Abstract
Myeloid-derived suppressor cells (MDSC) are induced during active TB disease to restore immune homeostasis but instead exacerbate disease outcome due to chronic inflammation. Autophagy, in conventional phagocytes, ensures successful clearance of M.tb. However, autophagy has been demonstrated to induce prolonged MDSC survival. Here we investigate the relationship between autophagy mediators and MDSC in the context of active TB disease and during anti-TB therapy. We demonstrate a significant increase in MDSC frequencies in untreated active TB cases with these MDSC expressing TLR4 and significantly more mTOR and IL-6 than healthy controls, with mTOR levels decreasing during anti-TB therapy. Finally, we show that HMGB1 serum concentrations decrease in parallel with mTOR. These findings suggest a complex interplay between MDSC and autophagic mediators, potentially dependent on cellular localisation and M.tb infection state.1
Keywords: myeloid-derived suppressor cells, Mycobacterium tuberculosis, Tuberculosis, Autophagy, high mobility group box protein 1
1. Introduction
Immune protection against tuberculosis (TB) disease is associated with induction of T helper cell-1 (Th1) and pro-inflammatory immune responses [1]. Endogenous amounts of proinflammatory cytokines produced locally in the lungs benefit the host by activating antimicrobial pathways and stimulating tissue repair, whereas uncontrolled release of large amounts of these mediators, signal the onset of tissue injury [2], [3]. This is a potentially disastrous scenario that is normally prevented by counter-regulatory mechanisms, evolved to inhibit cytokine overproduction. One mechanism of innate immune regulation in TB involves the induction of myeloid-derived suppressor cells (MDSC) [4].
MDSC are classified as immunosuppressive cells of monocytic (M-MDSC; Lin−/HLA-DRlow/−/CD33+/CD11b+/CD14+/CD15−) and granulocytic/polymorphonuclear (PMN-MDSC; Lin−/HLA-DRlow/−/CD33+/CD11b+/CD14−/CD15+) phenotypes [5], [6], arising from the immature precursor “early-stage” MDSC subset (e-MDSC; Lin−(CD3/14/15/19/56)/HLA-DR−/CD33+) [6]. More recently, a new subset resembling immature eosinophils (Eo-MDSC) were identified in mice infected with Staphylococcus aureus, and further expanded our knowledge of suppressive granulocytes [7]. These cells accumulate in response to chronic or excessive inflammatory conditions, such as cancer, mediating tumour escape through their potent T-cell suppressive functions in the tumour microenvironment [8], [9].
Detrimental immunosuppressive effects of MDSC are also recognised in infectious diseases, potentially restricting protective anti-pathogen responses [10]–[13]. We and others have previously shown that MDSC from individuals with untreated active TB disease exhibit a potent ability to impair CD4 T-cell homing by targeting their L-selectin expression, reducing CD4 T-cell cytokine production and reducing CD8 T-cell activation and proliferation [12], [13]. In cancer, MDSC suppression occurs through production of high levels of arginase-1 (Arg1) and inducible nitric oxide synthase (iNOS), resulting in the depletion of L-arginine which is essential for T-cell proliferation [14]–[16]. Other suppressive mechanisms reported from the cancer field include production of indoleamine-2,3 dioxygenase (IDO) and TGF-β-mediated immunosuppression [17]–[21]. In the context of TB, our previous investigations suggested minor roles for Arg-1, iNOS and IDO [12], [22].
MDSC-induced suppression in malignancy, through regulation of L-selectin on T-cells, is mediated by MDSC plasma membrane expression of the enzyme, A Disintegrin And Metalloproteinase Domain-17 (ADAM-17) [23], [24]. ADAM-17 cleaves the ectodomain of L-selectin on T-cells, decreasing their ability to home to tumour sites where they would be activated [23]. Tumor-derived MDSC have demonstrated ADAM-17 expression, and this expression may be further sustained by the extracellular, damage-associated molecular pattern (DAMP) and alarmin, high mobility group box 1 (HMGB1), which contributes to the downregulation of L-selectin on T-cells [23], [25]. HMGB1 may also act as a binding partner, inducer and/or chaperone for multiple pro-inflammatory molecules that drive MDSC expansion, like interleukin (IL)-6 [26], [27]. HMGB1 is secreted extracellularly during cellular stress responses induced through hypoxia or nutrient starvation, or after exposure of immunocompetent cells to products of pathogenic bacteria, binding to either the receptor for advanced glycation end products (RAGE) [28], or the toll-like receptors (TLR)-2 and/or −4 [27]; all shown to be highly expressed on the surface of tumor-derived MDSC. Binding of HMGB1 to these receptors activates the NF-κB signalling pathway and the subsequent production of IL-10 from the induced MDSC. HMGB1 has been demonstrated to further promote the development of MDSC, and their ability to suppress antigen-driven activation of T-cells by inducing MDSC secretion of IL-10 and IL-1β [25]. In addition to regulating MDSC immunosuppressive potential, HMGB1 also prolongs MDSC survival through the induction of autophagy under conditions of stress, as described in cancer by many [29]–[37], and is particularly beneficial for the human host during M.tb infection owing to its mechanism of virulent bacterial clearance [38].
The autophagic response is regulated by the mammalian target of rapamycin (mTOR) pathway a member of the phosphatidylinositol-3 (PI-3)-kinase-related family [28], [39], [40]. mTOR is regulated by the binding of HMGB1 to its receptors (RAGE, TLR2/4), with binding of HMGB1 facilitating the inhibition of mTOR and subsequently preventing the inhibition of the autophagic response [41], [42]. Recently, mTOR has been identified as a master regulator in cancer cells, driving the expression of tumor-promoting G-CSF and PMN-MDSC accumulation [35]. Direct MDSC treatment with rapamycin showed opposing effects on the two MDSC subsets: while PMN-MDSC generation was promoted by rapamycin [36], M-MDSC generation and function were inhibited [43] likely due to inhibition of their glycolytic metabolism [41]. In contrast, inhibition of mTOR has been shown to promote autophagy and increase the viability of bulk CD11b+ GR1+ MDSC [36]. In addition to this, autophagy has promoted the suppressive capacity of M-MDSC [30], while in PMN-MDSC autophagy reduced their accumulation and suppression [31]. Thus, the interplay between mTOR and autophagy and its impact on MDSC shows conflicting results [44].
The investigation of the mTOR signalling pathway and autophagy has, to our knowledge, not been done in the context of MDSC during active TB disease. Interestingly, studies investigating autophagy during M.tb infection in general have shown evidence that both mTOR activation and inhibition may occur depending on the species or strain virulence in question [45]. In this study we aimed to provide a M.tb infection-specific description of kinetic changes of circulating MDSC during TB treatment, and specifically investigate whether the HMBG1-mTOR signalling pathway is linked to MDSC induction or function. Here we demonstrate the multi-faceted role of the HMGB1 pathway in the immunosuppressive functioning of MDSC as a means by which their immunosuppressive functions may be exacerbated/lessened and host control of bacterial invasion inhibited. Extracellular HMGB1 produced systemically in response to infection has the potential to bind to one of its receptors, in this case TLR4, which we have demonstrated is expressed on MDSC. This may subsequently induce the production of proinflammatory cytokines like IL-6 to further induce MDSC expansion from the bone marrow of the host. Further research into this pathway in the context of M.tb infection and MDSC specifically may hold promise as unique targets for host-directed therapies.
2. Materials and methods
2.1. Subject enrolment and sample collection
In this study, eligible adult participants (>18 years of age) were selected from two existing longitudinal multi-centre studies in Cape Town, South Africa – the Concurrent Tuberculosis and Diabetes: Clinical Monitoring, Microbiological and Immunological Effects of Diabetes During TB Treatment (TANDEM) study and the ScreenTB study. Newly diagnosed, untreated active TB disease participants were recruited, and followed-up for 18 months. At the baseline visit, medical history and clinical examination occurred, followed by routine TB diagnostic tests: three independent samples of 2ml smear, GeneXpert and MGIT culture, as well as Chest X-ray and HIV rapid test (finger prick). At month 2 and month 6 follow up, all participants and only TB-case participants respectively, had repeat diagnostic assessment (medical history and examination, sputum smear, GeneXpert and culture, CXR). Each participant’s outcome was classified using harmonised case definitions (Table 1), with treatment response monitored by sputum culture conversion at the month 2 and month 6 time points.
Table 1:
Classification criteria of participants based on combinations of clinical and microbiological findings.
| Classification: | Definition: |
|---|---|
| Definite TB | 2 positive sputum cultures, or 1 positive culture and CXR suggestive of TB. |
| Probable TB | 2 positive smears or 1 culture, without documented CXR |
| evidence, but symptoms responding to TB treatment, or 1 positive smear with CXR consistent with PTB. | |
| Questionable diagnosis | Positive smear(s), but no other supporting evidence, or CXR suggestive of TB, but no other supporting evidence. |
| No TB | Negative cultures, negative smears, negative CXR leading to an alternate diagnosis. |
Select participants recruited at diagnosis also had peripheral blood drawn at additional time points, including two weeks- (W2), one month- (M1), two months- (M2), and six months (M6) after the initiation of 2HRZE/4HR anti-TB treatment. Non-TB (otherwise healthy) controls were also recruited, and peripheral blood collected only at the initial visit. Peripheral blood was collected in either two 9mL sodium heparin tubes (BD Biosciences), or one 5mL EDTA (BD Biosciences) anti-coagulant tube where available.
2.2. Ethics considerations
Ethics approval (S12/06/163) was obtained from the Health Research Ethics Committee (HREC) of the Faculty of Medicine and Health Sciences of Stellenbosch University, Cape Town, South Africa, and the research was performed in accordance with the amended Declaration of Helsinki. City of Cape Town regional approval and HREC ethics approval were obtained previously for each of the source cohort studies, and written informed consent was provided by all participants.
