Abstract
Background
Comprehensive pathogen genomic surveillance represents a powerful tool to complement and advance precision vaccinology. The emergence of the Alpha variant in December 2020 and the resulting efforts to track the spread of this and other severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern led to an expansion of genomic sequencing activities in Germany.
Methods
At Robert Koch Institute (RKI), the German National Institute of Public Health, we established the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) network to perform SARS-CoV-2 genomic surveillance at the national scale, SARS-CoV-2–positive samples from laboratories distributed across Germany regularly undergo whole-genome sequencing at RKI.
Results
We report analyses of 3623 SARS-CoV-2 genomes collected between December 2020 and December 2021, of which 3282 were randomly sampled. All variants of concern were identified in the sequenced sample set, at ratios equivalent to those in the 100-fold larger German GISAID sequence dataset from the same time period. Phylogenetic analysis confirmed variant assignments. Multiple mutations of concern emerged during the observation period. To model vaccine effectiveness in vitro, we employed authentic-virus neutralization assays, confirming that both the Beta and Zeta variants are capable of immune evasion. The IMS-SC2 sequence dataset facilitated an estimate of the SARS-CoV-2 incidence based on genetic evolution rates. Together with modeled vaccine efficacies, Delta-specific incidence estimation indicated that the German vaccination campaign contributed substantially to a deceleration of the nascent German Delta wave.
Conclusions
SARS-CoV-2 molecular and genomic surveillance may inform public health policies including vaccination strategies and enable a proactive approach to controlling coronavirus disease 2019 spread as the virus evolves.
Keywords: SARS-CoV-2, pathogen genomics, molecular surveillance, variant of concern, incidence estimation
For integrated molecular surveillance of SARS-CoV-2, a robust, medium-scale national surveillance system was established. It enables identification of variants and mutations of concern, vaccine efficacy assessment in vitro, and genome-based incidence estimates, thus informing public health policies, including vaccination strategies.
At the beginning of the third year of the coronavirus disease 2019 (COVID-19) pandemic, it is becoming evident that its causative agent, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), will not be eradicated. Reasons include viral persistence in some individuals [1–3]; multiple indications that zoonotic reservoirs can be, or have already been established [4–8]; and the emergence of variants of concern (VOCs), SARS-CoV-2 strains that have evolved to, for instance, transmit more effectively [9–11]. Thus, it seems that SARS-CoV-2 may continue to circulate for decades to come. Although the burden on the acute healthcare sector may lessen over time, there is considerable evidence that SARS-CoV-2 is more virulent than other respiratory viruses, including influenza viruses [12–14]; moreover, SARS-CoV-2 displays broad tissue tropism, frequently causing a multisystemic illness that is not limited to the respiratory tract and may have long-term consequences [15–18]. Therefore, the effective use of COVID-19 vaccines may remain a long-term public health imperative. While postpandemic circulation patterns cannot be predicted with absolute certainty, once high population immunity is achieved, COVID-19 could become an endemic disease with high impact and recurrent epidemic peaks [19, 20]. Depending on the quality and the duration of immune protection, these epidemic peaks may occur regularly during the winter season, similar to influenza. Additionally, they might be triggered, for example, by zoonotic reintroductions or the emergence of novel VOCs capable of immune escape [19, 20].
Pathogen surveillance can substantially contribute to maximizing vaccine effectiveness at the population level. Relevant factors include not only the timing of epidemic peaks but also pathogen genetic variation: For example, the Omicron surge of the COVID-19 pandemic is caused by a genetically distinct SARS-CoV-2 variant with pronounced immune-evasive properties [21–23]. The Omicron variant has a substantial transmission advantage and has therefore prompted the development of novel Omicron-specific vaccines. Given the severity and anticipated long-term epidemiological persistence of COVID-19, as well as SARS-CoV-2’s ability to evolve and adapt to immune selection pressure, there is an urgent need to establish adequate and robust surveillance instruments for this virus. Among these instruments, pathogen molecular and genomic surveillance, which also allow the tracking of viral evolution, may serve as powerful tools to complement and advance precision vaccinology.
In response to the emergence of the Alpha VOC in Europe in late 2020, SARS-CoV-2 sequencing efforts in Germany were ramped up substantially by a statutory order for diagnostic laboratories to submit viral genome information from a fraction of positive diagnostic specimens to a central hub [24] against financial compensation. Since then, a vast amount of complete genome sequences has been deposited in public repositories such as the GISAID database [25]. However, such large-scale, resource-intensive efforts will become difficult to sustain, especially in the postpandemic period. Thus, it is crucial to establish molecular surveillance approaches that are not only informative but also cost-effective.
In 2020, Robert Koch Institute (RKI, the German National Institute of Public Health) established the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) network with the explicit goal to complement the existing epidemiological surveillance with a national-scale, representative SARS-CoV-2 genomic surveillance that is also resource-efficient. Here, we present the results of 13 months of SARS-CoV-2 molecular and genomic surveillance in Germany, spanning the time period from the first emergence of Alpha to the rise of Omicron. We also demonstrate how genomic surveillance as part of a comprehensive surveillance system can be used as an unbiased and independent data source that allows researchers not only to gauge the overall dynamics of SARS-CoV-2 spread, but also to retrospectively assess the impact of the vaccination campaign on the pandemic trajectory of the Delta VOC in Germany.
METHODS
Sample Selection
IMS-SC2 network laboratories are distributed across Germany and include large regional laboratories involved in routine testing of samples from ambulatory healthcare centers and hospitals, academic diagnostic laboratories, and 2 public health laboratories (1 running diagnostics for local health departments and another running the national sentinel for acute respiratory infections [26]). The Supplementary Methods provide details on sampling approaches.
Ethics Statement
Assessment of vaccine-induced humoral immunity via neutralization assays was approved by the ethics committee of the Hesse Medical Association (2020-1664_2-evBO) and written informed consent was obtained from all participants. All investigations were conducted according to the principles laid down in the Helsinki Declaration. All analyses were based on anonymized data.
RNA Extraction, Sequencing, and Genome Reconstruction
At RKI, total RNA was extracted using established protocols [26, 27] as described in the Supplementary Methods, which also provide details on the sequencing and genome reconstruction procedures used.
Plaque Reduction Neutralization Test
The plaque reduction neutralization test was performed in duplicate in the biosafety level 3 (BSL-3) facility at RKI as described elsewhere [28] and outlined in the Supplementary Methods.
Sequence Data Set Preparation
The IMS-SC2 data set was prepared for downstream analysis employing poreCov v0.11.7 [29], Pangolin v3.1.17 [30], and covSonar v1.1.0 (https://gitlab.com/s.fuchs/covsonar) and restricted to the time period 1 December 2020 until 31 December 2021, as outlined in the Supplementary Methods, which also explains how the German GISAID dataset was prepared.
Geographical Distribution Analysis
A crucial component of national-level molecular surveillance is collecting a steady flow of samples that approximates the total population of a pathogen, which renders key importance to adequate geographical coverage. Therefore, we conducted geographical distribution analysis as described in the Supplementary Methods.
