Abstract
SARS-CoV-2, the agent of the COVID-19 pandemic, emerged in late 2019 in China, and rapidly spread throughout the world to reach all continents. As the virus expanded in its novel human host, viral lineages diversified through the accumulation of around two mutations a month on average. Different viral lineages have replaced each other since the start of the pandemic, with the most successful Alpha, Delta and Omicron variants of concern (VoCs) sequentially sweeping through the world to reach high global prevalence. Neither Alpha nor Delta was characterized by strong immune escape, with their success coming mainly from their higher transmissibility. Omicron is far more prone to immune evasion and spread primarily due to its increased ability to (re-)infect hosts with prior immunity. As host immunity reaches high levels globally through vaccination and prior infection, the epidemic is expected to transition from a pandemic regime to an endemic one where seasonality and waning host immunization are anticipated to become the primary forces shaping future SARS-CoV-2 lineage dynamics. In this review, we consider a body of evidence on the origins, host tropism, epidemiology, genomic and immunogenetic evolution of SARS-CoV-2 including an assessment of other coronaviruses infecting humans. Considering what is known so far, we conclude by delineating scenarios for the future dynamic of SARS-CoV-2, ranging from the good—circulation of a fifth endemic ‘common cold’ coronavirus of potentially low virulence, the bad—a situation roughly comparable with seasonal flu, and the ugly—extensive diversification into serotypes with long-term high-level endemicity.
Keywords: coronaviruses, phylogenetics, host range, specific immunity, epidemic dynamic, endemicity
INTRODUCTION
On New Year’s Eve of 2019, a cluster of cases of pneumonia was reported in Wuhan, China. The causative agent was identified as a novel coronavirus and the first genome sequence was made available to the scientific community by mid-January 2020 [1]. Soon thereafter, an additional five complete genomes collected from patients infected during the early stages of the outbreak were reported. Sequences were near-identical, a result in line with a recent host jump into humans from a single source [2]. The newly identified coronavirus, initially called 2019-nCoV, was renamed SARS-CoV-2 due to its relatedness to SARS-CoV-1, the agent of the 2003 SARS outbreak. These early efforts were followed by a remarkable and unprecedented undertaking by the international scientific community to sequence large numbers of SARS-CoV-2 genomes. Over 500 genomes from 39 countries and 9 continents were available by the time the WHO declared the COVID-19 pandemic on 11 March 2020, and the milestone of 1 million SARS-CoV-2 genomes was reached a year later in March 2021 [3]. The international sequencing effort intensified as the pandemic progressed and by March 2022 over 10 million SARS-CoV-2 genomes had been deposited on the GISAID database [4, 5].
Phylogenetic analyses point to a Most Recent Common Ancestor (MRCA) of all sequenced SARS-CoV-2 dating back to late 2019 [6, 7]. When the WHO announced that the outbreak of pneumonia in Wuhan was caused by a novel coronavirus on 9 January 2020, 59 cases had been reported in Wuhan, China. While some of the earliest cases have epidemiological links to the Huanan Seafood Wholesale Market [8], the lack of direct isolates from animals at the market [9] leaves the proximal origin and early stages of the COVID-19 pandemic timeline largely unclear. The first confirmed cases outside China were identified in Thailand and Japan on 20 January, and a day later, a Washington state resident who had returned from Wuhan on 15 January tested positive [10]. The first confirmed cases in Europe were reported in France on 24 January 2020 [11]. Athough it is suspected SARS-CoV-2 was present in Europe earlier, in particular with some evidence the virus may have been circulating in Northern Italy already in December 2019 [12].
Genome sequencing can shed only limited light on the early timeline of the pandemic due to the small number of genomes available from that period. Athough from March 2020 onwards, the global spread and genetic evolution of SARS-CoV-2 can be exquisitely documented. Over the first half of 2020, the global population of SARS-CoV-2 remained largely unstructured geographically, with the entire viral genetic diversity distributed in most regions of the world [6, 13], with the notable exception of China, which rapidly contained the initial outbreak. As different countries implemented travel bans and other mitigations measures, the global SARS-CoV-2 population became more geographically structured with different viral lineages attaining high prevalence locally from mid-to-late 2020 onwards [14, 15].
From the start of the pandemic, multiple lineages emerged while others went extinct, thereby keeping the genetic diversity of the SARS-CoV-2 population in circulation at a largely constant level during the first 2 years. Until the near-simultaneous characterization of four variants of concern (VoCs) towards the end of 2020 and the Omicron VoC in late 2021, this lineage-replacement dynamic remained mostly unnoticed with the exception of the emergence and global spread of the B.1 lineage carrying the D614G mutation that swept the world during the early stages of the pandemic [16, 17]. The more transmissible Alpha VoC, which emerged in the UK, spread globally [18, 19]. However, before it reached worldwide prevalence, it was replaced by the even more transmissible Delta VoC and daughter lineages, first identified in India, in 2021 [20, 21]. Delta was subsequently displaced by the Omicron VoC, identified in late November 2021 in South Africa [22], and which rapidly established global prevalence. Other designated VoCs of the pandemic include Beta, first detected in South Africa [23], and Gamma, first identified in cases linked to Brazil [24]. The emergence of VoCs represented a sharp turn in the evolutionary picture of SARS-CoV-2 and prompted the need for the early definition (variants under investigation/monitoring), detection and monitoring of lineages that pose an increased risk to global health.
With the pandemic in its third year, it is becoming increasingly urgent to understand the long-term evolutionary dynamics of SARS-CoV-2. Here, we first overview key aspects of SARS-CoV-2 genetic and immunogenetic evolution and draw parallels with what is known for other coronaviruses infecting humans. We conclude the review by delineating possible scenarios for the future.
AN OVERVIEW OF CORONAVIRUSES
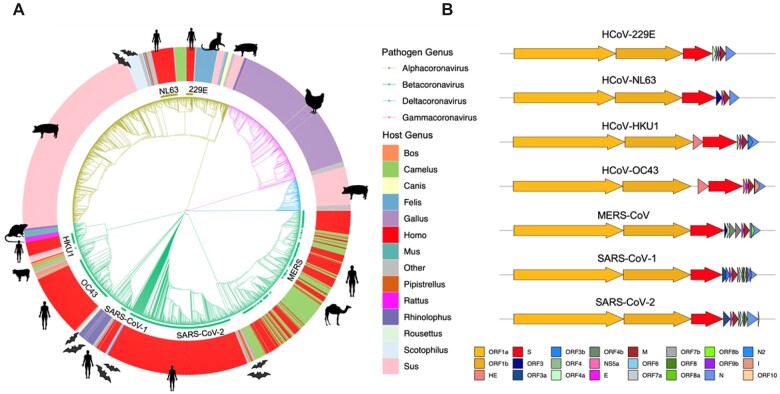
Coronaviruses belong to the viral family Coronaviridae, which comprises three subfamilies: Orthocoronavirinae, Pitovirinae and Letovirinae. The Orthocoronavirinae subfamily comprises the four genera: Alpha-, Beta-, Gamma- and Deltacoronavirus [25]. SARS-CoV-2 falls within the Sarbecovirus subgenus in the genus Betacoronavirus, together with SARS-CoV-1, the agent of the SARS 2002–04 epidemic, and related lineages circulating in populations of horseshoe bats (Rhinolophus sp.). Betacoronavirus also include MERS-CoV and two seasonal human endemic coronaviruses: HCoV-OC43 and HCoV-HKU1 (Fig. 1A). Lineages within Gammacoronavirus are restricted to birds but Alpha- and Deltacoronaviruses infect mammals [26]. Deltacoronaviruses have been primarily isolated from domestic pigs, but Alphacoronaviruses infect a broad range of mammals, including humans, with the genus comprising the two other seasonal human endemic coronaviruses HCoV-229E and HCoV-NL63 (Fig. 1A).
Figure 1:
Overview of the Orthocoronavirinae. Notes: (A) Phylogenetic representation of 2529 coronavirus genomes. The branch colour indicates the assignment into the four genera (Alpha-, Beta-, Gamma- and Deltacoronavirus). The coloured outer circle and icons provide the host the strains were isolated from. Each of the seven human-associated coronaviruses (HCoVs) are highlighted on the inner ring. (B) Genome structure of major human associated coronaviruses. Gene annotations are provided for the coding RNA sequences of seven representative human-associated coronaviruses (HCoV-229E NC_002645.1; HCoV-NL63 JX504050.1; HCoV-HKU1 NC_006577.2; HCoV-OC43 KX344031.1; MERS-CoV NC_019843.3; SARS-CoV-1 NC_004718.3; and SARS-CoV-2 NC_045512.2). Gene annotations are provided as per the colour legend at bottom, aligned to the coronavirus spike protein. Figure made using the R package gggenes (https://cran.r-project.org/web/packages/gggenes).
Coronaviruses are enveloped viruses with a single-strand, positive-sense RNA genome which, at ∼26–32 kilobases, is among the largest known genomes for RNA viruses (Fig. 1B). The basic organization of the genome is conserved across members of the Orthocoronavirinae. The bulk of the genome (around two-thirds) extending from the 5′-end comprises the replicase open-reading frame (ORF) 1a/b which encode two polyproteins which are cleaved into a suite of non-structural proteins (NSPs; ORF1a: NSP1-11, ORF1b: NSP12-16) involved in proteolytic processing, transcription and genome replication [27, 28]. These include the NSP12 RNA-dependent RNA polymerase that is essential for RNA synthesis and associated exoribonuclease NSP14 that exhibits proof-reading activity [29]. The remaining third of the genome encodes the structural proteins in 5′- to 3′-order; the surface or spike glycoprotein (S), the membrane glycoprotein (M), the envelope (E) and the nucleocapsid (N).
Of all ORFs identified in the Orthocoronavirinae, only ORF1ab, S, M and N can be considered homologues in >99% of sequenced coronavirus genomes (i.e. ‘core’ genes). Additionally, only ∼43% of the average coronavirus genome can be meaningfully aligned at the nucleotide level [30]. Some Betacoronaviruses, most notably HCoV-HKU1 and HCoV-OC43, harbour an additional hemagglutinin-esterase (HE) glycoprotein which can act as a secondary viral attachment protein [31]. Additionally, coronavirus genomes typically harbour a variable complement of accessory proteins depending on the genus, or even the individual species [32] (Fig. 1B).
Viral characterization has particularly focused on the spike glycoprotein, whose club-shaped surface projections from the lipid bilayer envelope give coronavirus virions their appearance [33]—reminiscent of the sun’s corona or halo during an eclipse [34]. The spike protein plays a key role in viral entry in the host cell, mediating host receptor recognition, cell attachment and membrane fusion [35, 36]. Notably, the host receptors recognized and used by the spike for viral entry vary between different coronavirus species. SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2), as does SARS-CoV-1 and HCoV-NL63, whereas other species within the same genus (including MERS-CoV) recognize a cell-surface serine peptidase, dipeptidyl peptidase 4 (DDP4), and others bind with 9-O-acetylated sialic acid (HCoV-OC43 and HCoV-HKU1) [32, 37–39]. Finally, HCoV-229E uses the human aminopeptidase N (hAPN) receptor [40]. All four host receptors are expressed in a wide range of human cell types, including those in the respiratory and gastrointestinal tracts [29–31], which may explain the broad tissue tropism and hence the diverse symptoms and transmission routes of coronavirus infections in humans [35].
The spike protein comprises two subunits, known as S1 and S2, with the former including important regions such as the N-terminal domain (NTD) and receptor binding domain (RBD) that directly binds host receptors during viral entry [38, 41]. The S2 domain additionally supports the fusion and eventual entry of the virus into host cells. Cell entry by SARS-CoV-2 is dependent on the activation of the spike protein by proteases. TMPRSS2 and cathepsin L are the two main proteases activating the spike protein [42]. TMPRSS2 is co-expressed together with ACE2 on the surface of most cells of the gastrointestinal, respiratory and urogenital epithelium [43]. If a virus–ACE2 complex fails to bind TMPRSS2, it can still enter cells through endocytosis and S2 is then activated by cathepsins [42]. Given its critical role in receptor binding, membrane fusion and immunogenicity, the spike protein is subject to natural selection for enhanced infectivity and antigenicity [44]. The S1 and S2 regions as well as the S1/S2 junction are the only targets of neutralizing antibodies (antibodies precluding pathogen entry into host cells). Due to this property, the spike protein was selected as the sole source of antigens in the majority of COVID-19 vaccines developed so far, including those based on mRNA (Pfizer/BioNTech/Moderna) and adenovirus recombinant technologies (Oxford/AstraZeneca/Sputnik V/Johnson and Johnson).
Coronavirus accessory proteins are more poorly characterized. While these proteins are generally not considered essential for viral replication [45], they play a role in pathogenesis, host interactions and transmissibility [46, 47]. ORF3a, the largest accessory protein in SARS-CoV-2, has been shown to interact strongly with the host immune system, activating the NLRP3 inflammasome and contributing to the generation of a cytokine storm [48]. Its functional role in cell apoptosis has been documented for both SARS-CoV-1 [49] and SARS-CoV-2 [50]. Consistently, in a SARS-CoV-1 animal model, deletion of ORF3a was shown to reduce viral replication [51]. The accessory proteins ORF6, ORF7a and ORF8 act as interferon antagonists in SARS-CoV-2 [46]. Of these, SARS-CoV-2 ORF8 is particularly interesting in that its presence as an intact gene (rather than fragmented into ORF8a/b) is unique among human-associated coronaviruses, with ORF8 being identified as highly immunogenic among the accessory genes [52]. A large deletion in ORF8 (Δ382) in SARS-CoV-2 circulating in a cohort of patients in Singapore in early 2020 was identified as being potentially associated with milder disease [53], further reinforcing its possible role in pathogenesis. While coronavirus accessory proteins have been studied most closely in SARS-CoV-2, many uncertainties remain about the exact role of different ORFs. For instance, to date no function has been associated to ORF10 and it has been questioned whether it should be considered a gene [54].
THE EVOLVING HOST RANGE OF SARS-CoV-2
Despite remarkable global genome sequencing and characterization efforts, the proximal phylogenetic origin of SARS-CoV-2 and its mode of introduction into human circulation remain unclear. There is no evidence for SARS-CoV-2 having been ‘engineered’ in a lab. Conversely, the escape of a strain from a lab or an accidental contamination during field work cannot formally be ruled out at this stage. However, a zoonotic spillover event in nature is considered as the most plausible scenario in the scientific community [2, 55–58]. A diversity of viruses closely related to SARS-CoV-2 have been isolated from multiple species of horseshoe bats (Rhinolophus sp.) sampled across East and Southeast Asia [2, 56–61]. Thus far, RaTG13 isolated from Rhinolophusaffinis in Yunnan in 2013 shares the highest whole-genome sequence identity with SARS-CoV-2 at 96.2% [2], followed by RpYN06 from Rhinolophuspusillus at 94.5% [57]. However, identity along the genome is highly variable. For example, phylogenetic analysis identified RmYN02 (Rhinolophusmalayanus; 93.3% identity) as the closest relative over the ORF1ab region (97.2%) [58]. The lower whole-genome sequence identity relative to that for RaTG13 is largely due to the sequences in the spike (71.9% sequence identity) [58]. However, at the region critical for host receptor binding—the RBD—RaTG13 has a low genetic similarity to SARS-CoV-2.
