ABSTRACT
Porcine epidemic diarrhea virus (PEDV) is the globally distributed alphacoronavirus that can cause lethal watery diarrhea in piglets, causing substantial economic damage. However, the current commercial vaccines cannot effectively the existing diseases. Thus, it is of great necessity to identify the host antiviral factors and the mechanism by which the host immune system responds against PEDV infection required to be explored. The current work demonstrated that the host protein, the far upstream element-binding protein 3 (FUBP3), could be controlled by the transcription factor TCFL5, which could suppress PEDV replication through targeting and degrading the nucleocapsid (N) protein of the virus based on selective autophagy. For the ubiquitination of the N protein, FUBP3 was found to recruit the E3 ubiquitin ligase MARCH8/MARCHF8, which was then identified, transported to, and degraded in autolysosomes via NDP52/CALCOCO2 (cargo receptors), resulting in impaired viral proliferation. Additionally, FUBP3 was found to positively regulate type-I interferon (IFN-I) signaling and activate the IFN-I signaling pathway by interacting and increasing the expression of tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3). Collectively, this study showed a novel mechanism of FUBP3-mediated virus restriction, where FUBP3 was found to degrade the viral N protein and induce IFN-I production, aiming to hinder the replication of PEDV.
IMPORTANCE PEDV refers to the alphacoronavirus that is found globally and has re-emerged recently, causing severe financial losses. In PEDV infection, the host activates various host restriction factors to maintain innate antiviral responses to suppress virus replication. Here, FUBP3 was detected as a new host restriction factor. FUBP3 was found to suppress PEDV replication via the degradation of the PEDV-encoded nucleocapsid (N) protein via E3 ubiquitin ligase MARCH8 as well as the cargo receptor NDP52/CALCOCO2. Additionally, FUBP3 upregulated the IFN-I signaling pathway by interacting with and increasing tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3) expression. This study further demonstrated that another layer of complexity could be added to the selective autophagy and innate immune response against PEDV infection are complicated.
KEYWORDS: FUBP3, IFN-I, MARCHF8, N protein, NDP52, PEDV, selective autophagy
INTRODUCTION
Porcine epidemic diarrhea (PED), which results from porcine epidemic diarrhea virus (PEDV), is an acute and extremely contagious intestinal infection characterized by vomiting, watery diarrhea, anorexia, and a high mortality rate in suckling pigs (1, 2). PED was originally discovered in 1971 in England and was successively discovered in other wine-producing nations in Europe and Asia within the early 1980s (3, 4). A novel variant strain of PEDV occurred in October 2010 and spread rapidly across China, causing 80 to 100% of deaths of suckling piglets (5, 6). This strain then quickly spread to other major pig-raising countries globally, causing substantial economic losses to the farms (7, 8). Vaccination is a vital way to effectively control infectious diseases, but recently PEDV outbreaks in large-scale pig farms frequently took place, indicating that the current commercially available vaccines may be insufficient to resist the epidemic strains (9). Identifying host antiviral factors and investigating the mechanism of host immune responses against PEDV infection generate a vital function in controlling viral epidemics and developing novel therapeutic targets.
As a positive-sense, single-stranded RNA virus, PEDV is a member of the genus Alphacoronavirus, Coronaviridae. Its genome (28 kb) can encode two polyproteins (pp1a, pp1ab), an accessory protein (ORF3), and four structural proteins (SPs; containing an envelope, E; nucleocapsid, N; membrane, M; spike, S) (10, 11). Of these, the conservation of the N protein is made among members of the genus Alphacoronavirus and can be abundantly expressed in infected cells (12). The coronavirus N protein, as a multifunctional protein, can strongly affect viral transcription, translation, immune evasions, and viral assembly (13), such as inducing endoplasmic reticulum (ER) stress to promote viral replication (14) and interacting with TANK-binding kinase (TBK1) for blocking IRF3 nuclear transport and phosphorylation, aiming to suppress the production of type-I interferon (IFN-I) (15). Therefore, the PEDV N protein plays a vital function in the proliferation of viral and can act as a beneficial therapeutic target for viral infection.
A continuous arms race between the virus and the host has given rise to different strategies to antagonize each other. During viral infection, viruses have evolved a variety of strategies for counteracting, evading, and even hijacking host defense. Conversely, the host induces a diverse array of antiviral reactions, such as intrinsic antiviral proteins (also named restriction factors), to resist virus infection (16, 17). In PEDV, 11 or more viral proteins (VPs) suppress IFN induction, thus counteracting congenital immune response in the host (18). The host, in turn, employed a lot of antiviral proteins independently induced by IFN to inhibit PEDV replication. For example, bone marrow stromal cell antigen 2 (BST2, which was named tetherin), induced by IRF1, reduces PEDV replication via targeting and degrading the virus N protein with the use of selective autophagy (19). Early growth response gene 1 (EGR1) can upregulate the expression of IRAV for degrading PEDV N protein via MARCHF8 and can subsequently inhibit PEDV replication (20). Cytoplasmic poly(A)-binding protein 4 (PABPC4) can be modulated by the transcription factor (TF) SP1, causing the reduction in PEDV replication (21). Investigating the continuous competition between the host and PEDV can provide new strategies for preventing and treating PEDV.
Far upstream element-binding protein 3 (FUBP3, also called MARTA2) is a member of the regulatory gene family, comprising FUBP1, FUBP2, and FUBP3 (22). FUBP, as an RNA-binding protein, strongly influences RNA translation and gene stability (23). It has been demonstrated that FUBP3 binds to the MAP2 mRNA 3′untranslated region (3′UTR) in rat neurons to regulate dendritic targeting and combines with 3′UTR of β-actin to regulate β-actin mRNA localization (24, 25). FUBP3 can also regulate the expression of fibroblast growth factor 9 (FGF9) post-transcriptionally through mRNA stabilization or by increasing the translation rate (26). FUBP3 has been discovered to interact with enterovirus 71 (EV71) IRES and play the role of a positive regulator of EV71 replication (27). According to the obtained results, the expression of FUBP3 could be controlled by the TF TCFL5 during PEDV infection. Additionally, FUBP3 suppressed PEDV replication through selective autophagy by targeting and degrading the viral N protein and activating the IFN-I signaling pathway via contact with the tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3). The current work described a novel antiviral mechanism for FUBP3, through which PEDV replication can be suppressed, and the IFN signaling pathway can be activated.
RESULTS
PEDV infection downregulated the expression of FUBP3 through the transcription factor of TCFL5.
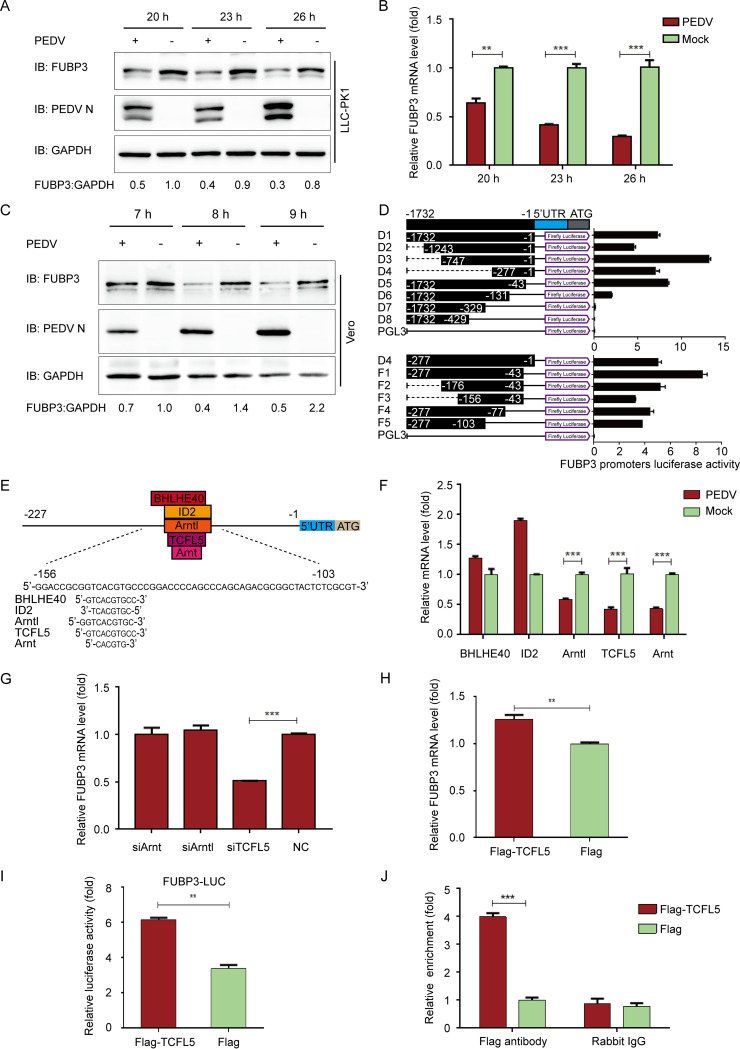
To screen the potential host antiviral proteins against PEDV, we performed a method of mass spectrometry (MS), aiming to identify the host factors interacting with the PEDV N protein. Meanwhile, the host factor FUBP3 was identified in the MS result. PEDV (strain JS-2013) infected porcine kidney cells (LLC-PK1) at the multiplicity of infection (MOI) of 1 to identify the correlation between FUBP3 and PEDV infection and the potential antiviral function of FUBP3. The endogenous expression level of FUBP3 was investigated by evaluating qRT-PCR and Western blot (WB), respectively. FUBP3 expression was found to decline in the cells infected by PEDV at 20/23/26 h postinfection (hpi) (Fig. 1A). Consistent with the FUBP3 protein expression level, its mRNA levels were demonstrated to decrease in the cells infected by PEDV, in comparison with the levels in uninfected cells (Fig. 1B). Similar to the results obtained with LLC-PK1 cells, PEDV infection also decreased the expression of FUBP3 in Vero cells (Fig. 1C). Based on the obtained results, the expression of FUBP3 is reduced by PEDV infection.
