Abstract
Background
The epidemiological advantage of Omicron variant is evidenced by its rapid spread and the ability to outcompete prior variants. Among Omicron sublineages, early outbreaks were dominated by BA.1, while BA.2 has gained dominance since February 2022. The relative pathogenicity and transmissibility of BA.1 and BA.2 have not been fully defined.
Methods
We compared viral loads and clinical signs in Syrian hamsters after infection with BA.1, BA.2, or D614G variant. A competitive transmission model and next-generation sequencing were used to compare the relative transmission potential of BA.1 and BA.2.
Results
BA.1 and BA.2 caused no apparent clinical signs, while D614G caused more than 10% weight loss. Higher viral loads were detected in nasal wash samples and nasal turbinate and lung tissues from BA.1-inoculated hamsters compared with BA.2-inoculated hamsters. No aerosol transmission was observed for BA.1 or BA.2 under the experimental condition in which D614G transmitted efficiently. BA.1 and BA.2 were able to transmit among hamsters via direct contact; however, BA.1 transmitted more efficiently than BA.2 under the competitive transmission model. No recombination was detected from direct contacts exposed simultaneously to BA.1 and BA.2.
Conclusions
Omicron BA.1 and BA.2 demonstrated attenuated pathogenicity and reduced transmission potential in hamsters compared with early SARS-CoV-2 strains.
Keywords: SARS-CoV-2, Omicron, pathogenicity, transmission, Syrian hamsters
The Syrian hamster model has been useful to evaluate the pathogenicity and transmissibility of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here we report that the Omicron BA.1 and BA.2 sublineages showed attenuated pathogenicity and reduced transmissibility in hamsters when compared with early SARS-CoV-2 strains.
Since the first report of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in December 2019, the ongoing coronavirus disease 2019 (COVID-19) pandemic has been sustained by the emergence of multiple variants of concern (VOC) that differ in their capacity to spread [1, 2], cause severe disease [3], or evade vaccine-induced immunity [4–8]. While the dynamics and the factors driving the emergence of VOC are not fully understood, it is anticipated that novel VOC will continue challenging our existing countermeasures [9–11] and diagnostic methods [12].
VOC Omicron (Pango lineage B.1.1.529) was first detected in Southern Africa in November 2021 and swept through the world within weeks [13]. Omicron has evolved into several major sublineages, with BA.1 as the initial dominant Omicron sublineage. The detection frequency of BA.2 has increased since early 2022 [14]. Omicron is genetically distinct from the prototype SARS-CoV-2 virus or other VOC that have previously spread regionally or globally. Compared with the prototype SARS-CoV-2 virus, the Omicron variant is characterized by >30 amino acid differences in the spike (S) glycoprotein and additional changes in the structural proteins (E, M, N), nonstructural proteins (NSPs; NSP3, NSP4, NSP5, NSP6, NSP12, and NSP14), and accessory protein (open reading frame [ORF] 8) [14]. Detailed mutagenesis study using pseudoviruses has confirmed that multiple amino acid changes located at the receptor-binding domain and the N-terminal domain in Omicron may confer resistance to neutralizing monoclonal antibodies [15] and evasion to infection or vaccine-induced immunity [16, 17]. However, the effect of changes in receptor-binding domain on viral attachment to human angiotensin-converting enzyme 2 has been under debate [18, 19].
Epidemiological data showed that infection with Omicron BA.1 was associated with reduced risk of hospitalization in South Africa, where prior infections by SARS-CoV-2 were prevalent [20]. Experimental data showed that Omicron BA.1 exhibit reduced susceptibility to transmembrane protease serine 2 (TMPRSS2) cleavage, which reduced viral entry via fusion at plasma membrane [21]. Comparable replication of Delta and BA.1 was reported in human nasal epithelial cells [21], while BA.1 showed reduced replication in human lung ex vivo culture when compared to the prototype or Delta viruses [22]. It is not clear whether the differential expression of TMPRSS2 in nasal and lung tissue or additional host factors have affected BA.1 replication efficiency at upper and lower airway. Omicron BA.1 also caused attenuated replication in mice and Syrian hamsters compared with other VOC, with reduced viral replication detected in the upper (mouse nasal turbinate) and lower respiratory tissues (mouse and hamster lungs) [23, 24].
While BA.2 is genetically related to BA.1, there are substantial differences in the S, M, and N structural proteins, NSPs (NSP1, NSP3, NSP4, NSP6, NSP13, and NSP15), and accessory proteins (ORF3a and ORF6). Clinical severity and the risk of hospitalization were found to be comparable after BA.1 and BA.2 infections [25]. Prior vaccination and infection history may confound the transmission dynamics of VOC in the human population, and it is not clear whether the increased detection frequency of BA.2 since February 2022 is due to an intrinsic fitness advantage of BA.2 over BA.1. Syrian hamsters have been reported to be an suitable animal model to study the pathogenicity and transmissibility of SARS-CoV-2 [26–29]. Prototype SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract and infected Syrian hamsters may lose 10%–15% body weight [26–29]. In addition, SARS-CoV-2 may transmit efficiently among hamsters via multiple routes, among which transmission was more efficient via aerosols than via fomites [28, 29]. To study whether there is an intrinsic fitness advantage of BA.2 over BA.1, we compared the pathogenicity and transmission potential of the 2 subvariants in Syrian hamsters.
