Abstract
pCD4, a small, highly stable theta-replicating lactococcal plasmid, was used to develop a food-grade cloning system. Sequence analysis revealed five open reading frames and two putative cis-acting regions. None appears to code for undesirable phenotypes with regard to food applications. Functional analysis of the replication module showed that only the cis-acting ori region and the repB gene coding for the replication initiator protein were needed for the stable replication and maintenance of pCD4 derivatives in Lactococcus lactis. A two-component food-grade cloning system was derived from the pCD4 replicon. The vector pVEC1, which carries the functional pCD4 replicon, is entirely made up of L. lactis DNA and has no selection marker. The companion pCOM1 is a repB-deficient pCD4 derivative that carries an erythromycin resistance gene as a dominant selection marker. The pCOM1 construct can only replicate in L. lactis if trans complemented by the RepB initiator provided by pVEC1. Since only the cotransformants that carry both pVEC1 and pCOM1 can survive on plates containing erythromycin, pCOM1 can be used transiently to select cells that have acquired pVEC1. Due to the intrinsic incompatibility between these plasmids, pCOM1 can be readily cured from the cells grown on an antibiotic-free medium after the selection step. The system was used to introduce a phage resistance mechanism into the laboratory strain MG1363 of L. lactis and two industrial strains. The introduction of the antiphage barrier did not alter the wild-type plasmid profile of the industrial strains. The phenotype was stable after 100 generations and conferred an effective resistance phenotype against phages of the 936 and c2 species.
Lactococcus lactis is a gram-positive bacterium used to manufacture a variety of fermented dairy products. Over the past 15 years, considerable effort has been put into characterizing industrially important traits of lactococci (i.e., lactose utilization, proteolytic activity, citrate metabolism, exopolysaccharide production, bacteriocinogenic activity, phage resistance, etc.). Consequently, a variety of gene cassettes have been isolated and characterized, some of which can be used to improve the lactococcal cultures of traditional fermentations or to develop novel and innovative products.
Strain improvement can be accomplished using plasmid vectors to transfer and maintain specific traits in bacteria. The selection of appropriate transformants or transconjugants relies on one or more genes coding for selection markers on the vector. Traditionally, antibiotic resistance genes have been used as versatile and efficient dominant selection markers in laboratory research vectors. On legal and ethical grounds, however, transferable genes that confer resistance to substances used in human drug therapy are unacceptable in food applications.
Consequently, alternate selection markers must be used and indeed have been proposed to construct food-grade vectors for L. lactis. Depending on the type of selection, they can be classified into two groups as either dominant or complementation markers (7). Dominant markers do not rely on host-expressed genes and can be used in most wild-type strains of L. lactis. For instance, lactococcal genes that confer immunity to nisin have been used for dominant selection (14, 15, 25, 35, 56). Since nisin is considered a food-grade molecule, it can be used throughout the fermentation processes to promote plasmid maintenance, as long as this bacteriocin is compatible with specific applications. Resistance to heavy metals (cadmium, copper, tin [33–35]) represents a second type of dominant marker. Although useful in the selection step, heavy metals are toxic and obviously cannot be used in food fermentation to maintain plasmids. However, maintenance of the selection pressure throughout the fermentation process may not be necessary, as the vector using cadmium resistance replicates through a theta mechanism (34), which is expected to be stable (30). Unfortunately in most of these dominant markers, the phenotypes used for the selection process occur naturally in numerous lactococcal strains, which consequently limit their host spectrum.
Complementation markers are based on mutations in the host chromosome that can be complemented by plasmid-expressed markers. One clear advantage of complementation systems is that the selection pressure, with carefully chosen genes, can be applied during industrial fermentation to ensure plasmid maintenance. For example, food-grade complementation marker systems have been based on the lacF gene coding for the essential enzyme IIA of the lactose phosphotransferase system of L. lactis (8, 37, 44). Lactose-deficient hosts with mutations or deletions of the lacF gene can be complemented by a plasmid that carries the wild-type enzyme IIA gene. The plasmid can be kept stable in medium having lactose as the main source of carbon (7). Furthermore, two auxotrophic complementation markers are based on altered tRNA genes that suppress nonsense mutations in genes involved in the biosynthetic pathway of purines and pyrimidines. In the first system, an ochre suppressor (supB) complements purine auxotrophic mutants of L. lactis grown in purine-free medium (9). However, supB suppresses both amber and ochre codons (82% of Lactococcus genes), which therefore causes major pleiotropic effects that alter bacterial growth. Alternatively, supD was used to complement pyrimidine auxotrophic hosts constructed by introducing an amber codon into the chromosomal pyrF gene (52). Since supD only suppresses amber codons (10% of Lactococcus genes), pleiotropic effects are significantly reduced. An auxotrophic supD-carrying mutant derived from an industrial strain showed an acidification rate in milk that is comparable to that of the parental strain. The major disadvantage of such complementation systems is that specific mutations must first be introduced, in a food-grade manner, into every recipient host before the complementing plasmids can be applied.
In this study, we present a third and novel strategy for a food-grade cloning system that has the advantages of both the dominant and the complementation strategies. The design consists of two plasmids that enable the dissociation of the dominant antibiotic marker from the vector plasmid. The vector pVEC1 consists exclusively of L. lactis DNA and complements the companion plasmid pCOM1, which bears the dominant marker. Both plasmids are required for the gene transfer step to L. lactis, but only the vector pVEC1 remains in the transformed cells afterward, resulting in a food-grade microorganism. The effectiveness of the design is ascertained by the stable transfer of a phage resistance mechanism to laboratory and industrial strains. This versatile and efficient vector system is potentially applicable to a large number of lactococcal hosts.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, and media.
The bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani medium (46) with shaking. L. lactis was grown at 30°C in M17 (54). For strain MG1363 and its derivatives and strain SMQ-481, M17 was supplemented with 0.5% glucose. All of the other lactococcal strains were grown in M17 supplemented with 0.5% lactose. Calcium chloride at a final concentration of 10 mM was added for the propagation of lactococcal bacteriophage. When appropriate, ampicillin was added at 50 μg/ml for E. coli. Chloramphenicol and erythromycin were used at a concentration of 5 μg/ml for L. lactis.