FIG 1.
PEDV infection reduces the expression of FUBP3 by the transcription factor of TCFL5 in LLC-PK1 cells. (A and B) After the infection of PEDV (MOI = 1), we harvested the cells at specific time points. Endogenous FUBP3 expression was determined by performing qRT-PCR and WB; DAPDH was used as the endogenous reference. (C) After the infection of PEDV (MOI = 1), we harvested the Vero cells at specific time points. Endogenous FUBP3 expression was determined by WB. (D) 293T cells were transfected with the truncated FUBP3 promoter constructs (D1 to D8, and F1 to F5) and Renilla luciferase reporter vector (pRL-TK-Luc) (D1 to D8, F1 to F5). The luciferase activity was measured in lysates. (E) The JASPAR vertebrate database (http://jaspar.genereg.net) was used to determine the cis-acting elements of FUBP3. (F) Arntl, BHLHE40, TCFL5, ID2, and Arnt mRNA levels in PEDV infected-PK1 cells were detected through qRT-PCR. (G) Arntl siRNA, TCFL5 siRNA, or Arnt siRNA was transfected into LLC-PK1 cells, and FUBP3 transcription was determined by performing qRT-PCR. (H) The plasmid that encoded Flag-TCFL5 was transfected in LLC-PK1 cells for a whole day, and qRT-PCR was conducted for analyzing the lysate. (I) 293T cells were subjected to transfection with FUBP3 promoter-driven luciferase vector, plasmids encoding Flag-TCFL5, and pRL-TK-Luc vector for 24 h. By analyzing the luciferase activity, the cells were collected. (J) An empty vector or a Flag-TCFL5 plasmid was transfected into LLC-PK1 cells for a day, and later, the cells were collected to conduct a ChIP assay. The normal rabbit IgG or anti-Flag antibody was adopted for precipitating chromatin-bound TCFL5. The findings are denoted as the mean ±SD from triplicate samples; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed Student's t test).
To detect the transcription factors of FUBP3, this study cloned the long FUBP3 promoter (1732 bp) and the truncated FUBP3 promoters (designated D1 to D8) into a luciferase vector (pGL3-Basic) and examined their capability of inducing luciferase expression in HEK 293T cells. According to our results, the promoter fragment that contained the nucleotides −277 to −43 induced luciferase activities in comparison with that induced by the full-length promoter, and the constructs lacking the sequence –277 to –43 triggered undetectable or low luciferase activities. Therefore, the core promoter of FUBP3 was identified to be in the −277 to −43 positions (Fig. 1D). The luciferase vector was inserted with different truncated promoters (denoted F1-5) with the above-mentioned sequence to verify the boundaries of the minimal FUBP3 promoter. In addition, based on the sequence from –277 to –43, several truncated promoters (designated F1-5) were cloned into the luciferase vector to deeply verify the boundaries of the minimal FUBP3 promoter. According to the results, the minimum FUBP3 core promoter was from the –156 to –103 positions (Fig. 1D).
To explore the transcriptional modulation of FUBP3, the JASPAR vertebrate database (http://jaspar.genereg.net/) was used to identify the potential TF-binding sites (TFBSs) in the FUBP3 promoter (19). The results showed that the minimal FUBP3 core promoter region contains several TFBSs, including Arntl-, BHLHE40-, TCFL5-, ID2-, and Arnt-binding sites (Fig. 1E). Subsequently, we identified the mRNA levels of the putative transcription factors with qRT-PCR, finding that the downregulation could merely be observed in Arntl, TCFL5, and Arnt, which conformed to FUBP3 during PEDV infection (Fig. 1F). Next, small interfering RNAs (siRNAs) that targeted putative transcription factors were synthesized to assess the transcription factors associated with the regulation of FUBP3 expression. In accordance with the results of the qRT-PCR analysis, the mRNA level of FUBP3 was considerably lowered in cells transfected with TCFL5, while it was almost unchanged in cells with the transfection of Arntl or Arnt siRNA (Fig. 1G). With the TCFL5 being overexpressed to explore the inducible expression of endogenous FUBP3, it could be found that the mRNA level and luciferase activity of FUBP3 were increased in cells transfected with plasmid encoding Flag-TCFL5 (Fig. 1H and I). The chromatin immunoprecipitation (ChIP) assay also verified that TCFL5 could directly bind to the FUBP3 core promoter region (Fig. 1J). It is suggested that TCFL5 is essential for triggering FUBP3 expression through the combination with the FUBP3 promoter.
FUBP3 suppressed PEDV replication.
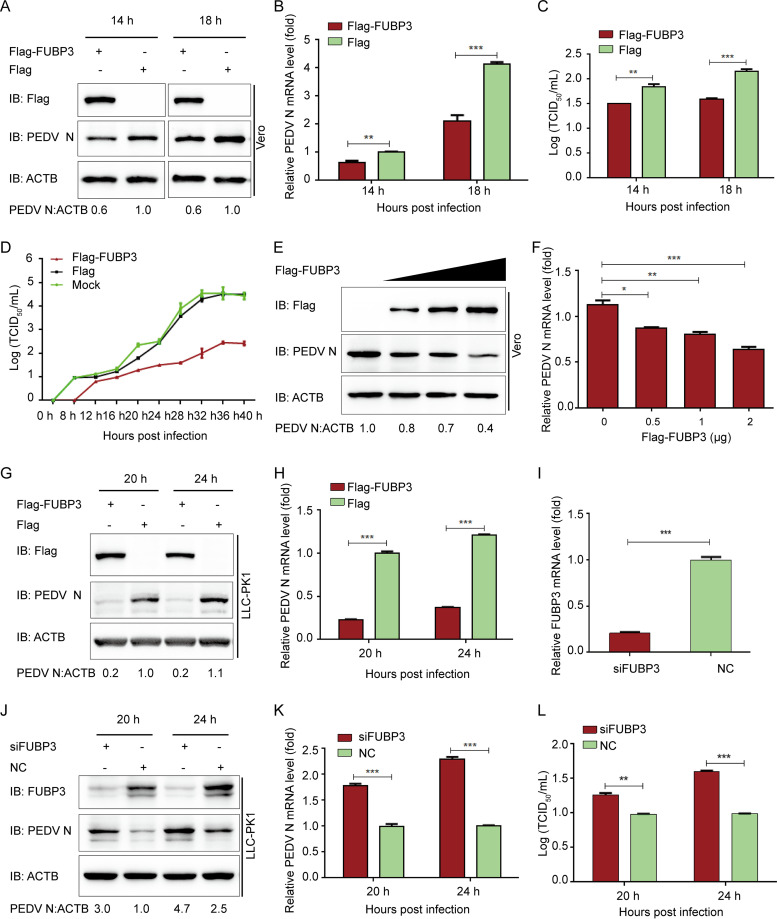
To further assess the function of FUBP3 in the proliferation of PEDV, Vero cells were subjected to the transfection with Flag-FUBP3 for a whole day and were subsequently exposed to PEDV (MOI = 0.01). Meanwhile, the viral loads were assayed in the cells and supernatants of the infected cell cultures. According to the results of the WB, during PEDV infection, FUBP3 overexpression reduced PEDV N protein in the Vero cells (Fig. 2A). Similarly, the results of the qRT-PCR analysis revealed significantly lower genomic mRNA expression in the Flag-FUBP3 group (Fig. 2B). The viral titers in the supernatants of the cells overexpressing FUBP3 were lower compared with those from cells that were subjected to the transfection with the control vector (Fig. 2C and D). Furthermore, an increasing amount of Flag-FUBP3 was ectopically expressed. We found that the viral loads of PEDV were reduced with the enhancement in the expression of FUBP3 (Fig. 2E and F). Moreover, high expressions of FUBP3 suppressed PEDV replication (Fig. 2G and H) while increasing PEDV replication by FUBP3 silencing in LLC-PK1 cells (Fig. 2I to L). As suggested by the findings, FUBP3 could significantly inhibit PEDV replication.
FIG 2.