2.3. Whole Blood Lysis
The peripheral blood samples were collected in sodium heparin tubes (BD Biosciences) and divided for whole blood lysis (1mL), and peripheral blood mononuclear cell (PBMC) isolations (remainder ~16mL). Erythrocytes were lysed using FACS lysing solution (BD Biosciences, San Jose, CA) to enumerate whole blood cell frequencies. Briefly, 1mL of peripheral blood was lysed with 9mL FACS lysing solution (1:10 as per instructions) for 20 minutes in the dark at room temperature (RT). Cells were then pelleted at 400 × g for 10 minutes and washed twice with 10mL 1X phosphate buffered saline (PBS). Washed cells were pelleted, suspended in 200μl 1X PBS and stored at 4°C prior to analysis by flow cytometry.
2.4. Peripheral Blood Mononuclear Cell Isolation and Cell enrichments
PBMC were isolated from peripheral blood by centrifugation using density gradient medium (Histopaque-1077; Sigma Chemical Co., St. Louis, MO). Cells were counted and the viability determined to be >90% by Trypan Blue exclusion method. PBMC were then used to enrich for MDSC using the MACS® (magnetically activated cell sorting) MicroBead Isolation technique (Miltenyi Biotec, Cologne, Germany) according to the manufacturer’s instructions. Briefly, PBMC were pelleted by centrifugation and stained with CD3 Microbeads (130–050101) by adding 20μl of Microbeads and 80μl (per 1×107 cells) MACS buffer (containing 1XPBS, 2mM EDTA and 0.5% bovine serum albumin (BSA)) for 15 minutes (4°C). Cells were then washed with 1ml (per 1×107 cells) MACS buffer and centrifuged at 300 × g for 10 minutes. Resuspended cells (500μl MACS buffer) were then separated using a column surrounded by a magnetic field, with the CD3 negative (unlabelled) and CD3 positive (labelled) fractions being collected into separate sterile 15ml Falcon tubes. The CD3 negative fraction was then used to enrich for HLA-DR negative cells using anti-HLA-DR Microbeads (130–046-101) and the method described above. The HLA-DR negative (unlabelled) fraction was collected in a sterile 15ml Falcon tube and used to enrich for CD33 positive cells using CD33 MicroBeads (130–045-501) and the same method described above. The CD33 positive (labelled) fraction was collected and represent the enriched MDSC population, with a final enriched phenotype of CD3−HLA-DR−CD33+. Enriched MDSC were then either used for stimulation assays or cryopreserved in cryomedia consisting of 90% fetal bovine serum (FBS; HyClone, GE Healthcare Life Sciences, Illinois, USA) and 10% dimethyl sulfoxide (DMSO; Sigma-Aldrich Co., St. Louis, MO) and stored in liquid nitrogen until later batch-analysis of cellular phenotypes by flow cytometry.
2.5. MDSC Stimulation with purified protein derivative (PPD)
The cytokine profile of MDSC stimulated with M.tb-specific proteins was assessed in culture using PPD. Enriched MDSC (2×105 MDSC per well) from each participant were cultured in a 96-well round-bottom culture plate for 48 hours under two stimulatory conditions: (i) unstimulated and (ii) stimulated with PPD (10μg/ml) in a total volume of 200μl complete RPMI-1640 supplemented with 1% L-glutamine (Sigma-Aldrich, St Louis, Missouri, USA), 2% FBS. Following the 48-hour incubation, supernatants were harvested and stored at −80°C until use in Luminex immunoassays, and the cells cryopreserved in cryomedia and stored in liquid nitrogen.
2.6. Flow cytometry
Phenotypes of enriched MDSC were analysed by flow cytometry in batches. For this, cells were thawed in a water bath set to 37°C and transferred to 10ml pre-warmed complete RPMI-1640. Cells were washed twice in 10ml pre-warmed complete RPMI-1640, centrifuging at 250 × g for 10 minutes between washes. Cell counts and viability were determined using a haemocytometer and the trypan blue exclusion method, and then fixed with 4% paraformaldehyde (PFA) for 10 minutes in the dark (RT) and washed with 1X PBS. Cells were then pelleted and stained with conjugated antibodies (volume per reaction: 50μl), purchased from BD Biosciences, for 30 minutes in the dark (RT). The surface marker antibodies included: CD3 APC-Cy7 (clone: SK7), CD14 PE (clone: MᵠP9), CD15 BV510 (clone: W6D3), CD11b PerCP-Cy5.5 (clone: M1/70), CD33 PE-Cy7 (clone: P67.7), HLA-DR APC (clone: L243), and TLR4 BV421 (clone: TF901).
To determine the number of MDSC expressing mTOR, fixed and lysed whole blood samples were stained with antibodies against intracellular and surface proteins for 30 minutes in the dark (RT) and washed as per manufacturer’s instructions. Cells were permeabilised using the eBioscience™ Intracellular Fixation and Permeabilization Buffer set (catalogue number: 88–8824; eBioscience, Thermo Fisher Scientific, Massachusetts, United States) according to the manufacturer’s instructions. Surface markers included: (1) LIN1-FITC (lineage marker 1 cocktail contains CD3, CD14, CD16, CD19, CD20, and CD56; used as exclusion marker), (2) CD33-PE (clone: P67.6), (3) HLA-DR-V500 (clone: G46–6), (4) CD15V450 (clone: MMA), and (5) CD11b-APC-Cy7 (clone: ICRF44); while (6) phosphorylated-mTOR (Ser2448)-PerCP (clone: MRRBY) was intracellular.
Cell acquisitions were performed on a FACS Canto II (BD Biosciences, New Jersey, USA). Instrument calibration was performed according to manufacturer’s instructions and compensation settings adjusted using antibody-capture beads (CompBeads, BD Biosciences). Data was analysed using FlowJo v10 software (Oregon, USA). After excluding doublet populations, lymphocytes were selected using the FSC-A vs SSC-A gates, followed by the selection of the CD3 and HLA-DR double negative population. CD33 and CD11b double positive (total MDSC; CD3−/HLA-DRlow/−/CD33+/CD11b+) populations were then selected, from which either CD14 (M-MDSC; CD3−/HLA-DRlow/−/CD33+/CD11b+/CD14+/CD15−) or CD15 (PMN-MDSC; CD3−/HLA-DRlow/−/CD33+/CD11b+/CD14−/CD15+) positive populations could be gated on. Single gates were then used to detect TLR4 expression on either the M-MDSC (CD14+/TLR4+) or PMN-MDSC (CD15+/TLR4+) populations. For the gating of lysed whole blood samples for the detection of mTOR expression, the same gating strategy was used, except for lymphocytes first being gated on the LIN1 negative population, instead of the CD3 negative population. Single gates were used to detect phosphorylated-mTOR expression in either the M-MDSC (CD14+/phospho-mTOR+) or PMN-MDSC (CD15+/phospho-mTOR+) populations.
2.7. HMGB1 and ADAM17/TACE ELISA assays
From the subset for which venous blood was collected into EDTA anti-coagulant tubes, the tubes were immediately centrifuged for 15 minutes at 1000 rpm (RT). The isolated plasma was then stored at −80°C until later batch analysis. The plasma samples from the patients and controls were routinely diluted 1:4 to assess the protein levels of ADAM17 (Abcam), and likewise diluted 1:50 to assess the protein levels of HMGB1 (Abcam) by commercial ELISA according to the manufacturer’s instructions. The OD values were measured using an automatic microplate reader at 450 nm. The concentration of samples was calculated using a log-log curve by regression/fit calculation program.
2.8. Multiplex cytokine assay
The Luminex immunoassay platform (Luminex, Bio Rad Laboratories, Hercules, CA, USA) using a MILLIPLEX® multiplex kit (MERCK MILLIPORE, Darmstadt, Germany) was used for detection and quantification of the following cytokines: IL-6, IL-8, IL-10, granulocyte-macrophage colony–stimulating factor (GM-CSF), macrophage inflammatory protein-1 alpha (MIP-1α), MIP-1β, monocyte chemotactic protein-1 (MCP-1), and interferon inducible protein (IP)-10 produced by MDSC in the presence/absence of PPD. Cytokine measurements were analysed using Bio-Plex Manager v.4.1.1. software (Bio-Rad, Richmond, CA) against internal quality controls and serial dilutions of cytokine standard curves prepared according to manufacturer’s instructions.
2.9. Statistical Analysis
All data were statistically analysed using GraphPad Prism version 8.0 (GraphPad Software, San Diego, CA, USA), and a p-value of < 0.05 was considered statistically significant. The D’Agostino & Pearson Omnibus test for normality was used to determine the distribution of the datasets, after which the appropriate statistical tests were performed and outlined in the results.
3. Results
3.1. M-MDSC frequencies are elevated in PBMC from TB patients and return to frequencies comparable with healthy controls following successful TB treatment.
