Abstract
Background
Neurofibromatosis type 2 (NF2) is an autosomal dominant genetic disease characterized by development of schwannomas on the VIIIth (vestibular) cranial nerves. Bromodomain and extra-terminal domain (BET) proteins regulate gene transcription and their activity is required in a variety of cancers including malignant peripheral nerve sheath tumors. The use of BET inhibitors as a therapeutic option to treat NF2 schwannomas has not been explored and is the focus of this study.
Methods
A panel of normal and NF2-null Schwann and schwannoma cell lines were used to characterize the impact of the BET inhibitor JQ1 in vitro and in vivo. The mechanism of action was explored by chromatin immunoprecipitation of the BET BRD4, phospho-kinase arrays and immunohistochemistry (IHC) of BRD4 in vestibular schwannomas.
Results
JQ1 inhibited proliferation of NF2-null schwannoma and Schwann cell lines in vitro and in vivo. Further, loss of NF2 by CRISPR deletion or siRNA knockdown increased sensitivity of cells to JQ1. Loss of function experiments identified BRD4, and to a lesser extent BRD2, as BET family members mediating the majority of JQ1 effects. IHC demonstrated elevated levels of BRD4 protein in human vestibular schwannomas. Analysis of signaling pathways effected by JQ1 treatment suggests that the effects of JQ1 treatment are mediated, at least in part, via inhibition of PI3K/Akt signaling.
Conclusions
NF2-deficient Schwann and schwannoma cells are sensitive to BET inhibition, primarily mediated by BRD4, which is overexpressed in human vestibular schwannomas. Our results suggest BRD4 regulates PI3K signaling and likely impedes NF2 schwannoma growth via this inhibition. These findings implicate BET inhibition as a therapeutic option for NF2-deficient schwannomas.
Keywords: BET proteins, BRD4, neurofibromatosis type 2, NF2
Key Points.
NF2-deficient Schwann and schwannoma cells are sensitive to BET inhibition (BETi).
BETi effects are mediated by BRD4, which is overexpressed in vestibular schwannomas.
The findings implicate BETi as a therapeutic option for NF2.
Importance of the Study.
This study identifies a new class of drugs, BET protein inhibitors, as potential therapeutic options for neurofibromatosis type 2 (NF2)-deficient schwannomas. There currently are no FDA approved therapeutic options for NF2 patients and treatment is mostly limited to surgical intervention associated with significant morbidity. Our studies identified that BET inhibition, via the prototypical inhibitor JQ1, can inhibit proliferation of NF2-deficient Schwann cells and schwannoma cells in culture and in vivo. These findings, combined with the finding that BET inhibition appears to be selective toward NF2-deficient cells and that schwannomas express elevated levels of BRD4, the likely mediator of BET activity, support the idea of BET inhibitors as a therapeutic option for NF2-deficient schwannoma, as a single agent or in combination with other drugs.
Neurofibromatosis type 2 (NF2) is a dominantly inherited autosomal disease (affecting 1 in 30 000), attributed to the loss-of-heterozygosity (LOH) of the NF2 gene. It is characterized by development of multiple benign tumors of Schwann cell origin (schwannomas), most typically affecting both VIIIth cranial nerves and also by meningiomas. In addition, mutations and LOH of the NF2 locus are detected at high frequency in various tumors of the nervous system, including sporadic schwannomas, meningiomas, and ependymomas.1 The NF2 tumor suppressor gene encodes a 69-kDa protein called Merlin (Moesin, ezrin, and radixin like protein). Merlin, which has been shown to function at the cell membrane and in the nucleus, has been implicated as a regulator of several signaling pathways that are influenced by changes in conditions such as cell density. Several studies suggest that as cells experience increased cell–cell contact, Merlin is recruited to cell junctions where it is thought to coordinate the establishment of intercellular contacts with concomitant inhibition of growth factor signaling through a number of pathways including those regulated by the small G-proteins Ras/Rac and the Hippo-YAP pathway.2–5 The loss of Merlin leads to aberrant activation of these signaling pathways and tumor development.
Bromodomain and extra-terminal domain (BET) proteins are a protein family comprised of 4 members in mammals including BRD2, BRD3, BRD4, and BRDT. The family is characterized by the presence of 2 tandem bromodomains that bind to acetyl-lysine, an extra-terminal domain (involved in protein–protein interactions) and a C-terminal domain (in BRD4 and BRDT).6,7 The BET proteins have been mostly characterized as epigenetic “readers” for histone acetylation marks.8 However, they can also recognize other acetylated proteins including transcription factors.9 The BET proteins are implicated in a number of pathological conditions including cancer.9 Direct evidence stems from NUT carcinoma, a rare squamous cell carcinoma characterized by chromosomal translocations involving the NUT gene on chromosome 15q13 to the BRD4 (localized to 19q13) or BRD3 (localized to 9q34.2) genes resulting in a BRD-NUT oncogenic fusion protein that drives tumorigenesis.10 In addition, BRD2 and BRD4 are upregulated in various cancers and required for survival of neoplastic cells.11–15 As positive regulators of gene expression, and especially given the association with super-enhancers, the BET proteins have been implicated in the regulation of genes that drive tumorigenesis including MYC, FOSL1, ILR, TERT, BCL2, and CDK6 among others.16 These findings led to significant attention to the BET proteins as potential therapeutic targets. One of the first BET inhibitors, JQ1, is a small molecule that inhibits binding of the bromodomain to acetylated histone H4.17 Promisingly, there seems to be high selectivity of BET inhibition toward transformed cells.9
Recent studies with BET inhibition have shown promising results in models of neurofibromatosis type 1 (NF1). Specifically, inhibition of BRD4 triggered cell death in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs). This appears to be mediated through regulation of the apoptotic effectors Bim and Bcl-2.18 In this report, we assess the impact of BET inhibition in NF2, in vitro and in vivo. Our findings suggest that BET inhibition suppresses the proliferation of NF2-deficient Schwann cells and schwannoma cells in culture and tumor growth in vivo, and that this is mediated through inhibition of bromodomain protein 4 (BRD4). In addition, we find that BRD4 is significantly upregulated in a majority of analyzed NF2-associated schwannomas. These findings implicate BET inhibition as a potential therapeutic modality for the treatment of NF2.
