Abstract
High-risk neuroblastoma, a pediatric tumor originating from the sympathetic nervous system, has a low mutation load but highly recurrent somatic DNA copy number variants. Previously, segmental gains and/or amplifications allowed identification of drivers for neuroblastoma development. Using this approach, combined with gene dosage impact on expression and survival, we identified ribonucleotide reductase subunit M2 (RRM2) as a candidate dependency factor further supported by growth inhibition upon in vitro knockdown and accelerated tumor formation in a neuroblastoma zebrafish model coexpressing human RRM2 with MYCN. Forced RRM2 induction alleviates excessive replicative stress induced by CHK1 inhibition, while high RRM2 expression in human neuroblastomas correlates with high CHK1 activity. MYCN-driven zebrafish tumors with RRM2 co-overexpression exhibit differentially expressed DNA repair genes in keeping with enhanced ATR-CHK1 signaling activity. In vitro, RRM2 inhibition enhances intrinsic replication stress checkpoint addiction. Last, combinatorial RRM2-CHK1 inhibition acts synergistic in high-risk neuroblastoma cell lines and patient-derived xenograft models, illustrating the therapeutic potential.
RRM2 is characterized as a dependency factor and entry point for CHK1 combination treatment in neuroblastoma.
INTRODUCTION
Neuroblastoma is a pediatric tumor arising from immature sympathetic neuroblasts (1, 2), with current survival rates for high-risk cases still being disappointingly low despite intensive multimodal therapy. Given that the number of mutations in newly diagnosed cases is typically very low, the search space for mutated druggable targets is limited except for ALK mutations. In contrast to the low mutational burden, DNA copy number changes are highly recurrent with 2p and 17q gains occurring in both MYCN-amplified and nonamplified high-risk cases, while 1p and 11q deletions are predominantly found in MYCN-amplified and MYCN-nonamplified high-risk cases, respectively. We and others previously showed that focal gains and amplifications can highlight candidate genes implicated in neuroblastoma initiation and/or maintenance, potentially expanding the current number of available druggable targets (3, 4). Recent whole-genome sequencing efforts have uncovered recurrent complex rearrangements, including chromothripsis affecting the chromosome 2 short arm. These rearrangements are often accompanied by amplicon formation encompassing the MYCN locus and regulatory sequences driving MYCN expression, as well as additional genes that have been proposed to have a tumor-promoting role distinct from MYCN activity itself (5), such as ODC1 (6), the gene encoding for the rate-limiting enzyme in polyamine biosynthesis; the ALK gene implicated in neuronal development and codriver of MYCN-driven neuroblastoma formation (7, 8); and SOX11, encoding a presumed lineage dependency transcription factor with functions distinct from the core regulatory transcription factor circuitry (9).
Here, we report on the further dissection of focal chromosome 2p imbalances and identified the “ribonucleotide reductase subunit M2” (RRM2) gene, located on 2p25.1, which encodes the small regulatory subunit of the ribonucleotide reductase (RNR) complex. RRM2 is the catalytic component of the RNR enzyme and is essential for the maintenance of deoxynucleotide triphosphate (dNTP) pool homeostasis required for DNA replication and repair. Synthesis of RRM2 protein is regulated in a cell cycle–dependent fashion, increasing to maximal levels during S phase of the cell cycle (10–12). Depletion causes G1-S phase arrest and rapidly leads to increased levels of replicative stress due to DNA replication fork stalling and decreased cell proliferation. Replicative stress activates the ATR (Ataxia Telangiectasia and Rad3-Related Protein)-CHK1 (Checkpoint Kinase 1) DNA damage response to control cell cycle checkpoints, origin firing, and replication fork stability to ensure genomic stability. This response includes RRM2 up-regulation through CHK1-E2F Transcription Factor 1 (E2F1) transcriptional regulation and ATR-controlled cyclin F inactivation to block RRM2 protein degradation (13). MYC and MYCN (V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma) proteins cause replicative stress, among others, through increased replication origin use and elevated global transcriptional activity (14–17). This can explain the up-regulation of many DNA damage response genes including RRM2 to cope with toxic replicative stress levels and increased DNA damage (18). Evidence for a more direct role of RRM2 in cancer formation or maintenance comes from mouse models in which increased RRM2 expression promotes lung (19), breast (20), and prostate cancer (21). Furthermore, RRM2 dependency was demonstrated in BRAFV600E-driven melanoma (22) and Ewing sarcoma (23). Of further interest, RRM2 was shown to be the target of synthetic lethal interaction with G2 Checkpoint Kinase (WEE1) inhibition in H3K36me3-deficient cancers (12). Together, these studies underline the potential importance of RRM2 levels in oncogenesis and cancer cell survival, thus marking RRM2 as a potential drug target.
Here, we provide evidence for enhanced RRM2 levels resulting from increased RRM2 copy numbers through large segmental or focal chromosome 2p gains or high-level amplification. Elevated RRM2 expression levels correlated with decreased overall and event-free survival in patients with neuroblastoma, and RRM2 expression levels were up-regulated during MYCN-driven neuroblastoma formation in mice. In vitro experiments support a role for RRM2 as a dependency factor in both neuroblastoma cell lines and three-dimensional (3D) neuroblastoma spheroid cultures. Further evidence for a direct cooperative role of RRM2 in neuroblastoma formation was provided through combined overexpression with MYCN in the sympathetic neuronal lineage in zebrafish, which led to increased tumor penetrance from 16% up to more than 80%, while overexpression of RRM2 alone did not cause tumor formation. High-risk neuroblastoma cells are addicted to the replicative stress-response ATR-CHK1 signaling pathway, with RRM2 as a key downstream factor of this signaling pathway. In vitro forced overexpression of RRM2 in neuroblastoma alleviates replicative stress, as monitored by pRPA32 (phosphorylated Replication Protein A 32 KDa Subunit) levels. This is further supported in vivo by our double transgenic MYCN-RRM2 zebrafish neuroblastoma model, with tumor cells exhibiting reduced S345pCHK1 and γH2AX (gamma H2A Histone Family Member X) protein levels. Last, RRM2 inhibition synergistically enhances sensitivity of neuroblastoma cells to pharmacological targeting of ATR-CHK1 pathway addiction, thus suggesting the therapeutic potential for this drug combination in high-risk CHK1-addicted, primary neuroblastomas.
RESULTS
RRM2 is a target for focal gains and amplifications affecting gene dosage and neuroblastoma patient survival
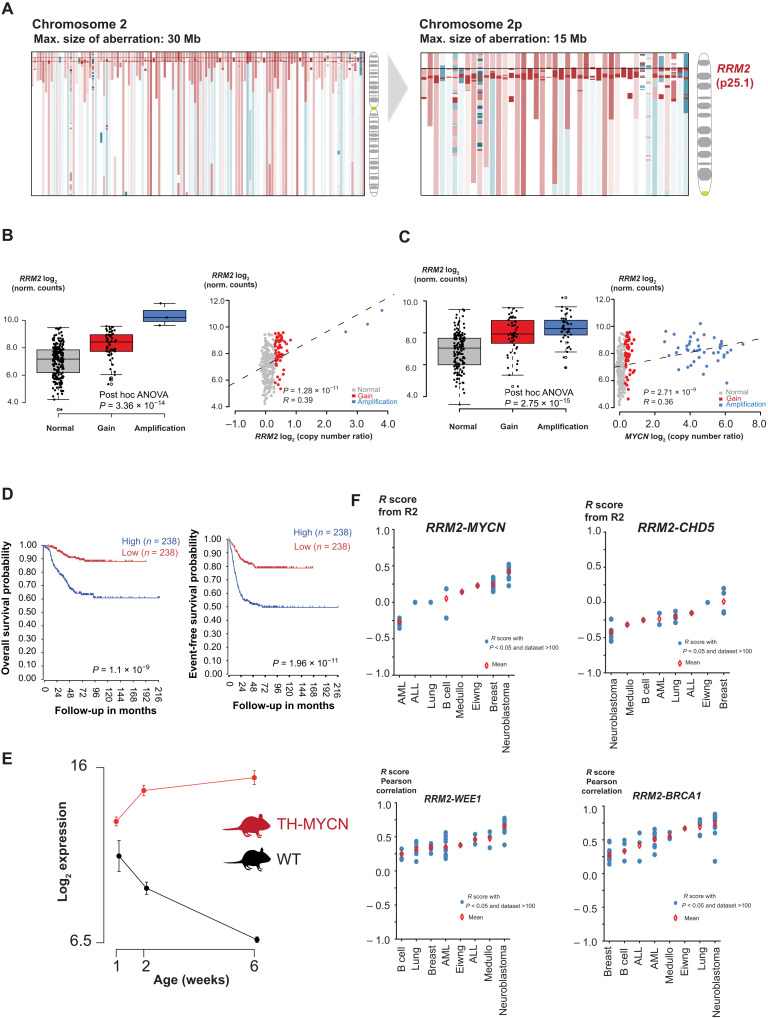
DNA copy number profiles of 556 primary high-risk neuroblastoma cases (24) were analyzed for recurrent small segmental gains or amplifications affecting chromosome 2p loci. In addition to known amplicons implicating MYCN, ODC1, ALK, and SOX11, we identified a previously unknown smallest region of overlap encompassing RRM2, encoded on 2p25.1 (Fig. 1A and fig. S1A). Subsequent additional analysis of high-resolution whole-genome data (63 cases; EGAS00001001308), whole-exome data (156 cases; EGAS00001003244), and low-resolution DNA copy number data (200 cases; GSE45480) revealed an additional 60 of 419 cases with 2p gains or amplifications (fig. S1B), with amplification defined as >4-fold increase of RRM2 signal in relation to the number of chromosomes 2 and gains with a 1.5- to 4-fold copy number in accordance with the European Neuroblastoma Quality Assessment group. In some cases, the RRM2 locus was involved in more complex amplicons, as illustrated by case “WGS-4” resulting from chromothripsis involving chromosome 2p encompassing the RRM2 locus, case “WGS-12” with the RRM2 locus being part of a complex amplification involving multiple loci across the entire chromosome 2, and case “WES-17” with the RRM2 gene involved in complex amplicon on 2p also involving MYCN and case “WES-19” displaying a similar pattern as observed for WES-17, but displaying an additional copy number jump within RRM2.
Fig. 1. In silico analysis of genomic and transcriptomic data of primary neuroblastoma converges toward RRM2 as a top-ranked 2p codriver in high-risk neuroblastoma.
(A) Array CGH (comparative genomic hybridization) profiles of >200 high-risk neuroblastoma cases converge toward the RRM2 gene (2p25.1) as recurrently gained on 2p (red: gained/amplified region and blue: deleted region). (B) Left: Boxplot indicating the gene dosage effect for RRM2 expression in relation to the RRM2 copy number status. Right: Correlation analysis of RRM2 expression with RRM2 copy number data [National Research Council (NRC) neuroblastoma cohort (n = 283); hgserver2.amc.nl]. ANOVA, analysis of variance. (C) Left: Boxplot indicating the gene dosage effect for RRM2 expression in relation to the MYCN copy number status. Right: Correlation analysis of RRM2 expression with MYCN copy number data [NRC neuroblastoma cohort (n = 283); hgserver2.amc.nl]. (D) High RRM2 expression levels correlate to a poor overall and event-free neuroblastoma patient survival [Kocak cohort (n = 283); hgserver2.amc.nl]. (E) Rrm2 expression is strongly up-regulated during TH-MYCN–driven neuroblastoma tumor development. (F) Pearson correlation of RRM2 and its upstream regulators (MYCN, WEE1, BRCA1, and CHD5) in expression data from various cancer entities available (hgserver2.amc.nl). AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia.
Given the crucial role of RRM2 in nucleotide metabolism and replicative stress control and its recent established role as (co)driver in various cancer subtypes, we performed further data mining to find support for a functional role for RRM2 in neuroblastoma. First, we evaluated the effects of RRM2 gene copy number increase on expression levels and observed a positive correlation (R = 0.39, P = 1.28 × 10−11) (Fig. 1B). We also observed a positive correlation between MYCN copy number status and RRM2 expression levels (Fig. 1C), in keeping with a direct regulatory role for MYCN, based on publicly available chromatin immunoprecipitation sequencing (ChIP-seq) data indicating direct binding to the RRM2 promotor (fig. S1C). MYC/MYCN also regulates the transcription of the E2F1, E2F2, and E2F3 genes (25), which directly regulate RRM2 expression (26). The transcriptional consequences of MYCN knockdown have been extensively documented by previous studies and enabled to verify whether modulated MYCN levels indeed affect RRM2 expression. First, Valentijn et al. (18) performed time course experiments in IMR-32 neuroblastoma cells (MYCN amplified) upon MYCN knockdown (fig. S1D), and our team executed similar experiments in IMR-5/75 neuroblastoma cells (fig. S1E) (also MYCN amplified) (27). In addition, Zeid et al. (28) characterized the kinetic effects of dynamic modulation of MYCN (at 0, 2, and 24 hours after inactivation) to profile changes in states with high, medium, and low levels of MYCN in Tet-OFF SHEP-21N neuroblastoma cells (fig. S1F). Collectively, these experiments consistently show strong down-regulation of RRM2 upon MYCN inactivation/depletion. Second, Kaplan-Meier analysis showed that high RRM2 expression levels predict both adverse overall and event-free survival probability of patients in three large primary tumor cohorts. These results are in keeping with our previously reported four-gene prognostic signature in neuroblastoma, which included RRM2 (Fig. 1D and fig. S1G) (29). Third, we looked into the gene expression dataset of early (hyperplastic) lesions at weeks 1 and 2 after birth and established tumors at week 6 from a murine model of MYCN-driven neuroblastoma (TH-MYCN) and expression data from normal matching sympathetic mouse ganglia. This allowed monitoring of the dynamic regulation of gene expression during the tumor formation process, and we observed strong up-regulation of Rrm2 expression levels in comparison to wild-type mice sympathetic ganglia (Fig. 1E) (30).
We also investigated whether RRM2 expression levels could be enhanced through DNA copy number gains, affecting RRM2 upstream regulators (Fig. 1F). Besides MYCN, a second bona fide oncogene and target of amplification, LIN28B (31), also regulates RRM2 through down-regulation of let-7 (32). In addition, CHD5, a known tumor suppressor and commonly deleted gene in the critical 1p36 chromosome region in neuroblastoma (33), is a transcriptional repressor of WEE1 (34), which itself controls cyclin-dependent kinase 2 (CDK2) (35), ensuring RRM2 degradation during S-G2-M transition. Last, BRCA1 (Breast Cancer gene 1) (encoded on the recurrently gained 17q region in neuroblastoma) was shown to be recruited by MYCN to promoter-proximal regions to prevent MYCN-dependent accumulation of stalled RNA polymerase II (RNAPII) (36) and has also been reported to up-regulate RRM2 expression (37). Together, these data suggest that, in neuroblastoma, an integrated gene regulatory network controls RRM2 expression levels, which is further enhanced by recurrent increased DNA copy numbers affecting these loci (fig. S1H).
Functional in vitro and in vivo validation of RRM2 as a novel dependency factor in neuroblastoma
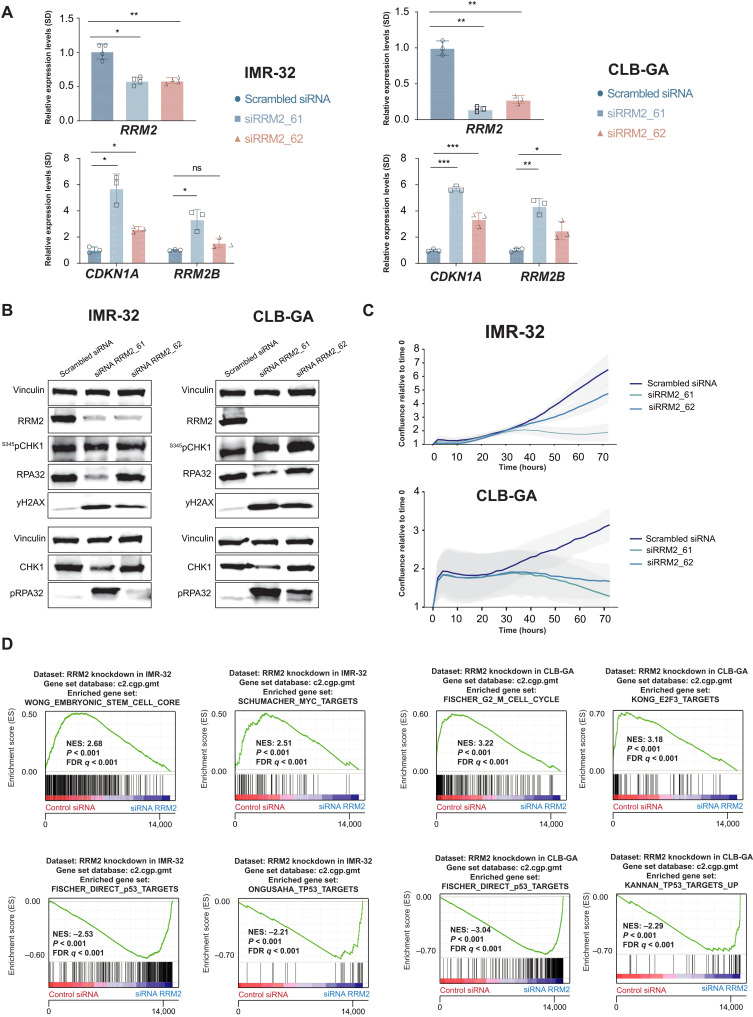
To assess the functional impact of RRM2 down-regulation, we performed a transient RRM2 knockdown using two independent small interfering RNAs (siRNAs) (Fig. 2A, left) in two high-risk neuroblastoma-derived cell lines, the MYCN-amplified IMR-32 cells and nonamplified CLB-GA cells. Transient RRM2 down-regulation induced the expected elevated levels of pRPA32 and 345pCHK1, reflecting increased replicative stress. In addition, RRM2 knockdown also led to increased DNA damage, evidenced by yH2AX induction known to result from replication fork collapse (Fig. 2B and fig. S2A) and reduced proliferation (Fig. 2C). In keeping with these findings, we also found evidence for p53 pathway activation illustrated by up-regulation of CDKN1A (encoding the p21 response gene) and RRM2B encoding the p53-controlled RNR (p53R2) (Fig. 2A, right).
Fig. 2. Transient in vitro RRM2 knockdown in neuroblastoma cells results in an increased DNA damage and p53 pathway response, supporting its putative dependency role in neuroblastoma.