Phylogenetic Tree Inference and Data Visualization
Phylogenetic tree inference and data visualization procedures are explained in detail in the Supplementary Methods. In brief, phylogenetic analysis was performed on the randomly sampled IMS-SC2 dataset employing MAFFT [31], IQ-TREE v2.1.4-beta [32], 1000 ultra-fast bootstrap replicates [33], and Iroki [34] for visualization; Sankey and UpSet graphs were generated using a custom script (https://github.com/hoelzer/sankey) inspired by the Pavian package [35] and the UpSetR package [36], respectively.
Testing Statistical Increase for a VOC Count
Significant increase in a VOC proportion was assessed using a Fisher exact test on sample counts between 2 successive calendar weeks (as detailed in [37]).
Genome-Based Incidence Estimation and Case Ascertainment
We used the newly established genome-based incidence estimation pipeline GInPipe [38] to assess the “true” number of SARS-CoV-2 infections and the case ascertainment (detection) rates in Germany over time. These analyses were based on the random IMS-SC2 sequence dataset (3282 sequences) and were validated using a sequence dataset from GISAID (226 316 sequences) covering the same time frame. Details on this analysis are provided in the Supplementary Methods.
Vaccine Efficacy and Impact of Vaccinations
Analyses on vaccine efficacy and impact of vaccination on Delta cases are based on mathematical modeling that is outlined in detail in the Supplementary Methods.
RESULTS
Integrated Molecular Surveillance for SARS-CoV-2
After completion of preparatory work, the IMS-SC2 network was established at RKI in November 2020 as a scalable surveillance instrument with the aim to guide and inform public health policies by providing insights into SARS-CoV-2 genomic epidemiology at national level.
The IMS-SC2 employs 2 complementary sampling strategies: (1) representative sampling, where SARS-CoV-2 reverse-transcription polymerase chain reaction–positive specimens are arbitrarily selected for whole-genome sequencing and (2) targeted sampling of SARS-CoV-2 cases occurring in settings of clinical or epidemiological interest, such as vaccine breakthrough infections, reinfections, outbreaks, and cases of travelers returning from countries with high VOC prevalence [39].
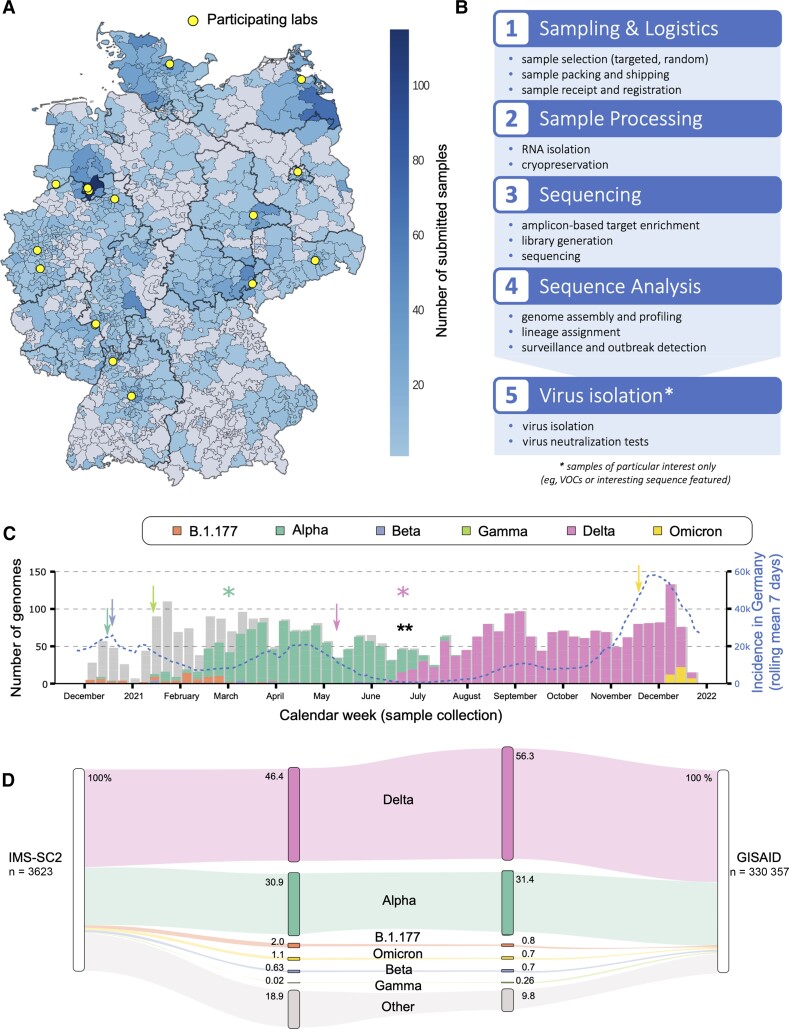
The IMS-SC2 laboratory network currently comprises 16 diagnostic laboratories distributed across Germany (Figure 1A). For representative sampling, the laboratories randomly select a total average of 58 ± 22 SARS-CoV-2–positive remainder specimens and send them to RKI for whole-genome sequencing on a weekly basis. In addition, a limited number of specimens fulfilling the targeted sampling criteria (see above) are sent to RKI on an as-needed basis. Thus, while the IMS-SC2 is primarily based on an unbiased sample set of SARS-CoV-2 cases in Germany, emerging genetic variants may be captured at an early timepoint, while they are still rare.
Figure 1.
A, Map visualizing the distribution and amount of submitted samples to the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) project in Germany based on 3-digit zip codes. The locations of participating laboratories are highlighted in yellow. The map is based on random samples (n = 3282), suspected samples (n = 194), and unknown samples (n = 147) of which 3.42% (n = 124) were excluded due to missing or incorrect geographical data. B, IMS-SC2 workflow. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive samples from multiple locations are transported to the Robert Koch Institute wet laboratory where they undergo processing for whole-genome sequencing. Sequence data are analyzed using standardized pipelines, which are updated at regular intervals to adjust for changing wet-lab protocols as well as sequence variation and evolution. All IMS-SC2 samples are cryopreserved, enabling isolation and further in vitro evaluation (eg, via neutralization assays) of SARS-CoV-2 strains displaying sequence features of interest, for example, amino acid substitutions in the spike gene that may be associated with immune evasion. C, SARS-CoV-2 lineage counts over time as captured by the IMS-SC2 network. Variants of concern (VOCs) shown are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529). Lineage B.1.177 is also shown as an early variant that emerged in Europe in early summer 2020 [41]. Information on sublineages (such as AY sublineages for Delta) are summarized in their parent lineage. Shadow bars indicate total sequence numbers determined. Blue line displays national SARS-CoV-2 incidences (right y-axis) at the corresponding time points. Arrows denote the time points that each VOC was declared as such by the World Health Organization (Alpha: mint; Beta: blue; Gamma: light green; Delta: pink; Omicron: yellow); a black double-asterisk shows the time point at which the weekly count of Delta genomes increased significantly (Fisher exact test, P <.01); colored asterisks denote the time points when Alpha and Delta were declared predominant variants in Germany, based on sequencing data, registered case counts, and polymerase chain reaction genotyping efforts. Additional information is shown in Supplementary Table 1. D, SARS-CoV-2 lineage and corresponding sublineage proportions as captured by the IMS-SC2, compared to German genome proportions in GISAID in the same time frame. VOCs shown here are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529), including their corresponding sublineages. Lineage B.1.177 is also shown as an early variant that emerged in Europe in early summer 2020. All remaining SARS-CoV-2 lineages are pooled into “Other.”