Materials and Methods
Cell Culture and Gene Editing or Knockdown
SC4 and SC4-Luc Nf2-null mouse Schwann cells and HEI-193 human NF2-mutant schwannoma cells were previously described.19–21 hSC2λ cells were obtained from the laboratory of Dr. Margret Wallace.22 The generation of the hSC2λ-shNF2 clones was previously described.23 For generation of the hSC2λ-NF2−/− cells, both NF2 alleles were targeted by CRISPR/Cas9 genome editing. hSC2λ cells were transfected with 2 independent pX459 plasmids containing guide sequences targeting exon 1 of NF2. gRNA-1 = GAACTCCATCTCGGCGTCCA, gRNA-2 = GATCCTCACGGTGAACGTCT. Cells were selected with puromycin (0.25 µg/mL) for 48 h and single cell clones picked. Multiple clones were expanded and analyzed by sequencing and western blotting for loss of NF2 protein expression (Supplementary Figure S1A).
For knockdown of target genes the following siRNA were used:
- BRD3 (ON-TARGETplus, Dharmacon Inc., Cat # LQ-004936),
- BRD2 (FlexiTube GeneSolution, Qiagen, Inc., Cat # GS6046 and ON-TARGETplus, Dharmacon Inc., Cat # LQ-004935)
- BRD4 (FlexiTube GeneSolution, Qiagen, Inc., Cat # GS23476 and ON-TARGETplus, Dharmacon Inc., Cat # LQ-004937)
- NF2 (ON-TARGETplus, Dharmacon Inc., Cat # LQ-003917)
The siRNA were reverse transfected into cells (10 pmol/2 × 105 cell) using Lipofectamine RNAiMax (Life Technologies Inc.). Twenty-four hours after transfection cells were trypsinized and replated for proliferation and/or JQ1 dose–response assays.
SC4, HEI-193, and hSC2λ cells were authenticated by short tandem repeat DNA profiling (DDC Medical). All cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin.
BRD4 Immunohistochemistry and Evaluation
Samples of human vestibular schwannomas and of normal peripheral nerve were obtained with IRB consent at the Mt Sinai Medical Center and were fixed in formalin and processed routinely through a graded series of alcohols, and then dehydrated and embedded in paraffin. Sections of these tissues were cut from the formalin fixed, paraffin embedded tissue. Immunohistochemical stains for BRD4 were made with antihuman BRD4 antibody (A301-985A50, Bethyl Laboratories, Inc.) and H&E stains were made using adjacent sections. Slides were evaluated for presence or absence of tumor, confirmation of the diagnosis as (benign) schwannoma, levels (intensity and number of cells) of BRD4 signal and cellular distribution by standard light microscopy.
BRD4 Chromatin Immunoprecipitation
BRD4 chromatin immunoprecipitation (ChIP) was carried out as described previously.24 Briefly, hSC2λ-shNF2 cells at ~75% confluency (1 × 107 cells/condition) were treated with DMSO or JQ1 (1 µM) for 12 h at 37°C. Cells were fixed with 1% formaldehyde for 10′, RT. Cells were incubated with 125 mmol/L glycine for 5′ then washed (2×) with cold PBS, centrifuged and lysed in 2 mL ChIP buffer (150 mmol/L NaCl, 1% Triton X-100, 5 mmol/L Ethylenediaminetetraacetic acid, 10 mmol/L Tris–Cl pH 7.5, 0.5 mmol/L Dithiothreitol, 0.5% sodium dodecyl sulfate [SDS]) by rocking for 10′ at 4°C. DNA was fragmented by sonication to 0.3–0.5 kb DNA fragments (Misonix S-3000, 4°C water bath, 30 min (30 s on, 30 s off, output 8). Input DNA (7 × 104 cell equivalent) was removed from fragmented DNA and stored at −80°C. Anti-BRD4 antibody (3 µg/per condition) (BL-149-2H5, Bethyl Laboratories) and Dynabeads Protein G (90 µl/ per condition) were added to lysate and incubated overnight at 4°C with rocking. Beads were washed with buffer 1 (150 mmol/L NaCl, 20 mmol/L Tris–Cl pH 8.0, 5 mmol/L EDTA, 65% (w/v) sucrose, 1% Triton X-100, and 0.2% SDS) and then washed with Tris-EDTA buffer. ChIP and input DNA were resuspended in TE/1% SDS and incubating at 65°C for 18 h. SDS was diluted with TE and treated with RNase A (0.2 mg/mL) at 37°C for 2 h, treated with proteinase K (0.2 mg/mL) 65°C for 2 h, then DNA was extracted with phenol chloroform isoamyl alcohol and resuspended in TE.
Data Availability
ChIP-seq data are stored in the Gene Expression Omnibus under accession number GSE167560.
ChIP-seq Data Analysis
Quality control of reads was done by trimming of exogenous adapter sequences, removing low-quality bases from the ends of reads, and removing the whole reads if average quality was too low. High-quality reads were mapped to the human genome (human-USCS: H.sapiens-UCSC-GRCh38hg38: downloaded March 22, 2016) using bowtie version 1.1.2 aligner and peak abundance estimated with macs version 2.1.0. Replicate BRD4 ChIP experiments (±JQ1 treatment) were processed and significant peaks were called in pairwise manner from BRD4_ChIP_DMSO versus input and BRD4_ChIP_DMSO versus BRD4_ChIP_JQ1. PAVIS (peak annotation visualization) (https://manticore.niehs.nih.gov/pavis2/) was used to annotate peaks and visualize relative enrichment levels to genomic regions. Panther website (http://pantherdb.org/about.jsp) was used to classify identified genes by GO term biological process. Genes lists were further classified into functionally related groups of genes by GO term or KEGG pathway using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/home.jsp).
Phospho-kinase Array
Human phospho-kinase array kit (ARY003B, R&D Systems) was used to detect phosphorylated kinases in hSC2λ-NF2−/− cells treated with DMSO or JQ1 (1 µM) for 15 h. Blots were imaged using ChemiDoc MP Imaging System and quantitated using Image Lab 6.1 software (Bio-Rad).
