ABSTRACT
The antibody response against the HIV-1 envelope glycoproteins (Envs) guides evolution of this protein within each host. Whether antibodies with similar target specificities are elicited in different individuals and affect the population-level evolution of Env is poorly understood. To address this question, we analyzed properties of emerging variants in the gp41 fusion peptide-proximal region (FPPR) that exhibit distinct evolutionary patterns in HIV-1 clade B. For positions 534, 536, and 539 in the FPPR, alanine was the major emerging variant. However, 534A and 536A show a constant frequency in the population between 1979 and 2016, whereas 539A is gradually increasing. To understand the basis for these differences, we introduced alanine substitutions in the FPPR of primary HIV-1 strains and examined their functional and antigenic properties. Evolutionary patterns could not be explained by fusion competence or structural stability of the emerging variants. Instead, 534A and 536A exhibited modest but significant increases in sensitivity to antibodies against the membrane-proximal external region (MPER) and gp120-gp41 interface. These Envs were also more sensitive to poorly neutralizing sera from HIV-1-infected individuals than the clade ancestral form or 539A variant. Competition binding assays confirmed for all sera tested the presence of antibodies against the base of the Env trimer that compete with monoclonal antibodies targeting the MPER and gp120-gp41 interface. Our findings suggest that weakly neutralizing antibodies against the trimer base are commonly elicited; they do not exert catastrophic population size reduction effects on emerging variants but, instead, determine their set point frequencies in the population and historical patterns of change.
IMPORTANCE Infection by HIV-1 elicits formation of antibodies that target the viral Env proteins and can inactivate the virus. The specific targets of these antibodies vary among infected individuals. It is unclear whether some target specificities are shared among the antibody responses of different individuals. We observed that antibodies against the base of the Env protein are commonly elicited during infection. The selective pressure applied by such antibodies is weak. As a result, they do not completely eliminate the sensitive forms of the virus from the population, but maintain their frequency at a low level that has not increased since the beginning of the AIDS pandemic. Interestingly, the changes in Env do not occur at the sites targeted by the antibodies, but at a distinct region of Env, the fusion peptide-proximal region, which regulates their exposure.
KEYWORDS: HIV-1, envelope glycoproteins, fusion peptide-proximal region, membrane-proximal external region, population-level evolution, antibody neutralization, immune selection, virus evolution
INTRODUCTION
The envelope glycoproteins (Envs) of HIV-1 are primary targets in AIDS vaccine design (1, 2). However, the Envs are not “stagnant” targets. Antigenic properties of these proteins have gradually changed during the AIDS pandemic among strains that circulate in the population (3–6). As a result, epitopes targeted by several broadly neutralizing antibodies (BNAbs) are found in decreasing proportions of strains (3). Changes in Env are caused by errors that occur during reverse transcription of the viral genome, which continuously introduce new variants in the host (7, 8). Persistence of the variants and their fixation in the population are determined by the selective pressures applied to them, including (i) fitness pressures, (ii) immune pressures, and (iii) bottlenecks that act during transmission (9–12). The relative roles of these forces in determining the population-level changes in Env are unclear. Nevertheless, some evidence suggests that their combined effects are similar in different populations. A recent study by Han and colleagues examined the amino acid diversity at different positions of Env (13). They observed that in distinct geographic regions, each Env position has evolved toward a specific frequency distribution of amino acids that replaced the regional ancestral form. The frequency distribution is specific to the HIV-1 clade and is similar in monophyletic and paraphyletic lineages, suggesting that, at a population level, Env encounters similar forces that guide its evolution (13).
Fitness pressures play a major role in determining properties of the emerging variants that can persist in the host (14); they guide Env toward structurally stable states that effectively recognize the entry receptors and mediate fusion with target cells. Fitness pressures are likely similar in different individuals during the same stage of infection (14, 15). In contrast, the target specificity of immune pressures applied in different hosts varies considerably (16, 17). Whether some targets are shared between antibody (Ab) responses elicited in different hosts and alter the evolutionary course of Env structure in the population is unclear (16, 18). That such common immune pressures may exist is suggested by the differences between conformational properties of Envs from lab-adapted strains and primary HIV-1 isolates. The Envs of primary isolates exhibit closed conformations that conceal immunogenic gp120 epitopes overlapping the coreceptor-binding site (CoR-BS) and CD4-BS (19–22). Lab adaptation of HIV-1 is often associated with changes to more open forms that expose these epitopes, which can exhibit reduced requirements for CD4 (22, 23). Subsequent passaging of the lab-adapted viruses in animal models of HIV-1 infection causes reversion to more closed conformations of the trimer (24). Such changes suggest that immune pressures reduce the frequency of forms that expose immunogenic epitopes on gp120. Whether similar pressures also affect the gp41 subunit and their impacts on conformation of the trimer are still unknown.
To better understand the contribution of immune pressures to the population-level changes in Env, we analyzed properties of emerging Env variants in a region of gp41 that exhibits a complex pattern of evolution. The fusion peptide-proximal region (FPPR), also designated the polar region, is located C terminal to the fusion peptide, extending from Env positions 528 to 540 (25, 26). Analyses of soluble Env trimers suggest that the FPPR is conformationally flexible (27, 28). Nevertheless, substitutions at some FPPR positions increase detachment of gp120 from virions (26, 29, 30), supporting the notion that this region contributes to structural stability of the trimer. We analyzed the historical changes in amino acid sequence of the FPPR among clade B viruses and tested the emerging variants for their function, stability, and antigenicity. We found that emerging FPPR variant 539A, which is increasing in its frequency, exhibits modestly higher fusion competence than the clade ancestral form and a similar neutralization phenotype. In contrast, FPPR variants 534A and 536A, which show a historically constant frequency, exhibit a unique “open-at-the-base” trimer conformation that exposes partially cryptic gp41 epitopes. Analysis of serum samples with low neutralization efficacy showed that Abs against the base of the Env trimer are commonly elicited in HIV-infected individuals. Our findings suggest that population-level changes in the FPPR can be explained by weak Ab pressures applied to the base of the Env trimer.
RESULTS
FPPR positions show distinct patterns of change during the 40 years of the AIDS pandemic.
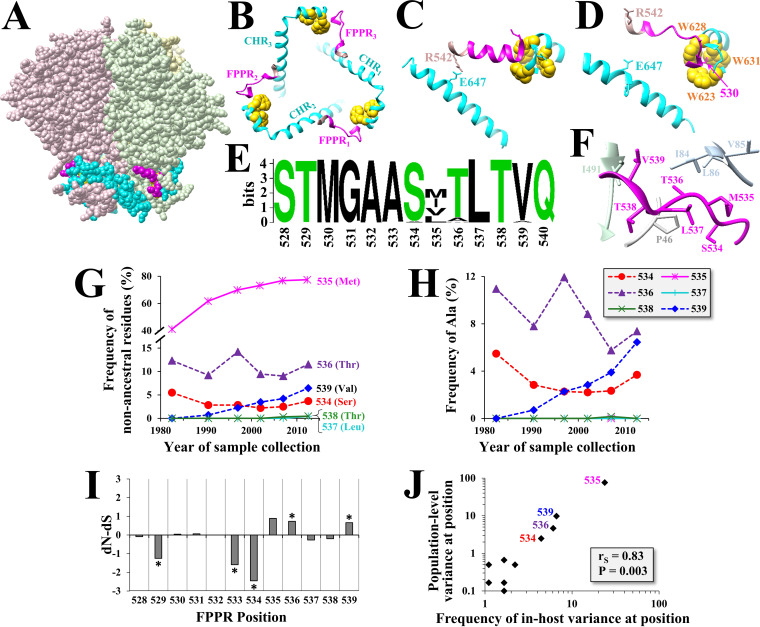
Cryo-electron microscopy (Cryo-EM) reconstructions of soluble gp140 trimers suggest that the FPPR resides at the base of the trimer and is only partially exposed (Fig. 1A) (31–34). The FPPR and C-terminal heptad repeat (CHR) of the three gp41 protomers appear to form a ring structure at the base of the trimer, which is supported by interactions between the two flanks of each FPPR and the CHRs of the same and adjacent protomers (Fig. 1B). At the N terminus of the FPPR, Met530 resides in a clasp-like structure formed by three Trp residues from the CHR of the same protomer (28, 32). Positioning of the C terminus of the FPPR and secondary structure of its midsegment appear to vary between the cryo-EM structures obtained in the presence and absence of CD4 (Fig. 1C and D, respectively). In the presence of CD4, Arg542, which flanks the FPPR, is positioned 2.9 Å from Glu647 in the CHR of the adjacent protomer and “seals” the ring at the base of the trimer. In the absence of CD4, the FPPR forms a helix-turn-helix structure in which Arg542 is more distantly positioned from Glu647. Consistent with this arrangement, the FPPR midsegment (residues 531 to 539) was suggested to be conformationally flexible in the unliganded form of Env (27, 28). The contribution of this region to the structure and function of Env is still poorly understood.
FIG 1.
Positions in the FPPR midsegment show distinct patterns of evolution. (A) Cryo-EM structure of the HIV-1 Env trimer strain B41 (PDB ID 6U59). The FPPR is colored in pink and the CHR in cyan. (B) Top view of the ring structure at the base of the trimer. The three residues of the Trp clasp are colored in yellow. (C) Relationship between the FPPR and CHR region in the B41 Env trimer bound to CD4 and antibody 17b (PDB ID 5VN3). (D) Relationship between the FPPR and CHR in B41 Env bound to antibody 13B (PDB ID 6U59). (E) Web logo of amino acid distribution in the FPPR among 1,576 clade B isolates circulating worldwide between 1979 and 2016. Polar and hydrophobic residues are colored in green and black, respectively. (F) Side view of the FPPR in the unliganded Env (PDB ID 6U59). Residues analyzed in this study are labeled in magenta. Other Env residues at distances smaller than 4 Å are shown. (G) Historical changes in amino acid sequence diversity at FPPR positions 534 to 539 among clade B isolates. Each data point describes the percentage of isolates with nonancestral residues for the indicated 5- to 7-year period (as a percentage of all strains from that period). Clade ancestral residues are shown in the 3-letter code. (H) Historical changes in frequency of Ala in clade B isolates at positions 534 to 539. (I) Estimated rates of nonsynonymous (dN) and synonymous (dS) substitutions were calculated for each codon using 6,285 clade B sequences. The test statistic indicates the difference between dN and dS values. Asterisks indicate the P value for rejecting the null hypothesis of neutral evolution: *, P < 0.05. (J) Correlation between in-host variance and population-level diversity at FPPR positions. A panel of 4,252 Env sequences from 181 patients infected by HIV-1 clade B was analyzed (10 to 80 sequences per patient). The percentage of patients that contain amino acid variability at each position is shown and compared with the frequency of nonancestral residues at the same position among 1,576 clade B Envs isolated from samples collected worldwide.