Once received at RKI, samples undergo RNA extraction, library preparation, sequencing, and bioinformatic analysis to generate a consensus genome for each sample, using standardized operating procedures (Figure 1B). Samples are sequenced with either Illumina or Nanopore instruments; the reads generated undergo sequence quality control and are subsequently subjected to specialized pipelines for reference-based genome reconstruction [40, 29]. The resulting consensus genomes are then forwarded to a pipeline that automatically performs the following steps on a daily basis: consensus and metadata quality control, lineage assignment using the most up-to-date software versions of pangolin [30], generation of mutation profiles and storage in a harmonized database, cluster detection, and generation of summaries for reporting and subsequent data analyses. Analysis protocols and bioinformatic algorithms undergo regular benchmarking and adjustment for optimal performance in the face of ongoing viral evolution. To enable subsequent analyses that are meaningful from a public health standpoint, the sequencing data are complemented with the corresponding metadata that hold information on the date, approximate location, sampling indication, and additional circumstances of each sequenced specimen.
Genome Sequencing
Of a total 4249 samples collected from 1 December 2020 to 31 December 2021, 3623 underwent sequencing via Nanopore (92.72%) or Illumina (7.27%) and passed quality control with at least 90% sequence identity to NC_045512.2 and allowing not more than 10% of ambiguous calls with not more than 5% of uninformative N calls. The consensus sequences reconstructed from Nanopore data were covered by 116k reads and comprised 1.62% N bases on average. The reconstructed SARS-CoV-2 genomes underwent downstream analyses as outlined below.
Total Sample: Timely Detection of VOCs at Representative Proportions
Of the total 3623 SARS-CoV-2–positive specimens with genomes successfully reconstructed, 90.6% (3282) were sampled randomly, whereas 5.4% (194) were collected via targeted selection (remaining unknown). The study samples originated from almost all regions in Germany (Figure 1A). The sampling period covered the second, the third, and the ongoing fourth wave of the COVID-19 pandemic in Germany (Figure 1C, incidence in Germany).
All World Health Organization (WHO)–declared VOCs were present in the IMS-SC2 sample set, at frequencies that varied over time (Figure 1C, Figure 2A; and section “Random sample: Variant, phylogenetic, and mutation analysis” below). VOCs relevant to COVID-19 epidemiology in Germany were detected in a timely manner: for example, Alpha (B.1.1.7), Beta (B.1.351), and Delta (B.1.617.2 and sublineages) were detected in IMS-SC2 specimens sampled over 1 week (Alpha, Beta) and over 3 weeks (Delta) before these variants were declared VOCs by the WHO (Figure 1C and Supplementary Table 1). To assess how adequately the IMS-SC2 data set captured relevant variants, we used the GISAID subset of sequences submitted by German laboratories from the same time period as reference and compared the relative proportions of selected SARS-CoV-2 lineages. Although the IMS-SC2 dataset was 2 orders of magnitude smaller than the total GISAID dataset, VOC proportions were well-correlated with the reference dataset (Figure 1D). As expected, non-VOC sequences were present in the IMS-SC2 dataset at a higher proportion (18.9%) than in the German GISAID dataset (9.8%), reflecting differences in the underlying sample acquisition system: While a considerable part of sequences submitted to GISAID originate from targeted samples that were sequenced because a VOC infection was suspected, the majority of IMS-SC2 samples (90%) have been obtained via random sampling as outlined above.
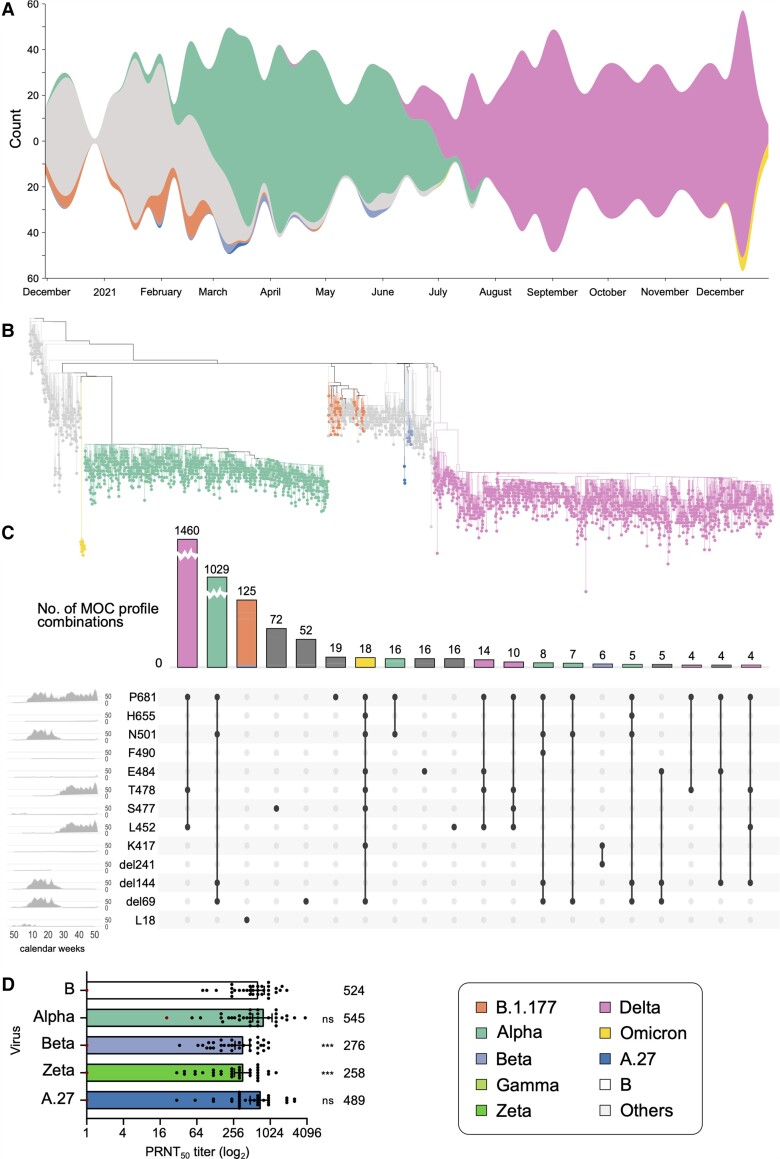
Figure 2.