Animal Experiments
All animal experiments complied with NIH guidelines and were approved by The Scripps Research Institute Animal Care and Use Committee. 2 × 105 SC4-Luc cells were harvested, washed twice and resuspended in sterile PBS and injected intraneurally into the sciatic nerves of NOD/SCID mice (6–8 weeks old). Tumor progression was monitored by bioluminescence imaging (BLI) on an IVIS-200 system (Xenogen) and treatment was commenced after detection of signal that correlates with an average tumor size of 1–2 mm3 (total flux ≥106 photons/s). For JQ1 treatment assessment, JQ1 was used at 50 mg/kg/daily. For OTX-015 the dose was 50 mg/kg/daily.
Statistical Analysis
Cell counting studies were analyzed by Student’s t-test (Graphpad QuickCalcs) in order to determine the significance of results and obtain 2-tailed P values. Two-tailed unpaired Student’s t-tests were used to assess the significance of tumor–body ratio results. The mean and SD were used to assess significance, unless otherwise noted. Statistical analyses were performed using Prism version 6.0e for Mac (GraphPad Software, www.graphpad.com).
Results
BET Inhibition Inhibits Proliferation and Tumorigenic Capacity of NF2-Null Schwann Cells
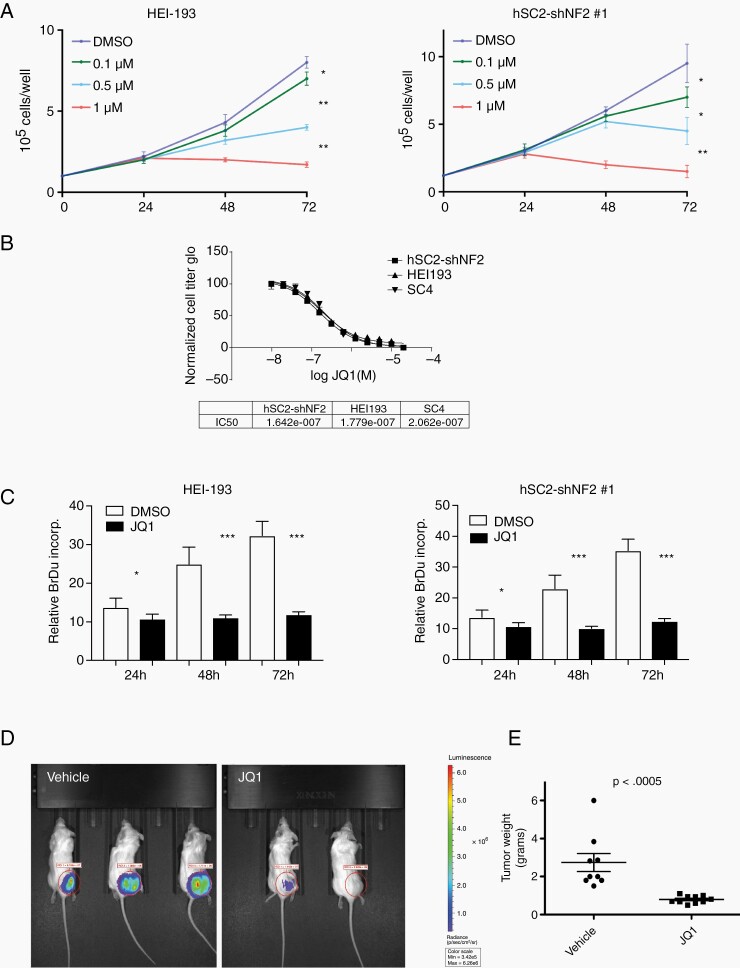
The involvement of BET bromodomain proteins in many different types of malignancies, including NF1-associated MPNSTs, prompted us to examine the sensitivity of NF2-null schwannoma cells to the BET inhibitor JQ1. To examine the role and requirement of BET proteins for growth of NF2-deficient Schwann cells we employed the human schwannoma cell line HEI-193,20 human Schwann cells with stable shRNA knockdown of NF2 (hSC2λ-shNF2 clones #1 and #2)23 and human Schwann cells in which the NF2 allele was inactivated by CRISPR mediated genome editing (hSC2λ-NF2−/− clones #5 and #9) (Supplementary Figure S1A). Cells were treated with increasing concentrations of JQ1 and counted over 3 days, which resulted in a dose-dependent inhibition of cell growth over time (Figure 1A and Supplementary Figure S1B and D). To determine the IC50 for JQ1 we assessed the impact of JQ1 treatment on cells in a dose–response format using the HEI-193, hSC2λ-shNF2, and mouse schwannoma SC4 cells. The IC50 was similar in all 3 cell lines, ranging from 160 to 210 nM (Figure 1B). To assess the impact of BET inhibition on cell proliferation we examined the effects of JQ1 treatment on BrdU incorporation in the HEI-193, SC4, and hSC2λ-shNF2 cells. BrdU incorporation was slightly reduced at 24 h but significantly reduced at 48 and 72 h post-treatment (Figure 1C and Supplementary Figure S1C). The pattern of reduced BrdU incorporation followed the pattern of cell growth inhibition after JQ1 treatment. To assess whether JQ1 treatment increased the rate of cell death in treated cells HEI-193, hSC2λ, hSC2λ-shNF2, and mouse schwannoma SC4 cells were treated with JQ1 and the impact caspase-3 status was assessed by western blotting. This analysis indicated that JQ1 treatment did not lead to a detectable change in the levels of activated caspase-3 (Supplementary Figure S1E).
Figure 1.