METHODS
Cells and Viruses
Vero E6 cells overexpressing transmembrane serine protease 2 (Vero-E6/TMPRSS2) were purchased from the Centre For AIDS Reagents (National Institute for Biological Standards and Control) and was maintained under conditions described elsewhere [30, 31]. Omicron BA.1 (hCoV-19/Hong Kong/VM21044713_ HKUVOC0195P3/2021) [16] and BA.2 (hCoV-19/Hong Kong/VM22000135_ HKUVOC0588P2/2022) were isolated from pooled nasopharyngeal and throat swab samples from imported cases. Viruses were isolated and propagated 2–3 times in Vero-E6/TMPRSS2. The consensus sequences of the stock viruses were validated to be identical to the original clinical specimens by next-generation sequencing.
Transmission Experiments in Syrian Hamsters
Male Syrian hamsters (AURA) 4–8 weeks old were obtained from the Laboratory Animal Services Centre, Chinese University of Hong Kong, and from the Centre for Comparative Medicine Research, University of Hong Kong. All experiments were performed at a biosafety level 3 core facility, the Li Ka Shing Faculty of Medicine at the University of Hong Kong. The study protocol has been approved by the Committee on the Use of Live Animals in Teaching and Research, University of Hong Kong (CULATR no. 5323-20).
To evaluate aerosol and contact transmission potential of D614G, Syrian hamsters were anesthetized with ketamine and xylazine and inoculated intranasally with 105 times the median tissue culture infective dose (TCID50) of SARS-CoV-2 in 80 µL phosphate-buffered saline (PBS). On day 1 after inoculation, 1 naive hamster was exposed to an inoculated donor in an individually ventilated cage for 8 hours (eg, aerosol transmission model), with the animals separated by stainless-steel wires to prevent physical contact. After exposure, the D614G-inoculated donors were each cohoused with 1 naive hamster for 24 hours (eg, from 32 to 56 hours after inoculation; direct contact transmission model). To evaluate the competitive transmissibility of BA.1 and BA.2 via aerosols, donors were anesthetized and separately inoculated with 1 × 105 TCID50 of BA.1 or BA.2 in 80 µL of PBS.
Aerosol and direct contact transmission experiments were performed as described above, except that each naive animal was simultaneously exposed to 2 donors separately inoculated with BA.1 or BA.2. The aerosol and contact transmission experiments were independently performed in duplicate or triplicate. After exposure, all hamsters were housed singly in separate individually ventilated cages until the end of the experiment. Weight changes and clinical signs were monitored daily. Nasal wash samples were harvested every other day from donors and exposed naive hamsters. On day 4 after inoculation, donors were euthanized to monitor viral replication in respiratory tissues. Nasal turbinate and lung tissues were homogenized in 1 mL of PBS for viral load determination.
Quantitative Real-time Reverse-Transcription Polymerase Chain Reaction
Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen). RNA was extracted from 70 µL from nasal wash samples and eluted with 60 µL of elution buffer containing poly(A) carrier (AVE buffer) or from 140 µL of tissue homogenate and eluted with 60 µL of AVE buffer. TaqManTM Fast Virus 1-Step Master Mix was used to determine N gene copy number with 4 µL of extracted RNA, following the conditions described elsewhere [32].
Virus Titration
Confluent Vero-E6/TMPRSS2 cells in 96-well culture plates were inoculated with 35 µL of serially half-log diluted samples in quadruplicate. Cytopathic effect of SARS-CoV-2 infection was observed under light microscopy 3 days after inoculation, and the TCID50 was calculated using the Reed-Muench method [33].
Histopathology and Immunohistochemistry
Respiratory tissues, including tissues from the head (sagittal cut), trachea, and right lung lobes were fixed in 10% neutral buffered formalin and processed for paraffin embedding. For immunohistochemistry, SARS-CoV-2 viral proteins were detected using rabbit anti-N polyclonal antibody (no. 40143-T62; Sinobiological), mouse anti-N monoclonal antibody (no. 40143-MM05; Sinobiological), or rabbit anti-S1 monoclonal antibody (no. 99423; Cell Signaling) and counterstained with hematoxylin. Images were captured using a Nanozoomer slide scanner and viewed using NDP View software (NDP.view2; Hamamatsu Photonics). To quantitatively compare the area with N protein detection, the percentage of positive pixels against the negative ones were calculated under ×2.5 magnification using Aperio ImageScope software (version 12.4.3).
Plaque Reduction Neutralization Assay
Postinfection or postexposure serum samples were heat inactivated at 56°C for 30 minutes and were initially diluted 1:10, followed by serially 2-fold dilution with Dulbecco's modified Eagle's medium. Diluted serum samples were incubated with 30 plaque-forming units of SARS-CoV-2 for 1 hour at 37°C. The virus-serum mixtures were added to confluent Vero-E6/TMPRSS2 cells in 24-well plates and incubated for 1 hour at 37°C. The inoculum was removed, and the plates were overlaid with 0.5% agarose in culture medium and incubated for 3 days. The plates were fixed with 10% formalin and stained with 0.3% crystal violet to determine the number of plaques formed in the presence and absence of hamster serum. The serum dilution that resulted in >50% reduction in the number of plaques was recorded as the 50% plaque reduction neutralization test (PRNT50) titer.
Next-Generation Sequencing and Data Analysis
The virus genome was reverse-transcribed with multiple gene-specific primers targeting different regions of the viral genome. The synthesized complementary DNA was then subjected to multiple overlapping 2-kb polymerase chain reactions (PCRs) for full-genome amplification. PCR amplicons obtained from the same specimen were pooled and sequenced using iSeq 100 sequencing platform (Illumina). The sequencing library was prepared by Nextera DNA flex. The sequencing reads generated were trimmed using fastp, a FASTQ data processing tool [34] with adapter and quality filters and mapped to a reference virus genome (GenBank accession no. MN908947.3) with BWA-MEM2 alignment tool (https://github.com/bwa-mem2/bwa-mem2). Potential PCR duplicates were identified and removed by means of samtools markup (https://www.htslib.org/doc/samtools-markdup.html).