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Bacterial strain, phage, or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| L. lactis strains | ||
| GLP31 | MG1363(pSA3; pCD4); Emr | This work |
| GLP148 | MG1363(pCD4) | This work |
| GLP265 | MG1363(pVEC1; pCOM1), Abi+, Emr | This work |
| GLP267 | MG1363(pVEC1), Abi+ | This work |
| MG1363 | Plasmid-free derivative of NCDO 712, host of phages p2 and c21, Lac− | 17 |
| MJC15 | Multiple plasmids including pCD4; Lac+, Bac+ | 2 |
| SMQ-481 | Strain used as a source for recA gene internal fragment | J. Bouchardb |
| SMQ-562 | Industrial strain, host of phage Q58 | This work |
| SMQ-652 | Industrial strain, host of phage Q60 | This work |
| E. coli strain | ||
| DH5α | supE 44 Δlac U169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA 96 thi-1 relA1 | Life Technologies |
| Bacteriophages | ||
| p2 | Small-isometric-headed, 936 species, infects L. lactis MG1363 | 42 |
| c21 | Prolate-headed, c2 species, infects L. lactis MG1363 | 11 |
| Q58 | Small-isometric-headed, 936 species, infects L. lactis SMQ-562 | This work |
| Q60 | Small-isometric-headed, 936 species, infects L. lactis SMQ-652 | This work |
| Plasmids | ||
| pCD4 | Theta-replicating cryptic plasmid from L. lactis MJC15, 6.1 kb | This work |
| pGK12 | 4.4-kb vector, source of chloramphenicol resistance gene, Cmr Emr | 31 |
| pIL252 | 4.6-kb vector, source of erythromycin resistance gene, Cmr Emr | 51 |
| pNC1 | Replicon screening vector, cat gene (0.9 kb) from pGK12 cloned into unique AatII site of pUC18, Apr Cmr | 3 |
| pNC1e | Replicon screening vector, ery gene (1.3 kb) from pIL252 cloned into unique AatII site of pUC18, Apr Emr | This work |
| pMIG3 | 5.5-kb shuttle vector, Cmr | 57 |
| pRL1 | 6.1-kb HindIII fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL2 | 3.4-kb EcoRI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL5 | 2.4-kb NspV/EcoRI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL6 | 3.0-kb NcoI/EcoRI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL7 | 2.2-kb EcoRI/NcoI fragment from pSRQ928 cloned into pVEC1, Apr Cmr, Abi+ | This work |
| pRL8 | 302-kb PCR product filled in with Klenow and cloned into the SmaI site of pRL2, Apr Cmr | This work |
| pRL10 | 2.1-kb Sau3A1/EcoRI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL11 | 1.8-kb ClaI/EcoRI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL12 | 1.8-kb HhaI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL20 | pRL5 cut with ClaI and self ligated, resulting in deletion of two tandem repeats of the iteron, Apr Cmr | This work |
| pRL30 | 3.5-kb BglII fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL32 | 2.1-kb NspV/AseI fragment from pCD4 cloned into pNC1, Apr Cmr | This work |
| pRL32e | 2.1-kb NspV/AseI fragment from pCD4 cloned into pNC1e, Apr Emr | This work |
| pRL33 | 0.7-kb EcoRI/BspMI fragment from pCD4 cloned into pNC1, ori, Apr Cmr | This work |
| pRL33e/pCOM1 | 0.7-kb NspV/BspMI fragment from pCD4 cloned into pNC1, ori, Apr Emr | This work |
| pSA3 | Shuttle vector, Cmr Tcr Emr, 10.2 kb | 4 |
| pSRQ900 | 10.8-kb wild-type plasmid from L. lactis W37, Abi+ | 11 |
| pSRQ928 | 2.2-kb EcoRI/HindIII fragment from pSRQ925 cloned into pNZ123; Abi+, Cmr | 11 |
| pUC18 | Cloning vector, Apr | 60 |
| pVEC1 | Self-ligated 3.4-kb EcoRI fragment from pCD4; food-grade vector for L. lactis | This work |
Abi+, phage resistance mechanism; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Cms, sensitivity to chloramphenicol; Emr, erythromycin resistance; Tcr, tetracycline resistance; Lac, lactose-fermenting ability; Bac+, bacteriocin production.
Department of Biochemistry and Microbiology, Université Laval, Québec, Canada.
Bacteriophage propagation and microbial assays.
Phage propagation, titration, and the assay for efficiency of plaquing (EOP) have been described elsewhere (26, 41, 47). One-step growth curves and centers of infection (COI) were assayed as described previously by Moineau et al. (40). The phage burst size was estimated as the ratio between the phage titer at two consecutive latent phases on the growth curves. The efficiency at which COI formed (ECOI) was calculated by dividing the number of COI on resistant strains by the number of COI on wild-type strains. The data presented for each assay represent the average of at least three independent experiments.
DNA isolation, manipulation, and sequencing.
Large-scale plasmid DNA purification from E. coli was carried out using the Qiagen kit (Chatsworth, Calif.), and the silica method (J. D. Brown, http://research.bmn.com/tto [T01281]) was used for small-scale preparations. To obtain L. lactis plasmid DNA, the silica method was modified as follows: 1.5 ml of an overnight culture of L. lactis was harvested and suspended in 200 μl of 25% sucrose containing 30 mg of lysozyme/ml. After incubation at 37°C for 15 min, 400 μl of lysis solution (3% sodium dodecyl sulfate [SDS], 0.2 N NaOH) was added, mixed, and incubated at room temperature for 5 min. Three hundred microliters of 3 M CH3COOK (pH 4.8) was added, mixed by inversion, and incubated on ice for 10 min. The samples were centrifuged for 10 min, and the aqueous phase was transferred to a new tube, avoiding white particles. Two hundred microliters of silica suspension was added and incubated for 5 min at room temperature. After a brief centrifugation, the silica was harvested and the aqueous phase was discarded. The pellet was suspended in an ethanol wash solution (50% ethanol, 0.1 M NaCl, 1 mM EDTA, 10 mM Tris-HCl [pH 7.5]) and centrifuged, and the solution was discarded. The washing step was repeated, and the silica pellet was air dried for 5 min. The pellet was suspended in 50 μl of 10 mM Tris-HCl, pH 8.0, incubated at 55°C for 5 min, and briefly centrifuged. The aqueous phase containing DNA was collected and transferred to a new tube.
For determination of the relative copy number of pCD4 derivatives (see below), the method of O'Sullivan and Klaenhammer (43) was used to ensure quantitative recovery of plasmid DNA. Total DNA from L. lactis was isolated by the method of Hill et al. (22), omitting the phage infection step. Restriction and modifying enzymes were used according to the supplier's instructions (Roche Diagnostics, Laval, Québec, Canada). Preparation and electroporation of E. coli and L. lactis have been described elsewhere (11, 23). DNA manipulations were essentially carried out as described previously by Sambrook et al. (46). Details on DNA constructions are presented in Table 1. DNA was sequenced on both strands by the dideoxy chain termination method (Nucleic Acid Analysis and Synthesis Units of the Life and Health Science Pavilion, Université Laval). The reactions were performed with the Dye Deoxy terminator Taq sequencing kit, and products were separated on the model 373A automated DNA sequencing system (Applied Biosystems, Foster City, Calif.). Sequence reactions began at previously known regions and continued by primer walking. DNA sequences were analyzed with the GCG package (version 10.1) and were compared to the databases using basic local alignment algorithms (6).
Southern transfer and hybridization.
Purified DNA was digested with restriction enzymes and separated by electrophoresis on 0.8% agarose gels. DNA fragments were stained with ethidium bromide, photographed under UV illumination, and then transferred onto positively charged nylon membranes (Roche Diagnostics) by capillary blotting (53). Probes were prepared by labeling DNA fragments with the DIG High-Prime labeling kit (Roche Diagnostics). Prehybridization, hybridization, and posthybridization washes as well as detection were performed as suggested in the DIG System User's Guide for Filter Hybridization (Roche Diagnostics). The DIG Easy Hyb buffer and CSPD were used for the hybridization steps and chemiluminescent detection, respectively.
Plasmid stability.