Impact of BET inhibition on NF2-null Schwann cell proliferation and tumor formation capacity. (A) HEI-193 and hSC2λ-shNF2 cells were cultured with DMSO or JQ1 (0.1, 0.5, or 1 µM) and counted daily, for 3 days. The data shown are the mean of 3 independent experiments. Error bars = SD, *P < .05, **P < .01. (B) Ten-point dose–response curves assessing viability of HEI-193, hSC2λ-shNF2, and SC4 cells treated with JQ1 at the indicated concentrations, for 48 h. Cell viability was measured by luminescent ATP-dependent assay (Cell Titer Glo), in quadruplicate. Nonlinear regression of log JQ1 versus normalized luminescence with variable slope is plotted and the IC50 calculated. The data shown are the mean of 3 independent experiments. Error bars = SD. (C) Bromodeoxyuridine (BrdU) incorporation was measured in HEI-193 and hSC-shNF2 cells cultured in DMSO or JQ1 (1 µM) over 3 days, in triplicate. The data shown are the mean of 3 independent experiments. Error bars = SD, *P < .05, ***P < .001. (D) JQ1 treatment inhibits tumor growth in vivo. Representative BLI images of NOD/SCID carrying orthotopic tumors originating from SC4-Luc cells, at day 14 of treatment with vehicle or JQ1 (50 mg/kg/daily). (E) Distribution of tumor weight from vehicle or JQ1-treated cohorts. The results of t-test with equal variances show the JQ1-treated cohort has significant lower average tumor weight compared to control group. N = 10 in each cohort.
To assess impact of BET inhibition on NF2-null Schwann cell tumorigenesis in vivo, we used an orthotopic model based on SC4-luciferase cells (SC4-Luc), which have been employed extensively to assess the antineoplastic activity of various drugs and small molecules,23–26 The SC4-Luc cells were orthotopically implanted into the sciatic nerves of NOD/SCID mice and tumor growth was monitored via BLI. At 10 days postsurgery similar flux readings for all animals were validated and animals were enrolled into control (vehicle only) JQ1-treated cohorts (50 mg/kg, IP, once daily) for a period of 14 days. Upon completion of treatment the mice were imaged, euthanized, and tumors removed and weighed. Comparison of tumor weights demonstrated a significantly lower average tumor weight in the JQ1-treated groups compared to control groups (Figure 1D and E). Similar results were obtained with OTX-015 (Birabresib), an improved BET inhibitor that is currently in clinical trials.16 Tracking the BLI signal in OTX-015-treated animals suggests that treatment partially impairs tumor growth rates compared to the control group (Supplementary Figure S1E). Like in the case of JQ1 treatment, comparison of tumor weights at the conclusion of the study demonstrated a significantly lower average tumor weight in the OTX-015-treated cohort compared to control group (Supplementary Figure S1F). Taken together, these data demonstrate that BETi has significant antiproliferative activity against human and mouse NF2-null schwannoma cells in vitro and antitumor activity in vivo.
The Effects of JQ1 Are Mediated by BRD4 and to a Lesser Extent by BRD2
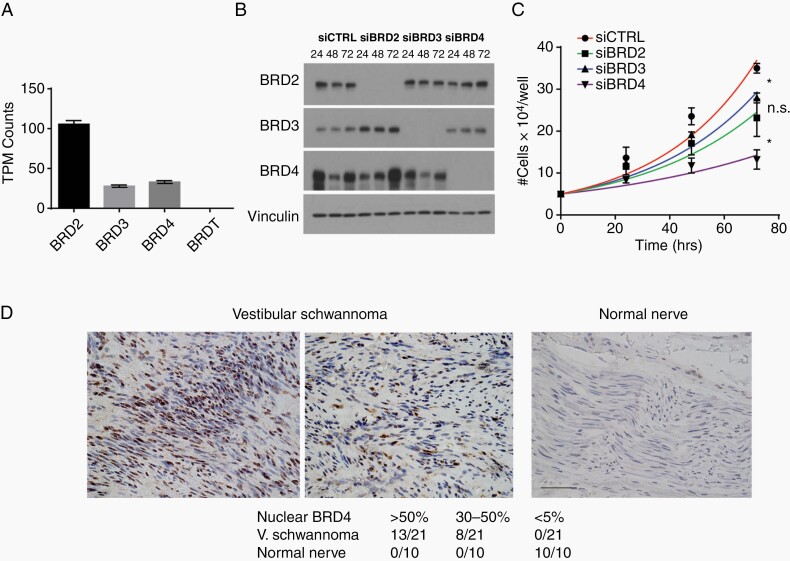
To directly determine the requirement for individual BET proteins in NF2-deficient Schwann cells, we first examined the expression profile of BET proteins in hSC2λ-shNF2 cells. While the level of BRDT mRNA was below levels of detection, BRD2, 3, and 4 are expressed and detected (Figure 2A). Similarly, when examining the expression of the BET proteins in these cells, BRD2, BRD3, and BRD4 proteins were readily detected by western blotting, while BRDT was below the level of detection (Figure 2B). To assess any requirement for BRD2, 3, or 4, we knocked down expression of the individual proteins in hSC2λ-shNF2, hSC2λ-NF2−/−, hSC2λ-wt, and HEI-193 cells (Figure 2B and Supplementary Figure S2A–C) and assessed the impact on cell proliferation. A significant decrease in cell numbers was observed in response to BRD4 knockdown in all cell lines tested, and to a lesser extent in the case of BRD2 or BRD3 knockdown (Figure 2C and Supplementary Figure S2A–C). These findings suggest that the effects of JQ1 are predominantly mediated through inhibition of BRD4 in NF2-deficient Schwann cells.
Figure 2.
Characterization of BET proteins expression and function in NF2-null Schwann cells and human schwannomas. (A) Relative expression levels of BRD family proteins in NF2-null Schwann cells, based on analysis of available data from GSE61528.23 (B) Confirmation of BRD protein knockdown. Cell lysates were western blotted for BRD2, BRD3, and BRD4 to confirm knockdown of protein by transfected siRNA. Vinculin used as a loading standard). (C) Proliferation of hSC2λ-shNF2 cells transfected with siRNA pools targeting BRD2 (squares), BRD3 (triangles), BRD4 (inverted triangles), and nontargeting siRNA (circles). Cells were counted daily, for 3 days, in triplicate. The data shown are the mean of 3 independent experiments. Error bars = SD, *P < .05, n.s. = nonsignificant. (D) Representative images of BRD4 immunostains of human vestibular schwannomas and normal nerve. Left panel shows schwannoma with >50% BRD4 immunopositive nuclei, middle panel shows schwannoma with 30%–50% BRD4 immunopositive nuclei, right panel shows normal nerve with <5% BRD4-positive nuclei. Scale bar = 100 µm. Bottom table shows number of tumors with the indicated level of nuclear BRD4 signal.