The genome consensus sequence was generated by the iVar tool with the PCR primer trimming protocol (minimum sequence depth, >10; minimum Q value, 30) [35]. The variants in the samples were identified using bcftools (version 1.14) (https://samtools.github.io/bcftools/howtos/variant-calling.html). The number and frequency of different mutations were deduced from results obtained with pysamstats software (https://github.com/alimanfoo/pysamstats). The mutation frequencies at different genomic sites were curated and visualized using custom R scripts (available on Github: https://github.com/Leo-Poon-Lab/BA.1-BA.2-competitive-transmission-hamster).
Statistical Analyses
Two-sided Mann-Whitney tests were performed to compare mean values between 2 groups, and Kruskal-Wallis test Dunn multiple comparisons post hoc tests to compare the mean values among multiple groups. Spearman rank correlation coefficient analysis was performed to evaluate the correlation between infectious virus load in oral swab and nasal wash samples. Data were analyzed using Microsoft Excel for Mac software, version 16.28, and GraphPad Prism software, version 8.4.1 for Windows (GraphPad Software).
RESULTS
Reduced Pathogenicity of Omicron BA.1 and BA.2 Compared to the D614G Variant
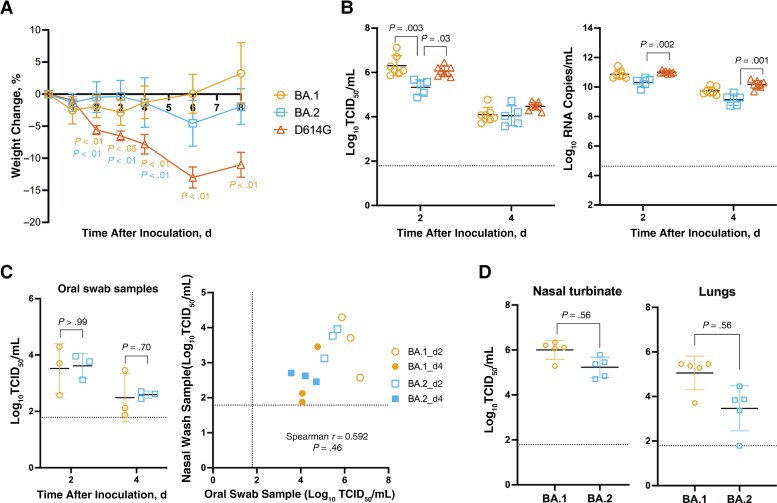
To compare the pathogenicity of Omicron BA.1 and BA.2 with an early D614G variant of SARS-CoV-2 virus (hCoV-19/Hong Kong/VM20109236/2020, EPI_ISL_2477019), hamsters were inoculated intranasally with 1 × 105 TCID50 of viruses. While D614G-inoculated hamsters showed the maximal average weight change of −13.00% on day 6 after inoculation, hamsters inoculated with BA.1 or BA.2 showed minimal weight loss, with maximal average weight changes of −3.90% at 1 day and −4.55% at 6 days after inoculation (Figure 1A). On day 2 after inoculation, comparable infectious viruses or viral RNA were detected from the nasal wash samples of BA.1- and D614G-inoculated hamsters, which were significantly higher than those detected from BA.2-inoculated hamsters (Figure 1B). On day 4 after inoculation, the infectious viral loads in the nasal wash samples were comparable. In addition, there was a positive correlation between infectious viral load detected in the nasal wash and oral swab samples (Spearman r = 0.592; P = .046), although the virus titers detected in the oral swab samples were about 2 logs lower than those in the nasal wash samples (Figure 1C). The mean infectious viral load detected in the nasal turbinate and lungs of BA.1-inoculated hamsters were higher than those in BA.2-inoculated hamsters, although the differences were not significant (Figure 1D).
Figure 1.
Pathogenicity of Omicron BA.1 and BA.2 in Syrian hamsters inoculated intranasally with BA.1 (n = 7), BA.2 (n = 5), or D614G (n= 8). A, Weight changes, shown as means with standard deviations. B, Infectious viral load and viral RNA detected in nasal wash samples on days 2 and 4 after inoculation. Abbreviation: TCID50, median tissue culture infective dose. C, Infectious viral load detected in oral swab samples from inoculated donors on days 2 and 4 after inoculation. D, Infectious viral load detected in the nasal turbinate and lungs of inoculated donors on day 4 after inoculation. A, B, Kruskal-Wallis test and Dunn multiple comparisons were performed to compare weight changes and viral loads of BA.1-, BA.2-, and D614G-inoculated hamsters at each time point. C, D, Mann-Whitney test was performed to compare viral loads of BA.1- and BA.2-inoculated hamsters. Spearman rank correlation coefficient analysis (right) was performed to evaluate the correlation between infectious virus load in oral swab (x-axis) and nasal wash (y-axis) samples. Experiments were independently performed 2–3 times. Dashed lines represent limits of detection.
