Abstract
The mlaA gene encodes a lipoprotein to maintain an outer membrane lipid asymmetry in gram-negative bacteria. Although the role of mlaA in bacterial virulence has been studied in several bacterial species, there are no reports of its role in E. coli virulence. In this study, we found that knockout of mlaA in E. coli increased its virulence against silkworms. The mlaA-knockout mutant was sensitive to several antibiotics and detergents, but resistant to vancomycin and chlorhexidine. The mlaA-knockout mutant grew faster than the parent strain in the presence of silkworm hemolymph. The mlaA-knockout mutant also produced a larger amount of outer membrane vesicles than the parent strain. These findings suggest that mlaA knockout causes E. coli resistance to specific antimicrobial substances and increases outer membrane vesicle production, thereby enhancing E. coli virulence properties in the silkworm infection model.
Introduction
The outer membrane of gram-negative bacteria, including E. coli, has a lipid asymmetry, in which the outer leaflet comprises lipopolysaccharide (LPS) and the inner leaflet comprises phospholipids [1]. The outer leaflet has low fluidity and contributes to inhibit the invasion of various antibiotics and foreign chemicals into bacterial cells [2]. The outer membrane lipid asymmetry is maintained by the Mla system, phospholipase A (PldA), and LPS palmitoyltransferase (PagP) [3]. PldA and PagP contribute to maintain the lipid asymmetry by degrading phospholipids in the outer leaflet of the outer membrane. The Mla system consists of 6 proteins, MlaA, MlaB, MlaC, MlaD, MlaE, and MlaF, that transport phospholipids from the outer leaflet of the outer membrane into the inner membrane. MlaA is a lipoprotein locating at the outer membrane that transfers phospholipids to MlaC, a periplasmic protein [4]. MlaC transfers phospholipids to the MlaFEDB complex in the inner membrane, and MlaFEDB then inserts the phospholipids into the inner membrane.
Knockout of the Mla system destroys the lipid asymmetry of the outer membrane and increases the sensitivity to various antimicrobial substances in many gram-negative bacteria. Knockout of MlaA causes bacterial sensitivity to various antimicrobial substances; sodium dodecyl sulfate (SDS)/EDTA in E. coli [4,5], hydrophobic antibiotics such as erythromycin, rifampicin, azithromycin, and polymyxin E in Haemophilus influenzae [6]; polymyxin B and vancomycin in Neisseria gonorrhoeae [7]; and tetracycline, ciprofloxacin, chloramphenicol, cathelicidin, and LL-37 in P. aeruginosa [8,9]. Knockout of MlaC also causes sensitivity to macrolides and fluoroquinolones in Burkholderia species [10], and gentamycin, novobiocin, and rifampicin in Acinetobacter baumanii [11]. In contrast, however, the MlaA-knockout mutant of E. coli shows resistance to chlorhexidine [12]. E. coli mutants resistant to the antimicrobial peptide aenicin-3 carry mutations in the mlaA gene or the mlaBCDEF operon [13]. The characteristics of antibiotics for which knockout of the Mla system causes bacterial sensitivity or resistance, however, are unknown.
Knockout of MlaA exerts both negative and positive effects on the virulence properties of various bacterial species. In Shigella flexneri, knockout of MlaA decreases bacterial spreading among host cells [14,15]. In H. influenzae and H. parasuis, knockout of MlaA attenuates bacterial infectivity in host epithelial cells and decreases the bacterial burden in a mouse model [6,16]. In Burkholderia pseudomallei, knockout of MlaA decreases bacterial persistence in mouse spleen [17]. On the other hand, in N. gonorrhoeae and Vibrio cholerae, knockout of MlaA increases the bacterial number in the mouse genital tract or gut [7,18]. In H. influenzae, A. baumannii, N. gonorrhoeae, and V. cholerae, knockout of MlaA increases the production of outer membrane vesicles (OMV), which have various functions in infectious processes, such as absorbing antimicrobial molecules and transporting toxins to host cells [7,18–21]. In P. aeruginosa, knockout of MlaA decreases bacterial virulence in a mouse lung infection model [9], but increases bacterial virulence in a fruit fly infection model [8]. How MlaA affects E. coli virulence, however, is unclear.
We previously investigated gene mutations that upregulate E. coli virulence properties using a silkworm infection model [22,23]. In the present study, we searched E. coli mutants with high virulence from among E. coli mutants resistant to vancomycin and found that knockout of MlaA upregulates E. coli virulence properties.
Results
Knockout of mlaA increases E. coli killing activity against silkworms
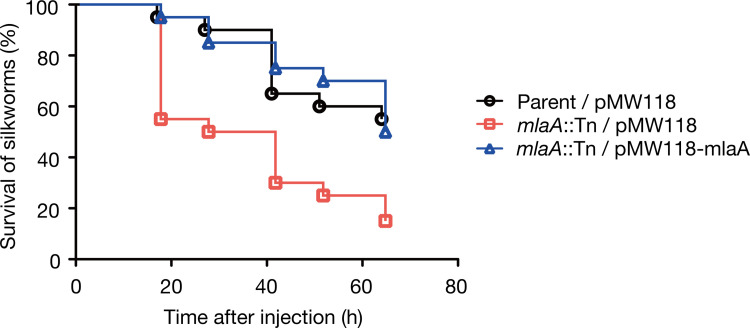
We previously revealed that amino acid substitutions in LptD or LptE, the LPS transporter subunits, as well as knockout of OpgG or OpgH, the synthetases for osmoregulated periplasmic glucan, cause E. coli resistance against vancomycin and increase E. coli killing activity against silkworms [22,23]. Based on this finding, we hypothesized the existence of E. coli genes whose knockout could lead to high vancomycin resistance as well as high killing activity against silkworms. By searching E. coli gene knockout mutants showing vancomycin resistance from a transposon mutant library, we identified 50 gene knockout mutants exhibiting vancomycin resistance (Table 1). We evaluated the killing activity of the vancomycin-resistant mutants against silkworms, and found that the mlaA-knockout mutant killed silkworms faster than the parent strain (Table 1, Fig 1). The increased killing activity of the mlaA-knockout mutant was blocked by introducing the intact mlaA gene (Fig 1). In addition, the mlaA-knockout mutant exhibited better growth than the parent strain in the presence of vancomycin, and the vancomycin resistance was decreased to the parent level by introducing the intact mlaA gene (Fig 2A). These results suggest that mlaA knockout leads to vancomycin resistance and increases E. coli virulence against silkworms.