Other bat coronaviruses, most notably those recently recovered from R. malayanus, R.marshalli and R. pusillus in Northern Laos, Indonesia, harbour RBD motifs far closer to SARS-CoV-2 and have been demonstrated to efficiently bind to human ACE2 [59]. The genetic similarity between RaTG13 and SARS-CoV-2 is largely comparable to that of the viral lineages most closely related to SARS-CoV-1 found in horseshoe bats (Rhinolophus spp.) [62, 63]. As such, it may be argued that the progenitor of SARS-CoV-1 has never been identified. Although in contrast to the situation for SARS-CoV-2, viral strains with near-perfect whole-genome sequence identity have been isolated from captive Himalayan palm civets (Paguma larvata) and a raccoon dog (Nyctereutes procyonoides) [64]. These observations led to the hypothesis that a carnivore may have acted as an intermediate host in the jump of SARS-CoV-1 into humans [65]. Early in the COVID-19 pandemic, there were suggestions that pangolins (Manis javanica) could have acted as an intermediate host for SARS-CoV-2 [66]. However, current evidence does not indicate that an established reservoir outside of bats may have been required for the host jump into humans [67] (Fig. 2).
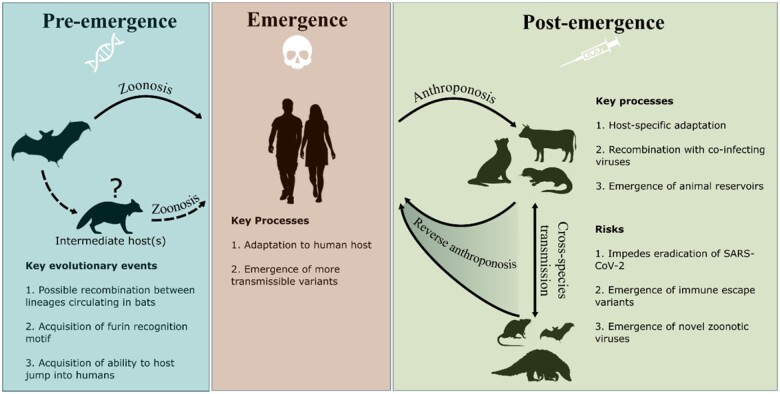
Figure 2:
SARS-CoV-2 host range evolution before, during and after the COVID-19 pandemic. Notes: The box on the left summarizes possible scenarios predating the host jump into humans, followed by key processes during the establishment of human-to-human transmission (central box). The box at the right displays the processes and risks associated with secondary spillovers from humans to wild and domestic animals. Icon choice is emblematic, sourced from flaticon.com.
Studies of the proximal origins of SARS-CoV-2 have focused on the Arginine-Arginine-Alanine-Arginine (RRAR) furin recognition motif at the S1/S2 spike subunits junction. Pre-activation of the SARS-CoV-2 spike by host proprotein convertase furin at this motif is crucial for cellular entry [68], viral replication and pathogenesis [69]. The presence of the RRAR motif in SARS-CoV-2 and its absence in close relatives has been suggested as a sign of bioengineering. However, while close relatives such as RmYN02 lack the exact furin recognition motif, they share a genomic region homologous to that in SARS-CoV-2 (positions 23603–23615) within the spike protein [70]. This observation suggests that recombination with a yet unsampled coronavirus may have played a role in the emergence of the furin recognition motif in SARS-CoV-2. Further, furin recognition motifs (RXXR) have emerged independently multiple times in the evolution of Betacoronaviruses [71], with MERS-CoV, HCoV-OC43 and HCoV-HKU1 also carrying furin cleavage sites. Hence, the natural emergence of the RRAR motif in SARS-CoV-2 through recombination and/or other evolutionary processes (point mutations and indels) represents a parsimonious explanation (Fig. 2).
As with other coronaviruses, SARS-CoV-2 can infect and transmit efficiently within different populations of mammals (Fig. 1A). This is evidenced by the plethora of studies that have probed the host tropism of SARS-CoV-2, in vitro (cell lines), in vivo (live inoculation) and through wildlife surveillance (Table 1). Human-to-animal spillover (i.e. anthroponosis) of SARS-CoV-2 into multiple wild, captive and domestic mammalian species has been observed repeatedly and is particularly well-documented in zoo animals [72], farmed mink [73, 74] and wild white-tailed deer [75, 76] (Table 1). Notably, evidence for natural or experimental infection may not necessarily entail efficient animal–animal transmission. For example, porcine cell lines are permissive to infection [77, 78] but studies have failed to experimentally infect pigs in vivo [78, 79]. Further, while animal hosts such as dogs and cattle have been shown to be susceptible to infection, transmission is poor or non-existent [80, 81], indicating that SARS-CoV-2 is not yet adapted for efficient transmission in these species. The broad host tropism of SARS-CoV-2 may be in part due to the usage of the ACE2 receptors for viral entry. ACE2 is fairly conserved across vertebrates [82], which entails that the accumulation of only few mutations may be required to evolve efficient binding to receptors in a novel host species. To exploit this, several studies have suggested that bioinformatic screening of key residues on animal ACE2 that govern binding affinity may be useful in assessing potential animal reservoirs [82–85].
Table 1:
Reports of susceptibility of different animal hosts to SARS-CoV-2
| Host | Species | In vitro susceptibility | In vivo susceptibility | Transmission | Detected in wild | Anthroponosis | Reverse anthroponosis |
|---|---|---|---|---|---|---|---|
| White-tailed deer | Odocoileus virginianus | Yes | Yes [86, 87] | Yes [86, 87] | Yes [75] | Yes [76, 88] | Probable [89] |
| American mink/ European mink | Neogale vison/ Mustela lutreola | Yes | Yes | Yes [73, 74] | Yes [90] | Yes [74] | Yes [74] |
| Other mustelids |
Mustela putorius furo
Martes martes Meles meles |
Yes | Yes [91] | Yes [79, 91–93] | Yes [94] | Yes [94] | |
| Domestic cats | Felis catus | Yes [77] | Yes [95, 96] | Yes [95, 97] | Yes [98, 99] | ||
| Dogs | Canis lupus familiaris | Yes | Yes [95, 96] | No [95] | Yes [81, 99] | ||
| Big cats |
Panthera spp. Puma concolor |
Yesa | Yes [72] | Yes [72] | |||
| Spotted hyenas | Crocuta crocuta | Yesa | Yesa | Yesa | |||
| Bearcat | Arctictis binturong | Yesa | Yesa | Yesa | |||
| Fishing cat | Prionailurus viverrinus | Yesa | Yesa | Yesa | |||
| Canada Lynx | Lynx canadensis | Yesa | Yesa | Yesa | |||
| Coati | Nasua nasua | Yesa | Yesa | Yesa | |||
| Cattle | Bos Taurus | Yes | Yes [80] | No [80] | |||
| Pigs | Sus scrofa | Yes [77, 78] | No [78, 79] | ||||
| Chickens | Gallus gallus | No [79] | |||||
| Fruit bats | Rousettus aegyptiacus | Yes | Yes [79] | ||||
| Horseshoe bats | Rhinolophus sinicus | Yes [77, 100, 101] | |||||
| Vesper bats | Pipistrellus abramus | Low [101] | |||||
| Rats/mice |
Rattus norvegicus
Mus musculus |
Variants only [102] | Variants only [102] | No [102] | |||
| Hamsters | Mesocricetus auratus | Yes | Yes [103] | Yes [104] | Yes [104] | ||
| Non-human primates |
Macaca mulatta
Macaca fascicularis Chlorocebus aethiops |
Yes | Yes [77, 103] |
https://www.aphis.usda.gov/aphis/dashboards/tableau/sars-dashboard; green: confirmed positive; red: confirmed negative; orange: unconfirmed; yellow: some variants only.
The host range of SARS-CoV-2 is likely dynamic and the virus may further expand its host repertoire in the future. For example, mice and rats were not susceptible to the first strains of SARS-CoV-2 circulating in humans [102]. However, following the global spread of the Alpha VoC, which carries the N501Y mutation facilitating infection in those species, they have become susceptible hosts [105]. Given the broad host tropism of SARS-CoV-2, there is a risk that viral mutations may emerge while circulating in novel animal host species following anthroponotic spillover. This entails a further risk that the transmission of animal-adapted SARS-CoV-2 back into humans (i.e. reverse anthroponosis) may alter the evolutionary trajectory of the virus, potentially leading to the emergence of more transmissible or immune-escape variants (Fig. 2). There is evidence for the emergence of host-adaptive mutations in SARS-CoV-2 strains circulating in farmed minks and wild white-tailed deer that have arisen recurrently and have reached a high frequency [106, 107]. For example, spike mutations, Y453F and F486L, are good candidates for adaptation in mink in addition to dampening human humoral and T cell-mediated immunity [108–111]. Similarly, following introduction in a research colony of cats [95], spike mutation H655Y rapidly reached fixation; a substitution that has been associated with immune escape in humans [112]. Reverse-anthroponotic transmission of SARS-CoV-2 has been documented for farmed minks [74, 106], white-tailed deer [89] and pet-shop hamsters [104] and it is to be expected that SARS-CoV-2 can also jump from other animal species back into humans. Putatively, mink-adaptive mutations are currently only maintained at a low frequency in human-associated SARS-CoV-2, suggesting that they do not confer a significant advantage in human circulation [106, 111]. As yet, the circulation of SARS-CoV-2 in mink and deer has not resulted in a significant alteration in extant genomic diversity [106].
The emergence of reservoirs in wildlife is of further concern since it is more difficult to assess and control anthroponotic or reverse-anthroponotic movement of SARS-CoV-2 between humans and wild animal populations than domestic species. Such reservoirs may emerge due to direct anthroponotic transmission from humans living in close proximity to wild animals, or indirect transmission via domesticated animals or contaminated water sources. SARS-CoV-2 has already established itself in wild white-tailed deer populations with a detected seroprevalence of 7–67% across different states in the USA [75, 76]. Further, there have been reports of deer-associated viral sequences that are considerably divergent from the human-associated sequences sampled within the same geographic region and time period [89, 113]. Given the broad host tropism of SARS-CoV-2, the emergence of other wild or farmed animal reservoirs remains a possibility. This may be particularly problematic in animals that already harbour a vast diversity of coronaviruses such as bats [56, 114, 115], since the propensity for recombination in coronaviruses [116] might lead to the emergence of novel recombinant viruses with the ability to transmit efficiently in humans (Fig. 2).
SARS-CoV-2 IN THE CONTEXT OF OTHER HUMAN CORONAVIRUSES
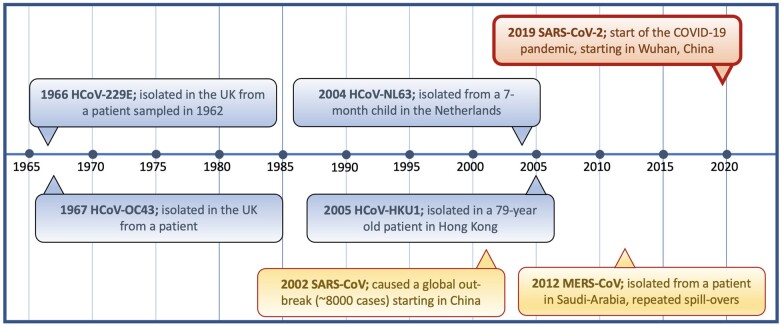
Prior to the emergence of SARS-CoV-2, six other coronaviruses were known to commonly infect humans (Fig. 1). HCoVs were first observed in the 1960s in samples at the Common Cold Unit in Salisbury, England [117] (Fig. 3). However, they were not considered highly pathogenic until the outbreak of SARS-CoV-1 in 2002 [118]. SARS-CoV-1 affected at least 8000 individuals causing severe respiratory disease with a case fatality rate of ∼10% [119]. Approximately 10 years later, MERS-CoV was identified in Saudi Arabia, thought to have also originated in bats [120] (Fig. 3). MERS-CoV also causes severe-to-fatal respiratory disease in humans and since its discovery over 2500 MERS-CoV cases have been reported with >850 associated deaths [121]. MERS-CoV is not well adapted to transmission in humans and is believed to spread through zoonotic reservoirs (mainly dromedary camels) with frequent ongoing spillovers into susceptible human populations [122]. MERS-CoV has been circulating in dromedary camels for decades as evidenced by the presence of MERS neutralizing antibodies in samples from East Africa dating back to 1983 [123].
Figure 3:
Timeline of the age of first observation for the seven HCoVs. Notes: Endemic coronaviruses are shown in blue, epidemic coronaviruses in yellow and pandemic SARS-CoV-2 is represented in red. Additionally, in the 1960s, several strains including B814, 692 and HCoV-OC16, -OC37 and -OC48 were observed, but lost for follow-up studies. It is unclear whether these strains may have been later identified or represent yet uncharacterized HCoVs. The figure is informed by the following sources [2, 117, 118, 124–126].
The four endemic respiratory seasonal coronaviruses (HCoV-229E, -OC43, -NL63 and -HKU1) circulate in all age groups with the first exposure generally happening in childhood. HCoVs tend to cause mild self-limiting upper respiratory or gastrointestinal disease in the immunocompetent accounting for ∼10–30% of common cold cases [127–129]. HCoVs tend to induce only relatively short-term protection against reinfection, which occurs repeatedly throughout life [130, 131]. This is likely due to fairly rapid waning of post-infection antibody titres to coronaviruses but there is also evidence that HCoV-OC43 and -229E undergo constant adaptive evolution to escape host immune recognition [132]. Both HCoVs tend to constantly acquire mutations in the S1 region of the spike protein and their genetic diversity over three decades falls into ladder-like phylogenetic trees compatible with antigenic drift, similar to those observed for seasonal flu (influenza A) [132]. Moreover, it has been demonstrated that historic sera able to potently neutralize virions pseudotyped with contemporary HCoV-229E spike proteins had little to no activity against spike proteins from HCoV-229E strains isolated 8–17 years later [133]. In the same study, modern adult sera were found to neutralize spike proteins from a variety of historical viruses, whereas modern sera from children best neutralized spike proteins from recent viruses that would have circulated in their lifetime [133]. These patterns provide evidence for antigenic evolution of the CoV spike protein in HCoV-229E, particularly in the spike RBD permitting the escape of neutralization by polyclonal sera within one or two decades [133]. No consistent evidence for antigenic drift has been found to date for HCoV-NL63 and -HKU1, but this may simply reflect the lack of sufficient longitudinal sequencing data to identify adaptive evolution [134].