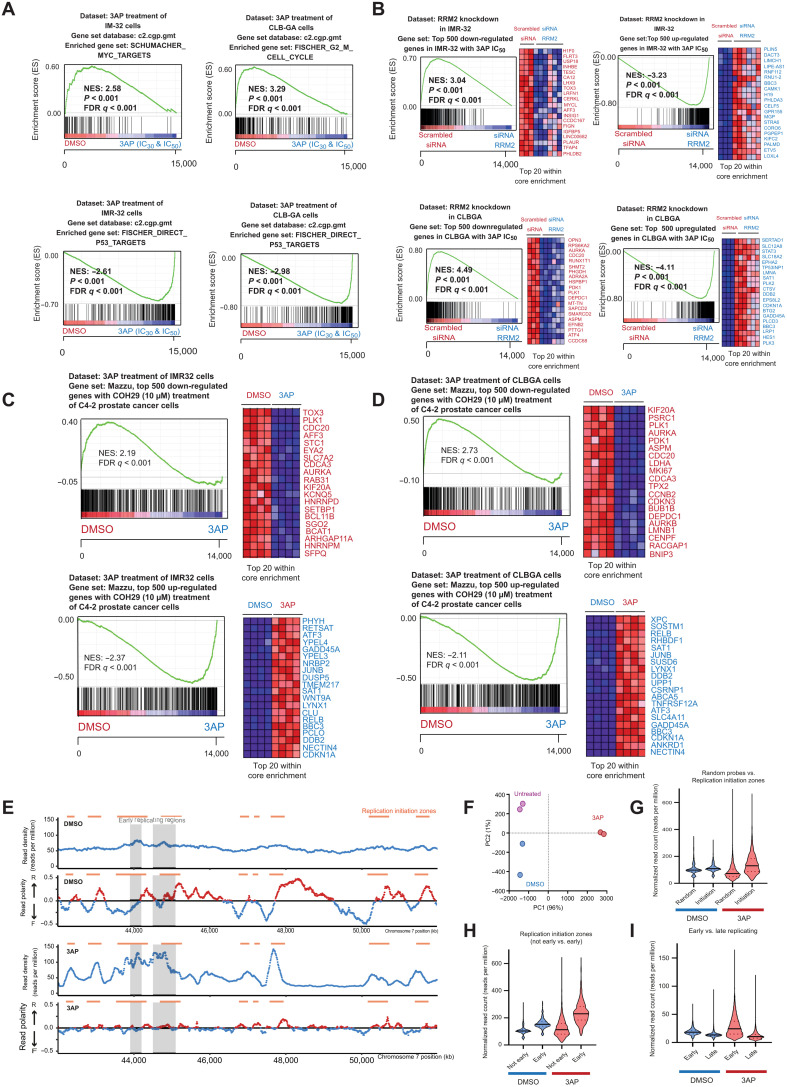
(A) Transient RRM2 knockdown in IMR-32 and CLB-GA neuroblastoma cells using two different RRM2-targeting siRNAs (denoted as si61 and si62) significantly down-regulates the expression of RRM2 and up-regulates the expression of the p53 target gene CDKN1A and the p53-inducible RRM2B gene, as shown by reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis. ns, not significant. (B) Immunblotting confirms that RRM2-targeting siRNA transfection in IMR-32 and CLB-GA cells strongly down-regulates RRM2 protein levels accompanied by DNA damage induction (increased pRPA32 and yH2AX signal) and checkpoint activation (increased pCHK1 levels) (see quantification in fig. S2). (C) IncuCyte live cell imaging analysis shows a strong reduced confluency upon siRRM2_61 transfection and, to a lesser extent, with siRRM2_62 in IMR-32 and CLB-GA cells. (D) GSEA following RNA sequencing (RNA-seq)–based transcriptome profiling of IMR-32 and CLB-GA cells transfected with RRM2-targeting siRNAs indicates a significant reduced expression of MYC targets and G2-M phase markers (top) and up-regulation of p53 target genes (bottom). FDR, false discovery rate; NES, normalized enrichment score.
Previous studies revealed strong effects on the cellular transcriptome under conditions of nucleotide stress. We explored this in IMR-32 and CLB-GA neuroblastoma cells by whole-transcriptome profiling followed by “gene set enrichment analysis” (GSEA) before and after RRM2 knockdown. This revealed a significant down-regulation of MYC and E2F targets (Fig. 2D, top) and up-regulation of p53 targets (Fig. 2D, bottom) compared to control cells, among others.
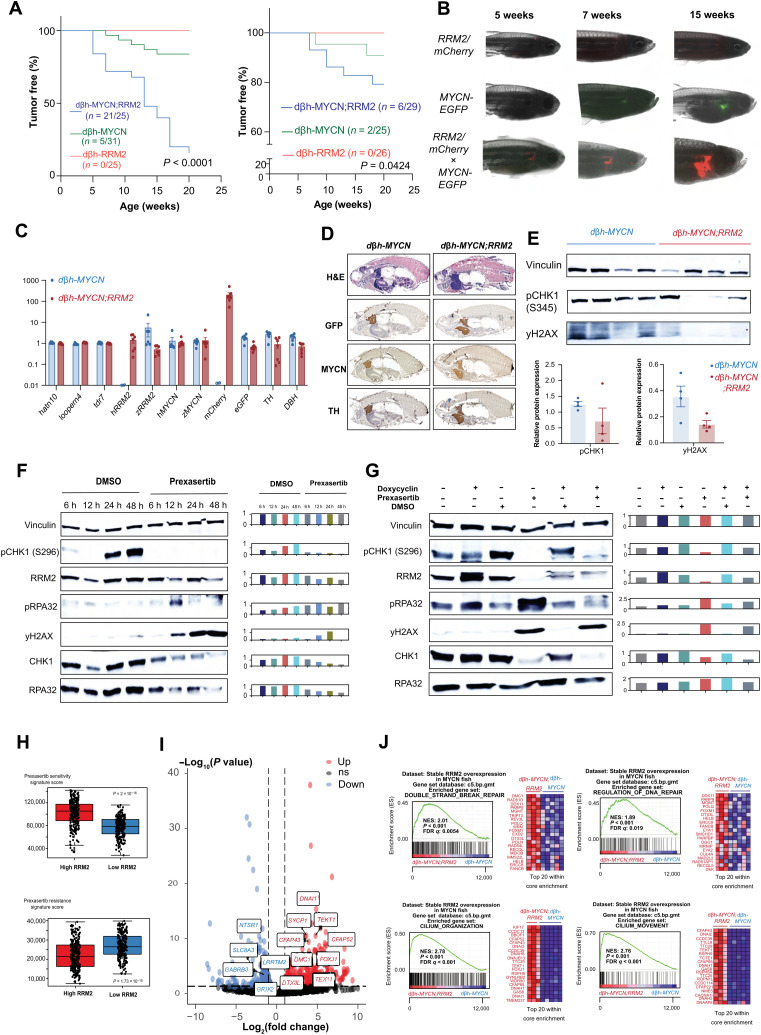
To investigate the role of RRM2 on MYCN-driven neuroblastoma formation in vivo, we evaluated the impact of increased RRM2 expression on a MYCN-driven neuroblastoma zebrafish model. Therefore, a stable Tg(dβh:hRRM2; dβh:mCherry) zebrafish line (further designated as RRM2 line) was generated and crossed with Tg(dβh:eGFP-MYCN)–overexpressing zebrafish, designated as MYCN (8). RRM2 overexpression in MYCN;RRM2 double transgenic zebrafish markedly increased tumor penetrance from 16 to 84% and accelerated in vivo neuroblastoma formation, which started already as early as 5 weeks of age (P < 0.0001) (Fig. 3A, left). To confirm these results and exclude an off-target effect of the integration site, we generated a mosaic model using the Tol2 transposase system (which induces random integration of overexpression constructs) to express cmlc2:eGFP/dβh:RRM2 in the MYCN zebrafish, whereby the integration of the transgene is tracked by a green fluorescent signal in the heart of the zebrafish driven by the cmlc2 promotor. The mosaic model supports the data from the stable lines with significant (P = 0.0424) acceleration of tumor formation in MYCN zebrafish expressing RRM2 (Fig. 3A, right). Tumor penetrance is less marked as compared with the stable line experiments, which is expected given that constructs are not integrated into all the sympathetic lineage precursors in all evaluated embryos for the mosaic approach (38). We used fluorescence microscopy to follow up tumor formation in both MYCN-only and MYCN-RRM2 double transgenic zebrafish over time, with human MYCN and RRM2 being coexpressed with green fluorescent protein (GFP) and mCherry, respectively. By reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis, we could show specific human RRM2 overexpression in the established stable MYCN-RRM2 double transgenic zebrafish compared to MYCN-only zebrafish (Fig. 3C). Next, we performed hematoxylin and eosin (H&E) staining and immunohistochemistry analysis for the markers GFP, TH, and MYCN, both on sections (×10 magnification) of MYCN and MYCN;RRM2 zebrafish (Fig. 3D), confirming that the tumors are indeed neuroblastomas in both model systems.
Fig. 3. Combined MYCN-RRM2 overexpression in zebrafish sympathetic neuroprogenitor cells results in accelerated neuroblastoma development and increased tumor penetrance versus MYCN-only fish.
(A) Kaplan-Meier analysis of dβh-MYCN;RRM2 double transgenic zebrafish (left) and the mosaic model expressing cmlc2-eGFP/dβh-RRM2 in dβh-MYCN zebrafish (right) both show significant accelerated neuroblastoma formation and strongly increased tumor penetrance compared to dβh-MYCN fish. (B) Fluorescence microscopy images show the development of neuroblastoma tumors over time in MYCN-only (GFP) or MYCN-RRM2 double transgenic fish (mCherry). (C) RT-qPCR analysis showing human RRM2 overexpression in the dβh-MYCN;RRM2 double transgenic zebrafish compared to MYCN-only fish. Tdr7, looprn4, and hatn10 were used as housekeeping genes in this analysis. (D) H&E staining and immunofluorescent staining for the markers GFP, TH, and MYCN (×10 magnification). (E) Immunoblotting for S345pCHK1 and yH2AX for protein samples derived from MYCN and MYCN-RRM2 zebrafish. (F) Left: Time course analysis of replication stress markers in CLB-GA neuroblastoma cells upon prexasertib exposure by immunoblotting. Right: Quantification of the immunoblotting relative to vinculin. (G) Left: Time course analysis of replication stress markers in CLB-GA neuroblastoma cells before and after doxycyclin-inducible RRM2 overexpression by immunoblotting. Right: Quantification of immunoblotting relative to vinculin. (H) Signature score analysis of publicly available prexasertib sensitivity and resistance gene signatures in a large primary cohort of neuroblastoma cases (GSE62564). (I) Volcano plot showing the set of significantly up-regulated (red) and down-regulated (blue) genes in dβh-MYCN;RRM2 double transgenic versus dβh-MYCN fish. (J) GSEA of the gene expression profiles using the C5 curated MSigDB gene sets of dβh-MYCN;RRM2 double transgenic versus dβh-MYCN fish shows a significant up-regulation of DNA repair and genes related to cilium organization and movement, while down-regulated gene sets were predominantly related to synapse transmission gene sets [see volcano plot in (I) (blue)].
Increased RRM2 levels coincide with a CHK1-driven gene response indicative of enhanced replicative stress resistance
Previous studies showed that MYC(N) executes both transcriptional and nontranscriptional mechanisms to facilitate progression through the cell cycle. In cancer cells with weakened G1-S control, enhanced MYC/MYCN levels increase the number of origins of replication to accelerate DNA replication, which subsequently causes increased need for nucleotide supply, leading to nucleotide stress sensed by the ATR-CHK1 replication stress checkpoint (14). During early S phase, RRM2 levels are still low, and dividing cells are particularly vulnerable to fork stalling. MYC(N) also indirectly anticipates to increase dNTP supply among others through activating E2Fs, which directly regulate RRM2 transcription (fig. S1H) (25, 39). To gain further insight into the functional impact of RRM2 overexpression on the neuroblastoma phenotype, we first performed immunoblotting for S345pCHK1 and yH2AX from protein extracts of both MYCN-only and MYCN-RRM2 double transgenic zebrafish. We could observe a down-regulation of S345pCHK1 and yH2AX in the MYCN-RRM2 double transgenic zebrafish (Fig. 3E), hinting toward enhanced replication stress resistance upon RRM2 overexpression in vivo. Next, we conducted an in vitro time series experiment in CLB-GA neuroblastoma cells exposed to pharmacological CHK1 inhibition using prexasertib as a stressor over a 6- to 48-hour time frame and monitored replicative stress marker induction (Fig. 3F). We observed increased down-regulation of CHK1 autophosphorylation (S296pCHK1) together with reduction in total CHK1 and RRM2 protein levels as well as up-regulated yH2AX (double-strand DNA breaks) and pRPA32 levels (binds single-stranded DNA and is a marker for enhanced form stalling). Next, we evaluated the presumed rescue effect of doxycyclin-inducible overexpression of RRM2 in CLB-GA neuroblastoma cells under conditions of prexasertib exposure (Fig. 3G). Combining prexasertib exposure with RRM2 overexpression strongly attenuated pRPA32 levels in keeping with our proposed hypothesis that enhanced RRM2 expression can suppress replicative stress. In addition to the direct and E2F-driven up-regulation of RRM2 through MYCN and other copy number–driven genes (see above), RRM2 levels are also tightly regulated through the ATR-CHK1 pathway (see fig. S1H). Moreover, ATR also dampens origin use to keep excessive effects of supraphysiological MYCN levels under control (40). In light of these observations, we further evaluated whether elevated RRM2 levels facilitate replication stress resistance in neuroblastoma cells. To this end, we used the unique dataset of Blosser et al. (41) to score gene signatures related to sensitivity or resistance to pharmacological CHK1 inhibition by prexasertib in a large cohort of primary neuroblastomas (GSE62564). We observed that high RRM2 expression indeed confers stronger sensitivity to prexasertib treatment, indicative of high ATR-CHK1 activity (Fig. 3H). The top-ranking gene sets in this transcriptional sensitivity signature include E2F transcriptional targets, G2-M cell cycle, and spindle-associated checkpoint genes (41).
To investigate the relation of RRM2 levels to replicative stress and ATR-CHK1 signaling activity, we also performed bulk RNA sequencing (RNA-seq) on emerging tumor from 6-week-old single and double transgenic fish. The volcano plot (Fig. 3I) shows top-scoring down- and up-regulated genes between the two groups [Padj < 0.05; log fold change < −1 (down) or log fold change > 1 (up)]. GSEA (Fig. 3J) for the differentially expressed genes between the double MYCN;RRM2 and MYCN-only transgenic fish shows up-regulated enrichment in the double transgenic group for DNA repair genes. Ten (DDX11, TRIP13, POLQ, FOXM1, RAD41L, MMS22L, ERCC8, FANCB, PARPBP, and RAD51AP1) of 31 highest-ranked genes from both DNA repair gene sets overlapped with the top-ranked CHK1-correlated genes in the Kocak neuroblastoma patient cohort (n = 649; GSE45545), suggesting that MYCN-RRM2–driven neuroblastomas in the double transgenic zebrafish model are also marked by enhanced ATR-CHK1 signaling activity. This includes FOXM1, a critical regulator of S-G2 transition and DNA repair; POLQ implicated in replication stress response (42); and the DNA helicase DDX11, which interacts with the fork protection complex to preserve replication fork integrity in stressful conditions (43). Of further interest, the differentially expressed RAD51D (44) plays a noncanonical role in sensing dNTP pool changes. We also find positive enrichment of genes related to cilium organization and movement, with 53 of previously reported cilia genes among the top 200 up-regulated genes from our analysis. In addition, the FOXJ1 (Forkhead Box J1) master regulator transcription factor controlling the expression of cilia genes was also among the top 50 regulated genes (log2 FC of 4.5, Padj of 2.15 × 10−8). It is highly intriguing that human ciliopathies have been brought into context of replicative stress, thus further pointing toward a functional role of increased RRM2 levels in MYCN-driven neuroblastoma in relation to replicative stress in our zebrafish model (45). In addition, recent studies point toward an association between primary cilia and cancer, as they play a role in the interactions between cancer cells and the tumor microenvironment (46). In addition, a direct link between ciliary signaling and regulation of tumor growth and response to treatment has been described, with an impact on core cancer signaling pathways, including DNA damage response (47). FOXJ1 and concomitant reduction of the ciliogenesis program have been previously linked to aggressive ependymoma tumor development (48). Further studies are warranted to clarify the possible role of FOXJ1 and cilia genes in connection to RRM2-enhanced MYCN-driven neuroblastoma formation.