To establish the relevance of BRD4, we used immunohistochemistry (IHC) to assess expression of BRD4 in schwannomas (N = 21) and compared this to normal nerve (N = 10). The IHC analysis found that in normal nerve sections (10/10) BRD4 levels are low and nuclear localization of BRD4 was observed in <5% of cells. In contrast, schwannomas displayed significantly elevated levels of BRD4 and extensive nuclear localization in 13/21 tumors (>50% of nuclei with immunopositivity) and moderate nuclear immunoreactivity in the remaining 8/21 tumors (between 30% and 50% of nuclei with immunopositivity). Examples are shown in Figure 2D. These findings suggest elevated BRD4 expression and nuclear localization, and thus activation, is exhibited by a majority of schwannomas.
NF2-Deficient Schwann Cells Are More Sensitive to JQ1 Treatment
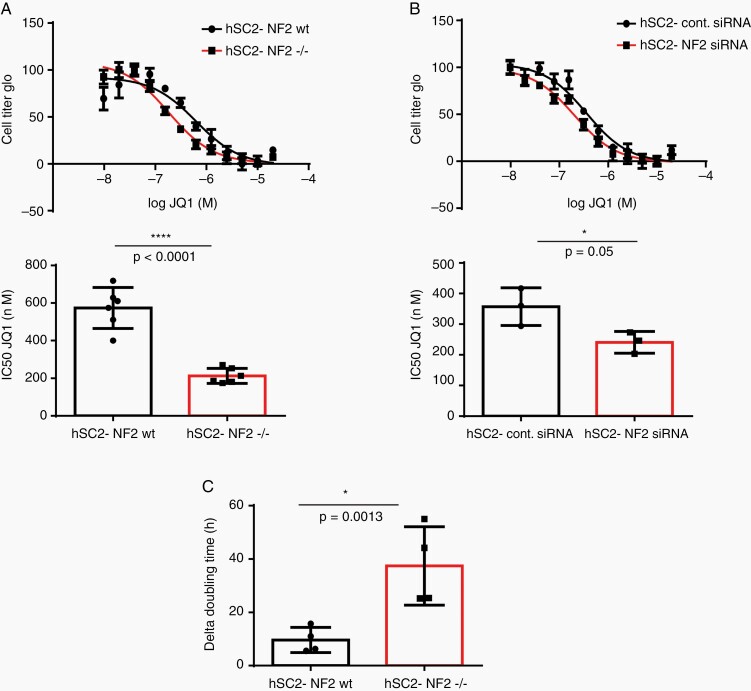
To determine if the sensitivity of Schwann cells to JQ1 was impacted by the loss of NF2 we employed the hSC2λ-NF2−/− cells (Figure 3A), and hSC2λ cells with siRNA knockdown of NF2 (Figure 3B, confirmation of knockdown shown in Supplementary Figure S3A), and compared their response to JQ1 to their wildtype counterparts. Dose–response analysis showed a significant and reproducible decrease in the IC50 of JQ1, in cells deficient for NF2, compared to their controls (Figure 3A and B). This was not due to changes in the levels of BET protein expression as these remain consistent between the hSC2λ and hSC2λ-NF2−/− cells (Supplementary Figure S3B). To further assess the impact of JQ1 treatment on the proliferation of the human Schwann cells, we examined the doubling time of hSC2λ-NF2−/− and hSC2λ cells, with or without JQ1 treatment (200 nM). This analysis demonstrated that the NF2-deficient cells have a significant increase in doubling time in the presence of JQ1 compared to cells wildtype for NF2, thus indicating that cellular proliferation was inhibited to a greater extent in NF2-deficient cells (Figure 3C). Together these data suggest that loss of NF2 sensitizes Schwann cells to inhibition by JQ1 and supports further exploration of its use as a potential therapeutic agent for NF2-deficient cancers.
Figure 3.
Enhanced sensitivity of NF2-null Schwann cells to BET inhibition. (A) Representative JQ1 dose–response curve for hSC2λ (black) and hSC2λ-NF2−/− (red) cells (top) and average IC50 as determined by of 6 biologic replicates of the dose–response curves (bottom). (B) Representative JQ1 dose–response curve for hSC2λ cells treated with control siRNA (circles) or siRNA against NF2 (squares) (top) and average IC50 as determined by of 3 biologic replicates of the dose–response curves (bottom). All concentration points were done in quadruplicate. (C) The differences in doubling time for hSC2λ or hSC2λ-NF2−/− cells were determined in the presence or absence of JQ1 (200 nM) and calculated (dt DMSO − dt JQ1) from 4 independent biologic replicates. Error bars = SD, where indicated P values are unpaired, 2-tailed, Student’s t-test.