Table 1. Silkworm-killing activities of transposon mutants exhibiting resistance against vancomycin.
| Strain ID | Gene | Product | Survival (%) |
|---|---|---|---|
| JD20172 | insH1 | Transposase InsH for insertion seguence element IS5A | 100 |
| JD20241 | carB | Carbamoyl-phosphate synthase large chain | 60 |
| JD20379 | acnB | Aconitate hydratase B | 20 |
| JD20391 | hpt | Hypoxanthine phosphoribosyltransferase | 40 |
| JD20727 | ybbO | Uncharacterized oxidoreductase | 20 |
| JD20913 | segA | Endonuclease segA | 80 |
| JD21066 | galU | UTP-glucose-1-phosphate uridylyltransferase | 100 |
| JD21400 | psuT | Putative pseudouridine transporter | 60 |
| JD21547 | crr | PTS system glucose-specific EIIA component | 40 |
| JD21607 | guaB | Inosine-5’-monophosphate dehydrogenase | 100 |
| JD21620 | ndk | Nucleoside diphosphate kinase | 80 |
| JD21662 | mltA | Membrane-bound lytic murein transglycosylase A | 60 |
| JD21673 | nlpI | Lipoprotein NlpI | 60 |
| JD21732 | yibB | Protein HtrL | 100 |
| JD21864 | ybiS | Probable L, D-transpeptidase YbiS | 100 |
| JD22094 | opgG | Glucans biosynthesis protein G | ND |
| JD22095 | opgH | Glucans biosynthesis glucosyltransferase H | ND |
| JD22143 | yceG | Endolytic murein transglycosylase | 80 |
| JD22152 | ycfL | Uncharacterized protein YcfL | 100 |
| JD22156 | lpoB | Penicillin-binding protein activator LpoB | 80 |
| JD23323 | mlaA | Intermembrane phospholipid transport system lipoprotein MlaA | 0 |
| JD23420 | uraA | Uracil permease | 100 |
| JD23606 | proX | Glycine betaine/proline betain-binding periplasmic protein | 100 |
| JD23607 | ygaZ | Inner membrane protein YgaZ | 100 |
| JD23873 | gshB | Glutathione synthetase | 100 |
| JD23934 | yghQ | Inner membrane protein YghQ | 100 |
| JD23935 | yghR | Uncharacterized ATP-binding protein YghR | 80 |
| JD23938 | yghS | Uncharacterized ATP-binding protein YghS | 100 |
| JD23939 | yghT | Uncharacterized ATP-binding protein YghT | 100 |
| JD24024 | tolC | Outer membrane protein TolC | 100 |
| JD24230 | mlaC | Intermembrane phospholipid transport system binding protein MlaC | 60 |
| JD24231 | mlaD | Intermembrane phospholipid transport system binding protein MlaD | 40 |
| JD24242 | yhbJ | Rnase adapter protein RapZ | 80 |
| JD24280 | sspA | Glutamyl endopeptidase | 100 |
| JD24335 | dusB | tRNA-dihydrouridine syntaseB | 100 |
| JD24462 | waaY | Lipopolysaccharide core heptose(Ⅱ) kinase RfaY | 80 |
| JD24466 | waaR | Lipopolysaccharide 1,2-glucosyltransferase | 100 |
| JD24468 | waaB | Lipopolysaccharide 1,6-galactosyltransferase | 100 |
| JD24476 | waaQ | Lipopolysaccharide core heptosyltransferase RfaQ | 100 |
| JD24492 | yicC | UPF0701 protein YicC | 100 |
| JD24700 | wecA | Undecaprenyl-phosphate alpha-N-acetylglucosaminly 1-phosphate transferase | 60 |
| JD25777 | slt | Soluble lytic murein transglycosylase | 20 |
| JD26183 | bioB | Biotin synthase | 80 |
| JD26764 | glf | UDP-galactopyranose mutase | 60 |
| JD27118 | mltB | Membrane-bound lytic murein transglycosylase B | 100 |
| JD27649 | hycB | Formate hydrogenlyase subunit 2 | 100 |
| JD27708 | mlaE | Intermembrane phospholipid transport system permease protein MlaE | 60 |
| JD27710 | mlaF | Intermembrane phospholipid transport system ATP-binding protein MlaF | 80 |
| JD27958 | citC | [Citrate[pro-3S]-lyase]ligase | 80 |
Transposon mutants exhibiting resistance to vancomycin are listed. Bacterial solutions (2 x 108 CFU) were injected into silkworms (n = 5) and percent survival at 2 days post infection was measured and is presented as “Survival”. Percent survival of the parent strain was 100%. ND, not determined.
Fig 1. Knockout of mlaA increases E. coli virulence against silkworms.