Pharmacological RRM2 inhibition suppresses growth of high-risk neuroblastoma-derived cell lines
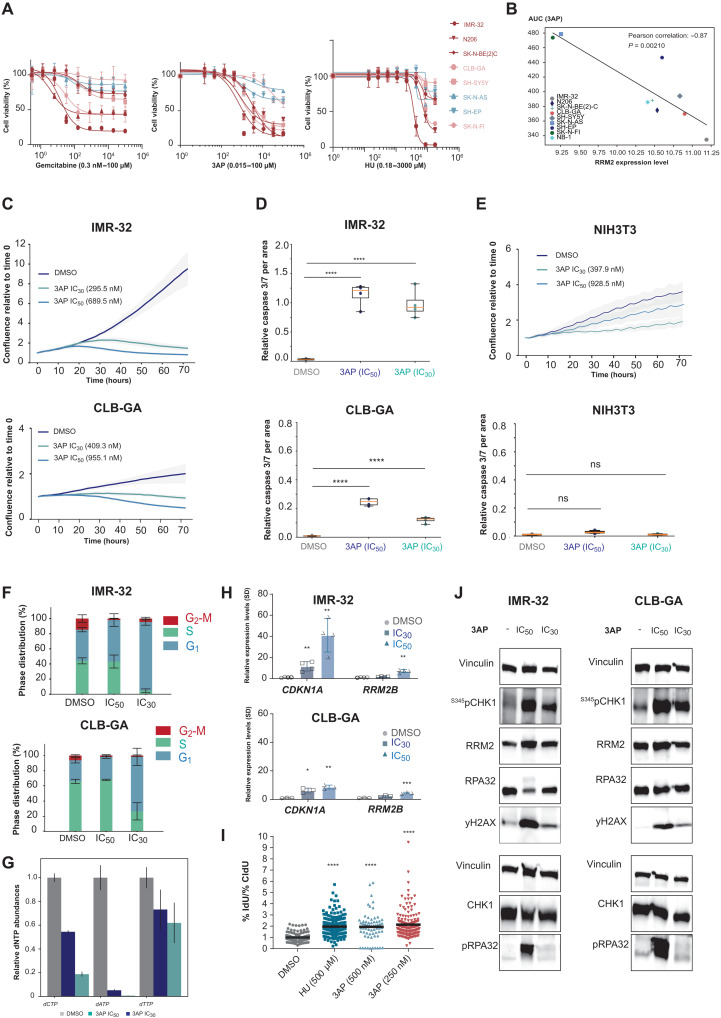
Several compounds targeting RRM2 or RNR activity have been reported, including the iron chelator triapine (further referred to as 3AP) for which positive safety and tolerability data are available from several clinical trials (49–51). First, to assess 3AP sensitivity, we determined average inhibitory concentration (IC50) values in a panel of eight neuroblastoma cell lines and compared the effects with the deoxycytosine analog and the RRM1 inhibitor gemcitabine (52), a commonly used chemotherapeutic in cancer treatment, as well as the effects of hydroxyurea (further denoted as HU) (53), a well-established RRM2 inhibitor in the same cell line panel. Cell viability was most effectively reduced with 3AP compared to gemcitabine or HU (Fig. 4A). MYCN-amplified cell lines and the nonamplified CLB-GA cell line responded well in the nanomolar range, while the other MYCN-nonamplified cell lines were poor responders to 3AP treatment. Correlation analysis of RRM2 expression levels with the area under the curve (AUC) values as obtained for 3AP (Fig. 4A) indicated that cell lines with high RRM2 expression display reduced sensitivity to 3AP (Fig. 4B). We next selected MYCN-amplified IMR-32 and MYCN-nonamplified CLB-GA neuroblastoma cells to study further the phenotypic and molecular effects of 3AP treatment. A significant reduction in cell confluence (Fig. 4C) and increased apoptosis could be observed for both cell lines (Fig. 4D) when exposed to respective IC50 and IC30 of 3AP. Notably, in nonmalignant murine NIH3T3 fibroblasts, no apoptotic effects were observed under IC50 drug conditions in the presence of the expected reduction in proliferation (Fig. 4E). Flow cytometry analyses revealed an almost complete G1-S phase arrest and increase of cells in G1 upon treatment with 3AP at IC30 (Fig. 4F). Both IC30 and IC50 could impose a reduction in the available dNTP pools (Fig. 4G), underscoring the on-target effect of 3AP treatment. In addition, a significant up-regulation of the p53 target genes CDKN1A and RRM2B was induced, as measured by RT-qPCR (Fig. 4H). RRM2 inhibition at IC50 and IC30 3AP concentrations caused enhanced RPA32 phosphorylation (marker for increased single-stranded DNA) and elevated yH2AX levels. In keeping with these observations, using DNA combing, we also found significant increased levels of stalled forks upon 3AP treatment versus controls (Fig. 4I), likely due to increased double-stranded DNA breaks (presumably reflecting increased fork collapse) (54). As expected, elevated fork stalling and accompanying increased pRPA32 and yH2AX levels led to S345pCHK1 activation (measured through pCHK1 levels) (Fig. 4J and fig. S2B). Given that p53 mutations can confer a more aggressive (drug-resistant) neuroblastoma phenotype, we also evaluated the phenotypic response to pharmacological RRM2 inhibition of the p53-mutant neuroblastoma cell line SK-N-BE(2)-C. We observed reduced cell confluence following 3AP treatment, with both IC30 and IC50 (fig. S3A) and concomitant cell death (fig. S3B). Immunoblotting following 3AP treatment of SK-N-BE(2)-C cells treated with 3AP showed up-regulated S345pCHK1 and pRPA32 levels. In addition, as observed in IMR-32 and CLB-GA cells, total CHK1 levels were down-regulated following 3AP exposure at IC50 (fig. S3C).
Fig. 4. Comparative RNR inhibitor analysis to kill neuroblastoma cells.
(A) 3AP treatment can establish lower half-maximal inhibitory concentrations than gemcitabine and HU in a panel of neuroblastoma cell lines, with MYCN-amplified cell lines and the nonamplified CLB-GA cell line being more sensitive than MYCN-nonamplified cell lines (red: MYCN amplified, adrenergic; orange: MYCN nonamplified, adrenergic; blue: MYCN nonamplified, mesenchymal). (B) 3AP sensitivity (AUC) is negatively correlated to RRM2 mRNA expression levels in a panel of neuroblastoma cell lines. (C) Treatment of IMR-32 and CLB-GA neuroblastoma cells with 3AP at their respective half-maximal inhibitory concentration significantly reduces cell confluence and (D) induces cell death. (E) Nonmalignant murine NIH3T3 fibroblasts did not show reduced confluence or apoptosis induction upon 3AP exposure. (F) IC30 values for IMR-32 and CLB-GA neuroblastoma cells impose a significant S phase cell cycle arrest. (G) Endogenous dNTP pools are reduced upon exposure of IMR-32 neuroblastoma cells to 3AP. (H) 3AP (IC50) treatment leads to a significant increased CDKN1A and RRM2B expression. (I) DNA combing following exposure of IMR-32 neuroblastoma cells to HU or 3AP shows a significant increased levels of stalled forks upon 3AP treatment versus controls. IdU, 5′-iododeoxyuridine. (J) Immunoblotting for DNA damage response markers (pRPA32 and yH2A) and CHK1/S345pCHK1 in protein extracts of neuroblastoma cells treated with fixed IC30 and IC50 of 3AP (for quantification, see fig. S2). CIdU, 5-chloro-2’-deoxyuridine.
Last, we evaluated the transcriptional responses of IMR-32 and CLB-GA cells after 48 hours of exposure to their respective IC30 and IC50 3AP concentrations. All four cell lines tested showed a p53 gene signature response induction (Fig. 5A). Furthermore, using GSEA, a strong overlap between 3AP and RRM2-targeting siRNAs induced gene signatures and was notable in both IMR-32 and CLB-GA cells, supporting the on-target effect of RRM2 pharmacological inhibition using 3AP (Fig. 5B). In further support of the on-target activity of 3AP as demonstrated by dNTP measurements (Fig. 4G), we also compared the transcriptome profiles after 3AP exposure of IMR-32 and CLB-GA neuroblastoma cells to published transcriptome profiling data of prostate cancer cells (cell line C4-2) following exposure to COH29, a small-molecule RRM2 inhibitor (21). From this analysis, we could show that 3AP-induced transcriptome changes in both IMR-32 and CLB-GA robustly overlap with those imposed upon COH29 exposure (Fig. 5, C and D).
Fig. 5. 3AP leads to dormant origin activation following replication fork stalling at early firing origins, as measured by transferase-activated end ligation sequencing.
(A) GSEA of RNA-seq–based transcriptome profiling using the C2 curated MSigDB gene sets for 3AP-treated IMR-32 and CLB-GA neuroblastoma cells. (B) GSEA shows a strongly significant overlap between up- and down-regulated gene signatures upon RRM2 knockdown and 3AP (IC50) treatment of IMR-32 and CLB-GA neuroblastoma cells. (C) GSEA shows a significant enrichment of publicly available transcriptome profiles of C4-2 prostate cancer cells upon exposure with the RRM2 inhibitor COH29 (10 μM) in the transcriptomes of IMR-32 neuroblastoma cells treated with the RRM2 inhibitor 3AP. (D) GSEA shows a significant enrichment of publicly available transcriptome profiles of C4-2 prostate cancer cells upon exposure with the RRM2 inhibitor COH29 (10 μM) in the transcriptomes of CLB-GA neuroblastoma cells treated with the RRM2 inhibitor 3AP. (E) TrAEL-seq read density and read polarity plots for IMR-32 cells treated for 24 hours with 3AP IC50 or DMSO alone. Read polarity was quantified by (R − F)/(R + F); data shown is an average of two biological replicates. Orange bars represent regions replication IZs called from DMSO-only control samples, and gray boxes represent early replicating regions based on published Repli-Seq data (56). (F) PCA for the TrAEL-seq libraries. (G to I) Violin plots of TrAEL-seq read count distributions (corrected for probe length) from DMSO- and 3AP-treated IMR-32 cells and solid and dotted lines denote median, upper quartile, and lower quartile, respectively. (G) Comparison of replication IZs to a set of 8760 random regions of equivalent average size. (H) Comparison of replication IZs that do or do not overlap with early replicating regions [defined in (E)]. (I) Read counts for early versus late replicating genomic regions defined on the basis of Repli-Seq data (56).
Pharmacological RRM2 inhibition imposes dormant origin activation following replication fork stalling at early firing origins
In addition, to monitor the impact of 3AP on individual replication forks by DNA combing, we applied the novel sequencing method “transferase-activated end ligation” sequencing (TrAEL-seq) (55) to establish the detailed genome-wide landscape of DNA replication changes imposed on neuroblastoma cells by 3AP exposure (IC50). Using this method on IMR-32 cells, we could measure replication fork stalling events as well as the impact on replication fork directionality and origin usage. In the “read density plot” for 3AP-treated cells, which shows the distribution of replication forks across the genome (Fig. 5E, panel 3), large and defined peaks appear in comparison to the relatively uniform profile of dimethyl sulfoxide (DMSO)–treated cells, indicative of increased replication fork stalling at these sites. Notably, these peaks line up with replication initiation zones (IZs) containing replication origins active in untreated cells, defined on the basis of the DMSO “read polarity plot” that displays the average direction of replication fork movement (Fig. 5E, panel 2; regions transitioning from negative to positive polarity), indicating that most forks initiated at normal S-phase IZs rapidly stall upon 3AP exposure. In addition, multiple dormant origins become active upon 3AP treatment (Fig. 5E, panel 4; regions transitioning from negative to positive polarity). The read polarity plot also shows that the replication profile of 3AP-treated cells becomes more disordered, as the reduced bias in read polarity results from the direction of replication fork movement at any given genomic location becoming less well defined (Fig. 5E). These substantial changes and reproducibility of the data could be shown by means of a principal components analysis (PCA) (Fig. 5F). Genome-wide, replication forks are uniformly distributed between IZs and random locations in DMSO-treated cells but accumulate at IZs and are depleted from random locations in 3AP-treated IMR-32 cells (Fig. 5G), with a particularly strong enrichment at early IZs (Fig. 5H) (56). Last, we observed an enrichment of 3AP-induced fork stalling at all early replicating regions and a depletion at late replicating regions relative to DMSO controls (Fig. 5I). Together, this shows that 3AP treatment induces significant stalling of replication forks initiated at commonly used and particularly early IZs, in keeping with the above-described cell cycle and gene expression data. Consequently, it appears that numerous typically dormant replication origins become activated, attempting to complete replication, resulting in the disordered replication profiles seen in 3AP-treated cells.
Combined pharmacological RRM2-CHK1 inhibition as a novel therapeutic strategy in high-risk neuroblastoma
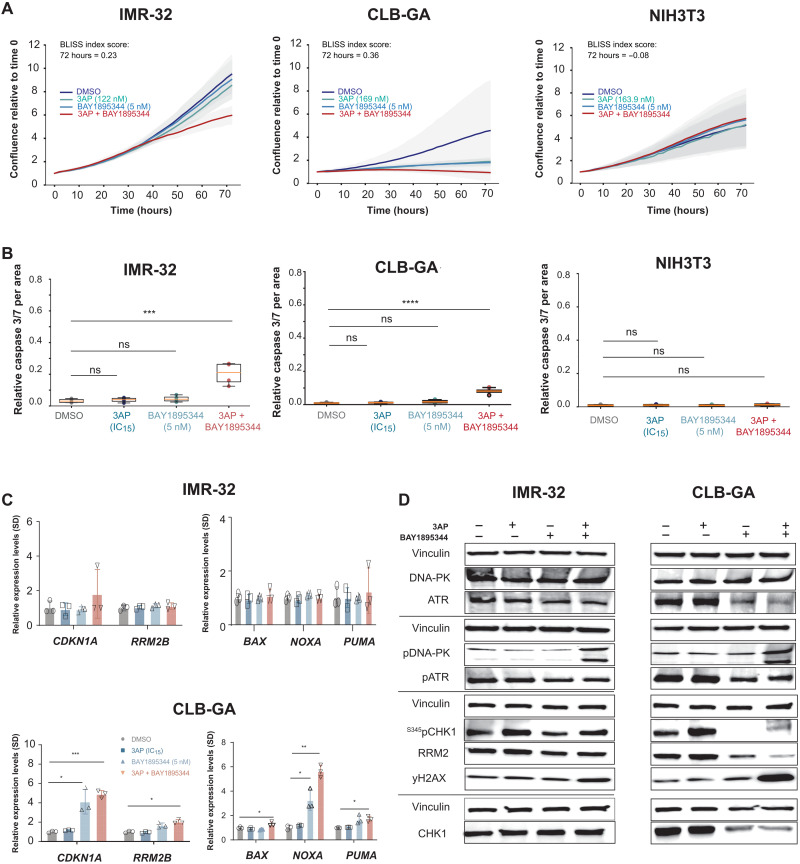
The ATR-CHK1 signaling axis plays a critical role on the control of replication fork stability and origin firing. CHK1 has been reported as a synthetic lethal target in MYCN-amplified neuroblastoma, and neuroblastoma was ranked as the most sensitive pediatric cancer for CHK1 inhibitor prexasertib (57). ATR inhibition has been shown to cause replication catastrophe under conditions of high replicative stress (58). Gemcitabine or HU has been used to enhance replicative stress and sensitize cells for CHK1 inhibition (59, 60). However, gemcitabine acts both directly on RRM1 (not RRM2) and through disruption of DNA synthesis through incorporation of dNTP analogs, while HU acts on RRM2 but only in the higher micromolar range. In view of this and the above data showing potent effects of 3AP, we selected this drug to further test its effect in combination with either ATR or CHK1 inhibitors. We first evaluated pharmacological ATR (BAY1895344) compared to CHK1 (prexasertib) inhibition in combination with 3AP at low-dose concentrations (IC15). Cell confluency was measured by means of IncuCyte live cell imaging for human IMR-32 and CLB-GA neuroblastoma cells under conditions of low single-dose RRM2 or ATR pharmacological inhibition in comparison to combined treatment. Drug synergism was observed at 72 hours after treatment with respective Bliss indices of 0.47 (IMR-32) and 0.36 (CLB-GA), concomitant with reduced cell confluence (Fig. 6A) and significant induction of apoptosis (Fig. 6B), the latter supported by induction of CDKN1A and PUMA gene expression in CLB-GA cells (Fig. 6C). Treatment of nonmalignant NIH3T3 fibroblast cells did not affect cell growth or survival, showing that neither cell confluence nor cell death was significantly changed in the combined treatment compared to single-agent or control treatment, indicating that the combination treatment can be tolerated by normal cells (Fig. 6, A and B). Increased apoptosis was also accompanied by up-regulated yH2AX levels (Fig. 6D and fig. S2C). Notably, the DNA-dependent protein kinase (DNA-PK) salvage pathway, as measured by pDNA-PK, was activated upon combined RRM2 and ATR inhibition (reducing p-ATR levels), in line with previous observations (40).
Fig. 6. Phenotypic effects of combined RRM2-ATR pharmacological inhibition are opposed through the activation of a DNA-PK salvage pathway.
(A) IncuCyte live cell imaging indicates a drug synergism between RRM2 and the ATR inhibitor BAY1895543 that could not be shown in normal fibroblast cells (NIH3T3). (B) Combined RRM2-ATR inhibition leads to a significant induction of apoptosis compared to control or single compound–treated neuroblastoma cells while leaving NIH3T3 cells unaffected. (C) RT-qPCR analysis for CDKN1A, RRM2B, NOXA, PUMA, and BAX in IMR-32 and CLB-GA neuroblastoma cells following control (DMSO), single (3AP or BAY1895543), or combined (3AP and BAY1895543) drug treatment. (D) Immunoblotting for various DNA damage markers in IMR-32 and CLB-GA cells upon treatment with DMSO, 3AP, or BAY1895344 as a single agent or combined treatment of 3AP with BAY1895344.