Analysis of 1,576 Envs from HIV-1 clade B showed limited variation in amino acid sequence at all FPPR positions except 535 (Fig. 1E). However, closer analysis of the historical changes in amino acid diversity in the FPPR midsegment (positions 534 to 539, Fig. 1F) revealed interesting patterns of change. Position 535 showed a rapid increase in the frequency of emerging variants during the pandemic and appears to have reached a plateau in the early 2000s (Fig. 1G). Position 539 showed a more gradual increase; the clade ancestral Val was mainly replaced by Ala (Fig. 1G and H). At positions 534 and 536, the ancestral residues (Ser and Thr, respectively) were also replaced by Ala; however, diversity and Ala frequency remained at a constant level (or decreased) during the pandemic (Fig. 1H).
To better understand these patterns, we examined FPPR codons for evidence of positive or negative selection. Nonsynonymous (dN) and synonymous (dS) substitution rates were calculated for 6,285 HIV-1 clade B isolates. The dN-dS statistic was used as an indicator of positive selection (dN-dS > 0) or purifying selection (dN-dS < 0). Positions 536 and 539 showed high positive dN-dS values. The dN-dS value of position 535, which rapidly diversified in the population, was also high, albeit not statistically significant. In contrast, dN-dS values for positions 529, 533, and 534 were negative, suggestive of purifying selection (Fig. 1I). Phylogenetic analysis of the distribution of Ala variants at position 534, 536, or 539 among clade B Envs revealed they do not localize to specific sublineages of this clade (data not shown).
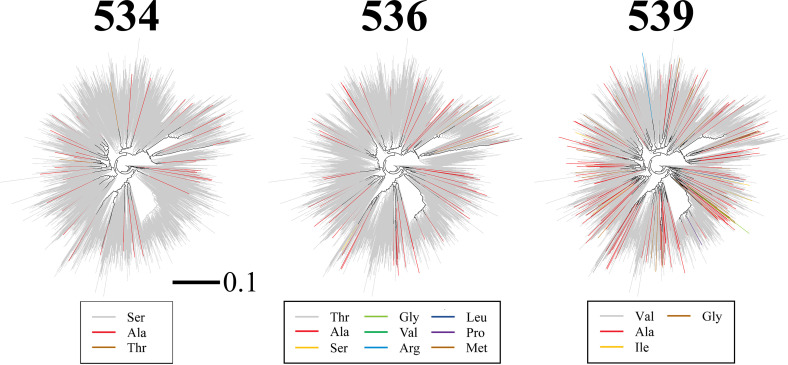
To further investigate the selective pressures applied on FPPR positions, we examined the in-host variance in amino acid sequence at each position of this domain. We previously showed that the level of variance in each Env feature within the host is conserved among hosts and correlates well with the population-level diversity of the feature (3). The in-host variance in amino acid sequence at all FPPR positions was analyzed using a panel of 4,252 Env sequences isolated from blood samples of 181 individuals (10 to 80 Env sequences per sample). For each position of the FPPR, we determined the percentage of patients that contain variance in amino acid sequence. This frequency was compared with the diversity at each position in clade B, as measured by the percentage of strains that did not contain the clade ancestral residue. A strong correlation was observed between the frequency of in-host variance at each position and its population-level diversity (Fig. 1J). This finding suggests that the selective pressures applied in the host on FPPR positions determine their patterns of change in the population. We note that the emerging Ala variants at positions 534, 536, and 539 did not localize to a specific lineage of clade B (see annotated phylogenetic trees in Fig. 2), indicating they represent independent substitution events.
FIG 2.
Envs with alanine substitutions at position 534, 536, or 539 do not localize to a specific lineage of HIV-1 clade B. Shown is a phylogenetic tree describing evolutionary relationships between Envs of 1,576 HIV-1 isolates from distinct individuals. Sequences are derived from samples collected worldwide between 1979 and 2016. The tree was reconstructed from nucleotide sequences of the entire env gene using the maximum likelihood method. Branches are colored by the amino acid contained at position 534, 536, or 539. The scale bar describes the number of substitutions per site.
Therefore, S534, T536, and V539 in the clade B ancestor were replaced primarily by Ala. However, the three positions show different patterns of historical changes in Ala frequency; 539A is gradually increasing, whereas 534A and 536A have remained constant (or decreased). Positions 536 and 539 show evidence for positive selection, whereas position 534 shows evidence for negative selection.
Population-level changes in amino acid sequence of the FPPR are not fully explained by fusion competence or stability of the emerging variants.
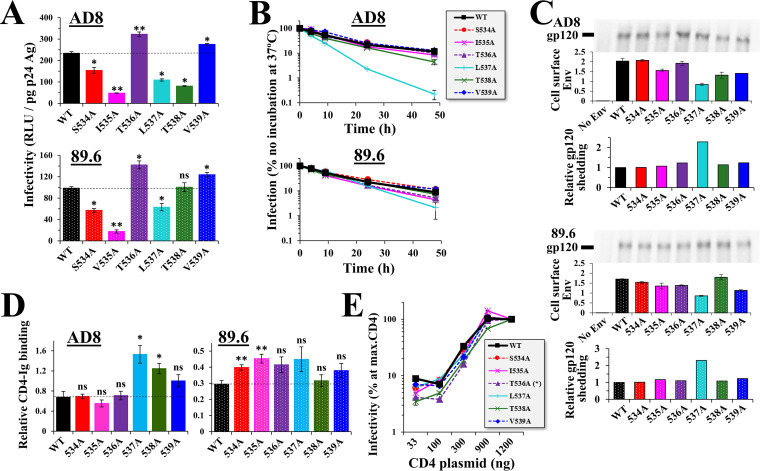
To better understand the selective pressures applied on the FPPR, we examined the effects of Ala substitutions at positions 534 to 539 on fusion competence of Env. The changes were first introduced in the Env of HIV-1 strain AD8, which occupies a “closed” conformation and exhibits a neutralization-resistant profile of a tier-2 virus (35–37). We also introduced the changes in the Env of HIV-1 strain 89.6, which occupies a partially open conformation and exhibits a neutralization-sensitive profile between tiers 1 and 2 (35). Replication-defective pseudoviruses that contain the Env variants and express the luciferase protein were generated and tested for their infectivity. In both AD8 and 89.6 Envs, Ala substitutions at positions 536 and 539 resulted in modest but significant increases in infection relative to the wild-type (WT) forms that contain the clade B ancestral residues (Fig. 3A). The change at position 534 caused a modest but significant decrease in infectivity. Other positions, which less frequently contain Ala in primary HIV-1 isolates, showed various levels of decrease in infectivity relative to the WT Envs.
FIG 3.
Fusion competence and stability of FPPR variants does not explain their emergence patterns in the population. (A) Infectivity of viruses that contain Envs with Ala substitutions at positions 534 to 539. Replication-defective viruses that express the luciferase protein and contain the indicated variants of Envs AD8 or 89.6 were used to infect Cf2Th-CD4+ CCR5+ cells. Infectivity was measured by luciferase activity (in relative light units [RLU]) and is expressed as a fraction of p24 antigen content in each sample. Error bars, standard error of the means (SEM). Statistical significance of the differences between infectivity of WT Env and the Ala variants was calculated using the results of 4 to 6 independent experiments by an unpaired t test: *, P < 0.05; **, P < 0.005. (B) Stability of FPPR variants at 37°C. Viruses were incubated at 37°C for different time periods and added to Cf2Th-CD4+ CCR5+ cells. Infectivity values are expressed as the percentage of infection measured in samples not preincubated at 37°C. (C) Shedding of gp120 from cells that express WT and mutant Envs. HOS cells were transfected by the indicated Env variants. Three days later, the supernatant was collected and the gp120 content was immunoprecipitated using protein A beads and MAbs 2G12, VRC03, and PGT121. Samples were analyzed by SDS-PAGE, and the blot was probed with goat anti-gp120 IgG. Values are expressed relative to the binding of the above MAb mixture to HOS cells that express the Env variants, as measured by ELISA. (D) Binding efficiency of CD4 to HOS cells expressing the FPPR variants. Env-expressing cells were incubated with the CD4-Ig probe or with MAb 2G12 (both at 0.5 μg/mL). CD4-Ig binding is expressed as a fraction of 2G12 binding. (E) CD4 usage efficiency. HEK 293T cells were transfected with 900 ng of plasmid that expresses human CCR5 and the indicated amounts of plasmid that expresses human CD4. Two days later, cells were seeded in 96-well plates, and viruses were added. Infectivity measured for each condition is expressed as a percentage of that measured at the maximal amount of CD4 plasmid. The amount of CD4 plasmid required for half-maximal infectivity was calculated by linear regression, and replicate values were compared between the WT and variants using an unpaired t test: *, P < 0.05.
