Abstract
Soluble angiotensin-converting enzyme 2 (ACE2) constitutes an attractive antiviral capable of targeting a wide range of coronaviruses using ACE2 as their receptor. Using structure-guided approaches, we developed a series of bivalent ACE2-Fcs harboring functionally and structurally validated mutations that enhance severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor binding domain recognition by up to ~12-fold and remove angiotensin enzymatic activity. The lead variant M81 potently cross-neutralized SARS-CoV-2 variants of concern (VOCs), including Omicron, at subnanomolar half-maximal inhibitory concentration and was capable of robust Fc-effector functions, including antibody-dependent cellular cytotoxicity, phagocytosis, and complement deposition. When tested in a stringent K18-hACE2 mouse model, Fc-enhanced ACE2-Fc delayed death by 3 to 5 days or effectively resolved lethal SARS-CoV-2 infection in both prophylactic and therapeutic settings via the combined effects of neutralization and Fc-effector functions. These data add to the demonstrated utility of soluble ACE2 as a valuable SARS-CoV-2 antiviral and indicate that Fc-effector functions may constitute an important component of ACE2-Fc therapeutic activity.
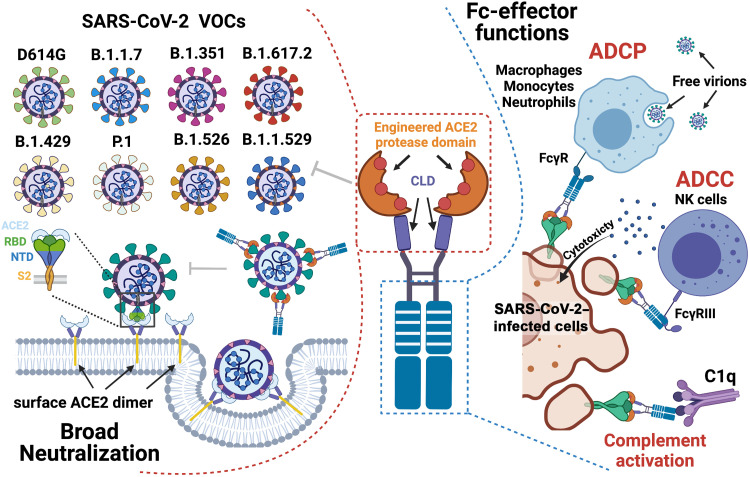
ACE2-based therapeutics prevents and treats SARS-CoV-2 infection by bridging neutralization with host innate immunity.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a betacoronavirus closely related to SARS-CoV-1, is the ninth documented coronavirus capable of infecting humans (1) and has led to a devastating ongoing pandemic, resulting in more than 6 million deaths worldwide since it first emerged in the Chinese city of Wuhan in late 2019. This highly transmissible airborne pathogen is an enveloped virus with a large, single-stranded, positive-sense RNA genome. Since the genetic sequence became available in January 2020, the development of both traditional vaccines (e.g., inactivated virus, recombinant proteins, viral vectors, etc.) and novel RNA/DNA strategies has moved at an unprecedented pace (2). The worldwide emergency rollout of vaccines aided in the suppression of viral circulation and reduced the risk of severe illnesses; however, continuous viral evolution and the resulting variants of concern (VOCs) have the potential to circumvent immunity conferred by both natural infection and vaccination (3). In preparation for the inevitable SARS-CoV-2 VOCs and any future potential pandemic or zoonotic spillovers, it is important that additional interventions and therapies effective against the vast natural CoV reservoirs are developed and stockpiled.
A major antigenic site on the SARS-CoV-2 virion surface is the spike (S) trimer, which mediates virus-host membrane fusion and subsequent entry via the primary host cell receptor angiotensin-converting enzyme 2 (ACE2) (4). Viral entry is initiated by specific interaction of the S1 subunit receptor binding domain (RBD) to ACE2, followed by S2-directed membrane fusion (5). Most neutralizing antibodies (nAbs) elicited through natural infection and vaccination act by disrupting this interaction; however, selection pressure results in viral escape mutations, in many cases generating VOCs with an enhanced ability to bind host receptors (3).
Full-length ACE2 consists of an N-terminal protease domain (PD; residues 18 to 615) that directly engages SARS-CoV-2 RBD, a collectrin-like domain (CLD; residues 616 to 740), a single transmembrane helix (residues 741 to 765), and a ~40–amino acid intracellular C-terminal domain (6). ACE2 is an essential zinc-dependent carboxypeptidase and critical regulator of the renin-angiotensin system (RAS). ACE2 PD converts angiotensin II (Ang II) to Ang 1-7, relieving the vasoconstriction, inflammation, and oxidative stress effect of Ang II (7).
Recombinant soluble ACE2 decoys were proposed and tested as potential SARS-CoV-2 therapies since the early onset of the coronavirus disease 2019 (COVID-19) pandemic (8, 9). A pilot clinical trial of human recombinant soluble ACE2 administered intravenously (0.4 mg/kg) in a patient severely infected with SARS-CoV-2 showed rapid viral clearance in sera, followed by nasal cavity and lung clearance at a later time (10). Concomitant with the viral load reduction was a profound decrease of Ang II and a proportional increase of the ACE2 products Ang 1-7 and Ang 1-9 in the plasma. Although compelling evidence confirms ACE2 enzymatic activity as an important component of the therapeutic activity of ACE2-Fc, due to its protective action against SARS-CoV-1– and SARS-CoV-2–induced acute lung injury (11, 12), ACE2-inactivated mutants with enhanced affinities for SARS-CoV-2 RBD and increased neutralization potencies have been developed (13) as possible alternative options.
Because monomeric ACE2 binds to nascent SARS-CoV-2 RBD (WA-1/2020 isolate) with only moderate affinity [dissociation constant (Kd) ~ 20 to 30 nM], engineered ACE2 derivatives with improved affinity to SARS-CoV-2 were extensively developed as antiviral therapeutics by several approaches. These include deep mutagenesis coupled with flow cytometry–based screening (14–16), computation-aided design and yeast display (13, 17), multimerization of ACE2 (18–20), de novo design of ACE2-derived miniprotein and peptides (21), and ACE2-decorated vesicles (22). Recently, bivalent ACE2-Fc [i.e., ACE2 extracellular domain (ECD) grafted onto an immunoglobulin G1 (IgG1) backbone] molecules have gained considerable attention as they are able to avidly bind SARS-CoV-2 S with increased affinity and potently neutralize VOCs, including those resistant to common nAbs (15, 16). As most currently investigated ACE2-Fc–based therapeutic approaches focus on direct neutralization, the potential of ACE2-Fcs as agents capable of Fc-mediated effector functions, including Ab-dependent cellular cytotoxicity (ADCC), cellular phagocytosis (ADCP), and complement deposition (ADCD), is largely unknown and has not been evaluated in vivo or in vivo in SARS-CoV-2 infection models.