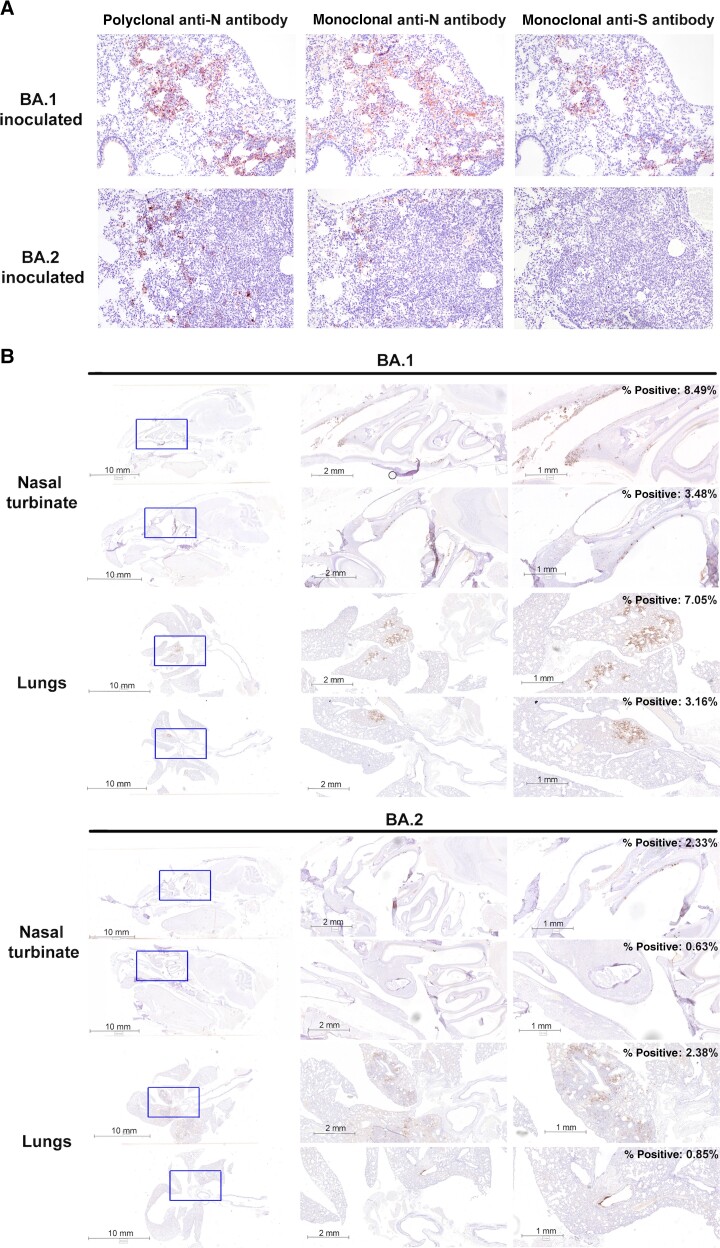
Histopathological examination and immunohistochemistry were performed on nasal and lung tissues collected on day 4 after inoculation. Because BA.1 and BA.2 differ by amino acid changes in the S and N protein, we compared the use of rabbit anti-N polyclonal antibody, mouse anti-N monoclonal antibody, and mouse anti-S monoclonal antibody in detecting BA.1 and BA.2 viral antigens in the lungs. Importantly, the mouse anti-S monoclonal antibody was able to detect BA.1 but not BA.2 viral antigen, highlighting the potential antigenic difference between the associated S proteins. Rabbit anti-N polyclonal antibody, which demonstrated the best sensitivity for both BA.1- and BA.2-infected lungs, was selected for subsequent analysis (Figure 2A).
Figure 2.
Detection of BA.1 and BA.2 viral antigens in the lungs and nasal cavity of inoculated Syrian hamsters by immunohistochemistry. A, Lung tissues collected on day 4 after inoculation from BA.1- or BA.2-inoculated hamsters were stained with rabbit anti-N polyclonal antibody, mouse anti-N monoclonal antibody, or mouse anti–spike (S) monoclonal antibody and counterstained with hematoxylin. B, Detection of N protein in nasal turbinates and lungs collected on day 4 after inoculation in BA.1-inoculated (n = 3) or BA.2-inoculated (n = 3) hamsters using the rabbit anti-N polyclonal antibody and counterstained with hematoxylin. Percentages of positive versus negative pixels were calculated under ×2.5 magnification using Aperio ImageScope software (version 12.4.3).
Viral antigens were detected in the nasal epithelial cells from BA.1- and BA.2-inoculated hamsters with minimal inflammation (Figure 2B). In the lungs, SARS-CoV-2 N protein was detected in focal bronchial epithelial cells and pneumocytes, accompanied by infiltration of mononuclear cells and lung consolidation. By calculating the percentage of positive versus negative pixels, higher percentages were detected in BA.1- than in B A.2-infected nasal turbinate and lung tissues (Figure 2B), which is consistent with the detection of more infectious virus in the nasal turbinates and lungs of BA.1-inoculated hamsters (Figure 1D). Overall, these results showed that BA.1 and BA.2 caused nonapparent clinical signs in Syrian hamsters compared with the early SARS-CoV-2 D614G variant. BA.1 replicated more efficiently than BA.2 in both upper and lower respiratory tract tissues.
BA.1 and BA.2 Showed Reduced Aerosol Transmissibility in Hamsters When Compared to the D614G Variant
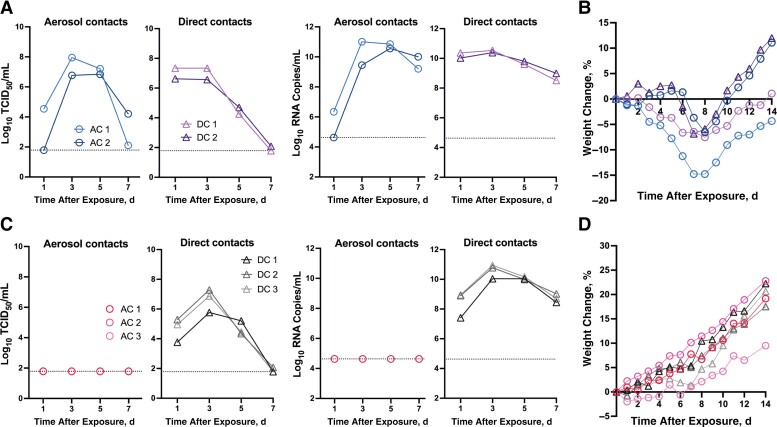
The D614G variant transmitted efficiently to exposed hamsters under the aerosol or direct contact transmission models (Figure 3A). Infectious virus and viral RNA were detected in nasal wash samples from 2 of 2 hamsters after aerosol contact (“aerosol contact hamsters”) 1 and 3 days after exposure and 2 of 2 hamsters after direct contact (“direct contact hamsters”) 1 day after exposure. The maximal weight loss (range of weight change, −6.5% to −14.8%) occurred in aerosol and direct contact hamsters on 7–8 days after exposure (Figure 3B). With the same experimental setting, we further evaluated the transmission potential of Omicron BA.1 and BA.2 under a competitive condition. Each naive hamster was simultaneously exposed to 2 donor hamsters separately inoculated with 1 × 105 TCID50 of BA.1 or BA.2.