Estimates for the age of emergence of the four endemic HCoVs remain under debate, though it is widely accepted that they were in circulation a good deal prior to first observations. For example, while HCoV-NL63 was first identified in 2004 (Fig. 3), the earliest available genome was later generated from a sample collected in 1983 [135] and the inferred MRCA of HCoV-NL63 strains currently in circulation has been estimated to date back to the ∼1920s [136]. HCoV-229E and -NL63 likely evolved in bats [137, 138], with camelids having possibly acted as an intermediatory for HCoV-229E transmission to humans [139]; a scenario similar to that of MERS-CoV. Both HCoV-OC43 and -HKU1 are thought to have originated in rodents and there is debate over whether cattle, pigs or other animals may have acted as an intermediate host for HCoV-OC43, with bovine coronavirus suggested as a possible ancestor [140]. It has also been hypothesized that the Russian flu pandemic (1889–1890) may have been caused by HCoV-OC43 [141, 142]. This hypothesis is primarily based on the list of respiratory, gastrointestinal and neurological symptoms including loss of taste and smell recorded at the time, which are more reminiscent of COVID-19 than influenza infections. Early phylogenetic analysis of the HCoV-OC43 spike protein sequence pointed to a time to the MRCA (tMRCA) compatible with a HCoV-OC43 host jump into humans in the late 19th century [140]. However, this estimated date does not seem supported by analyses of additional genomes, which point to a more recent common ancestor for HCoV-OC43 currently circulating in humans [134, 143].
Additional coronavirus spillovers into humans have been documented for species outside of the seven generally categorized as ‘human coronaviruses’. There is evidence of multiple separate cases of cattle strains being transmitted to humans. For instance, two samples of human enteric coronavirus 4408 strains have been documented, collected in 1988 and 2009 [135]. Further, there are clinical, epidemiological and serological observations that bovine coronaviruses can infect human subjects causing diarrhoea [144] and a recent study indicated that bovine coronavirus can be carried in human nasal mucosa after exposure to virus-shedding calves [145]. Coronavirus RNA identified as a novel canine–feline recombinant alphacoronavirus (since named CCoV-HuPn-2018) has been detected in nasopharyngeal swab samples from eight patients during 2017–18 in Sarawak, Malaysia [146]. Additionally, isolates with high identity to feline enteric coronavirus (Alphacoronavirus) were detected in three nasal swab samples collected in Arkansas in 2010, from patients exhibiting acute respiratory symptoms. One of these samples contained a 400-base pair fragment in the S region, which revealed an OC43-like spike protein, suggestive of feline coronavirus and HCoV-OC43 coinfection [147]. Porcine deltacoronavirus strains were also recently identified in blood plasma samples from three children in Haiti with acute undifferentiated febrile illness [148]. Prior to this, documented human coronavirus infections had been limited to Alpha- and Betacoronavirus strains; therefore, this example provides the first known case of Deltacoronavirus adaptation to human transmission [148].
These recently documented spillover events point to a wider diversity of coronaviruses circulating in wild and domestic animal populations being able to infect humans than previously recognized. Together with the emergence of three pandemic/epidemic human coronaviruses in the last two decades (SARS-CoV-1, SARS-CoV-2 and MERS-CoV), this observation illustrates the relative ease with which coronaviruses can jump into humans and sometimes subsequently adapt and spread. Coronaviruses infecting humans cover the entire epidemiological spectrum, ranging from rare to common spillover events, outbreaks, epidemics, a pandemic to a global endemic quasi-equilibrium state achieved by the four seasonal HCoVs.
GENETIC EVOLUTION OF SARS-CoV-2
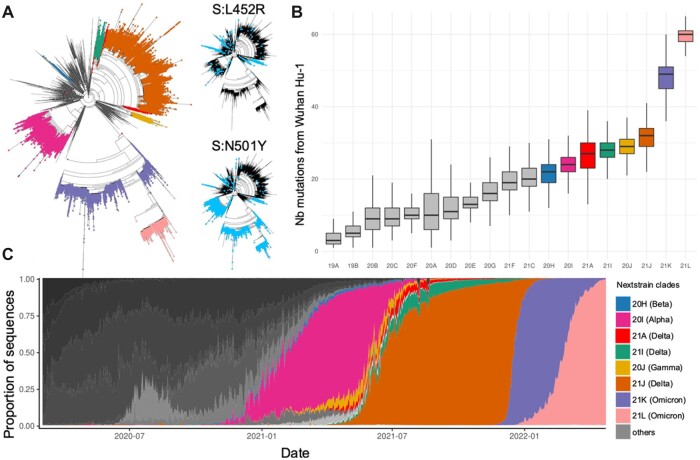
The genetic diversity of SARS-CoV-2 was initially very low and mutations accumulated steadily over the first 2 years of the pandemic at a rate of approximately 25–30 mutations per lineage, per year (Fig. 4A) [3]. Interestingly the same mutations can often be observed at a low frequency in many different lineages (Fig. 4A insets). As mutations accumulated over time, this led to more recently emerging lineages being a larger number of mutations away from the hypothetical ancestor of all SARS-CoV-2 strains circulating in humans (Fig. 4B). Even before the emergence of the Alpha, Delta and Omicron VoCs that successively swept the world, the dynamic of the SARS-CoV-2 population was characterized by a birth and death dynamic of lineages replacing each other (Fig. 4C). Interestingly, despite all three variants having achieved global prevalence, they do not derive from each other (Fig. 4A). Both the Delta and then the Alpha variant are descendants of early pandemic lineages that had been displaced globally first by Alpha and then Delta. It remains to be seen whether this pattern will persist, or whether successful variants of the future will derive from each other as is typical for seasonal flu or HCoVs.
Figure 4:
Lineage dynamic of the SARS-CoV-2 population. Notes: (A) Phylogenetic tree of ∼200,000 SARS-CoV-2 strains coloured by their Nextstrain clade assignment with the inset providing the presence of the Spike L452R and N501Y mutations highlighting the recurrence of independent emergence of mutations in multiple lineages. (B) Distributions of distances (in number of mutations) from the root of the tree (Wuhan-Hu-1) of Nextstrain clades (ordered by emergence). (C) Daily prevalence of each NextStrain clade estimated as the proportion of uploads to the GISAID genome database highlighting the dynamic of lineage replacement in SARS-CoV-2. Only Nextrain clades corresponding to VoC lineages were attributed a color, others were represented in grey.
Direct competition between lineages can only occur when two lineages simultaneously infect the same host leading to only one lineage being successfully transmitted. Even when SARS-CoV-2 is at high prevalence, such mixed infections are expected to be rare. Thus, the dynamic of lineage replacement is likely primarily fuelled by the emergence of new strains that are more transmissible and/or more able to (re-)infect hosts immunized through vaccination or prior infection. Additionally, fitness decay of lineages in circulation through the accumulation of slightly deleterious mutations (Muller’s ratchet [149]) may also play a role in lineage extinction [150, 151].
The mutation rate of SARS-CoV-2 is fairly unremarkable among RNA viruses [152–154] despite the proof-reading activity of its replication mechanism [155]. While genetic diversity of the SARS-CoV-2 population in circulation remains low 2 years into the pandemic (Fig. 4A and B), mutations can at this stage be found at every single base of the SARS-CoV-2 genome [4, 5]. The majority of these mutations have independently emerged many times in unrelated lineages [6, 15] (Fig. 4A). The bulk of the segregating diversity in SARS-CoV-2 is expected to be adaptively neutral or slightly deleterious. However, early selective sweeps of mutations essential for transmissibility in humans may have been missed [156]. Indeed, the earliest available SARS-CoV-2 genomes were sampled in late December 2019, by which time the virus had probably been in human circulation for several weeks [6].
Host antiviral defence mechanisms inducing mutations at specific sites of the viral genome [15, 157, 158] represent an additional selective force shaping the genetic diversity segregating in the SARS-CoV-2 population. Host-mediated editing of SARS-CoV-2 includes the activity of proteins form the APOlipoprotein B Editing Complex (APOBEC) and the Reactive Oxygen Species and Adenosine Deaminase Acting on RNA (ADAR), all suspected to introduce mutations in a nucleotide and sequence-context specific manner. For instance, the mutation spectrum of SARS-CoV-2 is largely dominated by C-to-U and G-to-U substitutions [15, 159]. These processes, coupled with ongoing selective reduction of CpGs [157, 159], create complex mutational dynamics [159, 160]. Host antiviral defence likely provides a potent selective pressure following spillover to a new host. As such, with the establishment of widespread human-to-human transmission, the rate of evolution in SARS-CoV-2 might slow down, with increasing fixation of advantageous mutations and depletion of sites susceptible to host-induced mutagenesis.
The accumulation of diversity in SARS-CoV-2 motivated the development of classification schemes to designate distinct lineages (Fig. 4A and C). Among the two most widely used SARS-CoV-2 classification schemes are the Nextstrain clade naming system, which provides an up-to-date nomenclature based on phylogeny, VoC status and global and regional frequency [161] and the PANGO nomenclature scheme which relies on the phylogeny and carriage of diagnostic mutations [162]. The latter classification includes many more sub-divisions (numbered labels). While such schemes are essential tools in scientific discourse, any taxonomic delineation into lineages is bound to remain somewhat arbitrary. Additionally, irrespective of the accuracy of the cut-off points separating lineages, any genetic-based taxonomy may struggle with the noise generated by recurrent and backwards mutations [6, 15], as well as lineage extinction, both prevailing properties of SARS-CoV-2 evolution (Fig. 4A and C).
SARS-CoV-2 GENOTYPE AND PHENOTYPE
While most mutations are expected to have no functional impact [163], some sites in the SARS-CoV-2 genome have been associated with changes in viral phenotype. Unsupervised assessment of the phenotypic effect of mutations has focused on characterizing proximal phenotypes such as binding affinity to the ACE2 host cell receptor or spike protein expression revealing a heterogenous phenotypic landscape over the spike protein [164]. Such an approach pinpointed spike mutation N501Y, characteristic of three of the five currently identified VoCs, as having a notable effect on receptor binding [164, 165]. Functional evidence for association of individual mutations with traits such as receptor binding affinity or gene expression provides useful mechanistic insight. However, more complex traits, such as virulence, transmissibility and immune escape, are more relevant epidemiologically. The genetic basis of complex traits is more challenging to infer mainly because they are affected by multiple mutations that do not necessarily act independently of each other. As such, a specific mutation may not always lead to the same phenotypic changes in different SARS-CoV-2 genetic backgrounds.
There is indeed evidence for extensive epistasis being involved in the evolution of SARS-CoV-2, with the fitness of viral lineages likely dependent on combinations of co-adapted mutations and deletions [166, 167]. More transmissible SARS-CoV-2 lineages did not primarily evolve through the sequential accumulation of mutations [15]. Instead, some VoCs seem to have emerged through the rapid acquisition of a set of mutations and deletions rarely observed together previously. For Alpha and Omicron in particular, it is suspected that new combinations of mutations arose during long-term infection in immunocompromised hosts [168, 169]. Rapid accumulation of rare mutations has been observed in chronic infections [170] and persistent infection in immunocompromised hosts may provide the virus with a possibility to explore a wider range of the fitness landscape. Favourable combinations of mutations that are deleterious in isolation may be unlikely to be acquired sequentially during transmission among immunocompetent hosts, as those less fit fitness lineages may rapidly go extinct. Persistent infections in immunocompromised hosts may allow such transient low fitness lineages to persist, with some eventually acquiring novel combinations of well co-adapted mutations. The evidence for the emergence in an immunocompromised host is particularly strong for the Omicron VoC, which acquired a large number of non-synonymous mutations in the immunodominant regions of the spike RBD region [22]. Additionally, Omicron-associated mutations confer significant escape from neutralizing antibodies but not T cell immunity [171, 172]. The unusual mutation pattern of Omicron might have arisen in an untreated HIV/AIDS patient with advanced CD4+ T-cell lymphopenia, who was unable to clear the infection [173]. Interestingly, Omicron comprises two deeply diverging lineages, BA.1 and BA.2 (nextstrain clades 21K and 21L). It is possible that both emerged in the same host before independently spreading in the community.
Recombination is also key for viral evolution as it allows for the combination, within a single genome, of mutations that arose independently in different genetic backgrounds. It is generally accepted that the emergence of all three human epidemic coronaviruses: SARS-CoV-1 [174–176], MERS-CoV [177] and SARS-CoV-2 [1, 178, 179] involved recombination events. Recombination also provides a mechanism for the generation of new antigenic combinations within species [180]. This can occur through the exchange of discrete segments of genetic material of different origins, well exemplified by antigenic shift in influenza [181, 182]. Recombination, however, requires co-infection of the same host by distinct lineages [183]. So far, relatively few recombinant SARS-CoV-2 genomes have been identified [184–188]. The relatively small number of recombinant SARS-CoV-2 genomes detected to date is intriguing given the large number of genomes available and epidemiological settings likely favouring co-infections by distinct lineages, for example, during mid 2021 (when prevalence was high and Alpha and Delta VoC were in co-circulation in multiple regions of the world). This may in part stem from difficulty in identifying recombinants because of low levels of SARS-CoV-2 genetic diversity in circulation at this stage. Low levels of genetic recombination in SARS-CoV-2 would however be in variance with the high recombination rates reported for other coronaviruses, including in human [116, 189–192] and animal-associated lineages [193–195], as inferred from genomic approaches [196], observed in cell culture [197, 198] and in vivo [199]. The low proportion of recombinant genomes detected to date might also in part be attributable to most of them having lower fitness, with recombinant lineages becoming extinct before they can be sampled.
Any replicating entity will be present in the future at a higher frequency if it makes more copies of itself [200]. Viruses are no exception to this fundamental rule of evolution. As such, through whatever means, viral lineages will evolve towards higher possible transmissibility. Efforts to computationally assess and predict transmissibility have focused on approaches based on logistic regression models of lineage growth rates [201] or phylogeny-based quantification of the association between carriage of specific mutations leading to an increase or decrease in the number of inferred descendants [150]. The latter approach capitalizes on the high rate of recurrent substitutions and deletions in SARS-CoV-2 [6, 15, 150, 202, 203] (Fig. 4A insets). Both those approaches to estimate transmissibility (viral fitness) highlight the propensity for a plethora of mutations, not only those within the critical spike RBD, to be implicated in increasing viral fitness [150, 201]. This is not entirely surprising since transmissibility reflects multiple underlying mechanisms such as the binding affinity to the human receptor ACE2 (i.e. fewer viral particles needed to generate a new infection) [164, 204], or viral replication rates [205, 206], or the ability to (re-)infect a larger fraction of the human population through immune escape.