The antiviral effect of FUBP3 against PEDV. (A and B) Vero cells were transfected with plasmid encoding Flag-FUBP3 and infected with PEDV (MOI = 0.01) and harvested at indicated times. qRT-PCR and WB were carried out to analyze cell lysates; ACTB served as the sample loading control. (C and D) Flag-FUBP3 was transfected into Vero cells, with the cells being infected with PEDV (MOI = 0.01). The culture supernatant was harvested at specific time points, with the viral titers being measured as TCID50 (E and F). The enhancing contents of a vector expressing Flag-FUBP3 (wedge) with the infection of PEDV (MOI = 0.01). qRT-PCR and a WB were conducted to analyze cell lysates. (G and H) LLC-PK1 cells were transfected with the plasmid that encoded Flag-FUBP3. PEDV (MOI = 1) was injected into the cells, which were collected at specific time points. Later, qRT-PCR and WB were conducted to analyze the cell lysates. (I) The knockdown efficiency of the FUBP3 siRNA in LLC-PK1 cells was analyzed by real-time PCR. (J, K, and L) LLC-PK1 cells were transfected using FUBP3 siRNA or negative control siRNA, followed by infection using PEDV (MOI = 1) and harvest the cells and culture supernatant at specific time points. PEDV N was examined by performing qRT-PCR and WB, and the viral titer was measured as TCID50. The obtained data are indicated as means ± SD of triplicate samples; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed Student's t test).
FUBP3 interacted with the N protein of PEDV.
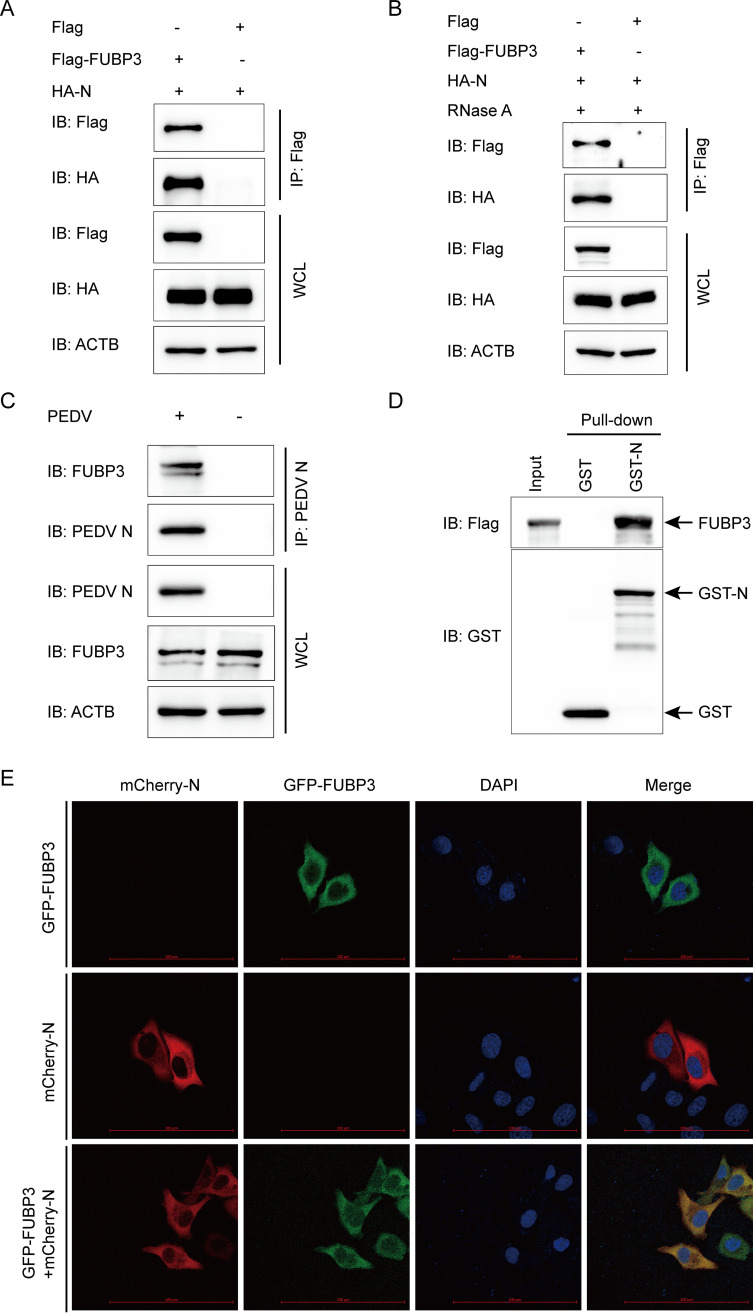
FUBP3 was identified as a protein that interacted with the PEDV N protein by the iTRAQ method. Thus, we hypothesized that FUBP3 might interact with the structural protein N of PEDV to inhibit the replication of PEDV. To confirm this, cells were subjected to cotransfection with HA-N and Flag-FUBP3 plasmids, and coimmunoprecipitation (Co-IP) was used to detect the interaction. We found that FUBP3 efficiently coimmunoprecipitated with N protein (Fig. 3A). Both FUBP3 and the PEDV N protein are discovered to be RNA binding proteins (23, 28). To exclude the possibility that the interaction between FUBP3 and N was connected by viral or host RNA, we treated cell lysates with RNase and found that RNase did not interrupt the association of FUBP3 with the N protein (Fig. 3B), implying that the interaction of FUBP3 with the PEDV N protein was RNA-independent. Additionally, the PEDV N protein contributed to the efficient coimmunoprecipitation with the endogenous FUBP3 protein (Fig. 3C). In the Co-IP assay, we conducted a glutathione S-transferase (GST) pulldown assay for confirming whether FUBP3 could bind to N. GST-fused N (GST-N) combined with FUBP3, which was not observed between GST and FUBO3 (Fig. 3D). Based on the obtained results, N could directly bind to FUBP3. Next, we performed confocal microscopy to determine whether FUBP3 could colocalize with N and found cytoplasmic colocalization of the N protein with FUBP3 (Fig. 3E). Thus, it was shown that FUBP3 could directly interact with the N protein of PEDV.
FIG 3.
FUBP3 can make interaction with the PEDV N protein. (A) Plasmids that encoded Flag-FUBP3 and HA-N were exposed to transfection into HEK 293T cells. Subsequently, a Co-IP assay was carried out by adopting anti-Flag-bound beads, and WB was conducted for analysis; ACTB was employed to be a sample loading control. (B) The cells cotransfected with Flag-FUBP3 and HA-N for 24 h were collected. RNase was used to incubate the lysate, and the relationship between FUBP3 and the N protein was analyzed through Co-IP assays. (C) Vero cells were mock-infected or infected with PEDV (MOI = 0.01) and were harvested 24 h after being infected. Co-IP assays were conducted using the anti-PEDV N protein antibody. (D) FUBP3 and GST-N expression was induced within the bacterial strain BL21(DE3), with the relationship between FUBP3 and the N protein being analyzed by performing a GST pulldown analysis. (E) N-mCherry and FUBP3-GFP were cotransfected into HeLa cells for a whole day. In addition, the cell nuclei were stained with DAPI, and the colocalization of FUBP3 and N was observed through confocal immunofluorescence microscopy; scale bars: 100 μm.
FUBP3 stimulated the autophagic degradation of N by recruiting MARCHF8 to ubiquitinate the N protein.
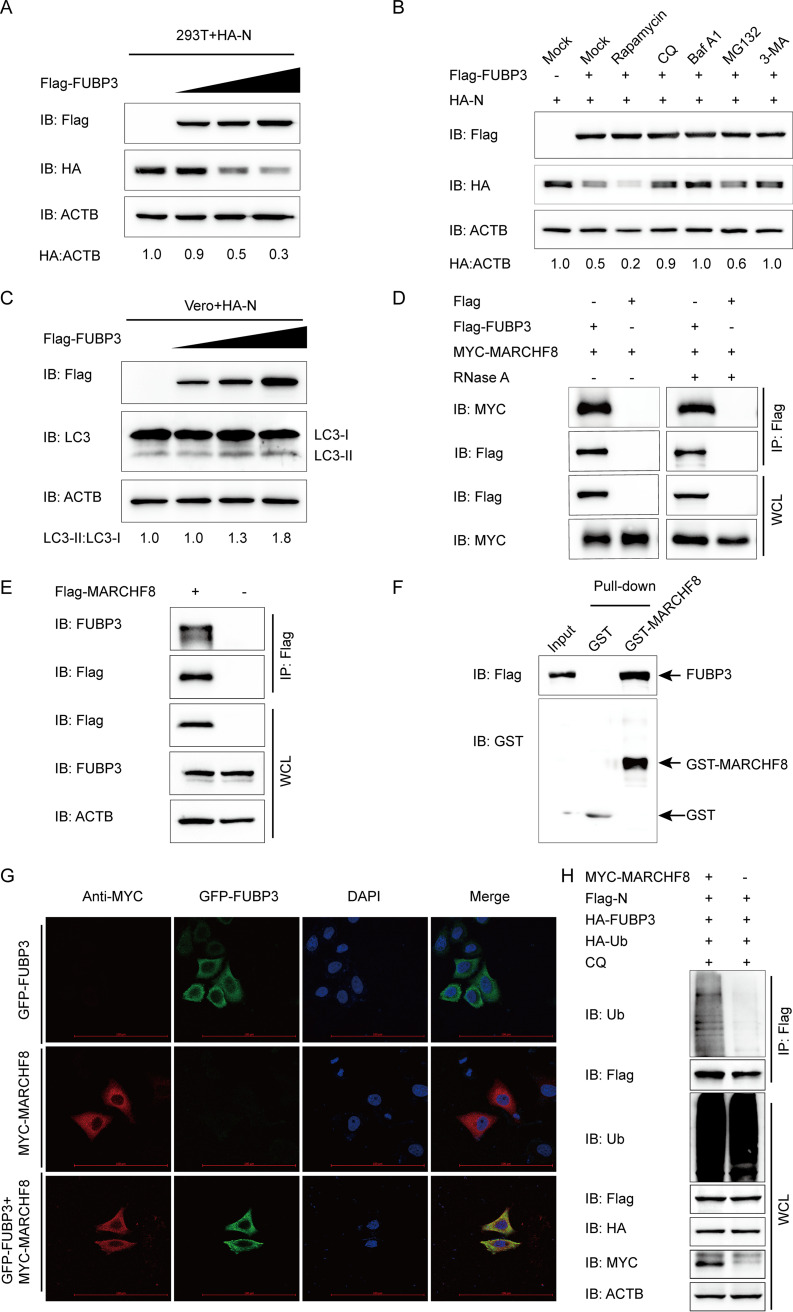
To elucidate the mechanisms through which FUBP3 hinders PEDV replication, the effect of FUBP3 on the stability of the N protein was examined in this study. HEK 293T cells were subjected to cotransfection with Flag-FUBP3 and HA-N eukaryotic expression plasmids. At the same time, to analyze the expression of the N protein abundances, a WB was carried out. N expression decreased significantly in a dose-dependent manner after being subjected to FUBP3 (Fig. 4A). Initially, the cellular proteins degenerated via the ubiquitin-proteasome and autolysosome pathways (29). To determine the pathway via which FUBP3 degraded N, HEK 293T cells were subjected to cotransfection with Flag-FUBP3 and HA-N as well as treated with autophagy inhibitors bafilomycin A1 (Baf A1), chloroquine (CQ), or 3-methyladenine (3-MA) or the proteasome inhibitor MG132. Based on the obtained results of the WB, autophagy inhibitors 3-MA, CQ, and Baf A1 suppressed the FUBP3-induced N protein degradation, while MG132 did not generate such an effect (Fig. 4B), suggesting that FUBP3 could promote autophagic degradation of N. Additionally, FUBP3 considerably enhanced N degradation during autophagic activation mediated by the autophagy inducer Rapamycin (Fig. 4B). The above results indicated that FUBP3 promoted N degradation via autophagy.