Here, we used a structure-guided approach to develop a series of ACE2-Fc variants, using human IgG1 or IgG3 backbones. Our variants were engineered to have (i) substantially increased affinity to SARS-CoV-2 RBD derived from the WA-1/2020 strain, B.1.1.7, and B.1.351; (ii) enhanced affinity for Fcγ receptors (FcγR) involved in Fc-effector mechanisms; and (iii) mutations to abrogate the angiotensin enzymatic activity of ACE2. All introduced mutations were validated for mechanism at the molecular level by structural biology approaches. Our best variant cross-neutralized nine SARS-CoV-2 VOCs, including Delta and Omicron, in a pseudo-viral assay with inhibitory potencies comparable to broad and potent anti–SARS-CoV-2 nAbs and was able mediate an array of Fc-effector activities (Fig. 1). When tested in humanized K18-hACE2 mice under prophylactic or single-dose therapeutic settings, the lead ACE2-Fc variant prevented or significantly delayed lethal SARS-CoV-2 infection. In both prophylaxis and therapy, in vivo protection was dependent on the combined effects of direct neutralization and Fc-effector functions of ACE2-Fc.
Fig. 1. Combined mechanism of direct neutralization and Fc-effector functions by engineered ACE2-Fcs.
The figure was generated in BioRender. NTD, N-terminal domain.
RESULTS
Structure-based engineering of ACE2-Fc variants with enhanced binding affinities to SARS-CoV-2 RBD
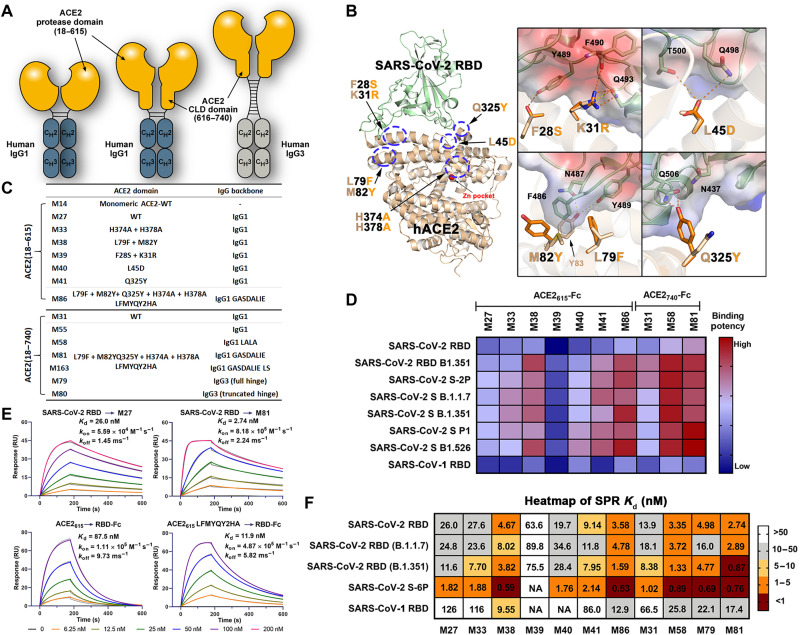
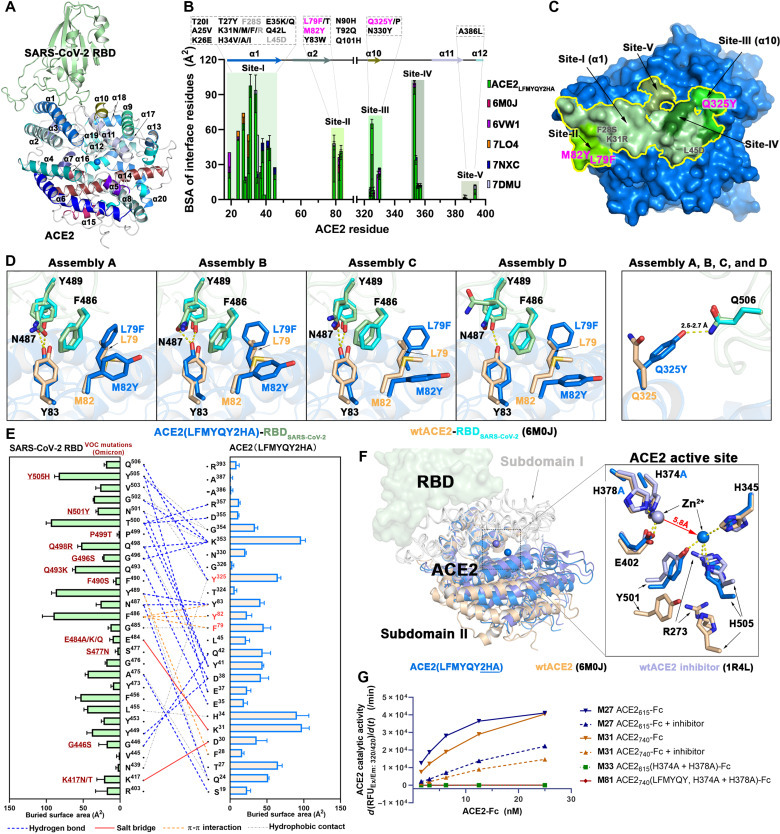
A series of hybrid molecules (ACE2-Fc fusion proteins) were generated by replacing the antigen binding fragment of human IgG1 or IgG3 with the ACE2 PD only (residues 18 to 615; hereafter referred to as ACE2615) or the whole extracellular ACE2 domain, consisting of both PD and CLD (residues 18 to 740; referred to as ACE2740) (Fig. 2A). These ACE2-Fc scaffolds were incorporated with mutations to (i) increase affinity for the SARS-CoV-2 RBD, (ii) abrogate the angiotensin cleavage activity of ACE2, and (iii) enhance the affinity for Fcγ receptors involved in Fc-mediated effector mechanisms. ACE2 mutation sites with the potential to increase affinity for SARS-CoV-2 RBD were identified using structure-based approaches (Fig. 2B). We started by analyzing the receptor-antigen interface of two high-resolution ACE2-RBD structures [6M0J (23) and 6VW1 (24)] to identify interface contacts that could be strengthened and/or optimized, while avoiding key interactions for interface stability, e.g., hydrogen bonds with distances of <3.0 Å or salt bridges. Interface residues were then analyzed on the basis of their electrostatic potential, and ACE2 point mutations with the potential to enhance charge complementarity with the RBD were introduced (e.g., K31R and L45D; Fig. 2B). Hydrophobic contacts within the flexible regions were also redesigned to improve binary packing and reduce steric repulsion (e.g., F28S), fill empty cavities (e.g., L79F), or improve aromatic interactions (e.g., M82Y). Point mutations to possibly facilitate the hydrogen bonding network (Q325Y) were also introduced. A list of the ACE2-Fc variants that were generated, expressed, and purified to homogeneity is shown in Fig. 2C; the size exclusion chromatography (SEC) profiles are shown in fig. S1. The enzymatically inactive ACE2-Fc variant was generated by introducing mutations to two Zn2+ binding histidines (H374A and H378A; Fig. 2B). In addition, to enhance binding to Fcγ receptors present on the effector cell surface, the well-known GASDALIE (G236A/S239D/A330L/I332E) and half-life–extending LS mutations (M428L/N434S) were added to the Fc region of the best-performing variants generated with the human IgG1 backbone (Fig. 2, A and C). The best-performing ACE2 variant was also prepared using human IgG3 backbone to test whether an equivalent IgG3 isotype, featuring by an elongated hinge, would display greater Fc-effector activities, as observed for anti–HIV-1 Abs (25, 26). Last, an “Fc-effector–null” (L234A/L235A and LALA) mutant was generated from the best-performing variant to assess the contribution of Fc-effector functions to antiviral activity.