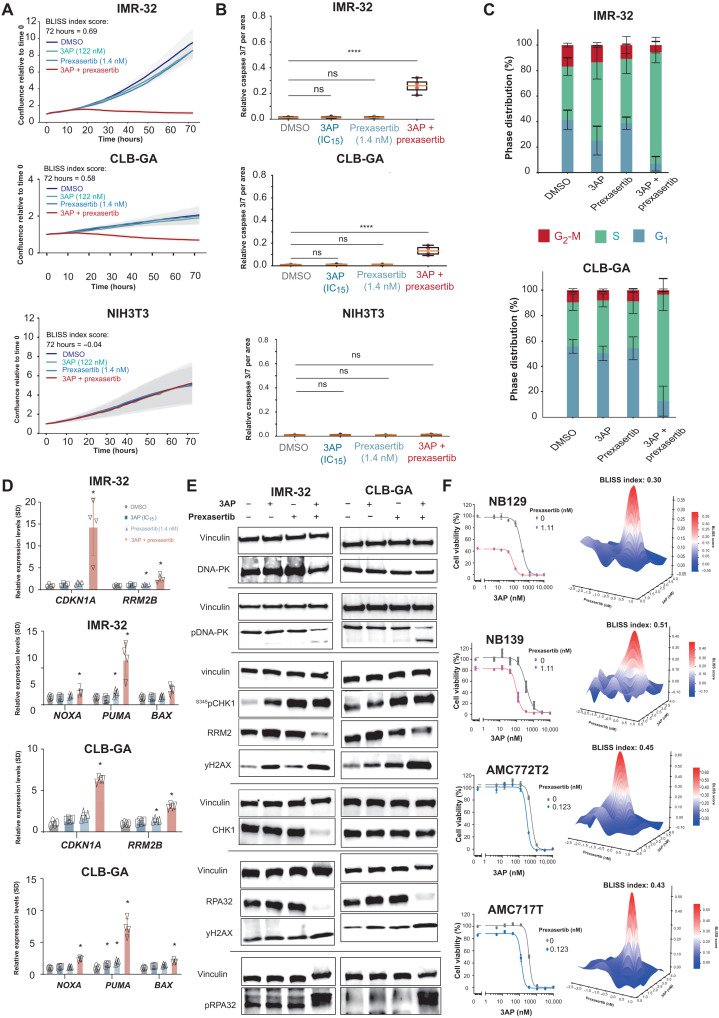
In a second step, using a similar approach, we dissected the phenotypic and molecular consequences of combined 3AP and prexasertib treatment. In comparison to combined RRM2 and ATR inhibition, higher BLISS indices were observed for this drug combination of 0.69 (IMR-32) and 0.58 (CLB-GA), respectively, and significant reduction of cell confluence (Fig. 7A). The drug combination also increased the caspase 3/7 signal (Fig. 7B) compared to single-drug and control-treated (DMSO) cells. In contrast, NIH3T3 fibroblast cells did not exhibit any measurable effects of a similar combined pharmacological RRM2-CHK1 inhibition (Fig. 7, A and B), thus suggesting low or no toxicity in normal cells. Reduced cell confluence was concomitant with a clear induction of an S phase arrest in the combination treatment versus controls (Fig. 7C). The induction of apoptosis was further confirmed by RT-qPCR analysis, indicating both p53 pathway activation (up-regulated CDKN1A and RRM2B expression) and increased expression of proapoptotic markers BAX, NOXA, and PUMA (Fig. 7D). Furthermore, 3AP-prexasertib synergism increased pRPA32 and yH2AX levels, and marked reduction in RRM2 protein levels was noted (Fig. 7E), suggesting that RRM2 could represent the target for synthetic lethal interaction. In contrast to combined RRM2-ATR pharmacological inhibition, we could no longer observe increased pDNA-PK levels upon combined RRM2-CHK1 inhibition, indicating that this drug combination can circumvent this rescue pathway while efficiently imposing DNA damage, as evidenced by up-regulated yH2AX levels (Fig. 7E and fig. S2D). Similar as for the 3AP single treatment, we also evaluated the phenotypic consequences of combined 3AP-prexasertib treatment in p53-mutant SK-N-BE(2)-C cells. Exposure of SK-N-BE(2)-C cells to 3AP-prexasertib combination treatment shows a synergistic effect on attenuated cell confluence, as measured by IncuCyte live cell imaging (fig. S3D), as well as cell death compared to control or (low-dose) single treatments (fig. S3E). Immunoblotting following combined 3AP-prexasertib treatment showed up-regulation of S345pCHK1 and yH2AX levels, together with reduced RRM2 and total CHK1 protein levels (fig. S3F), in line with the observations in IMR-32 and CLB-GA neuroblastoma cells. The effects of nucleotide inhibition by RNR inhibitors or RRM2 knockdown on CHK1 levels have been reported by several teams (61, 62). Particularly instructive is the recent work of Ohmura et al. in Ewing sarcoma (23), who demonstrated that, upon exposure to the gemcitabine RNR inhibitor, total CHK1 is down-regulated together with down-regulation of global protein levels. These authors also showed that, upon RRM2 knockdown, similar results are observed. Moreover, we could show that, upon combined 3AP-prexasertib treatment, S296pCHK1 autophosphorylation is effectively reduced, concomitant with an increase in S345pCHK1 signal (fig. S4A). It has been demonstrated that functional inhibition or knockdown of RRM2 directly affects protein translation through inhibition of the translational inhibitor 4E-BP1. We therefore also investigated whether the loss of the inhibitory phosphorylation of 4E-BP1 also occurred in neuroblastoma cells upon combined pharmacological RRM2-CHK1 inhibition. Our data indeed show a decrease in p4E-BP1 levels, which indicates activation of this translational inhibitor, in keeping with the observed reduction of both CHK1 and RRM2 protein levels (fig. S4B).
Fig. 7. Identification of 3AP-prexasertib as a synergistic drug combination in neuroblastoma.
(A) IncuCyte live cell imaging indicates a drug synergism between RRM2 and CHK1 pharmacological inhibition resulting in reduced cell confluence in IMR-32 and CLB-GA neuroblastoma cells, while not affecting NIH3T3 confluence. (B) Combined 3AP-prexasertib treatment of IMR-32 and CLB-GA neuroblastoma cells leads to a significant induction of apoptosis compared to a single compound treatment or DMSO-treated cells, while NIH3T3 cells did not show any apoptotic response. (C) Combined 3AP-prexasertib treatment of IMR-32 and CLB-GA neuroblastoma cells results in a strong S phase arrest compared to a single compound treatment or DMSO-treated cells. (D) RT-qPCR analysis for the p53 targets CDKN1A and RRM2B as well as the proapoptotic genes BAX, NOXA, and PUMA upon combined 3AP-prexasertib treatment. (E) Immunoblotting for various DNA damage markers in IMR-32 and CLB-GA cells upon treatment with DMSO or 3AP or prexasertib as a single agent or combined 3AP and prexasertib (see quantification in fig. S2). (F) 3AP-prexasertib combined treatment synergistically affected neuroblastoma spheroid cell viability 120 hours after treatment.
In a next step, we further explored the 3AP-prexasertib drug synergism using four different primary human neuroblastoma-derived 3D spheroid cultures, representative for three major adrenergic neuroblastoma subtypes: MYCN amplified (NB129; ALK mutated and AMC717T, ALK wild-type), MYCN nonamplified (NB139; ALK wild-type), and MYCN nonamplified and ATRX deleted (AMC772T2; ALK wild-type). The 3AP-prexasertib synergism could be achieved in all tested spheroids (Fig. 7F).
Combined 3AP-prexasertib drugging induces expression of the nucleotide stress–induced transcriptional regulator and tumor suppressor HEXIM1 (Hexamethylene Bis-Acetamide-Inducible Protein 1)
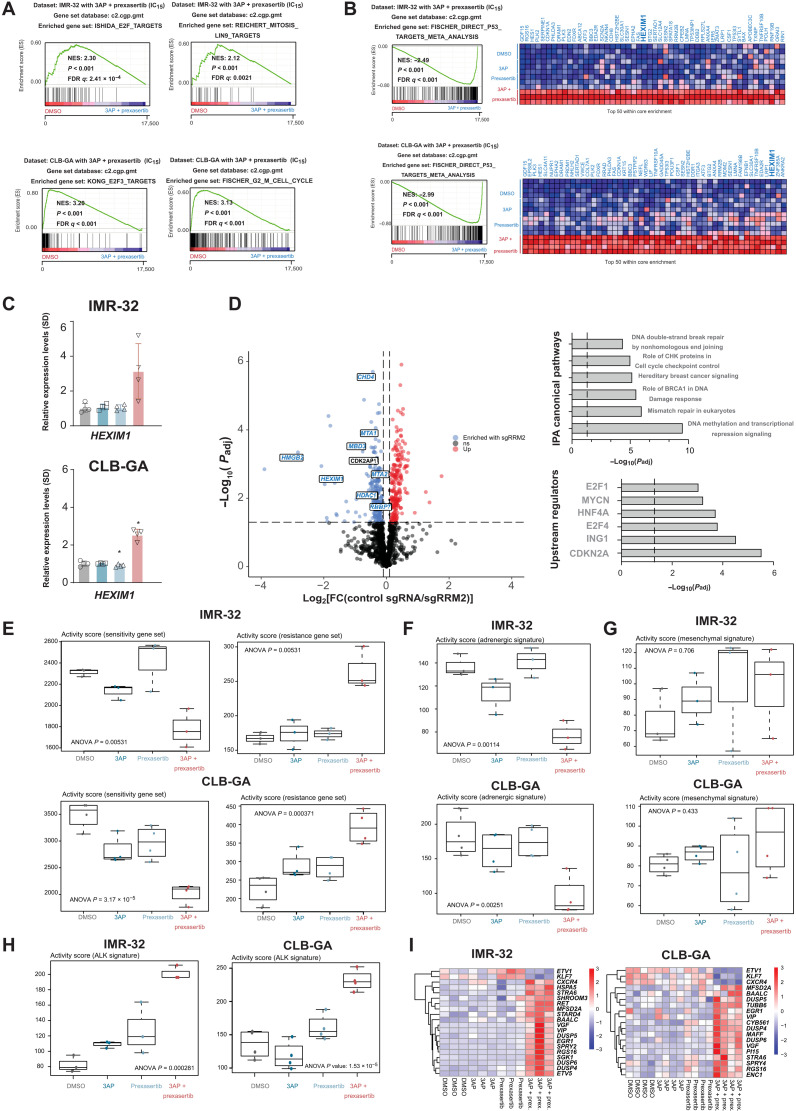
To gain further mechanistic insight into the observed drug synergism, we performed gene signature analysis following transcriptome profiling by RNA-seq. GSEA revealed a significant down-regulation of G2-M cell cycle genes and E2F targets (Fig. 8A). The set of up-regulated genes was also strongly enriched for p53 target genes and revealed that combined 3AP-prexasertib treatment significantly reduced the expression of various oncogenes with an established role in neuroblastoma, including TWIST1 (a direct MYCN target and interaction partner) (28, 63) and PBK (a converging target gene of LIN28B/let-7 and MYCN) together with a strong induction of expression of tumor suppressor HEXIM1, a negative regulator of the transcriptional regulator pTEFb (Fig. 8B). Transcriptional up-regulation of HEXIM1 was confirmed by RT-qPCR (Fig. 8C). To gain further insight into the upstream regulators of RRM2 expression, we performed an unbiased landscaping of RRM2 upstream regulators using CasID as a proximity-based labeling approach for RRM2 promotor interactome mapping (64). A total of four different single guide RNAs (sgRNAs) were designed that cover the RRM2 promotor in a tiling approach [300 base pairs (bp) upstream to transcription start site] versus a control sgRNA against lacZ. Following biotin-streptavidin affinity purification in SK-N-B(E)2-C cells over four biological replicates, high-confidence RRM2 promotor interactors [false discovery rate (FDR) ≤ 0.05] were identified (Fig. 8D and table S1), including HMGB2 and HEXIM1, as well as the NurD, PAF, and COMPASS chromatin modifier complexes. HEXIM1 is a negative regulator of the positive transcription elongation factor P-TEFb complex (consisting of CDK9 and cyclin T1). Recent data suggest that HEXIM1 could act as a tumor suppressor to block transcription under conditions of nucleotide stress by sequestration of P-TEFb (65). In addition, the P-TEFb component CDK9 itself was significantly enriched in the performed CasID experiment (FDR < 0.05). We also scrutinized this hit list using the “Ingenuity Pathway Analysis” (IPA) tool, indicating a clear enrichment of DNA damage and checkpoint proteins (Fig. 8D). Although we could not detect MYCN itself in our assay, IPA analysis indicated, among others, MYCN, E2F4, and CDKN2A/p16 as putative key upstream regulators of the enriched RRM2 regulator pool, and publicly available MYCN ChIP-seq data in neuroblastoma cell lines showed direct MYCN binding on the RRM2 promotor region (Fig. 1E). Next, we also evaluated the activity scores of various publicly available gene signatures in the transcriptome profiles obtained in IMR-32 and CLB-GA cells upon exposure to single and combined 3AP/prexasertib: (i) Scoring of prexasertib response gene sets established by Blosser et al. (41) showed a significant down-regulation of genes that are negatively correlated to prexasertib sensitivity and a significant up-regulation of genes that are correlated to prexasertib resistance (Fig. 8E); (ii) the adrenergic neuroblastoma gene set established by Van Groningen et al. (66) was significantly down-regulated upon combined 3AP-prexasertib treatment both in IMR-32 and CLB-GA cells, while the mesenchymal signature was not significantly altered (Fig. 8, F and G); and (iii) we determined the activity score of AKL (Anaplastic Lymphoma Receptor Tyrosine Kinase) signaling and observed that the set of genes activated downstream of the ALK receptor (67) was significantly up-regulated upon exposure of IMR-32 and CLB-GA cells to combined 3AP-prexasertib treatment compared to single-agent and control (DMSO) treatment (Fig. 8, H and I).
Fig. 8. Scrutinizing putative RRM2 upstream regulators by CasID.
(A) GSEA of transcriptome data generated following combined 3AP-prexasertib treatment of IMR-32 and CLB-GA neuroblastoma cells shows significantly reduced expression of E2F and G2-M cell cycle–controlled genes compared to control (DMSO)– or single compound–treated cells. (B) GSEA of transcriptome data generated following combined 3AP-prexasertib treatment of IMR-32 and CLB-GA neuroblastoma cells shows significantly up-regulated expression of p53 target genes compared to control (DMSO)– or single compound–treated cells. (C) RT-qPCR confirms significantly up-regulated HEXIM1 expression upon combined 3AP and prexasertib treatment compared to control treatment (DMSO). (D) Left: Volcano plot of significantly enriched hits from a proximity-based and biotin-dependent CasID approach for the identification of RRM2 upstream regulatory factors in SK-N-BE(2)-C cells (FDR < 0.05). Right: Ingenuity Pathway Analysis (IPA) for the identification of enriched pathways and putative upstream regulators of the putative RRM2 regulators as identified by CasID (overview of all hits can be found in table S1). (E) Boxplots depicting the activity score in IMR-32 and CLB-GA cells of a prexasertib “sensitivity” (left) and “resistance” (right) gene signature as defined in neuroblastoma patient-derived xenografts (PDX) (65). (F) Boxplots depicting the activity score in IMR-32 and CLB-GA cells of an adrenergic gene signature (66). (G) Boxplots depicting the activity score in IMR-32 and CLB-GA cells of a mesenchymal gene signature (66). (H) Boxplots depicting the activity score in IMR-32 and CLB-GA cells of an ALK signaling signature (67). (I) Heatmaps depicting the significantly differentially expressed genes of the ALK signaling signature as scored in (H).
In vivo validation of synergistic RRM2-CHK1 inhibition
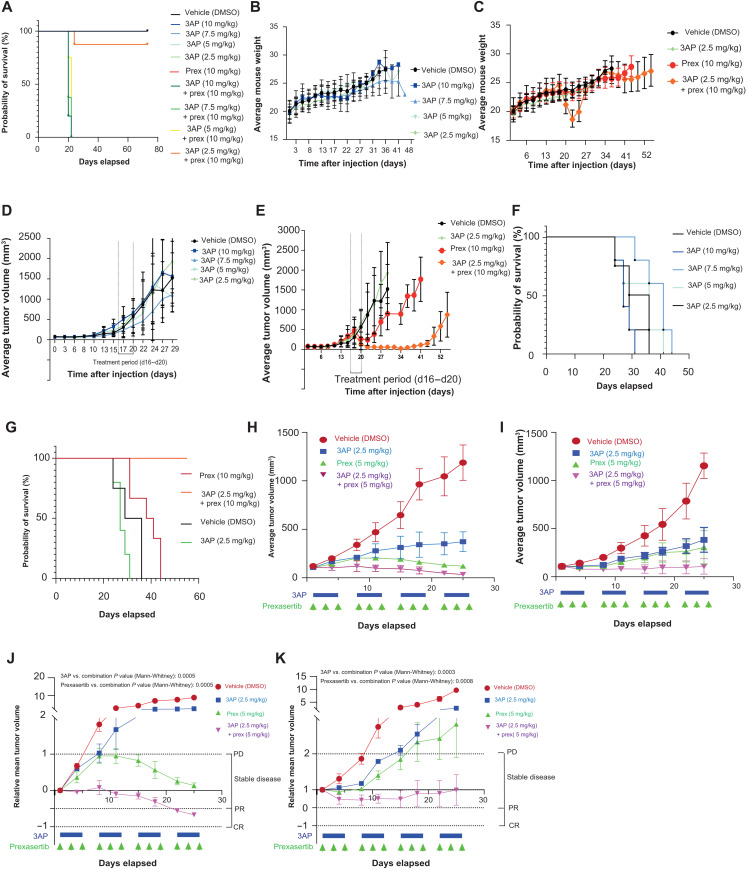
Next, we aimed to validate the observed in vitro synergism between 3AP and prexasertib using an in vivo murine xenograft model in immunodeficient mice. Here, subcutaneous xenografted tumors from IMR-32 neuroblastoma cells were treated with vehicle, 3AP only (10, 7.5, 5, or 2.5 mg/kg), prexasertib only (10 mg/kg), or a combination. Mice treated with 3AP or prexasertib monotherapy displayed no obvious signs of toxicity based on their body weight and animal behavior (Fig. 9, A and B). However, in the combination therapy, mice treated with higher 3AP doses (5, 7.5, or 10 mg/kg) and prexasertib (10 mg/kg) suffered from severe drug toxicities (Fig. 9A) and were euthanized before the end of the treatment regime as they reached the preset humane end points of this in vivo study. Mice treated with the lowest 3AP dose (2.5 mg/kg) in combination with prexasertib (10 mg/kg) also presented mild (seven of eight) to severe (one of eight) signs of toxicity, including a significant drop in body weight at the end of the treatment regime (P = 0.004) (Fig. 9C). Upon treatment discontinuation, these mice regained body weight and fully recovered.
Fig. 9. In vivo validation of 3AP-prexasertib synergism.
(A) Survival probabilities were measured over time of control-treated, 3AP single compound–treated, and prexasertib-treated mice and mice treated with different concentration combinations of 3AP and prexasertib. Statistical analyses were performed using the log-rank (Mantel-Cox) test. (B) Time course analysis of the average mouse weight per 3AP treatment group included in this murine cell line xenograft study. (C) Time course analysis of the average mouse weight per 3AP, prexasertib, and combination treatment groups included in this murine cell line xenograft study. (D) Average tumor volume (TV) of the different 3AP treatment groups included in this murine cell line xenograft study. (E) Average TV of the 3AP, prexasertib, or combination treatment group included in this murine cell line xenograft study. (F) Time course analysis of the survival probabilities of the different 3AP treatment groups included in this murine cell line xenograft study. (G) Time course analysis of the survival probabilities of the 3AP, prexassertib, and combination treatment group included in the experiment. (H) Average TV of the 3AP, prexasertib, or combination treatment group included for the treatment schedule of a MYNC-nonamplified (p53 wild type) neuroblastoma PDX model. (I) Average TV of the 3AP, prexasertib, or combination treatment group included for the treatment schedule of a MYNC-amplified (p53 wild type, ALK R1275Q mutant) neuroblastoma PDX model. (J) Relative mean TV of the 3AP, prexasertib, or combination treatment group included for the treatment schedule of a MYNC-nonamplified (p53 wild type) neuroblastoma PDX model. (K) Relative mean TV of the 3AP, prexasertib, or combination treatment group included for the treatment schedule of a MYNC-amplified (p53 wild type, ALK R1275Q mutant) neuroblastoma PDX model. CR, complete response; PR, partial response; PD, progressive disease.
