Abstract
Lignin is the main factor limiting the enzymatic conversion of lignocellulosic biomass into fermentable sugars. To reduce the recalcitrance engendered by the lignin polymer, the coumarin scopoletin was incorporated into the lignin polymer through the simultaneous expression of FERULOYL-CoA 6′-HYDROXYLASE 1 (F6′H1) and COUMARIN SYNTHASE (COSY) in lignifying cells in Arabidopsis. The transgenic lines overproduced scopoletin and incorporated it into the lignin polymer, without adversely affecting plant growth. About 3.3% of the lignin units in the transgenic lines were derived from scopoletin, thereby exceeding the levels of the traditional p-hydroxyphenyl units. Saccharification efficiency of alkali-pretreated scopoletin-overproducing lines was 40% higher than for wild type.
Biosynthesis of scopoletin in lignifying cells increases biomass processing efficiency.
INTRODUCTION
Lignin is an essential polymer in vascular plants that embeds cellulose and hemicelluloses in the secondary-thickened cell wall. Lignin provides strength to plant tissues, allows the transport of water through the vessels, and forms a physical barrier against pathogens (1–3). In dicots, lignin primarily forms by radical coupling of three monolignols—p-coumaryl, coniferyl, and sinapyl alcohols—and, depending on the plant species, from acetate, p-hydroxybenzoate, p-coumarate, and ferulate esters and the cinnamaldehyde precursors of these monolignols (4–9). Upon polymerization in the cell wall, these monolignol moieties give rise to the p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units in the lignin polymer, respectively (4, 6, 10).
The presence of lignin hinders the enzymatic conversion of lignocellulosic biomass into fermentable sugars (11–13). One strategy to overcome the recalcitrance of lignocellulosic biomass builds on the observation that plants can tolerate large shifts in their lignin compositions, often without adversely affecting plant growth and development (14–20). Moreover, plants have the remarkable property to allow the incorporation of novel building blocks into their lignin polymer (15, 21–25). In some cases, this results in a lignin polymer easier to extract or degrade, thereby improving the processing efficiency of plant biomass (26–29). To date, there are only a few examples of the successful use of alternative monomers in lignin engineering.
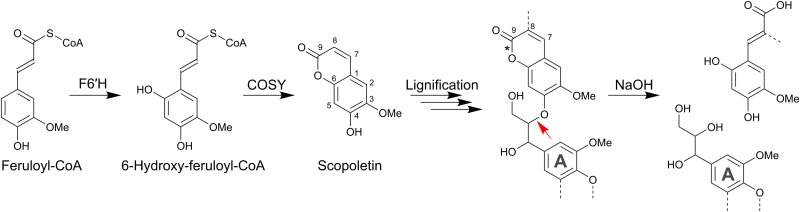
Here, we selected scopoletin (6-methoxy-7-hydroxycoumarin, which we number as a 3-methoxy-4-hydroxy aromatic to be in line with lignin nomenclature) as a candidate alternative monomer for lignin engineering (Fig. 1). We reasoned that incorporation of scopoletin via its O-4 position into the lignin polymer would result in β-aryl ethers with a conjugated carbonyl function (Fig. 1). This carbonyl functionality allows the delocalization of electrons over a larger part of the molecule and was previously proven to strongly reduce the hydrolysis temperature required to cleave aryl ethers in an alkaline environment (30–34). In addition, alkaline hydrolysis of scopoletin’s ester functionality results in the formation of a carboxylic acid and an additional free phenolic group. This hydrolysis would thus increase the hydrophilicity and the alkaline solubility of the pretreated polymer, benefiting the extraction of lignin in aqueous solvents (35, 36). Earlier studies already detected G(8-O-4)scopoletin structures in root exudates of Arabidopsis, demonstrating the coupling propensity of scopoletin with traditional monolignols (37). Building on this knowledge, we aimed to engineer Arabidopsis plants that incorporate scopoletin into their lignin polymer to improve the processing of biomass upon alkaline pretreatments.
Fig. 1. Biosynthesis and incorporation of scopoletin into the lignin polymer.
Feruloyl-CoA is biosynthesized via the phenylpropanoid pathway. F6′H1, FERULOYL-CoA 6′-HYDROXYLASE 1; COSY, COUMARIN SYNTHASE. It is hypothesized that, because of the presence of a conjugated carbonyl in the scopoletin monomer, the 8-O-4 bond between scopoletin and the A unit in the engineered lignin polymer becomes easier to cleave under alkaline conditions whether or not the 8-position is substituted. The red arrow indicates the bond with increased reactivity. Note that alkaline conditions will also hydrolyze the cyclic ester (lactone), thereby cleaving the bond indicated with an asterisk (*) and resulting in increased hydrophilicity and thus in a polymer that is easier to extract into aqueous solvents. Note that because scopoletin is shown as a lignin monomer, its numbering follows standard lignin conventions rather than formal coumarin numbering.
RESULTS AND DISCUSSION
Scopoletin is a natural metabolite in many plant species (38, 39). In Arabidopsis, scopoletin is biosynthesized from feruloyl-CoA, via FERULOYL-CoA 6′-HYDROXYLASE 1 (F6′H1) and the recently discovered COUMARIN SYNTHASE (COSY) (Fig. 1) (40, 41). In an attempt to engineer the scopoletin biosynthetic pathway into lignifying tissues of Arabidopsis, the coding sequences of F6′H1 and COSY were cloned as a bicistronic construct linked by a T2A sequence under transcriptional control of the promoter of the secondary cell wall–specific CELLULOSE SYNTHASE 4 (CesA4) gene (42, 43). Previously, we successfully used a similar strategy for the biosynthesis of curcumin in lignifying cells of Arabidopsis (27). The pCesA4:F6′H1_COSY (SCOP) construct was transferred into Arabidopsis via Agrobacterium-mediated transformation. Six independent homozygous single-locus transgenic lines that expressed the transgenes and produced high amounts of scopoletin (1) and its glucoside scopolin (2) were selected on the basis of quantitative reverse transcription polymerase chain reaction (qRT-PCR) and ultrahigh-performance liquid chromatography coupled to mass spectrometry (UHPLC-MS) (figs. S1 and S2). However, four of these lines (SCOP-A–D) suffered from silencing effects of the CesA4 gene that shared sequence similarity with the used promoter sequence (fig. S3). Therefore, all further analyses were performed with the two transgenic lines, named SCOP-1 and SCOP-2, that did not suffer from gene silencing. Detailed morphological analyses indicated that these lines were phenotypically similar to wild-type (WT) plants (fig. S4).