We previously showed that a Ser-to-Pro change at position 532 of the FPPR increases shedding of gp120 from Env trimers (26). We thus examined the effects of substitutions at positions 534 to 539 on Env trimer stability. We first determined functional stability of the variants, by measuring sensitivity of the viruses that contain them to incubation at 37°C. Primary strains of HIV-1 exhibit similar sensitivities to this treatment; half-lives at 37°C usually range between 7 and 10 h (38). In both strains, Ala substitutions at positions 534, 535, 536, 538, and 539 did not alter sensitivity to 37°C, whereas the 537A variant was considerably more sensitive than WT Env (Fig. 3B). We also examined the effects of the substitutions on structural stability of the Envs, by measuring spontaneous shedding of gp120 from Env trimers. For this purpose, human osteosarcoma (HOS) cells were transfected by the WT and mutant Envs. Shed gp120 was immunoprecipitated from the supernatant and quantified as a fraction of Env expressed on the surface of the cells. Consistent with the measurements of functional stability, only the 537A variants exhibited higher levels of gp120 shedding that the WT Envs (Fig. 3C).
To determine if differences in infectivity (Fig. 3A) result from changes in receptor recognition, we measured binding efficiency of the CD4 receptor to the Env variants expressed on the surface of HOS cells. In contrast to other cell types, HOS cells express only fully cleaved trimers on their surface (39). We measured binding of the Env mutants to the CD4-Ig probe, which is composed of two copies of domains 1 and 2 of the CD4 protein linked to the Fc region of human IgG1. To normalize for the cell surface expression level of each Env, we also measured in the same experiment binding of the 2G12 Ab, which recognizes an exposed epitope on the high-mannose patch of gp120 (40, 41). For Env AD8, the 537A and 538A variants exhibited modest but significantly higher binding of CD4-Ig than the WT Env, whereas other variants were similar to WT (Fig. 3D, left). For the partially open 89.6 Env, most variants showed higher binding efficiencies than the WT, with various levels of significance (Fig. 3D, right). Given that the 534A, 536A, and 539A variants of AD8 did not show significant changes in CD4-Ig binding efficiency, we examined the impact of these mutations on CD4 dependence for entry. To this end, we used target cells that transiently express high levels of CCR5 and a range of CD4 levels. The 534A and 539A variants exhibited a similar CD4 usage efficiency to the WT Env, whereas 536A exhibited a modest decrease (Fig. 3E). Therefore, CD4 binding and usage efficiency cannot account for the observed differences in infectivity.
The above results raised the following question: if fusion competence and stability of the 536A and 539A variants are higher than those of the ancestral form, why is the frequency of 539A increasing in clade B, whereas the frequency of 536A is constant or decreasing? We hypothesized that these patterns may be attributed to the differential sensitivity of these variants to immune selective pressures.
Substitutions in the FPPR midsegment enhance sensitivity to antibodies that target the trimer base.
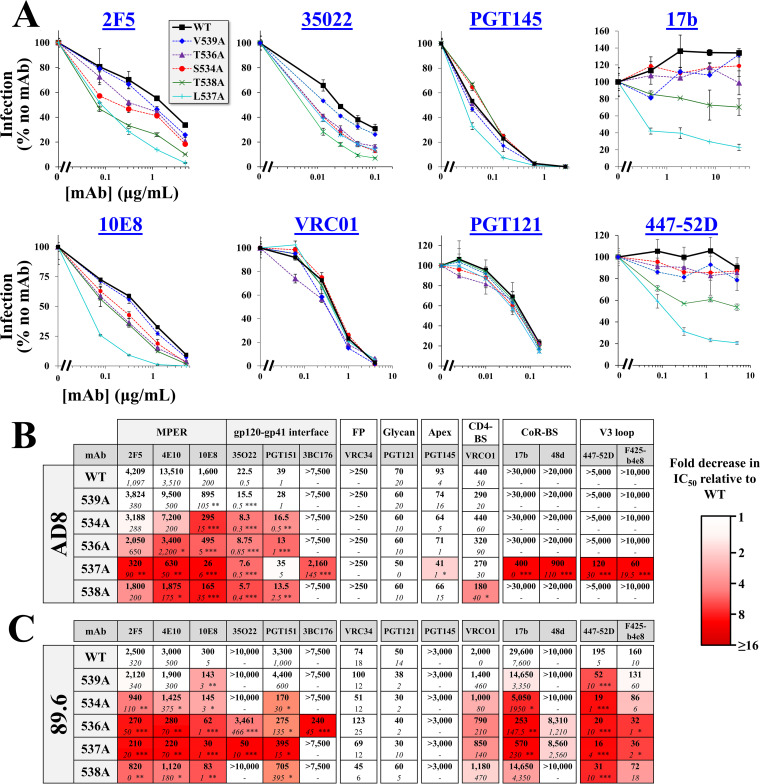
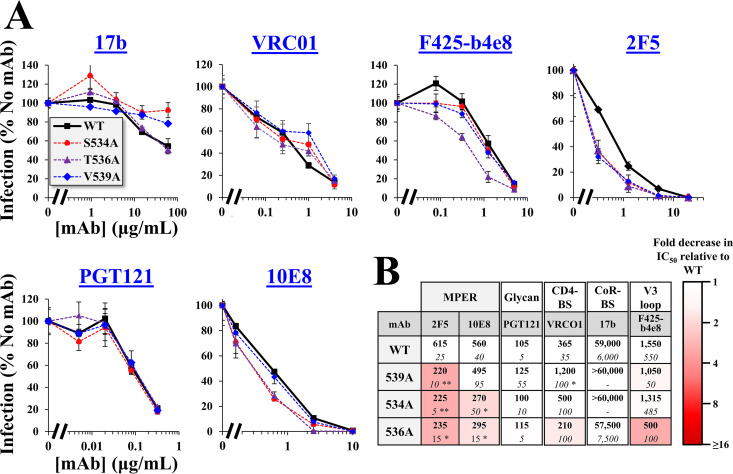
We examined whether the population-level changes in the FPPR can be explained by Ab pressure applied to the emerging variants. For this purpose, we analyzed the neutralization profiles of Ala variants at FPPR positions that exhibit intermediate and high levels of infectivity: 534, 536, 537, 538, and 539. Neutralization by monoclonal Abs (MAbs) that target distinct gp41 and gp120 epitopes was compared with that of the WT Envs. We first examined sensitivity of the Ala changes introduced in the tier-2-like AD8 Env. MAbs that target the membrane-proximal external region (MPER) of gp41 were tested, which recognize epitopes that are partially exposed on the unliganded form of Env (42). Ala substitutions at positions 534, 536, 537, and 538 increased sensitivity to MAbs 2F5, 4E10, and 10E8, with modest or no changes for the 539A variant (see Fig. 4A and all MAbs tested in Fig. 4B, which indicates statistical significance of the differences in 50% inhibitory concentration [IC50] values between the WT and all variants). We also examined sensitivity of the variants to MAbs that target epitopes overlapping the gp41-gp120 interface at the base of the trimer (43, 44). All FPPR variants were more sensitive than WT to MAbs 35O22 and PGT151, with only modest differences observed for variant 539A. In contrast to the higher sensitivity of the FPPR variants to MAbs that target the MPER and gp120-gp41 interface, their sensitivity to MAbs that target gp120 epitopes was generally similar to that of the WT AD8 Env, including (i) MAb PGT121, which targets an epitope that overlaps the high-mannose patch of gp120 (45), (ii) MAb PGT145, which targets a quaternary epitope at the apex of the trimer (46), and (iii) MAb VRC01, which targets the CD4-BS (47). We further examined sensitivity of the Envs to MAbs that target cryptic epitopes that overlap the CoR-BS, including 17b and 48d, (48) and the V3 loop, including 447-52D and F425-b4E8 (49, 50). For both groups of Abs, only the 537A variant showed 50% reduction of infectivity at the highest concentration of the MAbs.
FIG 4.
Alanine substitutions in the FPPR midsegment enhance HIV-1 sensitivity to antibodies that target the trimer base. (A) Neutralization sensitivity of virus containing AD8 Env with the indicated FPPR changes. Viruses were incubated with different concentrations of the MAbs for 1 h. Samples were then added to Cf2Th-CD4+ CCR5+ cells, and residual infectivity was measured 3 days later. Values describe the mean infection measured for each virus as a percentage of infection measured in the absence of MAbs. Error bars, SEM. (B, C) Summary of the effects of the FPPR substitutions on MAb sensitivity of viruses containing the WT or indicated variants of Env AD8 (B) or 89.6 (C). Mean IC50 values measured in three or more independent experiments are shown. Numbers in italics indicate SEMs calculated across experiments. Statistical significance of the differences between WT and mutant Envs was determined by an unpaired t test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Cells are shaded by the fold decrease in IC50 relative to WT Env.
We also tested the effects of the Ala mutations on neutralization sensitivity of the tier-1-like 89.6 Env. This Env exhibits a partially open conformation and, in contrast to tier-2 strains, is modestly sensitive to CoR-BS-targeting Abs (35). The sensitivity pattern of the 89.6 variants was similar to that of AD8 Env for MAbs that target the MPER and gp41-gp120 interface, as well as epitopes in the glycan patch and apex (Fig. 4C). No differences were observed for the fusion peptide targeting MAb VRC34 (51). However, the FPPR changes in Env 89.6 also increased sensitivity to CoR-BS and V3 loop Abs, suggesting that they further enhanced exposure of these domains. In addition, sensitivity to the CD4-BS MAb VRC01 was modestly increased. Since Envs of most primary HIV-1 strains occupy a more closed conformation than 89.6, we sought to determine whether introducing the FPPR substitutions in a closed form of this Env would also enhance exposure of the partially cryptic gp120 epitopes. For this purpose, we used a variant of 89.6 that contains two stabilizing changes in gp120: (i) Arg to Glu at position 305 of the V3 loop and (ii) Met to Ile at position 225 of the gp120 inner domain (52, 53). These changes were identified after serial passage in rhesus macaques of a chimeric simian-human immunodeficiency virus that contains the 89.6 Env (54). Previous analyses of the antigenic profile of 89.6(M225I, R305E) Env showed it occupies a more closed conformation than the parental 89.6 strain (35). We introduced substitution S534A, T536A, or V539A in the 89.6(M225I, R305E) Env and tested sensitivity to the different MAbs. As expected, 89.6(M225I, R305E) was more resistant than WT 89.6 to MAb 17b (IC50 values of >60 μg/mL and 25 μg/mL, respectively) (Fig. 5A). In contrast to their effects on WT 89.6, the 534A and 536A changes in 89.6(M225I, R305E) did not increase sensitivity to MAb 17b (Fig. 5B); a phenotype similar to that observed for AD8 Env. Furthermore, the 534A and 536A changes increased sensitivity of 89.6(M225I, R305E) to MPER MAbs 10E8 and 2F5. The 539A Env exhibited WT-like sensitivity to MAb 10E8 but higher sensitivity to 2F5. In addition, and in contrast to the WT 89.6 Env, the changes at positions 534 and 536 did not impact VRC01 neutralization of 89.6(M225I, R305E). Similarly, the 534A change in 89.6(M225I, R305E) did not affect sensitivity to MAb F425-b4e8, and the 536A change increased sensitivity to this MAb to a lesser extent than that observed for WT 89.6. Therefore, in the tier-2-like Env 89.6(M225, R305E), the 534A and 536A mutations induced limited changes in epitopes outside the base of the trimer.