Fig. 2. Structure-based development of ACE2-Fc variants with enhanced SARS-CoV-2 RBD affinity.
(A) Schematic overview of bivalent engineered ACE2-IgG Fc chimeras. (B) The structure-based design of ACE2 mutations with improved SARS-CoV-2 RBD binding affinity. Crystal structure of the ACE2 (light yellow) and RBD (pale green) complex (PDB: 6M0J) used for interface residue analysis (left), with residues selected for mutation shown as sticks. Blow-up views of mutation sites with wild-type (wt) and mutated residues shown as sticks (right). The RBD is shown as a semitransparent electrostatic potential surface (red for negatively charged residues and blue for positively charged residues), with a green cartoon for the polypeptide backbone. Wt and mutated ACE2 residues are colored as mustard and orange, respectively. (C) The developed ACE2-Fc variants. (D) Heatmap showing the binding efficacy of ACE2-Fc variants to SARS-CoV-2 RBDwt, RBDB.1.351, SARS-CoV RBD, and the selected SARS-CoV-2 VOCs. Binding was measured by enzyme-linked immunosorbent assay (ELISA) using SARS-CoV-2 antigens immobilized on the plate and ACE2-Fc in the concentration range of 0.05 to 125 nM. Area under the curve (AUC) for the unsaturated binding region (0.05 to 2.5 nM; fig. S2) was calculated and plotted as a heatmap. (E) SPR-based kinetic measurement of SARS-CoV-2 RBDwt binding to immobilized M27 or M81 (top) and monomeric wtACE2615 or ACE2615(LFMYQY2HA) to immobilized SARS-CoV-2 RBDwt-Fc (bottom). Experimental data are shown as colored curves overlapping the 1:1 Langmuir fit in gray. (F) The dissociation constants (Kd) for SARS-CoV-2 and SARS-CoV antigens binding to ACE2-Fc variants as measured by SPR. Experimental binding curves and the detailed kinetic constants are shown in fig. S3 and summarized in table S1.
The binding affinity of the ACE2-Fc variants to SARS-CoV-2 wild-type (wt; WA-1/2020 isolate) RBD and selected VOC RBDs (e.g., B.1.1.7 and B.1.351) were estimated by enzyme-linked immunosorbent assay (ELISA) (Fig. 2D and fig. S2) and surface plasmon resonance (SPR) (Fig. 2, E and F; fig. S3; and table S1). The wt ACE2615-Fc (M27; Fig. 2C) bound to RBDwt with a Kd of 26 nM, consistent with reported data (4, 14), and is around five times higher than the affinity of SARS-CoV-1 RBD binding. A slight enhancement (1.4- to 2.0-fold) to the binding affinity of all RBDs tested was observed for the ACE2740-Fc (M31) variant over the shorter ACE2615-Fc (M27) variant (Fig. 2F and figs. S2 and S3). Furthermore, the ACE2615-Fc variant with H374A/H378A mutations (M33; Fig. 2C) displayed RBD binding comparable to the wt (M27), indicating that the zinc site–disrupting substitutions do not interfere with the ACE2-RBD binding interface.
Among the interface mutations that could potentially facilitate RBD binding (Fig. 2, D and F), the dual L79F/M82Y mutant (M38 in Fig. 2C) and the single Q325Y mutant (M41) showed a 3- to 6-fold and 1.4- to 2.8-fold enhancement in binding affinity to the RBD, respectively, compared to the unmodified ACE2615-Fc (M27). In contrast, variants with F28S/K31R mutations (M39) or an L45D mutation (M40) showed reduced or unchanged binding affinity to SARS-CoV-2 RBDs. Combining the enhancing mutations (L79F/M82Y and Q325Y) generated variants with significantly increased affinity for RBDwt. Variants with the combined L79F/M82Y/Q325Y and H374A/H378A mutations (referred to as LFMYQY2HA) were fused to GASDALIE IgG1 Fc (M81 and M86), LALA IgG1 Fc (M58), or IgG3 Fc (M79 and M80). As shown in Fig. 2 (D and F), the best-performing variant, M81, showed increased affinity compared to the unmodified ACE2615-Fc (M27) by ~8.5- to 13.3-fold to RBDwt, RBDB.1.1.7, and RBDB1.351 (Kd range of 0.87 to 2.89 nM). This binding enhancement is likely a result of the faster RBD association (kon) (5- to 14-fold), while the dissociation constant (koff) was similar to wtACE2-Fc M27 (Fig. 2E). We observed decreased binding affinity for ACE2 variants tested in the SPR format, where SARS-CoV-2 RBD-Fc fusion was immobilized, probably because of the slower tumbling rate of monomeric ACE2615 (~75 kDa) when acting as the soluble analyte compared to the smaller ligand RBD (~26 kDa) (Fig. 2E). Together, our lead ACE2-Fc variant M81 showed comparable RBD affinity to the reported best-in-class engineered ACE2-Fcs (Kd below 1 nM) (13–16) and to many nAbs isolated from patients with SARS-CoV-2 (27).
Notably, the ACE2740-Fc showed enhanced binding affinity compared to ACE2615-Fc, while ACE2740 grafted onto an IgG3 backbone (M79) with an intact hinge (11 disulfides) displayed slightly decreased affinity to SARS-CoV-2 RBD (Fig. 2F). These data point toward the possibility that, while the extended CLD likely increases the structural plasticity of ACE2-Fc (28) and facilitates interaction with adjacent S trimers (13, 29), the elongated IgG3 hinge may partially restrict the ACE2 mobility required for optimal RBD recognition from separate spikes.
