S, Suthar M, Polonis VR, Matyas GR, Boritz EA, Douek DC, Seder RA, Daye SP, et al. 2021. A SARS-CoV-2 spike ferritin nanoparticle vaccine protects hamsters against Alpha and Beta virus variant challenge. NPJ Vaccines 6:129. 10.1038/s41541-021-00392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joyce MG, Chen W-H, Sankhala RS, Hajduczki A, Thomas PV, Choe M, Martinez EJ, Chang WC, Peterson CE, Morrison EB, Smith C, Chen RE, Ahmed A, Wieczorek L, Anderson A, Case JB, Li Y, Oertel T, Rosado L, Ganesh A, Whalen C, Carmen JM, Mendez-Rivera L, Karch CP, Gohain N, Villar Z, McCurdy D, Beck Z, Kim J, Shrivastava S, Jobe O, Dussupt V, Molnar S, Tran U, Kannadka CB, Soman S, Kuklis C, Zemil M, Khanh H, Wu W, Cole MA, Duso DK, Kummer LW, Lang TJ, Muncil SE, Currier JR, Krebs SJ, Polonis VR, Rajan S, McTamney PM, et al. 2021. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep 37:110143. 10.1016/j.celrep.2021.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geng Q, Tai W, Baxter VK, Shi J, Wan Y, Zhang X, Montgomery SA, Taft-Benz SA, Anderson EJ, Knight AC, Dinnon KH, III, Leist SR, Baric RS, Shang J, Hong SW, Drelich A, Tseng CK, Jenkins M, Heise M, Du L, Li F. 2021. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog 17:e1009897. 10.1371/journal.ppat.1009897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun C, Kang YF, Liu YT, Kong XW, Xu HQ, Xiong D, Xie C, Liu YH, Peng S, Feng GK, Liu Z, Zeng MS. 2022. Parallel profiling of antigenicity alteration and immune escape of SARS-CoV-2 Omicron and other variants. Signal Transduct Target Ther 7:42. 10.1038/s41392-022-00910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schultze V, D'Agosto V, Wack A, Novicki D, Zorn J, Hennig R. 2008. Safety of MF59 adjuvant. Vaccine 26:3209–3222. 10.1016/j.vaccine.2008.03.093. [DOI] [PubMed] [Google Scholar]