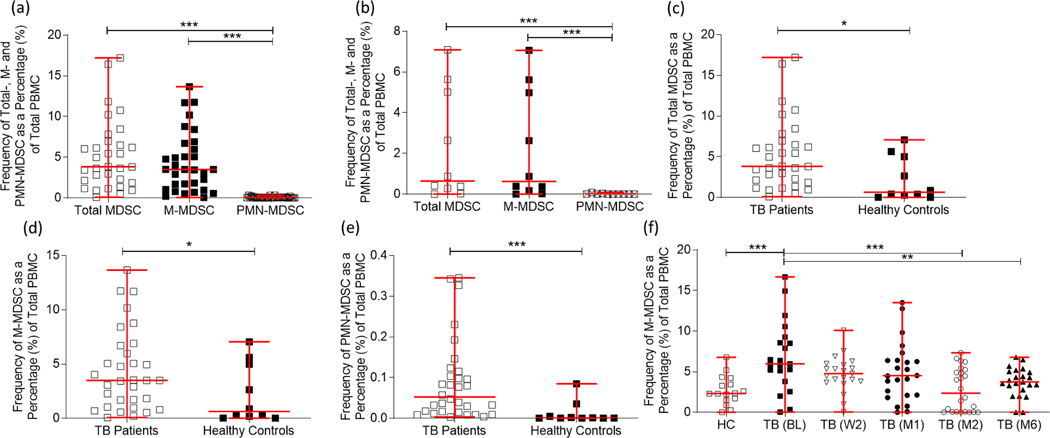
Total MDSC and major subtype frequencies in the PBMC of patients with active TB disease have previously been assessed in small sample populations with inconsistent means of characterising this innate population of myeloid cells [12]. In this study, the frequencies of total MDSC (CD3−/HLA-DRlow/−/CD33+/CD11b+), and the two dominant MDSC subtypes (MMDSC: CD3−/HLA-DRlow/−/CD33+/CD11b+/CD14+/CD15−; and PMN-MDSC: CD3−/HLADRlow/−/CD33+/CD11b+/CD14−/CD15+) were compared in PBMC from participants with active TB disease (n = 38) at the time of diagnosis (Figure 1a) and in healthy controls (n = 10; Figure 1b). No differences were observed between their total MDSC and M-MDSC frequencies (p = 0.3719 Figure 1a; p = 0.6842 Figure 1b), however there were significantly fewer PMN-MDSC compared to total MDSC (p < 0.0001), as well as M-MDSC (p < 0.0001) (Figure 1a and Figure 1b) demonstrating a potential M-MDSC dominance within the PBMC compartment of patients with active TB disease as well as in healthy controls. We then investigated the differences in frequencies of total MDSC and the major subtypes between patients with active TB disease and healthy controls. Total MDSC frequencies (Figure 1c; p = 0.0161), M-MDSC frequencies (Figure 1d; p = 0.0496), and PMN-MDSC frequencies (Figure 1e; p = 0.0003) in PBMC were observed to be upregulated in participants with active TB disease at the time of diagnosis compared to frequencies observed in healthy control participants. Based on these observations, M-MDSC are the dominant subset in the PBMC compartment.
Figure 1:
Myeloid-derived suppressor cells (MDSC) are enriched in peripheral blood mononuclear cells (PBMC) of participants with active TB disease compared to healthy controls, with the monocyte-like subset (M-MDSC) being the dominant subset compared to the granulocytic-type subset (PMN-MDSC). (a) The percentage of total PBMC for total MDSC, M-MDSC and PMN-MDSC subsets were compared in participants with active TB disease (n = 38) at the time of diagnosis and (b) in Healthy Controls (n = 10). (c) Total MDSC frequencies (p = 0.0161), (d) M-MDSC frequencies (p = 0.0496), and (e) PMN-MDSC frequencies (p = 0.0003) in PBMC were observed to be upregulated in participants with active TB disease (n = 38) at the time of diagnosis compared to frequencies observed in healthy control participants (n = 10). (f) Longitudinal M-MDSC frequencies in adult active TB disease cases. M-MDSC frequencies were investigated at the time of diagnosis/baseline (BL; n = 21), week two following treatment initiation (W2; n = 21), one month (M1; n = 24), two months (M2; n = 24), and six months (M6; n = 23) after treatment initiation and compared to healthy control participants (n= 16). Frequencies were determined using the FACS Canto II flow cytometer, and the third-party software FlowJo. D’Agostino & Pearson Omnibus test for normality was used to determine the distribution of the datasets: Kruskal-Wallis tests (a & b), Mann-Whitney tests (c-e), and a one-way ANOVA with Tukeys post-hoc test (f) were used where appropriate. Error bars represent the median and range. ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant.
MDSC induction and accumulation is known to be associated with chronic inflammation that occurs during active TB disease [12], [13]. However, no studies have assessed the change in MDSC frequencies over the course of anti-TB treatment. Owing to the previous observation that the M-MDSC subtype is dominant within the PBMC compartment, longitudinal analyses of this subtype were performed in adult active TB cases. Using paired samples where available, frequencies were investigated at the time of diagnosis/baseline (BL; n = 21), week two following treatment initiation (W2; n = 21), one month (M1; n = 24), two months (M2; n = 24), and six months (M6; n = 23) after treatment initiation and compared to healthy controls (HC; n = 16). The longitudinal analysis revealed that M-MDSC frequencies were significantly upregulated at the time of diagnosis compared to the frequencies observed in healthy controls (p = 0.001), and dropped significantly by the M2 time point (p = 0.0004), as well as by the M6 time point (p = 0.0028) to levels similar to those seen in healthy controls by the end of treatment (Figure 1f), strongly suggesting that the point at which these cells return to normal levels following the initiation of treatment is likely around the month 2 time point. Taken together, these data show that MDSC are elevated in TB patients at diagnosis and return to frequencies comparable with healthy donors at month 6, following successful treatment.
3.2. MDSC upregulate IL-6 following Purified Protein Derivative (PPD) stimulation.
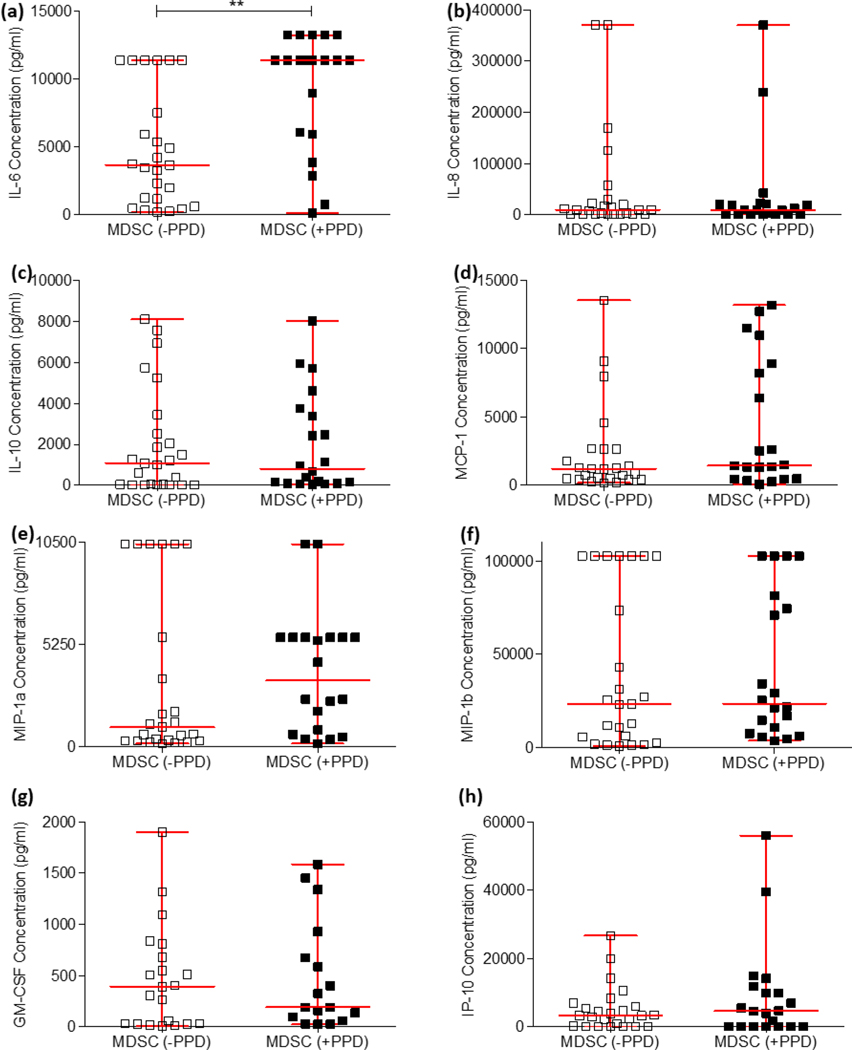
In cancer, various cytokines have been demonstrated to enhance MDSC expansion (IL6 and TNF-α [46]) and mediate their immunosuppressive functions (IL-10 and TGF-β) [47], [48]). These cytokine functions have recently been demonstrated to be synonymous with MDSC functions during active TB disease [10], [12]. To investigate which suppressive mediators are released by MDSC isolated from active TB patients, this study performed a stimulation experiment using PPD to determine whether mycobacterial proteins are sufficient to induce these mediators. Enriched MDSC isolated from the PBMC compartment of participants with active TB disease were cultured and stimulated with or without M.tb PPD. Supernatants were harvested after the 16 hour culture period, and the production of twelve extracellular cytokines were assessed using the Luminex immunoassay platform to compare differences in cytokine production between PPD-stimulated (n = 20) and unstimulated MDSC (n = 25); these included IL-6 (Figure 2a; p = 0.0011), IL-8 (Figure 2b; p = 0.8893), IL-10 (Figure 2c; p = 0.545), MCP-1 (Figure 2d; p = 0.2132), MIP-1a (Figure 2e; p = 0.6613), MIP-1b (Figure 2f; p = 0.6704), GM-CSF (Figure 2g; p = 0.8029), and IP-10 (Figure 2h; p = 0.4718). MDSC display no difference in cytokine responses following PPD-specific stimulation, except for IL-6, which was significantly upregulated (Figure 2c). It is generally accepted in literature that MDSC are unable to be isolated from healthy donors due to them not having a chronic inflammatory condition or are found in incredibly low numbers in healthy controls recently exposed to M.tb (tuberculin skin test positive) [4], [12]. For these reasons, this experiment was not conducted with MDSC from healthy volunteers. As observed in this study, the frequency of MDSC from healthy controls are significantly lower than that of TB patients (Fig 1c–e) and this posed a significant challenge in terms of adequate cell numbers when trying to conduct the above-mentioned experiment, as equal cell numbers were required for appropriate comparison, and is therefore a limitation of this study.