To examine the extent to which the combined expression of F6′H1 and COSY affected the phenolic metabolism of the plant, methanol-soluble metabolites were extracted from inflorescent stems and analyzed via UHPLC-MS. No statistical differences were found between the two independent SCOP lines. However, of all 1597 detected mass/charge ratio (m/z) features (LC-MS peaks characterized by a mass and a retention time), 181 features had a higher intensity in the extracts of both SCOP lines, whereas 128 features had a reduced intensity compared to control extracts. For the 181 features with higher intensity, we focused on the 40 features with the highest intensity in SCOP lines and that were at least fourfold higher in abundance as compared to WT plants. The 40 selected features corresponded to 26 compounds (because of in-source fragmentation and adduct formation, a single compound can give rise to multiple m/z features), of which 22 could be structurally characterized (Fig. 2 and table S1). Scopoletin (1) and its glucoside scopolin (2) accumulated up to 26- and 67-fold, respectively, in SCOP lines as compared to WT. Scopolin became the phenolic compound with the highest intensity in SCOP lines (fig. S5). In an independent experiment, scopoletin and scopolin were quantified to represent about 0.004 and 0.55% (w/w) of the stem dry weight, respectively (table S2). All of the other 20 (partially) characterized compounds also contained a coumarin moiety (Fig. 2 and table S1), of which 15 (3–17) containing a scopoletin moiety and one (18) a scopoletin derivative. In addition to scopoletin-containing metabolites, esculin (19) and hexosylated fraxetins (20, 21, and 22) were overproduced in the SCOP lines. Notably, except for scopoletin sulfate (3), all compounds contained a hexose moiety, suggesting the swift glycosylation of the accumulating coumarins in SCOP lines. Glycosylation of compounds by uridine 5′-diphosphate (UDP)–glucose–dependent glycosyltransferases (UGTs) and the storage of the resulting glycosides in the vacuole are common for phenolic metabolites in plants (44, 45).
Fig. 2. Phenolic profiling of SCOP lines and WT.
(A) Average ion intensities of scopoletin and its glucoside scopolin. For each compound, the average ion intensity was significantly higher in SCOP lines as compared to WT (P < 0.001, ANOVA followed by post hoc Tukey test; n = 9; error bars represent the SE). (B) Principal components analysis of the phenolic profile of SCOP-1, SCOP-2, and WT lines. PC, principal component. Venn diagrams represent the 95% confidence regions of all detected m/z features. (C) Overview of all (partially) characterized differential metabolites.
The 40 features with the largest relative decrease in abundance in SCOP lines as compared to WT corresponded to 33 compounds, of which 26 could be (partially) characterized (Fig. 2 and table S3). All compounds were derived from coniferyl alcohol, sinapyl alcohol, or both. A particularly affected metabolic class comprises the oligolignols (27–33), i.e., small oligomers of G and S units that have the potential to extend further to lignin polymers. Together, these observations suggest a redirection of the metabolic flux toward the biosynthesis of scopoletin, rather than toward the biosynthesis of monolignols, as both biosynthetic pathways share feruloyl-CoA as an intermediate.
As perturbations in the phenylpropanoid pathway often result in vessel collapse (46), we investigated the phenotypes of the SCOP lines at the cellular level. No structural abnormalities could be observed in the shape of the vessel cells in inflorescence stems of SCOP lines (Fig. 3). However, light microscopy experiments revealed a yellow color in the xylem vessels and interfascicular fibers of SCOP lines, closely resembling the yellow color of solutions that contain dissolved scopoletin. Stem sections stained with phloroglucinol-HCl showed a similar shift in coloration of the interfascicular fibers; the typical red staining, due mainly to hydroxycinnamaldehyde end groups in lignin, was conflated with a yellowish color (Fig. 3). This observation indicates a shift in lignin composition. To evaluate the presence of scopoletin in the cell wall, fluorescence microscopy was performed. Scopoletin fluoresces when excited at 405 nm (fig. S6). To account for lignin autofluorescence, the detector sensitivity was reduced until a weak emission signal was observed from WT inflorescence stem sections. Upon excitation at 405 nm, the emission observed in SCOP lines was much more intense (Fig. 3). The fluorescence signal in SCOP lines was observed in cell walls of lignifying tissues, i.e., vessels, xylary fibers, and interfascicular fibers, whereas no fluorescence was observed in the cell walls of nonlignifying tissues. These observations suggested a marked increase in lignin quantity or, more likely, the presence of scopoletin or scopoletin-related molecules in the secondary cell walls of SCOP lines.
Fig. 3. Bright-field and fluorescence microscopy of SCOP lines.
(Top) Bright-field microscopy revealed a yellow color in SCOP lines. (Middle) Phloroglucinol-HCl (Wiesner)–stained inflorescence stem sections were visualized using bright-field microscopy. (Bottom) Autofluorescence of inflorescence stem sections upon excitation at 405 nm. V, vessel; XF, xylary fiber; IF, interfascicular fiber; P, phloem. Scale bars, 100 μm. Representative for n = 3 biological replicates.
To investigate whether the increased fluorescence in lignified cell walls was caused by the accumulation of scopoletin or by an increase in lignin, fully senesced stems were analyzed for their lignin levels. Cell wall residue (CWR) was treated with sulfuric acid to remove polysaccharides and quantify lignin using the gravimetry-based Klason assay. Compared to WT, the SCOP-1 line showed a 9.9% reduction in Klason lignin, whereas no significant difference could be detected for the SCOP-2 line, although a trend toward lower lignin could be observed (Table 1). In addition, we investigated the ultraviolet-visible absorption properties of SCOP lignins upon solubilization in acetyl bromide (AcBr) solution. Analysis of the absorbance spectra at 280 nm indicated an 11.5% reduction in AcBr lignin amount for SCOP-1 (P = 0.01) and a 7.6% reduction for SCOP-2 (P = 0.09). As a potential shift in lignin composition in SCOP lines interferes with the spectrophotometry-based quantification procedure of this method, the Klason assay remains the preferred method for lignin quantification. The absorbance spectra of SCOP AcBr lignin had a shoulder peak between 320 and 370 nm, whereas this shoulder was absent in the spectra of the control (fig. S7A). Scopoletin has a distinctive absorption peak at 343 nm (fig. S7B). By subtracting the absorbance spectra of WT lines from those of SCOP lines, a difference spectrum was obtained with a defined absorption peak at 346 nm, almost perfectly matching that of the scopoletin standard (fig. S7B). The notable similarity between both spectra further suggests the incorporation of scopoletin or scopoletin-like moieties in the cell walls of SCOP lines.
Table 1. Lignin content and composition.
CWR was determined gravimetrically as a percentage of the dry weight. Lignin content was determined via the Klason + acid soluble lignin (ASL) assay and the AcBr method and expressed as a percentage of the CWR. The composition of the lignin polymer was determined using thioacidolysis followed by GC-MS. The relative proportions of the monomers were calculated as a percentage of the total thioacidolysis yield of H, G, and S units. S/G was calculated from the relative percentages of G and S. The values are the average (SD) of 10 biological replicates. Klason and ASL values are given as average (SD) of three independent pools of plants. *0.05 > P > 0.01; ***P < 0.001; ●, P = 0.09 (ANOVA with post hoc Tukey’s HSD test).