Molecular basis for the enhanced SARS-CoV-2 S binding and enzymatic inactivation of engineered ACE2-Fc
To dissect the molecular basis of the enhanced RBD affinity and abrogated enzymatic activity of our best ACE2 variant, we cocrystallized monomeric ACE2615 (with LFMYQY2HA mutations) with SARS-CoV-2 RBD, and the structure was determined at 3.5-Å resolution. Four ACE2-RBD complexes were presented in the asymmetric unit (ASU) of the crystal, and the final model was refined to an Rwork/Rfree of 0.24/0.29 (Fig. 3, fig. S4, and table S2). The overall interface and contact residues of ACE2615 LFMYQY2HA-RBD largely resemble ACE2wt-RBD (Fig. 3, B and E), with a slightly larger total buried surface area (BSA) (957.4 Å2) compared to the BSA of the ACE2wt-RBD complex (average of 869.1 Å2 from reported ACE2wt-RBD structures) and that of an engineered ACE2 [908.9 Å2; Protein Data Bank (PDB): 7DMU] (fig. S4B). More than 70% of the RBD contacts are mediated by the ACE2 α1 helix (residues 18 to 52), which contains many reported RBD binding–enhancing mutations (Fig. 3B) (13–15). To differentiate the engineered ACE2 mutations reported previously from those identified in this study, we divided the RBD contact surface on ACE2 into five subsites, designated as follows: Site-I (residues 18 to 45), Site-II (residues 79 to 83), Site-III (residues 324 to 330), Site-IV (residues 353 to 357), and Site-V (residues 386 to 393) (Fig. 3, B and C). The introduced affinity-enhancing mutants L79F/M82Y and Q325Y map to Site-II and Site-III, respectively, flanking the α1 helix/Site-I at the furthest edge of the RBD contact surface (Fig. 3C). In the ACE2wt-RBD structure (PDB: 6M0J), the RBD ridge (residues 473 to 490) is weakly associated with Site-II residues and represents the most mobile segment with the highest B-factors of residues across the interface. Contacts within this region, specifically with RBD ridge residue F486 that interacts with ACE2 residues Y83, L79, and M82, are significantly stabilized in complex with the mutant ACE2615 LFMYQY2HA. Specifically, L79F and M82Y are in face-to-face or face-to-edge stacking with F486RBD in two of the ASU copies (assemblies A and B), while only L79F is in face-to-edge stacking with F486RBD in assemblies C and D (Fig. 3D). As a result, residues (G485-F486-N487) of the RBD ridge with better hydrophobic packing in assemblies A and B have the lowest relative B-factor values, followed by those in assemblies C and D and in two ACE2wt-RBD crystal structures (fig. S4C). This observation supports the ACE2 L79F/M82Y-stabilizing effect on the RBD ridge; however, it is dependent on how well the three Site-II aromatic residues (L79F, M82Y, and Y83) pack against F486RBD. In contrast, in all four ASU assemblies, the other RBD affinity–enhancing mutant, Q325Y, uniformly forms a strong hydrogen bond with Q506RBD, which is not involved in receptor binding in the ACE2wt-RBD structure (Fig. 3D and fig. S4, A and D). The mutant’s binding affinity to SARS-CoV-2 RBD (M41 versus M27) is increased by 1.4- to 2.8-fold as a result of the extra hydrogen bond (Fig. 2E, fig. S3, and table S1). Notably, the two anchor RBD residues F486 and Q506, which interact with L79F/M82Y and Q325Y, respectively, are invariant across the SARS-CoV-2 VOCs identified thus far (Fig. 3E), suggesting that these RBD-enhancing mutations could be equally effective against these SARS-CoV-2 escape variants (fig. S5, A to C) as well as a wide range of CoVs that use ACE2 as a receptor (fig. S5D).
Fig. 3. Crystal structure of ACE2615 LFMYQY2HA mutant in complex with SARS-CoV-2 RBD.
(A) Overall structure of the mutant ACE2615-RBD complex. ACE2 helices (α1 to α20) are colored as defined in (31) in which helix α1 serves as the major element of RBD binding. (B to E) Properties of the mutant ACE2615-RBD interface. (B) BSA plots for individual ACE2 residues within the RBD interface of mutant ACE2615 and five reported ACE2-RBD crystal structures. The RBD binding residues are classified into five regions (Site-I to Site-V). ACE2 mutations from this study (pink for RBD binding–enhancing mutations and gray for null mutations) or reported by others are shown (black) above the plot. Notably, L79F has been currently reported to be favorable for RBD binding in (14, 67). (C) The RBD footprint (yellow contour) on mutant ACE2615 (blue surface). Shades of green are used to color Site-I to Site-V defined in (B). (D) Molecular interactions of the introduced L79F/M82Y and Q325Y mutations among four ASU copies of the mutant ACE2-RBD structure in comparison to ACE2wt-RBD (6M0J; see in fig. S4A). Hydrogen bonds are depicted as dashed lines. (E) Interaction network at mutant ACE2615-RBD interface. The antigen-receptor interactions as defined by 5-Å cutoff (e.g., H bonds and salt-bridge < 3.5 Å) are shown as lines with a diagram of BSA values for individual interface residues shown on the side. (F) Structural changes induced by H374A/H378A mutations. Mutant ACE2615-RBD, ACE2wt-RBD (6M0J), and an inhibitor-bound ACE2wt-RBD (1R4L) are aligned on the basis of ACE2 subdomain I (left) and a close-up view into of the ACE2 active site (right) [see also fig. S4 (B and C)]. (G) Angiotensin-converting activity of the ACE2-Fc variants. The slopes of the initial linear region of the reaction were plotted against the indicated ACE2-Fc concentrations (fig. S4G).
The second set of mutations, H374A and H378A, were introduced to disrupt the zinc-binding pocket and eliminate angiotensin-converting activity without altering ACE2-RBD recognition. Towler et al. (30) first described the ACE2 PD subdomains I and II that form the active site cleft and show ligand-dependent subdomain II closure. Figure 3F shows the structural alignment based on the ACE2 domain of the ACE2615 LFMYQY2HA-RBD complex, the ACE2wt-RBD “apo” complex (6M0J), and ACE2wt with a bound inhibitor (1R4L). There were no significant differences in the overall structure of subdomain I and the RBD binding regions, consistent with our finding that H374A/H378A does not interfere with binding to SARS-CoV-2 RBD (Fig. 2, D and F). In contrast, noticeable changes were observed in the overall conformation of subdomain II and the Zn2+ binding site (Fig. 3F). Subdomain II in the H374A/H378A mutant adopted a partially closed conformation, somewhere between the open unliganded and inhibitor-bound closed conformations, likely induced by catalytic Zn2+ relocation. In ACE2wt, the Zn2+ is coordinated by three subdomain I residues (H374, H378, and E402) that form a catalytic metal binding pocket. In the ACE2 mutant, we found this Zn2+ binding site vacant but observed a spherical additional electron density of ~5.8 Å away within the substrate binding pocket (fig. S4, E and F). As there were no divalent cations in the crystallization or protein buffers, we attributed this density to endogenous zinc. This new Zn2+ ion was tetrahedrally coordinated by R273, H345, Y501, and H505, which are responsible for angiotensin substrate recognition in ACE2wt (fig. S4, E and F). We speculate that this alternate Zn2+ coordination site is noncatalytic but of importance to the structural integrity of ACE2. A dual zinc coordination site is not uncommon in aminopeptidases, which remove N-terminal amino acids (31). Collectively, our data provide the structural basis for the ACE2 inactivation induced by our zinc coordination mutations. As predicted, the H374A/H378A mutations were sufficient to abrogate angiotensin-converting activity in ACE2-Fc as tested using a fluorescent peptide (Fig. 3G) and Ang II (fig. S4, G to I) as substrates.