Plasmid segregational stability was determined as the fraction of a culture that maintained the tested plasmid after exponential growth for 100 generations without selective pressure (19). Cultures were assayed for plasmid maintenance at 0 and 100 generations by testing 100 randomly selected colonies for the plasmid-borne antibiotic resistance phenotype. The strains SMQ-562(pRL7) and SMQ-652(pRL7) were evaluated for plasmid stability by testing for phage resistance with the cross-streaking method described by Moineau et al. (41). The correlation between phenotype and plasmid content was confirmed by analyzing the plasmid profile of resistant and sensitive colonies. The percentage of plasmid loss per generation was calculated using the formula of Roberts et al. (45). The results are an average of three independent determinations.
Plasmid copy number.
Plasmid DNA was linearized by digestion with restriction enzymes, separated by electrophoresis on a 0.8% agarose gel, and photographed under UV illumination with the Gel Doc 1000 photodocumentation system (Bio-Rad, Mississauga, Ontario, Canada) under nonsaturating conditions. The intensity of each plasmid DNA band was estimated using Molecular Analyst image analysis software (Bio-Rad). The relative copy number of each plasmid was evaluated by comparing the intensity of each DNA band and correcting for plasmid size. Experiments were done in triplicate.
For the estimation of pCD4 copy number per chromosome equivalent, a comparison was made between the hybridization signals of single-copy chromosomal and pRL8-encoded recA gene fragments in total DNA extracts from cultures of L. lactis MG1363 harboring pRL8. Restriction enzymes were selected to linearize plasmid DNA and to allow a clear discrimination between chromosome- and plasmid-borne signals. Exposure time was also calibrated to obtain unsaturated autoradiograms. The films were photographed with the Gel Doc 1000 photodocumentation system, and densitometric comparison was made using the Molecular Analyst image analysis software (Bio-Rad). The plasmid copy number corresponds to the signal generated by the plasmid band divided by that of the chromosomal band.
Nucleotide sequence accession number.
The complete sequence of pCD4 has been submitted to GenBank (accession no. AF306799).
RESULTS
Selection and isolation of a plasmid for food-grade vector construction.
L. lactis subsp. lactis MJC15 was isolated from raw milk cheese by Cardinal et al. (2). It carries five plasmids which range from 2.8 to 70 kb in size. To identify the plasmids that most likely replicate through the theta mechanism, a probe specific for the conserved repB gene of lactococcal theta plasmids (19, 49) was used. The probe was prepared by labeling an 896-bp PCR product obtained from the lactococcal theta plasmid pSRQ900 (accession no. AF001314) with primers SM3 (5′-CCTTTTTACCGTAGGTAGG) and SM11 (5′-GTCGTTTCAAAGAAGCGGTT). Four of the plasmids of strain MJC15 (pCD1, pCD2, pCD3, pCD4) hybridized with the probe. The fifth plasmid (pCD5; 2.8 kb) hybridized with a 1,251-bp RsaI fragment of pMG36ct corresponding to the leading-strand initiation and control region of pWV01, a lactococcal plasmid from the pMV158/pE194 family (28). This suggests that pCD5 replicates by the rolling circle mechanism. Two plasmids, pCD4 and pCD5, were found to be highly stable, as neither one could be cured from the host. Plasmid pCD4 was selected for food-grade vector construction for the following reasons. Firstly, vectors derived from theta replicons have shown higher intrinsic structural stability. Secondly, host range is limited, and thirdly, they are most often mutually compatible, which means that they can coexist in relatively high numbers in a single host (29, 30, 36).
Plasmid pCD4 was transferred by cotransformation with the marker plasmid pSA3 into plasmid-free strain MG1363, as previously described (12). Plasmid pSA3 was subsequently cured by subculturing in the absence of selective pressure. One derivative carrying only pCD4 was then selected and named L. lactis GLP148.
Sequence analysis of pCD4.
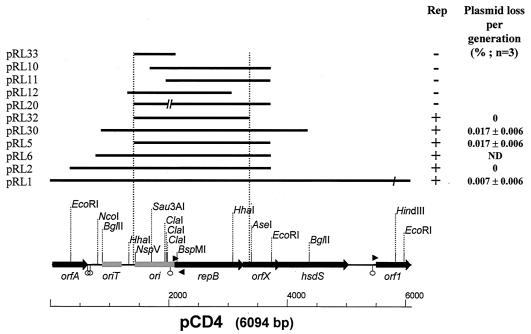
The plasmid pCD4 contained 6,094 bp and had a G+C content of 33.5% (Fig. 1). Five open reading frames (ORFs) and two cis-acting regions were identified in the sequence of pCD4 (Table 2). The first ORF (orfA) encodes a putative protein of 196 amino acids that bears strong similarities with hypothetical proteins found on a number of lactococcal plasmids. In a recent study (1a), bioinformatic evidence was presented suggesting that these proteins could be integrases belonging to a family of tyrosine recombinases that rearrange DNA duplexes by site-specific recombination. Two putative rho-independent terminators were identified within the first 80 bp downstream of orfA (Fig. 1).
FIG. 1.
Functional analysis of the replication region of the lactococcal plasmid pCD4. A linear restriction map is shown at the bottom of the diagram. Putative ORFs (black arrows), cis-acting regions (light grey boxes), promoters (arrowheads), and terminators (stem-loops) shown on the restriction map were inferred from sequence analysis. Thick lines above the linear map represent the fragments cloned into pNC1. The names of recombinant plasmids containing these inserts are indicated on the left. The single break in pRL1 corresponds to the HindIII site used for the cloning of the complete pCD4, and the double breaks in pRL20 correspond to the deletion of ClaI fragments. Only those restriction sites used to generate pCD4 derivatives are indicated on the map. The replication phenotype (Rep) and the calculation of the stability of pCD4 derivatives in strain MG1363 (average percent plasmid loss per generation ± standard deviation) are indicated on the right. The vertical lines delimit the core replicon. ND, not determined.
TABLE 2.
General features of the theta-replicating lactococcal plasmid pCD4
| Feature | Start position (codon) | End position (stop codon) | Length (amino acids) | pI | Mol wt | RBS (AGAAAGGAGGT)a | Similarity, motif, proposed function | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| orfA | 1 (AUG) | 591 (UAA) | 196 | 10.39 | 22.5 | AAAGGAGG | DNA recombinase (tyrosine recombinase family) | |
| oriT | 844 | 1179 | Transfer origin, homologous to oriT1 and oriT2 of pNZ4000; oriT of pCI528, pSRQ800, and pSRQ900 | 36, 55 | ||||
| ori | 1806 | 2074 | Origin of replication of L. lactis pCD4 | 19, 49 | ||||
| repB | 2075 (AUG) | 3235 (UGA) | 386 | 10.04 | 45.3 | AAAGGAG | Replication initiator protein of pCD4 | 19, 49 |
| orfX | 3232 (AUG) | 3861 (UAG) | 209 | 8.34 | 24.8 | AAAGG | DNA binding protein; unknown function | 49 |
| hsdS | 3837 (AUG) | 5021 (UGA) | 394 | 9.39 | 45.8 | AGAAA | S subunit of type IC R/M system | 48 |
| orf1 | 5498 (AUG) | 6079 (UAA) | 193 | 10.81 | 22.2 | AAAGGA | Similar to orf1 of pIL2614; motif for short chain alcohol dehydrogenase; unknown function | 27 |
See reference 36a.