Figure 3.
Transmissibility of D614G, BA.1, and BA.2 in Syrian hamsters. Donors were intranasally inoculated with 1 × 105 the median tissue culture infective dose (TCID50) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. On day 1 after inoculation, naive hamster was exposed to donors for 8 hours without physical contact (eg, aerosol transmission). Subsequently, donors were cohoused with another naive hamster for 24 hours (direct contact transmission). A, Infectious virus and viral RNA were detected in nasal wash samples from D614G-exposed hamsters (aerosol and direct contact). B, Weight changes in D614G-exposed hamsters (aerosol and direct contact). C, Under a competitive transmission model, infectious virus or viral RNA was detected in nasal wash samples from BA.1/BA.2-exposed hamsters (direct contact). D, Weight changes in BA.1/BA.2-exposed hamsters (aerosol and direct contact).
Under the experimental condition that supports efficient aerosol transmission of D614G variant, we observed no transmission of BA.1 or BA.2. Specifically, no viral RNA was detected in the nasal wash samples (Figure 3C), and no seroconversion was detected in the aerosol contacts. Instead, transmission by direct contact was observed in 3 of 3 replicates, with infectious virus and viral RNA detected in nasal wash samples from direct contact hamsters on day 1 after exposure (Figure 3C). No apparent weight loss was observed from the direct contact hamsters infected with BA.1/BA.2 viruses (Figure 3D). Seroconversion was detected 18 days after exposure, with all direct contact hamsters having higher neutralizing antibody titers against BA.1 than against BA.2 (PRNT50 geometric mean titer, 179.3 vs 57.7, respectively).
Infectious viral loads, approximated by calculating the area under the receiver operating characteristic curve (AUC), were higher in the D614G-exposed than in the BA.1/BA.2-exposed direct contacts (mean AUC [standard deviation], 21.1 [0.8] vs 18.4 [1.6], respectively) (Mann-Whitney test, P = .20). Taken together, findings show that no aerosol transmission was observed for BA.1 and BA.2 viruses under the experimental condition in which D614G virus transmitted efficiently to naive hamsters.
BA.1 was Transmitted More Efficiently Than BA.2 in Hamsters by Direct Contact
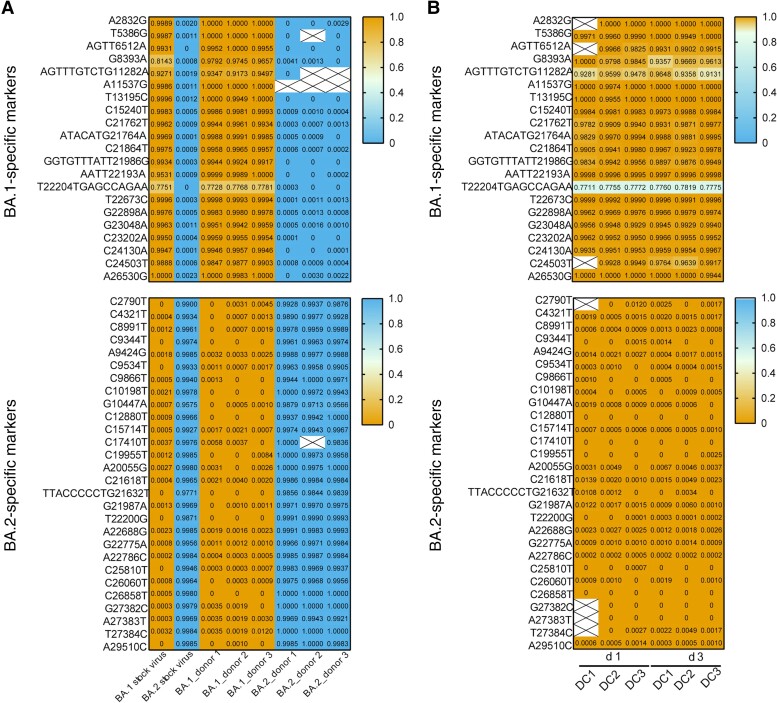
Next-generation sequencing was performed to delineate the competitive transmission potential of BA.1 and BA.2 in direct contact hamsters. Using the SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank no. MN908947.3) as the reference, a total of 40 unique molecular markers were identified that differentiate BA.1 (n = 14) and BA.2 (n = 26) viruses after exclusion of mutations shared in common between BA.1 and BA.2. We first analyzed the nasal wash samples collected from BA.1- or BA.2-inoculated donors on day 2 after inoculation (Figure 4A). High frequencies (≥91.73%) of BA.1-specific and low frequencies (≤1%) of BA.2-specific mutations were detected in BA.1-inoculated donors. In contrast, high frequencies (≥95.66%) of BA.2-specific mutations and low frequencies (≤1%) of BA.1-specific mutations were detected in BA.2-inoculated donors.