Genetic determinants of virulence are difficult to assess not least because virulence is not a property of a pathogenic strain alone, but rather of an interaction with its host. Moreover, there are currently no large datasets available with the necessary information to associate individual viral mutations with disease severity and outcomes, while controlling for key host factors such as age, sex or health. It is sometimes assumed that as a pathogen adapts to its host, it becomes intrinsically less virulent [207, 208]. However, such a prediction is only true under two conditions. The first is when transmission is primarily vertical (e.g. from mother to child) as in human cytomegalovirus where fitness of the virus and the host are closely correlated, so that the pathogen would reduce its transmissibility by harming its host. The second is for extremely lethal pathogens where mortality of the host upon infection is so high that it precludes transmission of the pathogen. The textbook example for pathogen attenuation is myxomatosis in rabbit populations with initial mortalities of 99% upon initial introduction of the pathogen going down after a few years of coevolution [209]. Neither of those conditions are fulfilled by SARS-CoV-2. Despite its mortality and morbidity burden, SARS-CoV-2 remains a moderately lethal pathogen. Moreover, about half of transmissions are from pre- and asymptomatic carriers [210], which further reduce the selective pressure on the virus to spare its host.
The morbidity and mortality resulting from SARS-CoV-2 infection have however reduced significantly since the start of the pandemic and will do so further as an increasing proportion of the human population is immunized through natural infection and vaccination. However, the evolution of the intrinsic virulence of SARS-CoV-2 is difficult to predict. The Alpha and Delta variants were both associated with increased virulence, with a higher proportion of people infected requiring hospitalization [18, 211–214]. The higher virulence of the Alpha and Delta VoCs is likely a secondary (pleiotropic) consequence of their increased replication rate and transmissibility. Conversely, all major Omicron lineages in circulation at the time of writing (BA.1, BA.2, BA.4 and BA.5) are intrinsically less virulent than the Alpha and Delta VoCs [215] and their severity falls below that of early pandemic strains. One explanation for Omicron’s reduced pathogenicity has been its altered use of TMPRSS2. In particular, it has been suggested that the Omicron spike protein tends to use the alternative cathepsin-mediated activation rather than the canonical TMPRSS2 pathway [216–218]. This property simultaneously results in a shift in tissue tropism away from TMPRSS2 expressing cells (common to the lungs) to the upper respiratory tract, resulting in lower intrinsic virulence [217, 218].
A lower frequency of severe COVID-19 and fatalities from Omicron infection appears to be the result of both intrinsic differences in the virus pathogenicity and widespread cross-reactive immunity [219]. This is mirrored in vaccine breakthrough data from South Africa that shows reduced frequency of severe disease with Omicron relative to previous VoCs [220]. Relatively low case fatality rates were recorded during Omicron waves both in countries with high vaccination rates (e.g. New Zealand) and high levels of prior infection (e.g. South Africa). Very high levels of neutralizing antibodies may be able to overcome partial escape to provide some cross-neutralization, while non-neutralizing immunity through antibody-dependent cellular cytotoxicity and T cells is postulated to provide protection against disease when breakthrough Omicron infection occurs [221–224]. However, in populations with hardly any prior infection and low immunization rates among the elderly, Omicron still caused devastating outbreaks as was observed in Hong Kong, which recorded the highest per capita death rate of any country, at any time throughout the pandemic in March 2022.
HOW ‘SPECIFIC’ IS SARS-CoV-2-SPECIFIC IMMUNITY
A major focus of SARS-CoV-2 research has been to understand the extent to which immunity generated by infection or vaccination can cross-recognize and cross-protect against different lineages of SARS-CoV-2 and other members of the Coronaviridae family. This is particularly relevant now that the global population consists of a mixture of individuals with different histories of exposure to SARS-CoV-2 antigens [225].
The majority of immunocompetent individuals infected with SARS-CoV-2 develop both T and B cell responses and these remain detectable in the circulation for at least 9–12 months [226, 227]. Previous infection has been shown to offer protection from reinfection with the same SARS-CoV-2 variant for at least 6 months [228–230], although this may be reduced in the elderly, or when exposed to a heterologous viral strain. Likewise, vaccine-induced immunity against SARS-CoV-2 has shown impressive efficacy against severe disease, hospitalization and death, at least in the short term [231]. Accumulating data suggest that T cells play an important role in the resolution of SARS-CoV-2 infection [232, 233] and probably against disease following breakthrough infection post-vaccination. However, it is likely that vaccine efficacy against infection is mediated predominantly by antibody responses. For example, titres of binding antibodies to spike and RBD are predictive of breakthrough Delta infection following mRNA-1273 vaccination, supporting their utility as correlates of protection (COP) [231].
Waning of effective immune protection over time can occur through a combination of changes in the circulating virus itself and subsequent loss of recognition by the immune response (escape), and/or by reduction in the number of immune effectors over time, such as antibody levels (reflective of plasma cells), memory B cells (able to generate new affinity-matured antibodies upon re-infection) and memory T cells. SARS-CoV-2 binding and neutralizing antibodies show measurable waning post-infection [226, 234, 235], which may correlate with risk of breakthrough infection [236]. However, memory B cells can persist after SARS-CoV-2 infection and vaccination [226, 237] even after neutralizing antibodies become undetectable in the blood [238].
Many mechanisms can be used by viruses to evade adaptive immunity (e.g. shielding epitopes with glycans/lipoproteins, cell–cell spread, induction of interfering antibodies and disrupting antigen presentation). For SARS-CoV-2, much attention has been paid to mutational changes in the spike protein [239]. Point mutations within the spike can lead to loss of recognition by monoclonal antibodies, including those used as therapeutics [164, 240], however, single mutations do not significantly evade polyclonal responses (that recognize many regions of spike) present in convalescent serum. Combinations of these point mutations are, however, common to certain variants and when occurring together result in a significant reduction in the ability of antibody responses to recognize and neutralize variants. This has been well demonstrated by the emergence of the Omicron BA.1 variant with 30 spike substitutions, 6 deletions and 3 insertions, which together lead to significantly reduced neutralization in vitro by convalescent and post-vaccination serum relative to other strains [223, 224, 241] and a higher rate of breakthrough infection.
Prior to the emergence of the Omicron variant, the rise and decline of VoCs did not broadly correlate with immune escape. Alpha emerged with a comparatively small number of spike mutations but was relatively well neutralized by convalescent and post-vaccination serum [242]. The Beta and Gamma variants demonstrated much more evidence of antibody escape, for instance evading 12 of 17 monoclonal tested and convalescent and post-vaccination serum [166, 242, 243], however, they failed to outcompete the Alpha VoC that was globally dominant at the time. Sufficient cross-reactive immunity may have been retained despite the changes in Beta and Gamma spike proteins [225]. Subsequently, Delta outcompeted all other strains, accounting for up to 99% of infections globally in October 2021 despite showing modest evidence of immune escape relative to ancestral strains [244–246], which suggests that infectivity and transmissibility may have been the dominant factors determining VoC in the earlier stages of the pandemic. For example, S: E484K is the individual mutation most strongly implicated in partial immune escape yet the Beta and Gamma VOCs, each carrying E484K, failed to dominate in the same manner as Alpha, Delta and Omicron. The E484K mutation has also been acquired repeatedly by strains belonging to the Alpha and Delta VoCs (referred to as ‘Alpha+’ and ‘Delta+’) and significantly reduces neutralization in those genetic backgrounds [247, 248]. Yet, Alpha+ and Delta+ lineages have not been particularly successful. With a large proportion of the human population having been vaccinated and/or previously exposed to SARS-CoV-2 from early 2022, the major selective pressure likely shifted from higher infectiousness to an increased ability to (re-)infect already immunized hosts. The Omicron strains broke pre-conceptions of how much room SARS-CoV-2 spike had to evolve; Omicron spike contains a complex pattern of mutations that retain RBD ACE2 binding while changing the shape of the RBD sufficiently to escape a significant proportion of neutralizing antibodies [241].
As with antibody recognition, mutations within epitopes can also lead to escape from the cellular arm of the adaptive immune response, whereby T cells fail to recognize infected cells, for instance due to the mutant epitope failing to be processed and presented effectively, or due to loss of recognition of the MHC-peptide complex by the T cells receptor (TCR) [91, 249–252]. Studies to date suggest the majority of T cells induced by vaccination or infection retain their ability to recognize epitopes within Omicron and other variants [253, 254]. T cell responses to SARS-CoV-2 post-infection are highly multispecific, targeting multiple potential human leukocyte antigens (HLA)-restricted epitopes within all viral proteins, not just the structural proteins and surface epitopes available to antibodies [225, 255–258]. Notably, all widely used SARS-CoV-2 vaccines also induced T cells responses which are polyclonal and multispecific in most individuals, however, these are restricted to the proteins contained within the vaccine, currently largely the spike [231, 259–261]. HLA have evolved to be highly polymorphic, meaning that the epitopes recognized vary substantially between individuals, implying that T cell escape variants are less likely to contribute to transmission within a population unless representing a highly immunodominant response restricted by a frequent HLA allele. TCRs, the receptors by which infected cells are recognized, are also randomly generated meaning each individual recognizes epitopes with a unique combination of TCRs; therefore, viral mutations will impact recognition in subtly different ways for each individual. Overall, these mechanisms reduce the selection pressure for individual escape mutations at the population level, in particular for an acute resolving infection. In other words, the advantage gained by mutating a single epitope would only be advantageous in another host if they share the same HLA restriction.
Related to SARS-CoV-2 lineage cross-immunity, pre-exposure to non-SARS-CoV-2 viruses can mediate cross-immunity. For instance, T cell responses that can cross-recognize SARS-CoV-2 can be detected in pre-pandemic samples taken before it circulated in humans in as many as 80% of individuals, depending on the sensitivity of the assay used [227, 262–265]. Due to sequence conservation with SARS-CoV-2, many groups have started to investigate the possibility that these pre-existing T cells were induced by universal exposure to HCoVs [256, 257, 262, 266, 267]. While it is unlikely that HCoVs are the sole source of pre-existing T cell responses [30], it has been demonstrated that T cells can cross-recognize SARS-CoV-2 and all four HCoVs in vitro at certain epitopes [256, 262, 266–268] and that T cells transduced with TCRs from convalescent samples could recognize HCoVs and SARS-CoV-2 with similar affinities [269].
Prior immunity would be expected to limit disease on exposure to SARS-CoV-2 as pre-existing memory T cells are imbued with characteristics which make them more efficient at viral control than naïve T cells. As an example, pre-existing T cells that target the highly conserved core replication transcription complex (NSP7 polymerase cofactor, NSP12 polymerase and NSP13 helicase) were shown to be enriched in health care workers that showed signs of exposure to SARS-CoV-2 but who appeared to control infection before it was detectable by PCR or induced an antibody response [262]. This early control of viral replication, before an infection could be established, may be explained by the rapid response of pre-existing T cells that target the earliest expressed proteins of the viral lifecycle in ORF1ab. Pre-existing cross-reactive T cells expand in vivo upon infection and vaccination [266] and have been associated with protection from infection [262, 270] and severe disease [271], but epidemiological data are still limited to support [272] or refute [273] that recent HCoV infection and associated immune responses are directly associated with protection from severe COVID-19.
Cross-reactive antibodies are less common in pre-pandemic samples, likely due to their targeting of less conserved structural viral proteins and greater sensitivity to point mutations altering structural epitopes [274, 275] and they have not been associated with protection from disease [276]. Nonetheless, it has been suggested that pre-existing cross-reactive immunity is the reason we have so few endemic coronaviruses, rather than lack of opportunity or lack of identification of coronavirus outbreaks in humans [277]. For another coronavirus to establish itself in the human population like SARS-CoV-2 did, it would have to circumvent significant pre-existing immunity generated by previous coronaviruses in circulation.
The biggest unknown in SARS-CoV-2 immunity is what correlates with protection from disease and from infection. Attempts have been made to identify antibody COP for SARS-CoV-2 by integrating efficacy and antibody data from vaccine trials [278, 279]; however, a single COP may be hard to identify for all SARS-CoV-2 VoCs without measuring variant-specific neutralizing antibodies, as well as non-neutralizing antibodies, and cellular immunity. A greater understanding of the immune responses at mucosal airway surfaces, the site of viral control, will likely be required [280]. Despite difficulties in accessing samples, compartmentalized immunity within the airway mucosa in the form of tissue-resident T cells [281–283] and local IgA, memory B cells [284, 285] have been described and associated with protection from severe disease and infection with SARS-CoV-2 [286].
A growing body of evidence suggests that vaccines for global health priorities should be focused on inducing both antibodies and T cells [287, 288]. Due to their longevity, multispecificity and propensity to target conserved regions, T cells could be particularly effective at mediating long-term protection against SARS-CoV-2. Since most T cell targets lie outside of the spike protein [255, 256, 262], it may be prudent to include non-spike antigens in vaccines, possibly targeting regions that are conserved across the wider Coronaviridae. Clinical trials are already underway to test second-generation vaccines to improve durability, infection and transmission blocking and the potential for inducing cross-reactive immunity that can protect against future SARS-CoV-2 variants and acquisition of novel coronaviruses in the future.
THE FOUR FUNDAMENTAL FORCES OF AN EPIDEMIC
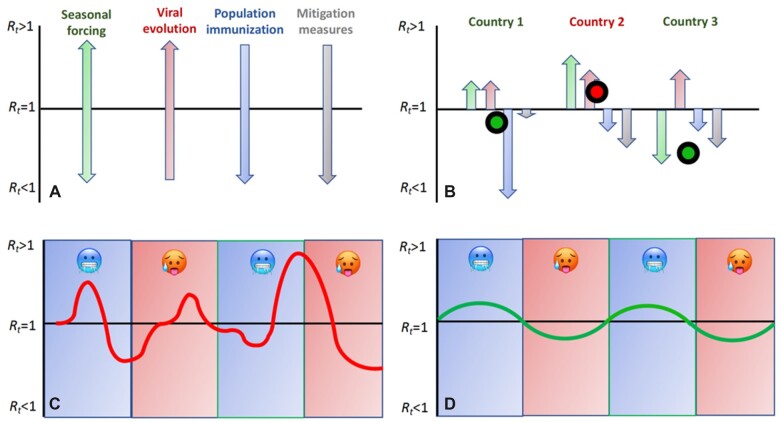
The epidemic dynamic of respiratory viruses such as SARS-CoV-2 is driven by four main factors, namely, seasonality, viral evolution, population immunization rates and mitigation measures affecting host behaviour (Fig. 5A). Seasonal forcing expresses the higher transmissibility of respiratory viruses during winter than summer in both hemispheres. This is in part due to physical conditions, such as viruses remaining infectious for longer in cold, dry air [289] and under low sunlight (UV) exposure [290]. However, there is also a host behaviour component with humans tending to spend more time indoors in poorly ventilated conditions during the winter [291, 292]. Viral evolution will always tend to push transmissibility upwards but it is expected to tend towards a fitness maximum. Population immunization reduces viral transmissibility by removing susceptible hosts from the population, but is constantly refuelled by viral antigenic drift, waning of immunization and introduction into the population of unexposed newborn hosts, thus eventually reaching a dynamic quasi-equilibrium. Finally, mitigation measures, which encompass interventions such as aiming to increase social distancing, will reduce transmission but for the most part are unlikely to stay in place indefinitely.