Figure 2:
The production of twelve extracellular cytokines were assessed using the Luminex immunoassay platform to compare differences in production between PPD-stimulated (n = 20) and unstimulated MDSC (n = 25). D’Agostino & Pearson Omnibus test for normality was used to determine the distribution of the datasets, after which the statistics used for the cytokines IL-6 and MIP-1a were parametric (unpaired t-test), and the statistics used for the remaining cytokines were non-parametric (MannWhitney test). Error bars represent the median and range. ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant.
3.3. HMGB1, but not ADAM17, is significantly downregulated following anti-TB therapy.
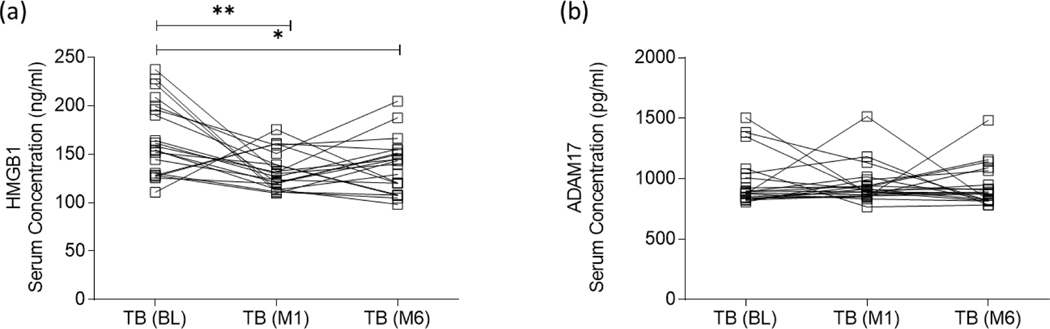
HMGB1, a secreted DAMP, has been demonstrated to drive the differentiation of MDSC within the bone marrow, and enhance their immunosuppressive effects through the induction of IL-10 [25]. This study, and others, was performed in the context of cancer, while no previous research has specifically investigated M.tb infection-specific production of HMGB1 and its role in MDSC [25], [36], [49], [50]. While the expression of ADAM17 on the surface of MDSC is well known, again it has not been investigated in the context of active TB disease, although its function is expected to be synonymous with that of cancer. Expression levels of HMGB1 and ADAM17 were therefore evaluated in serum of people with active TB disease (n = 20) at the time of diagnosis/baseline (BL), one month (M1) and six months (M6) after the initiation of treatment. While high levels of both proteins were measured at diagnosis, baseline levels of only HMGB1 protein (Figure 3a) diminished at the M1 (p = 0.0124) and M6 (p = 0.0278) time points. Longitudinal analysis of ADAM17 protein (Figure 3b) showed no difference between any of the investigated time points following TB treatment initiation. This downregulation of HMGB1 production corresponds with the downregulation of MDSC frequencies longitudinally over the course of anti-TB treatment (Figure 1f).
Figure 3:
Expression levels of (a) HMGB1 and (b) ADAM17 were evaluated using the ELISA platform in serum of participants with active TB disease (n = 20) at the time of diagnosis/baseline (BL), one month (M1) and six months (M6) after the initiation of treatment. D’Agostino & Pearson Omnibus test for normality was used to determine the distribution of the protein data, after which the statistics used for HMGB1 were parametric (repeated measures one-way ANOVA and Tukey’s test for multiple comparisons), while the statistics used for ADAM17 were non-parametric (Friedman test and Dunn’s test for multiple comparisons). Error bars represent the median and range. ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant.
3.4. TLR4 expression on MDSC is significantly upregulated at TB diagnosis.
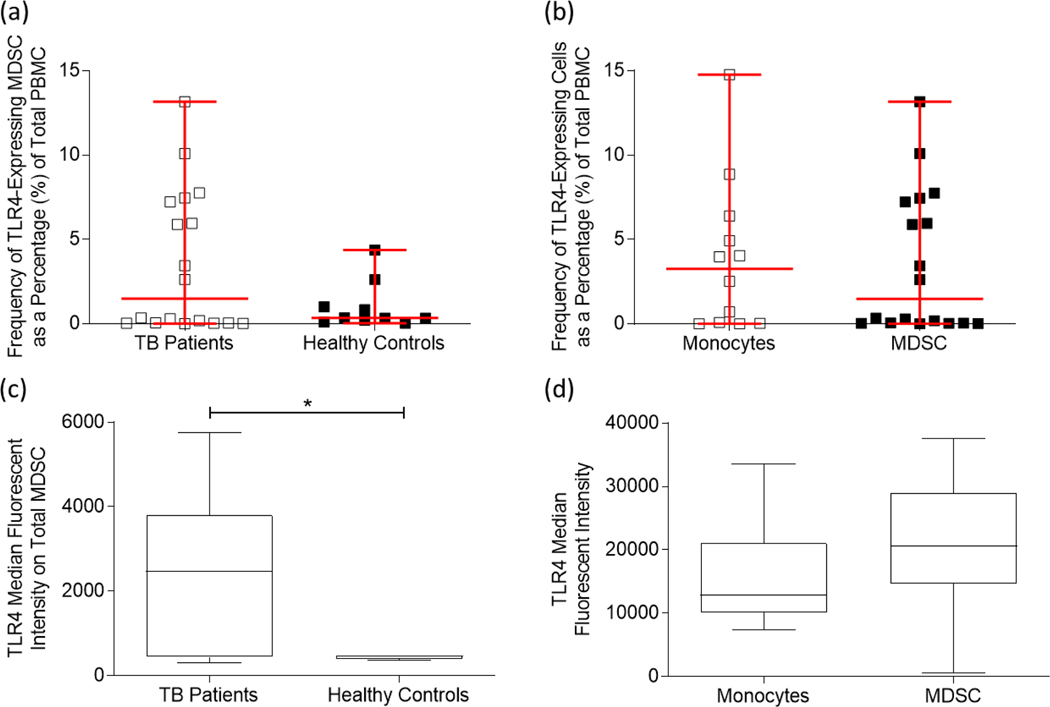
HMGB1 stimulates pro-inflammatory cytokine production in one of three ways: (1) through binding to the RAGE receptor, (2) binding to the TLR2 and/or TLR4 receptors, and (3) through DNA transcription in the nucleus. As mentioned previously, both RAGE [28] and TLR2/4 [37], [51] have been demonstrated to be expressed on the surface of MDSC. This study therefore aimed to investigate the expression of TLR4 specifically, in the hopes of delineating the relationship between HMGB1 and MDSC in the context of active TB disease. In this study, the frequency of TLR4-expressing MDSC was investigated in patients with active TB disease (n = 18) and in healthy controls (n = 10), and isolated CD14+ monocytes (n = 12) were used as a control cell population owing to the phenotypic similarities between conventional monocytes and M-MDSC, and the observation that the total MDSC population consisted mostly of MMDSC. The frequency of TLR4-expressing MDSC did not significantly differ between the two patient groups (Figure 4a; p = 0.0735), nor was there a significant difference between the monocyte control population and total MDSC (Figure 4b; p = 0.8667). There was, however, a significant upregulation of the median fluorescent intensity (MFI) of TLR4-expressing MDSC isolated from patients with active TB disease compared to healthy controls (Figure 4c; p = 0.0439). Unfortunately, the MFI of TLR4-expressing total MDSC and control monocytes showed no significant changes (Figure 4d; p = 0.2028). Where frequencies of MDSC expressing TLR4 demonstrate the number of MDSC expressing TLR4, the MFI is a measure of how many receptors are on a given cell. Therefore, while TLR4-MDSC and –monocyte control population frequencies do not significantly differ, it is reasonable to suggest that the MDSC cellular compartment expresses more of these receptors on their surface, thereby allowing for enhanced binding of ligands such as HMGB1.
Figure 4:
Myeloid-derived suppressor cells derived from the peripheral blood mononuclear cells of adult participants with active TB disease express Toll-like Receptor 4 (TLR4). (a) The frequency of TLR4-expression on MDSC from active TB disease participants (n = 18) and healthy control participants (n = 10). (b) TLR4 expression on total MDSC (n = 18) and control monocyte populations (n = 12). (c) The median fluorescent intensity (MFI) of TLR4 expression on MDSC from participants with active TB disease compared to healthy control participants; as well as (d) the MFI of TLR4 expression on control monocytes and total MDSC. Frequencies were determined using the FACS Canto II flow cytometer, and the third-party software FlowJo. Error bars represent the median and range, while the whiskers represent the 5th-95th percentiles. ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant.