Silkworm killing activity of the parent strain transformed with an empty vector (Parent/pMW118), the mlaA-knockout mutant transformed with an empty vector (mlaA::Tn/pMW118), and the mlaA-knockout mutant transformed with pMW118-mlaA (mlaA::Tn/pMW118-mlaA) was examined. Silkworms were injected with bacterial cells (2 x 108 CFU) and survival was monitored. The experiment was performed twice and the data were pooled (n = 20). Log-rank test p value was less than 0.05 between Parent/pMW118 and mlaA::Tn/pMW118, or between mlaA::Tn/pMW118 and mlaA::Tn/pMW118-mlaA.
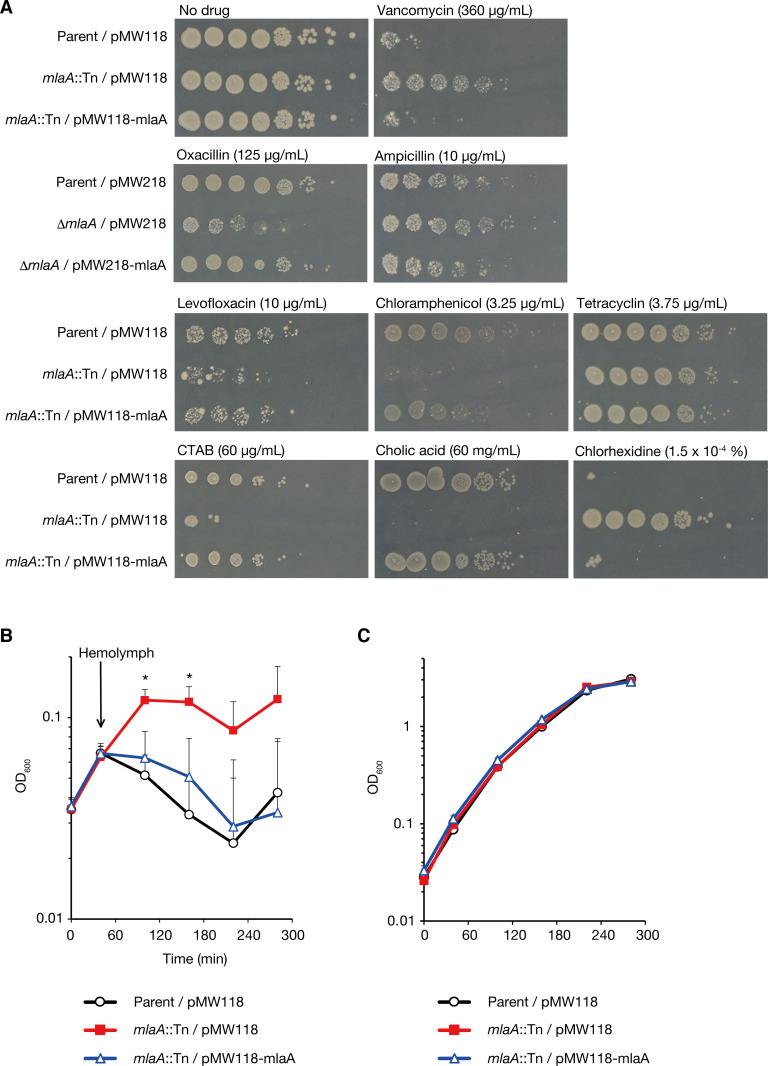
Fig 2. The mlaA-knockout mutant exhibits various sensitivities to antibiotics and resistance to silkworm antimicrobial substances.
(A) E. coli overnight culture of the Parent/pMW118, mlaA::Tn/pMW118, or mlaA::Tn/pMW118-mlaA strain was 10-fold serially diluted; spotted onto LB agar plates supplemented with or without vancomycin, levofloxacin, chloramphenicol, tetracycline, CTAB, cholic acid, or chlorhexidine; and incubated at 37˚C. To examine the sensitivity to oxacillin and ampicillin, the mlaA deletion mutant (markerless deletion mutant of mlaA) transformed with pMW218 (ΔmlaA/pMW218) and the mlaA deletion mutant transformed with pMW218-mlaA (ΔmlaA/pMW218-mlaA) were used. (B) E. coli strains of the Parent/pMW118, mlaA::Tn/pMW118, or mlaA::Tn/pMW118-mlaA were aerobically cultured in LB medium and silkworm hemolymph was added to the bacterial culture at 40 min after the bacterial inoculation. The vertical axis represents the OD600 value of the bacterial culture, and the horizontal axis represents the culture time. The means ± standard errors from 5 independent experiments are shown. Star indicates Student t-test p value less than 0.05 between the Parent/pMW118 vs. mlaA::Tn/pMW118, and between mlaA::Tn/pMW118 vs. mlaA::Tn/pMW118-mlaA. (C) E. coli strains of the Parent/pMW118, mlaA::Tn/pMW118, or mlaA::Tn/pMW118-mlaA were aerobically cultured in LB medium and the OD600 values of the cultures were measured.
Knockout of mlaA alters E. coli sensitivity to various antimicrobial molecules
Next, we examined whether mlaA knockout alters E. coli sensitivity to various antimicrobial molecules. The mlaA-knockout mutant showed less growth than the parent strain in the presence of antibiotics such as levofloxacin, chloramphenicol, and oxacillin, and detergents such as cetyltrimethylammonium bromide (CTAB) and cholic acid (Fig 2A). The growth of the mlaA-knockout mutant was indistinguishable from that of the parent strain in the presence of tetracycline (Fig 2A). Consistent with a previous report [12], the mlaA-knockout mutant showed better growth than the parent strain in the presence of chlorhexidine (Fig 2A). The alteration of drug sensitivity in the mlaA-knockout mutant was restored to the parent strain level by introducing the intact mlaA gene (Fig 2A). These results suggest that mlaA knockout alters E. coli sensitivity to various antimicrobial molecules.