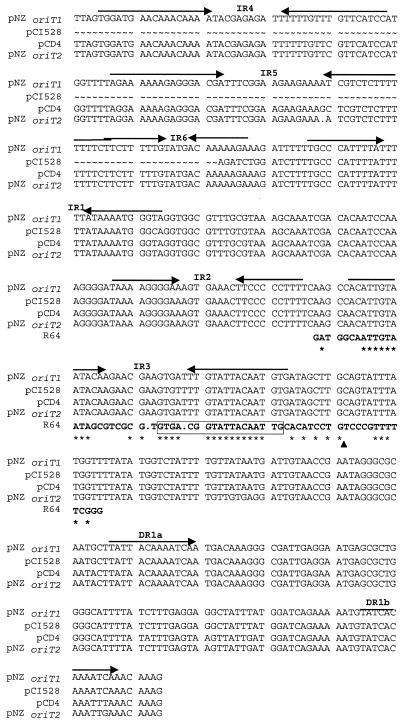
DNA sequences similar (>90% identical) to the functional transfer origin of the lactococcal plasmids pCI528 and pNZ4000 were identified downstream of orfA. This region includes three inverted repeats and one direct repeat (IR1, IR2, IR3, DR1) (Fig. 2). As reported for the two transfer origins of pNZ4000 (1998), oriT of pCD4 contains a region that shares significant sequence identity with the core region of the transfer origin of the E. coli plasmid R64 (Fig. 2). This core region covers the R64 repeat A, which contains the binding site for the R64 NikA protein, and the nick sequence (16). Sequence similarity with pNZ4000 extends further upstream of the functional transfer origin defined for pCI528 (36), with three extra conserved inverted repeats (IR4, IR5, IR6) (Fig. 2).
FIG. 2.
Sequence alignment of the oriT locus of lactococcal plasmids. Inverted and direct repeats are indicated above the sequence. IR1, IR2, and IR3 correspond to the inverted repeats that were identified on the functional transfer origins of pCI528 and pNZ4000 (36, 55). Inverted repeats IR4, IR5, and IR6 are present on the functional transfer origin of oriT1 and oriT2 (IR5 and IR6) defined on pNZ4000. The bold letters represent the core region of the R64 oriT locus. The NikA-binding site is boxed, and the nick site is indicated by the arrowhead.
A typical replication module of plasmids belonging to the lactococcal theta family was identified downstream of oriT (19, 49). It includes a presumptive ori region and a gene coding for a replication initiator protein (Table 2; Fig. 1). The cis-acting ori region includes a 41-bp AT-rich box flanked by short GC-rich clusters (Fig. 3A). The AT-rich box encompasses a conserved 10-bp direct repeat and is located upstream of an iteron consisting of a 22-bp sequence repeated three and one-half times. In addition, two sets of perfect inverted repeats, a putative promoter with both −35 and −10 consensus sequences, and an appropriate ribosome binding site (RBS) located 9 bp upstream of the repB start codon were identified. The first inverted repeat overlaps the iteron and the −35 box of the repB promoter, while the second is located between the −10 box of the repB promoter and the RBS. As described for pND324 (10), a putative promoter with an extended −10 box, located on the lagging strand within the RepB coding sequence, could direct the expression of a counter-transcript RNA of approximately 80 nucleotides, ending at the most proximal inverted repeat of the repB promoter region (Fig. 3A). The antisense RNA could pair with the leader sequence of repB mRNA, thus causing either premature termination of repB transcription or an inhibition of the repB mRNA translation (5).
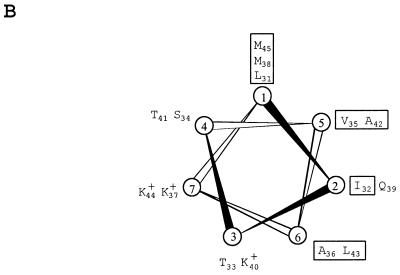
FIG. 3.
Sequence analysis of the minimal replicon of pCD4. (A) The restriction sites used to generate pCD4 derivatives are highlighted on the DNA sequence. The −35 and −10 boxes of repB promoter and RBSs are underlined. The reverse-complement sequence of the −35 and −10 boxes of the counter-transcript RNA promoter is double underlined. The AT-rich region is boxed, and the GC clusters are indicated with asterisks. Direct repeats within the AT-rich stretch are represented by double arrows, and the 22-bp iterated sequences repeated three and one-half times (iteron) are indicated by thick arrows above the sequence. Inverted repeats are represented by dashed arrows over the sequence. Protein translation of repB and of the truncated orfX is given below the DNA sequence. Amino acids in reverse video in the RepB sequence correspond to consensual residues with a plurality of at least 30 from an alignment of the amino acid sequence of 35 replication initiator proteins of lactococcal plasmids. The sequences used in the alignment were taken from GenBank. Amino acid motifs for the leucine zipper (positions 31 to 45), copy number control (129 to 144), and DNA binding (positions 215 to 241) are indicated with dashed underlines. The numbers on the left correspond to the nucleotide numbering of pCD4 as submitted to GenBank. The amino acid numbering of RepB is indicated on the right. (B) Helical wheel representation of the leucine motif of RepB. Hydrophobic residues are boxed, charged residues are indicated by a + sign, and the other residues correspond to uncharged polar residues.
The repB gene encodes a protein of 386 amino acids that shares a high level of sequence identity to initiator proteins (RepB) of lactococcal theta-replicating plasmids (Fig. 3A). As for many replication initiators, a motif was identified in the N-terminal region of RepB (amino acid positions 31 to 45) that can form an amphipathic alpha helix with a spine composed of periodically repeated leucine and methionine residues (Fig. 3B). This structure is in good agreement with the model for a leucine zipper (18, 32) and could be involved in protein dimerization (5). Another conserved domain (amino acid positions 129 to 144) (Fig. 3A) overlaps a region believed to be involved in copy number control (38, 59). Finally, a 27-amino-acid domain (positions 215 to 241) overlaps the region defined as the DNA binding site of several initiators (38). It also encompasses a 13-amino-acid stretch (positions 216 to 228) that has been proposed as the region governing ori-specific interactions for lactococcal theta plasmids (20). This suggestion is further supported by the poor amino acid conservation within the 13-amino-acid stretch observed for an alignment of 35 different lactococcal initiator proteins (Fig. 3A).
The theta replication modules of lactococcal plasmids are often associated with one or two additional ORFs that appear to be part of the same transcriptional unit as repB (48, 50). This is indeed the case for pCD4, where two genes (orfX and hsdS) were identified with proper RBSs, no promoter sequence, and overlapping coding sequences between repB, orfX, and hsdS (Table 2; Fig. 1). The orfX gene encodes a polypeptide of 209 amino acids containing a helix-turn-helix motif, which suggests that it could be a member of a family of DNA binding proteins. A role in plasmid copy number control and segregational stability has been shown for a homolog of OrfX coded by pUCL287, a Tetragenococcus halophilus plasmid (1). However, no such function has been demonstrated for other homologs in L. lactis plasmids (13, 19, 49). The last gene of this transcription unit encodes HsdS, the specificity subunit of a type I restriction/modification (R/M) system (58). Despite the presence of hsdS genes in several replication modules of lactococcal theta plasmids, no evidence has so far related this gene to plasmid replication. However, its presence on plasmids can expand the R/M specificity in lactococcal cells carrying a type I R/M system (48, 50).