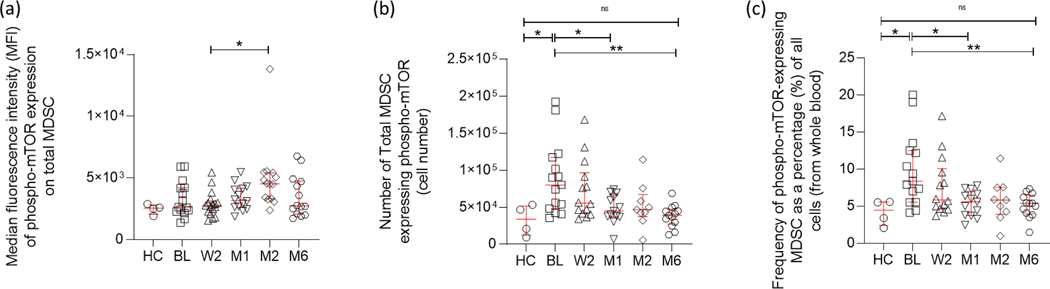
3.5. MDSC showing mTOR phosphorylation are enriched at TB diagnosis and decrease with TB treatment.
HMGB1 is known to enhance MDSC development and function, and may prolong MDSC survival by driving the autophagy process within these cells [36]. Under nutrient-starved conditions, HMGB1 binds to its receptors (RAGE, TLR2/4) signalling the inhibition of mTOR which subsequently initiates the autophagic response [39], [52]–[54]. Contrastingly, under nutrient-rich conditions, mTOR remains active and inhibits autophagy. Additionally, increased mTOR phosphorylation has been described as a marker for M-MDSC in the blood of tumor patients [43]. For this reason, the expression of intracellular phospho-mTOR was investigated in MDSC using flow cytometry.
Longitudinal analysis of phospho-mTOR+ total MDSC from the peripheral blood of patients with active TB disease at the time of diagnosis/baseline (BL; n = 15), week two following treatment initiation (W2; n = 14), one month (M1; n = 13), two months (M2; n = 9), and six months (M6; n = 13) after treatment initiation, were compared to healthy controls (HC; n= 4) using intracellular staining and flow cytometry. The MFI of phosphor-mTOR expression on total MDSC was assessed and no significant differences were observed across the period of anti-TB treatment from baseline to end of treatment. There was a single significant difference, however, between the week 2 and month 2 after treatment initiation time points (Figure 5a; p = 0.0109). In contrast, the absolute number of total MDSC expressing phospho-mTOR demonstrated a significant upregulation at the time of active TB disease diagnosis compared to healthy controls (Figure 5b; p = 0.0477 and Figure 5c; p = 0.039), and longitudinal analyses demonstrated the same downregulation over the course of anti-TB treatment as seen in total MDSC frequencies (Figure 1f) and total HMGB1 serum concentration (Figure 3a). Interestingly, after one month of anti-TB treatment, the number of phospho-mTOR+ MDSC showed a significant decrease compared to baseline (Figure 5b; p = 0.0257 and Figure 5c; p = 0.0097), raising the potential of this measurement for downstream investigations into biomarkers of treatment response. By the end of anti-TB treatment, the number of phospho-mTOR+ MDSC were significantly reduced compared to baseline (Figure 5b; p = 0.0026 and Figure 5c; p = 0.0048) and were similar to levels observed for healthy controls. These observations were consistent with the overall frequency of phosphor-mTOR-expressing MDSC as a percentage of all cells analysed in whole blood (Figure 5c). Considering these results, MDSC isolated from patients with active TB disease prior to the initiation of anti-TB therapy express significantly more phospho-mTOR, indicative of a non-autophagic phenotype, possibly promoting bacterial survival.
Figure 5:
Longitudinal phospho-mTOR+ MDSC cell numbers measured in the peripheral blood (whole blood) of adult, active TB disease cases. mTOR+ total MDSC cell numbers were investigated at the time of diagnosis/baseline (BL; n = 15), week two following treatment initiation (W2; n = 14), one month (M1; n = 13), two months (M2; n = 9), and six months (M6; n = 13) after treatment initiation and compared to healthy control participants (n= 4). (a) The median fluorescent intensity of phosphor-mTOR, (b) absolute cell numbers, and (c) frequency of phosphor-mTOR-expressing MDSC as a percentage of all cells (from whole blood) were assessed and determined using the FACS Canto II flow cytometer, and the third-party software FlowJo. D’Agostino & Pearson Omnibus test for normality was used to determine the distribution of the flow cytometry data, after which the statistics used were parametric (one-way ANOVA and Tukey’s multiple comparison test). Error bars represent the median and range. ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant.
4. Discussion
MDSC are a heterogeneous population of myeloid cells with strong immunomodulatory suppressive functions driven by excessive inflammation. Largely studied in cancer models, MDSC research in infectious diseases is showing great promise for host-directed therapeutic targets, as well as for improving our understanding of host responses to M.tb infection. In this study, the frequencies of MDSC from the PBMC compartment were investigated in patients with confirmed active TB disease and compared to otherwise healthy participants. Consistent with the literature, we observed a significantly larger population of MDSC in patients with active TB disease compared to healthy controls, with the predominant subset being M-MDSC in both participant groups [12], [22]. Recent advances in the cancer field have demonstrated that MDSC are more accurately measured in PBMC rather than whole blood and that phenotypic characterization using whole blood can lead to a gross overestimation of the PMN-MDSC subset particularly due to the shared CD15 marker with conventional neutrophils [55]. As such, previous research using active TB patient samples investigated the frequencies of MDSC in whole blood and not PBMC, making our study a unique investigation [12], [22], [56]. We observed the M-MDSC subset to be dominant, with little-to-no detection of PMN-MDSC, both in the context of active disease and healthy controls. But, this observation is likely not an accurate reflection of PMN-MDSC frequencies as it has been shown that PMN-MDSC do not readily survive the process of cryopreservation which was necessary post-isolation for the storage and batch analysis of these samples, and this is considered a limitation of the study [57]. Future studies should evaluate these frequencies using fresh samples which have not been cryopreserved.
The point at which MDSC frequencies change over the course of anti-TB treatment has not been characterised. We have previously demonstrated a reduction in MDSC frequencies between the time of diagnosis and the end of successful anti-TB treatment [12] however, this study used multiple longitudinal timepoints to further characterise the point at which there is a reduction in MDSC frequencies. We demonstrated that while there is a non-significant reduction in frequencies soon after treatment initiation (week two and month one), a significant reduction can be observed in the second month (Figure 1f). This significant decrease prior to the six-month treatment endpoint holds promise for mechanistic insights into disease resolution within TB patients.
MDSC also hold promise as targets for host-directed therapies owing to the immunomodulatory properties of these cells. Knaul et al. demonstrated in a murine model of M.tb infection that MDSC can phagocytose M.tb and subsequently secrete the cytokines IL-10, IL-6, and IL-1α to elicit their immunosuppressive functions [10]. Toll-like receptors −2 [51] and −4 [58], [59] appear to play a crucial role in MDSC functions during M.tb infection, likely during the uptake of M.tb into the cytosol of MDSC and also facilitating the activation of molecular pathways leading to the secretion of cytokines, like IL-6 and IL-10, and immunosuppressive effector functions. While the production of IL-10 in this study was not significant in PPD stimulated MDSC, the production of IL-6 was significantly increased in PPD stimulated MDSC, however, this was not a convincing incremental increase (Figure 2). This suggests that the MDSC investigated during this study had lower immunosuppressive potential than MDSC studied for example at the tumour site. In cancer it has been suggested that tumour infiltrating MDSC demonstrate a stronger immunosuppressive capacity than their circulatory counterparts owing to their drive to differentiate into tumour-associated macrophages (TAMs) which are highly immunosuppressive [60], thereby suggesting that those MDSC found in circulation during active TB disease may also display lessened immunosuppressive potentials compared to MDSC from the site of disease.
A known regulator of MDSC function both in the circulation and at the tumour site, HMGB1, was demonstrated in this study to be upregulated in patients with active TB disease suggesting the potential activation of autophagic pathways within host phagocytes associated with chronic M.tb infection. This upregulation was longitudinally downregulated during anti-TB therapy, suggesting that as the host environment becomes less inflammatory in response to a lower bacterial burden, so too does the release of DAMPs like HMGB1 lessen. This would, by definition, reduce the expansion of the MDSC subset within the host, as observed in Figure 1f. Mechanistically, HMGB1 has a multifaceted role in the regulation of MDSC: (1) HMGB1 can induce the production of pro-inflammatory cytokines like IL-10 and IL-6 after binding with the RAGE or TLR2/4 receptors, thereby inducing the NF-κB signalling pathway specifically to enhance the immunosuppressive activities of MDSC, significantly reducing T cell activation and expansion [25], [61], [62]; and contrastingly (2) HMGB1 can induce the autophagic pathway in MDSC to ensure prolonged survival while reducing immunosuppressive activity on T cell functions [36], [63][25], [36]. Based on our evidence, we propose that HMGB1 plays a crucial role as a modulator of MDSC function depending on the environmental conditions present and other contributing factors like the receptor being bound on the surface of MDSC. We have demonstrated in this study that MDSC enriched from active TB patients do indeed express TLR4, one of the receptors to which HMGB1 binds to initiate the NF-κB signalling pathway. Considering the knowledge that HMGB1 may bind multiple different receptors and thus initiate various pathways as a result, we speculate that there may be a difference in the immunosuppressive functions of MDSC in circulation (a more autophagy-driven response) compared to MDSC at the site of disease (a more immunosuppressive response), possibly driven by specific HMGB1 receptor binding. With our results at hand, it is possible to speculate that the MDSC investigated during this study specifically may well have adopted such an autophagic phenotype considering their origin and the lack of convincing pro-inflammatory cytokine upregulation. Future work should consider the investigation and comparison of MDSC from the periphery and the site of disease to establish the possibility of this being true.