Figure 5:
Schematic representation of the four main forces acting on pathogen transmission. Notes: (A) Effect of the four main forces on transmission. (B) Cumulative effect of the forces in three hypothetical countries. In countries 1 and 3, the cumulative effects of the four forces keep the epidemic from growing (R < 1), whereas in country 2, the epidemic is growing. (C) Epidemic behaviour over a 2-year period under a continental climate with blue area depicting colder months in the year and red warmer months. (D) Seasonal dynamic of endemic pathogen with smaller amplitude of cases and regular peaks during the colder months.
Together these four forces will jointly affect the effective reproduction number of a pathogen (Rt), the average number of new infections caused by a single infected individual at time t. When Rt is above 1.0, case numbers increase and when it falls below 1.0, they go down (Fig. 5B). In an epidemic dynamic, Rt, and hence case numbers, can fluctuate wildly leading to marked epidemic waves (Fig. 5C). Herd immunity is reached when Rt falls below 1.0, but for viruses such as SARS-CoV-2, where immunization against re-infection is relatively short lived, this endemic state is only transient. The only force in the system that remains unaltered is seasonal forcing, which ultimately becomes a major driver of the system, leading to higher case numbers in winter than in summer in the temperate zone (Fig. 5D).
Essentially all ∼200 endemic human respiratory viruses are seasonal in temperate regions of the world. Many endemic respiratory viruses including the four endemic HCoVs also tend to exhibit a biannual dynamic, with case number higher every second year [293]. This might in part stem from a non-equilibrium state caused by immune protection from reinfection following prior exposure lasting on average for longer than 1 year, but less than 2. SARS-CoV-2 is no exception to the general pattern of seasonality, and its transmissibility, at the time of writing, is already higher in winter. Seasonal variation in the transmissibility of SARS-CoV-2 had little impact during the early stages of the pandemic. This is because as long as immunization rates remained limited, SARS-CoV-2 could readily transmit at any time of year as long as conditions were otherwise favourable, and case rates were mainly driven by variation in the stringency of mitigation measures and the emergence of more transmissible lineages. For instance, India experienced a major epidemic peak outside the winter season, during the spread of the Delta variant in late spring 2021. As immunization levels increase further, SARS-CoV-2 is expected to adopt a seasonal endemic dynamic, with outbreaks and epidemics mostly happening in winter.
THE GOOD, THE BAD AND THE UGLY
While there is little doubt that SARS-CoV-2 will eventually become a seasonal endemic pathogen, there is limited consensus in the scientific community about what ‘endemic SARS-CoV-2’ entails in terms of future morbidity and mortality, both upon infection and through long COVID/PASC (post acute sequelae of SARS-CoV-2 infection). However, during the endemic phase of COVID-19, morbidity and mortality will primarily be dictated by the ability of SARS-CoV-2 to bypass global host immunity and the intrinsic virulence of future SARS-CoV-2 variants.
Worst case scenarios include high case numbers and circulation of a diversity of serotypes with limited protection across strains, possibly fuelled by reverse anthroponosis and recombination between human and animal coronaviruses. For example, infectious bronchitis virus (IBV), a Gammacoronavirus infecting chicken, circulates in the form of multiple serotypes with highly divergent sequences in their spike protein and minimal cross-immunization between certain strains in circulation [294]. This would present a highly challenging situation, leading to constantly high case numbers and a need for continual vaccine updates to match at least a subset of dominant serotypes in circulation. While such a scenario of diversification into multiple serotypes cannot be formally ruled out as the future for endemic SARS-CoV-2, it remains relatively unlikely. Humans, as long-lived hosts, are typically exposed multiple times in their lives to the four HCoVs and will in all likelihood be to SARS-CoV-2 in the future. Repeated exposure to the five coronaviruses in circulation is expected to restrict the immunological space that SARS-CoV-2 lineages can explore at any time [277]. The extent to which immune-escape mutations in the spike RBD act independently from each other will be critical for the emergence of new serotypes through previously unexplored epistatic combinations. There is currently no consensus on the evolutionary potential of SARS-CoV-2 to generate new serotypes through novel epistatic combinations of mutations [295, 296]. However, at the least, we can anticipate that SARS-CoV-2 will undergo antigenic drift in common with other HCoVs [133].
A situation where SARS-CoV-2 will circulate in the future as one or two serotypes at any time, constantly evolving through antigenic drift and with sequential lineage replacement represents the most likely scenario. This is the situation we currently experience with the four HCoVs and seasonal influenza A and B. People are generally infected for the first time to both HCoVs and seasonal influenza early in life and build immunity through multiple exposures throughout life. The majority of influenza infections are asymptomatic and people tend to get exposed to influenza multiple times [297]; the same is true for HCoVs [127–129]. In terms of epidemiology and public health, one major difference between HCoVs and influenza is the higher virulence of seasonal influenza. HCoVs are generally considered to exert only a minor burden on human health, and are often tellingly categorized among ‘common colds’, even if they are not always harmless, in particularly in the elderly. While also often underascertained, the burden of influenza tends to be more widely recognized. A recent study estimated the number of yearly deaths associated with seasonal influenza to around 400 000 globally (with considerable year on year variation), with around two-thirds of these among people over the age of 65 years old [298]. Additionally, influenza A undergoes rare events of antigenic shifts, which have not been documented in HCoVs to date. An antigenic shift corresponds to the emergence of a new lineage, to which the population has no or limited immunity, and which leads to an influenza pandemic as in 1918, 1957, 1968 or 2009. The emergence of the Omicron variant could be considered as largely analogous to an influenza A antigenic shift. The extent to which the emergence of Omicron was an exceptional event or part of reoccurring future pattern will be determinant for the future dynamic of SARS-CoV-2.
COVID-19 has been the worst respiratory disease pandemic since the ‘Spanish flu’ in 1918/20 when influenza H1N1 may have killed up to 50 million. The 1918–20 flu pandemic came to an end with H1N1 becoming the agent of endemic seasonal flu until it got displaced by the 1957–58 H2N2 pandemic lineage. The 1918 H1N1 lineage then made a comeback in the 1970s, again contributing to the yearly burden of seasonal flu until it got likely displaced in humans by the 2009 pandemic H1N1pdm09 lineage [299]. While pandemics of respiratory viruses end, the causal agents remain in circulation for decades. As such, whether SARS-CoV-2 will in time become a fifth ‘common cold HCoV’, or exert a more significant burden on human health comparable to, or even higher than, seasonal influenza will largely depend on the intrinsic virulence of future viral lineages. We have essentially no control over the global evolution of the virus and the trajectory in terms of virulence of future SARS-CoV-2 variants is unknown, if not unknowable. However, what we have considerable control over is the morbidity and mortality associated with endemic SARS-CoV-2 in the future. By far the best tool is global vaccination of the elderly and those otherwise most at risk, which needs to be scaled up and maintained globally if we hope to live with a fifth HCoV, rather than an additional seasonal flu-type virus.
ACKNOWLEDGEMENTS
We gratefully acknowledge all contributing and submitting laboratories around the globe including the COG-UK consortium (https://www.cogconsortium.uk) who have openly shared large numbers of UK SARS-CoV-2 assemblies. We are also grateful to developers and maintainers of CoVariants (https://covariants.org) and Outbreak.info (https://outbreak.info) for information on mutation and deletion carriage in VoCs and VoIs.
FUNDING
L.v.D. is supported by a UCL Excellence Fellowship. D.R. was supported by an NIHR Precision AMR award. D.R., L.v.D. and F.B. acknowledge funding from Horizon Europe for the ‘END-VoC’ consortium grant. F.B. acknowledges support from the BBSRC (equipment grant BB/R01356X/1). M.M. is supported by the UKRI UK-CIC grant and Wellcome Trust Investigator Award (214191/Z/18/Z) and L.S. by a Medical Research Foundation fellowship (044-0001).
DATA AVAILABILITY
Figure 4 was enabled by the GISAID EpiCoV™ database (https://www.gisaid.org) and regular Audacity releases. We gratefully acknowledge all authors of the data used herein, and their originating and submitting laboratories. Detailed acknowledgments are available at https://www.gisaid.org using the Data Acknowledgement Locator with the GISAID EPI_SET_20220509xa accession number.
CONFLICT OF INTEREST STATEMENT
None declared.
Contributor Information
François Balloux, UCL Genetics Institute, University College London, London WC1E 6BT, UK.
Cedric Tan, UCL Genetics Institute, University College London, London WC1E 6BT, UK; Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), 138672 Singapore, Singapore.
Leo Swadling, Division of Infection and Immunity, University College London, London NW3 2PP, UK.
Damien Richard, UCL Genetics Institute, University College London, London WC1E 6BT, UK; Division of Infection and Immunity, University College London, London NW3 2PP, UK.
Charlotte Jenner, UCL Genetics Institute, University College London, London WC1E 6BT, UK.
Mala Maini, Division of Infection and Immunity, University College London, London NW3 2PP, UK.
Lucy van Dorp, UCL Genetics Institute, University College London, London WC1E 6BT, UK.
References
- 1. Wu F, Zhao S, Yu B. et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9. 10.1038/s41586-020-2008-3. PubMed PMID: WOS:000519378900029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270. 10.1038/s41586-020-2012-7. PubMed PMID: WOS:000519378900030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Dorp L, Houldcroft CJ, Richard D. et al. COVID-19, the first pandemic in the post-genomic era. Curr Opin Virol 2021;50:40–8. 10.1016/j.coviro.2021.07.002. PubMed PMID: WOS:000700783300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elbe S, Buckland-Merrett G.. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 2017;1:33–46. 10.1002/gch2.1018. PubMed PMID: WOS:000412860800011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shu YL, McCauley J.. GISAID: Global initiative on sharing all influenza data—from vision to reality. Eurosurveillance 2017;22:2–4. 10.2807/1560-7917.Es.2017.22.13.30494. PubMed PMID: WOS:000398153900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Dorp L, Acman M, Richard D. et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 2020;83:104351. 10.1016/j.meegid.2020.104351. PubMed PMID: MEDLINE:32387564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumar S, Tao QQ, Weaver S. et al. An evolutionary portrait of the progenitor SARS-CoV-2 and its dominant offshoots in COVID-19 pandemic. Mol Biol Evol 2021;38:2021–3059. 10.1093/molbev/msab118. PubMed PMID: WOS:000693740300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Worobey M. Dissecting the early COVID-19 cases in Wuhan. Science 2021;374:1202–4. 10.1126/science.abm4454. [DOI] [PubMed] [Google Scholar]
- 9. Gao G, Liu W, Liu P. et al. Surveillance of SARS-CoV-2 in the environment and animal samples of the Huanan Seafood Market. Research Square2022. 10.21203/rs.3.rs-1370392/v1. [DOI]
- 10. Newsroom C. First Travel-related Case of 2019 Novel Coronavirus Detected in United States, 2020. https://www.cdc.gov/media/releases/2020/p0121-novel-coronavirus-travel-case.html.