The last gene identified on pCD4 codes for Orf1 (Table 2; Fig. 1), a polypeptide of 193 amino acids that contains a motif typical of proteins belonging to the family of short-chain dehydrogenase/reductase. This very large family of enzymes includes mainly NAD- or NADP-dependent oxidoreductases (27). A putative promoter with a stem-loop structure encompassing the −35 region of the promoter was identified upstream of orf1 (Fig. 1). Since this promoter is located just upstream of the orfA RBS, both genes could be transcriptionally coupled.
Functional analysis of the replicon of pCD4.
DNA fragments containing the functional replicon may be identified by the replication ability that they confer on L. lactis. Various fragments of pCD4 were cloned into the replicon screening vector pNC1 (Table 2) and were transferred to L. lactis MG1363 by electroporation. Only clones carrying the functional replicon of pCD4 produced chloramphenicol-resistant colonies (Fig. 1). The core replication module of pCD4 was localized to the 1,980-bp NspV/AseI fragment carried by the recombinant plasmid pRL32 (Fig. 3). This fragment includes the ori region and a complete copy of repB. Only the first 29 amino acids of OrfX are coded on this fragment, and the gene for HsdS is absent, showing that OrfX and HsdS are not essential for replication. All pCD4 derivatives tested (Fig. 1) proved to be very stable, as the loss rate was less than 0.02% per generation in L. lactis MG1363 over 100 generations without selection. Although these were found to be segregationally stable, it is still possible that partial or complete deletion of auxiliary factors could affect copy number (19). Measurement of the relative copy number of pCD4, pRL2, pRL5, pRL30, and pRL32 revealed no significant differences, further demonstrating that the absence of such auxiliary factors does not affect copy number.
The essential role of the iterated sequence in plasmid replication was shown by constructing pRL20, a deletion derivative of pRL5 that carries only one and one-half direct repeat sequences from the iteron (Fig. 1) and does not replicate in L. lactis. Another deletion derivative, pRL10, did not replicate in L. lactis MG1363 (Fig. 1), which is surprising since it lacks only a 273-bp NspV/Sau3A1 fragment located 127 bp upstream of the ori region defined for other theta lactococcal plasmids (49). To determine if the region comprised between NspV and Sau3A1 codes for essential cis- or trans-acting elements, trans complementation was tested by cotransforming pRL10 and pRL32e in L. lactis MG1363. Selection for pRL10 did not produce viable colonies, while cotransformants carrying pRL32 and pRL32e were repeatedly obtained at a relatively high frequency (>104/μg) when selecting for both plasmids (Table 3). These results show that the proposed cis-acting ori gene of pCD4 (Table 2) should be extended further upstream to include the NspV site at nucleotide 1399 (Fig. 3).
TABLE 3.
Transformation frequencies per microgram of DNA of pCD4 derivatives to L. lactis MG1363 in single and cotransformation assays
| Tested plasmida | Complementing plasmida
|
||
|---|---|---|---|
| None | pRL32e | pRL33e/pCOM1 | |
| pMIG3 | 9.5 × 105 | 4.6 × 104 | |
| pRL32 | 1.9 × 106 | 8.0 × 104 | |
| pRL10 | 0 | 0 | |
| pRL12 | 0 | 0 | |
| pRL33 | 0 | 8.3 × 104 | |
| pVEC1 | 6.1 × 104 | ||
| pRL7 | 5.0 × 104 | ||
A 1/1 molar ratio of plasmid DNA was used in all assays.
The ability of the cis-acting ori region to support plasmid replication when RepB is supplied in trans was investigated. Two repB-deficient derivatives (pRL12 and pRL33) (Fig. 1) harboring the complete ori locus and truncated repB gave no transformants when electroporated alone. However, chloramphenicol-resistant colonies were readily obtained when pRL33 was transformed along with pRL32e (Table 3). This indicates that RepB supplied by pRL32e can trans complement the ori region of pRL33. The frequency of cotransformation was comparable to that of the control experiment, where pairs of compatible (pMIG3 and pRL32e) and incompatible (pRL32 and pRL32e) plasmids were double selected with erythromycin and chloramphenicol. The rate of cotransformation was reduced by only 1 to 2 logs compared to the rate of lone transformation with repB-proficient plasmids (Table 3). The second repB-deficient plasmid, pRL12, did not produce cotransformants with pRL32e, even at a lower concentration of chloramphenicol (2.5 μg/ml). Plasmid pRL12 encodes the first 328 amino acids of RepB and harbors the domains for protein dimerization, copy number control, and DNA binding (Fig. 3). This truncated RepB possibly competes with the wild-type RepB for the DNA binding sites within the origin, thus interfering with the normal replication process.
Altogether, these results demonstrate that the replication module carried by pRL32 is fully functional in L. lactis and can be used to develop a stable food-grade vector.
Construction of the food-grade vector system.
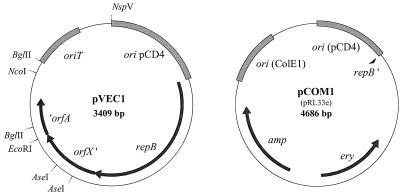
Our criteria for the construction of the two-component vector system were as follows: (i) the vector plasmid pVEC1 must be relatively small, stable without selective pressure, and compatible with wild-type lactococcal plasmids and must consist entirely of L. lactis DNA. (ii) The companion plasmid pCOM1 must contain a dominant selection marker gene, and its replication must be pVEC1 dependent although segregationally incompatible with the latter. The plasmid pRL33 has all the characteristics required for the companion plasmid pCOM1. For the convenience of cloning, the functions needed for propagation and selection in E. coli (ampicillin resistance gene, ColE1 origin of replication) were also included (Fig. 4). For pCOM1, chloramphenicol resistance was replaced by erythromycin (pRL33e) to avoid the potential repression of chloramphenicol gene expression by the replication initiator protein (13).
FIG. 4.
Circular maps of pVEC1 and pCOM1. Relevant features of the plasmids and restriction sites useful for cloning are indicated. Truncation in genes is indicated by apostrophes.
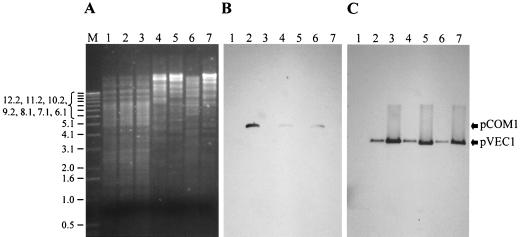
The plasmid pVEC1 consists of the 3.4-kb EcoRI fragment of pCD4, including the functional replicon of pCD4, the oriT region, and the truncated orfX and orfA genes. Although the minimal replicon is smaller, the loci for oriT, orfX, and orfA were retained for their useful cloning sites (Fig. 4), thus precluding the insertion of foreign or synthetic DNA. For the construction of pVEC1, the 3.4-kb EcoRI fragment of pCD4 was self ligated and cotransformed with pCOM1 in L. lactis MG1363, resulting in 103 erythromycin-resistant colonies. Plasmid analysis showed that all erythromycin-resistant isolates tested carried both pVEC1 and pCOM1. One isolate, GLP265, was selected and grown in GM17 for about 10 generations and was then scored for resistance to erythromycin. Over 90% of colonies reverted to an Erys phenotype (26% plasmid loss per generation), suggesting the loss of pCOM1, a loss that was later confirmed by plasmid analysis. One strain carrying only pVEC1 was selected and named GLP267. When the supercoiled vector DNA was used instead of the ligation mixture, the cotransformation frequency of pVEC1 and pCOM1 increased by almost 2 logs (Table 3). Retention of pVEC1 and loss of pCOM1 after the curing step was confirmed by Southern analysis of total DNA from GLP265 and GLP267 (Fig. 5). The loss of pCOM1 restored the copy number of pVEC1 in GLP267 (Fig. 5) to that of the other derivatives of pCD4 tested.