| Line | CWR (%) | Klason | ASL | AcBr | H (%) | G (%) | S (%) | S/G |
| WT | 80.7 (2.0) | 19.0 (0.5) | 2.4 (0.04) | 13.1 (0.7) | 0.99 (0.08) | 79.36 (0.74) | 19.21 (0.64) | 0.24 (0.01) |
| SCOP-1 | 81.2 (1.9) | 17.1 (1.3)* | 2.5 (0.3) | 11.6 (1.1)* | 1.53 (0.44)* | 74.67 (1.34)*** | 19.96 (0.79) | 0.27 (0.01) |
| SCOP-2 | 77.9 (1.9)*** | 17.9 (1.2) | 2.3 (0.3) | 12.1 (1.3)• | 1.36 (0.14) | 74.53 (1.57)*** | 20.15 (1.29) | 0.27 (0.02)* |
To unambiguously demonstrate the incorporation of scopoletin into the lignin polymer, we subjected enzyme lignins (ELs) isolated from the two SCOP lines to two-dimensional heteronuclear single-quantum coherence (2D HSQC) nuclear magnetic resonance (NMR). This technique allows the detection of monomers independent of their linkage type in the aromatic region and, in addition, the determination of the relative frequency of each interunit linkage type. Using a comparative analysis of SCOP lines and a scopoletin standard, 3.2 and 2.7% of the lignin polymer in SCOP-1 and SCOP-2 lines could be attributed to the incorporation of scopoletin (Fig. 4). Compared to the traditional H units, which commonly represent approximately 0.5% of the lignin polymer in Arabidopsis, scopoletin incorporated at sixfold higher levels. Analyses of the NMR spectra indicated that the coupling of scopoletin to traditional monolignols occurred, at least in large part, via the O-4 position, as illustrated by the peak contours originating from correlations of C2-H, C5-H, C7-H, and C8-H, suggesting that scopoletin mainly acts as a starting unit of the lignin polymer. Moreover, the splitting of the scopoletin resonance peaks indicates that scopoletin is involved in at least two different structures in the lignin-dominated samples. Furthermore, the remaining peaks in the difference spectrum might suggest the coupling of scopoletin via a variety of coupling methods (e.g., 8-8, 8-5, or 5-8). However, at this point, we could not characterize these structures. No scopoletin-related signals could be detected in the NMR spectra of WT lignin. In addition, spectral analyses illustrated that the relative amount of G units had slightly decreased, and that of S units had increased in SCOP lignin compared to those in WT (table S4). In line with the relative increase in S units, the relative frequency of β-aryl ether linkages (8-O-4) had increased in SCOP lines compared to WT, whereas the relative frequencies of condensed lignin bonds, i.e., phenylcoumaran (8–5) and resinol (8–8), were reduced. In addition, the frequency of cinnamyl alcohol end groups was reduced in the lignin of SCOP lines (table S5 and fig. S8). Because dimerization reactions favor the formation of condensed bonds and cinnamyl alcohol end groups, these data are consistent with the main role of scopoletin as a chain initiator, as deduced from the aromatic region of the NMR spectra.
Fig. 4. NMR spectrum of the lignin aromatic region.
Partial short-range 1H–13C HSQC NMR spectra from isolated ELs from cell walls of the main stem of senesced Arabidopsis plants. WT (A) and SCOP (B and C) lines were analyzed. The colors of the contours correspond to the structures drawn. H peaks are indicated as trace amounts as their signals coincide with those of Phe3/5. In the difference spectrum (D), hot-colored contours represent positive, whereas cold (light cyan) contours represent negative differences after subtracting (A) from (B); the scopoletin peaks colored in solid magenta were also positive. A scopoletin standard (E) was used to identify resonance peaks related to scopoletin incorporation in the difference spectrum. ppm, parts per million (Hz/MHz).
To gain deeper insight into the lignin composition of SCOP lines, we used thioacidolysis, a lignin degradative method that cleaves β-aryl ether linkages while leaving carbon-carbon bonds intact, thereby releasing monomers, dimers, and higher-order oligomers. The released thioacidolysates were subsequently analyzed for H, G, and S monomers via gas chromatography (GC)–MS. In line with the NMR results, the GC-MS analysis showed that the S/G ratio was increased up to 12% in SCOP lines as compared to the S/G ratio in WT (Table 1 and table S6). As with other Arabidopsis mutants that have a reduction in lignin content without large phenotypical abnormalities [cinnamate 4-hydroxylase (c4h), 4-coumarate-CoA ligase 1 (4cl1), caffeoyl shikimate esterase (cse), caffeoyl-CoA O-methyltransferase 1 (ccoaomt1), and transaldolase 2 (tra2)], the higher S/G ratio may be explained by the fact that the remaining flux through the phenylpropanoid pathway preferentially proceeds toward sinapaldehyde when F5H1 remains fully active (47, 48). qRT-PCR showed that, similar to mutants with an increased S/G ratio, F5H1 gene expression levels were not affected in SCOP lines compared to WT (fig. S9). Further analysis of the thioacidolysates indicated an ~1.5-fold increase in the relative frequency of released H units in SCOP lines as compared to WT (Table 1). Principal components analysis (PCA) on 388 thioacidolysates detected via GC-MS separated samples from SCOP lines from those of WT (fig. S10). Untargeted analysis of the thioacidolysates yielded two significant features; scopoletin accumulated up to sixfold in SCOP lines as compared to WT, whereas G end groups were reduced (table S7). The results of the untargeted analysis are in line with the observations in the targeted analysis, in which the absolute quantity of released scopoletin and G units was found to be increased and reduced, respectively, in SCOP lines (table S6).
To better understand how scopoletin was incorporated into the lignin polymer, we isolated EL, i.e., lignocellulosic biomass was treated with polysaccharide-degrading enzymes to remove the majority of the carbohydrates and the EL was subjected to derivatization followed by reductive cleavage (DFRC). Similar to thioacidolysis, this method cleaves β-aryl ether linkages. The use of deuterated acetic anhydride in the final step of the reaction allows the discrimination of lignin units linked via their phenolic group (4-O etherified) from those units originally having a free phenolic function. The former units become derivatized with deuterated acetate in the final step (after the reductive cleavage of the β-ether units), whereas the latter become derivatized with normal (nondeuterated) acetate in the initial AcBr derivatization/dissolution step (Fig. 5 and fig. S11). The released DFRC degradation products were analyzed via GC-MS. Notably, roughly 50% of the standard G units were etherified, whereas about 75% of the S units were etherified. Similar etherification levels have been determined in lignin monomers released by analytical thioacidolysis (49). The abundance of ether-linked scopoletin in lignin was investigated via the presence of scopoletin acetate [weight-average molecular weight (Mw) 234] and its deuterated analog (Mw 237). Scopoletin acetates (both deuterated and nondeuterated acetates) were detected as 3.3 and 2.3% of the total released DFRC degradation products in SCOP-1 and SCOP-2 lines, respectively, and were not detected in the WT samples (fig. S5). Up to 55% of these released scopoletin units appeared as deuterated acetate products, indicating that scopoletin coupled via its 4-O position into the lignin polymer to a considerable extent (Fig. 5 and fig. S11).
Fig. 5. Incorporation of scopoletin into the lignin polymer.
GC-MS analysis of selected DFRC cleavage products from isolated EL of (A) WT, (B) SCOP-1, and (C) SCOP-2 lines. 4-O etherified scopoletin and 8-O etherified scopoletin were detected with an m/z of 193 and 192, respectively. (D) Putative routes of scopoletin incorporation together with the expected chemical markers after DFRC analysis. Scopoletin covalently couples into the lignin polymer via radical coupling, giving rise to scopoletin as a starting unit (route i), as an internalized unit (route ii), and as a phenolic end unit (route iii). Ar stands for aryl, e.g., a G or S unit, or a unit derived from a second scopoletin monomer. Mw, weight-average molecular weight.