- 11. Stoecklin SB, Rolland P, Silue Y. et al. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Eurosurveillance 2020;25:20–6. 10.2807/1560-7917.Es.2020.25.6.2000094. PubMed PMID: WOS:000514439600004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. La Rosa G, Mancini P, Ferraro GB. et al. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci Tot Environ 2021;750. 10.1016/j.scitotenv.2020.141711. PubMed PMID: WOS:000585694600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ldu Plessis, McCrone JT, Zarebski AE. et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021;371:708–12. 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodcroft EB, Zuber M, Nadeau S. et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021;595:707–12. 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 15. van Dorp L, Richard D, Tan CCS. et al. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun 2020;11:5986. 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korber B, Fischer W, Gnanakaran S. et al. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020:2020.04.29.069054. 10.1101/2020.04.29.069054. [DOI] [Google Scholar]
- 17. Volz E, Hill V, McCrone JT. et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021;184:64–75.e11. 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davies NG, Abbott S, Barnard RC. et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021;372:eabg3055. 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Volz E, Mishra S, Chand M. et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021;593:266–9. 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 20. Pung R, Mak TM, Kucharski AJ. et al. Serial intervals in SARS-CoV-2 B.1.617.2 variant cases. Lancet 2021;398:837–8. 10.1016/S0140-6736(21)01697-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott P, Haw D, Wang H. et al. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the Delta variant. Science 2021;374:eabl9551. 10.1126/science.abl9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viana R, Moyo S, Amoako DG. et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022;603:679–86. 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tegally H, Wilkinson E, Giovanetti M. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature2021;592:438–43. [DOI] [PubMed] [Google Scholar]
- 24. Faria NR, Mellan TA, Whittaker C. et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science2021;372:815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker PJ, Siddell SG, Lefkowitz EJ. et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch Virol 2020;165:2737–48. 10.1007/s00705-020-04752-x. [DOI] [PubMed] [Google Scholar]
- 26. Shaw LP, Wang AD, Dylus D. et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol Ecol 2020;29:3361–79. 10.1111/mec.15463. PubMed PMID: WOS:000537468900001. [DOI] [PubMed] [Google Scholar]
- 27. Brian DA, Baric RS.. Coronavirus genome structure and replication. Curr Top Microbiol Immunol 2005;287:1–30. 10.1007/3-540-26765-4_1. PubMed PMID: 15609507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snijder EJ, Decroly E, Ziebuhr J.. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res 2016;96:59–126. Epub 2016/09/14. 10.1016/bs.aivir.2016.08.008. PubMed PMID: 27712628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denison MR, Graham RL, Donaldson EF. et al. Coronaviruses. RNA Biol 2011;8:270–9. 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan CCS, Owen CJ, Tham CYL. et al. Pre-existing T cell-mediated cross-reactivity to SARS-CoV-2 cannot solely be explained by prior exposure to endemic human coronaviruses. Infect Genet Evol 2021;95:105075. 10.1016/j.meegid.2021.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vlasak R, Luytjes W, Spaan W. et al. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA 1988;85:4526–9. 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cui J, Li F, Shi Z-L.. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein S, Cortese M, Winter SL. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun 2020;11:5885. Epub 2020/11/20. 10.1038/s41467-020-19619-7. PubMed PMID: 33208793; PubMed Central PMCID: PMCPMC7676268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almeida JD, Berry DM, Cunningham CH. et al. Virology: Coronaviruses. Nature 1968;220:650. 10.1038/220650b0. [DOI] [Google Scholar]
- 35. Huang Y, Yang C, Xu X-F. et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 2020;41:1141–9. 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolf KA, Kwan JC, Kamil JP. et al. Structural dynamics and molecular evolution of the SARS-CoV-2 spike protein. mBio 2022;13:e02030–21. 10.1128/mbio.02030-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schultze B, Gross HJ, Brossmer R. et al. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol 1991;65:6232–7. 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Millet JK, Jaimes JA, Whittaker GR.. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol Rev 2020;45. 10.1093/femsre/fuaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hulswit RJG, Lang Y, Bakkers MJG. et al. Human coronaviruses OC43 and HKU1 bind to 9-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci USA 2019;116:2681–90. 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yeager CL, Ashmun RA, Williams RK. et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992;357:420–2. 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qing E, Li P, Cooper L. et al. Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry. Cell Rep 2022;39. 10.1016/j.celrep.2022.110786. PubMed PMID: 110786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson CB, Farzan M, Chen B. et al. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 2022;23:3–20. 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szabo R, Bugge TH.. Type II transmembrane serine proteases in development and disease. Int J Biochem Cell Biol 2008;40:1297–316. Epub 2008/01/15. 10.1016/j.biocel.2007.11.013. PubMed PMID: 18191610. [DOI] [PubMed] [Google Scholar]
- 44. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016;3:237–61. 10.1146/annurev-virology-110615-042301. PubMed PMID: 27578435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu DX, Fung TS, Chong KK-L. et al. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res 2014;109:97–109. 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Redondo N, Zaldívar-López S, Garrido JJ. et al. SARS-CoV-2 accessory proteins in viral pathogenesis: Knowns and unknowns. Front Immunol 2021;12. 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohaim MA, El Naggar RF, Clayton E. et al. Structural and functional insights into non-structural proteins of coronaviruses. Microb Pathog 2021;150:104641. 10.1016/j.micpath.2020.104641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siu KL, Yuen KS, Castaño-Rodriguez C. et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J 2019;33:8865–77. Epub 2019/04/30. 10.1096/fj.201802418R. PubMed PMID: 31034780; PubMed Central PMCID: PMCPMC6662968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Law PTW, Wong CH, Au TCC. et al. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol 2005;86:1921–30. Epub 2005/06/17. 10.1099/vir.0.80813-0. PubMed PMID: 15958670. [DOI] [PubMed] [Google Scholar]
- 50. Ren Y, Shu T, Wu D. et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol 2020;17:881–3. 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J. et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio 2018;9. Epub 2018/05/24. 10.1128/mBio.02325-17. PubMed PMID: 29789363; PubMed Central PMCID: PMCPMC5964350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gordon DE, Jang GM, Bouhaddou M. et al. A SARS-CoV-2-human protein–protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 2020:2020.03.22.002386. 10.1101/2020.03.22.002386. [DOI] [Google Scholar]
- 53. Young BE, Fong S-W, Chan Y-H. et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet 2020;396:603–11. 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pancer K, Milewska A, Owczarek K. et al. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog 2020;16:e1008959. Epub 2020/12/11. 10.1371/journal.ppat.1008959. PubMed PMID: 33301543; PubMed Central PMCID: PMCPMC7755277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Banerjee A, Doxey AC, Mossman K. et al. Unraveling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol Evol 2021;36:180–4. 10.1016/j.tree.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wacharapluesadee S, Tan CW, Maneeorn P. et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 2021;12:972. 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou H, Ji J, Chen X. et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 2021;184:4380–91.e14. 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou H, Chen X, Hu T. et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 2020;30:2196–203.e3. 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Temmam S, Vongphayloth K, Baquero, E et al. Coronaviruses with a SARS-CoV-2-like receptor-binding domain allowing ACE2-mediated entry into human cells isolated from bats of Indochinese peninsula. Nature 2022;604:330–6. 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 60. Murakami S, Kitamura T, Suzuki J. et al. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis J 2020;26:3025. 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Delaune D, Hul V, Karlsson EA. et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun 2021;12:6563. 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ge X-Y, Li J-L, Yang X-L. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503:535–8. 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lau SKP, Woo PCY, Li KSM. et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA 2005;102:14040–5. 10.1073/pnas.0506735102. PubMed PMID: WOS:000232231900059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guan Y, Zheng BJ, He YQ. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 2003;302:276–8. 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 65. Lytras S, Hughes J, Martin D. et al. Exploring the natural origins of SARS-CoV-2 in the light of recombination. bioRxiv. 2021:2021.01.22.427830. 10.1101/2021.01.22.427830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lam TT-Y, Jia N, Zhang Y-W. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020;583:282–5. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 67. MacLean OA, Lytras S, Weaver S. et al. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol 2021;19:e3001115–e. 10.1371/journal.pbio.3001115. PubMed PMID: 33711012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shang J, Wan Y, Luo C. et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA 2020;117:11727–34. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Johnson BA, Xie X, Bailey AL. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021;591:293–9. 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lytras S, MacLean OA, Robertson DL.. The Sarbecovirus origin of SARS-CoV-2's during cleavage site 2020. https://virological.org/t/the-sarbecovirus-origin-of-sars-cov-2-s-furin-cleavage-site/536.
- 71. Wu Y, Zhao S.. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res 2021;50:102115. 10.1016/j.scr.2020.102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McAloose D, Laverack M, Wang L. et al. From people to Panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. mBio 2020;11:e02220–20. 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Oreshkova N, Molenaar RJ, Vreman S. et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020;25:2001005. 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Munnink BBO, Sikkema RS, Nieuwenhuijse DF. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021;371:172–7. 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chandler JC, Bevins SN, Ellis JW. et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci USA 2021;118:e2114828118. 10.1073/pnas.2114828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kuchipudi SV, Surendran-Nair M, Ruden RM. et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc Natl Acad Sci USA 2022;119:e2121644119. 10.1073/pnas.2121644119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chu H, Chan JF-W, Yuen TT-T. et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 2020;1:e14–e23. 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Meekins DA, Morozov I, Trujillo JD. et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect 2020;9:2278–88. 10.1080/22221751.2020.1831405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schlottau K, Rissmann M, Graaf A. et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020;1:e218–e25. 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ulrich L, Wernike K, Hoffmann D. et al. Experimental infection of cattle with SARS-CoV-2. Emerg Infect Dis J 2020;26:2979. 10.3201/eid2612.203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sit THC, Brackman CJ, Ip SM. et al. Infection of dogs with SARS-CoV-2. Nature 2020;586:776–8. 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Damas J, Hughes GM, Keough KC. et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA 2020;117:22311–22. 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lam SD, Bordin N, Waman VP. et al. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci Rep 2020;10:16471. 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu L, Chen Q, Liu K. et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov 2020;6:68. 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu Y, Hu G, Wang Y. et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc Natl Acad Sci USA 2021;118:e2025373118. 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cool K, Gaudreault NN, Morozov I. et al. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. bioRxiv. 2021:2021.08.15.456341. 10.1101/2021.08.15.456341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Palmer MV, Martins M, Falkenberg S. et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J Virol 2021;95:e00083–21. 10.1128/JVI.00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hale VL, Dennis PM, McBride DS. et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022;602:481–6. 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pickering B, Lung O, Maguire F. et al. Highly divergent white-tailed deer SARS-CoV-2 with potential deer-to-human transmission. bioRxiv. 2022:2022.02.22.481551. 10.1101/2022.02.22.481551. [DOI] [Google Scholar]
- 90. Aguiló-Gisbert J, Padilla-Blanco M, Lizana V. et al. First description of SARS-CoV-2 infection in two feral American mink (Neovison vison) caught in the wild. Animals 2021;11:1422. PubMed PMID: 10.3390/ani11051422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kim Y-I, Kim S-G, Kim S-M. et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020;27:704–9.e2. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gortázar C, Barroso-Arévalo S, Ferreras-Colino E. et al. Natural SARS-CoV-2 infection in kept ferrets, Spain. Emerg Infect Dis J 2021;27:1994. 10.3201/eid2707.210096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Richard M, Kok A, de Meulder D. et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun 2020;11:3496. 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Davoust B, Guérin P, Orain N. et al. Evidence of antibodies against SARS-CoV-2 in wild mustelids from Brittany (France). bioRxiv. 2022:2022.01.20.477038. 10.1101/2022.01.20.477038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Braun KM, Moreno GK, Halfmann PJ. et al. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog 2021;17:e1009373. 10.1371/journal.ppat.1009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bosco-Lauth AM, Hartwig AE, Porter SM. et al. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA 2020;117:26382–8. 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Halfmann PJ, Hatta M, Chiba S. et al. Transmission of SARS-CoV-2 in domestic cats. N Engl J Med 2020;383:592–4. 10.1056/NEJMc2013400. PubMed PMID: 32402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Garigliany M, Van Laere A-S, Clercx C. et al. SARS-CoV-2 natural transmission from human to cat, Belgium, March 2020. Emerg Infect Dis J 2020;26:3069. 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Patterson EI, Elia G, Grassi A. et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat Commun 2020;11:6231. 10.1038/s41467-020-20097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhou J, Li C, Liu X. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med 2020;26:1077–83. 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 101. Lau SKP, Wong ACP, Luk HKH. et al. Differential tropism of SARS-CoV and SARS-CoV-2 in bat cells. Emerg Infect Dis J 2020;26:2961. 10.3201/eid2612.202308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Shuai H, Chan JF-W, Yuen TT-T. et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 2021;73. 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Muñoz-Fontela C, Dowling WE, Funnell SGP. et al. Animal models for COVID-19. Nature 2020;586:509–15. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kok K-H, Wong S-C, Chan W-M. et al. Co-circulation of two SARS-CoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg Microbes Infect 2022;11:689–98. 10.1080/22221751.2022.2040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huang H, Zhu Y, Niu Z. et al. SARS-CoV-2 N501Y variants of concern and their potential transmission by mouse. Cell Death Differ 2021;28:2840–2. 10.1038/s41418-021-00846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tan CCS, Lam SD, Richard D. et al. Transmission of SARS-CoV-2 from humans to animals and potential host adaptation. Nature Communications. 2022;13:2988. 10.1038/s41467-022-30698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lu L, Sikkema RS, Velkers FC. et al. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat Commun 2021;12:6802. 10.1038/s41467-021-27096-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ren W, Lan J, Ju X. et al. Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog 2021;17:e1010053. 10.1371/journal.ppat.1010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Motozono C, Toyoda M, Zahradnik J. et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021;29:1124–36.e11. 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang L, Li Q, Nie J. et al. Cellular tropism and antigenicity of mink-derived SARS-CoV-2 variants. Signal Transduct Target Therapy 2021;6:196. 10.1038/s41392-021-00617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhou J, Peacock TP, Brown JC. et al. Mutations that adapt SARS-CoV-2 to mink or ferret do not increase fitness in the human airway. Cell Rep 2022;38:110344. 10.1016/j.celrep.2022.110344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Baum A, Fulton BO, Wloga E. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020;369:1014–8. 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Marques AD, Sherrill-Mix S, Everett JK. et al. Evolutionary trajectories of SARS-CoV-2 alpha and delta variants in white-tailed deer in Pennsylvania. medRxiv. 2022:2022.02.17.22270679. 10.1101/2022.02.17.22270679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Maganga GD, Pinto A, Mombo IM. et al. Genetic diversity and ecology of coronaviruses hosted by cave-dwelling bats in Gabon. Sci Rep 2020;10:7314. 10.1038/s41598-020-64159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lin X-D, Wang W, Hao Z-Y. et al. Extensive diversity of coronaviruses in bats from China. Virology 2017;507:1–10. 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Graham RL, Baric RS.. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J Virol 2010;84:3134–46. 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hamre D, Procknow JJ.. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 1966;121:190–3. 10.3181/00379727-121-30734. PubMed PMID: 4285768. [DOI] [PubMed] [Google Scholar]
- 118. Drosten C, Günther S, Preiser W. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967–76. 10.1056/NEJMoa030747. PubMed PMID: 12690091. [DOI] [PubMed] [Google Scholar]
- 119. Cheng VCC, Lau SKP, Woo PCY. et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microb Rev 2007;20:660–94. 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ithete NL, Stoffberg S, Corman VM. et al. Close relative of human middle east respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis J 2013;19:1697. 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.World Health Organization. MERS Situation Update, January 2020. 2020.