FIG 4.
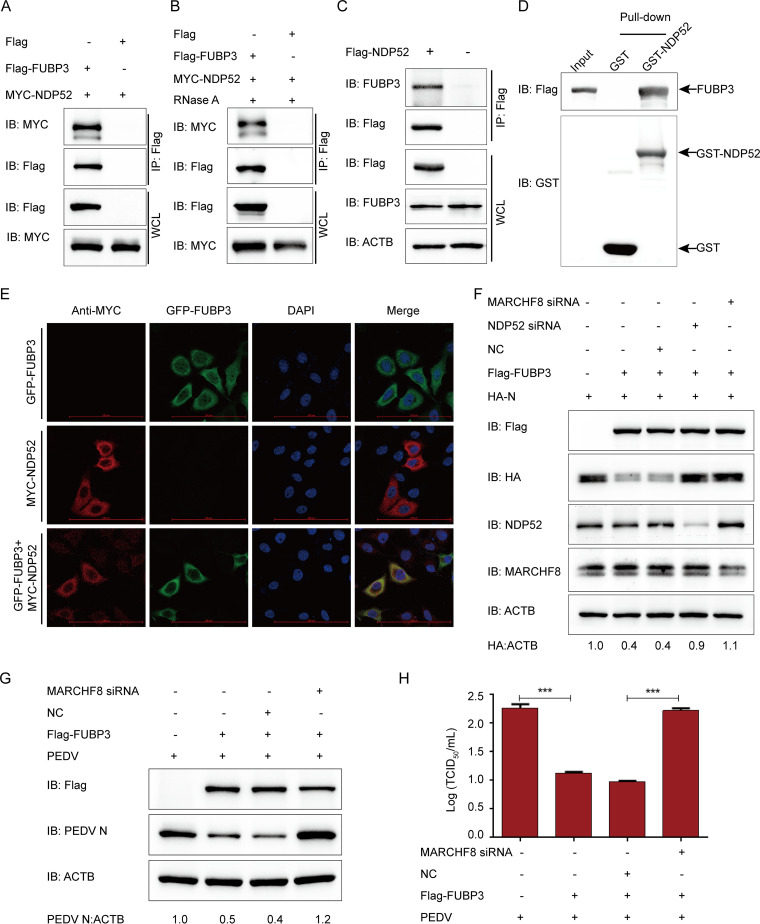
FUBP3 can enhance autophagic degradation of N by recruiting MARCHF8 to ubiquitinate the N protein. (A) The HA-N expression vector was transfected into HEK 293T cells, and subsequently, the Flag-FUBP3 expression vector (wedge) at increasing doses was also transfected into the cells. WB was conducted to analyze the cell lysates. ACTB functioned as a sample loading control. (B) The Flag-FUBP3 and HA-N expression vectors were transfected into the HEK 293T cells for 24 h, followed by treatment with Rapamycin (12.5 μM), CQ (50 μM), Baf A1 (50 μM), MG132 (5 μM), or 3-MA (1 mM) for 9 h. WB was conducted to analyze the cell lysates. (C) The HA-N expression vector was transfected into the HEK 293T cells along with the Flag-FUBP3 expression vector (wedge) at high doses. WB was conducted to analyze the cell lysates. (D) The plasmids that encoded MYC-MARCHF8 and Flag-FUBP3 were transfected into HEK 293T cells, and later, a Co-IP assay was carried out with the use of anti-Flag-bound beads, followed by WB analysis. (E) The plasmid that encoded Flag-MARCHF8 was transfected into HEK 293T cells for 24 h. Subsequently, a Co-IP assay was carried out with an anti-Flag antibody. (F) The expression of GST-MARCHF8 and FUBP3 was induced in the bacterial strain BL21(DE3), with the relationship between MARCHF8 and FUBP3 being analyzed by performing a GST pulldown assay. (G) Followed by incubation of the cells with anti-MYC MAb, FUBP3-GFP and MARCHF8-MYC were cotransfected into HeLa cells for a whole day. The colocalization of FUBP3 and MARCHF8 was found by confocal IF microscopy; scale bars: 100 μm. (H) The plasmids that encoded MYC-MARCHF8, Flag-N, HA-FUBP3, and HA-Ub were cotransfected into HEK 293T cells for 24 h, followed by treatment with CQ (50 μM) for 9 h. A Co-IP assay was carried out with an anti-Flag antibody. In the meanwhile, a WB assay was conducted to analyze the cell lysates with an anti-ubiquitin (Ub) antibody.
To determine whether FUBP3 could induce autophagy, the levels of microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3), a hallmark of autophagy (19), were checked in Flag-FUBP3 and HA-N coexpressed cells. The results showed that N and FUBP3 coexpression in the cells promoted a dose-dependent conversion of LC3-I to LC3-II (Fig. 4C), which suggested that FUBP3 promoted autophagy in the N protein-expressing cells. Ubiquitin can be modified to substrate proteins in the autophagy protein degradation pathways via the E3 ubiquitin ligase (30). The host factors BST2, IRAV, and PABPC4 recruit MARCHF8 for catalyzing PEDV N protein ubiquitination (2, 3, 19). HEK 293T cells were subjected to cotransfection with Flag-FUBP3 and MYC-MARCHF8 plasmids with the purpose of testing whether MARCHF8 can regulate the autophagy pathway mediated by FUBP3 to degrade N. The co-IP assay indicted that FUBP3 could bind to MARCHF8, and the interaction between FUBP3 and MARCHF8 was independent of RNA. (Fig. 4D). Furthermore, MARCHF8 facilitated efficient coimmunoprecipitation with FUBP3 (Fig. 4E). The results of a GST pulldown experiment confirmed that FUBP3 could interact with MARCHF8 (Fig. 4F). Moreover, the confocal immunofluorescence (IF) assay indicated that FUBP3 could colocalize with the MARCHF8 protein in the cytoplasm (Fig. 4G). To determine the function of MARCHF8 in FUBP3-induced autophagic degradation of the N protein, this study overexpressed MARCHF8 in the HA-FUBP3-and Flag-N-overexpressing HEK 293T cells. According to the results of the WB, the overexpressed MARCHF8 contributed to significantly enhanced ubiquitination of N, as decided based on IP and ubiquitination assays (Fig. 4H). According to the results, MARCHF8 increased the ubiquitination of N during FUBP3-mediated autophagic degradation.
N protein degraded by autophagy via the FUBP3-MARCHF8-NDP52-autophagosome pathway.
Using selective autophagy, cargo receptors affect substrate recognition and delivery into autophagosomes (31). The host factors BST2 and PABPC4 use the cargo receptor NDP52 to recognize and deliver the ubiquitinated N protein (19, 21). Therefore, FUBP3 was discovered to interact with NDP52. Our results showed that FUBP3 interacted with NDP52 (Fig. 5A), and this interaction did not depend on viral or host RNA (Fig. 5B). Additionally, we found that NDP52 contributed to the efficient coimmunoprecipitation with the endogenous FUBP3 protein (Fig. 5C). Furthermore, the GST pulldown assay confirmed that FUBP3 could bind to NDP52 (Fig. 5D). We cotransfected the FUBP3-GFP and NDP52-MYC plasmids to investigate the colocalization of FUBP3 and NDP52 by adopting confocal microscopy, finding the colocalization of FUBP3 and NDP52 in the cytoplasm. (Fig. 5E). This suggested that FUBP3 could directly interact with the cargo receptor NDP52.