After the successful engineering of scopoletin into the lignin of SCOP lines, we tested the hypothesis that the cell walls of SCOP lines would be easier to degrade under alkaline biomass pretreatment conditions before enzymatic hydrolysis of the cell wall carbohydrates into fermentable sugars. We therefore measured the enzymatic cellulose-to-glucose conversion of inflorescence stem segments of SCOP and WT lines after an alkaline pretreatment. CWRs of SCOP lines and WT plants had equal amounts of cellulose and hemicellulose (Table 2). After a 62.5 mM NaOH (3 hours at 90°C) pretreatment of stem biomass, the glucose release upon a cellulase incubation was followed over a period of 72 hours. At each time point, SCOP lines released significantly more glucose compared to WT. At the end of the saccharification, the cellulose-to-glucose conversion of SCOP-1 and SCOP-2 lines had increased by 40 and 39%, respectively, relative to the conversion of WT (Fig. 6). The increased enzymatic hydrolysis of cellulose was also visually obvious, as the structural integrity of the SCOP inflorescence stem segments was lost after 72 hours of cellulose treatment (Fig. 6B and fig. S13). In addition, the cellulose-to-glucose conversion of SCOP lines was evaluated after a low-concentration alkaline pretreatment, an acid pretreatment, and without the use of a pretreatment. After pretreatment with 1 M HCl for 2 hours at 80°C, cellulose-to-glucose conversion increased by 17% and 20% for SCOP-1 and SCOP-2 lines, respectively, as compared to WT. With a low alkaline concentration (1.91 mM NaOH for 3 hours at 90°C) or when no pretreatment was applied, the cellulose-to-glucose conversion of SCOP-2 was increased by only 14 and 11%, respectively, whereas the cellulose-to-glucose conversion of SCOP-1 was similar to that of WT (fig. S12). These observations are in line with our initial hypothesis that a certain alkaline load will hydrolyze scopoletin-derived β-aryl ethers, resulting in enhanced lignin degradation during alkaline pretreatment and an improved subsequent cellulose-to-glucose conversion (Fig. 1). However, we cannot exclude the possibility that changes to the lignin polymer—such as the increased S/G ratio, the lower lignin content, and the 1.5-fold relative increase in H-unit frequency—also contributed to the enhanced saccharification efficiency.
Table 2. Polysaccharide content and composition.
Cellulose content and matrix polysaccharides (MPSs) are expressed as a percentage of the CWR. The cellulose content was determined using an Updegraff assay, whereas the MPS content was determined gravimetrically. The composition of the MPS is expressed as mole percent of the total MPS content. The values are given as average (SD) of 10 biological replicates. **0.01 > P > 0.001 (ANOVA with post hoc Tukey’s HSD test).
| Line | Cellulose | MPS | Rhamnose | Fucose | Arabinose | Xylose | Mannose | Glucose | Galactose |
| WT | 42.70 (1.63) | 39.42 (1.54) | 2.61 (0.23) | 0.90 (0.07) | 18.62 (1.24) | 69.92 (3.22) | 1.21 (0.09) | 1.10 (0.30) | 5.63 (0.32) |
| SCOP-1 | 45.01 (1.83) | 42.14 (1.12)** | 2.60 (0.57) | 0.88 (0.08) | 18.94 (1.55) | 69.37 (3.54) | 1.34 (0.33) | 1.29 (0.44) | 5.59 (0.33) |
| SCOP-2 | 45.18 (3.14) | 39.92 (1.87) | 2.73 (0.47) | 0.93 (0.09) | 18.25 (1.37) | 69.70 (4.37) | 1.34 (0.18) | 1.30 (0.36) | 5.74 (0.31) |
Fig. 6. Enzymatic cellulose-to-glucose conversion of SCOP lines.
Before the saccharification, stem segments were pretreated with 62.5 mM NaOH at 90°C for 3 hours. (A) Released glucose was expressed as a percentage of the total cellulose in the respective lines. Saccharification was ended after 72 hours. At each time point, cellulose-to-glucose yield was significantly higher in both SCOP lines as compared to that of WT. Significance levels are indicated by ● and * for SCOP-1 and SCOP-2 lines, respectively, compared to WT [* and ●, 0.05 > P > 0.01; ** and ●●, 0.01 > P > 0.001; *** and ●●●, P < 0.001 (ANOVA followed by post hoc Tukey test; n = 10)]. (B) Biomass of SCOP lines and controls after 62.5 mM NaOH pretreatment and 72 hours of enzymatic hydrolysis. SCOP stem pieces are structurally degraded, whereas those of WT remain largely intact. Scale bars, 4 mm.
At the time that we initially submitted our findings on the SCOP lines, Sakamoto et al. (50) reported a screening method in Arabidopsis to discover lignin engineering strategies. From that screen, they found that overexpression of F6′H1 under control of the Populus sieboldii × Populus grandidentata C4H promoter resulted in incorporation of scopoletin into the alcohol-insoluble cell wall. However, lacking NMR- and DFRC-based evidence, it remained unclear whether scopoletin was an integral part of the lignin polymer in those transgenic plants. Furthermore, biomass from their plants did not show an improvement in saccharification efficiency upon alkaline pretreatment, possibly because they only expressed F6′H1, and not also COSY, in their screening strategy (40).
Our combined results show that the simultaneous expression of F6′H1 and COSY in lignifying tissues results in the production of scopoletin, thereby shifting the metabolism from the biosynthesis of traditional monolignols toward the biosynthesis of coumarins. In addition, scopoletin was successfully incorporated, via 4-O etherification, into the lignin polymer, exceeding the levels of the traditional H units. The integration of scopoletin into the lignin polymer is in line with recent in silico molecular modeling studies that predicted that a variety of lignin-related compounds can passively traverse the plasma membrane before their incorporation into the lignin polymer, rather than, but not excluding, the need for plasma membrane–located transporters (51). The corollary is that any aromatic compound that is made in lignifying cells, that meets the minimum requirement of having a phenolic function, and that has the right chemical properties to diffuse through a lipid bilayer may incorporate into the lignin polymer and can therefore be considered as a candidate alternative lignin monomer (8, 21, 23, 24, 35). Our concept study to enhance lignocellulosic biomass processing in the model Arabidopsis may serve as a basis for translation into biomass crops to engineer high-quality feedstock for the biorefinery.
MATERIALS AND METHODS
Plant material and vector construction
Arabidopsis thaliana (ecotype Col-0) was used as a control and for plant transformation. For the vector construction, we removed the stop codon of the coding sequence of F6′H1 (AT3G13610) and fused it to a T2A linker sequence, which was, on its turn, fused to the coding sequence of the COSY (AT1G28680). The resulting construct, flanked by AttL1 and AttL2 sites (AttL1-F6′H1:T2A:COSY-AttL2), was subsequently synthesized and cloned into the pBR332 vector by GenScript. The 4491–base pair (bp) promoter of Arabidopsis CesA4 (pCesA4) was available in-house as pEN-L4-pCesA4-R1 (27). The promoter was recombined with the pBR332-F6′H1:T2A:COSY vector into the destination vector pB7m24GW using a multisite Gateway reaction. The expression vector, pCesA4:F6′H1_COSY (SCOP), was introduced into Agrobacterium tumefaciens strain C58C1 PMP90 via electroporation. Positive colonies were selected via PCR using the following gene-specific primers: F6′H1_Fw (5′-CAACCCAATTCTCAAATCCAG-3′) and COSY_Rv: (5′-AGGGCAAATTCACCTTTGTG-3′). Five independent WT Arabidopsis plants were floral dipped (52) with Agrobacterium containing the expression vector. Of each floral-dipped plant, three transgenic seedlings were selected on Basta selective medium and subsequently planted in soil. In the T2 generation, the proportion of resistant seedlings was determined. Seed stocks that followed a 3:1 segregation, indicative for a single locus insertion, were selected and transferred to soil. Plants giving rise to a T3 seedstock of which all seeds germinated on the selective medium were selected as being homozygous.