Figure 4.
Detection of BA.1- and BA.2-specific markers by next-generation sequencing. A, Detection frequency of BA.1- and BA.2-specific markers in nasal wash samples from BA.1- or BA.2-inoculated donors on day 1 after inoculation. B, Detection frequency of BA.1- and BA.2-specific markers in the nasal wash samples from exposed direct contact hamsters 1 and 3 days after exposure (d 1 and d 3). BA.1- and BA.2-specific markers that differ from the prototype Wuhan-Hu-1 virus are shown. High allele frequency of BA.1-specific markers is shown in orange, and high allele frequency of BA.2-specific markers in blue. Boxes with a large X denote regions with <100 reads and were not included in the analysis.
The next-generation sequencing (NGS) result suggests there was minimal secondary infection of BA.1- and BA.2-inoculated donors after 32 hours of cohousing. In the nasal wash samples collected from direct contact hamsters 1 and 3 days after exposure, we detected high frequencies (≥91.31%) of BA.1-specific and low frequencies (≤1.39%) of BA.2-specific mutations, suggesting that the direct contact hamsters were preferentially infected by BA.1 virus.
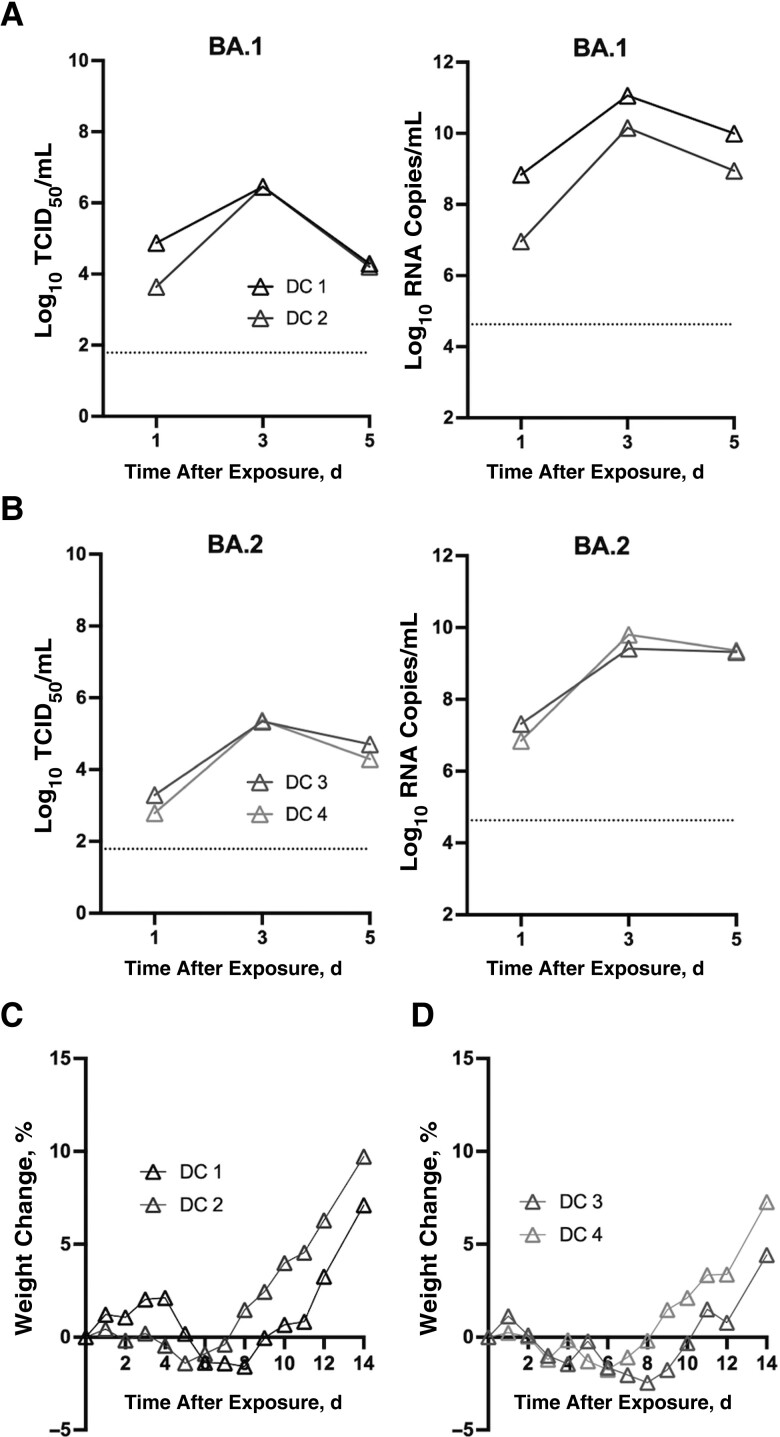
Because BA.1 was the dominant virus transmitted under the competitive model, it is important to further confirm whether BA.2 can transmit among hamsters via direct contact. After cohousing with donors inoculated with 1 × 105 TCID50 of BA.1 or BA.2 on day 1 after inoculation, infectious virus and viral RNA were detected from BA.1- or BA.2-exposed hamsters with direct contact 1 day after exposure (Figure 5A and 5B). The maximal weight losses were recorded in BA.1-exposed (maximal weight changes, −1.41% and −1.58%) (Figure 5C) and BA.2-exposed (−1.75% and −2.46%) hamsters (Figure 5D) 5–8 days after exposure. Higher viral loads were detected in nasal wash samples from BA.1-exposed contacts than in those from BA.2-exposed contacts (mean AUC [standard deviation], 16.3 [0.9] vs 11.1 [0.6], respectively) (Mann-Whitney test, P = .33). Taken together, the result demonstrates that both BA.1 and BA.2 are able to transmit among hamsters via direct contact; however, BA.1 transmits more efficiently than BA.2 in a competitive transmission model.