A, Variant proportions over time, as captured in the representative sampling subset of Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) laboratory network genome sequences. To visualize the dynamics in the virus population over time, virus lineages were determined with pangolin based on the randomly sampled genome sequences (n = 3282, see Materials and Methods). Lineage frequencies were aggregated based on the date of sampling relative to calendar weeks. Missing values have been interpolated. Visualization was performed using RAWGraphs. Please see Supplementary Figure 1 for a detailed visualization including non–variants of concern (VOCs). B, Phylogenetic tree highlighting VOC clades. Sequencing data presented here are based on all randomly selected severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive specimens from the IMS-SC2 network (n = 3282). Lineage B.1.177 is also shown as an early variant that emerged in Europe in early summer 2020 as well as 3 A.27 samples. Please see Supplementary Figure 2 for the full tree visualization, including the 2 long branch attractions described in the Supplementary Methods. C, Mutation of concern (MOC) proportions and combinations over time, as captured by the randomly sampled IMS-SC2 genomes. MOCs shown here highlight the spike amino acid positions, rather than the specific exchanged amino acid, as the selected positions can have >1 amino acid substitution. We constructed an UpSet diagram to visualize the 20 most common intersecting sets, ie, shared MOCs among the randomly selected IMS-SC2 sequences. For selected MOCs, the diagram shows all intersections (specific mutation profiles) and the number of IMS-SC2 sequences that harbor these profiles. On the leftmost panel, we show the frequencies of specific MOCs over time (x-axis: calendar weeks). For selected mutation profiles, we also show the distribution of SARS-CoV-2 lineages. For additional information on the selected mutations, please see Supplementary Figure 3 and Supplementary Table 3. D, Assessing the susceptibility of SARS-CoV-2 variants to neutralization. Thirty-four sera drawn from individuals vaccinated twice with the BNT162b2 vaccine were assessed for their capacity to neutralize different SARS-CoV-2 isolates in vitro. Bars represent the geometric mean plaque reduction neutralization test (PRNT50) titer and 95% confidence intervals. The red dot–marked patient in (A) is immunosuppressed and not included in the statistical analysis. The geometric mean titer is indicated above each bar. Significance was determined by 2-way analysis of variance. ***P < 0.001; ns, not significant.
Random Sample: Variant, Phylogenetic, and Mutation Analysis
The randomly selected sample set included 3282 SARS-CoV-2 specimens collected between December 2020 and December 2021, for which whole genomes were successfully reconstructed. The study population of this random sample was diverse with respect to their age distribution (Supplementary Table 2). Figure 2A and Supplementary Figure 1 depict the lineage composition over time in the sequence dataset derived from the random sample. Lineage composition was initially diverse and both the Alpha (B.1.1.7) and Beta (B.1.351) VOCs were detected less frequently than, for instance, B.1.177 (a variant prevalent in Europe during the 2020 summer [41]), and other non-VOC variants (Figure 2A, Supplementary Figure 1). Whereas Beta detections remained sporadic, Alpha rose to predominance in March 2021 and was frequently found throughout the first half of that year. A single sample, collected in July 2021, was classified as Gamma (P.1). Delta (B.1.617.2 and sublineages), first identified in a sample from April 2021, spread in mid-2021 to become the single most common VOC through December 2021, when the first Omicron (B.1.1.529) genomes were identified.
Phylogenetic analysis was performed to explore the genetic diversity within each VOC clade and between different VOCs (Figure 2B and Supplementary Figure 2). As expected, genomes belonging to the same VOC clustered together. The VOC topology in our phylogeny overall agrees with the one based on all global GISAID sequences, as shown in the Nextstrain website build (https://nextstrain.org/ncov/gisaid/global; accessed 17 January 2022): Omicron branching together with Alpha and B.1.177 branching together with Delta.
Mutation analyses focused on nonsynonymous spike mutations with previously characterized phenotypic effects (Supplementary Table 3 and Supplementary Figure 3). For several of these mutations of concern (MOCs), the time-dependent detection patterns appeared similar (Figure 2C, leftmost panel), indicating combined occurrence. We identified several MOC combination profiles, of which the most common ones were indeed specific to Delta, Alpha, and Omicron, the VOCs identified at the highest frequencies in the IMS-SC2 dataset (Figure 2C, upper and main panels). Figure 2C also reveals changes in single specific sites, for example, S:L18 for B.1.177 and Beta.
Assessing Variant Sensitivity to Vaccine-Induced Humoral Immunity
A major goal of SARS-CoV-2 molecular surveillance efforts is to predict phenotypical virus properties, such as immune evasion. For such genotype-to-phenotype predictions to be robust, sequencing data must be complemented with laboratory experimental data. Native swab samples submitted by IMS-SC2 network laboratories can be used for virus isolation in a dedicated high biosafety level (BSL-3) at RKI. To assess SARS-CoV-2 variants circulating in Germany for their susceptibility to vaccine-induced humoral immunity, we have established a protocol to measure the neutralizing activity of postvaccine plasma against different variants, using authentic viruses. Sera from 35 healthcare workers receiving routine COVID-19 vaccination with BNT162b2 [42] were tested against 1 of the first German SARS-CoV-2 isolates (reference, lineage B) and isolates obtained from the IMS-SC2 and classified as A.27 [37], Alpha, Beta, and Zeta (P.2). As shown in Figure 2D, sera from these vaccinated individuals displayed overall robust neutralization activity against each of these variants, with the exception of a serum sample originating from an immunocompromised study participant, which neutralized, at low titers, only 1 out of the 5 isolates tested. The mean neutralization titers were significantly higher against the reference strain, Alpha, and A.27 than they were against Beta and Zeta, confirming that both Beta and Zeta are capable of immune evasion [43, 44]. Thus, the IMS-SC2 infrastructure enables experimental studies to assess immune escape. This is a key priority with respect to relevant phenotype predictions [45].