A limitation of this study is that the continuous expression of ADAM17 over the course of anti-TB treatment did not correspond with the downregulation of MDSC frequencies over the course of anti-TB treatment as would be expected; however, it is prudent to consider that the responses seen in Figure 3a and Figure 3b, while M.tb infection-specific, are not MDSC-specific as these proteins were measured using whole blood plasma. It would be interesting for future studies to investigate the MDSC-specific production of ADAM17, as well as the release of its ligand CD62L from co-cultures of MDSC and T-cells in the context of active TB disease. While the infection of MDSC with live M.tb strains was not specifically evaluated in this study, they would be important for future works. In concordance with this, the HMGB1 responses seen in this study were not MDSC-specific, while the cytokine responses, like those for IL-6, were. For this reason, we were unable to draw parallels between the production of these two proteins in this study. We do suggest that this be an important consideration for future work to validate the proposed mechanism.
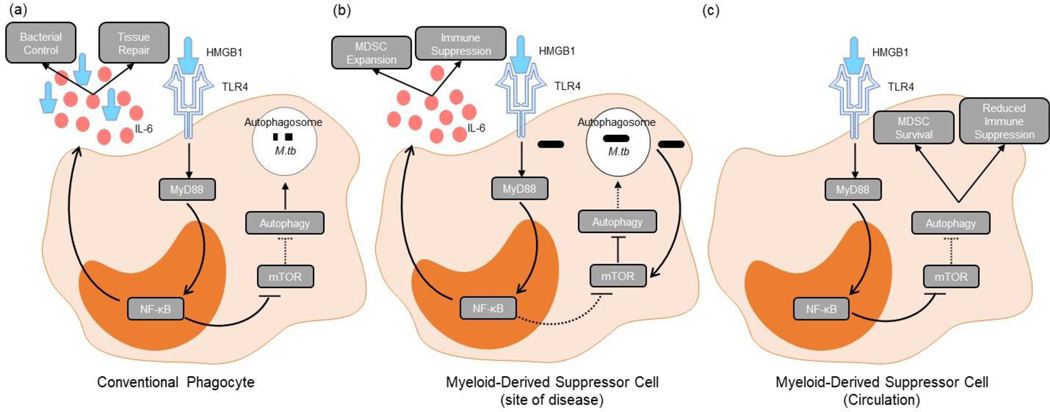
Lastly, our study also demonstrated increased numbers of MDSC expressing the phosphorylated autophagy inhibitor, mTOR, in active TB patients. The expression of this protein coincides with the upregulation of HMGB1 (autophagy activator) prior to the initiation of anti-TB treatment, with both protein levels significantly dropping over the course of treatment. While these results may seem somewhat contradictory, it is important to keep in mind that the population of MDSC investigated during this study may have contained subsets in various stages of their life cycle, i.e., early-stage MDSC which had just recently been recruited from the bone marrow, activated subsets which had entered the periphery after pathogen exposure and/or uptake at the site of disease, or long-lived subsets which may patrol the periphery as a result of excessive chronic inflammation. Based on the literature and the findings from this study, the results may represent a mixed population of MDSC at various stages in their life cycle. It is reasonable to suggest that the upregulation of HMGB1 may be representative of MDSC subsets which have adopted an autophagic phenotype, not producing the usual immunosuppressive cytokines like IL-10 and IL-6 (as evidenced by our results) and thereby promoting MDSC survival; while the upregulation of phospho-mTOR may be specific to immunosuppressive MDSC which may then be found to be infected with M.tb. Thus, we propose that during chronic infection with M.tb, host-protective anti-inflammatory pathways are modified by the bacteria to promote pathogen survival within the phagocytic MDSC (Figure 6). Specifically, M.tb promotes phospho-mTOR within M-MDSC, inhibiting autophagy and thereby enhancing the immunosuppressive properties of M-MDSC as shown before [43], and promoting pathogen survival. This hypothesis is further supported by previous work which has demonstrated that autophagy is important for restriction of intracellular bacterial growth, with subsequent knock-out experiments of the crucial autophagy-related protein, Atg5, resulting in significantly increased M.tb bacterial burdens, inflammation, and IL-1 levels in a murine model [64]–[67].
Figure 6:
Proposed mechanism for how MDSC utilise the regulatory factor, HMGB1, to control their effector functions depending on the environmental need after binding to the receptor TLR4. (a) Conventional phagocytes inhibit mTOR through the binding of HMGB1 to TLR4 (and other receptors like TLR2 and RAGE), allowing for the fusion of the lysosome to the autophagosome and subsequent killing of the bacteria. (b) M.tb-infected MDSC (likely found predominantly at the site of disease) on the other hand, increase the production of mTOR, thereby inhibiting the autophagy process and prohibiting the lysosome fusion to the autophagosome, harbouring the bacteria from killing mechanisms. Pro-inflammatory cytokines, like IL-6, produced during this process induce expansion of MDSC. (c) Uninfected MDSC (likely found predominantly in circulation) make use of HMGB1 in an alternate manner to induce autophagy within the cells. The induction of autophagy prolongs the survival of these cells while limiting their suppressive potential.
Conflicting evidence exists with regards to the role of autophagy, and even the HMGB1 signalling pathway, in MDSC function and control [36], [68], [69]. Disease-specific signalling pathways activated in response to the host environment should first be validated based on the disease being investigated, as data from this research has demonstrated that specific MDSC subsets and disease-specific effector functions can differentially control various induction and activation signals of MDSC. Autophagy, like other anti-inflammatory pathways including those mediated by MDSC, physiologically function to limit excessive pro-inflammatory responses in the host during infection to prevent tissue damage and pathology. The importance of a strictly balanced pro- and anti-inflammatory immune response to efficiently control, clear and recover from invading pathogens has long been stressed [70], and autophagy has been shown to have potential as a host-directed therapy (HDT) target for M.tb infection [71]. However, the necessity for strict immune control raises an important question for future studies, as well as for application in medical practice considerations: at what point during infection/the immune response should host-directed therapies be administered/activated?
To conclude, we have demonstrated that MDSC isolated from the PBMC compartment of the blood of patients with active TB disease are significantly elevated prior to the initiation of anti-TB therapy and return to frequencies comparable to those of healthy controls after two months of therapy. Prior to the initiation of anti-TB therapy, these MDSC produce IL-6 in response to PPD stimulation, albeit in low levels, and express the TLR4 receptor in significantly larger proportions to healthy controls. The DAMP, HMGB1, is downregulated during anti-TB therapy suggesting a reduction in autophagic responses in response to control of bacterial infection, preventing the prolonged survival of MDSC and further suggesting the restoration of immune homeostasis. The number of MDSC expressing phospho-mTOR was significantly upregulated prior to the initiation of anti-TB therapy, and declined over the course of treatment, suggesting an MDSC-specific inhibition of the autophagy pathway in M.tb-infected M-MDSC specifically and the possible exacerbation of MDSC suppressive functions as a direct result. Taken together, the PBMC compartment contains MDSC in various stages of the life cycle, with indications that prolonged MDSC survival is being driven and possibly prioritised in the periphery before the initiation of anti-TB treatment, and indications that autophagy is being inhibited in those cells that are potentially entering the periphery from the site of infection and which are infected with M.tb. In this way, we propose a mechanism by which M.tb bacilli may persist and evade the host’s protective immune responses, but future studies should be directed at validating these findings in a more functional manner using knockouts.
Highlights.
MDSC frequencies in TB patients are significantly increased before treatment.
MDSC frequencies returned to healthy control levels after two months of treatment.
MDSC from TB patients express significantly increased phospho-mTOR, TLR4, and IL-6.
The expression of phospho-mTOR on MDSC decreased during anti-TB treatment.
HMGB1 serum concentrations decrease in parallel with phospho-mTOR.
Acknowledgements
This work was supported by the European & Developing Countries Clinical Trials Partnership (EDCTP; CDF1546), National Institute of Health (NIH) International Collaborations in Infectious Disease Research (ICIDR): Biology and Biosignatures of anti-TB Treatment Response (5U01IA115619/03), DFG LU851/18-1 and South African National Research Foundation SA Research Chair Initiative (SARCHI; 86535).
1. Abbreviations:
- MDSC
myeloid-derived suppressor cell(s)
- mTOR
mammalian target of rapamycin
- HMGB1
high mobility group box protein 1
- ADAM17
A Disintegrin and Metalloproteinase Domain-17
- RAGE
receptor for advanced glycation end products
- TLR
Toll-like receptor
- DAMP
damage associated molecular protein
- IDO
indoleamine-2,3 dioxygenase
- iNOS
inducible nitric oxide synthase
- Arg1
arginase 1
Footnotes
CRediT authorship contribution statement
Leigh A. Kotze: Investigation, Methodology, Formal analysis, Writing-Original Draft. Vinzeigh N. Leukes: Investigation, Methodology, Writing – Review and Editing. Zhuo Fang: Investigation, Methodology, Writing – Reviewing and Editing. Manfred B. Lutz: Conceptualization, Writing – Reviewing and Editing. Bryna L. Fitzgerald: Writing – Reviewing and Editing, Funding acquisition, Resources. John Belisle: Writing – Reviewing and Editing, Funding acquisition, Resources. Andre G. Loxton: Writing – Reviewing and Editing. Gerhard Walzl: Resources, Writing – Reviewing and Editing, Funding acquisition. Nelita du Plessis: Conceptualization, Supervision, Methodology, Resources, Project Administration, Writing – Reviewing and Editing, Funding acquisition.