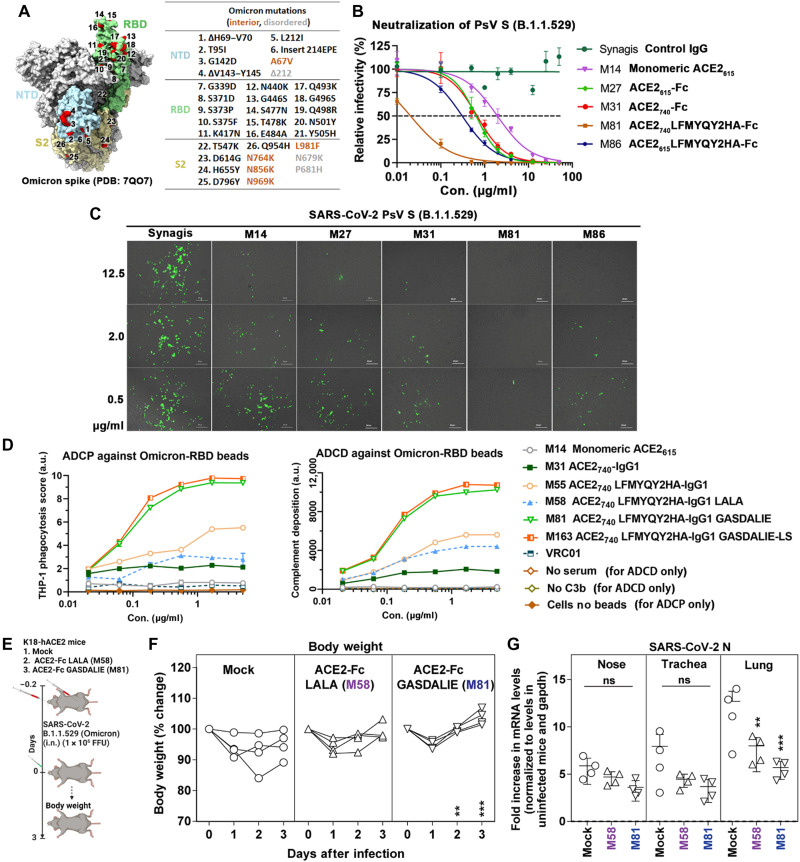
Engineered ACE2-Fcs show potent neutralization of SARS-CoV-2 VOCs in vitro
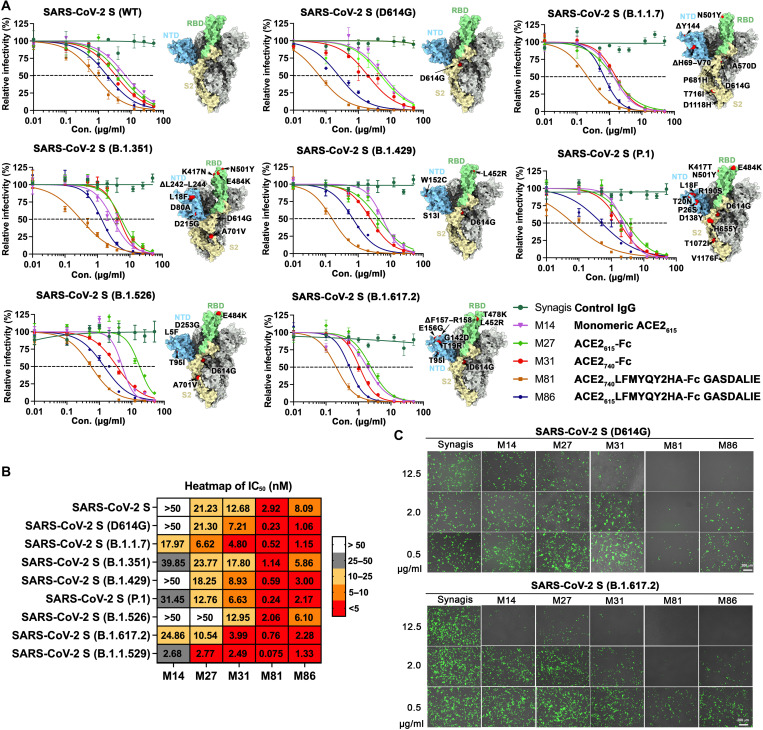
Our engineered ACE2-Fcs showed enhanced affinity for wt SARS-CoV-2 RBD and several VOCs (Fig. 2). To evaluate whether the increased S affinity translated to improved neutralizing activity, we tested the best-performing ACE2 LFMYQY2HA-Fc variants in an in vitro neutralization assay using a lentivirus pseudo-typed with the S from nine SARS-CoV-2 strains, including WA-1/2020 (wt), D614G, B.1.1.7 (Alpha), B.1.351 (Beta), P1 (Gamma), B.1.429 (Epsilon), B1.526 (Iota), B.1.617 (Delta), and the currently dominant B.1.1.529 (Omicron) (Figs. 4 and 8, C and D, and figs. S6 and S7). For all tested SARS-CoV-2 pseudotyped viruses (PsVs), neutralization by the bivalent ACE2-Fcs was >2-fold greater than monovalent ACE2615 (M14), as reflected in the half-maximal inhibitory concentrations (IC50) (molar units; Fig. 4B), highlighting the importance of multivalency in the mechanism of action of soluble ACE2-based therapeutics (32). The ACE2wt-Fcs (M27 and M31) neutralized SARS-CoV-2 PsVwt with an IC50 of 21.2 and 12.6 nM, respectively (Fig. 4, A and B), which concurs with their SPR Kd values (26 and 13.9 nM, respectively) for SARS-CoV-2 RBD (Fig. 2F) and with previous reports (13–15).
Fig. 4. Broad neutralization of engineered ACE2-Fcs against SARS-CoV-2 PsV.
(A) ACE2-Fc neutralization of SARS-CoV-2 lentivirus pseudo-typed with eight SAR2-CoV-2 S variants. hACE2-expressing 293T cells were infected with different variants of SARS-CoV-2 PsV in the presence of varying concentrations of monomeric ACE2615, selected ACE2-Fc variants, Synagis IgG (negative control), or phosphate-buffered saline (PBS). Infectivity was quantified by the cellular luciferase signal 48 hours after infection. Relative infectivity was normalized by the luciferase signal in infected cells without intervention (PBS). The spike graphics for individual VOCs were generated using PDB 7C2L with mutation sites colored in red. Data are shown as means ± SEM from three independent replicates. (B) Heatmap summary of neutralization IC50 values for the ACE2-Fc variants tested. The neutralization IC50 of Omicron (B.1.1.529) was derived from Fig. 8C. (C) Representative fluorescent imaging of hACE2-293T cells that were infected with SARS-CoV-2 S (D614G; top) or SARS-CoV-2 S (B.1.617.2; bottom) carrying ZsGreen1 reporter gene in the presence of the indicated concentrations of ACE2-Fc variants. Images obtained 48 hours after infection are shown as merged bright field (cell shape) and green field (ZsGreen signal). Scale bars, 200 μm. n = 3 replicates per group.
Fig. 8. In vitro and In vivo activities of ACE2-Fcs against B.1.1.529 VOC.
(A) Spike mutations in the B.1.1.529 (Omicron) variant. (B) Neutralization of S B.1.1.529–pseudo-typed lentivirus by ACE2-Fc (n = 3). hACE2-expressing 293T cells were infected with PsVB.1.1.529 (~106 PFU), which was preincubated with varied concentrations of selected ACE2-Fc 1 hour before. Luciferase readout was measured 48 hours after infection. The data were shown as means ± SEM. (C) Representative fluorescent imaging (n = 3) of hACE2-293T cells infected with PsVB.1.1.529 in the presence of varied concentrations of ACE2-Fc. Images were acquired 48 hours after infection. (D) ADCP and ADCD against RBDB.1.1.529-coated beads, measured as described in Materials and Methods. ADCP and ADCD data were the mean of four and two replicates, respectively. (E) Experimental design of the in vivo efficacy test of M58 (ACE2740 LFMYQY2HA-Fc LALA) and M81 (ACE2740 LFMYQY2HA-Fc GASDALIE) on K18 hACE2 transgenic mice challenged by authentic B.1.1.529 virus. ACE2-Fc was delivered at a dose of 12.5 mg/kg of body weight intranasally and 6.25 mg/kg of body weight intraperitoneally 5 hours before infection. PBS (n = 4) mice were used as control. (F) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in (E). (G) Fold changes in nucleocapsid mRNA expression in nose, trachea, and lung tissues after necropsy on day 3. Data were normalized to Gapdh mRNA in the same sample and that in noninfected mice after necropsy. Grouped data were analyzed by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests. **P < 0.01 and ***P < 0.001. Mean values ± SD are depicted. ns, not significant.