Next, we evaluated the effects of the treatment on the tumor progression. Mice treated with a dose range of 3AP monotherapy displayed little or no effect on the in vivo tumor progression rate compared to the vehicle-treated group (Fig. 9D). Single treatment with prexasertib (10 mg/kg) could slow down tumor growth (P = 0.04; Fig. 9E). Our results show that combined 3AP (2.5 mg/kg) and prexasertib (10 mg/kg) were strongly synergistic and could completely halt neuroblastoma tumor development in vivo (Fig. 9E). Again, a strong significant difference could be observed at day 24 (4 days after the treatment was stopped) between vehicle and combined 3AP (2.5 mg/kg) with prexasertib (10 mg/kg)–treated mice (P = 0.004). These observations were further confirmed in terms of progression-free survival of the different treatment groups included in this study (Fig. 9, F and G). No statistical difference (log-rank Mantel-Cox test) in tumor size could be observed between vehicle- and 3AP-treated mice for all tested 3AP concentrations including 10 mg/kg (P = 0.90), 7.5 mg/kg (P = 0.07), 5 mg/kg (P = 0.59), and 2.5 mg/kg (P = 0.18), while only slight significance was noted for the prexasertib-treated group (P = 0.05) (Fig. 9, F and G). In contrast, a clear significant difference (log-rank Mantel-Cox test) in tumor size–based survival could be shown between vehicle-treated mice and those that received the combination treatment of 3AP (2.5 mg/kg) and prexasertib (10 mg/kg) (P = 0.0007).
Last, we also evaluated 3AP-prexasertib synergism in patient-derived xenograft (PDX) models. First, three different concentration combinations of 3AP and prexasertib [3AP (2.5 mg/kg) and prexasertib (10 mg/kg), 3AP (2.5 mg/kg) and prexasertib (5 mg/kg), and 3AP (2 mg/kg) and prexasertib (5 mg/kg)] were evaluated in nontumor nonobese diabetic (NOD) severe combined immunodeficient (SCID) gamma (NSG) mice to evaluate toxicity effects. No mortality was observed for any of the dose levels tested. However, significant weight loss was observed in treated animals at the highest dose level [i.e., 3AP (2.5 mg/kg) + prexasertib (10 mg/kg)], and treatment was only tolerated for 1 week because of severe weight loss and poor clinical appearance. In contrast, animals treated with lower doses [i.e., 3AP (2.5 mg/kg) + prexasertib (5 mg/kg) and 3AP (2 mg/kg) + prexasertib (5 mg/kg)] had few to no dosing holidays and were tolerated for nearly two treatment cycles (fig. S5A). In addition, serial blood cell count measurements were performed, which show treatment-associated anemia and thrombocytopenia, but with consequent recovery (fig. S5B). On the basis of the tolerability data, the middle dosing [i.e., 3AP (2.5 mg/kg) + prexasertib (5 mg/kg)] was used for subsequent therapeutic study in which we evaluated responses in two PDX models [one MYCN nonamplified/p53 wild type and one MYCN amplified/p53 wild type/ALK mutant (R1275Q)] over a treatment period of 4 weeks [3AP (2.5 mg/kg) and prexasertib (5 mg/kg)]. For both PDX models, we could achieve significant differences when comparing the combination response to the other arms, both in terms of average (Fig. 9, H and I) and relative mean tumor volumes (Fig. 9, J and K). However, given that only three mice per treatment group were included, further experiments are warranted to learn whether the combination treatment is truly synergistic in these models compared to the single-treatment arms.
DISCUSSION
High-risk neuroblastoma genomes are predominantly marked by a landscape of highly recurrent large segmental and focal DNA copy imbalances, while mutations are sparse. Multiple loci located on the short arm of chromosome 2 frequently undergo segmental gains or chromothripsis events, causing focal high-level amplifications of which MYCN amplification is the most frequently altered target. Here, we identified RRM2 as a previously unidentified copy number–driven dependency gene, which is strongly associated with poor prognosis, up-regulated during MYCN-driven mouse neuroblastoma formation and causing increased penetrance and accelerated neuroblastoma formation under forced combined overexpression with MYCN in the developing zebrafish sympathetic neuronal lineage. In vitro knock down experiments and pharmacological inhibition of RRM2 showed strong dependency in adrenergic neuroblastoma cell lines and neuroblastoma patient tumor-derived spheroid cultures.
How do we explain the RRM2 dependency in neuroblastoma cells? First, high MYCN/MYC activity causes weakened G1-S control and activates dormant origins of replication, causing enhanced proliferation but at the cost of increased demand for nucleotides (among others), which causes enhanced replicative stress levels and ATR-CHK1 activation. Second, RRM2 is cell cycle–regulated, and while induced during G1-S transition, RRM2 levels are still low during early S phase. In view of this, ATR-CHK1 activation is critical in suppressing origin firing and enhancing RRM2 levels through control of CDK2-mediated E2F degradation during early S phase. In this context, the previously reported synthetic lethality for CHK1 in MYCN-amplified neuroblastomas (57) is not unexpected and further supported by the highest CHK1 inhibitor sensitivity for neuroblastoma across a wide range of tumor entities. Further evidence that elevated RRM2 levels facilitate replication stress resistance in neuroblastoma cells comes from the data from Blosser et al. (41), showing that high RRM2 expression is associated with enhanced sensitivity to the prexasertib CHK1 inhibitor, thus indicative of high ATR-CHK1 activity. Forced RRM2 overexpression in MYCN-nonamplified CLB-GA neuroblastoma cells suppresses excessive prexasertib-induced replicative stress and further supports the role of elevated RRM2 expression for suppression of excessive replicative stress. Moreover, GSEA for the differentially expressed genes for double MYCN-RRM2 versus MYCN-only transgenic zebrafish neuroblastomas also reveals enhanced ATR-CHK1 signaling activity in keeping with increased pCHK1 and yH2AX in MYCN- versus MYCN/RRM2-overexpressing zebrafish neuroblastomas. This dataset also revealed possible involvement of cilia genes and their master regulator FOXJ1. However, the exact significance of this intriguing finding remains to be unraveled by further studies.
Elevated MYC/MYCN activity is also considered to induce transcriptional amplification of actively transcribed loci, which may cause so-called transcription-dependent replicative stress because of the formation of paused transcriptional complexes and R loops, which pose a physical and topological challenge to the DNA replication machinery and subsequent slowing down and stalling of DNA replication forks (68). Evidence has emerged that several factors are regulated by MYC/MYCN that alleviate replicative stress. Among others, the work of Herold et al. (36) revealed a pronounced role for AURKA (Aurora Kinase A) in suppressing R loop formation and a novel role for BRCA1 (located on 17q and also often affected by gains) in limiting MYCN-driven accumulation of stalled RNAP (36). Further studies are warranted to investigate the role of ATR-CHK1 control of RRM2 levels in relation to transcription-replication conflicts and R loop suppression.
While monotherapies for RRM2 inhibition have not led to durable responses on tumor growth inhibition, previous effects of gemcitabine, which inhibits DNA replication through RRM1 inhibition and incorporation of toxic nucleotide analogs, showed the potential for CHK1 sensitization. Given the previously established strong dependency of neuroblastoma for CHK1-mediated checkpoint control and, thus far, poorly explored potential for synergistic drugging, we therefore decided to evaluate the use of RRM2 inhibition to enhance CHK1 sensitivity. We observed a strong synergism between the combined inhibition of RRM2 and CHK1, using 3AP and prexasertib, respectively. Both in a cell line xenograft and several initially treated PDXs, sensitivity was noted at the initially used high concentrations. In the cell line xenograft, a short period of 4 days of treatment already resulted in a complete regression of the engrafted tumor, both for prexasertib monotherapy and the 3AP-prexasertib combination. Upon drug withdrawal, tumors treated with “prexasertib only” rapidly recurred, while mice remained tumor free for almost 40 days in the combination treatment group, indicating the potential value of 3AP-prexasertib combination to treat patients with high-risk neuroblastoma. Next, we also evaluated the response to the 3AP-prexasertib combination in PDXs (MYCN nonamplified or MYCN amplified with ALK mutation) and observed similar strong growth reduction.
Using a combined transcriptome and proteome approach, we also sought for mechanistic insights into the underlying molecular mechanism driving the synthetic lethal drug interaction. In response to combined RRM2 and CHK1 inhibition, we consistently observed strong transcriptional HEXIM1 induction. HEXIM1 was reported to respond to nucleotide stress and acts as a negative regulator of transcription elongation regulator at oncogenic loci and stabilizes mRNAs of tumor suppressor genes in melanoma (69). Of further importance, we used programmable DNA binding of dCas9 with the promiscuous biotin ligase BirA* (CasID) for proximity biotinylation of proteins (table S1), and we identified HEXIM1 as one of the RRM2 promotor–bound proteins that, together with ATR-induced E2F1 degradation, may further attenuate RRM2 transcription. In addition, several epigenetic regulatory protein complexes were identified at the RRM2 promotor, including NurD, PAF, and COMPASS chromatin factors. Furthermore, CHD5 is encoded on the commonly deleted 1p36 chromosomal region in neuroblastoma and known to replace CHD4 in NurD complexes during neuronal differentiation (70), suggesting that tight RRM2 regulation may also be an important factor in normal differentiation of sympathoblasts. In addition, HMGB2 was one of the top enriched factors in this assay, recently described as a master regulator of the chromatin landscape during senescence (71), with loss of its nuclear expression being instructive to CTCF (CCCTC-binding factor) clustering (72), the latter also strongly enriched in our CasID assay.
RRM2 expression and function is tightly regulated by various factors. Several of these regulators are also affected by copy number changes, further suggesting that these highly recurrent chromosomal and focal genomic imbalances exert effects that support tumor initiation and/or maintenance. To the best of our knowledge, RRM2 is the first copy number–driven dependency gene for which direct impact of transcriptional up-regulation has been successfully modeled in the zebrafish MYCN-driven neuroblastoma model. We are currently also generating mouse and zebrafish models overexpressing RRM2-dTAG to allow to monitor in vivo the effect of RRM2 protein degradation on tumor maintenance as a prelude for previously unknown protein-degrading drugging approaches. At present, it is difficult to irrefutably prove the impact of a copy number gain for a given gene on neuroblastoma formation and behavior and, even more so, to establish the biological (and possible epistatic) effects of multiple genes implicated in large chromosomal gains or losses. One approach that has been tested is CRISPR-based chromosomal deletions in mouse neural crest cells before MYCN overexpression–mediated transformation to neuroblasts and injection in mice (73).
In conclusion, our results converge toward a key RRM2 dependency in neuroblastoma cells by RRM2-controlled modulation of checkpoint integrity and replication fork stability in response to MYCN-induced replicative stress. Co-overexpression of RRM2 induced enhanced neuroblastoma formation and increased tumor penetrance in the MYCN-driven zebrafish model. We provide evidence that elevated RRM2 levels facilitate neuroblastoma cells to cope with replicative stress along with evidence for enhanced activation of ATR-CHK1 response. We present preclinical evidence that selective and/or combinatorial targeting of the RRM2 axis opens perspectives for potent and tolerable previously unidentified targeted drug combinations for the clinic. The emerging role of RRM2 and CHK1 dependency in other tumor entities including Ewing sarcoma and glioblastoma may warrant to broaden clinical trial efforts to evaluate the potential of the proposed combination therapy. Given the critical role of RRM2 in high-risk neuroblastoma, we also hope that our work will trigger further investigation toward novel pharmacological compounds, such as protein degraders to target RRM2 in combination with previously unknown replication stress checkpoint signaling drugs.
MATERIALS AND METHODS
Cell culture
Human neuroblastoma cell lines SK-N-AS, SH-SY5Y, SK-N-BE(2)-C, IMR-32, CLB-GA, NB-1, SH-EP, and SK-N-FI were grown in RPMI 1640 supplemented with 10% fetal calf serum (FCS), penicillin/streptomycin (100 IU/ml), and 2 mM l-glutamine. NIH3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS, 10 nM β-mercaptoethanol, penicillin/streptomycin (100 IU/ml), and 1 mM nonessential amino acid. All cell lines used were cultured in 5% CO2 atmosphere at 37°C on plastic cultured plates.
Patient-derived neuroblastoma tumor organoids AMC717T (74) were grown in DMEM-GlutaMAX containing low glucose and supplemented with 25% (v/v) Ham’s F-12 nutrient mixture, B27 supplement minus vitamin A, penicillin (100 IU/ml), streptomycin (100 μg/ml), epidermal growth factor (EGF) (20 ng/ml), and fibroblast growth factor-basic (FGF-2) (40 ng/ml). Patient-derived neuroblastoma tumor organoids (NB129 and NB139) and AMC772T2 were grown in DMEM-GlutaMAX containing low glucose and supplemented with 20% (v/v) Ham’s F-12 nutrient mixture, B27 supplement minus vitamin A, N-2 supplement, penicillin (100 IU/ml), streptomycin (100 μg/ml), EGF (20 ng/ml), FGF-2 (40 ng/ml), insulin-like growth factor 1 (IGF-1) (200 ng/ml), platelet-derived growth factor–AA (PDGF-AA) (10 ng/ml), and PDGF-BB (10 ng/ml). EGF, FGF-2, PDGF-AA, and PDGF-BB were obtained from PeproTech, and IGF-1 was obtained from R&D Systems. B27 minus vitamin A and N-2 supplements were obtained from Thermo Fisher Scientific. The origin of the cell lines is provided in table S2, and all cell culture–related materials are provided on table S3.
Inducible RRM2 overexpression cell line
The RRM2 fragment was amplified by PCR (forward and reverse primer sequences are provided on table S6), and OriGene clone GC-Z9335-GS was used as a template. The obtained fragment was gel-purified and ligated into the opened Mlu I/Nde I sites of response vector pLVX-TRE3G-Zsgreen1 producing pLVX-TRE3G-Zsgreen1-IRES-RRM2. The sequence of the constructed plasmid was verified by Sanger DNA sequencing (GATC).
Lenti-X 293T cells were transfected with the regulator vector pLVX-pEF1a-Tet3G and Lenti-X Packaging Single Shots (VSV-G) according to the manufacturer’s instructions. The supernatant containing the lentivirus was collected, filtered through a 0.45-μm filter, and concentrated using PEG-it. CLB-GA cells were infected with the concentrated virus. After 48 hours of incubation, the transduced clones were obtained by limiting dilution. After clonal expansion, the TAT protein expression in each clone was checked by immunoblotting using TetR monoclonal antibody (clone 9G9). In addition, induction of each expressing clone was tested after transduction with the pLVX-TRE3G-Luc control vector. Selected clones were transduced with the lentivirus produced, as described above, from vector pLVX-TRE3G-Zsgreen1-IRES-hsRRM2 and subsequently selected with only 4 μg/mL of puromycin.
Compounds and chemicals
3AP, gemcitabine, and HU were obtained from Sigma-Aldrich; prexasertib and BAY1895344 were obtained from Bio-Connect.
Cell viability measurements for a single compound treatment
The adherent cell lines were plated in 96-well plates at a density of 2 × 103 to 1.5 × 104 cells per well, depending on the cell line. Cells were allowed to adhere overnight, after which different compounds—3AP, HU, and gemcitabine—were added in a range of concentrations. Cytotoxicity assays were performed at 48 and 72 hours after treatment with CellTiter-Glo reagent. In addition, the apoptosis levels were measured, at 72 hours after treatment, using the Caspase-Glo 3/7 Assay System. Both protocols were adapted, adding 50 μl of reagent for each assay. The results were normalized to vehicle (0.1% DMSO), and the different inhibitory concentration values and AUC were computed using GraphPad Prism Software (version 9.2). The dose-response curve analysis was performed through ECanything equation assuming a standard slope of −1.0. For the caspase analysis, each caspase signal was normalized to the area of occupancy, given by the IncuCyte Software. The error bars in figures represent the SD from three biological replicates.
Combination and synergism measurements
To find synergism, cells were seeded in 384-well plates at a density of 1.5 × 103 to 2 × 103 cells per well, depending on the cell line. Cells were allowed to adhere overnight, after which these were exposed in a range of concentration of different compounds, alone or in a combination matrix or in fixed combination (3AP, prexasertib, and BAY1895344). The treatment was performed using a D300 TECAN instrument. Cell proliferation was monitored for 72 hours, in which pictures were taken through IncuCyte Live Cell Imaging System. Each image was analyzed through the IncuCyte Software. Cell proliferation was monitored by analyzing the occupied area (percentage of confluence) of cell images over time. The synergism was computed according to the Bliss independent (BI) method (75) using the HTSplotter tool (75). Once the combinations with the highest BI score were selected, the adherent cell lines were plated in 96-well plates at a density of 2 × 103 to 1.5 × 104 cells per well, depending on the cell line. These ones were allowed to adhere overnight, following their exposure to a determined concentration of each different compound as mentioned above. The proliferation was monitored for 72 hours by the same system, as well as the analyses of each image. From the same plate, once the latest time point was scanned by the IncuCyte software, the apoptosis levels were measured using the Caspase-Glo 3/7 Assay; however, this protocol was adapted by adding 50 μl of reagent for each assay. The caspase analysis was performed as mentioned above. Proliferation plots were generated using our recently published HTSplotter tool (76), in which the SD from three or more biological replicates is represented as error bars. As for the caspase assay, the data was mean-centered and autoscaled.
Organoid cell viability screening
Patient-derived neuroblastoma tumor organoids were harvested using Accutase solution (Sigma-Aldrich), made single cell, filtered using a 70-μm nylon cell strainer (Falcon), and resuspended in an appropriate growth medium. Subsequently, cells were plated at densities ranging from 1000 to 6000 cells per well using the Multi-drop Combi Reagent Dispenser on repellent black 384-well plates (Corning). Following 24 hours of recovery time, cells were treated with 0 to 10 nM prexasertib and/or 0 to 10 μM 3AP or DMSO (negative control) using the Tecan D300e Digital Dispense (HP). Two technical replicates were included in each experiment, and two biological replicates were completed for each patient-derived neuroblastoma tumor organoid. After 5 days of treatment, adenosine triphosphate levels were measured using CellTiter-Glo 3D (Promega) according to the manufacturer’s instructions. The results were normalized to vehicle (0.1% DMSO), and data were analyzed with GraphPad Prism v7.04.
siRNA-mediated knockdown of RRM2
NB cells were transfected using the appropriate Neo kit (catalog no. MPK10096) with siRNA toward RRM2 [s12361 (siRRM2-61) and s12362 (siRRM2-62)] (Ambion, Life Technologies) and or scrambled siRNA (Ambion, #AM4635). After transfection, the cells of each condition were split and seeded in a 96-well plate, at a density of 1.5 × 104 to 3 × 104 cells per well, and in a T-25, at a density of 2.2 × 106 cells per flask. The cells seeded in a 96-well plate were monitored for proliferation during 72 hours in which pictures were taken through IncuCyte. Proliferation was analyzed as mentioned above. From the same plate, once the latest time point was scanned in IncuCyte, the apoptosis levels were measured using the Caspase-Glo 3/7 Assay System. The protocol was adapted, adding 50 μl of reagent for each assay. The caspase analysis was performed as mentioned above, and the HTSplotter tool (75) was used to generate the proliferation plots. The error bars from the proliferation assay represents the SD from three or more biological replicates, while the ones showed on the caspase assay were mean-centered and autoscaled. The cells seeded in a T-25 flask were collected for RNA and protein isolation, 72 hours after transfection. The knockdown was evaluated by RT-qPCR and immunoblotting.