To clarify the molecular mechanisms by which mlaA knockout increases E. coli virulence against silkworms, we examined bacterial growth in the presence of silkworm hemolymph in which antimicrobial peptides were induced by injection of heat-killed bacteria [24]. The mlaA-knockout mutant grew faster than the parent strain in the presence of silkworm hemolymph (Fig 2B). The faster growth of the mlaA-knockout mutant in the presence of silkworm hemolymph was abolished by introducing the intact mlaA gene (Fig 2B). In contrast, in the absence of silkworm hemolymph, the growth of the mlaA-knockout mutant was indistinguishable from that of the parent strain (Fig 2C). These results suggest that the mlaA-knockout confers E. coli resistance to silkworm immune mechanisms.
Knockout of mlaA increases OMV production
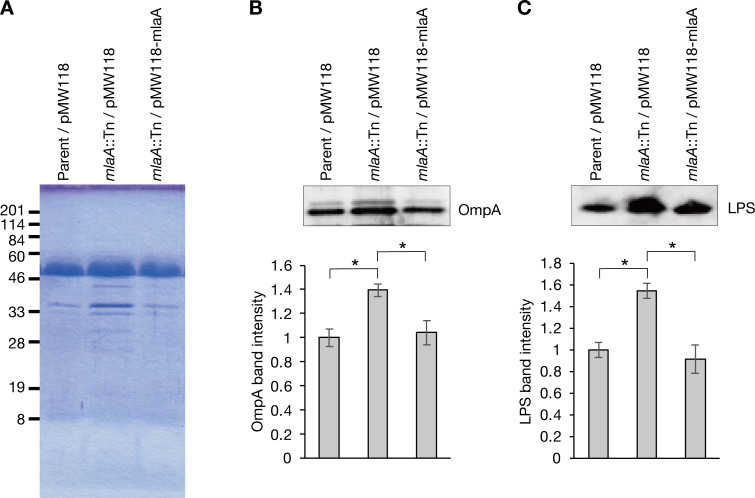
Knockout of mlaA leads to increased production of OMVs in N. gonorrhoeae, H. influenza, V. cholerae [7,20]. We examined whether mlaA knockout increases OMV production in E. coli. SDS-polyacrylamide gel electrophoresis analysis revealed that the amounts of proteins with about 33 kDa increased in the OMV fraction of the mlaA-knockout mutant compared with the parent strain (Fig 3A, S1 Raw images). Western blot analysis revealed that the amounts of OmpA and LPS, which are components of OMV, increased in the OMV fraction of the mlaA-knockout mutant compared with the parent strain (Fig 3B and 3C, S1 Raw images). The increase in OmpA and LPS was decreased by introducing the intact mlaA gene (Fig 3B and 3C, S1 Raw images). These results suggest that mlaA knockout increases OMV production.
Fig 3. Knockout of mlaA increases OMV production.
(A) Culture supernatants of the Parent/pMW118, mlaA::Tn/pMW118, or mlaA::Tn/pMW118-mlaA strains were ultracentrifuged and the precipitates were electrophoresed in a SDS polyacrylamide gel. The gel was stained by Coomassie brilliant blue. (B, C) The OMV fraction obtained in (A) was subjected to Western blot analysis using an anti-OmpA antibody (B) or an anti-LPS antibody (C). Lower graph indicate the relative band intensity compared with that in the parent strain. Data shown are means ± standard errors from three independent experiments. The asterisk represents a p value less than 0.05 (Student’s t test).
Knockout of pldA does not affect E. coli killing activity against silkworms
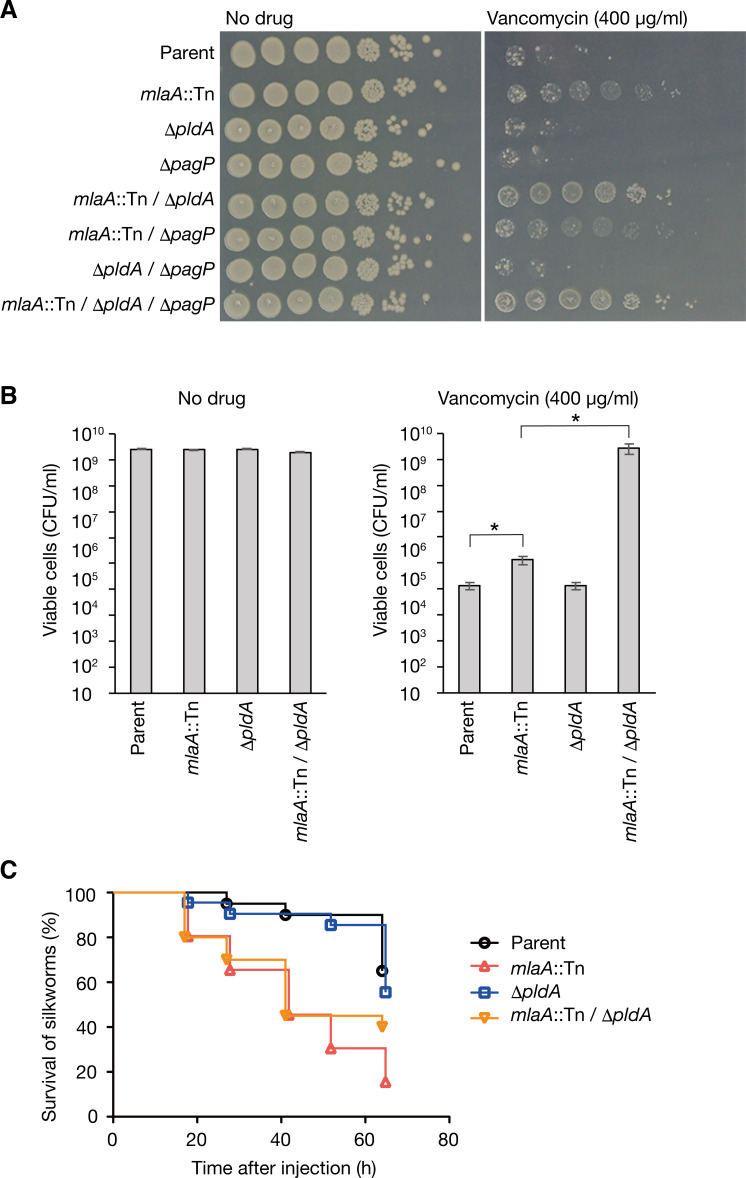
PldA and PagP degrade phospholipids of the outer membrane and contribute to maintain the lipid asymmetry of the outer membrane independently of the Mla system [3]. Double knockout of mlaA and pldA leads to a higher accumulation of phospholipids in the outer membrane than the respective single knockouts [25]. We examined whether double knockout of mlaA and pldA, or mlaA and pagP increases vancomycin resistance. The growth of single knockout mutants of pldA or pagP was indistinguishable from that of the parent strain in the presence of vancomycin (Fig 4A). Double knockout mutants of mlaA and pldA showed slightly better growth than the respective single knockout mutants in the presence of vancomycin (Fig 4A). Colony forming unit assay confirmed the different sensitivity to vancomycin between the mlaA-knockout mutant and the mlaA/pldA double knockout mutant (Fig 4B). The growth of double knockout mutants of mlaA and pagP was similar to that of the mlaA-knockout mutant in the presence of vancomycin (Fig 4A). These results suggest that knockouts of mlaA and pldA additively increase vancomycin resistance.