Both engineered ACE2-Fc variants, M81 (ACE2740 LFMYQY2HA-Fc) and its truncated version M86 (ACE2615 LFMYQY2HA-Fc), cross-neutralized eight SARS-Cov-2 PsV with low nanomolar IC50 values (Fig. 4, A and B). The best-performing variant, M81, which bound to SARS-CoV-2 RBD variants with a Kd of 0.87 to 2.89 nM, inhibited SARS-CoV-2 PsVwt with an IC50 of 2.92 nM. The CLD-containing M81 showed better neutralization than the CLD-lacking M86, as reflected by a 2.2- to 9.0-fold reduction of IC50 across the tested VOCs. Similar IC50 differences in the range of 1.6- to 3.0-fold were observed between M31 and M27 for VOC neutralization, which further supports the observation that the CLD promotes SARS-CoV-2 S/ACE2-Fc recognition (13, 14, 19, 28).
Notably, we observed significantly better neutralization with M81 and M86 toward VOCs containing the D614G mutation (Fig. 4, A and C). As the first recurrent S mutation present in all VOCs to date, D614G has been shown to shift the RBD conformational equilibrium to a wider range of open trimer states, facilitating enhanced receptor binding and virus transmission (33). The single substitution of D614G substantially enhanced PsV infectivity, as demonstrated by the higher ZsGreen signal (Fig. 4C). Consistent with a recent study (34), this allosteric mutation also makes VOCs more susceptible to RBD-specific nAbs and ACE2-Fc, as shown by the >10-fold reduction in the M81 IC50 to PsVD614G. Although other SARS-CoV-2 VOCs were less sensitive to M81 neutralization, IC50 values were still in the range of 0.24 to 2.06 nM, comparable to high-affinity Abs isolated from convalescent patients (35). Together, our PsV-based neutralization studies demonstrated that the best RBD binder M81 can neutralize SARS-CoV-2 PsV and VOCs that have D614G with enhanced potency, i.e., with a low nanomolar IC50 that is ~10- to 90-fold lower than wt ACE2615-Fc (M27).
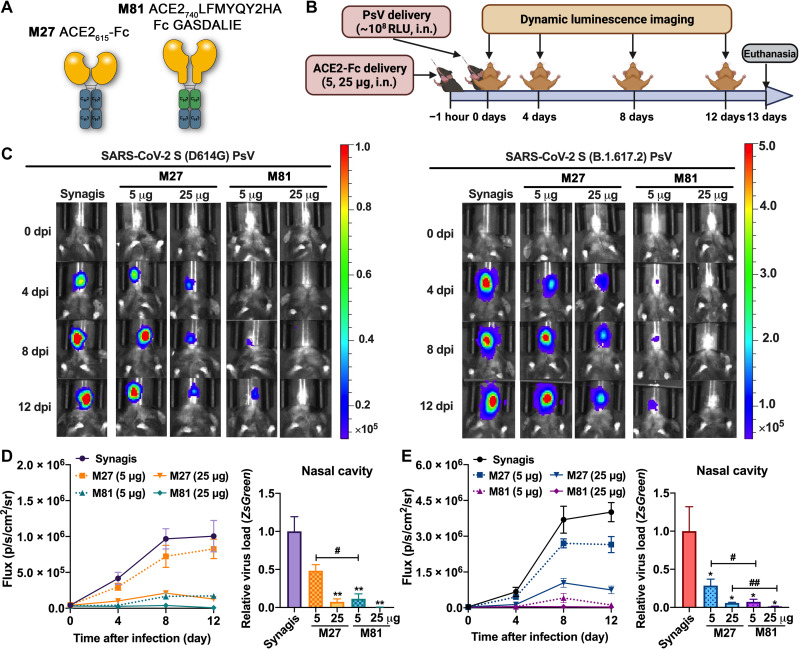
Engineered hACE2-Fc efficiently blocks SARS-CoV-2 PsV transduction in K18-hACE2 mice
Next, we tested the capacity of engineered hACE2-Fc to prevent SARS-CoV-2 viral transduction in vitro using an adapted pseudo-virus–based mouse infection protocol (36) that provides a safe alternative for evaluating antivirals in vitro under ABSL-2 (animal biosafety level 2) conditions (Fig. 5). Lentivirus pseudo-typed with S from two highly infective SARS-CoV-2 variants, D614G and B.1.617, were produced using the same protocol as above (37) and concentrated by PEG-8000 (polyethylene glycol, molecular weight 8000). In K18-hACE2 transgenic mice, 5 or 25 μg of ACE2-Fc (M27 or M81) or 25 μg of Synagis (control IgG) was delivered intranasally (i.n.) 1 hour before administration of replication-defective SARS-CoV-2 PsVD614G or PsVB.1.617 [~1× 108 plaque-forming units (PFU), i.n.] expressing Luc2 firefly luciferase. Longitudinal bioluminescence imaging (BLI) on live mice was performed at 4, 8, and 12 days post-infection (dpi) (Fig. 5, A and B). Because of the nonreplicative nature of the pseudo-virus and a suboptimal luciferase reporter for in vitro imaging, the BLI signal was only detected around the nasal cavity. In the control IgG–treated mice, the fluorescent signal increased by a factor of >1000 over the 12-day time course, demonstrating that SARS-CoV-2 PsV was capable of transducing cells in the nasal cavity of K18-hACE2 mice (Fig. 5, C to E, and fig. S8D). The luminescence intensity increased between 0 and 8 days and plateaued thereafter.
Fig. 5. In vivo efficacy of engineered ACE2-Fc in blocking SARS-CoV-2 PsV transduction in K18-hACE2 mice.
(A) Scheme of wt ACE2615-Fc (M27) and the engineered variant M81. (B) Experimental design of the PsV-challenged K18-hACE2 mouse model. M27, M81, or Synagis (a negative control) were intranasally administrated 1 hour before SARS-CoV-2 PsVD614G or PsVB.1.617.2 challenge (~108 RFU, i.n.), and noninvasive luminescence imaging was performed every 4 days. (C) Representative BLI images that indicate the luciferase signal for PsVD614G (left) and PsVB.1.617.2 (right). (D and E) Quantification of luciferase signal as flux (photons/s) computed noninvasively in the nasal area (left) and real-time polymerase chain reaction (PCR) quantification of SARS-CoV-2 PsV RNA loads (targeting ZsGreen) in the nasal cavity at the end point (13 dpi; right) for PsVD614G (D) (n = 3 to 4) and PsVB.1.617.2 (E) (n = 4 to 5). The data are shown as means ± SEM. Kruskal-Wallis test with Dunn’s post hoc test: *P < 0.05 and **P < 0.01 versus synagis and #P < 0.05 and ##P < 0.01.
Mice pretreated with 5 μg of wtACE2615-Fc (M27) before PsVD614G exposure showed a slight reduction in luminescent signal, whereas increasing M27 to 25 μg led to a viral inhibition of >85%. In contrast, only 5 μg of our lead variant ACE2 LFMYQY2HA-Fc (M81) was needed to reach >85% inhibition, while 25 μg of M81 nearly completely eradicated the BLI signal (Fig. 5C, left). End point analysis (13 dpi) after necropsy to estimate viral transduction levels (ZsGreen mRNA level) in the nasal cavity demonstrated that the low-dose M81 had equivalent antiviral activity to high-dose M27 (Fig. 5D).