DNA combing
Exponentially expanding neuroblastoma cells were pulse-labeled for 20 min with 25 μM thymidine analog 5-iodo-2′-deoxyuridine. Cells were washed with warm medium and pulse-labeled a second time for 20 min with 5-chloro-2′-deoxyuridine, and, depending on the condition, 500 μM HU and 500 or 250 nM 3AP were combined. Cells were refreshed three times with warm medium and harvested by trypsinization. The cell pellet was washed with ice-cold phosphate-buffered saline (PBS); cells were resuspended at a cell density of 1 × 106 cells/ml and placed on ice. In total, 2 μl of the cell suspension was spotted at one end of a glass slide. When the drop became opaque, 7 μl of the lysis buffer [50 mM EDTA, 200 mM tris (pH 7.4), and 0.6% SDS] was added. After 7 min of incubation, tilting the slide allowed the spreading of the DNA fibers. The air-dried slides were immersed in methanol/acetic acid (3:1), dried, and stored at −20°C until immunofluorescence staining. DNA fibers were acid-treated with 2.5 M HCl for 80 min, blocked in 5% bovine serum albumin (BSA) in PBS with Tween 20 and immunolabeled overnight at 4°C with mouse anti–5-bromo-2′-deoxyuridine (BrdU) B44 (1:100; BD347580) and rat anti-BrdU Bu1/75 (1:150; Ab6326). The secondary antibodies were goat anti-mouse AF647 (1:100; Life Technologies, A21241) and goat anti-rat AF488 (1:100; Life Technologies, A11006). Incubation time was 1 hour at room temperature. The slides were rinsed with PBS followed by an alcohol series (70 to 95% ethanol), dried, and mounted with 1% propyl gallate as an antifading reagent. Imaging was done on a Zeiss Axio Observer.Z1 epifluorescence microscope equipped with a Plan-Apochromat 63×/1.40 Oil DIC M27 lens and connected with an Axiocam 506 mono camera. The length of the fiber tracks was converted from pixels to micrometers and measured with Fiji (ImageJ) software. The measured fibers were further randomly selected the usingrandom.choice function from Python (version 3.9).
Cell culture for fluorescence-activated cell sorting, RNA, and protein collection
The neuroblastoma cell lines were seeded in a T-75 flask at a density of 2 × 106 to 2.25 × 106 cells per flask, depending on the cell line. As for the inducible CLB-GA RRM2 overexpressing cell line, doxycyclin was added at 1 μg/ml immediately upon seeding. Cells were allowed to adhere for 48 hours, after which the medium was replaced by fresh medium and the treatment was added. Upon the selected treatment time points, the cells were scraped and centrifuged for 5 min at 1200 rpm, and the pellet was washed twice with ice-cold PBS. During each wash, the cells were pelleted during 5 min at 1200 rpm. The samples were divided for RNA and protein isolation and/or for flow cytometric analysis for cell cycle measurements.
dNTP pool assay
Neuroblastoma cells were seeded in a six-well plate at a density of 3.0 × 105 cells per well, depending on the cell line. Cells were allowed to adhere overnight, after which the medium was replaced by fresh medium and treatment was added. After 48 hours of treatment, the medium of all conditions were collected for further analyses. The plates were put on ice, in which each well was washed with 1 ml of ice-cold washing solution (0.9% NaCl). Next, 300 μl of ice-cold extraction buffer was added and incubated for 2 to 3 min on ice. Then, the cells were scraped and collected.
Next, 20 μl of each sample was loaded into the Dionex UltiMate 3000 LC System (Thermo Fisher Scientific, Bremen, Germany) equipped with a C-18 column (Acquity UPLC-HSS T3, 1.8 μm; 2.1 mm by 150 mm; Waters) coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in negative ion mode. A step gradient was carried out using solvent A [10 mM tetrabutylammonium hydroxide (TBA) and 15 mM acetic acid] and solvent B (100% methanol). The gradient started with 5% of solvent B and 95% of solvent A and remained at 5% B until 2 min after injection. A linear gradient to 37% B was carried out until 7 min and increased to 41% until 14 min. Between 14 and 26 min, the gradient increased to 95% B and remained at 95% B for 4 min. At 30 min, the gradient returned to 5% B. The chromatography was stopped at 40 min. The flow was kept constant at 0.25 ml/min at the column that was placed at 40°C throughout the analysis. The mass spectrometry operated in full scan mode (mass/charge ratio range: [400,000 - 600,000]) using a pray voltage of 4.80 kV, capillary temperature of 300°C, sheath gas at 40.0, and auxiliary gas at 10.0. The AGC (Automatic Gain Control) target was set at 1.0E+006 using a resolution of 35,000, with a maximum IT (injection time) fill time of 200 ms. Data collection was performed using the Xcalibur software (Thermo Fisher Scientific). The data analyses were performed by integrating the peak areas (El-Maven Polly-Elucidata). This experiment was performed once with three technical replicates.
Cell culture for fluorescence-activated cell sorting, RNA, and protein collection
The neuroblastoma cell lines were seeded in a T-75 flask at a density of 2 × 106 to 2.25 × 106 cells per flask, depending on the cell line. Cells were allowed to adhere for 48 hours, after which the medium was replaced by fresh medium and the treatment was added. After 48 hours of treatment, the cells were scraped and centrifuged for 5 min at 1200 rpm, and the pellet was washed twice with ice-cold PBS. During each wash, the cells were pelleted during 5 min at 1200 rpm. The samples were divided for RNA and protein isolation and for flow cytometric analysis of cell cycle.
Cell cycle analysis
Cell pellet was fixed in cold 70% ethanol for at least 1 hour. After fixation, cells were centrifuged for 5 min at 1200 rpm. The pellet was washed twice in 1 ml of PBS. During each wash, the cells were pelleted during 5 min at 1200 rpm. Then, the pellet was resuspended, and ribonuclease A (RNase A) was added to a final concentration of 0.2 mg/ml in PBS. The samples were incubated for 1 hour at 37°C. Last, 59.8 μM propidium iodide (PI) was added to the solution. Samples were analyzed in PI/RNase A solution by Bio-Rad S3 fluorescence-activated cell sorting (FACS) cell sorter flow cytometer. All data were further analyzed by FlowJo software, following the cell cycle instructions.
Western blot analysis
Cells were lysed in cold radioimmunoprecipitation assay (RIPA) buffer [12.7 mM; 150 mM NaCl, 50 mM tris-HCl (pH 7.5), 0.01% SDS solution, and 0.1% NP-40] supplemented with protease and phosphatase inhibitors. Samples were rotated for 1 hour at 4°C to obtain more complete lysis. The cleared lysates were collected and centrifuged at 10,000 rpm in a microcentrifuge for 10 min at 4°C. Protein concentrations were determined using the Pierce BCA Protein Assay Kit. The lysates were denaturized before loading on a gel, through five times Laemli denaturation buffer supplemented with β-mercaptoethanol. Thirty micrograms of protein extracts was loaded on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels with 10× tris/glycine/SDS buffer and run for 1 hour at 130 V. Samples were blotted on nitrocellulose or polyvinylidene difluoride (PVDF) membranes in 10% of 10× tris/glycine buffer and 20% of methanol. The membranes were blocked during 1 hour in 5% milk or 5% BSA in Tris-buffered saline with 0.1% Tween® 20 detergent (TBST). Primary antibody incubations were done in blocking buffer overnight at 4°C. Blots were washed three times with TBST before the incubation for 1 hour of secondary antibodies. The immunoblots were visualized by using the enhanced chemiluminescent Femto (Bio-Rad). The protein quantification analysis of the generated blots was performed through ImageJ software, where the area from each protein was normalized to the loading protein in respect to each blot. Antibodies used were the following: RRM2, pCHK1(Ser345), pCHK1(Ser296), CHK1, pRPA32, RPA32, yH2AX, vinculin, tubulin, ATR, p-ATR, 4E-BP1 (53H11), and p-4E-BP1(Thr37/46). The material origin used for Western blot and the antibodies are provided on tables S4 and S5, respectively.
RNA isolation complementary DNA synthesis and real-time qPCR
RNA extraction was performed, practicing the manufacturer’s instructions of miRNeasy kit (QIAGEN) including on-column deoxyribonuclease treatment, and NanoDrop (Thermo Fisher Scientific) was used to determine the concentration. Complementary DNA (cDNA) synthesis was achieved, practicing the iScript Advanced cDNA synthesis kit instructions from Bio-Rad. The PCR mix contained 5 ng of cDNA, 2.5 μl of SsoAdvanced SYBR qPCR super mix (Bio-Rad), and 0.25 μl of forward and reverse primers (to a final concentration of 250 nM; Integrated DNA Technologies). The RT-qPCR cycling analysis was performed using a LC-480 device (Roche). qBasePlus software 3.2 (www.biogazelle.com) was used for the analysis of the gene expression levels. For the neuroblastoma cell lines, the following reference genes were used: B2M, HPRT1, TBP, and YHWAZ. The error bars in figures represent SD after error propagation, with mean centering and scaling to control. The primer designs are provided on table S6.
CasID
To identify the putative upstream regulators of RRM2 binding at its promotor region, we exploited the CasID technology. Briefly, SK-N-BE(2)-C cells were transduced with lentiviral constructs for stable expression of the different tested RRM2 promotor–targeting sgRNAs (Addgene) (MP-I-1142 sg86RRM2, MP-I-1143 sg150RRM2, MP-I-1144 sg403RRM2, and MP-I-1145 sg685RRM2) versus an sgRNA-targeting LacZ as control (MP-I-1104 sgLacZ). Infections were done with titers corresponding to a final multiplicity of infection of 15. Cells were seeded at a density of 7.5 × 106 cells 24 hours after infection and incubated with doxycycline (48 hours) and 50 μM biotin (18 to 24 hours) (1 μg/ml) and subsequently harvested. Harvested cells were washed twice with PBS and collected by scraping in urea lysis buffer [50 mM Hepes (pH 8) and 9 M urea]. The obtained lysates were cleared by centrifugation. To the supernatant, 1/278 volume of 1.25 M dithiothreitol was added and incubated for 30 min at 55°C. Next, 1/10 volume of iodoacetamide solution was added and incubated at room temperature for 15 min. Next, the sample is fourfold diluted with 20 mM Hepes (pH 8.0) to a final concentration of 2 M urea. Subsequently, 30 μl of prewashed GE Streptavidin Sepharose High Performance bead suspension was added to each sample and incubated for 2 hours with agitation at room temperature. Beads were washed three times with 20 mM Hepes (pH 8.0) + 2 M urea and resuspended in resuspend beads in 20 μl of 20 mM Hepes (pH 8.0) + 2 M urea. In a next step, 0.4 μg of LysC (Wako) was added to the beads/proteins [assume 100 μg; 1:250 (w/w)] and digested in an incubator, for 4 hours at 37°C. Then, 1 μg of trypsin (Promega) was added to the beads/proteins [assume 100 μg; 1:100 (w/w)] and digested in an incubator overnight at 37°C, and beads were removed by centrifugation at 500g for 2 min. Add trifluoroacetic acid to the digest for a final pH of 2 to 3. After acidification, precipitate was allowed to form by letting it stand for 15 min on ice. The acidified peptide solution was centrifuged for 15 min at full speed (room temperature) to remove the precipitate and was analyzed on a Q-HF standard gradient. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE repository with the dataset identifier PXD029630.
RNA-seq processing and GSEA
RNA-seq was carried out for siRNA-mediated knockdown of RRM2 in IMR-32 and CLB-GA cells, 3AP treatment (48 hours) with two different concentrations (IC30 and IC50; table S7) for IMR-32 and CLB-GA cells and combined 3AP-prexasertib treatment (48 hours) for IMR-32 and CLB-GA cells. Libraries for mRNA-seq were prepared using the QuantSeq 3′ mRNA library prep kit (Lexogen, Vienna, Austria) with umi barcoding according to the manufacturer’s protocol. The libraries were equimolarly pooled and sequenced on an Illumina NextSeq 500 high-throughput flow cell, generating single-end 75-bp reads. Sequencing quality was confirmed using FastQC (version 0.11.7). Trimmed reads were aligned to the human reference genome using the STAR alignment software. The resulting mapped files were inspected for quality using Qualimap BamQC. Feature counts were used to infer gene-level expressions from the mapped data, and expression levels per sample were aggregated using custom Python scripts. All subsequent analyses were conducted in R statistical language. BioMartR was used for obtaining gene annotation and references from BioMart. DESeq2 was used for differential gene expression analysis; data were additionally normalized using a regularized log transformation (rlog2) followed by log FC shrinkage to account for variability at extreme ranges (lfcShrink). As apparent batch effects between cell lines were observed in the data, Limma’s “batchEffectCorrection” method was used to account for batch effect to deeper investigate the distinct conditions. GSEA was done using either the c2 gene set collection from MSigDB (gsea-msigdb.org) or gene lists compiled from this study. All RNA-seq data are available through the Gene Expression Omnibus (GEO) repository (GSE161902).
TrAEL-seq library preparation and data processing
IMR-32 cells were seeded in a T-25 (2.2 × 106 cells per flask) and allowed to adhere for 48 hours, after which the medium was replaced by a fresh one with either 0.1% of DMSO (vehicle) or 3AP treatment (689.5 nM). After 24 hours of treatment, the supernatant of each flask was collected and added to the trypsinized cells and centrifuged (5 min at 1200 rpm), and the pellet was washed with ice-cold PBS. Cells were collected, 1 × 106 per condition, and washed with L buffer [100 mM EDTA (pH 8), 10 mM tris (pH 7.5), and 20 mM NaCl] and lastly resuspended in 60 μl of the buffer. The cell suspension was warmed for at least 2 min at 50°C before adding to 40 μl of melted and warm CleanCut agarose. The mixture was vortexed for 5 s and directly pipetted into a plug mold.
Agarose-embedded DNA from IMR-32 cells was generated and processed into libraries as previously described (55), and libraries were sequenced on an Illumina NextSeq 500 as high-output 75-bp single end by the Babraham Institute Next-Generation Sequencing facility.
Scripts used for unique molecular identifier (UMI) handling and more detailed information on data processing are available at https://github.com/FelixKrueger/TrAEL-seq and described in detail in (55). TrAEL-seq read structure is NNNNNNNN(T)nSEQUENCESPECIFIC, where NNNNNNNN represents the UMI and (T)n represents poly(T). Before TrAEL-seq preprocessing, sequences were deduplicated on the basis of the first 23 bp on their 5′ end (using the script TrAELseq_sequence_based_deduplication.py). This region contains both the UMI sequence and the first 15 bp of genomic sequence and identifies (and removes) PCR-amplified multicopy sequences that would, under normal conditions, survive the UMI-aware deduplication procedure by aligning to several different genomic regions at random. The script TrAELseq_preprocessing.py was then used to remove and store the first 8 bp (UMI). Up to 3 T (inclusive) at the start of the sequence were removed, and reads were subjected to adapter and quality trimming using Trim Galore (v0.6.5; default parameters; https://github.com/FelixKrueger/TrimGalore). UMI-preprocessed and adapter-/quality-trimmed files were then aligned to GRCh38 with Bowtie2 (v2.4.1; option: --local; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) using local alignments. Last, alignment results files were deduplicated using UmiBam (v0.2.0; https://github.com/FelixKrueger/Umi-Grinder). This procedure deduplicates alignments on the basis of the mapping position, read orientation, and the UMI sequence. Deduplicated mapped reads were imported into SeqMonk v1.47 (www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and immediately truncated to one nucleotide at the 5′ end, representing the last nucleotide 5′ of the DNA end. Reads were then summed in 200-kb running windows spaced every 10 kb. Read counts were normalized to reads per million mapped, exported from SeqMonk, and plotted in GraphPad Prism 8. Read polarity values were calculated and plotted for each quantification window using the formula read polarity = (R − F)/(R + F), where F and R relate to the total forward and reverse read counts, respectively. The R code to generate these plots can also be found here: https://github.com/FelixKrueger/TrAEL-seq. To determine regions where replication initiates, the ForkCall program was used using the read polarity data (the code for ForkCall can be found here: https://github.com/s-andrews/forkcall). The read polarity signal from 200-kb running windows tiled every 10 kb along the genome was smoothed using LOWESS regression to remove local noise. Filters were applied to remove poorly covered or particularly short areas, and remaining regions where the directionality index consistently increases were called as replication IZs and imported into SeqMonk v1.47. Early and late replicating regions were defined on the basis of Repli-Seq data from HCT116 cells (GSE137764) (56). Gaussian smoothed and normalized data from BrdU-labeled reads from the first three time points (S1 to S3; early replicating) and last three time points (S14 to S16; late replicating) in S phase were totaled. The 10% of regions with the highest values were imported into SeqMonk v1.47 and used to define replication timing. All sequencing data are available through the GEO repository (GSE186122, SuperSeries GSE161902).
Cell line xenograft
All animal experiments were performed according to the Guide for the Care and Use of Laboratory Animals (Eight Edition) following approval of the Committee on the Ethics of Animal Experiments of Ghent University (permit number: ECD 20/55). Persons who carried out the described experiment received appropriate training in animal care and handling. The mice were allowed to acclimatize for 1 week before the experimental procedure started and were randomly assigned to the different treatment groups. IMR-32 cells were cultured as described above to investigate the synergistic effect of the combinatorial 3AP and prexasertib treatment in vivo. A total of 2 × 106 cells were mixed with Matrigel (354230, Corning) and subsequently injected subcutaneously in the right flank of 5-week-old female Crl:NU-Foxn1nu nude mice (strain code 088, Charles River Laboratories). Compound treatment was started when tumors reached 300 mm3. For 3AP, a dose range of 10, 7.5, 5, and 2.5 mg/kg was tested, while the dose of prexasertib was fixed to 10 mg/kg. Treatment was continued for five consecutive days for 3AP (intraperitoneally; 10% DMSO in sterile H2O, twice a day with a minimum of 8 hours in between treatments) and three consecutive days for prexasertib (subcutaneously; 20% Captisol in sterile PBS, twice a day). The control group received both vehicles. Following treatment discontinuation, animals were observed for another 6 to 7 weeks to determine complete or partial response (PR) to the compound treatment (i.e., regrowth of tumors). TV was assessed using a caliper and calculated according to the spheroid formula: V = 0.5 × a × b2, with a as the largest and b as the smallest superficial perpendicular diameter. To control for systemic toxicity during the treatment period, the body weight and physical status of the animals were monitored by animal caretakers until they were judged to be in discomfort. When systemic toxicity (weight gain/loss of 20% or more) or maximum TV (2000 mm3) was reached, the animal was euthanized by cervical dislocation.