Fig 4. Knockout of pldA does not affect silkworm-killing activity.
(A) E. coli overnight culture of the Parent, mlaA::Tn, ΔpldA, ΔpagP, mlaA::Tn/ΔpldA, mlaA::Tn/ΔpagP, ΔpldA/ΔpagP, or mlaA::Tn/ΔpldA/ΔpagP strain was 10-fold serially diluted, spotted onto LB agar plates supplemented with or without vancomycin (400 μg/ml), and incubated at 37˚C. (B) E. coli overnight culture of the Parent, mlaA::Tn, ΔpldA, or mlaA::Tn/ΔpldA strain was 10-fold serially diluted, spread onto LB agar plates supplemented with or without vancomycin (400 μg/ml), and incubated at 37˚C. The appeared colonies were counted and CFU/ml was calculated. Data shown are means ± standard errors from three independent experiments. The asterisk represents a p value less than 0.05 (Student’s t test). (C) Silkworms were injected with bacterial cells (2 x 108 CFU) of the Parent, mlaA::Tn, ΔpldA, or mlaA::Tn/ΔpldA, and survival was monitored. The experiment was performed twice and the data were pooled (n = 20). Log-rank test p value was less than 0.05 between Parent and mlaA::Tn, or between ΔpldA and mlaA::Tn/ΔpldA.
Based on these observations, we examined the effect of pldA knockout on E. coli virulence against silkworms. The pldA single knockout mutant killed silkworms with a similar time course as the parent strain (Fig 4C). The mlaA/pldA double knockout mutant killed silkworms with a similar time course as the mlaA single knockout mutant (Fig 4C). Thus, the pldA knockout does not increase silkworm killing activity.
Discussion
The findings of the present study revealed that knockout of mlaA, which maintains the lipid asymmetry of the outer membrane leads to increased virulence of E. coli against silkworms. The mlaA-knockout mutant exhibited increased OMV production, resistance to vancomycin, and increased killing activity against silkworms. Thus, this study unveiled a novel function of mlaA in E. coli virulence properties.
Knockout of mlaA increases bacterial virulence not only in E. coli, but also in N. gonorrhoeae, V. cholerae, and P. aeruginosa [7,8,18]. In contrast, knockout of mlaA attenuates bacterial virulence in S. flexneri, H. influenzae, H. parasuis, and B. pseudomallei [6,14–16]. Thus, the effect of the mlaA knockout on bacterial virulence differs between bacterial species. A previous study reported that in N. gonorrhoeae, knockout of mlaA sensitizes bacteria to antimicrobial peptides (defensin and polymyxin), vancomycin, and ampicillin, but increases OMV production; the authors speculated that the increased OMV could be advantageous for bacteria to survive in the mouse lower genital tract [7]. In V. cholerae, knockout of mlaA increases OMV production and alters the lipid composition of the outer membrane, which could be beneficial for bacteria adaptation in host environment containing antimicrobial peptides and bile acids [18]. In P. aeruginosa, knockout of mlaA increases bacterial virulence via ZnuA, which functions in Zn incorporation [8]. The effect of the mlaA knockout on OMV production was not examined in P. aeruginosa. Thus, the increased OMV production in the mlaA-knockout mutant is conserved among E. coli, N. gonorrhoeae, and V. cholerae, and could underlie the increased virulence of the mlaA-knockout mutant. It should also be noted that the animal infection models were different among the studies of different bacterial species, and the host environment could be differently involved in the bacterial virulence phenotypes of these mlaA-knockout bacteria.
The mlaA-knockout mutant of E. coli exhibited sensitivity to many antimicrobial molecules, including levofloxacin, chloramphenicol, oxacillin, CTAB, and cholic acid, but was resistant to vancomycin (Fig 2), chlorhexidine [12], and arenicin-3 [13]. Because the conserved chemical structure or conserved target molecules between vancomycin, chlorhexidine, and arenicin-3 is not known, it is difficult to understand the mechanism by which the mlaA-knockout mutant exhibits resistance to these 3 antimicrobial molecules, while it exhibits sensitivity to other antimicrobial molecules. We speculate that there are 3 possible reasons, as follows: (i) Because OMV have the capacity to absorb various antimicrobial molecules, the increased OMV production in the mlaA-knockout mutant may contribute to the resistance to these antimicrobial molecules. (ii) The mlaA knockout increases the amount of phospholipids in the outer leaflet of the outer membrane, which could alter the permeability of antimicrobial molecules and affect bacterial sensitivity to antimicrobial molecules. (iii) MlaA may transport phospholipids bound to antimicrobial molecules from the outer leaflet of the outer membrane to the inner membrane. The knockout of mlaA may block the transport of antimicrobial molecules from the outer membrane to the inner membrane. These possibilities should be addressed in future studies.