In the PsVB.1.617 challenge model, which demonstrated considerably higher infectivity than PsVD614G (Fig. 5C, right, and fig. S8D), administration of 5 μg of M27 failed to inhibit viral transduction, and the 25-μg M27–treated group maintained ~20% luciferase signal at 12 dpi (Fig. 5E). Conversely, 5 μg of M81effectively protected mice, with only the basal luciferase signal detected at 12 dpi. Our SARS-CoV-2 PsV-challenged mouse model thus demonstrated that engineered ACE2-Fc M81 has the potential to effectively inhibit SARS-CoV-2 infection when administered prophylactically.
Engineered ACE2-Fc variants mediate potent Fc-effector functions
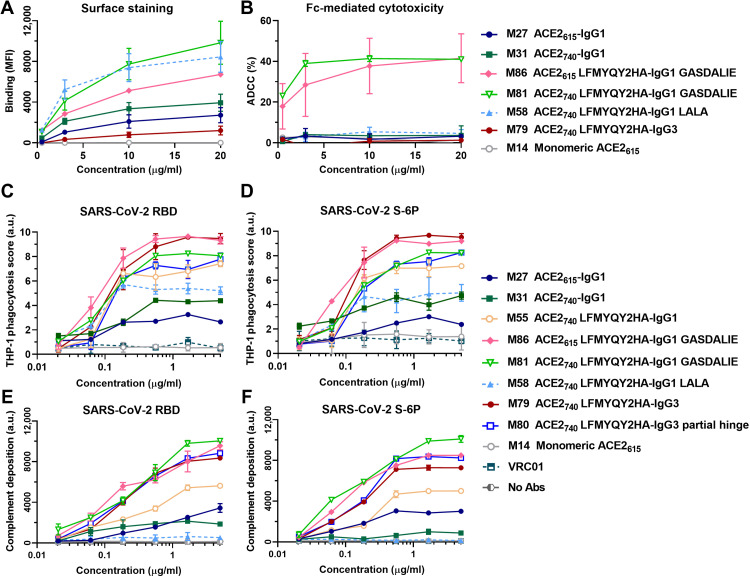
To further dissect the mechanism of action for protection observed in mice, engineered ACE2-Fc variants with different Fc modifications and subclasses were assessed for Fc-mediated effector activities in several in vitro assays that quantify ADCC, ADCP, and ADCD, as previously described (38, 39). As shown in Fig. 6A, engineered ACE2 LFMYQY2HA-Fc variants (M58, M81, and M86) showed higher surface binding to target cells expressing SARS-CoV-2 S compared to ACE2-Fcs without RBD binding–enhancing mutations (M27 and M31). We also observed low-binding efficiency of M79, ACE2740 LFMYQY2HA grafted to the IgG3 backbone, most likely due to the mismatch of the anti-IgG1 secondary Ab used for fluorescent quantification (Materials and Methods). Although all ACE2-Fcs tested showed binding to the target cells (Fig. 6A), only variants with Fc GASDALIE (M81 and M86) were able to stimulate robust cytotoxic responses, leading to the killing of ~40% of target cells expressing S protein (Fig. 6B). No ADCC activity was observed for the equivalent ACE2 variants with either Fc LALA or IgG3 backbones.
Fig. 6. Fc-effector function activities of engineered ACE2-Fcs.
(A) Mean fluorescence intensity (MFI) of CEM.NKr cells expressing SARS-CoV-2 S (CEM.NKr-S) stained with varying concentrations of ACE2-Fcs or monomeric ACE2. The background MFI signal obtained on parental CEM.NKr CCR5+ cells was subtracted to the signal on CEM.NKr-S cells. (B) Percentage of ADCC in the presence of titrated amounts of ACE2-Fcs or monomeric ACE2 as in (A) using 1:1 ratio of parental CEM.NKr CCR5+ cells and CEM.NKr-S cells as targets. Peripheral blood mononuclear cells from healthy donors were used as effector cells. (C and D) Fc-dependent cellular phagocytosis by THP-1 effector cells against the fluorescent microspheres (1 μm) coated with SARS-CoV-2 RBD (C) or S-6P (D) in the presence of varying concentrations (0.02 to 5 μg/ml) of ACE2-Fcs, monomeric ACE2, or VRC01. Data were means ± SD from at least two technical replicates. a.u., arbitrary units. (E and F) Fc-mediated complement deposition. Multiplex assay microspheres coated with SARS-CoV-2 RBD (E) or S-6P (F) were incubated with fourfold serial diluted ACE2-Fcs, monomeric ACE2 (0.02 to 5 μg/ml), or blank buffer control (no Abs) before incubation with guinea pig complement. Anti–guinea pig C3 IgG (conjugated with a red pigment) was used to detect the bound C3 on immune complexes. Data were means ± SD of four replicates.
Unlike ADCC, all ACE2-Fc variants showed dose-dependent ADCP activity against beads coated with both viral antigens (Fig. 6, C and D). M86 (ACE2615 LFMYQY2HA-Fc GASDALIE) and M79 (ACE2740 LFMYQY2HA-Fc in the IgG3 backbone with full hinge) stimulated the highest phagocytic responses, followed by M81 (ACE2740 LFMYQY2HA-Fc GASDALIE) and M80 (ACE2740 LFMYQY2HA in the IgG3 backbone but with a truncated hinge). This suggests that FcγR-Fc engagement of ACE2-Fc variants is partially impaired by increased flexibility in the CLD (M86 versus M81) and shorter hinge length (M79 versus M80). The latter observation is in line with a recent study that shows that several anti–HIV-1 Abs with extended hinges mediated enhanced ADCP against gp140-coated beads (25). Notably, residual ADCP activity for M58, the Fc LALA variant, was observed against antigen-coated beads (Fig. 6, C and D) and S-expressing CEM.NKr target cells (fig. S9). This finding is consistent with previous studies that LALA mutations do not completely abrogate binding to FcγRII, a primary receptor in ADCP signaling (40, 41).
In the bead-based ADCD assay (Fig. 6, E and F), Fc-mediated complement activation was affected by both ACE2-Fc binding capacity to antigens coupled on the beads and Fc subclasses. The LFMYQY2HA bearing M81 and M86 with Fc GASDALIE elicited the most potent deposition of complement C3, followed by the IgG3 subtypes M79 and M80. M55 with LFMYQY2HA and wt IgG1 Fc showed significantly weaker ADCD activity, and its Fc LALA (M58) exhibited activity only slightly above baseline. The ACE2-Fc with no RBD binding–enhancing mutations (M27 and M3) displayed only ~30% of ADCD efficiency as compared to M81 and M86. In summary, the ADCD efficiency of ACE-Fc was positively correlated with its binding affinity to the antigen-coated beads, as well as Fc composition, with a C1q recruitment efficiency rank of IgG1 Fc GASDALIE > IgG3-Fc > IgG1-Fc > IgG1 LALA-Fc.