Declaration of Competing Interest
The authors declare no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, and Berry MPR, “The Immune Response in Tuberculosis,” Annu. Rev. Immunol, vol. 31, no. 1, pp. 475–527, Mar. 2013, doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- [2].Van Crevel R, Ottenhoff THM, and van der Meer JWM, “Innate Immunity to Mycobacterium tuberculosis,” Clin. Microbiol. Rev, vol. 15, no. 2, pp. 294–309, Apr. 2002, doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Flynn JL and Chan J, “Immunology of tuberculosis,” Annu. Rev. Immunol, vol. 19, pp. 93–129, 2001, doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- [4].Gabrilovich DI and Nagaraj S, “Myeloid-derived suppressor cells as regulators of the immune system,” Nat. Rev. Immunol, vol. 9, no. 3, pp. 162–174, Mar. 2009, doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Talmadge JE, Cole K, Britton H, Dafferner A, and Warkentin P, “Human myeloid derived suppressor cell (MDSC) subset phenotypes,” J. Immunol, vol. 198, no. 1 Supplement, p. 211.2–211.2, May 2017. [Google Scholar]
- [6].Bronte V. et al. , “Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards,” Nat. Commun, vol. 7, p. 12150, 06 2016, doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Goldmann O, Beineke A, and Medina E, “Identification of a Novel Subset of Myeloid-Derived Suppressor Cells During Chronic Staphylococcal Infection That Resembles Immature Eosinophils,” J. Infect. Dis, vol. 216, no. 11, pp. 1444–1451, 12 2017, doi: 10.1093/infdis/jix494. [DOI] [PubMed] [Google Scholar]
- [8].Nagaraj S. and Gabrilovich DI, “Regulation of suppressive function of myeloidderived suppressor cells by CD4+ T cells MDSC and CD4+ T cells,” Semin. Cancer Biol, vol. 22, no. 4, pp. 282–288, Aug. 2012, doi: 10.1016/j.semcancer.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Condamine T, Ramachandran I, Youn J-I, and Gabrilovich DI, “Regulation of Tumor Metastasis by Myeloid-Derived Suppressor Cells,” Annu. Rev. Med, vol. 66, no. 1, pp. 97–110, Jan. 2015, doi: 10.1146/annurev-med-051013-052304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Knaul JK et al., “Lung-residing myeloid-derived suppressors display dual functionality in murine pulmonary tuberculosis,” Am. J. Respir. Crit. Care Med., vol. 190, no. 9, pp. 1053–1066, Nov. 2014, doi: 10.1164/rccm.201405-0828OC. [DOI] [PubMed] [Google Scholar]
- [11].Agrawal N. et al. , “Human Monocytic Suppressive Cells Promote Replication of Mycobacterium tuberculosis and Alter Stability of in vitro Generated Granulomas,” Front. Immunol, vol. 9, 2018, doi: 10.3389/fimmu.2018.02417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].du Plessis N. et al. , “Increased Frequency of Myeloid-derived Suppressor Cells during Active Tuberculosis and after Recent Mycobacterium tuberculosis Infection Suppresses T-Cell Function,” Am. J. Respir. Crit. Care Med, vol. 188, no. 6, pp. 724–732, Jul. 2013, doi: 10.1164/rccm.201302-0249OC. [DOI] [PubMed] [Google Scholar]
- [13].El Daker S. et al. , “Granulocytic Myeloid Derived Suppressor Cells Expansion during Active Pulmonary Tuberculosis Is Associated with High Nitric Oxide Plasma Level,” PLoS ONE, vol. 10, no. 4, Apr. 2015, doi: 10.1371/journal.pone.0123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miret JJ et al. , “Suppression of Myeloid Cell Arginase Activity leads to Therapeutic Response in a NSCLC Mouse Model by Activating Anti-Tumor Immunity,” J. Immunother. Cancer, vol. 7, no. 1, p. 32, Feb. 2019, doi: 10.1186/s40425-019-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Movahedi K. et al. , “Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity,” Blood, vol. 111, no. 8, pp. 4233–4244, Apr. 2008, doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- [16].Dolcetti L. et al. , “Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF,” Eur. J. Immunol, vol. 40, no. 1, pp. 22–35, Jan. 2010, doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- [17].Novitskiy SV et al. , “Adenosine receptors in regulation of dendritic cell differentiation and function,” Blood, vol. 112, no. 5, pp. 1822–1831, Sep. 2008, doi: 10.1182/blood-2008-02136325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Baban B. et al. , “IDO activates regulatory T cells and blocks their conversion into Th17like T cells,” J. Immunol. Baltim. Md 1950, vol. 183, no. 4, pp. 2475–2483, Aug. 2009, doi: 10.4049/jimmunol.0900986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang B. et al. , “Gr-1+CD115+ Immature Myeloid Suppressor Cells Mediate the Development of Tumor-Induced T Regulatory Cells and T-Cell Anergy in Tumor-Bearing Host,” Cancer Res., vol. 66, no. 2, pp. 1123–1131, Jan. 2006, doi: 10.1158/0008-5472.CAN05-1299. [DOI] [PubMed] [Google Scholar]
- [20].Torroella-Kouri M. et al. , “Identification of a Subpopulation of Macrophages in Mammary Tumor–Bearing Mice That Are Neither M1 nor M2 and Are Less Differentiated,” Cancer Res., vol. 69, no. 11, pp. 4800–4809, Jun. 2009, doi: 10.1158/0008-5472.CAN-083427. [DOI] [PubMed] [Google Scholar]
- [21].Hoechst B, Gamrekelashvili J, Manns MP, Greten TF, and Korangy F, “Plasticity of human Th17 cells and iTregs is orchestrated by different subsets of myeloid cells,” Blood, vol. 117, no. 24, pp. 6532–6541, Jun. 2011, doi: 10.1182/blood-2010-11-317321. [DOI] [PubMed] [Google Scholar]
- [22].Du Plessis N. et al. , “Phenotypically resembling myeloid derived suppressor cells are increased in children with HIV and exposed/infected with Mycobacterium tuberculosis.,” Eur. J. Immunol, vol. 47, no. 1, pp. 107–118, Jan. 2017, doi: 10.1002/eji.201646658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hanson EM, Clements VK, Sinha P, Ilkovitch D, and Ostrand-Rosenberg S, “Myeloid-Derived Suppressor Cells Down-Regulate L-Selectin Expression on CD4+ and CD8+ T Cells,” J. Immunol., vol. 183, no. 2, pp. 937–944, Jul. 2009, doi: 10.4049/jimmunol.0804253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gabrilovich DI, Ostrand-Rosenberg S, and Bronte V, “Coordinated regulation of myeloid cells by tumours,” Nat. Rev. Immunol, vol. 12, no. 4, pp. 253–268, Mar. 2012, doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Parker KH et al. , “HMGB1 Enhances Immune Suppression by Facilitating the Differentiation and Suppressive Activity of Myeloid-Derived Suppressor Cells,” Cancer Res., vol. 74, no. 20, pp. 5723–5733, Oct. 2014, doi: 10.1158/0008-5472.CAN-13-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sims GP, Rowe DC, Rietdijk ST, Herbst R, and Coyle AJ, “HMGB1 and RAGE in inflammation and cancer,” Annu. Rev. Immunol, vol. 28, pp. 367–388, 2010, doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- [27].Lotze MT and Tracey KJ, “High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal,” Nat. Rev. Immunol, vol. 5, no. 4, pp. 331–342, Apr. 2005, doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- [28].Vernon PJ et al. , “The receptor for advanced glycation end products promotes pancreatic carcinogenesis and accumulation of myeloid-derived suppressor cells,” J. Immunol. Baltim. Md 1950, vol. 190, no. 3, pp. 1372–1379, Feb. 2013, doi: 10.4049/jimmunol.1201151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li W. et al. , “Aerobic Glycolysis Controls Myeloid-Derived Suppressor Cells and Tumor Immunity via a Specific CEBPB Isoform in Triple-Negative Breast Cancer,” Cell Metab., vol. 28, no. 1, pp. 87–103.e6, Jul. 2018, doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Alissafi T. et al. , “Autophagy orchestrates the regulatory program of tumor-associated myeloid-derived suppressor cells,” J. Clin. Invest, vol. 128, no. 9, pp. 3840–3852, 31 2018, doi: 10.1172/JCI120888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dong G. et al. , “Autophagy regulates accumulation and functional activity of granulocytic myeloid-derived suppressor cells via STAT3 signaling in endotoxin shock,” Biochim. Biophys. Acta BBA - Mol. Basis Dis, vol. 1863, no. 11, pp. 2796–2807, Nov. 2017, doi: 10.1016/j.bbadis.2017.08.005. [DOI] [PubMed] [Google Scholar]
- [32].Ostrand-Rosenberg S, Beury DW, Parker KH, and Horn LA, “Survival of the fittest: how myeloid-derived suppressor cells survive in the inhospitable tumor microenvironment,” Cancer Immunol. Immunother., vol. 69, no. 2, pp. 215–221, Feb. 2020, doi: 10.1007/s00262-019-02388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Welte T. et al. , “Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation,” Nat. Cell Biol, vol. 18, no. 6, pp. 632–644, 2016, doi: 10.1038/ncb3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang C. et al. , “The mTOR signal regulates myeloid-derived suppressor cells differentiation and immunosuppressive function in acute kidney injury,” Cell Death Dis., vol. 8, no. 3, Art. no. 3, Mar. 2017, doi: 10.1038/cddis.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen X. et al. , “mTOR signaling disruption from myeloid-derived suppressive cells protects against immune-mediated hepatic injury through the HIF1α-dependent glycolytic pathway,” J. Leukoc. Biol, vol. 100, no. 6, pp. 1349–1362, 2016, doi: 10.1189/jlb.2A1115492R. [DOI] [PubMed] [Google Scholar]