- 122. Corman VM, Muth D, Niemeyer D. et al. Chapter 8—Hosts and sources of endemic human coronaviruses. In: Kielian M, Mettenleiter TC, Roossinck MJ (eds), Advances in Virus Research, Vol. 100, pp. 163–88. Academic Press, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Müller MA, Corman VM, Jores J. et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis 2014;20:2093–5. 10.3201/eid2012.141026. PubMed PMID: 25425139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. McIntosh K, Dees JH, Becker WB. et al. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA 1967;57:933–40. 10.1073/pnas.57.4.933. PubMed PMID: 5231356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. van der Hoek L, Pyrc K, Jebbink MF. et al. Identification of a new human coronavirus. Nat Med 2004;10:368–73. 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zaki AM, van Boheemen S, Bestebroer TM. et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814–20. 10.1056/NEJMoa1211721. PubMed PMID: 23075143. [DOI] [PubMed] [Google Scholar]
- 127. Su S, Wong G, Shi W. et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24:490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nickbakhsh S, Ho A, Marques DFP. et al. Epidemiology of seasonal coronaviruses: Establishing the context for the emergence of coronavirus disease 2019. J Infect Dis 2020;222:17–25. 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Aldridge R, Lewer D, Beale S. et al. Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the Flu Watch cohort study [version 2; peer review: 2 approved]. Wellcome Open Res 2020;5. 10.12688/wellcomeopenres.15812.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Edridge AWD, Kaczorowska J, Hoste ACR. et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med 2020;26:1691–3. 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 131. Callow KA, Parry HF, Sergeant M. et al. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990;105:435–46. Epub 2009/05/15. 10.1017/S0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Jo WK, Drosten C, Drexler JF.. The evolutionary dynamics of endemic human coronaviruses. Virus Evol 2021;7. 10.1093/ve/veab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Eguia RT, Crawford KHD, Stevens-Ayers T. et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog 2021;17:e1009453. 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kistler KE, Bedford T.. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. eLife 2021;10:e64509. 10.7554/eLife.64509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hatcher EL, Zhdanov SA, Bao Y. et al. Virus variation resource—improved response to emergent viral outbreaks. Nucleic Acids Res 2016;45(D1):D482–D90. 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Al-Khannaq MN, Ng KT, Oong XY. et al. Diversity and evolutionary histories of human coronaviruses NL63 and 229E associated with acute upper respiratory tract symptoms in Kuala Lumpur, Malaysia. Am Soc Trop Med Hyg 2016;94:1058–64. 10.4269/ajtmh.15-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Corman VM, Baldwin HJ, Tateno AF. et al. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 2015;89:11858–70. 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Huynh J, Li S, Yount B. et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 2012;86:12816–25. Epub 2012/09/19. 10.1128/JVI.00906-12. PubMed PMID: 22993147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Corman VM, Eckerle I, Memish ZA. et al. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci USA 2016;113:9864–9. 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Vijgen L, Keyaerts E, Moës E. et al. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 2005;79:1595–604. 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Christakis NA. Apollo’s Arrow: The Profound and Enduring Impact of Coronavirus on the Way We Live. New York: Little, Brown Spark/Hachette Book Group, 2020. [Google Scholar]
- 142. Brussow H, Brussow L.. Clinical evidence that the pandemic from 1889 to 1891 commonly called the Russian flu might have been an earlier coronavirus pandemic. Microb Biotechnol 2021;14:1860–70. 10.1111/1751-7915.13889. PubMed PMID: WOS:000672472900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pollett S, Conte MA, Sanborn M. et al. A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic. Sci Rep 2021;11:17365. 10.1038/s41598-021-96626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Storz J, Rott R.. Reactivity of antibodies in human serum with antigens of an enteropathogenic bovine coronavirus. Med Microbiol Immunol 1981;169:169–78. 10.1007/BF02123590. PubMed PMID: 6265746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Oma VS, Klem T, Tråvén M. et al. Temporary carriage of bovine coronavirus and bovine respiratory syncytial virus by fomites and human nasal mucosa after exposure to infected calves. BMC Vet Res 2018;14:22. 10.1186/s12917-018-1335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Vlasova AN, Diaz A, Damtie D. et al. Novel canine coronavirus isolated from a hospitalized patient with pneumonia in East Malaysia. Clin Infect Dis 2021. 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Silva CS, Mullis LB, Pereira O Jr. et al. Human respiratory coronaviruses detected in patients with influenza-like illness in Arkansas, USA. Virol Mycol 2014;2014:004. Epub 2014/03/26. 10.4172/2161-0517.S2-004. PubMed PMID: 27588218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lednicky JA, Tagliamonte MS, White SK. et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv. 2021:2021.03.19.21253391. 10.1101/2021.03.19.21253391. [DOI] [Google Scholar]
- 149. Muller HJ. The relation of recombination to mutational advance. Mutat Res 1964;1:2–9. [DOI] [PubMed] [Google Scholar]
- 150. Richard D, Shaw LP, Lanfear R. et al. A phylogeny-based metric for estimating changes in transmissibility from recurrent mutations in SARS-CoV-2. bioRxiv. 2021:2021.05.06.442903. 10.1101/2021.05.06.442903. [DOI] [Google Scholar]
- 151. Brufsky A, Lotze MT.. Ratcheting down the virulence of SARS-CoV-2 in the COVID-19 pandemic. J Med Virol 2020;92:2379–80. Epub 2020/06/03. 10.1002/jmv.26067. PubMed PMID: 32458475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Peck KM, Lauring AS, Sullivan CS.. Complexities of viral mutation rates. J Virol 2018;92:e01031–17. 10.1128/JVI.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Domingo-Calap P, Schubert B, Joly M. et al. An unusually high substitution rate in transplant-associated BK polyomavirus in vivo is further concentrated in HLA-C-bound viral peptides. PLoS Pathog 2018;14:18. 10.1371/journal.ppat.1007368. PubMed PMID: WOS:000448978700042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Holmes EC, Dudas G, Rambaut A. et al. The evolution of Ebola virus: Insights from the 2013–2016 epidemic. Nature 2016;538:193–200. 10.1038/nature19790. PubMed PMID: WOS:000386671000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Robson F, Khan KS, Le TK. et al. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol Cell 2020;79:710–27. 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. van Dorp L, Tan CC, Lam SD. et al. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv. 2020:2020.11.16.384743. 10.1101/2020.11.16.384743. [DOI] [Google Scholar]
- 157. Di Giorgio S, Martignano F, Torcia MG. et al. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. bioRxiv. 2020:2020.03.02.973255. 10.1101/2020.03.02.973255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Simmonds P, Schwemmle M.. Rampant C->U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: Causes and consequences for their short- and long-term evolutionary trajectories. mSphere 2020;5:e00408–20. 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Graudenzi A, Maspero D, Angaroni F. et al. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. iScience 2021;24:102116. 10.1016/j.isci.2021.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Mourier T, Sadykov M, Carr MJ. et al. Host-directed editing of the SARS-CoV-2 genome. Biochem Biophys Res Commun 2021;538:35–9. 10.1016/j.bbrc.2020.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Bedford T, Hodcroft EB, Neher RA.. Updated Nextstrain SARS-CoV-2 clade naming strategy 2021. https://nextstrain.org/blog/2021-01-06-updated-SARS-CoV-2-clade-naming.
- 162. Rambaut A, Holmes EC, Hill V. et al. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv. 2020:2020.04.17.046086. 10.1101/2020.04.17.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Frost SDW, Magalis BR, Kosakovsky Pond SL.. Neutral theory and rapidly evolving viral pathogens. Mol Biol Evol 2018;35:1348–54. 10.1093/molbev/msy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Starr TN, Greaney AJ, Hilton SK. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020;182:1295–310.e20. 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Teruel N, Mailhot O, Najmanovich RJ.. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 spike protein variants. PLoS Comput Biol 2021;17:e1009286. 10.1371/journal.pcbi.1009286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Li Q, Wu J, Nie J. et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020;182:1284–94.e9. Epub 2020/07/17. 10.1016/j.cell.2020.07.012. PubMed PMID: 32730807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Rochman ND, Wolf YI, Faure G. et al. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc Natl Acad Sci USA 2021;118:e2104241118. 10.1073/pnas.2104241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Corey L, Beyrer C, Cohen MS. et al. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med 2021;385:562–6. 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Baang JH, Smith C, Mirabelli C. et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2020;223:23–7. 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kemp SA, Collier DA, Datir RP. et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021;592:277–82. 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Cao Y, Wang J, Jian F. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022;602:657–63. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Naranbhai V, Nathan A, Kaseke C. et al. T cell reactivity to the SARS-CoV-2 omicron variant is preserved in most but not all individuals. Cell 2022;185:1041–51.e6. 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Spinicci M, Mazzoni A, Borchi B. et al. AIDS patient with severe T cell depletion achieved control but not clearance of SARS-CoV-2 infection. Eur J Immunol 2022;52:352–5. Epub 2021/11/26. 10.1002/eji.202149574. PubMed PMID: 34822185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Hon C-C, Lam T-Y, Shi Z-L. et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol 2008;82:1819–26. 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hu B, Zeng L-P, Yang X-L. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 2017;13:e1006698. 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lau SKP, Li KSM, Huang Y. et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol 2010;84:2808–19. 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Corman VM, Ithete NL, Richards LR. et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 2014;88:11297–303. Epub 2014/07/18. 10.1128/jvi.01498-14. PubMed PMID: 25031349; PubMed Central PMCID: PMCPMC4178802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Andersen KG, Rambaut A, Lipkin WI. et al. The proximal origin of SARS-CoV-2. Nat Med 2020;26:450–2. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Boni MF, Lemey P, Jiang X. et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 2020;5:1408–17. 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 180. Pérez-Losada M, Arenas M, Galán JC. et al. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol 2015;30:296–307. 10.1016/j.meegid.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Nelson MI, Viboud C, Simonsen L. et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 2008;4:e1000012. 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Smith G, Vijaykrishna D, Bahl J. et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009;459:1122–5. PubMed PMID: citeulike:4832143. [DOI] [PubMed] [Google Scholar]
- 183. Lythgoe KA, Hall M, Ferretti L. et al. SARS-CoV-2 within-host diversity and transmission. Science 2021;372:eabg0821. 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. VanInsberghe D, Neish AS, Lowen AC. et al. Recombinant SARS-CoV-2 genomes are currently circulating at low levels. bioRxiv. 2021:2020.08.05.238386. 10.1101/2020.08.05.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Gutierrez B, Castelan HG, da Silva Candido D. et al. Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America. medRxiv. 2021:2021.11.19.21266601. 10.1101/2021.11.19.21266601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Colson P, Delerce J, Burel E. et al. Emergence in Southern France of a new SARS-CoV-2 variant of probably Cameroonian origin harbouring both substitutions N501Y and E484K in the spike protein. medRxiv. 2021:2021.12.24.21268174. 10.1101/2021.12.24.21268174. [DOI] [Google Scholar]
- 187. Lacek KA, Rambo-Martin BL, Batra D. et al. Identification of a novel SARS-CoV-2 delta-omicron recombinant virus in the United States. bioRxiv. 2022:2022.03.19.484981. 10.1101/2022.03.19.484981. [DOI] [Google Scholar]
- 188. Jackson B, Boni MF, Bull MJ. et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell 2021;184:5179–88.e8. 10.1016/j.cell.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Lau SKP, Lee P, Tsang AKL. et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol 2011;85(21):11325–37. 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kin N, Miszczak F, Lin W. et al. Genomic analysis of 15 human coronaviruses OC43 (HCoV-OC43s) circulating in France from 2001 to 2013 reveals a high intra-specific diversity with new recombinant genotypes. Viruses 2015;7:2358–77. 10.3390/v7052358. PubMed PMID: 26008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Pyrc K, Dijkman R, Deng L. et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 2006;364:964–73. 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Woo PCY, Lau SKP, Yip CCY. et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol 2006;80:7136–45. 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Terada Y, Matsui N, Noguchi K. et al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE 2014;9:e106534. 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Decaro N, Mari V, Campolo M. et al. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J Virol 2009;83:1532–7. 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Tian P-F, Jin Y-L, Xing G. et al. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis 2014;20:1735–8. 10.3201/eid2010.140338. PubMed PMID: 25272273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Herrewegh AA, Smeenk I, Horzinek MC. et al. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol 1998;72:4508–14. Epub 1998/04/29. 10.1128/jvi.72.5.4508-4514.1998. PubMed PMID: 9557750; PubMed Central PMCID: PMCPMC109693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Lai MM, Baric RS, Makino S. et al. Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol 1985;56:449–56. Epub 1985/11/01. 10.1128/jvi.56.2.449-456.1985. PubMed PMID: 2997467; PubMed Central PMCID: PMCPMC252599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Makino S, Keck JG, Stohlman SA. et al. High-frequency RNA recombination of murine coronaviruses. J Virol 1986;57:729–37. Epub 1986/03/01. 10.1128/jvi.57.3.729-737.1986. PubMed PMID: 3005623; PubMed Central PMCID: PMCPMC252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Keck JG, Matsushima GK, Makino S. et al. In vivo RNA–RNA recombination of coronavirus in mouse brain. J Virol 1988;62:1810–3. Epub 1988/05/01. 10.1128/jvi.62.5.1810-1813.1988. PubMed PMID: 2833625; PubMed Central PMCID: PMCPMC253235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Darwin C. On the Origin of Species by Means of Natural Selection, or, the Preservation of Favoured Races in the Struggle for Life. London, UK: John Murray, 1859. [PMC free article] [PubMed] [Google Scholar]
- 201. Obermeyer F, Schaffner SF, Jankowiak M. et al. Analysis of 2.1 million SARS-CoV-2 genomes identifies mutations associated with transmissibility. medRxiv. 2021:2021.09.07.21263228. 10.1101/2021.09.07.21263228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. De Maio N, Walker CR, Turakhia Y. et al. Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol Evol 2021;13. 10.1093/gbe/evab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. McCarthy KR, Rennick LJ, Nambulli S. et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021;371:1139–42. 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Chan KK, Tan TJC, Narayanan KK. et al. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci Adv 2021;7:eabf1738. 10.1126/sciadv.abf1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Lista MJ, Winstone H, Wilson HD. et al. The P681H mutation in the Spike glycoprotein confers Type I interferon resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. bioRxiv. 2021:2021.11.09.467693. 10.1101/2021.11.09.467693. [DOI] [Google Scholar]
- 206. Syed AM, Taha TY, Tabata T. et al. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science 2021;374:1626–32. 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Anderson RM, May RM.. Infectious Diseases of Humans. Dynamics and Control. Oxford: Oxford University Press, 1991. [Google Scholar]
- 208. Balloux F, van Dorp L.. Q&A: What are pathogens, and what have they done to and for us? BMC Biol 2017;15:6. 10.1186/s12915-017-0433-z. PubMed PMID: WOS:000413251900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Kerr PJ, Liu J, Cattadori I. et al. Myxoma virus and the leporipoxviruses: An evolutionary paradigm. Viruses Basel 2015;7:1020–61. 10.3390/v7031020. PubMed PMID: WOS:000351943000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Qiu X, Nergiz AI, Maraolo AE. et al. The role of asymptomatic and pre-symptomatic infection in SARS-CoV-2 transmission—a living systematic review. Clin Microbiol Infect 2021;27:511–9. 10.1016/j.cmi.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Bager P, Wohlfahrt J, Rasmussen M. et al. Hospitalisation associated with SARS-CoV-2 delta variant in Denmark. Lancet Infect Dis 2021;21:1351. 10.1016/S1473-3099(21)00580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Graham MS, Sudre CH, May A. et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Pub Health 2021;6:e335–e45. 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Nyberg T, Twohig KA, Harris RJ. et al. Risk of hospital admission for patients with SARS-CoV-2 variant B.1.1.7: cohort analysis. BMJ 2021;373:n1412. 10.1136/bmj.n1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Twohig KA, Nyberg T, Zaidi A. et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22:35–42. 10.1016/S1473-3099(21)00475-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Nyberg T, Ferguson NM, Nash SG. et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399:1303–12. 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Willett BJ, Grove J, MacLean OA. et al. The hyper-transmissible SARS-CoV-2 omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. 2022:2022.01.03.21268111. 10.1101/2022.01.03.21268111. [DOI] [Google Scholar]