By constructing multiple gene deletion mutants of pldA, pagP, and mlaA, which respectively maintains the lipid asymmetry of the outer membrane, we revealed that the mlaA/pldA double knockout mutant, compared with the mlaA-knockout mutant, had increased resistance to vancomycin. We also revealed that, compared with the mlaA-knockout mutant, the mlaA/pldA double knockout mutant did not have increased killing activity in silkworms. Thus, pldA has a role with mlaA in vancomycin resistance, but no role in the silkworm-killing activity. Our analysis suggests the mlaA is the main factor among pldA, pagP, and mlaA that affects the vancomycin resistance and virulence of E. coli.
MlaA is attracting attention as a drug target, because mlaA knockout increases the sensitivity of many bacteria to various antibiotics. The present study, however, demonstrated that the mlaA knockout confers E. coli resistance to vancomycin and exhibits high virulence in silkworms. Investigating the molecular mechanisms of how the mlaA knockout upregulates bacterial virulence will help to elucidate the biological significance of the Mla system, and contribute to the evaluation of MlaA as a drug target.
Materials and methods
Bacteria and culture condition
E. coli KP7600 strain and its gene-knockout mutants were cultured on LB agar medium and the bacterial colonies were aerobically cultured in LB liquid medium at 37˚C. E. coli strains transformed with pCP20 or pMW118 were cultured on LB agar plates containing 100 μg/ml ampicillin. The details of the bacterial strains and plasmids used in this study are provided in Table 2.
Table 2. List of bacterial strains and plasmids used.
| Strain or plasmid | Genotypes or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| KP7600 | W3110 type-A, F-, lacIq, lacZΔM15, λ-, galK2, galT22, IN(rrnD-rrnE)1 | NBRP |
| JD strains | Transposon library using mini-Tn10; Kanr | NBRP |
| JD23323 | KP7600 mlaA::mini-Tn10; Kanr | NBRP |
| JW3794 | BW25113 ΔpldA::kan; Kanr | NBRP |
| JW0617 | BW25113 ΔpagP::kan; Kanr | NBRP |
| JW2343 | BW25113ΔmlaA:: kan; Kanr | NBRP |
| N0001 | KP7600 ΔpldA:: kan; Kanr (transduced from Keio collection JW3794) | This study |
| N0002 | KP7600 ΔpagP:: kan; Kanr (transduced from Keio collection JW0617) | This study |
| N0003 | KP7600 mlaA::mini-Tn10; Kanr, ΔpldA::markerless | This study |
| N0004 | KP7600 mlaA::mini-Tn10; Kanr, ΔpagP::markerless | This study |
| N0005 | KP7600 mlaA::mini-Tn10; Kanr, ΔpldA::markerless, ΔpagP::markerless | This study |
| N0006 | KP7600 ΔmlaA:: kan; Kanr (transduced from Keio collection JW2343) | This study |
| N0006 | KP7600 mlaA::markerless | This study |
| Plasmids | ||
| pMW118 | A low copy plasmid, Ampr | Nippongene |
| pMW218 | A low copy plasmid, Kanr | Nippongene |
| pMW118-mlaA | pMW118 with mlaA, Ampr | This study |
| pMW218-mlaA | pMW218 with mlaA, Kanr | This study |
| pCP20 | FLP recombinase, Ampr | [26] |
Kan: Kanamycin, Cm: Chloramphenicol, Amp: Ampicillin.
Silkworm infection experiment
Third instar silkworms (Fu/Yo X Tsukuba/Ne) were purchased from Ehime Sansyu (Ehime, Japan). The silkworms were fed an artificial diet (Silkmate, Nosan, Japan) and maintained at 27˚C. Fifth instar silkworms were fed an antibiotic-free artificial diet (Sysmex) for 1 day and used for the infection experiment. E. coli overnight culture was centrifuged at 4050 g for 10 min, and the precipitated cells were suspended in 0.9% NaCl. Silkworms were injected with the bacterial solution using 1-ml syringes equipped with a 27-gauge needle via the intra-hemolymph route [27] and maintained at 37˚C. Silkworm survival was measured every ~12 h after the injection. The OD600 values of the bacterial solutions were measured before the injection to confirm that the number of bacteria did not differ between samples. The number of bacteria was determined by plating bacterial solution on LB agar plates.
Genetic manipulation
To construct gene-knockout mutants of mlaA, pldA, and pagP, transduction using a phage P1 vir was performed. First, the pldA knockout strain (N0001) or pagP knockout strain (N0002) were constructed by transduction from JW3794 or JW0617 to KP7600 strain. After the transduction step, the kanamycin resistance marker was removed by transformation with pCP20 expressing FLP recombinase, and pCP20 was removed by culturing the bacteria at 43˚C. Next, the pldA/mlaA double knockout strain (N0003) or pagP/mlaA double knockout strain (N0004) were constructed by transduction from JD23323 to the pldA or pagP markerless knockout strains. Third, the pldA/pagP double knockout strain was constructed by transduction from JW0617 to the pldA markerless knockout strain. The kanamycin resistance marker was removed from the pldA/pagP double knockout strain by transformation with pCP20, and the transduction was performed from JD23323 to the markerless pldA/pagP double knockout strain, resulting in the mlaA/pldA/pagP triple knockout strain. The gene knockouts in the mutant strains were confirmed by PCR. To construct a plasmid carrying the mlaA gene, the DNA fragment encoding the mlaA gene was amplified by PCR using primer pairs (forward, 5’-TCTTCTAGACCGCAGTCACGGTATTTTC-3’, reverse, 5’-GGTGGTACCTGGTTCCGATCATCAGGTT-3’) from genomic DNA of KP7600 as a template. The amplified DNA fragment was cloned into XbaI and KpnI sites of pMW118 or pMW218, resulting in pMW118-mlaA or pMW218-mlaA.