Figure 5.
Transmissibility of BA.1 and BA.2 via direct contact. A, B, Infectious and viral RNA were detected in nasal wash samples from BA.1-exposed (A) and BA.2-exposed (B) hamsters (direct contact). Abbreviation: TCID50, median tissue culture infective dose. C, D, Weight changes in BA.1-exposed (C) and BA.2-exposed (D) hamsters (direct contact).
DISCUSSION
Since its first detection in November 2021, the Omicron SARS-CoV-2 variant, that is capable of evading neutralizing antibodies elicited after prior infection or vaccination, has rapidly replaced previously circulating VOC. In the present study, we evaluated the pathogenicity and transmission potential of Omicron sublineages BA.1 and BA.2 in Syrian hamsters. In inoculated hamsters, compared with the early D614G virus that caused >10% weight loss, both BA.1 and BA.2 caused minimal weight changes and no apparent clinical signs. Lower viral load was detected in nasal wash samples from BA.2-inoculated hamsters than in those from BA.1- or D614G-inoculated hamsters.
Under the experimental conditions in which D614G transmitted efficiently among Syrian hamsters via aerosol or direct contact transmission models, BA.1 and BA.2 were able to transmit among hamsters via direct contact alone. Furthermore, BA.1 transmitted more efficiently than BA.2 via direct contact under a competitive transmission model. Our result is consistent with previous studies that reported attenuated pathogenicity of BA.1 in hamsters compared with previous SARS-CoV-2 strains [23] and reported that BA.2 is no more pathogenic than BA.1 in the Syrian hamster model [36]. The differential detection of BA.1 and BA.2 viral antigens by the anti-S monoclonal antibody highlights potential antigenic differences in the S protein. Caution may be needed on the use of reagents to detect antigens of Omicron, and the use of polyclonal anti-N antibodies may provide less biased results across different SARS-CoV-2 variants.
Syrian hamsters have been a useful model for studying SARS-CoV-2 transmission via multiple transmission routes [28, 29]. Interestingly, Omicron BA.1 and BA.2 lacked the ability to transmit in hamsters via aerosols under the experimental condition in which the DD614G variant transmitted efficiently. The furin cleavage site at the S1/S2 junction was previously demonstrated to be required for SARS-CoV-2 transmission in the ferret model [37], and amino acid changes in proximity to the furin cleavage site may enhance viral fusion and pathogenicity [38]. Both Omicron BA.1 and BA.2 contained N679K and P681H amino acid changes near the furin cleavage site.
These changes were previously identified in other VOC and were associated with increased prevalence of Gamma sublineages in Brazil [39]. Omicron differs from previous SARS-CoV-2 variants in its reduced dependence on TMPRSS2 cleavage, which facilitates fusion and entry at plasma membrane [21]. Instead, Omicron uses an endosomal entry pathway. In addition, removing the furin cleavage site from SARS-CoV-2 or introducing it showed that the furin cleavage site of S protein determines viral dependence on TMPRSS2 [40]. This furin-TMPRSS2 interdependence is also supported by the detection of variants with deletion in the furin cleavage site after passaging in Vero-E6 cells that lack TMPRSS2 [41]. Further studies are required to understand whether the reduced TMPRSS2 dependence of Omicron has any effect on S1/S2 cleavage by furin in humans and in different animal models.
In a competitive transmission model, we observed that BA.1 transmitted more efficiently than BA.2 by direct contact. This may be due to less efficient viral replication of BA.2 in Syrian hamsters, as noted in the viral loads detected in nasal wash samples and nasal turbinate and lung tissues. While both BA.1 and BA.2 may separately transmit to naive direct contacts, NGS data suggest that coinfection of BA.1 and BA.2 was infrequent under the competitive model. Specifically, high frequencies (≥91.31%) of BA.1-specific and low frequencies (≤1.39%) of BA.2-specific mutations were detected in the nasal wash samples of direct contact hamsters 1 and 3 days after exposure, and no recombination of BA.1 and BA.2 was noted. Our experiments are limited by a small sample size, which may not fully recapitulate the viral dynamics in humans.
While animal models provide helpful insights on SARS-CoV-2 pathogenicity and transmissibility, the absence of aerosol transmission in hamsters appears to differ from what is observed epidemiologically in humans. This may limit the ability to extrapolate transmission data in hamsters to humans. However, the reduced pathogenicity of Omicron in Syrian hamsters is concordant with data observed in humans. It remains to be investigated whether Omicron with multiple changes in the S protein has changed the ability to infect a wide range of mammalian species [42], although Omicron viral RNA was recently detected in white-tailed deer [43].
In conclusion, Omicron BA.1 and BA.2 showed reduced pathogenicity and transmission potential compared with prototype or D614G SARS-CoV-2 viruses in Syrian hamsters. Furthermore, BA.1 demonstrated a transmission advantage over BA.2 via direct contact in hamsters. The reduced transmission potential of BA.1 and BA.2 in Syrian hamsters, compared with previous SARS-CoV-2 variants, provides a research opportunity for the study of host factors and virus-host interaction critical to SARS-CoV-2 transmission.
Contributor Information
Wen Su, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Ka Tim Choy, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Haogao Gu, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Sin Fun Sia, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Ka Man Cheng, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Sarea Islam Nuha Nizami, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Pavithra Krishnan, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Yuet Mai Ng, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Lydia Dai Jia Chang, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Yingzhi Liu, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Samuel M S Cheng, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Malik Peiris, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Leo L M Poon, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
John M Nicholls, Department of Pathology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Hui-Ling Yen, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Notes
Financial support. This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant U01AI151810 and 75N93021C00016), and Research Grant Council Theme-based Research Schemes, Hong Kong, China (grants T11-712/19-N and T11-705/21-N).