The Genomic Landscape of BRD4 Binding in NF2-Null Schwann Cells
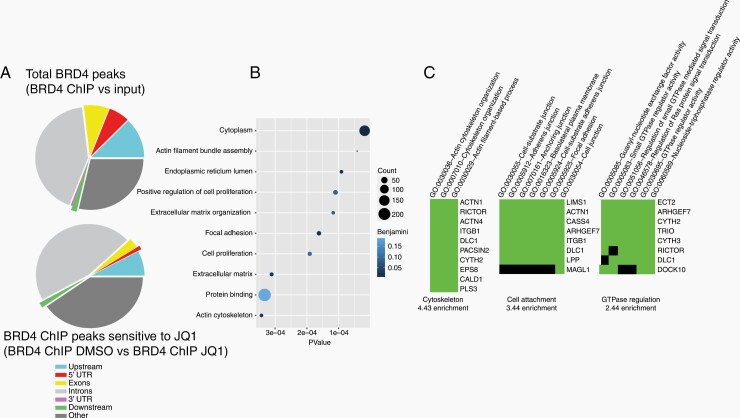
To identify genes directly engaged by BRD4 in the NF2-null Schwann cells that are sensitive to JQ1 treatment, we carried out BRD4 ChIPs in hSC2λ-shNF2 cells with or without JQ1 treatment. A total of 55 568 BRD4 ChIP peaks were called in control-treated cells compared to input DNA (BRD4-ChIP DMSO vs input) and a total of 436 JQ1-sensitive BRD4 peaks were called in control-treated cells compared to JQ1-treated cells (BRD4 ChIP DMSO vs BRD4 ChIP JQ1). The peaks were then mapped, with 71% (39 105) of total BRD peaks and 60% (259) of JQ1-sensitive BRD4 ChIP peaks in close proximity to known genes or regulatory elements (Figure 4A). BRD4 appeared to bind predominantly in intronic regions but could also be found in all other genomic regions. 12 145 unique genes were identified from the total BRD4 peaks and 164 unique genes were identified from the JQ1-sensitive BRD4 peaks. The genes associated with the JQ1-sensitive BRD4 peaks were sorted according to GO biological process and plotted by number of genes in group and P value. Several genes were associated with positive regulation of cell proliferation, thus supporting the antiproliferative activity of JQ1 in these NF2 depleted cells (Figure 4B). To further explore the biology of these genes we looked for enrichment of functional-related gene groups (Figure 4C). The top 3 clusters enriched in the JQ1-sensitive BRD4 peaks are associated with genes regulating cytoskeletal organization, cell attachment and GTPase regulation, highlighting processes that are regulated by Merlin.27–29
Figure 4.
BRD4 genomic distribution in NF2-null Schwann cells. BRD4 was chromatin immunoprecipitated from hSC2λ-shNF2 Schwann cells treated with DMSO or JQ1 (1 µM) and total BRD4 peaks mapped to GRCh38/hg38. (A) Distribution of BRD4 peaks (BRD4 ChIP vs input) (top) and BRD4 peaks sensitive to JQ1 treatment (BRD4 ChIP DMSO vs BRD4 ChIP JQ1) (bottom), relative to gene bodies. Shown are pie charts of peak enrichment relative to genomic location (peak annotation visualization, PAVIS) (B) GO annotation of biological process of genes associated with JQ1-sensitive BRD4 peaks. (C) Gene list of JQ1-sensitive BRD4 binding peaks, classified into functionally related groups by GO term annotation.
JQ1 Inhibits PI3K/AKT Signaling in NF2-Null Schwann Cells
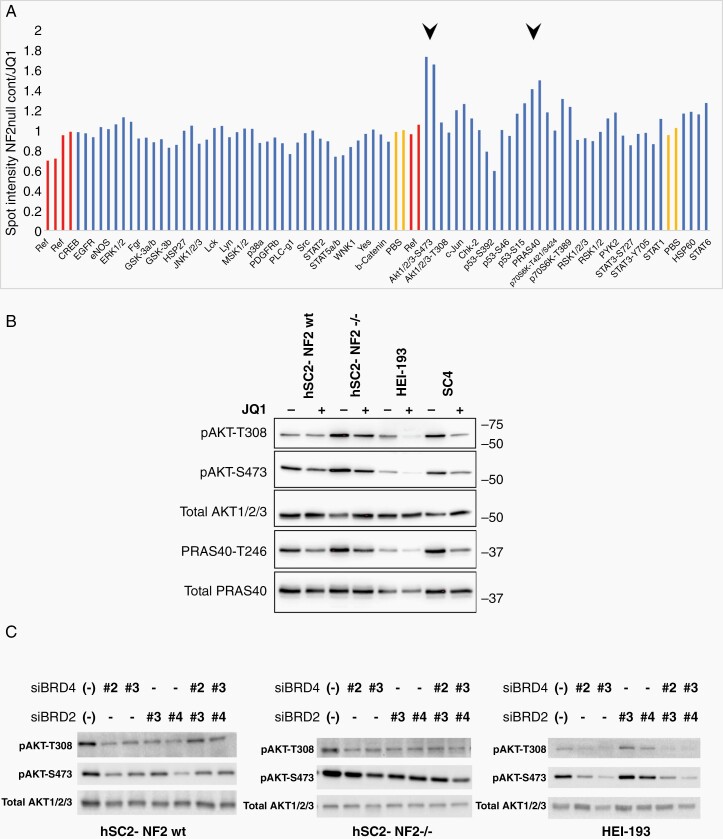
To explore signaling pathways impacted by JQ1 we used a phospho-kinase array to detect changes in phosphorylation of key signaling effectors after JQ1 treatment in hSC2λ-NF2−/− cells. JQ1-treated cells showed a significant decrease in phosphorylation of AKT1/2/3-S473, and PRAS40-T246. In addition, a significant increase in P53-S392 phosphorylation was observed (Figure 5A and Supplementary Figure S5A). To confirm and expand the array results, we validated the results of the phospho-kinase array for the individual phosphorylation sites in hSC2λ-NF2−/−, hSC2λ-NF2wt, HEI-193, and SC4 cells, using western blot analysis. Indeed, pAKT-S473 showed significant decreases in all cell lines after JQ1 treatment (Figure 5B and Supplementary Figure S5B), while pAKT-T308 showed significant decreases in all cell lines besides the hSC2λ-NF2wt cells (Figure 5B and Supplementary Figure S5C). These results indicate that JQ1 inhibits PI3K/Akt signaling and suggests an additional potential mechanism for inhibition of proliferation. Using western blot analysis of pPRAS40-T246 and pP53-S392 phosphorylation, we were unable to confirm these sites are consistently differentially phosphorylated as a consequence of JQ1 treatment (Supplementary Figure S5D and E).
Figure 5.