Evaluation of bacterial resistance to antimicrobial substances
To measure bacterial resistance to antibiotics and detergents, autoclaved LB agar medium was mixed with antibiotics or detergents and poured into square plastic dishes (Eiken Chemical, Tokyo, Japan). E. coli overnight cultures were serially diluted 10-fold in a 96-well microplate and 5 μl of the diluted bacterial solution was spotted onto the LB agar plates supplemented with drugs. The plates were incubated at 37˚C for 1 day and colonies were photographed using a digital camera.
Bacterial resistance to silkworm hemolymph was measured according to our previous method [24]. Briefly, fifth instar silkworms were injected with heat-killed E. coli KP7600 cells and the hemolymph was collected at 1 day after inoculation. The collected hemolymph was frozen in liquid nitrogen and stored at -80˚C. Overnight cultures (10 μl) of E. coli strains (Parent/pMW118, mlaA::Tn/pMW118, or mlaA::Tn/pMW118-mlaA) were inoculated into fresh LB medium (1 ml) and aerobically cultured at 37˚C. The silkworm hemolymph (27 μl) was added to the culture at 40 min after the inoculation and the OD600 was measured every 1 h.
Preparation of OMV
E. coli overnight culture (1 ml) was inoculated into 100 ml of LB medium in a flask and aerobically cultured at 37 ˚C for 24 h. The culture was centrifuged at 4450 g for 10 min, and the supernatant was filtered through a 0.22-μm polyvinylidene difluoride membrane (Millipore). The supernatant was centrifuged at 45,000 g for 3 h and the precipitate was dissolved with SDS sample buffer. The sample was electrophoresed in 15% SDS polyacrylamide gel and the gel was stained with Coomassie brilliant blue.
Western blot analysis
The OMV samples were electrophoresed in 15% SDS polyacrylamide gel, and blotted to a PVDF membrane (Immobilon-P, Millipore). The membrane was treated with TBST (20 mM Tris-HCl [pH7.6], 150 mM NaCl, 0.12% Tween20) containing 5% skim milk for 1 h. The membrane was treated with a TBST buffer containing 1:5000 anti-OmpA IgG (111120, Antibody research corp., MO, USA) or 1:10000 anti-LPS core (WN1 222–5, Hycult Biotech, Uden, The Netherlands) for 1 h at room temperature. After washing with TBST, the membrane was treated with TBST containing 1:5000 anti-rabbit IgG conjugated with horseradish peroxidase HRP or 1:5000 anti-mouse IgG conjugated with HRP for 1 h at room temperature. After washing with TBST, the membrane was treated with a HRP substrate (Western Lightning Plus-ECL, Perkin Elmer). The signal was visualized using ImageQuant LAS 4000 (Fujifilm, Tokyo, Japan). The band intensity was measured by Image J software [28].
Statistical analysis
Differences of the growth curves in the presence of silkworm hemolymph were assessed using the Student t test in Microsoft Excel for Mac (version 16.56). Statistical analyses of the survival curves of silkworms were performed using the log rank test with GraphPad PRISM software (version 5.0c).
Supporting information
(PDF)
Acknowledgments
We thank the National BioResource Project-E. coli (National Institute of Genetics, Japan) for providing the mini-Tn10 library and the Keio Collection.
Data Availability
All relevant data are within the article.
Funding Statement
This study was supported by JSPS Grants-in-Aid for Scientific Research (grants 19H03466 and 19K22523 [CK]), the Takeda Science Foundation (CK), the Ichiro Kanehara Foundation (CK), and the Ryobi Teien Memory Foundation (CK).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nikaido H. Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim Biophys Acta. 1976;433(1):118–32. Epub 1976/04/16. doi: 10.1016/0005-2736(76)90182-6 . [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. Epub 2003/12/11. doi: 10.1128/MMBR.67.4.593-656.2003 ; PubMed Central PMCID: PMC309051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lundstedt E, Kahne D, Ruiz N. Assembly and Maintenance of Lipids at the Bacterial Outer Membrane. Chem Rev. 2021;121(9):5098–123. Epub 2020/09/22. doi: 10.1021/acs.chemrev.0c00587 ; PubMed Central PMCID: PMC7981291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abellon-Ruiz J, Kaptan SS, Basle A, Claudi B, Bumann D, Kleinekathofer U, et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol. 2017;2(12):1616–23. Epub 2017/10/19. doi: 10.1038/s41564-017-0046-x . [DOI] [PubMed] [Google Scholar]
- 5.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A. 2009;106(19):8009–14. Epub 2009/04/23. doi: 10.1073/pnas.0903229106 ; PubMed Central PMCID: PMC2683108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Calvet A, Rodriguez-Arce I, Almagro G, Moleres J, Euba B, Caballero L, et al. Modulation of Haemophilus influenzae interaction with hydrophobic molecules by the VacJ/MlaA lipoprotein impacts strongly on its interplay with the airways. Sci Rep. 2018;8(1):6872. Epub 2018/05/04. doi: 10.1038/s41598-018-25232-y ; PubMed Central PMCID: PMC5932069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baarda BI, Zielke RA, Le Van A, Jerse AE, Sikora AE. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog. 2019;15(3):e1007385. Epub 2019/03/08. doi: 10.1371/journal.ppat.1007385 ; PubMed Central PMCID: PMC6424457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen L, Gao X, Wei J, Chen L, Zhao X, Li B, et al. PA2800 plays an important role in both antibiotic susceptibility and virulence in Pseudomonas aeruginosa. Curr Microbiol. 2012;65(5):601–9. Epub 2012/08/11. doi: 10.1007/s00284-012-0196-2 . [DOI] [PubMed] [Google Scholar]
