Abstract
We isolated a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) BA.2 variant from a person with coronavirus disease 2019 recrudescence after nirmatrelvir/ritonavir treatment. Antiviral sensitivity and neutralizing antibody testing were performed with both parental SARS-CoV-2 and multiple variants of concern. We found that neither nirmatrelvir resistance nor absence of neutralizing immunity was a likely cause of the recrudescence.
Keywords: COVID-19, treatment rebound, COVID-19 recrudescence, nirmatrelvir, Omicron
Early administration of the oral protease inhibitor nirmatrelvir combined with ritonavir (NM/r) (Paxlovid) can reduce severe disease due to coronavirus disease 2019 (COVID-19) [1]. Nirmatrelvir inhibits the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease, thus blocking viral replication, but virologic and symptomatic rebound after NM/r treatment was recently reported [2]. We evaluated if NM resistance or impaired humoral immunity contributed to a case of COVID-19 recrudescence after NM/r treatment.
RESULTS
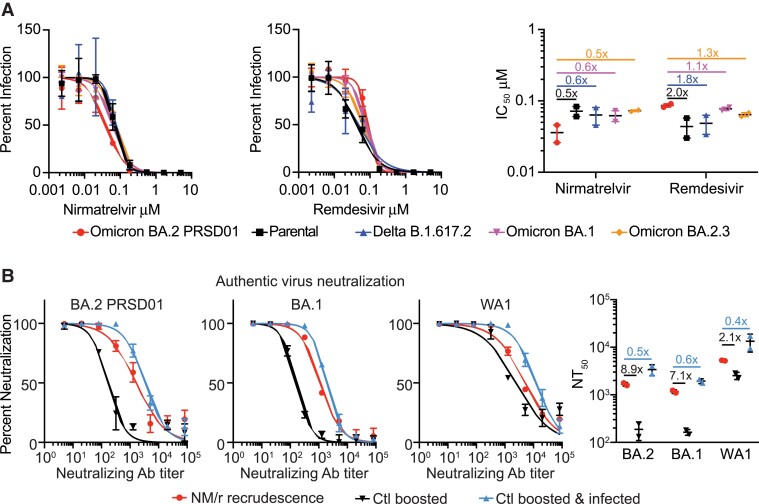
Three boosted adult travelers acquired COVID-19 after returning to the United States from South Africa and were treated with NM/r 300/100 mg taken orally twice daily for 5 days (Supplementary Table 1). All cases resolved quickly except in 1 traveler who experienced initial improvement with NM/r followed by rebounding symptoms. Progressively worsening fatigue, sore throat, nasal congestion, and diarrhea were associated with high viral shedding (cycle threshold = 21.7, FluxErgy, Inc) and culturable virus 5 days after the NM/r course. We isolated SARS-CoV-2 from a nasopharyngeal swab following development of worsening symptoms (Supplementary Methods). SARS-CoV-2 full genome sequences isolated from the nasopharyngeal swab sample and culture isolate were assigned as SARS-CoV2 BA.2 lineage (Phylogenetic Assignment of Named Global Outbreak) strain PRSD01 [3]. Sequence comparison to the BA.2 reference showed no amino acid differences in any coding region, including ORF1a and spike protein. Phenotypic analyses of the antiviral susceptibility of BA.2 PRSD01 (isolate), WA1/2020 (parental), B.1.617.2 (Delta), and BA.1 and BA.2.3 variants to NM and remdesivir were conducted in Calu-3 human lung epithelial cells. The half-maximal inhibitory concentrations (IC50) of NM against BA.2 PRSD01 were 2.0-, 1.8-, 1.7-, and 2.0-fold lower than the parental, Delta, BA.1, and BA.2.3 strains, respectively (Figure 1A). As a control, we determined the IC50 for remdesivir, a drug to which this individual was not exposed, for each strain. The remdesivir IC50 for BA.2 PRSD01 was 2.0, 1.8, 1.1, and 1.3 times higher than the parental, Delta, BA.1, and BA.2.3 strains, respectively (Figure 1A).
Figure 1.
Comparison of antiviral activity (A) and neutralization (B) against BA.2 PRSD01 after nirmatrelvir combined with ritonavir recrudescence. Dose response curves, half-maximal inhibitory concentrations, and half-maximal authentic virus-neutralizing antibody concentrations show the averages ± standard deviation from 2 independent experiments with 2 biological replicates. Abbreviations: Ab, antibody; Ctl, control; IC50, half-maximal inhibitory concentration; NM/r, nirmatrelvir combined with ritonavir; NT50, half-maximal authentic virus-neutralizing antibody concentration.