FIG 5.
Alanine substitutions in the FPPR of a tier-2-like variant of 89.6 Env enhance sensitivity to MPER Abs but not to CoR-BS Abs. (A) Effects of Ala substitutions at positions 534, 536, and 539 on neutralization sensitivity of Env 89.6(M225I, R305E). Representative results from at least three independent experiments are shown. Error bars, SEM. (B) Summary of MAb sensitivities of the FPPR variants. Mean IC50 values measured in the experiments are shown. Numbers in italics indicate SEMs calculated across experiments. Statistical significance of the differences between WT and mutant Envs was determined by an unpaired t test: *, P < 0.05; **, P < 0.005.
The above data suggest that the FPPR regulates exposure of epitopes at the trimer base. In primary tier-2-like viruses such as AD8 and 89.6(M225I, R305E), Ala substitutions at FPPR positions 534 and 536 induce isolated exposure of epitopes in the MPER and gp120-gp41 interface. Ala substitutions at FPPR positions 537 and 538 (or in partially open forms, such as 89.6 Env) can induce more drastic changes in trimer conformation and enhance exposure of epitopes that overlap the CoR-BS, V3 loop, and CD4-BS. The V539A variants exhibited considerably lower sensitivity to the MPER and gp120-gp41 interface Abs, a pattern that more closely resembled that of the WT Env.
FPPR substitutions that induce an open-at-the-base conformation sensitize HIV-1 to antibodies commonly elicited during infection.
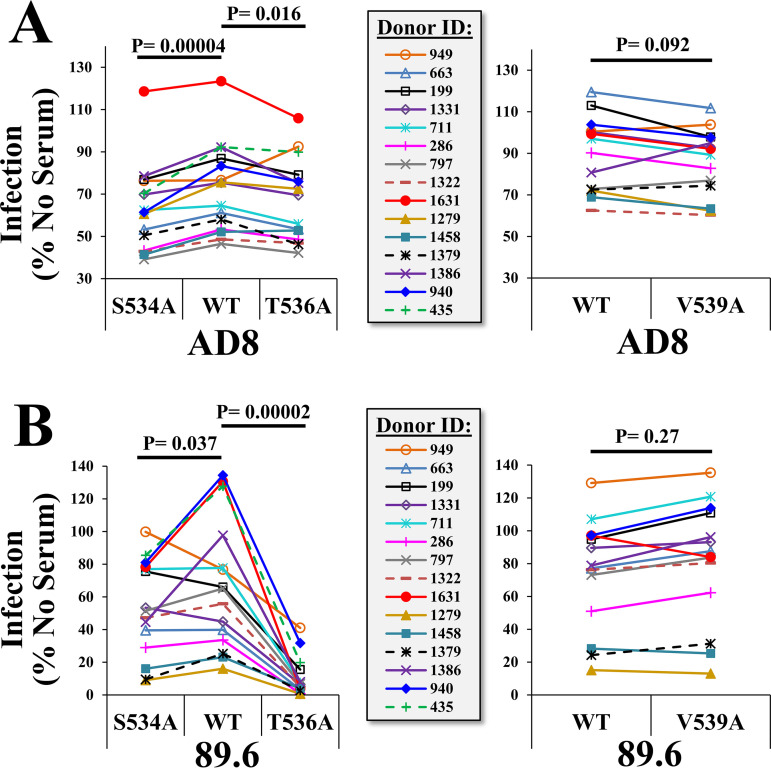
The emerging variants 534A and 536A show a historically constant frequency in the population and exhibit an open-at-the-base conformation of the trimer. In contrast, the 539A variant that is increasing in frequency exhibits a more closed conformation. We asked whether common Ab pressures applied on the open-at-the-base forms may account for their lack of increase in the population. To address this question, we first tested the effects of the substitutions on sensitivity to serum samples from HIV-infected individuals. Samples from 14 donors that exhibit low potency (IC90 values at a dilution of 1:10 or lower, as measured for virus containing the WT AD8 Env) were tested for their neutralization of pseudovirus containing the WT Envs, as well as variants 534A, 536A, and 539A (Fig. 6). The 534A and 536A variants of AD8 were modestly but significantly more sensitive to the sera than the WT form (see results of paired t tests in Fig. 6A). Greater enhancement of sensitivity was observed for the 534A and 536A variants of 89.6 (Fig. 6B). In contrast, sensitivity of the 539A variant did not differ from that of the WT Env for both strains.
FIG 6.
Alanine substitutions in the FPPR enhance sensitivity to weakly neutralizing serum from HIV-infected individuals. Samples from 14 individuals that exhibit low neutralization efficacy (IC90 value at a 1:10 dilution or lower for HIV-1AD8) were incubated with the WT, S534A, T536A, or V539A variants of Env AD8 (A) or Env 89.6 (B). All serum samples were used at a final dilution of 1:10. The virus-serum mixture was incubated for 1 h at 37°C and added to Cf2Th-CD4+ CCR5+ cells to measure residual infectivity. P values describe the results of a paired t test that compares infectivity of the WT and mutant Envs in the presence of each serum.
These data suggested that Abs targeting the open conformation of the 534A and 536A variants might be commonly elicited in HIV-1-infected individuals and contained in samples with low neutralization efficacy. To directly test this hypothesis, we sought to identify the Ab specificities in the samples by competition enzyme-linked immunosorbent assay (ELISA). For this purpose, we conjugated horseradish peroxidase (HRP) to MAbs 10E8 and 35O22. In addition, since patients frequently contain Abs that target the V3 loop and CD4-BS (55–57), we generated HRP conjugates for MAbs 447-52D and VRC01. As a control for an epitope that is less frequently targeted in patients, we used the glycan-targeting MAb PGT121 (45). HRP-conjugated MAbs were incubated with HOS cells that express Env in the presence of serum from 3 HIV-1-negative and 6 HIV-1-positive individuals: 3 with low neutralizing activity (IC50 of >1:50) and 3 with high neutralizing activity (IC50 of <1:50), as measured using virus containing WT 89.6 Env. Properties of the HIV-1-positive donor samples are shown in Table 1. To maximize recognition of serum-derived Abs that target the trimer base, we used the open 89.6(536A) Env as the capture antigen on the cells.
TABLE 1.
Serum samples from HIV-1-infected individuals used in this study
| Patient ID | Viral load (copies/mL)a | CD4 count (cells/mm3) | Antiretroviral treatmentb | Time since seroconversion (yr) |
|---|---|---|---|---|
| 1614 | 31,700 | 547 | None | >1 |
| 1631 | 79,300 | 415 | None | >4 |
| 1508 | 11,700 | 701 | None | >5 |
| 1279 | 509 | 120 | Nelfinavir, emtricitabine, TDF | >3 |
| 1235 | 321 | 616 | Stavudine, efavirenz | >7 |
| 1458 | 4,300 | 79 | Lopinavir, ritonavir, emtricitabine, TDF | >5 |
The viral load and CD4 counts shown describe values measured at the closest time point available to the collection time of the serum sample used in our tests (interval less than 3 months).
Treatment(s) received at the collection time of the samples used in our tests. TDF, tenofovir disoproxil fumarate.
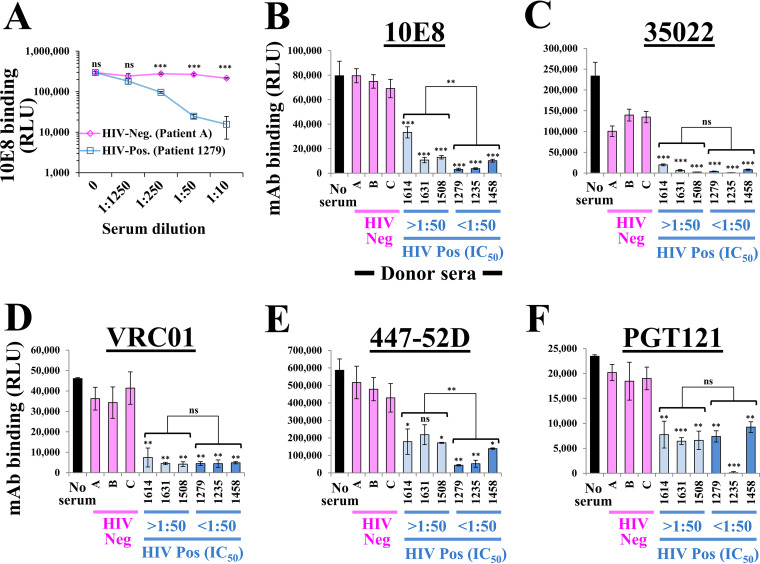
We first examined the ability of serum from HIV-positive and HIV-negative individuals to compete with binding of the HRP-conjugated MAb 10E8. As shown in Fig. 7A, the HIV-1-positive serum significantly reduced binding of the MAbs at dilutions of 1:10 to 1:250. We proceeded to test the ability of the HIV-positive and HIV-negative sera to reduce binding of the different MAbs. All sera were used at a dilution of 1:10. Binding of MAb 10E8 to the cell-surface Envs was considerably reduced by all HIV-positive sera, whereas the HIV-negative sera had no impact relative to the buffer (no-serum) control (Fig. 7B). Greater effects were observed for the sera that contained higher neutralizing activity. The HIV-positive sera also completely abrogated binding of the HRP-tagged MAb 35O22 to Env (Fig. 7C). Interestingly, binding of MAb 35O22 was modestly reduced by the HIV-1-negative sera; such 2-fold changes were observed in four independent experiments and may be attributed to cross-reactivity with a common human Ab. Considerable effects on binding of MAb VRC01 were also observed, with more modest effects on binding of MAbs 447-52D and PGT121 (Fig. 7D to F).