IMS-SC2 Facilitates Genome-Based Incidence Estimation to Assess Unreported Case Numbers
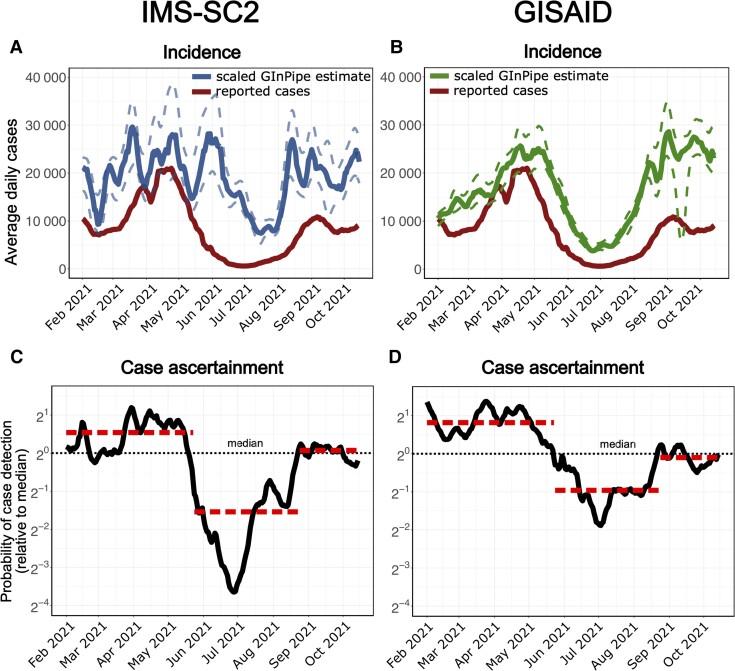
To assess the epidemic activity and spread of COVID-19, it is important to know the size of the population infected per time, that is, the incidence of SARS-CoV-2 infections. This key epidemiologic parameter is commonly gauged by monitoring the number of confirmed cases. However, number of confirmed cases may considerably underestimate the true number of infections, for example, due to varying test capacities or test strategies [46–48, 38]. We have previously established a bioinformatic method that enabled an unbiased estimate of SARS-CoV-2 incidence, based on genome sequencing data [38]. We employed this novel approach to retrospectively assess SARS-CoV-2 incidences in Germany, based on the IMS-SC2 genome sequencing dataset. To this end, sequences are assigned to consecutive subsets according to their sampling dates (temporal bins) and for each temporal bin, the number of sequences different from a reference, as well as the number of unique sequences, is computed in order to infer the incidence correlate, which can then be used to estimate the “true” number of infections (ie, independent of the testing strategies, which are biased; see Materials and Methods). In Figure 3A, we show the sequence-based incidence estimation for the randomly selected IMS-SC2 data (3282 sequences). For validation purposes, we performed the same calculation using the German sequence data (226 316 sequences) available on GISAID, which were generated during a corresponding time frame, that is, starting in February 2021, when the IMS-SC2 began to generate a steady data influx. This validation showed comparable results for the GISIAD sequence data, although confidence intervals were narrower compared to the smaller IMS-SC2 data set. Based on the sequence-based incidence estimate, it is also possible to predict the case ascertainment probability over time (the probability that an infected case is reported). This analysis suggests that SARS-CoV-2 was underreported between end of May and end of August 2021 (Figure 3C), similar to findings observed in other European countries in the summer of 2020 [38]. Convincingly, case ascertainment analysis with the entire GISAID dataset largely confirmed these trends (Figure 3D). These calculations suggest that it is possible to perform genome-based incidence estimation and accurate case ascertainment analysis using a well-designed national-scale genomic surveillance dataset.
Figure 3.
A and B, Reported severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cases in Germany (rolling average, red line) and genome-based incidence estimation using GInPipe [39]. A, The solid and dashed blue lines depict the median trajectory and its 5th to 95th percentile of GInPipe’s incidence estimate using only Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) laboratory network data (3282 sequences, random set). B, The solid and dashed green lines depict the corresponding estimates using all available German sequences deposited in GISAID (226 316 sequences). C and D, Predicted changes in case ascertainment using GInPipe with the IMS-SC2 data (C), as well as all available German sequences deposited in GISAID (D). Case ascertainment is centered on the median case ascertainment probability.
The German Vaccination Campaign Decelerated and Delayed the Delta Wave
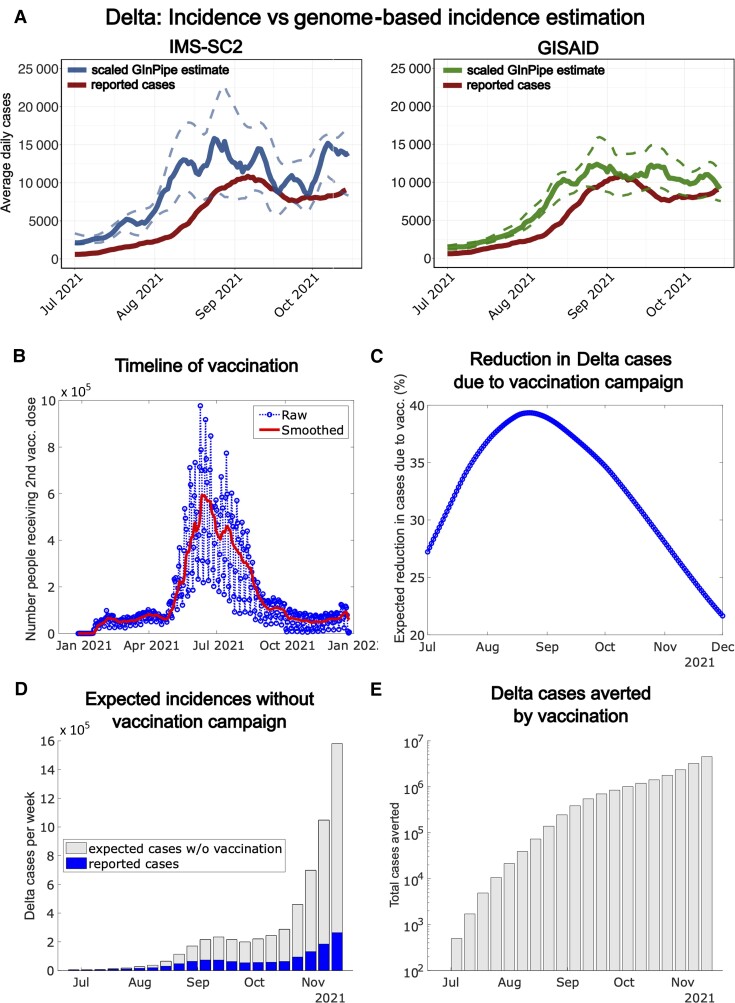
The number of reported cases in Germany increased exponentially from July to late August 2021 (Figures 3A and 4A, red line). We hypothesized that this pronounced increase was borne by the Delta VOC, which had emerged in India in March 2021, rapidly spread around the world, and became predominant in Germany in late June 2021 (Figure 1C). To confirm this hypothesis, we inferred the “true” Delta incidences by genome-based estimations focusing on Delta genomes from the IMS-SC2 random sample set (1497 sequences, blue line in Figure 4A). Indeed, this Delta-specific subanalysis confirmed that the increase of total reported cases coincided with an exponential rise in cases (Figure 4A and 4B); this finding was validated by analysis based on the GISAID data set (132 610 sequences, green line in Figure 4B). Thus, it is reasonable to assume that the rising case counts in July and August represented the nascent Delta wave.
Figure 4.
A, Reported cases (rolling average) during the onset of the Delta wave in Germany (July–November 2021, solid red line) and genome-based incidence estimation to infer the “true” Delta incidences for the considered time window, using an established in-house bioinformatic method [38] using the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) sequence set (solid blue line = median estimate; dashed blue lines = 5th and 95th percentile, 1497 sequences) vs the GISAID sequence set (solid green line = median estimate; dashed green lines = 5th and 95th percentile, 132 610 sequences). B, Daily number of individuals receiving the second vaccine shot (blue line) in Germany and smoothed 7-day average (red line). C, Expected reduction of new Delta cases in Germany resulting from the timeline of vaccination and the waning dynamics of vaccine efficacy. D, Reported weekly severe acute respiratory syndrome coronavirus 2 cases during the onset of the Delta wave in Germany (blue bars) and expected additional weekly Delta cases if the German vaccine campaign had not been rolled out (gray bars; computed using the susceptible-infected-recovered [SIR] model outlined in the Supplementary Methods). E, Expected total Delta cases averted by the German vaccination campaign until December 2021 (computed using the SIR model outlined in the Supplementary Methods).