BET inhibition impacts PI3K/AKT signaling in NF2-null Schwann cells. (A) Lysates from hSC2λ-NF2−/− cells treated with DMSO or JQ1 (1 µM) for 15 h were used to interrogate an antibody array targeting phospho-kinases on the indicated effectors. The arrays were imaged and signal intensity of control treated versus to JQ1 treated were calculated. Duplicate spots are shown for the individual effectors (blue), control references (red), and negative control (no protein) (yellow) are shown. Spots for pAKT1/2/3-S473 and pPRAS40 showed a >1.5-fold increase in signal intensity in the control-treated cells (black arrowheads). (B) Representative western blot shown for assessment of pAKT1/2/3-S473, pAKT1/2/3-S308, and pPRAS40 phosphorylation in hSC2λ, hSC2λ-NF2−/−, HEI-193, and SC4 cells treated with JQ1 (1 µM). (C) Western blot analysis of pAkt-T308, pAkt-S473, and total Akt levels of expression in hSC2λ, hSC2λ-NF2−/−, and HEI-193 cells following BRD4 and BRD2 knockdown. Cells were transfected with individual siRNAs targeting BRD4, BRD2, the combination, or a nontargeting control (−). Protein was collected 48 h later. Images are representative of 3 individual experiments.
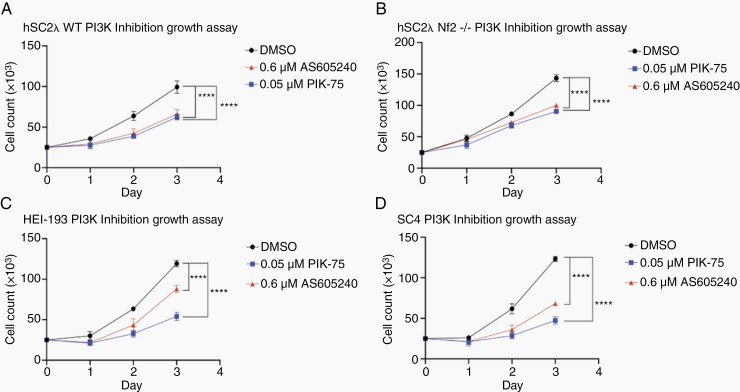
To determine whether the effects of JQ1 on Akt phosphorylation were mediated vis BRD4 or BRD2, we knocked the expression of the proteins down individually or in combination. In agreement with the results of JQ1 treatment, knockdown of BRD4 resulted in reduced levels of pAKT-p308 and pAKT-p473, while BRD2 knockdown did not result in a consistent reduction in the phosphorylation levels of these site on Akt (Figure 5C). This suggests that the effects of JQ1 on Akt are likely mediated via BRD4 inhibition. Finally, to confirm that inhibition of PI3K signaling impairs cell growth in the model systems we are using we treated the cells with the PI3K inhibitors PIK-75 and AS605240. As expected, this treatment significantly impaired cell growth (Figure 6).
Figure 6.
PI3K inhibition impairs cell growth. Growth assays were conducted using (A) hSC2λ cells; (B) hSC2λ-NF2−/− cells; (C) HEI-193 cells; and (D) SC4 cells. Cells were plated in triplicate in 24-well plates, 25 000 cells per well. Cells were treated with 0.05 µM PIK-75, 0.6 µM AS605240, or DMSO (0.5%) and counted for 3 days. Significance was assessed by 2-way ANOVA. Post hoc analysis was conducted when ANOVA determined the interaction of treatment was significant. ****P < .0001. Error bars = SD. Graphs representative of 3 individual experiments.
Discussion
Recent studies in a mouse model of NF1-associated MPNSTs demonstrated that BRD4 was highly expressed and tumors were sensitive to BET protein inhibition. In this model, inhibition of BRD4 triggered cell death in the MPNSTs and this appears to be mediated through regulation of the apoptotic effectors Bim and Bcl-2.18 Interestingly, subsequent studies offer conflicting results. While 1 report suggested that treatment of NF1-associated MPNST cell lines with BET inhibitors, either alone or in combination with other agents, impaired cellular proliferation, another report30 concluded that MPNST cells do not display an increased sensitivity to JQ1 treatment.31 These findings suggest that BET inhibition in this tumor type could be beneficial and illustrates the fact that further work is needed to understand the mechanisms of action of these drugs. To determine the utility of BET inhibition in the context of NF2, we carried out an extensive analysis of cell-based and animal models of this disease. Our findings demonstrate that NF2-deficient schwannoma cells are sensitive to the BET protein inhibitor JQ1, suggesting a novel therapeutic option for treating these tumors. NF2-deficient Schwann cells and schwannoma cell lines were arrested at low, nanomolar, concentrations of JQ1 in culture and in vivo, in an orthotopic model of NF2-associated schwannoma. Interestingly, the sensitivity to JQ1 appeared to be enhanced in the context of NF2 loss, as CRISPR knockout or siRNA knockdown of NF2 in human Schwann cells increased sensitivity to JQ1. In NF2-null Schwann cells, JQ1 appeared to be acting mostly through inhibition of BRD4 as siRNA knockdown of BRD4 arrested proliferation to a similar extent as JQ1 treatment and had similar effects on p-AKT. While we do not provide direct proof that the effects of JQ1 require BRD4, as these studies require tools allowing decoupling of BRD4 from the effects of JQ1 while retaining normal function, we believe the evidence provided supports this conclusion. Significantly, human vestibular schwannomas demonstrated high levels of BRD4 protein by IHC, not seen in normal peripheral nerve, implying a functional role for BRD4 in these tumors.
Although our studies indicate that BET protein inhibitors may be effective for inhibiting the slow growth of NF2 schwannomas, clinical studies have shown adverse effects of these inhibitors that could be exacerbated with long-term use. Indeed, because of their toxicity, many studies are now focusing on combination therapies that allow lower doses of BET protein inhibitors to be used.32 Examples include combining BET protein inhibitors with immune checkpoint inhibitors or chromatin modifying inhibitors such as HDAC inhibitors. Alternatively, BRD4 inhibitors that have lower toxicity are currently under development (NHWD-870).33 Our work demonstrating sensitivity and exploring the mechanism of action of BET inhibitors in NF2 schwannomas is an important first step in assessing combination therapies to treat this cancer.
Supplementary Material
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Library preparation and sequencing were performed at The Scripps Research Institute Florida Genomics Core.
Contributor Information
Joanne Doherty, Department of Molecular Medicine, The Scripps Research Institute, Jupiter, Florida, USA.
Vinay Mandati, Department of Molecular Medicine, The Scripps Research Institute, Jupiter, Florida, USA.