FIG 7.
Competition ELISA to detect antibodies against the trimer base in patient sera. (A) HOS cells that express 89.6(536A) Env were incubated with an HRP-tagged form of MAb 10E8 (at 4 μg/mL) and the indicated dilutions of serum from an HIV-negative or HIV-positive individual. Binding of the tagged MAb was detected by chemiluminescence. Values for the two samples were compared by an unpaired t test: ns, not significant; *, P < 0.05; **, P < 0.005; ***, P < 0.0005. (B to F) Competition of patient sera with MAb binding to Env. HOS cells expressing the 89.6(536A) Envs were incubated with the HRP-tagged MAb 10E8 (4 μg/mL), 35O22 (0.7 μg/mL), VRC01 (4 μg/mL), 447-52D (2 μg/mL), or PGT121 (4 μg/mL) in the presence of patient sera (all used at a 1:10 dilution). Patient sera were collected from three HIV-negative and six HIV-positive individuals. Samples are grouped by their IC50 values measured against virus containing the WT AD8 Env. Binding values for each serum (in four independent experiments) were compared with values measured for the HIV-negative samples using an unpaired t test, and significance is indicated by asterisks above the bars. A similar comparison between the sample groups with low and high IC50 values is also shown.
These findings suggest that patient sera commonly contain Abs against the trimer base that interfere with binding of MAbs against the MPER and gp120-gp41 interface. The 534A and 536A FPPR variants are more sensitive to the effects of such gp41-targeting Abs than the WT Env or 539A (Fig. 6), which likely explains the patterns of change in these emerging variants in the population during the course of the AIDS pandemic.
DISCUSSION
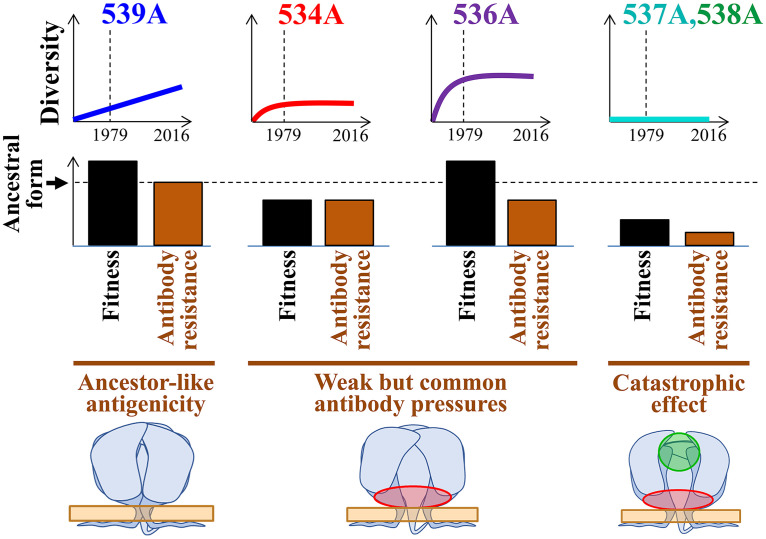
Many positions of HIV-1 Env show considerable diversity in amino acid sequence among strains that circulate in the population, whereas others are highly conserved. In many cases, conservation results from fitness pressures applied on the sites, allowing only specific Env variants to persist due to their functionality or structural stability (58, 59). Over the course of multiple replication cycles, even small reductions in fusion competence (as measured in vitro) can result in rapid loss of the variant from the host and thus limited representation in the population. However, in some cases, Env variants exhibit in vitro functionality levels that are similar to (or higher than) those of the ancestral form but are still not found among circulating strains. For example, several lab-adapted strains of HIV-1 exhibit higher fusion competence than their parental primary isolates, with a lower or no requirement for CD4 to mediate fusion (60, 61). The ability to surmount the necessity for CD4 would potentially allow more efficient replication in the host. Nevertheless, CD4-independent primary strains are rarely encountered; their absence is explained by the altered conformations of their Envs, which expose epitopes that overlap the CoR-BS (37, 62, 63). Abs that target the CoR-BS are commonly elicited in infected individuals (64) and likely apply strong selective pressures that account for the absence of CD4-independent strains in the population despite their potential fitness advantage. Our data suggest that a similar type of Ab selection pressure is applied to epitopes at the base of the trimer. However, in contrast to the CD4-independent strains, such effects do not cause catastrophic population size reductions: strains with an open-at-the-base conformation (e.g., 534A or 536A) are indeed isolated from infected individuals. Instead, their immune selection is incomplete, which has resulted in a lack of increase in their frequency during the past 4 decades. The balance between the rate of appearance of such variants in the host, their fitness, and sensitivity to immune pressures determines their “set point frequency” in the population and historical patterns of change (Fig. 8).
FIG 8.
Model of the population-level evolution of the FPPR. Evolutionary patterns of emerging FPPR variants can be explained by their fitness profiles and sensitivity to common antibody pressures. Fitness is inferred from the in vitro fusion competence of the Envs and their levels of structural and functional stability. The 539A variant exhibits modestly higher fusion competence than the clade B ancestral form and a similar closed conformation of the trimer that is relatively resistant to antibodies. Consequently, it is gradually increasing in its frequency. The 534A and 536A variants exhibit fitness levels that are lower than or higher than those of the clade ancestral form, respectively; their open-at-the-base conformation (highlighted in red) renders them sensitive to common but weakly neutralizing Abs against this region. Thus, following an initial increase in their frequency in the population, they reached steady states that reflect their relative infectivity levels. The 537A and 538A variants exhibit lower fitness, and a conformation that exposes immunogenic epitopes at multiple sites on Env targeted by frequently elicited antibodies (highlighted in green). As a consequence, such emerging variants are effectively eliminated in the host and poorly represented in the population.
Abs against Env subunit gp41 appear in patients at very early stages of infection (65). Many of these are nonneutralizing and target the immunodominant region of gp41, which is not exposed on the native trimer (66–68). In addition, Abs against exposed or partially exposed regions of gp41, which can neutralize the virus, are elicited in many patients. For example, serum profiling studies based on neutralization of chimeric Envs containing the MPER of HIV-1 or HIV-2 suggested that approximately 8% of patients contain 10E8-like Abs and 27% contain MPER-targeting Abs (69). Experiments based on identification of “neutralization fingerprints” of sera suggested that 50% of the sera contain 2F5- or 10E8-like Abs and 38% contain Abs with 35O22-like neutralization patterns (43). In our study, we detected Abs against the trimer base that interfere with binding of MAbs 10E8 and 35O22 in all 6 samples tested. Several reasons may explain the high proportion of sera with such Abs in our tests. First, our measurements are based on serum competition with MAbs for binding to Env (70). Thus, serum Abs that target partially overlapping epitopes and possibly conformationally related epitopes at the base of the trimer may sterically hinder binding of the MAbs (e.g., anti-MPER Abs that may interfere with binding of anti-gp120-gp41 interface Abs). Second, to maximize detection of the Abs, we used the 89.6(T536A) Env. The Env of strain 89.6 exhibits an open conformation that was further “opened” by the T536A change. Third, the above-cited studies tested for presence of the Abs by measuring neutralization activity of the sera, whereas our approach focuses on their ability to reduce binding of MAbs, thus also detecting Abs with low potency. Indeed, our results suggest that Abs against the trimer base exhibit low neutralizing activity, primarily against tier-2-like strains.
Structural integrity of the Env trimer is maintained by multiple interactions within and between subunits. Such sites control the overall architecture of the trimer and keep it in a functional closed form that does not expose immunogenic epitopes. Disruption of the interactions by mutations, chemical or physical treatments, or specific ligands can cause conformational changes to functional nonnative forms. A common feature of these perturbed forms is exposure of epitopes that overlap the CoR-BS (35, 37, 71). This common path reflects the natural propensity of the trimer to “spring” into an open form that can bind the CoR. During the course of their evolution, primate lentiviruses likely gained dependence upon CD4, a feature that limits exposure of the conserved immunogenic CoR-BS. Similar to the CoR-BS, the MPER is highly conserved (72) and is required for fusion (73–75). The MPER is thus maintained as less accessible to targeting Abs (76, 77); those BNAbs that target this domain generally have lower potencies than BNAbs that target more exposed epitopes on gp120. Here, we show that exposure of epitopes in the trimer base is controlled by the FPPR. Modestly perturbing substitutions in the FPPR (e.g., 534A and 536A) cause isolated exposure of these epitopes, whereas more perturbing changes (e.g., L537A) induce “fully open” trimer forms that also expose the CoR-BS. Interestingly, a similar phenotype is caused by disruption of the anchoring interaction between gp41 and membrane cholesterol. Low concentrations of cholesterol-depleting agents increase exposure of the MPER, whereas higher concentrations also increase exposure of the CoR-BS (78). We propose that the changes in the FPPR induce similar release of structural constraints. Based on the proximity of the FPPR to the CHR and MPER, is it likely that the FPPR substitutions affect the interaction with these domains (26). The nature of the association between the FPPR and MPER, which may explain the structural basis for our observations, remains to be clarified.