- [36].Parker KH, Horn LA, and Ostrand-Rosenberg S, “High-mobility group box protein 1 promotes the survival of myeloid-derived suppressor cells by inducing autophagy,” J. Leukoc. Biol, vol. 100, no. 3, pp. 463–470, 2016, doi: 10.1189/jlb.3HI0715-305R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Parker K. and Ostrand-Rosenberg S, “HMGB1 promotes MDSC survival through autophagy (TUM6P.956),” J. Immunol, vol. 194, no. 1 Supplement, p. 141.4–141.4, May 2015. [Google Scholar]
- [38].Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, and Deretic V, “Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages,” Cell, vol. 119, no. 6, pp. 753–766, Dec. 2004, doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- [39].Yorimitsu T. and Klionsky DJ, “Autophagy: molecular machinery for self-eating,” Cell Death Differ., vol. 12 Suppl 2, pp. 1542–1552, Nov. 2005, doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saxton RA and Sabatini DM, “mTOR Signaling in Growth, Metabolism, and Disease,” Cell, vol. 168, no. 6, pp. 960–976, Mar. 2017, doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meng L. et al. , “The protective effect of dexmedetomidine on LPS-induced acute lung injury through the HMGB1-mediated TLR4/NF-κB and PI3K/Akt/mTOR pathways,” Mol. Immunol, vol. 94, pp. 7–17, Feb. 2018, doi: 10.1016/j.molimm.2017.12.008. [DOI] [PubMed] [Google Scholar]
- [42].Yang Q-W et al. , “High-Mobility Group Protein Box-1 and its Relevance to Cerebral Ischemia,” J. Cereb. Blood Flow Metab, vol. 30, no. 2, pp. 243–254, Feb. 2010, doi: 10.1038/jcbfm.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ribechini E. et al. , “Novel GM-CSF signals via IFN-γR/IRF-1 and AKT/mTOR license monocytes for suppressor function,” Blood Adv., vol. 1, no. 14, pp. 947–960, Jun. 2017, doi: 10.1182/bloodadvances.2017006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Welte T. et al. , “Oncogenic mTOR signaling recruits myeloid-derived suppressor cells to promote tumor initiation,” Nat. Cell Biol, vol. 18, no. 6, pp. 632–644, Jun. 2016, doi: 10.1038/ncb3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zullo AJ and Lee S, “Mycobacterial induction of autophagy varies by species and occurs independently of mammalian target of rapamycin inhibition,” J. Biol. Chem, vol. 287, no. 16, pp. 12668–12678, Apr. 2012, doi: 10.1074/jbc.M111.320135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shipp C, Speigl L, Janssen N, Martens A, and Pawelec G, “A clinical and biological perspective of human myeloid-derived suppressor cells in cancer,” Cell. Mol. Life Sci. CMLS, vol. 73, no. 21, pp. 4043–4061, 2016, doi: 10.1007/s00018-016-2278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ostrand-Rosenberg S. and Sinha P, “Myeloid-derived suppressor cells: linking inflammation and cancer,” J. Immunol. Baltim. Md 1950, vol. 182, no. 8, pp. 4499–4506, Apr. 2009, doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kumar V, Patel S, Tcyganov E, and Gabrilovich DI, “The nature of myeloid-derived suppressor cells in the tumor microenvironment,” Trends Immunol., vol. 37, no. 3, pp. 208–220, Mar. 2016, doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li J. et al. , “HMGB1 promotes myeloid-derived suppressor cells and renal cell carcinoma immune escape,” Oncotarget, vol. 8, no. 38, pp. 63290–63298, Jun. 2017, doi: 10.18632/oncotarget.18796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gebhardt C. et al. , “Myeloid Cells and Related Chronic Inflammatory Factors as Novel Predictive Markers in Melanoma Treatment with Ipilimumab,” Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res, vol. 21, no. 24, pp. 5453–5459, Dec. 2015, doi: 10.1158/10780432.CCR-15-0676. [DOI] [PubMed] [Google Scholar]
- [51].John V, Kotze LA, Ribechini E, Walzl G, Du Plessis N, and Lutz MB, “Caveolin1 Controls Vesicular TLR2 Expression, p38 Signaling and T Cell Suppression in BCG Infected Murine Monocytic Myeloid-Derived Suppressor Cells,” Front. Immunol, vol. 10, 2019, doi: 10.3389/fimmu.2019.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xu X. et al. , “Exogenous High-Mobility Group Box 1 Inhibits Apoptosis and Promotes the Proliferation of Lewis Cells via RAGE/TLR4-Dependent Signal Pathways,” Scand. J. Immunol, vol. 79, no. 6, pp. 386–394, 2014, doi: 10.1111/sji.12174. [DOI] [PubMed] [Google Scholar]
- [53].Zhao C-B et al. , “Co-expression of RAGE and HMGB1 is associated with cancer progression and poor patient outcome of prostate cancer,” Am. J. Cancer Res, vol. 4, no. 4, pp. 369–377, Jul. 2014. [PMC free article] [PubMed] [Google Scholar]
- [54].Morgan TM, Koreckij TD, and Corey E, “Targeted Therapy for Advanced Prostate Cancer: Inhibition of the PI3K/Akt/mTOR Pathway,” Curr. Cancer Drug Targets, vol. 9, no. 2, pp. 237–249, Mar. 2009, doi: 10.2174/156800909787580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dorhoi A. et al. , “Therapies for tuberculosis and AIDS: myeloid-derived suppressor cells in focus,” J. Clin. Invest, May 2020, doi: 10.1172/JCI136288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Magcwebeba T, Dorhoi A, and du Plessis N, “The Emerging Role of Myeloid-Derived Suppressor Cells in Tuberculosis,” Front. Immunol, vol. 10, p. 917, 2019, doi: 10.3389/fimmu.2019.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Trellakis S. et al. , “Granulocytic myeloid-derived suppressor cells are cryosensitive and their frequency does not correlate with serum concentrations of colony-stimulating factors in head and neck cancer,” Innate Immun., vol. 19, no. 3, pp. 328–336, Jun. 2013, doi: 10.1177/1753425912463618. [DOI] [PubMed] [Google Scholar]
- [58].Garg AJ and Spector SA, “Myeloid derived suppressor cells (MDSC) from HIVinfected individuals are defective in innate immunity to Mycobacterium tuberculosis thus increasing the risk of tuberculosis,” J. Immunol., vol. 202, no. 1 Supplement, p. 62.13–62.13, May 2019. [Google Scholar]
- [59].Kotzé LA et al. , “Mycobacterium tuberculosis and myeloid-derived suppressor cells: Insights into caveolin rich lipid rafts,” EBioMedicine, vol. 53, p. 102670, Mar. 2020, doi: 10.1016/j.ebiom.2020.102670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yin Z, Li C, Wang J, and Xue L, “Myeloid-derived suppressor cells: Roles in the tumor microenvironment and tumor radiotherapy,” Int. J. Cancer, vol. 144, no. 5, pp. 933–946, 2019, doi: 10.1002/ijc.31744. [DOI] [PubMed] [Google Scholar]
- [61].Bunt SK, Clements VK, Hanson EM, Sinha P, and Ostrand-Rosenberg S, “Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through Tolllike receptor 4,” J. Leukoc. Biol, vol. 85, no. 6, pp. 996–1004, Jun. 2009, doi: 10.1189/jlb.0708446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Huang M. et al. , “S100A9 Regulates MDSCs-Mediated Immune Suppression via the RAGE and TLR4 Signaling Pathways in Colorectal Carcinoma,” Front. Immunol, vol. 10, p. 2243, 2019, doi: 10.3389/fimmu.2019.02243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tang D. et al. , “HMGB1 release and redox regulates autophagy and apoptosis in cancer cells,” Oncogene, vol. 29, no. 38, pp. 5299–5310, Sep. 2010, doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ye X, Zhou X-J, and Zhang H, “Exploring the Role of Autophagy-Related Gene 5 (ATG5) Yields Important Insights Into Autophagy in Autoimmune/Autoinflammatory Diseases,” Front. Immunol, vol. 9, 2018, doi: 10.3389/fimmu.2018.02334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Castillo EF et al. , “Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation,” Proc. Natl. Acad. Sci, vol. 109, no. 46, pp. E3168–E3176, Nov. 2012, doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Guo X-G, Ji T-X, Xia Y, and Ma Y-Y, “Autophagy protects type II alveolar epithelial cells from Mycobacterium tuberculosis infection,” Biochem. Biophys. Res. Commun, vol. 432, no. 2, pp. 308–313, Mar. 2013, doi: 10.1016/j.bbrc.2013.01.111. [DOI] [PubMed] [Google Scholar]
- [67].Harris J et al., “T Helper 2 Cytokines Inhibit Autophagic Control of Intracellular Mycobacterium tuberculosis,” Immunity, vol. 27, no. 3, pp. 505–517, Sep. 2007, doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- [68].Wu T. et al. , “mTOR masters monocytic myeloid-derived suppressor cells in mice with allografts or tumors,” Sci. Rep, vol. 6, no. 1, Art. no. 1, Feb. 2016, doi: 10.1038/srep20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Qu L. et al. , “High-Mobility Group Box 1 (HMGB1) and Autophagy in Acute Lung Injury (ALI): A Review,” Med. Sci. Monit. Int. Med. J. Exp. Clin. Res, vol. 25, pp. 1828–1837, Mar. 2019, doi: 10.12659/MSM.912867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Keane MP and Strieter RM, “The importance of balanced pro-inflammatory and anti-inflammatory mechanisms in diffuse lung disease,” Respir. Res, vol. 3, no. 1, p. 5, Oct. 2001, doi: 10.1186/rr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Deretic V, “Autophagy in Tuberculosis,” Cold Spring Harb. Perspect. Med, vol. 4, no. 11, Nov. 2014, doi: 10.1101/cshperspect.a018481. [DOI] [PMC free article] [PubMed] [Google Scholar]