Growth conditions
Vernalized seeds of WT and SCOP lines were germinated in soil and were grown in short-day conditions (8-hour light, 16-hour dark; 21°C, day; 18°C, night) for approximately 6 weeks, before being transferred to long-day conditions (16-hour light, 8-hour dark; 21°C, day; 18°C, night). Plants used for metabolomics and microscopy were grown until they were 30 and 20 cm tall before analysis, respectively. Plants grown for cell wall analyses and biomass experiments were grown until senescence. Root phenotypical analyses were performed on 10-day-old seedlings grown on vertical Murashige and Skoog plates, containing 0.6% (w/v) GELRITE, 1 × Murashige and Skoog medium [Murashige and Skoog basal salt mixture powder (4.31 g liter−1; Duchefa), sucrose (10 g liter−1), 2-morpholinoethanesulphonic acid (MES) monohydrate (0.5 g liter−1), myo-inositol (0.1 g liter−1), plant tissue GELRITE (6.0 g liter−1; Duchefa); pH 5.7]. After sowing, plates were incubated at 4°C for 2 days for vernalization, after which the plates were placed in vertical orientation in the tissue culture room under long-day growth conditions (16-hour light/8-hour dark) at 21°C.
Phenotyping experiments
Dry weight and height of the main stem of senesced Arabidopsis lines were measured on a precision balance (Mettler Toledo XP105 Delta Range) after removal of the side branches, cauline leaves, and siliques. Root phenotypic analyses were performed by counting the number of emerged lateral roots using a stereomicroscope (CETI Binocular Zoom Stereo) and by quantifying the root length using the ImageJ analysis software.
Phenolic profiling
The bottom 15 cm of Arabidopsis stems was harvested and immediately frozen in liquid nitrogen. Subsequently, the basal 1 cm was removed, and the following 10 cm was ground in 2-ml Eppendorf tubes with precooled 4-mm stainless steel beads using a Retsch ball milling machine (20 Hz). The phenolic metabolites were extracted with 1 ml of methanol for 15 min at 70°C in a thermomixer (1000 rpm). An 800 μl of aliquot of each phenolic extract was dried at reduced pressure in a SpeedVac and subsequently solubilized in 100 μl of cyclohexane and 100 μl of Milli-Q water. The water phase (15 μl) was injected on an UHPLC system (Waters ACQUITY UPLC) equipped with a BEH C18 column (2.1 mm by 150 mm, 1.7 μM; Waters) and coupled to a quadrupole time-of-flight mass spectrometer [Q-TOF mass spectrometer, Synapt Q-TOF (Waters Corp., Milford, MA, USA)]. A gradient of two buffers was used: buffer A (99:1:0.1, H2O/ACN/formic acid, pH 3) and buffer B (99:1:0.1, ACN/H2O/formic acid, pH 3); 99% buffer A for 0.1 min decreased to 50% buffer A in 30 min (350 μl/min, column temperature of 40°C). The flow was diverted to the mass spectrometer equipped with an electrospray ionization source and LockSpray interface for accurate mass measurements. The MS source parameters were as follows: capillary voltage of 2.5 kV, sampling cone of 37 V, extraction cone of 3.5 V, source temperature of 120°C, desolvation temperature of 400°C, cone gas flow of 50 liter/hour, and desolvation gas flow of 500 liter/hour. The collision energy for the trap and transfer cells was 4 and 3 V, respectively. For data acquisition, the dynamic range enhancement mode was activated. Full-scan data were recorded in negative centroid V-mode; the mass ranged between m/z 100 and 1200, with a scan speed of 0.2 s per scan, with MassLynx software v4.1 (Waters). Leucine-enkephalin [250 pg/μl solubilized in 1:1 (v/v) water/ACN, with 0.1% formic acid] was used for the lock mass calibration, with scanning every 10 s and with a scan time of 0.5 s. From the resulting chromatograms, 8035 deisotoped peaks were integrated and aligned via Progenesis QI v2.4 (Waters Corp., Milford, MA, USA), each peak having an m/z and a retention time. PCA (Pareto scaling, all 8035 peaks included) was performed using MetaboAnalyst v4 (53). The following filters were applied to analyze the data: average ion intensity within at least one group >1000, P value Student’s t test <0.001, and fold change >4 or <0.25 compared to WT. Statistical analyses were performed on ArcSinh-transformed ion intensities. For structural elucidation, MS/MS was used. For MS/MS, all settings were the same as in full MS, except that the collision energy was ramped from 10 to 20 eV for low mass and from 20 to 45 eV for high mass, in the trap, and the scan time was set at 0.3 s.
Scopoletin and scopolin quantification
Phenolic extracts were prepared as described above, except for dried phenolic extracts that were solubilized in 200 μl of cyclohexane and 200 μl of Milli-Q water. All samples were diluted 100 and 1000 times for quantification purposes. Three independent dilution series of scopoletin (Sigma-Aldrich) and scopolin [synthesized from scopoletin (54)] were used to generate calibration curves. Of each sample, 10 μl was injected onto a UHPLC system (Waters ACQUITY UPLC) equipped with a BEH C18 column (2.1 mm × 50 mm, 1.7 μm; Waters) coupled to a Q-TOF mass spectrometer [Vion IMS Q-TOF (Waters Corporation, Milford, MA, USA)]. For chromatographic separation, a gradient of two buffers was used: buffer A (99:1:0.1 H2O/ACN/formic acid, pH 3) and buffer B (99:1:0.1 ACN/H2O/formic acid, pH 3); 99% buffer A for 0.1 min decreased to 50% buffer A in 5 min followed by a decrease to 30% buffer A from 5 to 7 min and 0% buffer A from 7 to 10 min. The flow rate was set to 0.5 ml min−1 (column temperature of 40°C). The flow was diverted to the mass spectrometer equipped with an electrospray ionization source and LockSpray interface for accurate mass measurements. The MS source parameters were as follows: capillary voltage of 2.5 kV, sampling cone of 40 V, extraction cone of 3.5 V, source temperature of 120°C, desolvation temperature of 600°C, cone gas flow of 50 liter/hour, and desolvation gas flow of 1000 liter/hour. For data acquisition, Intelligent Data Capture Intensity threshold was set at 5. Full-scan data were recorded in negative ionization mode through Uni-Fi 2.0.0 Workstation (Waters). Profile data were recorded, and the mass range was set between m/z 100 and 1200 with a scan speed of 0.1 s per scan. Leucine-enkephalin [250 pg/μl solubilized in 1:1 (v/v) water/ACN, with 0.1% formic acid] was used for the lock mass calibration. Chromatogram alignment and peak detection were done with Progenesis QI v2.4 (Waters Corporation, Milford, MA, USA). The amounts of scopoletin and scopolin in the biological samples were calculated on the basis of the respective peak intensities and the calibration curve for each.