Maria A González Rodriguez, Department of Molecular Oncology, H. Lee Moffitt Cancer Center, Tampa, Florida, USA.
Scott Troutman, Department of Molecular Oncology, H. Lee Moffitt Cancer Center, Tampa, Florida, USA.
Alyssa Shepard, Department of Molecular Medicine, The Scripps Research Institute, Jupiter, Florida, USA.
David Harbaugh, Department of Molecular Medicine, The Scripps Research Institute, Jupiter, Florida, USA.
Rachel Brody, Department of Pathology, Molecular, and Cell-based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Douglas C Miller, Department of Pathology and Anatomical Sciences, University of Missouri School of Medicine, Columbia, Missouri, USA.
Michael S Kareta, Genetics and Genomics Group, Sanford Research, Sioux Falls, South Dakota, USA.
Joseph L Kissil, Department of Molecular Oncology, H. Lee Moffitt Cancer Center, Tampa, Florida, USA.
Funding
The work was supported by NS117926 (National Institute of Neurological Disorders and Stroke / National Institute of Health) to J.L.K. M.S.K is supported by the National Institute of Health / National Cancer Institute / National Institute of General Medicine grant R01CA233661 and the National Institute of Health / National Institute of General Medicine Center for Pediatric Research 5P20GM103620.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship Statement. J.R.D., V.M., M.S.K., and J.L.K.: conceptualization, investigation, methodology, and data analysis. M.A.G.-R., A.S., M.L.C., M.S.K., D.C.M., and R.B.: methodology, data analysis, and review of manuscript. S.T. and D.H.: investigation and methodology. J.R.D and J.L.K.: writing of manuscript.
References
- 1. Gusella JF, Ramesh V, MacCollin M, Jacoby LB. Merlin: the neurofibromatosis 2 tumor suppressor. Biochim Biophys Acta. 1999;1423(2):M29–M36. [DOI] [PubMed] [Google Scholar]
- 2. Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177(5):893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rangwala R, Banine F, Borg JP, Sherman LS. Erbin regulates mitogen-activated protein (MAP) kinase activation and MAP kinase-dependent interactions between Merlin and adherens junction protein complexes in Schwann cells. J Biol Chem. 2005;280(12):11790–11797. [DOI] [PubMed] [Google Scholar]
- 4. Curto M, McClatchey AI. Nf2/Merlin: a coordinator of receptor signalling and intercellular contact. Br J Cancer. 2008;98(2):256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17(9):1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. [DOI] [PubMed] [Google Scholar]
- 7. Filippakopoulos P, Picaud S, Mangos M, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moriniere J, Rousseaux S, Steuerwald U, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461(7264):664–668. [DOI] [PubMed] [Google Scholar]
- 9. Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem Sci. 2015;40(8):468–479. [DOI] [PubMed] [Google Scholar]
- 10. French CA, Ramirez CL, Kolmakova J, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237–2242. [DOI] [PubMed] [Google Scholar]
- 11. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcotte R, Sayad A, Brown KR, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164(1–2):293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baratta MG, Schinzel AC, Zwang Y, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci U S A. 2015;112(1):232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toyoshima M, Howie HL, Imakura M, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8(1):24–36. [DOI] [PubMed] [Google Scholar]
- 17. Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel AJ, Liao CP, Chen Z, Liu C, Wang Y, Le LQ. BET bromodomain inhibition triggers apoptosis of NF1-associated malignant peripheral nerve sheath tumors through Bim induction. Cell Rep. 2014;6(1):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morrison H, Sperka T, Manent J, Giovannini M, Ponta H, Herrlich P. Merlin/neurofibromatosis type 2 suppresses growth by inhibiting the activation of Ras and Rac. Cancer Res. 2007;67(2):520–527. [DOI] [PubMed] [Google Scholar]
- 20. Hung G, Faudoa R, Li X, et al. Establishment of primary vestibular schwannoma cultures from neurofibromatosis type-2 patients. Int J Oncol. 1999;14(3):409–415. [DOI] [PubMed] [Google Scholar]
- 21. Yi C, Troutman S, Fera D, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19(4):527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Chang LJ, Neubauer DR, Muir DF, Wallace MR. Immortalization of human normal and NF1 neurofibroma Schwann cells. Lab Invest. 2016;96(10):1105–1115. [DOI] [PubMed] [Google Scholar]
- 23. Guerrant W, Kota S, Troutman S, et al. YAP mediates tumorigenesis in neurofibromatosis type 2 by promoting cell survival and proliferation through a COX2-EGFR signaling axis. Cancer Res. 2016;76(12):3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoxha S, Shepard A, Troutman S, et al. YAP-mediated recruitment of YY1 and EZH2 represses transcription of key cell-cycle regulators. Cancer Res. 2020;80(12):2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Licciulli S, Maksimoska J, Zhou C, et al. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem. 2013;288(40):29105–29114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Troutman S, Moleirinho S, Kota S, et al. Crizotinib inhibits NF2-associated schwannoma through inhibition of focal adhesion kinase 1. Oncotarget. 2016;7(34):54515–54525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2015;35(5):537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett. 2014;588(16):2743–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yi C, Kissil JL. Merlin in organ size control and tumorigenesis: Hippo versus EGFR? Genes Dev. 2010;24(16):1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varin J, Poulain L, Hivelin M, et al. Dual mTORC1/2 inhibition induces anti-proliferative effect in NF1-associated plexiform neurofibroma and malignant peripheral nerve sheath tumor cells. Oncotarget. 2016;7(24):35753–35767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amirnasr A, Verdijk RM, van Kuijk PF, Taal W, Sleijfer S, Wiemer EAC. Expression and inhibition of BRD4, EZH2 and TOP2A in neurofibromas and malignant peripheral nerve sheath tumors. PLoS One. 2017;12(8):e0183155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol. 2017;28(8):1776–1787. [DOI] [PubMed] [Google Scholar]
- 33. Yin M, Guo Y, Hu R, et al. Potent BRD4 inhibitor suppresses cancer cell-macrophage interaction. Nat Commun. 2020;11(1):1833–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data are stored in the Gene Expression Omnibus under accession number GSE167560.