Interestingly, both reporting data and genome-based Delta incidence estimation revealed a plateau and decreasing trend during August and September (Figure 4A, red line), followed by rapid increase from mid-October onward (fourth wave in Germany). Underreporting might have affected these dynamics. However, not only the reporting data, but also genome-based incidence estimation point towards the plateau and decrease, which makes it unlikely that this represents a reporting artefact. Rather, there appears to be a “true” deceleration of the nascent Delta wave, which is related to a real-world decrease in Delta infections (Figure 4A).
A reasonable cause for the Delta wave deceleration (Figure 4A), in addition to containment measures, may in fact be the German vaccination campaign: As shown in Figure 4B, most Germans received their second vaccine dose during July and August 2021, that is, during a time period which immediately preceded the Delta plateau in August/September. To assess whether vaccination may have decelerated the Delta wave, we estimated the onset and waning dynamics of vaccine efficacy against Delta infections (Supplementary Methods). Based on these dynamics and the timeline of vaccine administration, we were able to estimate the expected reductions of infections solely attributed to vaccination (Figure 4C; Supplementary Methods). We then fitted a susceptible-infected-recovered (SIR) model to the case reporting data to derive an estimate of the time-varying “force of infection” (Supplementary Methods). By calculating the impact of the vaccination campaign on the force of infection (Supplementary Methods) and using the derived SIR model, we could simulate a hypothetical scenario in which vaccination had not slowed down the pandemic. The resulting weekly cases and the overall cases, if vaccination had not been rolled out in Germany, are depicted in Figure 4D and 4E. This analysis indicates that the German vaccination campaign considerably reduced the velocity of the Delta wave at its onset during July to October. Our model predictions indicate that if vaccination had not taken place, the nonaverted cases in the summer (ie, during the early phase of the Delta wave) would have led to an exponential rise in cases during late autumn and winter, with weekly incidences >10 times higher than those actually observed (Figure 4D). In conclusion, the German vaccination campaign contributed substantially to a true deceleration of the Delta wave during August–October 2021, and profoundly reduced the overall number of cases until December 2021 (Figure 4E).
DISCUSSION
Robust molecular and genomic SARS-CoV-2 surveillance is crucial for detection, monitoring, and evaluation of viral variants with potentially negative public health implications due to concerning characteristics [39, 45, 49–52]. In January 2021, Germany, through use of financial incentives, implemented a large-scale, mainly commercial-laboratory based molecular surveillance of SARS-CoV-2. In 2021, this surveillance collected >450 000 sequences. In the long term, maintaining genomic surveillance of such high intensity may prove financially and logistically challenging. Thus, molecular surveillance instruments that are efficient and sustainable are becoming increasingly important.
Here, we present the first results of the German IMS-SC2, a well-designed medium-scale molecular surveillance network at the national level that could be adopted as a postpandemic genomic surveillance infrastructure to monitor SARS-CoV-2 evolution. We show that this surveillance instrument allows for the detection and experimental laboratory study of important VOCs (Figures 1C and 2D). Moreover, the robust primary surveillance dataset captures the abundance of major variants quite accurately, when compared to the entire German sequence data set (Figures 1C, 1D, and 2A), thereby allowing the monitoring of concerning mutations (Figure 2C), to characterize outlier variants by phylogenetic analysis (Figure 2B), and to assess antigenic attributes of the humoral vaccine response against emerging variants; the latter is crucial to predicting immune evasion, that is, a highly relevant clinical phenotype (Figure 2D). Furthermore, these data enabled us to monitor infection dynamics through genome-based incidence estimation and to monitor case ascertainment rates over time (Figure 3). Sample size selection in molecular surveillance balances resource allocation against downstream analysis requirements. While large-scale SARS-CoV-2 genomic surveillance may be scaled down considerably in the future, our data demonstrate that a well-designed surveillance network using substantially smaller sample sizes allows to capture important aspects of SARS-CoV-2 viral evolution and infection dynamics. Limitations of smaller sample sizes include greater uncertainty, as illustrated by the wider 95% confidence intervals of the IMS-SC2–based incidence estimation (Figure 3A) compared to the GISAID-based estimation (Figure 3B); and potentially longer lag times to the detection of low-frequency (eg, emerging) variants. Depending on the certainty levels required for downstream applications, adjusting the numbers of sequenced samples may therefore be beneficial.
We show that the IMS-SC2 data can be used for genome-based incidence estimation [38] to obtain information on the “true” SARS-CoV-2 infection dynamics (Figure 3). In addition to laboratory-confirmed infections, this approach allows us to quantify unreported cases, that is, infections that do not find their way into official case counts. Using this method, we observed considerably higher incidences during the 2021 summer than were captured via registered cases; these high infection levels during the summer were setting the stage for the 2021 winter surge (Figure 3). Importantly, we were able to reliably predict changes in case ascertainment probabilities (ie, the likelihood that a SARS-CoV-2 case is detected and reported). Thus, the IMS-SC2 provides an additional data source that facilitates the assessment of testing policies for adequacy, and the surveillance of infection dynamics, even if these are hidden, for example, due to paucity of testing. Such capability will be particularly important in the postpandemic future, when testing may decrease substantially below the current levels. The IMS-SC2 data may then also serve to more precisely assess the impact of vaccine timing and vaccine coverage on infection dynamics at the population level (Figure 4), thereby helping to inform and optimize future vaccination campaigns.
Antigenic variation in SARS-CoV-2 can decrease vaccine efficacy substantially, thereby increasing virus circulation and leaving vulnerable groups unprotected. The Delta and, in particular, Omicron surges of the COVID-19 pandemic are caused by variants capable of substantial immune evasion [53, 54]. They indicate a need for SARS-CoV-2 surveillance that closely monitors virus evolution and antigenic drift to inform public health decisions and potentially vaccine strain adjustment in a timely manner. Current vaccines have induced robust protection toward SARS-CoV-2 circulating until summer 2021, but show a rapid waning towards more divergent variants such as Delta and Omicron, requiring booster immunizations to maintain high vaccine efficacy. The messenger RNA vaccine technologies enable swift designing and redesigning of vaccines, possibly including the option to produce bivalent or trivalent formulations if needed. The combination of these technologies with the monitoring of SARS-CoV-2 genomic diversity and evolution at adequate temporal and geographic resolution will open up the possibility to administer vaccines that have been optimally adjusted to pathogen genetic diversity, considering partial as well as total immune evasion that may emerge in the future.
In summary, the IMS-SC2 is an efficient surveillance instrument, complementing and enhancing existing pathogen surveillance mechanisms. Of particular value, IMS-SC2 data can inform vaccination strategies to maximize vaccine effectiveness at the population level.
Supplementary Material
Contributor Information
Djin Ye Oh, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Martin Hölzer, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Sofia Paraskevopoulou, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Maria Trofimova, Systems Medicine of Infectious Disease (P5), Robert Koch Institute, Berlin, Germany.
Felix Hartkopf, Methodology and Research Infrastructure, Genome Sequencing and Genomic Epidemiology (MF2), Robert Koch Institute, Berlin, Germany.
Matthias Budt, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Marianne Wedde, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Hugues Richard, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Berit Haldemann, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Teresa Domaszewska, Respiratory Infections (FG36), Robert Koch Institute, Berlin, Germany.