Light and fluorescence microscopy
Fresh Arabidopsis stems were harvested from WT and SCOP lines when they had an approximate height of 20 cm. SCOP lines that suffered from gene silencing effects (SCOP-A–D) were harvested at the same age with an average height of 12 cm. The bottom 2 cm was discarded, and the following 5 cm was embedded in 7% (w/v) agarose according to Mitra et al. (55). Slices of 100-μm thickness were made using a Leica VT 1200S vibratome. Stem sections were first stained using Wiesner (phloroglucinol-HCl) reagent according to Mitra et al. (55) and subsequently visualized using an OLYMPUS BX51 microscope with a 20× objective. Lignin autofluorescence was determined using a Zeiss LSM 710 confocal microscope with a Plan-Apochromat 20×/0.8 M27 objective. The fluorescence signal originating from the lignin polymer was obtained using an excitation wavelength of 405 nm and an emission window between 415 and 735 nm.
Cell wall preparation
CWR was prepared using a series of sequential extraction steps (27). In summary, after removal of the bottom 1 cm of the main stem, the basal 30 cm of senesced Arabidopsis stems was cut into 2-mm pieces and sequentially washed for 30 min in Milli-Q-water at 98°C, ethanol at 76°C, chloroform at 59°C, and acetone at 54°C. CWRs were dried under reduced pressure and determined gravimetrically as a percentage of the dry weight.
Lignin quantification
The lignin content was determined using the gravimetry-based Klason assay and the spectrophotometry-based AcBr lignin quantification assay. Klason lignin was determined on three pools of 100 mg of CWR and essentially performed according to National Renewable Energy Laboratory (56) with modifications as reported by Oyarce et al. (27). AcBr lignin was determined on 10 mg of CWR and essentially performed according to Foster et al. (57) with small modifications as reported by Van Acker et al. (11). The scopoletin standard (1.04 mg) was dissolved in 5 ml of AcBr solution and taken through the whole AcBr lignin quantification procedure, after which its absorbance spectrum was compared to those of biological samples.
Lignin composition via thioacidolysis
The composition of the lignin polymer was investigated using thioacidolysis of the isolated cell wall preparations (58–60). Released monomers, involved in β-O-4 linkages, were detected as their trimethylsilyl derivatives. GC-MS analysis was carried out using a 7890B GC system equipped with a 7693A automatic liquid sampler and a 7250 Accurate Mass Q-TOF MS system (Agilent Technologies, Santa Clara, CA, USA). One microliter of the sample was injected in splitless mode with the injector port set to 280°C. Separation was achieved with a VF-5ms column (30 m × 0.25 mm, 0.25 μm; Varian CP9013, Agilent Technologies) with helium carrier gas at a constant flow of 1.2 ml/min. The oven was held at 130°C for 3 min after injection, ramped to 200°C at 10°C/min, ramped to 250°C at 3°C/min, held at 250°C for 5 min, ramped to 320°C at 20°C/min, held at 320°C for 5 min, and, lastly, cooled to 130°C at 50°C/min at the end of the run. The mass selective detector (MSD) transfer line was set to 280°C, and the electron ionization energy was 70 eV. Full electron impact–MS spectra were recorded between m/z 50 and 800 at a resolution of >25,000 and with a solvent delay of 10.0 min. Peak integrations for quantification of the lignin monomers were carried out using the MassHunter Quantitative Analysis (for Q-TOF) software package (Agilent Technologies) using specific quantifier ions for each metabolite (table S8). Response factors for the lignin monomers were determined on the basis of the standard curves of each of the different monomers: Hm/z 205 (1.17), Gm/z 235 (2.48), Sm/z 265 (1.98), and scopoletinm/z 234 (0.19).
For untargeted analysis, the resulting GC-MS chromatograms were analyzed and deconvoluted with Analyzer Pro XD. In total, 388 thioacidolysates were detected and quantified. Statistical analysis was performed via analysis of variance (ANOVA) (P < 0.01) followed by a post hoc Student’s t test (P < 0.01).
Lignin composition via 2D HSQC NMR
After removal of the bottom 1 cm, the basal 30 cm of the main inflorescence stem of SCOP lines and control plants was chopped into 2 mm pieces. Stem pieces of multiple individual SCOP plants were pooled to finally obtain three independent pools of 300 mg each. Control plants were pooled similarly, but only two independent pools of 300 mg could be obtained. EL was prepared using a CELLULYSIN enzyme mixture, and 2D HSQC NMR was performed as previously reported (61).
Scopoletin incorporation via DFRC
EL was degraded by DFRC using previously described experimental conditions (62, 63). The DFRC degradation products were acetylated using deuterated acetic anhydride and pyridine [1:1 (v/v), 2 hours], allowing distinction of monomers deriving from originally etherified versus originally free phenolic units (26) before being analyzed on a Shimadzu GC/MS-QP2010 Plus instrument using previously described chromatographic conditions (62). Monomer determination was performed by monitoring the m/z as described in (64): 192, 222, and 252, the base peaks resulting from the loss of ketene (CH2-C=O) for nondeuterated H, G, and S acetates, and 193, 223, and 253 for the deuterated H, G, and S analogs, respectively. The nondeuterated acetate of scopoletin was monitored at m/z 192, and its deuterated analog was monitored at m/z 193. The response of each ion is depicted as an extracted ion chromatogram in Fig. 5.
Carbohydrate analyses
Prepared CWRs were used to determine the amount of matrix polysaccharides (MPSs), cellulose, and hemicellulose. The composition of the hemicellulose together with the amount of MPS was determined using previously described methods (11). The cellulose content was determined as reported in (65).
Saccharification assays
Main stems of senesced SCOP lines and WT plants were collected for saccharification experiments. Upon removal of the bottom 1 cm, the basal 30 cm was chopped into 2-mm pieces that were pooled per two individuals. The saccharification assay was performed according to Van Acker et al. (66). For alkaline biomass pretreatments, 1.91 mM NaOH and 62.5 mM NaOH were used. For the acid pretreatment, a 1 M HCl solution was used. The activity of the enzyme mixture was determined using a filter paper assay (67), and 0.016 filter paper units were added to each sample. The structural integrity of inflorescence stem pieces was imaged using a Leica M80 stereomicroscope equipped with an ICE90 E camera (Wetzlar, Germany).
Quantitative reverse transcription PCR
Transcript levels of CESA4 (AT5G44030), CESA7 (AT5G17420), F5H1 (AT4G36220), and the chimeric pCesA4:F6′H1_COSY gene were determined in 8-week-old Arabidopsis stems of transgenic lines and WT grown in long-day conditions (16-hour light, 8-hour dark). The bottom 2 cm of the main stem was harvested and immediately flash frozen in liquid nitrogen. RNA was extracted from four pools consisting of each three individual stem pieces using the RNeasy Plant Mini Kit (QIAGEN). An on-column treatment with deoxyribonuclease I (Promega) was performed to digest contaminating genomic DNA according to the manufacturer’s instructions. A total of 1 μg of RNA was used as a template for cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad). Relative transcript abundances were determined in triplicate on the LightCycler 480 II Real-Time SYBR Green PCR System (Roche) according to the manufacturer’s instructions. Obtained cycle threshold values were converted to relative expression values using the second derivative maximum method. PP2AA3 (AT1G13320) and MON1 (AT2G28390) were used as reference genes within the analysis. All primer sequences are listed in table S9.
Acknowledgments
We thank L. Goossens, E. Goethals, and A. Hermans for the help with the experiments, and A. Bleys for helping us prepare this manuscript.