FIG 5.
FUBP3 caused N protein degradation via the MARCHF8-NDP52-autophagosome pathway. (A) Plasmids that encoded MYC-NDP52 and Flag-FUBP3 were cotransfected into HEK 293T cells, then a Co-IP assay was conducted based on anti-Flag-bound beads, and a WB was conducted for the analysis. (B) The HEK 293T cells cotransfected with Flag-FUBP3 and MYC-NDP52 for 24 h were harvested. RNase was used for incubating the lysates, and the correlation between FUBP3 and NDP52 was analyzed by Co-IP assays. (C) The plasmids that encoded Flag-NDP52 were transfected into HEK 293T cells for a day. Co-IP assays were conducted using an anti-Flag antibody. (D) The expression of GST-NDP52 and FUBP3 was induced in the bacterial strain BL21(DE3). The relationship between FUBP3 and NDP52 was analyzed by performing a GST pulldown analysis. (E) HeLa cells were cotransfected with FUBP3-GFP and NDP52-MYC for 24 h based on the incubation with anti-MYC MAb. The colocalization of FUBP3 and NDP52 was found using confocal IF microscopy; scale bars: 100 μm. (F) NDP52 siRNA or MARCHF8 siRNA was transfected into HEK 293T cells with plasmids that encoded HA-N and Flag-FUBP3. WB was performed for analysis. (G and H) Vero cells were transfected with the plasmids encoding Flag-FUBP3 and MARCHF8 siRNA for 24 h, and subsequently, PEDV (MOI = 0.01) was used to infect cells. Western blot and TCID50 were analyzed at 20 h postinfection.
To determine the importance of MARCHF8 and NDP52 in FUBP3- induced degradation of N protein, HEK 293T cells were cotransfected with Flag-FUBP3 and HA-N plasmids, and later, the expression of MARCHF8 or NDP52 was inhibited by small interfering RNAs (MARCHF8 siRNA or NDP52 siRNA). The findings of the Western blot demonstrated that interrupting the expression of MARCHF8 or NDP52 expression could prevent the degradation of N protein by FUBP3 (Fig. 5F). Next, we investigated whether the MARCHF8-NDP52-autophagosome axis is required to hinder the replication of PEDV by FUBP3 and found that interfering with MARCHF8 protein expression prevented FUBP3-mediated inhibition of PEDV replication in Vero cells (Fig. 5G and H). Therefore, FUBP3 contributes to PEDV N protein degradation based on the MARCHF8-NDP52 autophagosome pathway.
FUBP3 activated the IFN signaling pathway by increasing the TRAF3 expression.
IFN and the IFN-induced cellular antiviral response are considered the leading constituents of the host to establish the antiviral status and offer the first line of defense to resist viral infection (32). To decide whether FUBP3 is engaged in the regulation of the IFN-I signaling pathway, IFN-β promoter and IFN-stimulated response element (ISRE)-driven luciferase reporter assay was performed in this study, showing that FUBP3 induced IFN expression through a dose-dependent manner (Fig. 6A and B). Apart from that, the mRNA level of IFN was also consistently enhanced in the FUBP3-transfected cells (Fig. 6C). As shown in the results, FUBP3 expression induced IFN-I production.
FIG 6.
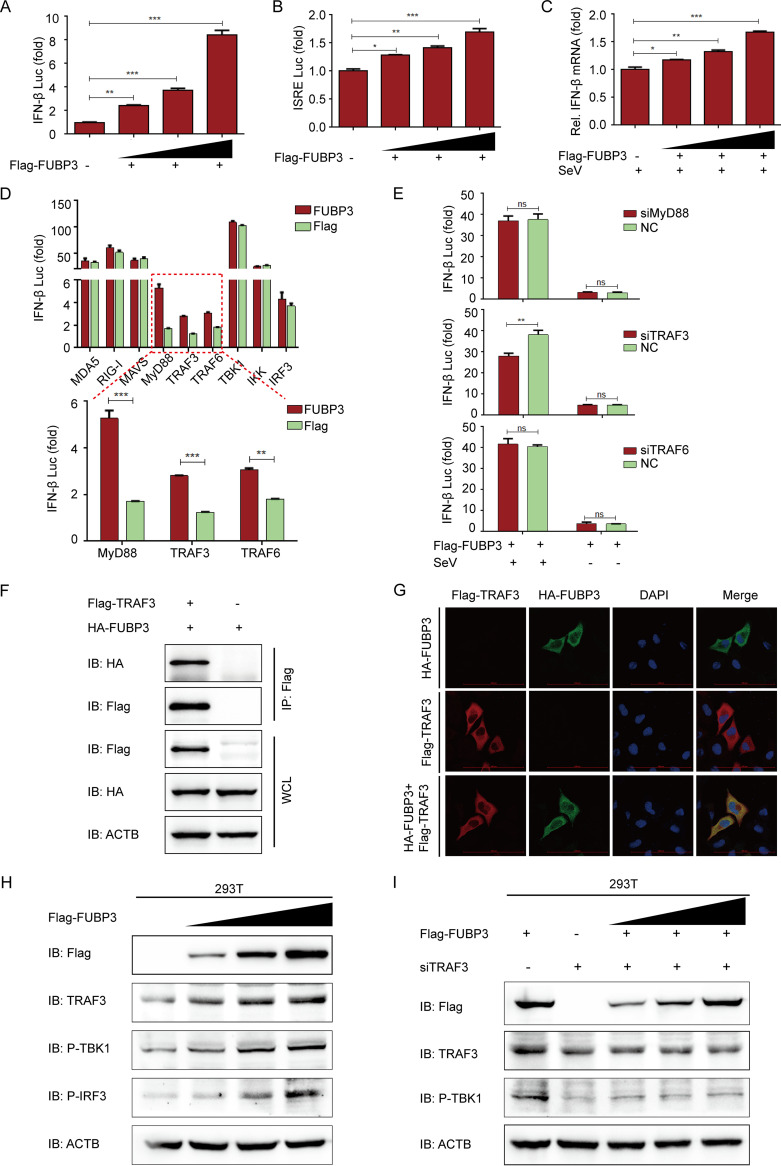
FUBP3 activated the IFN signaling pathway by increasing TRAF3 expression. (A and B). The ISRE or IFNB luciferase reporter was cotransfected into HEK 293T cells with the Flag-FUBP3 expression vector (wedge) at increasing doses. (C) The lysates of HEK 293T cells were subjected to transfection with the Flag-FUBP3 expression vector (wedge), and subsequently, the Sendai virus (SeV) was used to infect cells and then conduct the luciferase assay. (D) The plasmids that encoded the FUBP3 and IFNB luciferase reporter were transfected into HEK 293 T cells with plasmids encoding MDA5, RIG-I, MAVS, MyD88, TRAF3, TRAF6, TBK1, IKK, or IRF3. Luciferase activities were then measured. (E) The lysates of HEK 293T cells cotransfected with the Flag-FUBP3 expression vector and MyD88 siRNA, TRAF3 siRNA, or TRAF6 siRNA, and later infected with SeV, were selected for the luciferase assay. (F) The plasmids that encoded Flag-TRAF3 and HA-FUBP3 were cotransfected into HEK 293T cells. Subsequently, a Co-IP assay was conducted with the anti-Flag-bound beads, and WB was performed for the analysis. (G) Flag-TRAF3 and HA-FUBP3 were cotransfected into HeLa cells for a day, followed by incubation of the cells with anti-Flag and anti-HA MAbs. Confocal IF microscopy was performed to observe TRAF3-FUBP3 colocalization; scale bars: 100 μm. (H) The Flag-FUBP3 expression vector (wedge) at increasing doses was transfected into HEK 293T cells for a day. WB was carried out to analyze the cell lysates. (I) The Flag-FUBP3 expression vector (wedge) at increasing doses was cotransfected with TRAF3 siRNA into HEK 293T cells for 24 h. WB was performed to analyze the cell lysates.
Within viral infection, host pattern recognition receptors (PRRs) can sense cytoplasmic viral RNAs and interact with MAVS to activate the downstream tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3), causing the activation of the interferon regulatory factor 3 (IRF3). Phosphorylated IRF3 can translocate to the nucleus and trigger IFN-Is (15, 33, 34). To determine the molecular mechanisms by which FUBP3 activates IFN signal transduction, a luciferase reporter assay was conducted, and the results suggested that FUBP3 increased the luciferase reporter activity due to MyD88, TRAF3, and TRAF6 (Fig. 6D). Next, we synthesized and selected siRNAs that target MyD88, TRAF3, or TRAF6 and cotransfected them with FUBP3 into cells. According to the findings of the luciferase reporter assay, only the siRNA-mediated downregulation of TRAF3 expression contributed to interrupting the activation of IFN induced by FUBP3 (Fig. 6E). This suggested that FUBP3 can mediate the activation of TRAF3 to regulate the IFN signaling pathway. Moreover, based on the co-IP and confocal immunofluorescence assays, the FUBP3 protein showed interaction and colocalization with TRAF3 in the cytoplasm (Fig. 6F and G). We also transfected FUBP3 into cells to identify the regulation of the downstream signaling. Based on the findings of the Western blot, overexpression of FUBP3 could increase TRAF3 overexpression and activate TBK1 and IRF3 (Fig. 6H). Thus, the siRNA's inhibition of TRAF3 protein expression could effectively prevent the activation of TBK1 by FUBP3 (Fig. 6I). The obtained findings suggested that FUBP3 activated the IFN signaling pathway by increasing TRAF3 expression.