In different populations worldwide infected by virus from the same HIV-1 clade, each Env position exhibits a similar frequency distribution of amino acids (13). In many cases, founder effects of substitutions in monophyletic lineages are reduced by evolution of the position toward the clade-specific frequency distribution. That evolution is directed toward distributions rather than single amino acids suggests that a fine balance exists between the inherent propensity of the virus for diversification and the selective pressures that oppose it. In the absence of catastrophic effects of the selective pressures, no single factor dominates the pattern of circulating variants. For example, the emerging variant T536A shows modestly but significantly higher fusion competence than the clade ancestral form but also higher sensitivity to immune pressures normally elicited in the host. Such factors are balanced and likely explain the constant frequency of this variant in the population (Fig. 8). In contrast, other FPPR positions do suggest population size reduction effects. For example, position 538 contains the same nucleotide sequence in the clade B ancestor as position 536; however, 538A variants are not found among circulating strains, likely due to their lower infectivity and higher sensitivity to Abs normally elicited. Similarly, the L537A change reduces functionality of Env and also induces an open conformation of the trimer with high exposure of epitopes that overlap the CoR-BS, V3 loop, MPER, and gp120-gp41 interface. This Env form is also unstable, both structurally and functionally, as measured by the high propensity for shedding of gp120 and the loss of fusion competence after incubation at 37°C. Based on previous studies, it is likely that the L537A change disrupts the association between the FPPR and Trp at position 666 of the MPER (29, 79), thus releasing structural constraints and enhancing exposure of epitopes in both gp120 and gp41 and potentially reduce trimer stability. These factors likely account for the highly conserved nature of Leu at position 537. In this study, we show that in vitro analyses of the fusion competence, structural stability, and Ab sensitivity profiles can explain, at least in some cases, the distribution of emerging variants in the population and their historical patterns of change.
Broadly acting and potently neutralizing Abs against HIV-1 Env are rarely elicited in patients. A better understanding of the lower-efficacy neutralizing responses that are more frequently elicited may provide insights that can further enhance their potency. Our findings suggest that poorly neutralizing sera contain weak Ab pressures that target the trimer base. The high conservation and contiguous nature of the MPER render it an attractive target for immunogen design. However, previous attempts to elicit MPER-targeting Abs using MPER peptides have yielded mixed results (80, 81). The low neutralization efficacy of the elicited Abs is likely not caused by low immunogenicity of the MPER peptides used, but by the low exposure of the epitopes on the virus. A second limitation of MPER Abs is their propensity to associate nonspecifically with the plasma membrane, resulting in autoreactivity and polyreactivity (82, 83). Nevertheless, some MPER Abs have shown high levels of specificity, indicating that polyreactivity is not an inherent property of these Abs (84). That such Abs are commonly elicited in the host, as our data suggest, indicates that additional focus on the exposed segment of this domain (85) may further increase the neutralizing efficacy of the response. In the context of an immunogen composed of the complete ectodomain of the Env trimer, exposure of such epitopes can be controlled by the FPPR.
MATERIALS AND METHODS
HIV-1 Env sequence analyses.
To examine historical changes in the FPPR, HIV-1 env nucleotide sequences were downloaded from the Los Alamos National Lab (LANL) database using the sequence search interface (https://www.hiv.lanl.gov) and from the NCBI database (https://www.ncbi.nlm.nih.gov). Sequences tagged as nonfunctional Envs were removed, as were sequences with nucleotide ambiguities or large deletions in conserved regions. A single sequence from each patient and a single sequence from known transmission pairs were used. In addition, to avoid related sequences, we applied a minimal cutoff of 0.03 nucleotide substitution per site for sequence selection. The remaining 1,576 nucleotide sequences were aligned using a hidden Markov model with the HMMER3 software (86). Sequences were then translated, and positions were numbered according to the standard HXBc2 numbering system of the Env protein (25). To analyze the historical changes in amino acid frequency at Env positions, sequences were grouped according to the year of sample collection, and the frequency of each residue was calculated as a percentage of that of all isolates from the same time period. A maximum likelihood phylogenetic tree based on the 1,576 nucleotide sequences was constructed using FASTTREE (87, 88). The tree was rooted to the clade B consensus sequence (87).
To identify FPPR codons under selective pressure, we downloaded from the NCBI database nucleotide sequences of the FPPR midsegment (positions 7806 to 7841 of the HIV-1 genome, corresponding to amino acid positions 528 to 539 of Env). A panel of 6,285 clade B env sequences from samples collected between 1979 and 2018 was used to estimate the number of inferred synonymous and nonsynonymous changes. Analyses were performed using the main instance of Galaxy (88). To estimate maximum likelihood values, a tree topology was computed using a generalized time-reversible model with the CAT approximation (GTR-CAT) nucleotide evolution model with FASTTREE (89). To detect codons under selection, we calculated the number of synonymous substitutions per site (dS), and the number of nonsynonymous substitutions per site (dN). Maximum likelihood computations of dN and dS were conducted using HyPhy-SLAC (90). The dN-dS statistic was applied to detect codons under positive selection (dN-dS > 0) or negative selection (dN-dS < 0). P values for rejecting the null hypothesis of neutral evolution were calculated, whereby P values of <0.05 were considered significant.
To calculate within-host diversity at FPPR positions, we used nucleotide sequences of 4,252 clade B Envs from 181 different HIV-infected individuals. For each individual, 10 to 80 Env sequences were analyzed. Sequences were aligned using a hidden Markov model with the HMMER3 software (86). For each patient sample, we determined the presence or absence of in-host amino acid variation at each position of the FPPR.
Antibodies and cells.
The following MAbs were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. The MAb 35O22 that targets the gp120-gp41 interface was contributed by Jinghe Huang and Mark Connors (43). James Robinson provided MAbs 17b and 48d, which recognize epitopes that overlap the CoR-BS (48). Hermann Katinger provided MAb 2G12, which targets a carbohydrate-dependent gp120 epitope (40), and MAb 2F5, which recognizes the membrane-proximal external region (MPER) of gp41 (91). The MPER-targeting MAb 10E8 was contributed by Mark Connors (69), and MAb 4E10 was contributed by DAIDS/NIAID (92). John Mascola provided the CD4-BS MAb VRC01 and MAb VRC34 that targets the fusion peptide (51, 93). The International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium provided MAb PGT121, which recognizes a glycan-dependent gp120 epitope (45), and MAb PGT145, which recognizes a quaternary epitope at the trimer apex (45). The MAb 447-52D was contributed by Susan Zolla-Pazner (50). Plasmids encoding the heavy and light chains of MAb 3BC176 were kindly provided by Michel Nussenzweig; this MAb was purified as previously described (94).
Serum samples were obtained from HIV-1 chronically infected adult subjects (more than 12 months since seroconversion) who gave informed consent under clinical protocols approved by the human use review boards at the University of Washington at Seattle Center for AIDS Research (CFAR) and the University of Iowa (IRB no. 8807313, 200010008, and 2010101730). All samples were heat inactivated at 55°C for 30 min before use.
Human embryonic kidney 293T cells and human osteosarcoma (HOS) cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10 μg/mL penicillin-streptomycin and 10% fetal calf serum (DMEM/FCS). Canis familiaris thymus normal (Cf2Th) cells (kindly provided by Joseph Sodroski) were used to measure infection. These cells stably express human CD4 and CCR5 (Cf2Th-CD4+ CCR5+) and were cultured in DMEM/FCS supplemented with 200 μg/mL hygromycin and 400 μg/mL G418.
Envelope glycoprotein constructs.
The full-length Envs of HIV-1 strains AD8 (accession no. AF004394) and 89.6 (accession no. U39362) were expressed from the pSVIIIenv plasmid, as previously described (35). Amino acid numbering of Env positions is based on the HXBc2 reference system (25). Mutations were introduced into the pSVIIIenv vector by site-directed mutagenesis using the PrimeStar Max polymerase (TaKaRa), followed by DpnI digestion, and transformation of Stellar competent cells. The env genes of all variants generated were sequenced to verify that unwanted mutations were not introduced during this process.
Preparation of recombinant luciferase-expressing HIV-1.
Single-round, recombinant HIV-1 that expresses the luciferase gene was generated by transfection of 293T cells using JetPrime transfection reagent (Polyplus). Cells were seeded in 6-well plates (8.5 × 105 cells per well) and transfected the next day with 0.4 μg of the HIV-1 packaging construct pCMVΔP1ΔenvpA, 1.2 μg of the firefly luciferase-expressing construct pHIvec2.luc, 0.4 μg of plasmid expressing HIV-1 Env and 0.2 μg of plasmid expressing HIV-1 Rev and 4.2 μL of JetPrime reagent. The transfection medium was replaced the next day, and virus-containing supernatant was collected 24 h later. Supernatants were cleared of cell debris by centrifugation at 700 × g and filtered through 0.45-μm-pore-sized membranes. Samples were snap-frozen on dry ice immersed in ethanol for 15 min and stored at −80°C until use.
Measurements of infectivity, antibody neutralization, and functional stability.
Cf2Th-CD4+ CCR5+ cells were seeded in 96-well luminometer-compatible plates at a density of 2 × 104 cells per well and infected the next day. For neutralization assays, virus preparations were incubated with the indicated concentration of the MAbs for 1 h at 37°C. In all neutralization assays, sample input was adjusted so that infectivity of any two variants did not vary by a factor of more than 2-fold. Samples were then added to Cf2Th-CD4+ CCR5+ cells and incubated for 3 days to allow infection. To measure infection, the medium was removed, and the cells were lysed with 35 μL passive lysis buffer (Promega) and subjected to three freeze-thaw cycles. To measure luciferase activity, 100 μL of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.6], 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM d-luciferin potassium salt (Syd Labs, MA) were added to each sample. Luminescence was recorded using a Synergy H1 microplate reader (BioTek Instruments).
To measure virus sensitivity to inactivation at 37°C, recombinant viruses were generated as described above, diluted, and divided into aliquots (one sample for each prospective time point). All samples were then snap-frozen on dry ice immersed in ethanol for 15 min and stored at −80°C. At different time points, the samples were thawed in a 37°C water bath for 2 min and then further incubated at 37°C for different time periods. All samples were subsequently added to Cf2Th-CD4+ CCR5+ cells, and infectivity was measured 3 days later by luciferase activity.