Funding: This work was supported by the IWT-FISCH-SBO project ARBOREF (grant 140894) and iBOF project NextBioRef. L.H. and M.W. are indebted to the Research Foundation Flanders (FWO) for a predoctoral fellowship and R.V. for a postdoctoral fellowship. L.H. was additionally funded by FWO project number G011620N. B.D.M. was funded by the Institute for the promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) for a predoctoral fellowship and, in addition, funded by FWO project number G020618N. H.K. and J.R. received funding from the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-SC0018409).
Author contributions: L.H., B.D.M., R.V., H.K., J.R., and W.B. designed the experiments. L.H., M.W., H.K., and B.D.M. performed the experiments. L.H., R.V., M.W., H.K., B.D.M., S.V., K.M., J.P., S.D., G.G., and J.R. collected and analyzed the data. L.H., R.V., and W.B. wrote the manuscript with the help of all of the authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.Turner S. R., Somerville C. R., Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson R. L., Hammerschmidt R., Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389 (1992). [Google Scholar]
- 3.Zhong R., Taylor J. J., Ye Z. H., Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanholme R., De Meester B., Ralph J., Boerjan W., Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Del Río J. C., Marques G., Rencoret J., Martínez Á. T., Gutiérrez A., Occurrence of naturally acetylated lignin units. J. Agric. Food Chem. 55, 5461–5468 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Ralph J., Lapierre C., Boerjan W., Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Regner M., Bartuce A., Padmakshan D., Ralph J., Karlen S. D., Reductive cleavage method for quantitation of monolignols and low-abundance monolignol conjugates. ChemSusChem 11, 1600–1605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Río J. C., Rencoret J., Gutiérrez A., Elder T., Kim H., Ralph J., Lignin monomers from beyond the canonical monolignol biosynthetic pathway: Another brick in the wall. ACS Sustain. Chem. Eng. 8, 4997–5012 (2020). [Google Scholar]
- 9.Karlen S. D., Zhang C., Peck M. L., Smith R. A., Padmakshan D., Helmich K. E., Free H. C. A., Lee S., Smith B. G., Lu F., Sedbrook J. C., Sibout R., Grabber J. H., Runge T. M., Mysore K. S., Harris P. J., Bartley L. E., Ralph J., Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2, e1600393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. Freudenberg, A. C. Neish, Constitution and Biosynthesis of Lignin (Springer-Verlag, 1968), vol. 2. [Google Scholar]
- 11.Van Acker R., Vanholme R., Storme V., Mortimer J. C., Dupree P., Boerjan W., Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 6, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F., Dixon R. A., Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Himmel M. E., Ding S.-Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D., Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science 315, 804–807 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Franke R., McMichael C. M., Meyer K., Shirley A. M., Cusumano J. C., Chapple C., Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22, 223–234 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Stewart J. J., Akiyama T., Chapple C. C. S., Ralph J., Mansfield S. D., The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150, 621–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries L., Vanholme R., Van Acker R., De Meester B., Sundin L., Boerjan W., Stacking of a low-lignin trait with an increased guaiacyl and 5-hydroxyguaiacyl unit trait leads to additive and synergistic effects on saccharification efficiency in Arabidopsis thaliana. Biotechnol. Biofuels 11, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J. H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W., A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24, 3506–3529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson L. A., Shadle G. L., Zhou R., Nakashima J., Chen F., Dixon R. A., Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. Bioenergy Res. 1, 180–192 (2008). [Google Scholar]
- 19.Marita J. M., Ralph J., Hatfield R. D., Chapple C., NMR characterization of lignins in Arabidopsis altered in the activity of ferulate 5-hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 96, 12328–12332 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eudes A., Dutta T., Deng K., Jacquet N., Sinha A., Benites V. T., Baidoo E. E. K., Richel A., Sattler S. E., Northen T. R., Singh S., Simmons B. A., Loqué D., SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLOS ONE 12, e0178160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S. D., Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Sederoff R. R., MacKay J. J., Ralph J., Hatfield R. D., Unexpected variation in lignin. Curr. Opin. Plant Biol. 2, 145–152 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P. F., Marita J. M., Hatfield R. D., Ralph S. A., Christensen J. H., Boerjan W., Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, 29–60 (2004). [Google Scholar]
- 24.J. Ralph, G. Brunow, P. J. Harris, R. A. Dixon, P. F. Schatz, W. Boerjan, in Recent Advances in Polyphenol Research, F. Daayf, V. Lattanzio, Eds. (Wiley-Blackwell Publishing, 2008), vol. 1, pp. 36–66. [Google Scholar]
- 25.Vanholme R., Morreel K., Ralph J., Boerjan W., Lignin engineering. Curr. Opin. Plant Biol. 11, 278–285 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Wilkerson C. G., Mansfield S. D., Lu F., Withers S., Park J.-Y., Karlen S. D., Gonzales-Vigil E., Padmakshan D., Unda F., Rencoret J., Ralph J., Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344, 90–93 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Oyarce P., De Meester B., Fonseca F., de Vries L., Goeminne G., Pallidis A., De Rycke R., Tsuji Y., Li Y., Van den Bosch S., Sels B., Ralph J., Vanholme R., Boerjan W., Introducing curcumin biosynthesis in Arabidopsis enhances lignocellulosic biomass processing. Nat. Plants 5, 225–237 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Smith R. A., Gonzales-Vigil E., Karlen S. D., Park J.-Y., Lu F., Wilkerson C. G., Samuels L., Ralph J., Mansfield S. D., Engineering monolignol p-coumarate conjugates into poplar and Arabidopsis lignins. Plant Physiol. 169, 2992–3001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibout R., Le Bris P., Legée F., Cézard L., Renault H., Lapierre C., Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:monolignol transferase PMT. Plant Physiol. 170, 1358–1366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Criss D. L., Fisher T. H., Schultz T. P., Alkaline hydrolysis of nonphenolic α-carbonyl β-0-4 lignin dimers substituted on the leaving phenoxide ring: Comparison with benzylic hydroxyl analogues. Holzforschung 52, 57–60 (1998). [Google Scholar]
- 31.Imai A., Yokoyama T., Matsumoto Y., Meshitsuka G., Significant lability of guaiacylglycerol β-phenacyl ether under alkaline conditions. J. Agric. Food Chem. 55, 9043–9046 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Gierer J., Ljunggren S., The reactions of lignins during sulfate pulping. Part 16. The kinetics of the cleavage of β-aryl ether linkages in structures containing carbonyl groups. Sven. Papperstidn. 82, 71–81 (1979). [Google Scholar]
- 33.Gierer J., Ljunggren S., Ljungquist P., Norén I., The reactions of lignin during sulfate pulping. Part 18. The significance of α-carbonyl groups for the cleavage of β-aryl ether structures. Sven. Papperstidn. 83, 75–82 (1980). [Google Scholar]
- 34.Rahimi A., Ulbrich A., Coon J. J., Stahl S. S., Formic-acid-induced depolymerization of oxidized lignin to aromatics. Nature 515, 249–252 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Vanholme R., Morreel K., Darrah C., Oyarce P., Grabber J. H., Ralph J., Boerjan W., Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Lapierre C., Jouin D., Monties B., On the molecular origin of the alkali solubility of Gramineae lignins. Phytochemistry 28, 1401–1403 (1989). [Google Scholar]