DISCUSSION
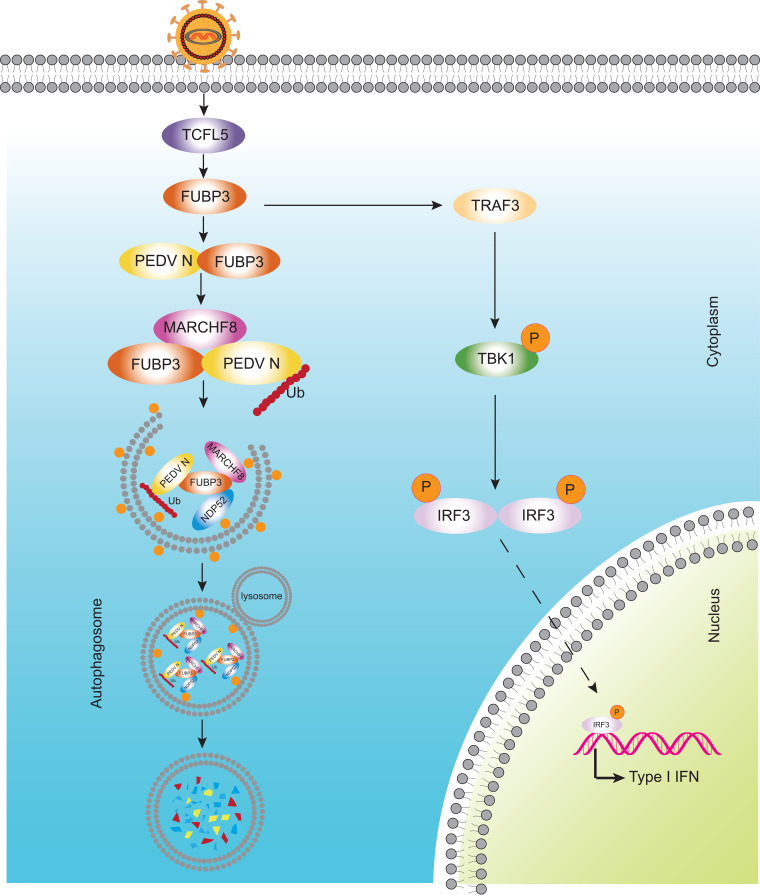
PEDV refers to a highly virulent re-emerging enteric coronavirus leading to epidemic infection in pig populations in many countries, resulting in substantial financial losses in the pig and swine industry (7, 34). Some vaccines for PEDV are commercially available, but they cannot completely resist the epidemic strains (9, 35). As a result, determining the association of PEDV with host antiviral factors can help elucidate and understand specific antiviral mechanisms and develop effective vaccines and new targets for treating PEDV infection. In this study, a new function of FUBP3 in resisting PEDV infection was found. It could be discovered that the transcription factor of TCFL5 could regulate the expression of FUBP3 through PEDV infection. According to the results, FUBP3 could suppress PEDV replication by degrading the virus N protein by promoting the FUBP3-MARCHF8-NDP52 autophagosome pathway. Additionally, FUBP3 activated the IFN signaling pathway by increasing TRAF3 expression (Fig. 7). Our results indicated that FUBP3 could suppress PEDV replication through the degradation of the viral N protein via the FUBP3-MARCHF8-NDP52 autophagosome pathway.
FIG 7.
The antiviral mechanism of FUBP3 can inhibit PEDV replication. During PEDV infection, host cells recognize the virus and activate TCFL5 to upregulate the abundance of FUBP3. FUBP3 interacts with E3 ubiquitin ligase MARCHF8 to ubiquitin N protein, then recruiting the cargo receptor NDP52 to recognize the ubiquitinated N protein and deliver it to autophagosome for degradation. Additionally, TARDBP activates type I IFN signaling by directly targeting TRAF3, which then induces the phosphorylation of TBK1.
The evolution of the host antiviral proteins has certainly been fostered by a continuous arms race between viruses and hosts. Thus, viral infections may drive proteins to evolve in various ways to resist viral replication (36). The physiological function of FUBP3 is primarily to make interactions with many single-stranded nucleic acids in the cells to regulate various biological processes, including mRNA transcription, mRNA degradation, and RNA transport (37–39). Host cells were recently shown to regulate the expression of FUBP3 to affect virus replication (37, 40). However, the regulatory mechanism of FUBP3 during virus infection is still poorly understood. We found that PEDV infection could effectively regulate FUBP3. To further identify the transcription factors regulating FUBP3 expression, the FUBP3 promoter sequence was amplified. We found that the minimum FUBP3 core promoter was in the range between −156 and −103 positions. Through analyzing the regulatory elements of FUBP3, we found that TCFL5 could be directly combined with the promoter of FUBP3 and regulate the expression of FUBP3.
FUBPs, which comprise FUBP1, FUBP2, and FUBP3, can bind to diverse RNA or DNA targets in cells to regulate RNA translation and gene stability (27, 39). The structure of FUBPs includes four regularly spaced K homology (KH) patterns, which are responsible for recognizing and interacting with similar sequences in single-stranded RNA and DNA targets (41). It is shown that FUBP3 is required to regulate the c-Myc proto-oncogene properly and regulate central cellular processes (42). Additionally, FUBP3 is found to positively regulate enterovirus 71 (EV-A71) replication by interacting with the viral 5′UTR (27, 33) and promotes Japanese encephalitis virus (JEV) replication through the interaction with the viral 3′UTR (37). However, our findings proved that FUBP3 could inhibit the replication of PEDV and reduce FUBP3 expression, thereby dramatically increasing PEDV replication. We also found that FUBP3 directly interacted with the PEDV N protein in an RNA-independent manner. N protein amount is negatively related to the FUBP3 protein amount. As a multifunctional structural protein, the PEDV N protein can facilitate immune escape and viral replication (12, 15). Thus, the role of FUBP3 in targeting and degrading the PEDV N protein indicated that there might be other antiviral functions of FUBP3 to regulate virus replication besides the interaction of FUBP3 with the viral 5′UTR or 3′UTR.
The autolysosome and proteasome pathways are the two main protein degradation pathways in eukaryotes (29). We showed that FUBP3 could enhance the degradation of N protein through autophagy. Autophagy is regarded as a vital cell catabolic process for keeping cellular homeostasis via degrading misfolded or long-lived cytoplasmic proteins and damaged organelles triggered by various cellular and environmental stresses like energy deficiency endoplasmic reticulum (ER) stress as well as pathogen infection (43, 44). Autophagy targets specific substrates and engulfs cargo very selectively (45). During selective autophagy, the specific substrates could be ubiquitinated by E3 ubiquitin ligase as well as identified by cargo receptors. The cargo receptors which target the ubiquitinated substrates interact with Atg8-family proteins and are sequestered within autophagosomes, fusing with lysosomes. The complexes of cargo receptors and specific substrates are discovered to degrade in the lysosome's proteolytic environment (45). Our previous study showed that the host antiviral proteins BST2 and PABPC4 promoted the selective autophagy process for degrading the N protein of PEDV and inhibited PEDV replication (19, 21). In this study, FUBP3 was demonstrated to recruit E3 ubiquitin ligase MARCHF8 for catalyzing PEDV N protein, and the ubiquitinated N protein was later detected and lowered by NDP52-dependent selective autophagy. Thus, FUBP3 promoted the degradation of the PEDV N protein through selective autophagy. In addition, after the selective autophagy pathway was disrupted, there existed an increase in PEDV replication.
The innate immune system serves as the first-line host defense against viral invasion, and the antiviral responses are controlled principally by the IFN, causing the production of hundreds of interferon-stimulating genes (ISGs) to create an antiviral state (12). During virus infection, the PRRs recruit MAVS and interact with TRAF3 through its proline enrichment region. The activated TRAF3 recruits and activates TANK-binding kinase 1 (TBK1) to induce IFN-I (15). According to recent studies, signaling pathways containing the PI3K-Akt, RLR, Rap1, TLR, JAK-STAT, and MAPK pathways, are differentially regulated by PEDV. This indicates that PEDV can modulate innate host immunity (46, 47). The current work discovered that FUBP3 could combine and enhance TRAF3 expression, which then regulates the phosphorylation of TBK1 and induces IFN-I production.