Patient-derived xenograft
Patient tumor tissue used to generate a PDX was obtained under the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board–approved protocols #17-387 and #06-107. Tumors were implanted in the subcutaneous flank of NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson Laboratory). Mice were monitored at least weekly until signs of tumor engraftment were observed. Tumor measurements were performed using manual calipers, and treatment was initiated when TV was ~100 mm3 (TV = 1/2 length × width2). Animals were assigned to four treatment arms through block randomization and included vehicle (30% polyethylene glycol, molecular weight 400/5% Tween 80/10% n-methyl-2-pyrrolidone in 5% dextrose water), prexasertib [5 mg/kg, intraperitoneally (IP); BID Monday/Wednesday/Friday], 3AP (2.5 mg/kg, IP; BID (twice a day) 5 days on/2 days off), and combination at equivalent single-agent doses for prexasertib and 3AP. Animals were treated for 4 weeks, with tumors measured twice weekly. Treatment cohorts were compared using Mann-Whitney-Wilcoxon test. Time to progression was estimated using a Kaplan-Meier estimator and compared using a log-rank test. Progressive disease was defined as doubling of TV relative to baseline, stable disease is <100% TV relative to baseline and <50% TV regression, PR is >50% TV regression, and complete response (CR) is >95% TV regression or no measurable tumor. Disease control rate is defined as the sum of stable disease + PR + CR. All animal studies were conducted under barrier conditions, and experiments were performed using protocol and conditions approved by the MSKCC Institutional Animal Care and Use Committee under protocol 16-08-011.
Zebrafish modeling
Zebrafish maintenance and generation of a stable line
Zebrafish were housed in a Zebtec semiclosed recirculation housing system (Techniplast, Italy) and maintained following standard procedures. All zebrafish studies and maintenance of the animals were in accordance with protocol #17/100, approved by the Ethics Committee for the use of animals in research of the Faculty of Medicine and Health Sciences at Ghent University. The dβh:hRRM2 DNA construct for the Tg(dβh:hRRM2; dβh:mCherry) transgenic line was generated using multisite Gateway cloning by combining three entry clones: dbh-pDONRP4-P1R (gift from the Look laboratory), RRM2-pDONR221 (GeneCopoeia, #GC-Z9335-GS) (see also table S8), and p3E-polyA (gift from the Look laboratory) into a modified destination vector containing I-SceI cleavage sites (gift from the Look laboratory) (8).
Generation of stable Tg(dβh:hRRM2; dβh:mCherry) transgenic fish
One-cell-stage wild-type zebrafish embryos were injected with mix containing dβh:RRM2 (12 ng/μl), dβh:mCherry (20 ng/μl) (gift from the Look laboratory), I-SceI enzyme (0.5 U/μl), and CutSmart buffer (0.5 U/μl). The injected fish were raised to adulthood and crossed to screen for founder fish. The offspring of the founder fish were raised, and DNA and RNA samples were collected to confirm the presence of the dβh-hRMM2 construct in the genome and expression of hRRM2 mRNA, respectively. For analysis, the following primer pairs were used: CCAACAGAAGTGGACCAACA (forward) and GGCAGCTGCTTTAGTTTTCG (reverse). A 472-bp fragment of the dβh:RRM2 transgene fragment was amplified and further confirmed by Fragment Analyzer (Applied Biosystems). RT-qCPR primers were the same as used for the in vitro experiments. The Tg(dβh:eGFP-MYCN) line was a gift from the Look laboratory.
Generation of mosaic dβh:MYCN;RRM2 fish
The dβh:RRM2 DNA construct for the mosaic injections was generated using the same multisite Gateway cloning strategy, but using a destination vector containing Tol2 transposase sites as well as a cmlc2-GFP sequence (gift from the Langenau laboratory). Each time, 20 ng/μl was injected together with transposon mRNA (35 ng/μl). Tumor watch was executed in the same way as done for the stable transgenic lines.
Screening and sample collection
From 5 weeks on, fish were screened every 2 weeks for neuroblastoma development by the use of a fluorescent microscope (Nikon, SMZ18) using the NIS Elements software. Statistical analysis was performed using GraphPad Prism software version 5.0 (La Jolla, CA). The method of Kaplan and Meier was used to estimate the rate of tumor development. Fish that died without evidence of EGFP+ or mCherry+ masses were censored. The log-rank test was used to assess differences in the cumulative frequency of neuroblastoma between MYCN-only transgenic fish and MYCN;RRM2 transgenic fish.
Western blot analysis
Cells were lysed using a tissue lyser and cold RIPA buffer [12.7 mM; 150 mM NaCl, 50 mM tris-HCl (pH 7.5), 0.01% SDS solution, and 0.1% NP-40] supplemented with protease and phosphatase inhibitors. Samples were rotated for 1 hour at 4°C to obtain more complete lysis. The cleared lysates were collected and centrifuged at 10,000 rpm in a microcentrifuge for 10 min at 4°C. Protein concentrations were determined using the Pierce BCA Protein Assay Kit. The lysates were denaturized before loading on a gel, through five times Laemli denaturation buffer supplemented with β-mercaptoethanol. Thirty micrograms of protein extracts was loaded on 10% SDS-PAGE gels with 10× tris/glycine/SDS buffer and run for 1 hour at 130 V. Samples were blotted on nitrocellulose or PVDF membranes in 10% of 10× tris/glycine buffer and 20% of methanol. The membranes were blocked during 1 hour in 5% milk or 5% BSA in TBST. Primary antibody incubations were done in blocking buffer overnight at 4°C. Blots were washed three times with TBST before the incubation for 1 hour of secondary antibodies. The immunoblots were visualized by using the enhanced chemiluminescent Femto (Bio-Rad). The protein quantification analysis of the generated blots was performed through ImageJ software, where the area from each protein was normalized to the loading protein in respect to each blot. Antibodies used were the following: pCHK1(Ser345), yH2AX, and vinculin. The material origin used for Western blot and the antibodies are provided on tables S4 and S5, respectively.
Bulk RNA-seq after FACS of the zebrafish lines
A pool of three to five 6-week-old fish per sample (for a total of three samples of MYCN;RRM2 and five samples of MYCN fish) was euthanized using an overdose of MS-222. After mechanical dissociation in resuspension solution (0.9× PBS and 10% FCS) using a scalpel, the cell suspension was passed through a 40μm mesh filter before cell sorting. The cell suspension was analyzed by imaging flow cytometry in collaboration with the Flow Cytometry Core Facility (VIB, Ghent University, Belgium). Forward and side scatters were used to gate for live, single cells, while sorting itself was based on enhanced GFP (eGFP)–positive cells and could also be evaluated through imaging on the sorter. Images were captured using a BD Biosciences FACS imaging–enabled prototype cell sorter that is equipped with an optical module, allowing multicolor fluorescence imaging of fast flowing cells in a stream enabled by BD CellView Image Technology based on fluorescence imaging using radiofrequency-tagged emission (77). No other markers were used. The sorted cells were collected in lysis buffer (RTL) of the RNeasy plus micro kit (QIAGEN, 74,034), supplemented with 2-mercaptoethanol (10 μl/1 ml of RTL, as recommended). The cells were kept on ice/cooled in the sorter during the whole procedure. Further RNA isolation was done using the RNeasy plus micro kit (QIAGEN, 74,034). Integrity and concentration of the RNA was evaluated using the Fragment Analyzer (Advanced Analytical Technologies) with the High Sensitivity RNA analysis kit (DNF-472-0500). The ProSize software version 3.0.1.5 was used to determine the RQN (RNA quality number) RNA integrity score considering the entire electropherogram. Full-length cDNA synthesis and amplification were done using the SMART-Seq v4 Ultra Low Input RNA Kit (Takara Biosystems, 634,890). An input of 1 ng of SMART-Seq v4 amplified cDNA was used for Nextera XT DNA library prep (Illumina, FC-131-1024), and the libraries were sequenced using a NextSeq 500 (Illumina). Quality control on fastq files was performed with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to Danio rerio reference genome GRCz10 with STAR v2.4.2a using a two-pass strategy. Genes were quantified using the Danio_rerio.GRCz10.91.gtf transcriptome. The DESeq2 R package was used for count normalization and differential gene expression analysis. The normalized read counts were used to generate PCA plots, heatmaps, and the correlation matrix. Pearson’s correlation coefficient was calculated on log-transformed normalized read counts. Preranked GSEA was preformed using GenePattern 2.0 (www.genepattern.org).
Immunohistochemistry analysis
For immunohistochemistry analysis, fish were euthanized with an overdose of MS-222, and their belly was cut open with a scalpel to increase impregnation. They were fixed overnight using modified Davidson fixation buffer (for 100 ml: 22 ml of 37% formaldehyde, 12 ml of glacial acetic acid, 33 ml of 95% EtOH, and 33 ml of distilled water). The fish were subsequently emerged overnight in 10% neutral buffered formalin. After fixation, the fish were decalcified with citric acid for several hours. Thereafter, the fish were dehydrated and brought to paraffin (70% EtOH overnight, 2 hours of 90% EtOH, 3× 1 hour of 100% EtOH, 3× 1 hour of xylene, and 2× 2 hours of paraffin at 65°C). Sagittal sections were made, and standard H&E staining was performed next to immunohistochemistry staining with the primary antibodies MYCN (Santa Cruz Biotechnology, sc-53993), TH (Pel-Freez Biologicals, P40101), and GFP (Cell Signaling Technology, #2956).
Quantification and statistical analyses
Expression datasets available in R2 (https://r2.amc.nl) were downloaded, and Pearson correlation from SciPy (version 1.6.2) from Python (version 3.9) was applied to evaluate the correlation between the selected genes. In vitro quantitative data were presented as means and SD from at least more than three biological replicates. Paired two-tailed Student’s t test was applied to the qPCR data using Microsoft Excel. As for the caspase 3/7 activity, the statistical analysis was applied to mean-centered and autoscaled data. As to determine the overall P value, Kruskal-Wallis test was applied, followed by a post hoc comparison by Dunn’s test. The analysis was done on Python (version 3.9) using SciPy (version 1.6.2) and scikit-posthocs library (version 0.6.7). One-way analysis of variance (ANOVA) and Holm-Šídák method for multiple comparison testing were applied to DNA combing fibers using GraphPad Prism 9.2.0 software.
For the in vivo xenograft, the Mantel-Cox method was applied for the survival analysis using GraphPad Prism 9.2.0 software. For all the analyses, the P value lower than 0.05 was considered as statistically significant. For the significance, P values lower than 0.05 were represented as “*,” P values lower than 0.01 as “**,” P values lower than 0.001 as “***,” and, lastly, P values lower than 0.0001 as “****.”
Acknowledgments
Funding: This work was supported by Research Foundation–Flanders 1197617N (to C.N.); Research Foundation–Flanders 11M1422N (to M.R.); Research Foundation–Flanders 3F014219 (to A.L.); Research Foundation–Flanders 1238420 (to B.D.); Bijzonder Onderzoeksfonds–ZAP (Ghent University) 202011/GE/ZAP/018 (to K.D.); Kom op Tegen Kanker (KOTK): “Preclinical evaluation of combined RRM2/DNA damage response signaling as therapy for high-risk neuroblastoma and entry point for sensitization to immunce checkpoint inhibition” (to F.S.); Stichting tegen Kanker (STK): “Novel combination therapies to combat therapy resistant neuroblastoma: Translating preclinical investigations to clinical trials” (to F.S.); Fight Kids Cancer: Replicative stress resistance modeling and therapeutic targeting in neuroblastoma (to F.S.); Innovative Training Networks (ITN): Validation of actionable genomic aberrations in a pediatric oncology network for doctorate students (to F.S.); and Olivia Hendrickx Research Fund (to F.S.). This research was funded, in part, through the “Geconcenteerde onderzoeksactie” (GOA), Ghent University: Replication fork protector dependency factors as novel targets for combination treatment and immunomodulation in neuroblastoma (BOF22/GOA/009) (to F.S. and S.E.). This research was funded, in part, through the GOA, Ghent University: Long non-coding RNAs in cancer: Deciphering the functional role of the dark matter of the human cancer genome (BOF16/GOA/023) (to F.S. and S.E.); Villa Joep (to F.S.); Wellcome Trust (110216) (to J.H. and A.W.); BBSRC (BI epigenetics ISP: BBS/E/B/000C0423) (to J.H.); MRC (iCASE studentship) (to N.K.); Artios Pharma (to N.K.); and UKRI-BBSCR (core capability grant) (to S.R.A.). This research was funded, in part, through NIH/NCI Cancer Center Support Grant P30 CA008748 (to S.S.R.). L.Del was funded by FWO (1S11817N) UGent BOF-GOA (BOF16/GOA/023). M.F. was supported by grants of the Deutsche Forschungsgemeinschaft (DFG; grant no. FI 1926/2-1 and as part of the SFB 1399) and the Förderverein für Krebskranke Kinder e.V. Köln (endowed chair).
Author contributions: Conceptualization: C.N., L.D., S.S.R., B.D.W., K.D., and F.S. Methodology: C.N., L.Dep., L.M., K.M.K., L.Del., F.D.V., C.T.C., S.D.C., E.D., C.B., F.D.V., E.S., A.E., S.-L.B., J.W.B., G.D., S.L., G.V.I., and B.G. Investigation: C.N., L.D., L.M., A.L., M.R., A.W., N.K., C.T.C., S.D.C., F.D., D.Y., A.S., E.D., S.L., L.T., F.V.N., and B.G. Visualization: C.N., L.Dep., K.M.K., L.Del., A.W., S.R.A., F.D., C.B., S.-L.B., W.V.L., B.D., L.T., C.V.N., and S.G. Supervision: C.N., L.D., V.V., S.G., S.E., K.D.P., M.F., J.H., J.M., B.D.W., S.S.R., K.D., and F.S. Writing—Original draft: C.N., L.Dep., L.M., L.Del., A.W., F.D., E.D., F.S., and K.D. Writing—Review and editing: C.N., L.D., B.D., K.D.P., M.F., J.H., J.M., B.D.W., S.S.R., K.D., and F.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. https://hgserver2.amc.nl/cgi-bin/r2/main.cgi?open_page=login:All data presented from primary neuroblastoma patient cohorts were extracted from the publicly available database at https://hgserver2.amc.nl/cgi-bin/r2/main.cgi?open_page=login. https://hgserver2.amc.nl/cgi-bin/r2/main.cgi?open_page=login:Publicly available MYCN ChIP-seq data were extracted from the database at https://hgserver2.amc.nl/cgi-bin/r2/main.cgi?open_page=login. https://hgserver2.amc.nl/cgi-bin/r2/main.cgi?open_page=login:RNA-seq and TrAEL-seq data generated for this manuscript are available through the GEO database through the SuperSeries with accession number GSE161902.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S1 to S9
REFERENCES AND NOTES
- 1.Jansky S., Sharma A. K., Korber V., Quintero A., Toprak U. H., Wecht E. M., Gartlgruber M., Greco A., Chomsky E., Grunewald T. G. P., Henrich K. O., Tanay A., Herrmann C., Hofer T., Westermann F., Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 53, 683–693 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Kameneva P., Artemov A. V., Kastriti M. E., Faure L., Olsen T. K., Otte J., Erickson A., Semsch B., Andersson E. R., Ratz M., Frisen J., Tischler A. S., de Krijger R. R., Bouderlique T., Akkuratova N., Vorontsova M., Gusev O., Fried K., Sundstrom E., Mei S., Kogner P., Baryawno N., Kharchenko P. V., Adameyko I., Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin. Nat. Genet. 53, 694–706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peifer M., Hertwig F., Roels F., Dreidax D., Gartlgruber M., Menon R., Krämer A., Roncaioli J. L., Sand F., Heuckmann J. M., Ikram F., Schmidt R., Ackermann S., Engesser A., Kahlert Y., Vogel W., Altmüller J., Nürnberg P., Thierry-Mieg J., Thierry-Mieg D., Mariappan A., Heynck S., Mariotti E., Henrich K.-O., Gloeckner C., Bosco G., Leuschner I., Schweiger M. R., Savelyeva L., Watkins S. C., Shao C., Bell E., Höfer T., Achter V., Lang U., Theissen J., Volland R., Saadati M., Eggert A., de Wilde B., Berthold F., Peng Z., Zhao C., Shi L., Ortmann M., Büttner R., Perner S., Hero B., Schramm A., Schulte J. H., Herrmann C., O’Sullivan R. J., Westermann F., Thomas R. K., Fischer M., Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526, 700–704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decaesteker B., Durinck K., Van Roy N., De Wilde B., Van Neste C., Van Haver S., Roberts S., De Preter K., Vermeirssen V., Speleman F., From DNA copy number gains and tumor dependencies to novel therapeutic targets for high-risk neuroblastoma. J. Pers. Med. 11, 1286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koche R. P., Rodriguez-Fos E., Helmsauer K., Burkert M., MacArthur I. C., Maag J., Chamorro R., Munoz-Perez N., Puiggròs M., Garcia H. D., Bei Y., Röefzaad C., Bardinet V., Szymansky A., Winkler A., Thole T., Timme N., Kasack K., Fuchs S., Klironomos F., Thiessen N., Blanc E., Schmelz K., Künkele A., Hundsdörfer P., Rosswog C., Theissen J., Beule D., Deubzer H., Sauer S., Toedling J., Fischer M., Hertwig F., Schwarz R. F., Eggert A., Torrents D., Schulte J. H., Henssen A. G., Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 52, 29–34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogarty M. D., Norris M. D., Davis K., Liu X., Evageliou N. F., Hayes C. S., Pawel B., Guo R., Zhao H., Sekyere E., Keating J., Thomas W., Cheng N. C., Murray J., Smith J., Sutton R., Venn N., London W. B., Buxton A., Gilmour S. K., Marshall G. M., Haber M., ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 68, 9735–9745 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosse Y. P., Laudenslager M., Longo L., Cole K. A., Wood A., Attiyeh E. F., Laquaglia M. J., Sennett R., Lynch J. E., Perri P., Laureys G., Speleman F., Kim C., Hou C., Hakonarson H., Torkamani A., Schork N. J., Brodeur G. M., Tonini G. P., Rappaport E., Devoto M., Maris J. M., Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S., Lee J. S., Guo F., Shin J., Perez-Atayde A. R., Kutok J. L., Rodig S. J., Neuberg D. S., Helman D., Feng H., Stewart R. A., Wang W., George R. E., Kanki J. P., Look A. T., Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. Decaesteker, A. Louwagie, S. Loontiens, F. De Vloed, J. Roels, S. Vanhauwaert, S. De Brouwer, E. Sanders, G. Denecker, E. D’haene, S. Van Haver, W. Van Loocke, J. Van Dorpe, D. Creytens, N. Van Roy, T. Pieters, C. Van Neste, M. Fischer, P. Van Vlierberghe, S. S. Roberts, J. Schulte, S. Ek, R. Versteeg, J. Koster, J. van Nes, K. De Preter, F. Speleman, SOX11 is a lineage-dependency factor and master epigenetic regulator in neuroblastoma. bioRxiv 2020.08.21.261131 [Preprint]. 21 August 2020. https://doi.org/10.1101/2020.08.21.261131.