FIG. 5.
Detection by hybridization of pVEC1 and pCOM1 in total DNA extracts from variants of L. lactis MG1363 harboring different combinations of plasmids by hybridization. (A) Agarose gel before Southern transfer. (B) Autoradiogram obtained after hybridization with a probe specific for pCOM1 prepared by labeling pNC1. (C) Autoradiogram obtained after hybridization with a probe specific for pVEC1 obtained by labeling a repB internal fragment of pSRQ900. Lanes: M, 1-kb DNA mass ladder (Gibco/BRL Life Technologies, Burlington, Ontario, Canada); 1, MG1363 total DNA/EcoRI; 2, MG1363(pCOM1, pVEC1)/EcoRI; 3, MG1363(pVEC1)/EcoRI; 4, MG1363(pCOM1, pVEC1)/ClaI; 5, MG1363(pVEC1)/NspV; 6, MG1363(pCOM1, pVEC1)/NcoI/XbaI; 7, MG1363(pVEC1)/NcoI. The molecular size of marker bands is indicated on the left in kilobases, and hybridization signals corresponding to pCOM1 and pVEC1 are indicated on the right.
Determination of the copy number of pCD4.
An internal fragment of 302 bp of the single-copy lactococcal gene recA was used to estimate the copy number of pCD4 and its derivatives per chromosome equivalent. The recA fragment was obtained by PCR amplification using genomic DNA of L. lactis SMQ-481 with primers Rec1 (5′-GACCCAGAATATGCAAA AGCACTCGGTG) and Rec2 (5′-CCAACTTTTTCACGCAATTGGTTGATG) and cloning into pRL2, giving pRL8 (Table 1). Plasmid pRL8 was introduced into L. lactis MG1363, and total DNA was submitted to Southern analysis. The recA-specific probe hybridized with two DNA bands, one corresponding to the chromosomal copy and the other to the pRL8-expressed copy of recA (data not shown). Comparison of the intensities of these bands established that pCD4 is a low-copy-number plasmid with 2.3 ± 1.1 (standard deviation) copies per genome equivalent in L. lactis MG1363.
Application of the two-component cloning system for the transfer of phage abortive resistance to industrial strains.
The L. lactis W-37 plasmid pSRQ900 contains the gene coding for AbiQ, a phage resistance mechanism effective against lactococcal phages belonging to the 936 and c2 species. A 2.2-kb fragment coding for AbiQ (Table 1) was subcloned into pVEC1 as an EcoRI/NcoI fragment and was introduced into L. lactis MG1363 by cotransformation with pCOM1. An erythromycin-resistant isolate carrying the proper construction (pRL7) was selected, cured of pCOM1, and tested for phage resistance. L. lactis MG1363 harboring pRL7 conferred high resistance against two lactococcal phages belonging to the 936 and c2 species (Table 4). The potential of pVEC1 as a food-grade vector was further demonstrated by transferring pRL7 into two industrial strains. Southern analysis was carried out to verify the total loss of pCOM1 from the industrial strains carrying pRL7. Only pVEC1 remained in the cells after the curing step, and none of the plasmids was integrated into the host chromosome or in the wild-type plasmids. In addition, resident wild-type plasmids were unaffected by the introduction of pRL7 as shown by 100 generations of coexistence. The plasmid pRL7 is highly stable in these hosts, as no loss of the phage resistance phenotype was recorded after growth for 100 generations. The effectiveness of pRL7 in controlling phage infection was demonstrated by a reduction of the burst size and of the ECOI (Table 4). The phenotype conferred by pRL7 also resulted in the severe reduction of EOP.
TABLE 4.
Microbiological impacts of pRL7 on different bacteriophages
| Phage | Assay type
|
||
|---|---|---|---|
| EOP (n = 3) | Burst sizea (n = 3) | ECOI (%)a (n = 3) | |
| p2 | |||
| MG1363 | 1 | ||
| MG1363(pRL7) | <10−8 | ||
| c21 | |||
| MG1363 | 1 | ||
| MG1363(pRL7) | <10−8 | ||
| Q58 | |||
| SMQ-562 | 1 | 18 ± 5 | 100 |
| SMQ-562(pRL7) | <10−8 | 0.4 ± 0.1 | 18 ± 7 |
| Q60 | |||
| SMQ-652 | 1 | 29 ± 7 | 100 |
| SMQ-652(pRL7) | <10−8 | 0.7 ± 0.2 | 1.0 ± 0.6 |
Multiplicity of infection = 0.15.
DISCUSSION
A two-component food-grade cloning system was developed for L. lactis, where the uncoupling of the selection marker and the vector enabled the transient use of a non-food-grade but versatile dominant erythromycin resistance marker that is efficient in virtually all lactococcal hosts. The designed system included (i) a replication-deficient companion plasmid (pCOM1) coding erythromycin resistance and harboring a replication origin and (ii) an autonomously replicating vector (pVEC1) composed of only L. lactis DNA and the trans-acting initiator needed to support replication of pCOM1. Cloning is accomplished by first recombining pVEC1 DNA with the desired genes, followed by its coelectroporation with pCOM1 in L. lactis. Media containing erythromycin allow only the growth of cells that contain both plasmids. Curing of the marker plasmid after selection of cotransformants is easily accomplished in antibiotic-free medium. Plasmid pVEC1 is highly stable and can be maintained without selective pressure for many generations.
As stability is a major requirement for this cloning system, pVEC1 and pCOM1 were based on a theta-type replicon. Plasmids using this mode of replication are structurally and segregationally more stable, especially when carrying long DNA inserts (30). In addition, lactococcal vectors of the theta family were shown to have a narrow host range (24, 29, 36, 56), thus limiting transfer to other microorganisms. Stable coexistence of pRL7 with wild-type plasmids for at least 100 generations in two lactococcal hosts agrees with previous observations that multiple theta-type plasmids can coexist in a single lactococcal host, even if they carry highly related replicons (13, 20, 21, 49).
The segregational stability of plasmids relies on control elements that adjust the rate of replication to maintain a constant copy number (reviewed by del Solar et al. [5]). Control of the plasmid copy number is achieved by modulating the intracellular concentration of the initiator protein with negative regulatory circuits that may include antisense RNA, both antisense RNA and proteins, and sites for binding initiator protein. The fact that all replication-proficient derivatives of pCD4 were as stable as the parent plasmid indicates that all of the control elements were contained within the defined minimal replicon.