This work shows a new effect of FUBP3 on the inhibition of PEDV replication via recruiting E3 ubiquitin ligase MARCHF8 to catalyze N protein, and the ubiquitinated N protein was identified and degraded by NDP52-dependent selective autophagy. Additionally, we found that FUBP3 could activate the IFN signaling pathway by increasing TRAF3 expression. Our findings collectively reveal a novel role of FUBP3-mediated viral restriction mechanisms against PEDV infection, contributing to strengthening the host innate immune system defense mechanism and providing a new effective strategy for the control of PEDV infection.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero cells; ATCC, CCL-81) and human embryonic kidney cells (HEK 293T cells; ATCC, CRL-11268) were kept in Dulbecco's modified Eagle's medium (Invitrogen, 12430054) by adding 10% fetal bovine serum (Gibco, 10099141). We obtained porcine kidney LLC-PK1 cells (LLC-PK1 cells) from Dr. Rui Luo (Huazhong Agricultural University, Wuhan, China) and kept them in the modified Eagle's medium (Invitrogen, 11095080). The cells were then inoculated at 37°C and 5% CO2. We isolated the variant PEDV JS-2013 strain and kept it in the laboratory (47). The propagation and titration of PEDV were performed with Vero cells, and Karber's method was used to determine the titers (47).
Antibodies and reagents.
The monoclonal anti-PEDV N antibody was stocked in our laboratory (48). Anti-Flag-tag antibody (F1804), chloroquine phosphate (CQ; PHR1258), 3-Methyladenine (3-MA; M9281), and MG132 (M7449) were obtained from Sigma-Aldrich. Antibodies against ACTB/β-actin (66009-1-lg), LC3 (14600-1-AP), NDP52 (12229-1-AP), MARCHF8 (14119-1-AP), FUBP3 (10623-1-AP), GST-tag (10000-0-AP), GAPDH (60004-1-lg), and TRAF3 (66310-1-lg), along with HRP-labeled anti-mouse (SA00001-1) and anti-rabbit (SA00001-2) IgG antibodies could be acquired from Proteintech Group. Bafilomycin A1 (Baf A1; 54645), anti-pIRF3 antibody (4947), anti-pTBK1 antibody (5483), anti-MYC-tag antibody (2278), and anti-HA-tag antibody (3724) were bought from Cell Signaling Technology. Human MARCHF8 siRNA (SC-90432), human NDP52 siRNA (SC-93738) as well as an anti-ubiquitin antibody (SC-8017) were acquired from Santa Cruz Biotechnology. In addition, Rapamycin (HY-10219) was bought from MedChemExpress.
Plasmids and transfection.
With the use of ClonExpress II one-step cloning kit (C112–02, Vazyme Biotech Co., Ltd.), amplification of target protein genes was completed, followed by clone into the eukaryotic expression vector using homologous recombination. The recombinant plasmids transfected into cells with Lipofectamine 3000 reagent (L3000015, Invitrogen) when the cells had grown to nearly 80% to 90% confluence and transfected the siRNAs into cells based on Lipofectamine RNAiMAX (13778150, Invitrogen) under the condition that the cells had grown to around 50% to 60% confluence in line with the required guidance.
Western blot (WB).
RIPA lysis and extraction buffer (89901, Thermo Fisher Scientific), including protease inhibitor cocktail (B14001, Bimake) as well as phosphatase inhibitor cocktail (B15001, Bimake) were used to lyse cell samples that were washed with cold PBS. We then boiled cell lysates with 5×SDS loading buffer for 10 min. SDS-PAGE was conducted to separate the proteins, followed by transfer onto nitrocellulose membrane (10600001, GE Healthcare). The membrane was treated with 5% skim milk powder (232100, BD Biosciences) for an hour. Incubation with primary antibodies was performed for an hour at ambient temperature. Subsequently, based on HRP-labeled secondary antibodies, the membrane was rinsed with 0.1% PBST and nurtured with HRP-conjugated secondary antibody for 60 min. In the end, enhanced chemiluminescence (ECL; Share-bio, SB-WB012) was used to measure the proteins.
Coimmunoprecipitation (Co-IP) assay.
As required by specified times, cells were subjected to transfection with the indicated plasmids or infected PEDV. With the addition of a protease inhibitor cocktail for an hour, the cells were rinsed with ice-cold PBS and lysed with NP-40 cell lysis buffer (FNN0021, Life Technologies). After centrifugation, the lysates were nurtured with affinity antibodies bound to protein G Dynabeads (10004D, Life Technologies) for half an hour at 25°C. After rinsing the Dynabeads in 0.02% PBST, they were boiled in the sample buffer. In addition, standardized immunoblotting procedures were used to examine the relationship between proteins.
RNA isolation and qRT-PCR.
Total RNA was extracted with the use of RNeasy Minikit (74104, Qiagen) and transcribed to cDNA based on the PrimeScript RT reagent kit (RRO47A, TaKaRa Bio) following the instructions of the manufacturer. The SYBR Premix Ex Taq (q711–03, Vazyme Biotech Co., Ltd.) was used to perform qRT-PCR for synthesized cDNA following the specific instruction. The ΔΔCt method was used for exploring the results. As shown in Table 1, the list of primer sequences employed in qRT-PCR is presented. GAPDH and ACTB were applied as the reference genes for normalization.
TABLE 1.
The sequences of the primers and siRNAs
| Purpose | Names | Sequence (5′–3′) |
|---|---|---|
| Real-time PCR Primers | PEDV N forward | GAGGGTGTTTTCTGGGTTG |
| PEDV N reverse | CGTGAAGTAGGAGGTGTGTTAG | |
| pFUBP3 forward | GCATAAACCAGCAGTCGGG | |
| pFUBP3 reverse | TGAAGGGGCTCTGTCCAAA | |
| TCFL5 forward | AGCAGGTTTGGATTAAAGTAGG | |
| TCFL5 reverse | AGGCTGGACTGCGAGGA | |
| ACTB forward | TCCCTGGAGAAGAGCTACGA | |
|
ACTB reverse pGAPDH forward pGAPDH reverse hIFN-β forward hIFN-β forward |
AGCACTGTGTTGGCGTACAG ATGGATGACGATATTGCTGCGCTC TTCTCACGGTTGGCTTTGG TCTTTCCATGAGCTACAACTTGCT GCAGTATTCAAGCCTCCCATTC |
|
| ChIP assay | ChIP primer forward | CAGAGACGGGACGGCAGCCTCACGC |
| ChIP primer reverse | GGACCCGTAGCCGGAAAGGGAAAGT | |
| siRNA sequences | si-FUBP3 sense si-FUBP3 antisense si-TCFL5 sense si-TCFL5 antisense si-MyD88 sense si-MyD88 antisense si-TRAF3 sense si-TARF3 antisense si-TRAF6 sense si-TRAF6 antisense NC sense NC antisense |
GCGUGAAGAUGGUCAUGAUTT
AUCAUGACCAUCUUCACGCTT CGACAUCCGUCCGAAUUAATT UUAAUUCGGACGGAUGUCGTT GUACAAGGCAAUGAAGAAATT UUUCUUCAUUGCCUUGUACTT GGCCGUUUAAGCAGAAAGUTT ACUUUCUGCUUAAACGGCCTT GCGCUGUGCAAACUAUAUATT UAUAUAGUUUGCACAGCGCTT UUCUCCGAACGUGUCACGUTT ACGUGACACGUUCGGAGAATT |
GST affinity-isolation assay.
PEDV N, FUBP3, MARCHF8, and NDP52 were cloned into prokaryotic expression vector pCold GST (3372, Clontech Laboratories, Inc) or pCold TF (3365, Clontech Laboratories, Inc). We expressed recombinant proteins in the BL21 competent cells (C504–03, Vazyme Biotech Co., Ltd.). In addition, the interaction was assessed using the GST Protein Interaction Pull-Down kit (21516, Thermo Fisher Scientific), following the specific instruction. The elution of the bound proteins was made using elution buffer (50 mM glycine, pH 2.8) and investigated by a Western blot using specific antibodies.
Dual-luciferase reporter assay.
In this study, 293T cells were seeded in 24-well plates, followed by transfection using the indicated plasmids, luciferase reporters, and pRL-TK by adopting Lipofectamine 3000. After transfection for 24 h, cell lysis was conducted, and then the dual-luciferase reporter assay (DL101–01, Vazyme Biotech Co., Ltd.) was carried out for measuring firefly luciferase and Renilla luciferase activities based on specific protocols. With Renilla luciferase activity as the internal control, luciferase values were normalized. In addition, three independent experiments were carried out, with each experiment in triplicate.
ChIP assay.
LLC-PK1 cells were exposed to transfection with Plasmid encoding Flag-TCFL5. After transfection for 24 h, cell lysis was conducted, and the SimpleChIP enzymatic ChIP kit (9003, Cell Signaling Technology) was applied to conduct the ChIP assay following the specific instructions. An anti-flag antibody was adopted for precipitating the TCFL5 promoter sequences, which were then amplified with the application of qRT-PCR.
Fluorescence microscopy.
HeLa cells were subjected to cotransfection with the indicated plasmids for the whole day. PBS was used to wash the cells, which were fixed in 4% paraformaldehyde (PFA, P6148, Sigma-Aldrich) for a quarter. They were then permeabilized using 0.1% Triton X-100 (T9284, Sigma-Aldrich) for 10 min at the ambient temperature. After being rinsed with PBS, the cells were incubated with the primary antibody for an hour at 37°C. The cells were rinsed with PBS again and nurtured with fluorescent-labeled secondary antibodies for an hour at 37°C. In the end, we stained the cells with DAPI (C1002, Beyotime Biotechnology) for 5 min at ambient temperature, and the confocal IF microscope (Carl Zeiss) was used.