Janine Reiche, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Kathrin Keeren, Gastroenteritis and Hepatitis Pathogens and Enteroviruses (FG15), Robert Koch Institute, Berlin, Germany.
Aleksandar Radonić, Methodology and Research Infrastructure, Genome Sequencing and Genomic Epidemiology (MF2), Robert Koch Institute, Berlin, Germany.
Julia Patricia Ramos Calderón, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Maureen Rebecca Smith, Systems Medicine of Infectious Disease (P5), Robert Koch Institute, Berlin, Germany.
Annika Brinkmann, Centre for Biological Threats and Special Pathogens, Highly Pathogenic Viruses (ZBS1), Robert Koch Institute, Berlin, Germany.
Kathrin Trappe, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Oliver Drechsel, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Kathleen Klaper, Methodology and Research Infrastructure, Genome Sequencing and Genomic Epidemiology (MF2), Robert Koch Institute, Berlin, Germany; Sexually Transmitted Bacterial Pathogens and HIV (FG18), Robert Koch Institute, Berlin, Germany.
Sascha Hein, Division of Virology, Paul Ehrlich Institute, Langen, Germany.
Eberhardt Hildt, Division of Virology, Paul Ehrlich Institute, Langen, Germany.
Walter Haas, Gastroenteritis and Hepatitis Pathogens and Enteroviruses (FG15), Robert Koch Institute, Berlin, Germany.
Sébastien Calvignac-Spencer, Epidemiology of Highly Pathogenic Microorganisms (P3), Viral Evolution, Robert Koch Institute, Berlin, Germany.
Torsten Semmler, Methodology and Research Infrastructure, Genome Sequencing and Genomic Epidemiology (MF2), Robert Koch Institute, Berlin, Germany.
Ralf Dürrwald, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
Andrea Thürmer, Methodology and Research Infrastructure, Genome Sequencing and Genomic Epidemiology (MF2), Robert Koch Institute, Berlin, Germany.
Christian Drosten, Institute of Virology, Charité-University Medicine Berlin, Berlin, Germany.
Stephan Fuchs, Methodology and Research Infrastructure, Bioinformatics and Systems Biology (MF1), Robert Koch Institute, Berlin, Germany.
Stefan Kröger, Gastroenteritis and Hepatitis Pathogens and Enteroviruses (FG15), Robert Koch Institute, Berlin, Germany.
Max von Kleist, Systems Medicine of Infectious Disease (P5), Robert Koch Institute, Berlin, Germany.
Thorsten Wolff, Influenza and Other Respiratory Viruses (FG17), Robert Koch Institute, Berlin, Germany.
for the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) Laboratory Network:
Barbara Biere, Konrad Bode, Victor Corman, Michael Erren, Patrick Finzer, Roger Grosser, Manuel Haffner, Beate Hermann, Christina Kiel, Andi Krumbholz, Kristian Meinck, Andreas Nitsche, Markus Petzold, Thomas Schwanz, Florian Szabados, Friedemann Tewald, and Carsten Tiemann
Data Availability
All Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) genome sequences used in this study were uploaded to the European Nucleotide Archive (project accession number: PRJEB50616).
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors are grateful to Essia Belarbi, Ariane Düx, Jan Gogarten, Kevin Merkel, Caroline Rothemeier, Andreas Sachse, Grit Schubert, Petra Kurzendörfer, and Tanja Pilz for outstanding sequencing support; and Heike Fischer, Ute Hopf-Guevara, and Gudrun Heins for excellent technical assistance. The authors thank Martin Mielke for critically reading the manuscript and his Working Group on SARS-CoV-2 Diagnostics at Robert Koch Institute (RKI) for valuable discussion of the results. The authors also appreciate Denis Beslic, Matt Huska, Sandra Kaiser, René Kmiecinski, Kunaphas Kongkitimanon, Marie Lataretu, and Paul Wolk for keeping the SARS-CoV-2 bioinformatics pipelines running day after day.
IMS-SC2 Laboratory Network consortium authors. Barbara Biere, RKI, FG17: National Influenza Centre, RKI, Berlin; Konrad Bode, MVZ Labor Dr Limbach, Heidelberg; Victor Corman, Institute of Virology, Charité-University Medicine, Berlin; Michael Erren, MVZ Laborzentrum Weser-Ems; Patrick Finzer, MVZ Düsseldorf-Centrum, Düsseldorf; Roger Grosser, Labor Dr Wisplinghoff, Köln; Manuel Haffner, MVZ Labor Dr Kirkamm, Mainz; Beate Hermann, MVZ Dianovis, Greiz; Christina Kiel, MVZ Labor Dessau, Dessau-Roßlau; Andi Krumbholz, Thomas Lorentz, Labor Dr Krause, Kiel; Kristian Meinck, IMD-Laborverbund, Greifswald; Andreas Nitsche, RKI, ZBS 1: Hochpathogene Viren; Markus Petzold, Institut für Medizinische Mikrobiologie und Hygiene, Institut für Virologie, TU Dresden; Thomas Schwanz, Institut für Medizinische Mikrobiologie und Hygiene, Universitätsmedizin Mainz; Florian Szabados, Laborarztpraxis Osnabrück, Georgsmarienhütte; Friedemann Tewald, Labor Enders, Stuttgart; and Carsten Tiemann, Labor Krone, Bad Salzuflen.
Financial support. This work was supported by the Bundesministerium für Bildung und Forschung (German Ministry for Science and Education) and Bundesministerium für Wirtschaft und Klima (German ministry for economic affairs and climate action), grant number 01MK21009H to M.v.K and M.R.S; COVID-19 emergency crisis funds provided by the German Ministry of Health (project number D81785 to T.W., W.H., T.S. and C.D.); RKI’s special research fund (dissertation grant to M. T.); and the European Centre for Disease Prevention and Control (grant number 2021/008 ECD.12222 to T. S., S. F., A. T., and T. W.).
Supplement sponsorship. This supplement is sponsored by the Precision Vaccines Program of Boston Children's Hospital.