CD4 dependence assay.
To determine the relative requirements of the Env variants for CD4, we infected 293T cells that transiently express human CCR5 and different levels of CD4. Briefly, 293T cells were seeded in 6-well plates (8.5 × 105 cells per well) and transfected the next day with a plasmid that expresses human CCR5 (0.9 μg in all wells) and different amounts of plasmid that expresses human CD4, using JetPrime reagent. Two days after transfection, cells were detached using phosphate-buffered saline (PBS) supplemented with 7.5 mM EDTA and seeded in 96-well plates (7 × 104 cells per well). Six hours later, viruses were added to the cells. Infectivity was measured after 2 days as described above.
p24 antigen assay to quantify virus particle content.
To normalize measured infectivity values by virus particle content in the samples, we quantified p24 antigen levels. For this purpose, human anti-HIV-1 p24 antigen MAb was incubated at 1 μg/mL in luminometer-compatible 96-well protein-binding plates overnight. The wells were then washed with blocking buffer, composed of 150 mM NaCl, 3 mM Tris (pH 8), 1.8 mM CaCl2, 1 mM MgCl2, and 2% bovine serum albumin (BSA). To lyse virions, samples were supplemented with 0.5% Triton X and incubated at 100°C for 5 min. Samples were then added to the 96-well plate and incubated for 2 h at room temperature. Wells were then washed 4 times with blocking buffer, and a rabbit anti-p24 antigen MAb suspended in blocking buffer was added. Binding of the latter was detected by a horseradish peroxidase (HRP)-conjugated goat anti-rabbit polyclonal Ab and measured by luminescence using SuperSignal West Pico enhanced chemiluminescence reagents and a Synergy H1 microplate reader.
Shedding of gp120 from Env-expressing cells.
To quantify the propensity of Env trimer to shed gp120, HOS cells were seeded in 6-well plates (2.75 × 105 cells per well) and transfected the next day with plasmids expressing Env, Tat, and Rev using 0.4, 0.33, and 0.2 μg of each plasmid per well, respectively, and 4.2 μL JetPrime transfection reagent. The next day, samples were washed twice with Tris-saline buffer (140 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, and 25 mM Tris [pH 7.5]), and then FreeStyle 293 expression medium (Gibco) was added. Two days later, the supernatant was collected, cleared of cell debris by centrifugation at 700 × g, and filtered through 0.45-μm-pore membranes. Envs were immunoprecipitated using protein A beads and a combination of MAbs 2G12, VRC01, and PGT121 (all at 1 μg/mL). Samples were then analyzed by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, probed with goat anti-gp120 and detected by an HRP-conjugated rabbit anti-goat Ab.
The amount of gp120 shed into the medium was normalized for the relative expression level of each Env. For this purpose, HOS cells were seeded in 96-well plates (1.4 × 104 cells per well) and transfected the next day with plasmids expressing Env, Tat, and Rev, using 60, 11, and 6 ng of each plasmid per well, respectively, and 0.18 μL per well of JetPrime reagent. In all experiments, we also used a negative-control plasmid that contains a stop codon at amino acid position 46 of Env to determine background binding to the cells. Three days later, cells were washed in Tris-saline buffer containing 2% BSA (TS/BSA) and incubated with the above combination of MAbs (all at 1 μg/mL) in TS/BSA for 1 h at room temperature. Cells were then washed twice with TS/BSA and once with TS. Binding was detected using HRP-conjugated goat anti-human IgG and measured by luminescence using SuperSignal West Pico reagents and a Synergy H1 microplate reader. Intensity of the gp120 band from the supernatant (quantified by densitometry) was compared with the expression level of each Env, and the ratio between the two values was used to quantify gp120 shedding.
Cell-based ELISA and serum competition assays.
Binding of the CD4-Ig probe (95) to the FPPR variants was measured by cell-based ELISA as described previously (3, 78). Briefly, HOS cells were seeded in 96-well plates and transfected the next day by Env-, Rev-, and Tat-expressing plasmids, as described above. Three days after transfection, cells were washed twice with TS/BSA and incubated with the CD4-Ig probe in blocking buffer for 45 min. To normalize for the cell surface expression level of each Env, we also measured binding of MAb 2G12, which recognizes an exposed epitope in the high-mannose patch of Env (40). Both probes were added at 0.5 μg/mL. Samples were then washed 4 times with blocking buffer and incubated with a horseradish peroxidase (HRP)-conjugated goat anti-human IgG polyclonal antibody preparation for 45 min. Cells were subsequently washed six times with TS/BSA and six times with TS buffer. HRP enzyme activity was determined after addition of 35 μL per well of a 1:1 mix of SuperSignal West Pico chemiluminescent peroxide and luminol enhancer solutions (Thermo Scientific) supplemented with 150 mM NaCl. Light emission was measured with a Synergy H1 microplate reader.
To map the target specificity of antibodies in patient sera, we performed a competition ELISA. First, MAbs 10E8, 35O22, PGT121, VRC01, and 447-52D were conjugated to HRP using EZ-Link Plus activated peroxidase (Thermo Scientific) as recommended by the manufacturer. The conjugated product was then purified using protein A beads, resuspended in PBS, and frozen at −20°C until use. To measure binding of the HRP-conjugated MAbs to Env, we transfected HOS cells (in 96-well plates) with Env 89.6(536A) as described above. Three days later, cells were incubated with serum from HIV-negative or HIV-positive individuals for 1 h at 37°C. All sera were suspended in blocking buffer at a 1:10 dilution. Media were then removed and replaced by blocking buffer containing the same serum dilution and supplemented with one of the HRP-conjugated MAbs 10E8, 35O22, PGT121, VRC01, and 447-52D at the indicated concentrations. Samples were then incubated with the cells for 1 h at 37°C and washed, and binding of the HRP-conjugated MAbs was detected by chemiluminescence as described above.
ACKNOWLEDGMENTS
This work was partly supported by amfAR grant 110028-67-RGRL to H.H. and by NIH grants AI141495 and AI150343 to L.W.
We declare no conflict of interest.
Contributor Information
Li Wu, Email: li-wu@uiowa.edu.
Hillel Haim, Email: hillel-haim@uiowa.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.de Taeye SW, Moore JP, Sanders RW. 2016. HIV-1 envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol 37:221–232. 10.1016/j.it.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Excler JL, Robb ML, Kim JH. 2015. Prospects for a globally effective HIV-1 vaccine. Am J Prev Med 49:S307–S318. 10.1016/j.amepre.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 3.DeLeon O, Hodis H, O'Malley Y, Johnson J, Salimi H, Zhai Y, Winter E, Remec C, Eichelberger N, Van Cleave B, Puliadi R, Harrington RD, Stapleton JT, Haim H. 2017. Accurate predictions of population-level changes in sequence and structural properties of HIV-1 Env using a volatility-controlled diffusion model. PLoS Biol 15:e2001549. 10.1371/journal.pbio.2001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunnik EM, Euler Z, Welkers MR, Boeser-Nunnink BD, Grijsen ML, Prins JM, Schuitemaker H. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat Med 16:995–997. 10.1038/nm.2203. [DOI] [PubMed] [Google Scholar]
- 5.Bouvin-Pley M, Morgand M, Moreau A, Jestin P, Simonnet C, Tran L, Goujard C, Meyer L, Barin F, Braibant M. 2013. Evidence for a continuous drift of the HIV-1 species towards higher resistance to neutralizing antibodies over the course of the epidemic. PLoS Pathog 9:e1003477. 10.1371/journal.ppat.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Euler Z, Bunnik EM, Burger JA, Boeser-Nunnink BD, Grijsen ML, Prins JM, Schuitemaker H. 2011. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J Virol 85:7236–7245. 10.1128/JVI.00196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu WS, Hughes SH. 2012. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2:a006882. 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preston BD, Poiesz BJ, Loeb LA. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168–1171. 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 9.Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, Moore JP, Shaw GM, Hahn BH, Doms RW. 2013. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol 87:2401–2411. 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci USA 110:6626–6633. 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rademeyer C, Korber B, Seaman MS, Giorgi EE, Thebus R, Robles A, Sheward DJ, Wagh K, Garrity J, Carey BR, Gao H, Greene KM, Tang H, Bandawe GP, Marais JC, Diphoko TE, Hraber P, Tumba N, Moore PL, Gray GE, Kublin J, McElrath MJ, Vermeulen M, Middelkoop K, Bekker LG, Hoelscher M, Maboko L, Makhema J, Robb ML, Abdool Karim S, Abdool Karim Q, Kim JH, Hahn BH, Gao F, Swanstrom R, Morris L, Montefiori DC, Williamson C. 2016. Features of recently transmitted HIV-1 clade C viruses that impact antibody recognition: implications for active and passive immunization. PLoS Pathog 12:e1005742. 10.1371/journal.ppat.1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, Montefiori DC, Tilton JC, Shaw GM, Hahn BH, Doms RW. 2011. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol 85:8514–8527. 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han C, Johnson J, Dong R, Kandula R, Kort A, Wong M, Yang T, Breheny PJ, Brown GD, Haim H. 2020. Key positions of HIV-1 Env and signatures of vaccine efficacy show gradual reduction of population founder effects at the clade and regional levels. mBio 11:e00126-20. 10.1128/mBio.00126-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddox HK, Dingens AS, Hilton SK, Overbaugh J, Bloom JD. 2018. Mapping mutational effects along the evolutionary landscape of HIV envelope. eLife 7:e34420. 10.7554/eLife.34420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanini F, Puller V, Brodin J, Albert J, Neher RA. 2017. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol 3:vex003. 10.1093/ve/vex003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hraber P, Korber BT, Lapedes AS, Bailer RT, Seaman MS, Gao H, Greene KM, McCutchan F, Williamson C, Kim JH, Tovanabutra S, Hahn BH, Swanstrom R, Thomson MM, Gao F, Harris L, Giorgi E, Hengartner N, Bhattacharya T, Mascola JR, Montefiori DC. 2014. Impact of clade, geography, and age of the epidemic on HIV-1 neutralization by antibodies. J Virol 88:12623–12643. 10.1128/JVI.01705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol 82:11651–11668. 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang G-Y, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. 2013. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340:751–756. 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol 69:4413–4422. 10.1128/JVI.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8–22. 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Spenlehauer C, Saragosti S, Fleury HJ, Kirn A, Aubertin AM, Moog C. 1998. Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol 72:9855–9864. 10.1128/JVI.72.12.9855-9864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrin T, Loh TP, Vennari JC, Schuitemaker H, Nunberg JH. 1995. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol 69:39–48. 