We next evaluated the susceptibility of the viral panel to the neutralizing antibody response in the plasma from the patient and 2 controls. While both controls were fully vaccinated and boosted with the BNT162b2 messenger RNA vaccine (Pfizer-BioNTech), one had been infected with and recovered from a SARS-CoV-2 infection with symptom onset 3 days prior to the individual who experienced NM/r recrudescence (Supplementary Table 1). The average half-maximal authentic virus-neutralizing antibody concentrations against BA.2 PRSD01, BA.1, and parental were 1668, 1170, and 5239, respectively, in the NM/r-treated patient. These levels were 8.9, 7.1, and 2.1 times higher than the boosted control but 2.0, 1.7, and 2.6 times lower than the boosted and infected control (Figure 1B).
DISCUSSION
This study suggests that neither development of NM resistance nor absence of neutralizing antibody were likely causes of the observed recrudescence. The phenomenon of relapse after NM/r has been described in a limited number of individuals, all of whom developed virologic rebound approximately 9–14 days after symptom onset and were likely infected with Omicron variants [2]. A small percentage of both NM/r- and placebo-treated individuals enrolled in the primary NM/r study, Evaluation of Protease Inhibition for COVID-19 in High-Risk Patients (EPIC-HR), which was mostly conducted before Omicron emergence, also experienced increasing SARS-CoV-2 RNA levels after stopping NM/r treatment at day 10 or 14 after infection [1, 4].
Our in vitro NM antiviral testing as well as sequencing, from our and other studies [2, 3], have not yet identified emergence of NM resistance. Since immune responses likely contribute to eradication of replication-competent virus during NM/r treatment, we evaluated neutralizing antibody responses but found no evidence of an impaired neutralizing antibody response in this immunocompetent individual, suggesting that impaired humoral immunity is not required for NM/r recrudescence. Although we were unable to measure T-cell responses or serum drug concentrations, we believe the most likely possibility for the observed recrudescence is insufficient drug exposure by individual pharmacokinetics or insufficient duration. Furthermore, the clinical significance of COVID-19 recrudescence after treatment remains unclear, so additional studies are needed to define the etiology, frequency and clinical consequences (eg, hospitalizations, deaths, transmissions) of such recrudescence.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the University of California, San Diego Center for Advanced Laboratory Medicine Microbiology Laboratory and EXCITE laboratory for assistance in SARS-CoV-2 isolation, and John Ayers for insightful discussions. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52281. The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate hCoV-19/USA/PHC658/2021 (lineage B.1.617.2; Delta variant), NR-55611, contributed by Dr Richard Webby and Dr Anami Patel.
Financial support. This work was supported by the NIH (grant numbers AI036214 and AI131385 to D. M. S.; CA177322, DA039562, DA046171, and AI125103 to T. M. R.); the San Diego Center for AIDS Research (grant number AI100665); the Department of Veterans Affairs; the John and Mary Tu Foundation; and the James B. Pendleton Charitable Trust. A. F. C. was supported by the NIH (grant number K08 AI130381) and a Career Award for Medical Scientists from the Burroughs Wellcome Fund. A. C. was supported by the NIH (grant numbers DA049644, AI145555, MH128153, AI106039, and DP2 CA051915).
Potential conflicts of interest. A. F. C. has received contract payments from Nurix Therapeutics and has options in Covicept Therapeutics. D. M. S. has served as a consultant for Bayer Healthcare, Kiadis Pharmaceuticals, and Signant Health, and has equity stake in Vx Biosciences, Model Medicines, Linear Therapies, and FluxErgy. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Supplementary Material
Contributor Information
Aaron F Carlin, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA.
Alex E Clark, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA.
Antoine Chaillon, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA.
Aaron F Garretson, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA.
William Bray, Department of Pediatrics, University of California, San Diego School of Medicine, La Jolla, California, USA.
Magali Porrachia, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA.
AsherLev T Santos, Department of Public Health, College of Education, Health and Human Services, California State University San Marcos, San Marcos, California, USA.
Tariq M Rana, Division of Genetics, Department of Pediatrics, Program in Immunology, Institute for Genomic Medicine, UCSD Center for AIDS Research, University of California, San Diego, La Jolla, California, USA.
Davey M Smith, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California, USA; Veterans Affairs San Diego Healthcare System, San Diego, California, USA.
References
- 1. Hammond J, Leister-Tebbe H, Gardner A, et al. . EPIC-HR Investigators. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med 2022; 386:1397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charness M, Gupta K, Stack G, et al. . Rapid relapse of symptomatic Omicron SARS-CoV-2 infection following early suppression with nirmatrelvir/ritonavir. BioRxiv 1588371 [Preprint]. May 23, 2022. Available at: 10.21203/rs.3.rs-1588371/v3. [DOI]
- 3. O’Toole A, Scher E, Underwood A, et al. . Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 2021; 7; veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. US Food and Drug Administration, Center for Drug Evaluation and Research. Emergency use authorization (EUA) for Paxlovid (nirmatrelvir tablets copackaged with ritonavir tablets). 2021. Available at: https://www.fda.gov/media/155194/download. Accessed 31 May 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.