- 10.Chabes A. L., Bjorklund S., Thelander L., S phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. J. Biol. Chem. 279, 10796–10807 (2004). [DOI] [PubMed] [Google Scholar]
- 11.D’Angiolella V., Donato V., Forrester F. M., Jeong Y. T., Pellacani C., Kudo Y., Saraf A., Florens L., Washburn M. P., Pagano M., Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149, 1023–1034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfister S. X., Markkanen E., Jiang Y., Sarkar S., Woodcock M., Orlando G., Mavrommati I., Pai C.-C., Zalmas L.-P., Drobnitzky N., Dianov G. L., Verrill C., Macaulay V. M., Ying S., La Thangue N. B., D’Angiolella V., Ryan A. J., Humphrey T. C., Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28, 557–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saldivar J. C., Cortez D., Cimprich K. A., The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold S., Herkert B., Eilers M., Facilitating replication under stress: An oncogenic function of MYC? Nat. Rev. Cancer 9, 441–444 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Kotsantis P., Silva L. M., Irmscher S., Jones R. M., Folkes L., Gromak N., Petermann E., Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 7, 13087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campaner S., Amati B., Two sides of the Myc-induced DNA damage response: From tumor suppression to tumor maintenance. Cell Div. 7, 6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petroni M., Sardina F., Infante P., Bartolazzi A., Locatelli E., Fabretti F., Di Giulio S., Capalbo C., Cardinali B., Coppa A., Tessitore A., Colicchia V., Sahun Roncero M., Belardinilli F., Di Marcotullio L., Soddu S., Comes Franchini M., Petricci E., Gulino A., Giannini G., MRE11 inhibition highlights a replication stress-dependent vulnerability of MYCN-driven tumors. Cell Death Dis. 9, 895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentijn L. J., Koster J., Haneveld F., Aissa R. A., van Sluis P., Broekmans M. E. C., Molenaar J. J., van Nes J., Versteeg R., Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. U.S.A. 109, 19190–19195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X., Li Y., Zhang N., Gao Y., Han L., Li S., Li J., Liu X., Gong Y., Xie C., RRM2 silencing suppresses malignant phenotype and enhances radiosensitivity via activating cGAS/STING signaling pathway in lung adenocarcinoma. Cell Biosci. 11, 74 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang S., Li L., Zang Y., Li G., Wang F., RRM2 elicits the metastatic potential of breast cancer cells by regulating cell invasion, migration and VEGF expression via the PI3K/AKT signaling. Oncol. Lett. 19, 3349–3355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzu Y. Z., Armenia J., Chakraborty G., Yoshikawa Y., Coggins S. A., Nandakumar S., Gerke T. A., Pomerantz M. M., Qiu X., Zhao H., Atiq M., Khan N., Komura K., Lee G.-S. M., Fine S. W., Bell C., O’Connor E., Long H. W., Freedman M. L., Kim B., Kantoff P. W., A novel mechanism driving poor-prognosis prostate cancer: Overexpression of the DNA repair gene, ribonucleotide reductase small subunit M2 (RRM2). Clin. Cancer Res. 25, 4480–4492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fatkhutdinov N., Sproesser K., Krepler C., Liu Q., Brafford P. A., Herlyn M., Aird K. M., Zhang R., Targeting RRM2 and mutant BRAF is a novel combinatorial strategy for melanoma. Mol. Cancer Res. 14, 767–775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohmura S., Marchetto A., Orth M. F., Li J., Jabar S., Ranft A., Vinca E., Ceranski K., Carreno-Gonzalez M. J., Romero-Perez L., Wehweck F. S., Musa J., Bestvater F., Knott M. M. L., Holting T. L. B., Hartmann W., Dirksen U., Kirchner T., Cidre-Aranaz F., Grunewald T. G. P., Translational evidence for RRM2 as a prognostic biomarker and therapeutic target in Ewing sarcoma. Mol. Cancer 20, 97 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depuydt P., Koster J., Boeva V., Hocking T. D., Speleman F., Schleiermacher G., De Preter K., Meta-mining of copy number profiles of high-risk neuroblastoma tumors. Sci. Data 5, 180240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leone G., Sears R., Huang E., Rempel R., Nuckolls F., Park C. H., Giangrande P., Wu L., Saavedra H. I., Field S. J., Thompson M. A., Yang H., Fujiwara Y., Greenberg M. E., Orkin S., Smith C., Nevins J. R., Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8, 105–113 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J. R., Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21, 4684–4699 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decaesteker B., Denecker G., Van Neste C., Dolman E. M., Van Loocke W., Gartlgruber M., Nunes C., De Vloed F., Depuydt P., Verboom K., Rombaut D., Loontiens S., De Wyn J., Kholosy W. M., Koopmans B., Essing A. H. W., Herrmann C., Dreidax D., Durinck K., Deforce D., Van Nieuwerburgh F., Henssen A., Versteeg R., Boeva V., Schleiermacher G., van Nes J., Mestdagh P., Vanhauwaert S., Schulte J. H., Westermann F., Molenaar J. J., De Preter K., Speleman F., TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat. Commun. 9, 4866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeid R., Lawlor M. A., Poon E., Reyes J. M., Fulciniti M., Lopez M. A., Scott T. G., Nabet B., Erb M. A., Winter G. E., Jacobson Z., Polaski D. R., Karlin K. L., Hirsch R. A., Munshi N. P., Westbrook T. F., Chesler L., Lin C. Y., Bradner J. E., Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbieri E., De Preter K., Capasso M., Johansson P., Man T. K., Chen Z., Stowers P., Tonini G. P., Speleman F., Shohet J. M., A p53 drug response signature identifies prognostic genes in high-risk neuroblastoma. PLOS ONE 8, e79843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss W. A., Aldape K., Mohapatra G., Feuerstein B. G., Bishop J. M., Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 16, 2985–2995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beckers A., Van Peer G., Carter D. R., Gartlgruber M., Herrmann C., Agarwal S., Helsmoortel H. H., Althoff K., Molenaar J. J., Cheung B. B., Schulte J. H., Benoit Y., Shohet J. M., Westermann F., Marshall G. M., Vandesompele J., De Preter K., Speleman F., MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett. 366, 123–132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhutia Y. D., Hung S. W., Krentz M., Patel D., Lovin D., Manoharan R., Thomson J. M., Govindarajan R., Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: Role of LIN-28 and SET oncoprotein. PLOS ONE 8, e53436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita T., Igarashi J., Okawa E. R., Gotoh T., Manne J., Kolla V., Kim J., Zhao H., Pawel B. R., London W. B., Maris J. M., White P. S., Brodeur G. M., CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J. Natl. Cancer Inst. 100, 940–949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quan J., Adelmant G., Marto J. A., Look A. T., Yusufzai T., The chromatin remodeling factor CHD5 is a transcriptional repressor of WEE1. PLOS ONE 9, e108066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppenhafer S. L., Goss K. L., Terry W. W., Gordon D. J., Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells causes DNA damage and apoptosis via the CDK2-mediated degradation of RRM2. Mol. Cancer Res. 18, 91–104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herold S., Kalb J., Buchel G., Ade C. P., Baluapuri A., Xu J., Koster J., Solvie D., Carstensen A., Klotz C., Rodewald S., Schulein-Volk C., Dobbelstein M., Wolf E., Molenaar J., Versteeg R., Walz S., Eilers M., Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567, 545–549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen R. D., Gajjar M. K., Tuckova L., Jensen K. E., Maya-Mendoza A., Holst C. B., Mollgaard K., Rasmussen J. S., Brennum J., Bartek J. Jr., Syrucek M., Sedlakova E., Andersen K. K., Frederiksen M. H., Bartek J., Hamerlik P., BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat. Commun. 7, 13398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ung C. Y., Guo F., Zhang X., Zhu Z., Zhu S., Mosaic zebrafish transgenesis for functional genomic analysis of candidate cooperative genes in tumor pathogenesis. J. Vis. Exp. 97, 52567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B., Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buisson R., Boisvert J. L., Benes C. H., Zou L., Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell 59, 1011–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blosser W. D., Dempsey J. A., McNulty A. M., Rao X., Ebert P. J., Lowery C. D., Iversen P. W., Webster Y. W., Donoho G. P., Gong X., Merzoug F. F., Buchanan S., Boehnke K., Yu C., You X. T., Beckmann R. P., Wu W., McNeely S. C., Lin A. B., Martinez R., A pan-cancer transcriptome analysis identifies replication fork and innate immunity genes as modifiers of response to the CHK1 inhibitor prexasertib. Oncotarget 11, 216–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelso A. A., Lopezcolorado F. W., Bhargava R., Stark J. M., Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLOS Genet. 15, e1008319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahtab M., Boavida A., Santos D., Pisani F. M., The genome stability maintenance DNA helicase DDX11 and its role in cancer. Genes (Basel) 12, 395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saxena S., Somyajit K., Nagaraju G., XRCC2 regulates replication fork progression during dNTP alterations. Cell Rep. 25, 3273–3282.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Slaats G. G., Giles R. H., Are renal ciliopathies (replication) stressed out? Trends Cell Biol. 25, 317–319 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Fabbri L., Bost F., Mazure N. M., Primary cilium in cancer hallmarks. Int. J. Mol. Sci. 20, 1336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H., Kiseleva A. A., Golemis E. A., Ciliary signalling in cancer. Nat. Rev. Cancer 18, 511–524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abedalthagafi M. S., Wu M. P., Merrill P. H., Du Z., Woo T., Sheu S. H., Hurwitz S., Ligon K. L., Santagata S., Decreased FOXJ1 expression and its ciliogenesis programme in aggressive ependymoma and choroid plexus tumours. J. Pathol. 238, 584–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knox J. J., Hotte S. J., Kollmannsberger C., Winquist E., Fisher B., Eisenhauer E. A., Phase II study of triapine in patients with metastatic renal cell carcinoma: A trial of the National Cancer Institute of Canada Clinical Trials Group (NCIC IND.161). Invest. New Drugs 25, 471–477 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Kunos C. A., Chu E., Makower D., Kaubisch A., Sznol M., Ivy S. P., Phase I trial of triapine-cisplatin-paclitaxel chemotherapy for advanced stage or metastatic solid tumor cancers. Front. Oncol. 7, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeidner J. F., Karp J. E., Blackford A. L., Smith B. D., Gojo I., Gore S. D., Levis M. J., Carraway H. E., Greer J. M., Ivy S. P., Pratz K. W., McDevitt M. A., A phase II trial of sequential ribonucleotide reductase inhibition in aggressive myeloproliferative neoplasms. Haematologica 99, 672–678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira S., Fernandes P. A., Ramos M. J., Mechanism for ribonucleotide reductase inactivation by the anticancer drug gemcitabine. J. Comput. Chem. 25, 1286–1294 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Singh A., Xu Y. J., The cell killing mechanisms of hydroxyurea. Genes (Basel) 7, 99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunos C. A., Ivy S. P., Triapine radiochemotherapy in advanced stage cervical cancer. Front. Oncol. 8, 149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kara N., Krueger F., Rugg-Gunn P., Houseley J., Genome-wide analysis of DNA replication and DNA double-strand breaks using TrAEL-seq. PLoS Biol. 19, e3000886 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao P. A., Sasaki T., Gilbert D. M., High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biol. 21, 76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole K. A., Huggins J., Laquaglia M., Hulderman C. E., Russell M. R., Bosse K., Diskin S. J., Attiyeh E. F., Sennett R., Norris G., Laudenslager M., Wood A. C., Mayes P. A., Jagannathan J., Winter C., Mosse Y. P., Maris J. M., RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 108, 3336–3341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toledo L. I., Altmeyer M., Rask M. B., Lukas C., Larsen D. H., Povlsen L. K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Liu Y., Li Y., Wang X., Liu F., Gao P., Quinn M. M., Li F., Merlino A. A., Benes C., Liu Q., Gray N. S., Wong K. K., Gemcitabine and Chk1 Inhibitor AZD7762 synergistically suppress the growth of Lkb1-deficient lung adenocarcinoma. Cancer Res. 77, 5068–5076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang M., Zhao T., Ma L., Guo Y., CHK1 inhibition sensitizes pancreatic cancer cells to gemcitabine via promoting CDK-dependent DNA damage and ribonucleotide reductase downregulation. Oncol. Rep. 39, 1322–1330 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Lowery C. D., VanWye A. B., Dowless M., Blosser W., Falcon B. L., Stewart J., Stephens J., Beckmann R. P., Bence Lin A., Stancato L. F., The checkpoint kinase 1 inhibitor prexasertib induces regression of preclinical models of human neuroblastoma. Clin. Cancer Res. 23, 4354–4363 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Morimoto Y., Takada K., Takeuchi O., Takagi A., Watanabe K., Hirohara M., Hamamoto T., Masuda Y., Prexasertib increases the sensitivity of pancreatic cancer cells to gemcitabine and S‑1. Oncol. Rep. 43, 689–699 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Selmi A., de Saint-Jean M., Jallas A. C., Garin E., Hogarty M. D., Benard J., Puisieux A., Marabelle A., Valsesia-Wittmann S., TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett. 357, 412–418 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Schmidtmann E., Anton T., Rombaut P., Herzog F., Leonhardt H., Determination of local chromatin composition by CasID. Nucleus 7, 476–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oscherwitz J., The promise and challenge of epitope-focused vaccines. Hum. Vaccin. Immunother. 12, 2113–2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Groningen T., Koster J., Valentijn L. J., Zwijnenburg D. A., Akogul N., Hasselt N. E., Broekmans M., Haneveld F., Nowakowska N. E., Bras J., van Noesel C. J. M., Jongejan A., van Kampen A. H., Koster L., Baas F., van Dijk-Kerkhoven L., Huizer-Smit M., Lecca M. C., Chan A., Lakeman A., Molenaar P., Volckmann R., Westerhout E. M., Hamdi M., van Sluis P. G., Ebus M. E., Molenaar J. J., Tytgat G. A., Westerman B. A., van Nes J., Versteeg R., Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Lambertz I., Kumps C., Claeys S., Lindner S., Beckers A., Janssens E., Carter D. R., Cazes A., Cheung B. B., De Mariano M., De Bondt A., De Brouwer S., Delattre O., Gibbons J., Janoueix-Lerosey I., Laureys G., Liang C., Marchall G. M., Porcu M., Takita J., Trujillo D. C., Van Den Wyngaert I., Van Roy N., Van Goethem A., Van Maerken T., Zabrocki P., Cools J., Schulte J. H., Vialard J., Speleman F., De Preter K., Upregulation of MAPK negative feedback regulators and RET in mutant ALK neuroblastoma: Implications for targeted treatment. Clin. Cancer Res. 21, 3327–3339 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J. L., Li X., R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan J. L., Fogley R. D., Flynn R. A., Ablain J., Yang S., Saint-André V., Fan Z. P., Do B. T., Laga A. C., Fujinaga K., Santoriello C., Greer C. B., Kim Y. J., Clohessy J. G., Bothmer A., Pandell N., Avagyan S., Brogie J. E., van Rooijen E., Hagedorn E. J., Shyh-Chang N., White R. M., Price D. H., Pandolfi P. P., Peterlin B. M., Zhou Y., Kim T. H., Asara J. M., Chang H. Y., Young R. A., Zon L. I., Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol. Cell 62, 34–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nitarska J., Smith J. G., Sherlock W. T., Hillege M. M. G., Nott A., Barshop W. D., Vashisht A. A., Wohlschlegel J. A., Mitter R., Riccio A., A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 17, 1683–1698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aird K. M., Iwasaki O., Kossenkov A. V., Tanizawa H., Fatkhutdinov N., Bitler B. G., Le L., Alicea G., Yang T. L., Johnson F. B., Noma K. I., Zhang R., HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J. Cell Biol. 215, 325–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zirkel A., Nikolic M., Sofiadis K., Mallm J.-P., Brackley C. A., Gothe H., Drechsel O., Becker C., Altmüller J., Josipovic N., Georgomanolis T., Brant L., Franzen J., Koker M., Gusmao E. G., Costa I. G., Ullrich R. T., Wagner W., Roukos V., Nürnberg P., Marenduzzo D., Rippe K., Papantonis A., HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell 70, 730–744.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Cohen M. A., Zhang S., Sengupta S., Ma H., Bell G. W., Horton B., Sharma B., George R. E., Spranger S., Jaenisch R., Formation of human neuroblastoma in mouse-human neural crest chimeras. Cell Stem Cell 26, 579–592.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bate-Eya L. T., Ebus M. E., Koster J., den Hartog I. J. M., Zwijnenburg D. A., Schild L., van der Ploeg I., Dolman M. E. M., Caron H. N., Versteeg R., Molenaar J. J., Newly-derived neuroblastoma cell lines propagated in serum-free media recapitulate the genotype and phenotype of primary neuroblastoma tumours. Eur. J. Cancer 50, 628–637 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Zhao W., Sachsenmeier K., Zhang L., Sult E., Hollingsworth R. E., Yang H., A new bliss independence model to analyze drug combination data. J. Biomol. Screen. 19, 817–821 (2014). [DOI] [PubMed] [Google Scholar]
- 76.C. Nunes, J. Anckaert, F. De Vloed, J. De Wyn, K. Durinck, J. Vandesompele, F. Speleman, V. Vermeirssen, Automatic end-to-end analysis of high-throughput in vitro cell culture screening by HTSplotter. bioRxiv 2021.09.06.459128 [Preprint]. 6 September 2021. https://doi.org/10.1101/2021.09.06.459128.
- 77.Diebold E. D., Buckley B. W., Gossett D. R., Jalali B., Digitally synthesized beat frequency multiplexing for sub-millisecond fluorescence microscopy. Nat. Photonics 7, 806–810 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S1 to S9