Plasmid copies that initiate a replication cycle are selected at random from a pool of identical plasmids within the cell (5). This has a major consequence for plasmids that share replication control functions, as they will compete for stable inheritance and are unable to coexist in a stable manner within a given cell in the absence of selective pressure. This incompatibility leads to the rapid segregation of plasmids within the host population. Deriving pVEC1 and pCOM1 from the same replicon ensures strong incompatibility between the plasmids and facilitates the loss of pCOM1. Incompatibility was also shown by the interference in the replication control of pVEC1 and the consequent reduction in copy number when accompanied by pCOM1 in L. lactis. Plasmid pCOM1 can be conveniently used to probe lactococcal hosts for incompatibility with pVEC1 because resident plasmids that support pCOM1 replication in trans would produce erythromycin-resistant colonies. In situations of incompatibility, a different set of two-component vector plasmids could be used. In a study of the lactococcal plasmid pJW563, complementation of a rep-deficient derivative with an incompatible plasmid led to rearrangement of the plasmids in about 50% of the isolates (19). Although a recA-negative derivative of MG1363 was not used in cotransformation assays, this phenomenon was not observed in this study.
The core replicon of pCD4 exceeds the boundaries generally defined for the minimal replicon of lactococcal theta-type plasmids (49). It was shown that the boundaries of the pSC101 minimal replicon are conditional to host and plasmid genes and to sites external to the core replicon (39). Further investigations should reveal binding of host factors needed for replication of pCD4 in L. lactis MG1363 in the region between the NspV and Sau3A1 sites of the pCD4 replicon.
Partial orfX and orfA as well as the oriT region were maintained in pVEC1 to provide convenient cloning sites. The presence of oriT in recombinant plasmids may result in increased transfer of the construction to other lactococci, if trans-acting conjugation factors are expressed by the host. Future generations of two-component vector systems will be constructed without the oriT locus, and a rho-independent terminator will be cloned downstream of repB.
Even though bacteriophages threaten the long-term use of lactococcal starter cultures in the food industry, the introduction of antiphage barriers in lactococcal strains constitutes a promising avenue for limiting their impact. We have shown the potential of the two-component cloning vector system for transferring AbiQ into industrial L. lactis strains and the stable maintenance of the phenotype under laboratory conditions. Field assays will have to be conducted to test the stability of the transferred characters under industrial processing conditions.
ACKNOWLEDGMENTS
We acknowledge A. Lucas, A. Pepin, and A. Cebron for technical assistance. We are also grateful to N. Corneau for kindly providing the replicon probe vector pNC1 prior to publication. We thank J. D. Bouchard, E. Lamiot, and R. St.-Laurent for critical reading of the manuscript and M. Parrot for helpful discussion.
We thank the Natural Sciences and Engineering Research Council of Canada (Research Partnership Program—Research Network on Lactic Acid Bacteria), Agriculture and Agri-Food Canada, Novalait Inc., Dairy Farmers of Canada, and Institut Rosell-Lallemand Inc. for financial support.
REFERENCES
- 1.Benachour A, Frère J, Flahaut S, Novel G, Auffray Y. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol Gen Genet. 1997;255:504–513. doi: 10.1007/s004380050523. [DOI] [PubMed] [Google Scholar]
- 1a.Boucher, I., E. Emond, M. Parrot, and S. Moineau. DNA sequence analysis of three Lactococcus lactis plasmids encoding phage resistance mechanisms. J. Dairy Sci., in press. [DOI] [PubMed]
- 2.Cardinal M-J, Meghrous J, Lacroix C, Simard R E. Isolation of Lactococcus lactis strains producing inhibitory activity against Listeria. Food Biotechnol. 1997;11:129–146. [Google Scholar]
- 3.Corneau N. M. Sc. thesis. Québec, Canada: Université Laval; 2000. [Google Scholar]
- 4.Dao M L, Ferretti J J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985;49:115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.del Solar G, Giraldo R, Ruiz-Echevarría M J, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos W M. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. Int Dairy J. 1999;9:3–10. [Google Scholar]
- 8.de Vos W M, Boerrigter I, van Roijen R J, Reiche B, Hengstenberg W. Characterization of lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1990;265:22554–22560. [PubMed] [Google Scholar]
- 9.Dickely F, Nillson D, Hansen E B, Johansen E. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol Microbiol. 1995;15:839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x. [DOI] [PubMed] [Google Scholar]
- 10.Duan K, Liu C-Q, Supple S, Dunn N W. Involvement of antisense RNA in replication control of the lactococcal plasmid pND324. FEMS Microbiol Lett. 1998;164:419–426. doi: 10.1111/j.1574-6968.1998.tb13118.x. [DOI] [PubMed] [Google Scholar]
- 11.Emond E, Dion E, Walker S A, Vedamuthu E R, Kondo J K, Moineau S. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl Environ Microbiol. 1998;64:4748–4756. doi: 10.1128/aem.64.12.4748-4756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emond E, Holler B J, Boucher I, Vandenbergh P A, Vedamuthu E R, Kondo J K, Moineau S. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl Environ Microbiol. 1997;63:1274–1283. doi: 10.1128/aem.63.4.1274-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frère J, Novel M, Novel G. Molecular analysis of the Lactococcus lactis subspecies lactis CNRZ270 bidirectional theta replicating lactose plasmid pUCL22. Mol Microbiol. 1993;10:1113–1124. doi: 10.1111/j.1365-2958.1993.tb00981.x. [DOI] [PubMed] [Google Scholar]
- 14.Froseth B R, Herman R E, McKay L L. Cloning of nisin resistance determinant and replication origin on 7.6-kilobase EcoRI fragment of pNP40 from Streptococcus lactis subsp. diacetylactis DRC3. Appl Environ Microbiol. 1988;54:2136–2139. doi: 10.1128/aem.54.8.2136-2139.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froseth B R, McKay L L. Development and application of pFM011 as a possible food-grade cloning vector. J Dairy Sci. 1991;74:1445–1453. [Google Scholar]
- 16.Furuya N, Komano T. Mutational analysis of the R64 oriT region: requirement for precise location of the NikA-binding sequence. J Bacteriol. 1997;179:7291–7297. doi: 10.1128/jb.179.23.7291-7297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasson M J. Plasmid complements of Streptococcus cremoris NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraldo R, Nieto C, Fernández-Teresguerres M E, Díaz R. Bacterial zipper. Nature. 1989;342:866. doi: 10.1038/342866a0. [DOI] [PubMed] [Google Scholar]
- 19.Gravesen A, Josephsen J, von Wright A, Vogensen F K. Characterization of the replicon from the lactococcal theta-replication plasmid pJW563. Plasmid. 1995;34:105–118. doi: 10.1006/plas.1995.9996. [DOI] [PubMed] [Google Scholar]
- 20.Gravesen A, von Wright A, Josephsen J, Vogensen F K. Replication of two pairs of incompatible lactococcal theta-replicating plasmids. Plasmid. 1997;38:115–127. doi: 10.1006/plas.1997.1302. [DOI] [PubMed] [Google Scholar]