- 217. Meng B, Abdullahi A, Ferreira IATM. et al. Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts infectivity and fusogenicity. Nature 2022;603:706–14. 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Hui KPY, Ho JCW, Cheung M-C. et al. SARS-CoV-2 omicron variant replication in human bronchus and lung ex vivo. Nature 2022;603:715–20. 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 219. Sigal A. Milder disease with omicron: is it the virus or the pre-existing immunity? Nat Rev Immunol 2022;22:69–71. 10.1038/s41577-022-00678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Goga A, Bekker L-G, Garrett N. et al. Breakthrough Covid-19 infections during periods of circulating beta, delta and omicron variants of concern, among health care workers in the Sisonke Ad26.COV2.S vaccine trial, South Africa. medRxiv. 2021:2021.12.21.21268171. 10.1101/2021.12.21.21268171. [DOI] [Google Scholar]
- 221. Kaplonek P, Fischinger S, Cizmeci D. et al. mRNA-1273 vaccine-induced antibodies maintain Fc effector functions across SARS-CoV-2 variants of concern. Immunity 2022;55:355–65.e4. Epub 2022/01/30. 10.1016/j.immuni.2022.01.001. PubMed PMID: 35090580; PubMed Central PMCID: PMCPMC8733218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Tai CG, Maragakis LL, Connolly S. et al. Booster protection against omicron infection in a highly vaccinated cohort. medRxiv. 2022:2022.02.24.22271347. 10.1101/2022.02.24.22271347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Cele S, Jackson L, Khoury DS. et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022;602:654–6. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Rössler A, Riepler L, Bante D. et al. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent persons. N Engl J Med 2022;386:698–700. 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Reynolds CJ, Pade C, Gibbons JM. et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science 2021. Epub 2021/05/02. 10.1126/science.abh1282. PubMed PMID: 33931567; PubMed Central PMCID: PMCPMC8168614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Dan JM, Mateus J, Kato Y. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021;371. Epub 2021/01/08. 10.1126/science.abf4063. PubMed PMID: 33408181; PubMed Central PMCID: PMCPMC7919858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Le Bert N, Tan AT, Kunasegaran K. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020;584:457–62. 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 228. Hansen CH, Michlmayr D, Gubbels SM. et al. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet 2021;397:1204–12. 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Lumley SF, O’Donnell D, Stoesser NE. et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med 2020;384:533–40. 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Townsend JP, Hassler HB, Wang Z. et al. The durability of immunity against reinfection by SARS-CoV-2: A comparative evolutionary study. Lancet Microbe 2021;2:e666–e75. 10.1016/S2666-5247(21)00219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. McDonald I, Murray SM, Reynolds CJ. et al. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. NPJ Vaccines 2021;6:74. 10.1038/s41541-021-00336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Sette A, Crotty S.. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021;184:861–80. Epub 2021/01/27. 10.1016/j.cell.2021.01.007. PubMed PMID: 33497610; PubMed Central PMCID: PMCPMC7803150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Bertoletti A, Tan AT, Le Bert N.. The T-cell response to SARS-CoV-2: Kinetic and quantitative aspects and the case for their protective role. Oxford Open Immunol 2021;2. 10.1093/oxfimm/iqab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Manisty C, Treibel TA, Jensen M. et al. Time series analysis and mechanistic modelling of heterogeneity and sero-reversion in antibody responses to mild SARS-CoV-2 infection. EBioMedicine 2021;65:103259. Epub 2021/03/05. 10.1016/j.ebiom.2021.103259. PubMed PMID: 33662833; PubMed Central PMCID: PMCPMC7920816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Bilich T, Nelde A, Heitmann JS. et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci Transl Med 2021;13:eabf7517. 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Mizrahi B, Shilo S, Rossman H. et al. Longitudinal symptom dynamics of COVID-19 infection. Nat Commun 2020;11:6208. 10.1038/s41467-020-20053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Goel RR, Apostolidis SA, Painter MM. et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol 2021;6. Epub 2021/04/17. 10.1126/sciimmunol.abi6950. PubMed PMID: 33858945; PubMed Central PMCID: PMCPMC8158969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Jeffery-Smith A, Burton AR, Lens S. et al. SARS-CoV-2-specific memory B cells can persist in the elderly who have lost detectable neutralizing antibodies. J Clin Invest 2022;132. 10.1172/JCI152042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Harvey WT, Carabelli AM, Jackson B. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021;19:409–24. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Rees-Spear C, Muir L, Griffith SA. et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep 2021;34:108890. Epub 2021/03/14. 10.1016/j.celrep.2021.108890. PubMed PMID: 33713594; PubMed Central PMCID: PMCPMC7936541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Dejnirattisai W, Huo J, Zhou D. et al. SARS-CoV-2 omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022;185:467–84.e15. Epub 2022/01/27. 10.1016/j.cell.2021.12.046. PubMed PMID: 35081335; PubMed Central PMCID: PMCPMC8723827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Wang P, Nair MS, Liu L. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021;593:130–5. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 243. Wu K, Werner AP, Koch M. et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N Engl J Med 2021;384:1468–70. 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Mlcochova P, Kemp SA, Dhar MS. et al. SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature 2021;599:114–9. 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Edara V-V, Pinsky BA, Suthar MS. et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N Engl J Med 2021;385:664–6. 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Planas D, Saunders N, Maes P. et al. Considerable escape of SARS-CoV-2 omicron to antibody neutralization. Nature 2022;602:671–5. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 247. Carreño JM, Alshammary H, Singh G. et al. Evidence for retained spike-binding and neutralizing activity against emerging SARS-CoV-2 variants in serum of COVID-19 mRNA vaccine recipients. EBioMedicine 2021;73. 10.1016/j.ebiom.2021.103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. UKHSA. SARS-CoV-2 variants of concern and variants under investigation in England. Technical Briefing 28, 2021.
- 249. Dolton G, Rius C, Hasan MS. et al. Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope. medRxiv. 2021:2021.06.21.21259010. 10.1101/2021.06.21.21259010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Agerer B, Koblischke M, Gudipati V. et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci Immunol 2021;6. Epub 2021/03/06. 10.1126/sciimmunol.abg6461. PubMed PMID: 33664060; PubMed Central PMCID: PMCPMC8224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. de Silva TI, Liu G, Lindsey BB. et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience 2021;24:103353–. Epub 2021/10/28. 10.1016/j.isci.2021.103353. PubMed PMID: 34729465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Skelly DT, Harding AC, Gilbert-Jaramillo J. et al. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat Commun 2021;12:5061. 10.1038/s41467-021-25167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Gao Y, Cai C, Grifoni A. et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med 2022;28:472–6. 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Tarke A, Coelho CH, Zhang Z. et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from alpha to omicron. Cell 2022;185:847–59.e11. 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Tarke A, Sidney J, Methot N. et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021;2:100355. Epub 2021/07/08. 10.1016/j.xcrm.2021.100355. PubMed PMID: 34230917; PubMed Central PMCID: PMCPMC8249675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Ferretti AP, Kula T, Wang Y. et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity 2020;53:1095–107.e3. 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Saini SK, Hersby DS, Tamhane T. et al. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci Immunol 2021;6:eabf7550. 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Grifoni A, Sidney J, Zhang Y. et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020;27:671–80.e2. 10.1016/j.chom.2020.03.002. PubMed PMID: 32183941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Sahin U, Muik A, Derhovanessian E. et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020;586:594–9. 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 260. Folegatti PM, Ewer KJ, Aley PK. et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020;396:467–78. 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Jackson LA, Anderson EJ, Rouphael NG. et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med 2020;383:1920–31. 10.1056/NEJMoa2022483. PubMed PMID: 32663912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Swadling L, Diniz MO, Schmidt NM. et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022;601:110–7. 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Mateus J, Grifoni A, Tarke A. et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020;370:89–94. Epub 2020/08/06. 10.1126/science.abd3871. PubMed PMID: 32753554; PubMed Central PMCID: PMCPMC7574914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Nelde A, Bilich T, Heitmann JS. et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol 2021;22:74–85. 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 265. Braun J, Loyal L, Frentsch M. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020;587:270–4. 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 266. Loyal L, Braun J, Henze L. et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science 2021;374:eabh1823. 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Low JS, Vaqueirinho D, Mele F. et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science 2021;372:1336–41. Epub 2021/05/20. 10.1126/science.abg8985. PubMed PMID: 34006597; PubMed Central PMCID: PMCPMC8168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Woldemeskel BA, Garliss CC, Blankson JN.. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J Clin Invest 2021;131. 10.1172/JCI149335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Francis JM, Leistritz-Edwards D, Dunn A. et al. Allelic variation in class I HLA determines CD8+ T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci Immunol 2022;7:eabk3070. 10.1126/sciimmunol.abk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Kundu R, Narean JS, Wang L. et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun 2022;13:80. 10.1038/s41467-021-27674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Mallajosyula V, Ganjavi C, Chakraborty S. et al. CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in patients with COVID-19. Sci Immunol 2021;6:eabg5669. 10.1126/sciimmunol.abg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Sagar M, Reifler K, Rossi M. et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest 2021;131. 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Ringlander J, Martner A, Nilsson S. et al. Verified infections with endemic common cold coronaviruses do not entail significant protection against SARS-CoV-2. J Infect Dis 2021;223:131–2. 10.1093/infdis/jiab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Ng KW, Faulkner N, Cornish GH. et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020;370:1339–43. Epub 2020/11/08. 10.1126/science.abe1107. PubMed PMID: 33159009; PubMed Central PMCID: PMCPMC7857411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Shrock E, Fujimura E, Kula T. et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020;370:eabd4250. 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Anderson EM, Goodwin EC, Verma A. et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 2021;184:1858–64.e10. Epub 2021/02/26. 10.1016/j.cell.2021.02.010. PubMed PMID: 33631096; PubMed Central PMCID: PMCPMC7871851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Rice BL, Douek DC, McDermott AB. et al. Why are there so few (or so many) circulating coronaviruses? Trends Immunol 2021;42:751–63. 10.1016/j.it.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Khoury DS, Cromer D, Reynaldi A. et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021;27:1205–11. 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 279. Feng S, Phillips DJ, White T. et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med 2021;27:2032–40. 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Mudgal R, Nehul S, Tomar S.. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum Vaccin Immunother 2020;16:2921–31. Epub 2020/09/16. 10.1080/21645515.2020.1805992. PubMed PMID: 32931361; PubMed Central PMCID: PMCPMC7544966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Poon MML, Rybkina K, Kato Y. et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci Immunol 2021;6:eabl9105. 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Niessl J, Sekine T, Lange J. et al. Identification of resident memory CD8+ T cells with functional specificity for SARS-CoV-2 in unexposed oropharyngeal lymphoid tissue. Sci Immunol 2021;6:eabk0894. 10.1126/sciimmunol.abk0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Grau-Expósito J, Sánchez-Gaona N, Massana N. et al. Peripheral and lung resident memory T cell responses against SARS-CoV-2. Nat Commun 2021;12:3010. Epub 2021/05/23. 10.1038/s41467-021-23333-3. PubMed PMID: 34021148; PubMed Central PMCID: PMCPMC8140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Allie SR, Bradley JE, Mudunuru U. et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol 2019;20:97–108. Epub 2018/12/05. 10.1038/s41590-018-0260-6. PubMed PMID: 30510223; PubMed Central PMCID: PMCPMC6392030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Mao T, Israelow B, Suberi A. et al. Unadjuvanted intranasal spike vaccine booster elicits robust protective mucosal immunity against sarbecoviruses. bioRxiv. 2022:2022.01.24.477597. 10.1101/2022.01.24.477597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Terreri S, Piano Mortari E, Vinci MR. et al. Persistent B cell memory after SARS-CoV-2 vaccination is functional during breakthrough infections. Cell Host Microbe 2022;30:400–8.e4. 10.1016/j.chom.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. King A. Vaccines beyond antibodies. EMBO Rep 2021;22:e54073. 10.15252/embr.202154073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Rappuoli R, Aderem A.. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature 2011;473:463–9. 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- 289. Morris DH, Yinda KC, Gamble A. et al. Mechanistic theory predicts the effects of temperature and humidity on inactivation of SARS-CoV-2 and other enveloped viruses. eLife 2021;10:e65902. 10.7554/eLife.65902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Schuit M, Ratnesar-Shumate S, Yolitz J. et al. Airborne SARS-CoV-2 is rapidly inactivated by simulated sunlight. J Infect Dis 2020;222:564–71. 10.1093/infdis/jiaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Fares A. Factors influencing the seasonal patterns of infectious diseases. Int J Prev Med 2013;4:128–32. PubMed PMID: 23543865. [PMC free article] [PubMed] [Google Scholar]
- 292. Martinez ME. The calendar of epidemics: Seasonal cycles of infectious diseases. PLoS Pathog 2018;14:e1007327. 10.1371/journal.ppat.1007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Dyrdak R, Hodcroft EB, Wahlund M. et al. Interactions between seasonal human coronaviruses and implications for the SARS-CoV-2 pandemic: A retrospective study in Stockholm, Sweden, 2009–2020. J Clin Virol 2021;136:104754. Epub 2021/02/19. 10.1016/j.jcv.2021.104754. PubMed PMID: 33601153; PubMed Central PMCID: PMCPMC7869750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Wickramasinghe INA, van Beurden SJ, Weerts EAWS. et al. The avian coronavirus spike protein. Virus Res 2014;194:37–48. Epub 2014/10/17. 10.1016/j.virusres.2014.10.009. PubMed PMID: 25451062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Rochman ND, Faure G, Wolf YI. et al. Epistasis at the SARS-CoV-2 receptor-binding domain interface and the propitiously boring implications for vaccine escape. mBio 2022;13:e00135–22. 10.1128/mbio.00135-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Starr TN, Greaney AJ, Hannon WW. et al. Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. bioRxiv. 2022:2022.02.24.481899. 10.1101/2022.02.24.481899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Hayward AC, Fragaszy EB, Bermingham A. et al. Comparative community burden and severity of seasonal and pandemic influenza: results of the Flu Watch cohort study. Lancet Respir Med 2014;2:445–54. 10.1016/S2213-2600(14)70034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Paget J, Spreeuwenberg P, Charu V. et al. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J Glob Health 2019;9:020421–. 10.7189/jogh.09.020421. PubMed PMID: 31673337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Rambo-Martin BL, Keller MW, Wilson MM. et al. Influenza A virus field surveillance at a swine–human interface. mSphere 2020;5:e00822–19. 10.1128/mSphere.00822-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Figure 4 was enabled by the GISAID EpiCoV™ database (https://www.gisaid.org) and regular Audacity releases. We gratefully acknowledge all authors of the data used herein, and their originating and submitting laboratories. Detailed acknowledgments are available at https://www.gisaid.org using the Data Acknowledgement Locator with the GISAID EPI_SET_20220509xa accession number.