- 9.Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, et al. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J Mol Med (Berl). 2017;95(10):1127–36. Epub 2017/08/28. doi: 10.1007/s00109-017-1579-4 ; PubMed Central PMCID: PMC5671890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernier SP, Son S, Surette MG. The Mla Pathway Plays an Essential Role in the Intrinsic Resistance of Burkholderia cepacia Complex Species to Antimicrobials and Host Innate Components. J Bacteriol. 2018;200(18). Epub 2018/07/11. doi: 10.1128/JB.00156-18 ; PubMed Central PMCID: PMC6112004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife. 2019;8. Epub 2019/01/15. doi: 10.7554/eLife.40171 ; PubMed Central PMCID: PMC6365058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregorchuk BSJ, Reimer SL, Green KAC, Cartwright NH, Beniac DR, Hiebert SL, et al. Phenotypic and Multi-Omics Characterization of Escherichia coli K-12 Adapted to Chlorhexidine Identifies the Role of MlaA and Other Cell Envelope Alterations Regulated by Stress Inducible Pathways in CHX Resistance. Front Mol Biosci. 2021;8:659058. Epub 2021/06/08. doi: 10.3389/fmolb.2021.659058 ; PubMed Central PMCID: PMC8170033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott AG, Huang JX, Neve S, Zuegg J, Edwards IA, Cain AK, et al. An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria. Nat Commun. 2020;11(1):3184. Epub 2020/06/25. doi: 10.1038/s41467-020-16950-x ; PubMed Central PMCID: PMC7311426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, et al. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun. 2014;82(2):660–9. Epub 2014/01/31. doi: 10.1128/IAI.01057-13 ; PubMed Central PMCID: PMC3911398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol. 1994;11(1):31–41. Epub 1994/01/01. doi: 10.1111/j.1365-2958.1994.tb00287.x [DOI] [PubMed] [Google Scholar]
- 16.Zhao L, Gao X, Liu C, Lv X, Jiang N, Zheng S. Deletion of the vacJ gene affects the biology and virulence in Haemophilus parasuis serovar 5. Gene. 2017;603:42–53. Epub 2016/12/19. doi: 10.1016/j.gene.2016.12.009 . [DOI] [PubMed] [Google Scholar]
- 17.Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, et al. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun. 2007;75(3):1186–95. Epub 2006/12/26. doi: 10.1128/IAI.01240-06 ; PubMed Central PMCID: PMC1828585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zingl FG, Kohl P, Cakar F, Leitner DR, Mitterer F, Bonnington KE, et al. Outer Membrane Vesiculation Facilitates Surface Exchange and In Vivo Adaptation of Vibrio cholerae. Cell Host Microbe. 2020;27(2):225–37 e8. Epub 2020/01/07. doi: 10.1016/j.chom.2019.12.002 ; PubMed Central PMCID: PMC7155939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powers MJ, Trent MS. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci U S A. 2018;115(36):E8518–E27. Epub 2018/08/09. doi: 10.1073/pnas.1806714115 ; PubMed Central PMCID: PMC6130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun. 2016;7:10515. Epub 2016/01/26. doi: 10.1038/ncomms10515 ; PubMed Central PMCID: PMC4737802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. Epub 2011/12/03. doi: 10.1186/1471-2180-11-258 ; PubMed Central PMCID: PMC3248377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaito C, Yoshikai H, Wakamatsu A, Miyashita A, Matsumoto Y, Fujiyuki T, et al. Non-pathogenic Escherichia coli acquires virulence by mutating a growth-essential LPS transporter. PLoS Pathog. 2020;16(4):e1008469. Epub 2020/04/24. doi: 10.1371/journal.ppat.1008469 ; PubMed Central PMCID: PMC7179839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami K, Nasu H, Fujiwara T, Takatsu N, Yoshida N, Furuta K, et al. The Absence of Osmoregulated Periplasmic Glucan Confers Antimicrobial Resistance and Increases Virulence in Escherichia coli. J Bacteriol. 2021;203(12):e0051520. Epub 2021/04/14. doi: 10.1128/JB.00515-20 ; PubMed Central PMCID: PMC8316038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashita A, Kizaki H, Kawasaki K, Sekimizu K, Kaito C. Primed immune responses to gram-negative peptidoglycans confer infection resistance in silkworms. J Biol Chem. 2014;289(20):14412–21. Epub 2014/04/08. doi: 10.1074/jbc.M113.525139 ; PubMed Central PMCID: PMC4022907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong ZS, Woo WF, Chng SS. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol. 2015;98(6):1133–46. Epub 2015/09/01. doi: 10.1111/mmi.13202 . [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. Epub 2000/06/01. doi: 10.1073/pnas.120163297 ; PubMed Central PMCID: PMC18686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaito C, Murakami K, Imai L, Furuta K. Animal infection models using non-mammals. Microbiol Immunol. 2020;64(9):585–92. Epub 2020/08/07. doi: 10.1111/1348-0421.12834 ; PubMed Central PMCID: PMC7590188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. Epub 2012/08/30. doi: 10.1038/nmeth.2089 ; PubMed Central PMCID: PMC5554542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the article.