- 21.Hayes F, Vos P, Fitzgerald G F, de Vos W M, Daly C. Molecular organization of the minimal replicon of novel narrow-host range, lactococcal plasmid pCI305. Plasmid. 1991;25:16–26. doi: 10.1016/0147-619x(91)90003-f. [DOI] [PubMed] [Google Scholar]
- 22.Hill C, Massey I J, Klaenhammer T R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991;57:283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horng J S, Polzin K M, McKay L L. Replication and temperature-sensitive maintenance functions of lactose plasmid pSK11L from Lactococcus lactis subsp. cremoris. J Bacteriol. 1991;173:7573–7581. doi: 10.1128/jb.173.23.7573-7581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes B F, McKay L L. Deriving phage-insensitive lactococci using a food-grade vector encoding phage and nisin resistance. J Dairy Sci. 1992;75:914–923. [Google Scholar]
- 26.Jarvis A W. Serological studies of a host range mutant of a lactic streptococcal bacteriophage. Appl Environ Microbiol. 1978;36:785–789. doi: 10.1128/aem.36.6.785-789.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jornvall, Persson H B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 28.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiewiet R, Bron S, Venema G, Seegers J F M L. Theta replication of the lactococcal plasmid pWV02. Mol Microbiol. 1993;10:319–327. [PubMed] [Google Scholar]
- 30.Kiewiet R, Kok J, Seegers J F M L, Venema G, Bron S. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl Environ Microbiol. 1993;59:358–364. doi: 10.1128/aem.59.2.358-364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kok J, van der Vossen J M, Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landschultz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 33.Leelawatcharamas V, Chia L G, Charoenchai P, Kunajakr N, Liu C-Q, Dunn N W. Plasmid-encoded copper resistance in Lactococcus lactis. Biotechnol Lett. 1997;19:639–643. [Google Scholar]
- 34.Liu C-Q, Khunajakr N, Chia L G, Deng Y-M, Charoenchai P, Dunn N W. Genetic analysis of regions involved in replication and cadmium resistance of the plasmid pND302 from Lactococcus lactis. Plasmid. 1997;38:79–90. doi: 10.1006/plas.1997.1301. [DOI] [PubMed] [Google Scholar]
- 35.Liu C-Q, Leelawatcharamas V, Harvey M L, Dunn N W. Cloning vectors for lactococci based on plasmid encoding resistance to cadmium. Curr Microbiol. 1996;33:35–39. doi: 10.1007/s002849900070. [DOI] [PubMed] [Google Scholar]
- 36.Lucey M, Daly C, Fitzgerald G. Identification and sequence analysis of the replication region of the phage resistance plasmid pCI528 from Lactococcus lactis subsp. cremoris UC503. FEMS Microbiol Lett. 1993;110:249–256. doi: 10.1111/j.1574-6968.1993.tb06330.x. [DOI] [PubMed] [Google Scholar]
- 36a.Ludwig W, Seewaldt E, Klipper-Balz R, Schleifer K H, Magnum L, Woese C R, Fox G E, Stackebrandt E. The phylogenetic position of Steptococcus and Enterococcus. J Gen Microbiol. 1985;131:543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 37.MacCormick C A, Griffin H G, Gasson M J. Construction of a food-grade host/vector system for Lactococcus lactis based on the lactose operon. FEMS Microbiol Lett. 1995;127:105–109. doi: 10.1111/j.1574-6968.1995.tb07457.x. [DOI] [PubMed] [Google Scholar]
- 38.Matsunaga F, Kawasaki Y, Ishiai M, Nishikawa K, Yura T, Wada C. DNA-binding domain of the RepE initiator protein of mini-F plasmid: involvement of the carboxyl-terminal region. J Bacteriol. 1995;177:1994–2001. doi: 10.1128/jb.177.8.1994-2001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller C A, Ingmer H, Cohen S N. Boundaries of pSC101 minimal replicon are conditional. J Bacteriol. 1995;177:4865–4871. doi: 10.1128/jb.177.17.4865-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moineau S, Durmaz E, Pandian S, Klaenhammer T R. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl Environ Microbiol. 1993;59:208–212. doi: 10.1128/aem.59.1.208-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moineau S, Fortier J, Ackermann H W, Pandian S. Characterization of lactococcal bacteriophages from Québec cheese plants. Can J Microbiol. 1992;38:875–882. [Google Scholar]
- 42.Moineau S, Walker S A, Vedamuthu E R, Vandenbergh P A. Cloning and sequencing of LlaDCHI restriction/modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae DpnII system. Appl Environ Microbiol. 1995;61:2193–2202. doi: 10.1128/aem.61.6.2193-2202.1995. . (Erratum, 61:3514.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Sullivan D J, Klaenhammer T R. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus lactis and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platteeuw C, van Alen-Boerrigter I, van Schalkwijk S, de Vos W M. Food-grade cloning and expression system for Lactococcus lactis. Appl Environ Microbiol. 1996;62:1008–1013. doi: 10.1128/aem.62.3.1008-1013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts R C, Burioni R, Helsinki D R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990;172:6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Sanders E M, Klaenhammer T R. Restriction/modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl Environ Microbiol. 1980;40:500–506. doi: 10.1128/aem.40.3.500-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schouler C, Gautier M, Ehrlich S D, Chopin M-C. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol Microbiol. 1998;28:169–178. doi: 10.1046/j.1365-2958.1998.00787.x. [DOI] [PubMed] [Google Scholar]
- 49.Seegers J F L M, Bron S, Franke C M, Venema G, Kiewiet R. The majority of lactococcal plasmids carry a highly related replicon. Microbiology. 1994;140:1291–1300. doi: 10.1099/00221287-140-6-1291. [DOI] [PubMed] [Google Scholar]
- 50.Seegers J F M, van Sinderen D, Fitzgerald G F. Molecular characterization of the lactococcal plasmid pCIS3: natural stacking of specificity subunits of a type I restriction/modification system in a single lactococcal strain. Microbiology. 2000;146:435–443. doi: 10.1099/00221287-146-2-435. [DOI] [PubMed] [Google Scholar]
- 51.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 52.Sørensen K I, Larsen R, Kibenich A, Junge M P, Johansen E. A food-grade cloning system for industrial strains of Lactococcus lactis. Appl Environ Microbiol. 2000;66:1253–1285. doi: 10.1128/aem.66.4.1253-1258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 54.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Kranenburg R, de Vos W M. Characterization of multiple regions involved in replication and mobilization of plasmid pNZ4000 coding for exopolysaccharide production in Lactococcus lactis. J Bacteriol. 1998;180:5285–5290. doi: 10.1128/jb.180.20.5285-5290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Wright A, Wessels S, Tynkkynen S, Saarela M. Isolation of a replicon region of a large lactococcal plasmid and use in cloning of a nisin resistance determinant. Appl Environ Microbiol. 1990;56:2029–2035. doi: 10.1128/aem.56.7.2029-2035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells J M, Wilson P W, LePage R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 58.Wilson G W. Restriction and modification systems. Annu Rev Genet. 1993;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 59.Xia G, Manen D, Yu Y, Caro L. In vivo and in vitro studies of a copy number mutation of the RepA replication protein of plasmid pSC101. J Bacteriol. 1993;175:4165–4175. doi: 10.1128/jb.175.13.4165-4175.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]