- 37.Strehmel N., Böttcher C., Schmidt S., Scheel D., Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 108, 35–46 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Clemens S., Weber M., The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav. 11, e1114197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stringlis I. A., de Jonge R., Pieterse C. M. J., The age of coumarins in plant–microbe interactions. Plant Cell Physiol. 60, 1405–1419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanholme R., Sundin L., Seetso K. C., Kim H., Liu X., Li J., De Meester B., Hoengenaert L., Goeminne G., Morreel K., Haustraete J., Tsai H.-H., Schmidt W., Vanholme B., Ralph J., Boerjan W., COSY catalyses trans–cis isomerization and lactonization in the biosynthesis of coumarins. Nat. Plants. 5, 1066–1075 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Kai K., Mizutani M., Kawamura N., Yamamoto R., Tamai M., Yamaguchi H., Sakata K., Shimizu B., Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 55, 989–999 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Halpin C., Cooke S. E., Barakate A., El Amrani A., Ryan M. D., Self-processing 2A-polyproteins - A system for co-ordinate expression of multiple proteins in transgenic plants. Plant J. 17, 453–459 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Eudes A., George A., Mukerjee P., Kim J. S., Pollet B., Benke P. I., Yang F., Mitra P., Sun L., Çetinkol Ö. P., Chabout S., Mouille G., Soubigou-Taconnat L., Balzergue S., Singh S., Holmes B. M., Mukhopadhyay A., Keasling J. D., Simmons B. A., Lapierre C., Ralph J., Loqué D., Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol. J. 10, 609–620 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Le Roy J., Huss B., Creach A., Hawkins S., Neutelings G., Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7, 735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dima O., Morreel K., Vanholme B., Kim H., Ralph J., Boerjan W., Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27, 695–710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones L., Ennos A. R., Turner S. R., Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Meyer K., Shirley A. M., Cusumano J. C., Bell-Lelong D. A., Chapple C., Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 6619–6623 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J. P., Naik P. P., Chen H.-C., Shi R., Lin C.-Y., Liu J., Shuford C. M., Li Q., Sun Y.-H., Tunlaya-Anukit S., Williams C. M., Muddiman D. C., Ducoste J. J., Sederoff R. R., Chiang V. L., Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26, 894–914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lapierre C., Rolando C., Thioacidolyses of pre-methylated lignin samples from pine compression and poplar woods. Holzforschung 42, 1–4 (1988). [Google Scholar]
- 50.Sakamoto S., Kamimura N., Tokue Y., Nakata M. T., Yamamoto M., Hu S., Masai E., Mitsuda N., Kajita S., Identification of enzymatic genes with the potential to reduce biomass recalcitrance through lignin manipulation in Arabidopsis. Biotechnol. Biofuels 13, 97 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vermaas J. V., Dixon R. A., Chen F., Mansfield S. D., Boerjan W., Ralph J., Crowley M. F., Beckham G. T., Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. U.S.A. 116, 23117–23123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., Xia J., MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaudhury D. N., Holland R. A., Robertson A., 338. The syntheses of glycosides. Part XII. Fabiatrin. J. Chem. Soc. 1948, 1671–1672 (1948). [DOI] [PubMed] [Google Scholar]
- 55.Pradhan Mitra P., Loqué D., Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp. , e51381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A. Sluiter, B. Hames, R. Ruiz, C. Scarlata, J. Sluiter, D. Templeton, D. Crocker, “Determination of structural carbohydrates and lignin in Biomass. Laboratory Analytical Procedure” (Technical Report NREL/TP-510–42618, National Renewable Energy Laboratory, 2012).
- 57.Foster C. E., Martin T. M., Pauly M., Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part I: Lignin. J. Vis. Exp. , e1745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson A. R., Mansfield S. D., Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J. 58, 706–714 (2009). [DOI] [PubMed] [Google Scholar]
- 59.C. Lapierre, in Forage Cell Wall Structure and Digestibility (American S., 2015), pp. 133–166. [Google Scholar]
- 60.Rolando C., Monties B., Lapierre C., Thioacidolysis. Methods Lignin Chem. , 334–349 (1992). [Google Scholar]
- 61.Ralph J., Hatfield R. D., Quideau S., Helm R. F., Grabber J. H., Jung H.-J. G., Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J. Am. Chem. Soc. 116, 9448–9456 (1994). [Google Scholar]
- 62.del Río J. C., Rencoret J., Prinsen P., Martínez Á. T., Ralph J., Gutiérrez A., Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60, 5922–5935 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Lu F., Ralph J., Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: Protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45, 2590–2592 (1997). [Google Scholar]
- 64.Martone P. T., Estevez J. M., Lu F., Ruel K., Denny M. W., Somerville C., Ralph J., Discovery of Lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19, 169–175 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Sundin L., Vanholme R., Geerinck J., Goeminne G., Höfer R., Kim H., Ralph J., Boerjan W., Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol. 166, 1956–1971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Acker R., Vanholme R., Piens K., Boerjan W., Saccharification protocol for small-scale lignocellulosic biomass samples to test processing of cellulose into glucose. Bio-Protocol 6, e1701 (2016). [Google Scholar]
- 67.Xiao Z., Storms R., Tsang A., Microplate-based filter paper assay to measure total cellulase activity. Biotechnol. Bioeng. 88, 832–837 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R., Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardiner J. C., Taylor N. G., Turner S. R., Control of cellulose synthase complex localization in developing xylem. Plant Cell 15, 1740–1748 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eudes A., Zhao N., Sathitsuksanoh N., Baidoo E. E., Lao J., Wang G., Yogiswara S., Lee T. S., Singh S., Mortimer J. C., Keasling J. D., Simmons B. A., Loqué D., Expression of S-adenosylmethionine hydrolase in tissues synthesizing secondary cell walls alters specific methylated cell wall fractions and improves biomass digestibility. Front. Bioeng. Biotechnol. 4, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skoog F., Montaldi E., Auxin-kinetin interaction regulating the scopoletin and scopolin levels in tobacco tissue cultures. Proc. Natl. Acad. Sci. U.S.A. 47, 36–49 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goodwin R. H., Kavanagh F., The isolation of scopoletin, a blue-fluorescing compound from oat roots. Bull. Torrey. Bot. Club 76, 255 (1949). [Google Scholar]
- 73.De Meester B., de Vries L., Özparpucu M., Gierlinger N., Corneillie S., Pallidis A., Goeminne G., Morreel K., De Bruyne M., De Rycke R., Vanholme R., Boerjan W., Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176, 611–633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morreel K., Dima O., Kim H., Lu F., Niculaes C., Vanholme R., Dauwe R., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W., Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 153, 1464–1478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morreel K., Kim H., Lu F., Dima O., Akiyama T., Vanholme R., Niculaes C., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W., Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal. Chem. 82, 8095–8105 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Tsuji Y., Vanholme R., Tobimatsu Y., Ishikawa Y., Foster C. E., Kamimura N., Hishiyama S., Hashimoto S., Shino A., Hara H., Sato-Izawa K., Oyarce P., Goeminne G., Morreel K., Kikuchi J., Takano T., Fukuda M., Katayama Y., Boerjan W., Ralph J., Masai E., Kajita S., Introduction of chemically labile substructures into Arabidopsis lignin through the use of LigD, the Cα-dehydrogenase from Sphingobium sp. strain SYK-6. Plant Biotechnol. J. 13, 821–832 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S9
References
Data S1 to S3