Statistical analysis.
All the findings stand for three independent experiments, and the Data are stood for as means ± standard deviations (SD). This study employed the GraphPad Prism 5 software (GraphPad Software, USA) to carry out Student's t test (two-sided) as well as to detect significant differences between groups. The importance of the different groups was identified using a two-tailed Student's t test with the GraphPad Prism 5 software (GraphPad Software, USA). P < 0.05 was shown to have statistical significance.
ACKNOWLEDGMENTS
The work was supported by the National Key Research and Development Programs of China (no. 2021YFD1801102) and the National Natural Science Foundation of China (no. 31872478 and 32102665).
We declare no conflict of interest.
Contributor Information
Guangzhi Tong, Email: gztong@shvri.ac.cn.
Tongling Shan, Email: shantongling@shvri.ac.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. 2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18:161–163. 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun D, Wang X, Wei S, Chen J, Feng L. 2016. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci 78:355–363. 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood EN. 1977. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec 100:243–244. 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Okada K, Ohshima K. 1983. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nihon Juigaku Zasshi 45:829–832. 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q. 2012. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis 18:1350–1353. 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XM, Niu BB, Yan H, Gao DS, Yang X, Chen L, Chang HT, Zhao J, Wang CQ. 2013. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch Virol 158:2487–2494. 10.1007/s00705-013-1767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. 2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol 52:234–243. 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 9.Gao Q, Zheng Z, Wang H, Yi S, Zhang G, Gong L. 2021. The new porcine epidemic diarrhea virus outbreak may mean that existing commercial vaccines are not enough to fully protect against the epidemic strains. Front Vet Sci 8:697839. 10.3389/fvets.2021.697839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte M, Gelfi J, Lambert P, Rasschaert D, Laude H. 1993. Genome organization of porcine epidemic diarrhoea virus. Adv Exp Med Biol 342:55–60. 10.1007/978-1-4615-2996-5_9. [DOI] [PubMed] [Google Scholar]
- 11.Kocherhans R, Bridgen A, Ackermann M, Tobler K. 2001. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23:137–144. 10.1023/a:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Yoo D. 2016. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res 226:128–141. 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride R, van Zyl M, Fielding BC. 2014. The coronavirus nucleocapsid is a multifunctional protein. Viruses 6:2991–3018. 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Zhang H, Zhang Q, Huang Y, Dong J, Liang Y, Liu HJ, Tong D. 2013. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol 164:212–221. 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia X, Zhao Q, Xiong Y. 2015. HIV suppression by host restriction factors and viral immune evasion. Curr Opin Struct Biol 31:106–114. 10.1016/j.sbi.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Yu M, Liu A, Bao Y, Qi X, Gao L, Chen Y, Liu P, Wang Y, Xing L, Meng L, Zhang Y, Fan L, Li X, Pan Q, Zhang Y, Cui H, Li K, Liu C, He X, Gao Y, Wang X. 2021. TRIM25 inhibits infectious bursal disease virus replication by targeting VP3 for ubiquitination and degradation. PLoS Pathog 17:e1009900. 10.1371/journal.ppat.1009900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Shi K, Yoo D. 2016. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489:252–268. 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, Tong W, Yu L, Jiang Y, Zhou Y, Li G, Gao F, Yu H, Zheng H, Tong G. 2020. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy 16:1737–1752. 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Kong N, Jiao Y, Dong S, Sun D, Chen X, Zheng H, Tong W, Yu H, Yu L, Zhang W, Tong G, Shan T. 2021. EGR1 suppresses porcine epidemic diarrhea virus replication by regulating IRAV to degrade viral nucleocapsid protein. J Virol 95:e0064521. 10.1128/JVI.00645-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, Kong N, Wang H, Sun D, Dong S, Chen X, Zheng H, Tong W, Yu H, Yu L, Huang Y, Wang H, Sui B, Zhao L, Liao Y, Zhang W, Tong G, Shan T. 2021. PABPC4 broadly inhibits coronavirus replication by degrading nucleocapsid protein through selective autophagy. Microbiol Spectr 9:e0090821. 10.1128/Spectrum.00908-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. 1996. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem 271:31679–31687. 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 23.Gao Q, Zhou R, Meng Y, Duan R, Wu L, Li R, Deng F, Lin C, Zhao L. 2020. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3-c-Myc axis. Oncogene 39:3926–3938. 10.1038/s41388-020-1266-8. [DOI] [PubMed] [Google Scholar]
- 24.Rehbein M, Kindler S, Horke S, Richter D. 2000. Two trans-acting rat-brain proteins, MARTA1 and MARTA2, interact specifically with the dendritic targeting element in MAP2 mRNAs. Brain Res Mol Brain Res 79:192–201. 10.1016/s0169-328x(00)00114-5. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee J, Hermesh O, Eliscovich C, Nalpas N, Franz-Wachtel M, Macek B, Jansen RP. 2019. beta-Actin mRNA interactome mapping by proximity biotinylation. Proc Natl Acad Sci USA 116:12863–12872. 10.1073/pnas.1820737116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gau BH, Chen TM, Shih YH, Sun HS. 2011. FUBP3 interacts with FGF9 3' microsatellite and positively regulates FGF9 translation. Nucleic Acids Res 39:3582–3593. 10.1093/nar/gkq1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang HI, Chang YY, Lin JY, Kuo RL, Liu HP, Shih SR, Wu CC. 2016. Interactome analysis of the EV71 5' untranslated region in differentiated neuronal cells SH-SY5Y and regulatory role of FBP3 in viral replication. Proteomics 16:2351–2362. 10.1002/pmic.201600098. [DOI] [PubMed] [Google Scholar]
- 28.Cong Y, Ulasli M, Schepers H, Mauthe M, V’kovski P, Kriegenburg F, Thiel V, de Haan CAM, Reggiori F. 2020. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol 94:e01925-19. 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korolchuk VI, Menzies FM, Rubinsztein DC. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584:1393–1398. 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 30.Deretic V, Saitoh T, Akira S. 2013. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13:722–737. 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green DR, Levine B. 2014. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157:65–75. 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. 2018. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J Virol 92:e01677-17. 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Luo J, Yu J, Mao X, Luo Y, Zheng P, He J, Yu B, Chen D. 2019. Manipulation of intestinal antiviral innate immunity and immune evasion strategies of porcine epidemic diarrhea virus. Biomed Res Int 2019:1862531. 10.1155/2019/1862531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Yang J, Zhu Z, Zheng H. 2020. Porcine epidemic diarrhea virus and the host innate immune response. Pathogens 9:367. 10.3390/pathogens9050367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford K, Lager KM, Kulshreshtha V, Miller LC, Faaberg KS. 2016. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res 226:108–116. 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P, Tong W, Chen YM. 2021. FUSE binding protein FUBP3 is a potent regulator in Japanese encephalitis virus infection. Virol J 18:224. 10.1186/s12985-021-01697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gherzi R, Chen CY, Ramos A, Briata P. 2014. KSRP controls pleiotropic cellular functions. Semin Cell Dev Biol 34:2–8. 10.1016/j.semcdb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Chung HJ, Liu J, Dundr M, Nie Z, Sanford S, Levens D. 2006. FBPs are calibrated molecular tools to adjust gene expression. Mol Cell Biol 26:6584–6597. 10.1128/MCB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai FJ, Lin CW, Lai CC, Lan YC, Lai CH, Hung CH, Hsueh KC, Lin TH, Chang HC, Wan L, Sheu JJ, Lin YJ. 2011. Kaempferol inhibits enterovirus 71 replication and internal ribosome entry site (IRES) activity through FUBP and HNRP proteins. Food Chem 128:312–322. 10.1016/j.foodchem.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Chen QM. 2013. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene 32:2907–2916. 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, May M, Meyer HA, Behnke S, Moch H, Kristiansen G. 2008. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer 8:369. 10.1186/1471-2407-8-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boya P, Reggiori F, Codogno P. 2013. Emerging regulation and functions of autophagy. Nat Cell Biol 15:713–720. 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King JS, Veltman DM, Insall RH. 2011. The induction of autophagy by mechanical stress. Autophagy 7:1490–1499. 10.4161/auto.7.12.17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul P, Munz C. 2016. Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond. Adv Virus Res 95:149–195. 10.1016/bs.aivir.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Sun D, Shi H, Guo D, Chen J, Shi D, Zhu Q, Zhang X, Feng L. 2015. Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J Virol Methods 218:27–39. 10.1016/j.jviromet.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong N, Wu Y, Meng Q, Wang Z, Zuo Y, Pan X, Tong W, Zheng H, Li G, Yang S, Yu H, Zhou EM, Shan T, Tong G. 2016. Suppression of virulent porcine epidemic diarrhea virus proliferation by the PI3K/Akt/GSK-3alpha/beta pathway. PLoS One 11:e0161508. 10.1371/journal.pone.0161508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan X, Kong N, Shan T, Zheng H, Tong W, Yang S, Li G, Zhou E, Tong G. 2015. Monoclonal antibody to N protein of porcine epidemic diarrhea virus. Monoclon Antib Immunodiagn Immunother 34:51–54. 10.1089/mab.2014.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]