References
- 1. Davies NG, Abbott S, Barnard RC, et al. . Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021; 372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hart WS, Miller E, Andrews NJ, et al. . Generation time of the Alpha and Delta SARS-CoV-2 variants: an epidemiological analysis. Lancet Infect Dis 2022; 22:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Twohig KA, Nyberg T, Zaidi A, et al. . Hospital admission and emergency care attendance risk for SARS-CoV-2 Delta (B.1.617.2) compared with Alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis 2022; 22:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao Y, Yisimayi A, Bai Y, et al. . Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res 2021; 31:732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cele S, Gazy I, Jackson L, et al. . Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 2021; 593:142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann M, Arora P, Gross R, et al. . SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021; 184:2384–93.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Planas D, Veyer D, Baidaliuk A, et al. . Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276–80. [DOI] [PubMed] [Google Scholar]
- 8. Wu J, Nie J, Zhang L, et al. . The antigenicity of SARS-CoV-2 Delta variants aggregated 10 high-frequency mutations in RBD has not changed sufficiently to replace the current vaccine strain. Signal Transduct Target Ther 2022; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emary KRW, Golubchik T, Aley PK, et al. . Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet 2021; 397:1351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eyre DW, Taylor D, Purver M, et al. . Effect of Covid-19 vaccination on transmission of Alpha and Delta variants. N Engl J Med 2022; 386:744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madhi SA, Baillie V, Cutland CL, et al. . Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med 2021; 384:1885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osorio NS, Correia-Neves M. Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays. Lancet Infect Dis 2021; 21:166–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Viana R, Moyo S, Amoako DG, et al. . Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022; 603:679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsueng G, Mullen JL, Alkuzweny M, et al. . Outbreak.info Research Library: a standardized, searchable platform to discover and explore COVID-19 resources. bioRxiv 477133 [Preprint: not peer reviewed]. 2 June 2022. Available from: https://www.biorxiv.org/content/10.1101/2022.01.20.477133v4. doi: 10.1101/2022.01.20.477133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu L, Iketani S, Guo Y, et al. . Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022; 602:676–81. [DOI] [PubMed] [Google Scholar]
- 16. Cheng SMS, Mok CKP, Leung YWY, et al. . Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med 2022; 28:486–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perez-Then E, Lucas C, Monteiro VS, et al. . Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med 2022; 28:481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cameroni E, Bowen JE, Rosen LE, et al. . Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022; 602:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han P, Li L, Liu S, et al. . Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2. Cell 2022; 185:630–40.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolter N, Jassat W, Walaza S, et al. . Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa: a data linkage study. Lancet 2022; 399:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meng B, Abdullahi A, Ferreira I, et al. . Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022; 603:706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hui KPY, Ho JCW, Cheung MC, et al. . SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022; 603:715–20. [DOI] [PubMed] [Google Scholar]
- 23. Halfmann PJ, Iida S, Iwatsuki-Horimoto K, et al. . SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022; 603:687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shuai H, Chan JF, Hu B, et al. . Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022; 603:693–9. [DOI] [PubMed] [Google Scholar]
- 25. Fonager J, Bennedbaek M, Bager P, et al. . Molecular epidemiology of the SARS-CoV-2 variant Omicron BA.2 sub-lineage in Denmark, 29 November 2021 to 2 January 2022. Euro Surveill 2022; 27:2200181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan JF, Zhang AJ, Yuan S, et al. . Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020; 71:2428–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imai M, Iwatsuki-Horimoto K, Hatta M, et al. . Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 2020; 117:16587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Port JR, Yinda CK, Owusu IO, et al. . SARS-CoV-2 disease severity and transmission efficiency is increased for airborne compared to fomite exposure in Syrian hamsters. Nat Commun 2021; 12:4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sia SF, Yan LM, Chin AWH, et al. . Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020; 583:834–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuyama S, Nao N, Shirato K, et al. . Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020; 117:7001–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shirogane Y, Takeda M, Iwasaki M, et al. . Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 2008; 82:8942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu DKW, Pan Y, Cheng SMS, et al. . Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 2020; 66:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493–7. [Google Scholar]
- 34. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grubaugh ND, Gangavarapu K, Quick J, et al. . An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 2019; 20:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uraki R, Kiso M, Iida S, et al. . Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Nature 2022; 607:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peacock TP, Goldhill DH, Zhou J, et al. . The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 2021; 6:899–909. [DOI] [PubMed] [Google Scholar]
- 38. Saito A, Irie T, Suzuki R, et al. . Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022; 602:300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naveca FG, Nascimento V, Souza V, et al. . Spread of Gamma (P.1) sub-lineages carrying spike mutations close to the furin cleavage site and deletions in the N-terminal domain drives ongoing transmission of SARS-CoV-2 in Amazonas, Brazil. Microbiol Spectr 2022; 10:e0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ou T, Mou H, Zhang L, Ojha A, Choe H, Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog 2021; 17:e1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sasaki M, Uemura K, Sato A, et al. . SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog 2021; 17:e1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hobbs EC, Reid TJ. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound Emerg Dis 2021; 68:1850–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vandegrift KJ, Yon M, Surendran-Nair M, et al. . Detection of SARS-CoV-2 Omicron variant (B.1.1.529) infection of white-tailed deer. bioRxiv 479189 [Preprint: not peer reviewed]. 7 February 2022. Available from:https://www.biorxiv.org/content/10.1101/2022.02.04.479189v1. doi: 10.1101/2022.02.04.479189 [DOI] [Google Scholar]