References
- 1. Avanzato VA, Matson MJ, Seifert SN, et al. . Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–1912.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tarhini H, Recoing A, Bridier-Nahmias A, et al. . Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis 2021; 223:1522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cele S, Karim F, Lustig G, et al. . SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 2022; 30:154–62.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chandler JC, Bevins SN, Ellis JW, et al. . SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci U S A 2021; 118:e2114828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Griffin BD, Chan M, Tailor N, et al. . SARS-CoV-2 infection and transmission in the North American deer mouse. Nat Commun 2021; 12:3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, et al. . Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021; 371:172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peacock TP, Brown JC, Zhou J, et al. . The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv [Preprint]. Posted online 3 January 2022. doi: 10.1101/2021.12.31.474653. [DOI] [Google Scholar]
- 8. Patterson EI, Elia G, Grassi A, et al. . Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat Commun 2020; 11:6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burki TK. Lifting of COVID-19 restrictions in the UK and the Delta variant. Lancet Respir Med 2021; 9:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishiura H, Ito K, Anzai A, Kobayashi T, Piantham C, Rodriguez-Morales AJ. Relative reproduction number of SARS-CoV-2 Omicron (B.1.1.529) compared with Delta variant in South Africa. J Clin Med 2021; 11:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ito K, Piantham C, Nishiura H. Relative instantaneous reproduction number of Omicron SARS-CoV-2 variant with respect to the Delta variant in Denmark. J Med Virol 2021; 94:2265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pormohammad A, Ghorbani S, Khatami A, et al. . Comparison of influenza type A and B with COVID-19: a global systematic review and meta-analysis on clinical, laboratory and radiographic findings. Rev Med Virol 2021; 31:e2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piroth L, Cottenet J, Mariet AS, et al. . Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: a nationwide, population-based retrospective cohort study. Lancet Respir Med 2021; 9:251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levin AT, Hanage WP, Owusu-Boaitey N, Cochran KB, Walsh SP, Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol 2020; 35:1123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV; World Health Organization Clinical Case Definition Working Group on Post–COVID-19 Condition . A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis 2021; 9:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spudich S, Nath A. Nervous system consequences of COVID-19. Science 2022; 375:267–9. [DOI] [PubMed] [Google Scholar]
- 17. Gupta A, Madhavan MV, Sehgal K, et al. . Extrapulmonary manifestations of COVID-19. Nat Med 2020; 26:1017–32. [DOI] [PubMed] [Google Scholar]
- 18. Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol 2020; 20:389–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murray CJL, Piot P. The potential future of the COVID-19 pandemic: will SARS-CoV-2 become a recurrent seasonal infection? JAMA 2021; 325:1249–50. [DOI] [PubMed] [Google Scholar]
- 20. Telenti A, Arvin A, Corey L, et al. . After the pandemic: perspectives on the future trajectory of COVID-19. Nature 2021; 596:495–504. [DOI] [PubMed] [Google Scholar]
- 21. Liu L, Iketani S, Guo Y, et al. . Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2021; 602:676–81. [DOI] [PubMed] [Google Scholar]
- 22. Cele S, Jackson L, Khoury DS, et al. . Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021; 602:654–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmidt F, Muecksch F, Weisblum Y, et al. . Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med 2022; 386:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robert Koch Institute German Electronic Sequence Data Hub (DESH) [in German]. 2022. Available at: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/DESH/DESH.html. Accessed 7 February 2022.
- 25. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data—from vision to reality. Euro Surveill 2017; 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oh DY, Buda S, Biere B, et al. . Trends in respiratory virus circulation following COVID-19-targeted nonpharmaceutical interventions in Germany, January—September 2020: analysis of national surveillance data. Lancet Reg Health Eur 2021; 6:100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oh DY, Biere B, Grenz M, et al. . Virological surveillance and molecular characterization of human parainfluenzavirus infection in children with acute respiratory illness: Germany, 2015–2019. Microorganisms 2021; 9:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 29. Brandt C, Krautwurst S, Spott R, et al. . poreCov—an easy to use, fast, and robust workflow for SARS-CoV-2 genome reconstruction via Nanopore sequencing. Front Genet 2021; 12:711437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Toole A, Scher E, Underwood A, et al. . Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 2021; 7:veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minh BQ, Schmidt HA, Chernomor O, et al. . IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020; 37:1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 2018; 35:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moore RM, Harrison AO, McAllister SM, Polson SW, Wommack KE. Iroki: automatic customization and visualization of phylogenetic trees. PeerJ 2020; 8:e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breitwieser FP, Salzberg SL. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 2020; 36:1303–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017; 33:2938–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calvignac-Spencer S, Budt M, Huska M, et al. . Rise and fall of SARS-CoV-2 lineage A.27 in Germany. Viruses 2021; 13:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith MR, Trofimova M, Weber A, Duport Y, Kuhnert D, von Kleist M. Rapid incidence estimation from SARS-CoV-2 genomes reveals decreased case detection in Europe during summer 2020. Nat Commun 2021; 12:6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. European Centre for Disease Prevention and Control . Guidance for representative and targeted genomic SARS-CoV-2 monitoring—3 May 2021. 2021. Available at: https://www.ecdc.europa.eu/en/publications-data/guidance-representative-and-targeted-genomic-sars-cov-2-monitoring. Accessed 7 February 2022.
- 40. Hufsky F, Lamkiewicz K, Almeida A, et al. . Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief Bioinform 2021; 22:642–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hodcroft EB, Zuber M, Nadeau S, et al. . Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021; 595:707–12. [DOI] [PubMed] [Google Scholar]
- 42. Hein S, Herrlein ML, Mhedhbi I, et al. . Analysis of BNT162b2- and CVnCoV-elicited sera and of convalescent sera toward SARS-CoV-2 viruses [manuscript published online ahead of print 25 November 2021]. Allergy 2021. doi: 10.1111/all.15189. [DOI] [PubMed] [Google Scholar]
- 43. Madhi SA, Baillie V, Cutland CL, et al. . Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med 2021; 384:1885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bekliz M, Adea K, Vetter P, et al. . Neutralization of ancestral SARS-CoV-2 and variants Alpha, Beta, Gamma, Delta, Zeta and Omicron by mRNA vaccination and infection-derived immunity through homologous and heterologous variants. medRxiv [Preprint]. Posted online 31 December 2022. doi: 10.1101/2021.12.28.21268491. [DOI] [Google Scholar]
- 45. Oude Munnink BB, Worp N, Nieuwenhuijse DF, et al. . The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med 2021; 27:1518–24. [DOI] [PubMed] [Google Scholar]
- 46. Irons NJ, Raftery AE. Estimating SARS-CoV-2 infections from deaths, confirmed cases, tests, and random surveys. Proc Natl Acad Sci U S A 2021; 118:e2103272118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schneble M, De Nicola G, Kauermann G, Berger U. A statistical model for the dynamics of COVID-19 infections and their case detection ratio in 2020. Biom J 2021; 63:1623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hippich M, Holthaus L, Assfalg R, et al. . A public health antibody screening indicates a 6-fold higher SARS-CoV-2 exposure rate than reported cases in children. Med (N Y) 2021; 2:149–63.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paul P, France AM, Aoki Y, et al. . Genomic surveillance for SARS-CoV-2 variants circulating in the United States, December 2020–May 2021. MMWR Morb Mortal Wkly Rep 2021; 70:846–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benslimane FM, Al Khatib HA, Al-Jamal O, et al. . One year of SARS-CoV-2: genomic characterization of COVID-19 outbreak in Qatar. Front Cell Infect Microbiol 2021; 11:768883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicholls SM, Poplawski R, Bull MJ, et al. . CLIMB-COVID: continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Genome Biol 2021; 22:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robishaw JD, Alter SM, Solano JJ, et al. . Genomic surveillance to combat COVID-19: challenges and opportunities. Lancet Microbe 2021; 2:e481–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mlcochova P, Kemp SA, Dhar MS, et al. . SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021; 599:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoffmann M, Kruger N, Schulz S, et al. . The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 2021; 185:447–56.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) genome sequences used in this study were uploaded to the European Nucleotide Archive (project accession number: PRJEB50616).