10.1128/JVI.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Si ZH, Moore JP, Sodroski J. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J Virol 74:11955–11962. 10.1128/jvi.74.24.11955-11962.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Si Z, Cayabyab M, Sodroski J. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J Virol 75:4208–4218. 10.1128/JVI.75.9.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber B, Foley B, Kuiken C, Pillai S, Sodroski J. 1998. Numbering positions in HIV relative to HXBc2, p III-102–III-111. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 26.Lu W, Chen S, Yu J, Behrens R, Wiggins J, Sherer N, Liu SL, Xiong Y, Xiang SH, Wu L. 2019. The polar region of the HIV-1 envelope protein determines viral fusion and infectivity by stabilizing the gp120-gp41 association. J Virol 93:e02128-18. 10.1128/JVI.02128-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, Bjorkman PJ. 2018. DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49:235–246.e4. 10.1016/j.immuni.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Sarkar A, Pugach P, Sanders RW, Moore JP, Ward AB, Wilson IA. 2019. Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide. Nat Commun 10:763. 10.1038/s41467-019-08738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellamy-McIntyre AK, Lay CS, Baar S, Maerz AL, Talbo GH, Drummer HE, Poumbourios P. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem 282:23104–23116. 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Li TW, Phillips S, Wu L. 2021. Reverted HIV-1 mutants in CD4(+) T-cells reveal critical residues in the polar region of viral envelope glycoprotein. Microbiol Spectr 9:e01653-21. 10.1128/spectrum.01653-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu K, Acharya P, Kong R, Cheng C, Chuang G-Y, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, Shen C-H, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai Y-T, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd J-P, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, et al. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–363. 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson J, Zhai Y, Salimi H, Espy N, Eichelberger N, DeLeon O, O’Malley Y, Courter J, Smith AB, III, Madani N, Sodroski J, Haim H. 2017. Induction of a tier-1-like phenotype in diverse tier-2 isolates by agents that guide HIV-1 Env to perturbation-sensitive, nonnative states. J Virol 91:e00174-17. 10.1128/JVI.00174-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal N, Leaman DP, Rowcliffe E, Kinkead H, Nohria R, Akagi J, Bauer K, Du SX, Whalen RG, Burton DR, Zwick MB. 2011. Functional stability of unliganded envelope glycoprotein spikes among isolates of human immunodeficiency virus type 1 (HIV-1). PLoS One 6:e21339. 10.1371/journal.pone.0021339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70:1100–1108. 10.1128/JVI.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murin CD, Julien JP, Sok D, Stanfield RL, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, Ward AB. 2014. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol 88:10177–10188. 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, Wyatt RT. 2011. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol 85:8217–8226. 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Leaman DP, Kim AS, Torrents de la Pena A, Sliepen K, Yasmeen A, Derking R, Ramos A, de Taeye SW, Ozorowski G, Klein F, Burton DR, Nussenzweig MC, Poignard P, Moore JP, Klasse PJ, Sanders RW, Zwick MB, Wilson IA, Ward AB. 2015. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat Commun 6:8167. 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P, Protocol G Principal Investigators. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-Ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavacini L, Duval M, Song L, Sangster R, Xiang SH, Sodroski J, Posner M. 2003. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17:685–689. 10.1097/00002030-200303280-00006. [DOI] [PubMed] [Google Scholar]
- 50.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol 66:7538–7542. 10.1128/JVI.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, Cale EM, Chen L, Choi CW, Chuang G-Y, Doria-Rose NA, Druz A, Georgiev IS, Gorman J, Huang J, Joyce MG, Louder MK, Ma X, McKee K, O'Dell S, Pancera M, Yang Y, Blanchard SC, Mothes W, Burton DR, Koff WC, Connors M, Ward AB, Kwong PD, Mascola JR. 2016. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352:828–833. 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cayabyab M, Karlsson GB, Etemad-Moghadam BA, Hofmann W, Steenbeke T, Halloran M, Fanton JW, Axthelm MK, Letvin NL, Sodroski JG. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J Virol 73:976–984. 10.1128/JVI.73.2.976-984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J Virol 75:5646–5655. 10.1128/JVI.75.12.5646-5655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol 70:6922–6928. 10.1128/JVI.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol 76:9035–9045. 10.1128/jvi.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pincus SH, Messer KG, Nara PL, Blattner WA, Colclough G, Reitz M. 1994. Temporal analysis of the antibody response to HIV envelope protein in HIV-infected laboratory workers. J Clin Invest 93:2505–2513. 10.1172/JCI117260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiber M, Wachsmuth C, Muller H, Odemuyiwa S, Schmitz H, Meyer S, Meyer B, Schneider-Mergener J. 1997. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J Virol 71:9198–9205. 10.1128/JVI.71.12.9198-9205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouyos RD, Leventhal GE, Hinkley T, Haddad M, Whitcomb JM, Petropoulos CJ, Bonhoeffer S. 2012. Exploring the complexity of the HIV-1 fitness landscape. PLoS Genet 8:e1002551. 10.1371/journal.pgen.1002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. 2005. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol 79:9006–9018. 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabat D, Kozak SL, Wehrly K, Chesebro B. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol 68:2570–2577. 10.1128/JVI.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozak SL, Platt EJ, Madani N, Ferro FE, Jr, Peden K, Kabat D. 1997. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol 71:873–882. 10.1128/JVI.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA 96:6359–6364. 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shakirzyanova M, Kong XP, Cheng-Mayer C. 2017. Determinants of HIV-1 CD4-independent brain adaptation. J Acquir Immune Defic Syndr 76:209–218. 10.1097/QAI.0000000000001478. [DOI] [PubMed] [Google Scholar]
- 64.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82:12449–12463. 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cook JD, Khondker A, Lee JE. 2022. Conformational plasticity of the HIV-1 gp41 immunodominant region is recognized by multiple non-neutralizing antibodies. Commun Biol 5:291. 10.1038/s42003-022-03235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frey G, Chen J, Rits-Volloch S, Freeman MM, Zolla-Pazner S, Chen B. 2010. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat Struct Mol Biol 17:1486–1491. 10.1038/nsmb.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol 65:4832–4838. 10.1128/JVI.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2016. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu Q, Shaik MM, Cai Y, Ghantous F, Piai A, Peng H, Rits-Volloch S, Liu Z, Harrison SC, Seaman MS, Chen B, Chou JJ. 2018. Structure of the membrane proximal external region of HIV-1 envelope glycoprotein. Proc Natl Acad Sci USA 115:E8892–E8899. 10.1073/pnas.1807259115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dimitrov AS, Rawat SS, Jiang S, Blumenthal R. 2003. Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion. Biochemistry 42:14150–14158. 10.1021/bi035154g. [DOI] [PubMed] [Google Scholar]
- 74.Salzwedel K, West JT, Hunter E. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol 73:2469–2480. 10.1128/JVI.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol 73:6089–6092. 10.1128/JVI.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46:1398–1401. 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- 77.Shen X, Dennison SM, Liu P, Gao F, Jaeger F, Montefiori DC, Verkoczy L, Haynes BF, Alam SM, Tomaras GD. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci USA 107:5972–5977. 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salimi H, Johnson J, Flores MG, Zhang MS, O'Malley Y, Houtman JC, Schlievert PM, Haim H. 2020. The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization. J Biol Chem 295:348–362. 10.1074/jbc.RA119.009481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. 2010. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog 6:e1000880. 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banerjee S, Shi H, Habte HH, Qin Y, Cho MW. 2016. Modulating immunogenic properties of HIV-1 gp41 membrane-proximal external region by destabilizing six-helix bundle structure. Virology 490:17–26. 10.1016/j.virol.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banerjee S, Shi H, Banasik M, Moon H, Lees W, Qin Y, Harley A, Shepherd A, Cho MW. 2017. Evaluation of a novel multi-immunogen vaccine strategy for targeting 4E10/10E8 neutralizing epitopes on HIV-1 gp41 membrane proximal external region. Virology 505:113–126. 10.1016/j.virol.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908. 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 83.Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, Spicer L, VandeBerg JL, Haynes BF, Kelsoe G. 2013. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med 210:241–256. 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donius LR, Cheng Y, Choi J, Sun ZY, Hanson M, Zhang M, Gierahn TM, Marquez S, Uduman M, Kleinstein SH, Irvine D, Love JC, Reinherz EL, Kim M. 2016. Generation of long-lived bone marrow plasma cells secreting antibodies specific for the HIV-1 gp41 membrane-proximal external region in the absence of polyreactivity. J Virol 90:8875–8890. 10.1128/JVI.01089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim M, Song L, Moon J, Sun ZY, Bershteyn A, Hanson M, Cain D, Goka S, Kelsoe G, Wagner G, Irvine D, Reinherz EL. 2013. Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J Biol Chem 288:31888–31901. 10.1074/jbc.M113.494609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaschen B, Kuiken C, Korber B, Foley B. 2001. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics 17:415–418. 10.1093/bioinformatics/17.5.415. [DOI] [PubMed] [Google Scholar]
- 87.Linchangco GV, Jr, Foley B, Leitner T. 2021. Updated HIV-1 consensus sequences change but stay within similar distance from worldwide samples. Front Microbiol 12:828765. 10.3389/fmicb.2021.828765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 91.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67:6642–6647. 10.1128/JVI.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 17:1757–1765. 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 93.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. 2011. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One 6:e24078. 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, III, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci USA 101:5036–5041. 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]