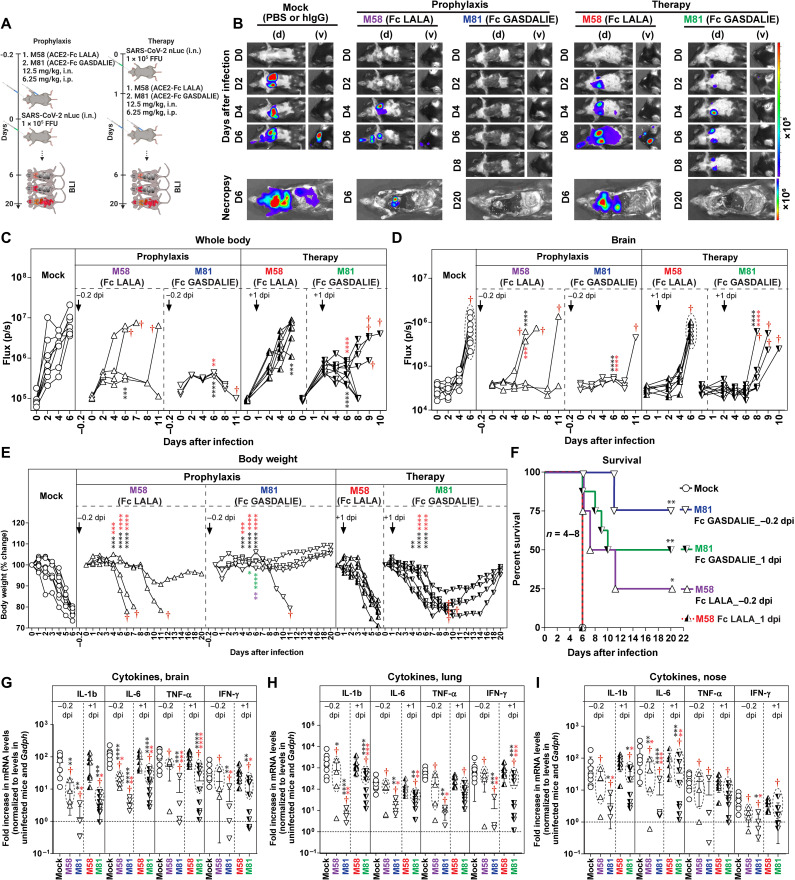
Engineered ACE2-Fc with enhanced Fc-effector functions protects or delays SARS-CoV-2 lethal infection in K18-hACE2 mice
We first confirmed that the Fc modifications (LALA and GASDALIE) did not significantly alter the pharmacokinetics in C57BL/6J mice before carrying out in vivo efficacy studies (fig. S8C). Next, we tested the in vivo efficacy and contribution of Fc-effector components to the overall functionality of engineered ACE2-Fcs under prophylactic and therapeutic settings, in K18-hACE2 mice challenged with SARS-CoV-2 nLuc (WA/2020). For delivery, we used a combination of intranasal and intraperitoneal routes to provide mucosal and systemic protection, respectively. K18-hACE2 mice were treated with two engineered ACE2-Fcs, i.e., ACE2740 LFMYQY2HA-Fc GASDALIE (M81) or ACE2740 LFMYQY2HA-Fc LALA (M58), both intranasally (12.5 mg/kg) and intraperitoneally (6.25 mg/kg), 5 hours before (prophylaxis) or 24 hours after (therapeutic) intranasal challenge with SARS-CoV-2 nLuc [1 × 105 focus-forming units (FFU)] (Fig. 7A). Longitudinal noninvasive BLI and terminal imaging revealed that prophylactic administration of M81 efficiently inhibited SARS-CoV-2 replication in the lungs and nose and prevented neuroinvasion compared to control mice (Fig. 7, B to D, and fig. S10, A and B). While control mice showed rapid weight loss and succumbed to infection at 6 dpi (Fig. 7, E and F), three of four mice treated with M81 (Fc GASDALIE) did not experience any weight loss and were completely protected, and the fourth mouse showed a significant delay in weight loss and survived until 11 dpi (Fig. 7, B and C). Although all mice pretreated with M58 (Fc LALA) showed delayed weight loss during the initial phase of infection, only one of four mice fully recovered (Fig. 7, B to F). These results revealed a significant difference in the prophylactic outcome for mice pretreated with the GASDALIE variant, engineered to have enhanced affinity for Fcγ receptors (Fig. 8), to those pretreated with the LALA variant that are capable of direct neutralization (Fig. 6) but impaired in Fc-effector functionality. Together, these data demonstrated that both direct neutralization and Fc-effector activities of ACE2-Fc contributed to protection from infection and mortality in a prophylactic setting (Fig. 7, B and C).
Fig. 7. In vivo efficacy of Fc-null/enhancing ACE2-Fcs in prevention from lethal SARS-CoV-2 infection in K18 hACE2 transgenic mice.
(A) Schemes showing the experimental design for testing the in vivo efficacy of M58 (ACE2740 LFMYQY2HA-Fc LALA) and M81 (ACE2740 LFMYQY2HA-Fc GASDALIE) delivered with a dose of 12.5 mg/kg, i.n. and 6.25 mg/kg, i.p. 5 hours before (−0.2 dpi, prophylaxis) or 1 day after (+1 dpi, therapy) challenge of K18-hACE2 mice with 1 × 105 FFU of SARS-CoV-2 nLuc. PBS (n = 4) or human IgG–treated (n = 4) mice were used as controls. (B) Representative BLI images of SARS-CoV-2 nLuc–infected mice in ventral (v) and dorsal (d) positions. The dorsal view shows images of the head to highlight virus neuroinvasion. (C and D) Temporal quantification of the nLuc signal as flux (photons/s) computed noninvasively. (E) Temporal changes in mouse body weight, with initial body weight set to 100%. Mice that succumbed to infection (cohorts not 100% mortality) are denoted with red daggers. (F) Kaplan-Meier survival curves of mice (n = 4 to 8 per group) statistically compared by log-rank (Mantel-Cox) test for the experiments shown in (A). (G to I) Fold changes of the end point cytokine mRNA expression in brain, lung, and nasal cavity tissues. Data were normalized to Gapdh mRNA in the same sample and that in noninfected mice after necropsy. Cytokines in indicated tissues were determined when they succumbed to infection (dashed ellipse with red dagger) and at 20 dpi for surviving mice. Statistical significance for group comparisons to control are shown in black, M58 (prophylaxis) in purple, M81 (prophylaxis) in blue, M58 (therapy) in red, and M81 (therapy) in green. Nonsignificant comparison is not shown. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Mean values ± SD are depicted.
Next, we tested whether M81 or M58 could resolve an established infection when administered 1 day after SARS-CoV-2 challenge (Fig. 7, A and B). While M58 (Fc LALA) failed to rescue SARS-CoV-2–infected mice, treatment with M81 (Fc GASDALIE) protected 50% of mice and significantly delayed weight loss and death in mice that succumbed to infection (Fig. 7B). The protection correlated with significantly reduced viral replication in the brain, lungs, and nose, as determined by imaging (nLuc activity in organs; flux) after terminal necropsy and by viral load analyses (fig. S10, A to D). Concomitant with the efficient viral clearance, substantial reductions in the mRNA level of several proinflammatory cytokines, including interleukin-1β (IL-1β), IL-6, tumor necrosis factor–α (TNFα), and interferon-γ (IFN-γ), in the nose, lung, and brain were observed in M81-treated mice compared to mock [phosphate-buffered saline (PBS) or IgG] and M58-treated cohorts (Fig. 7, G to I). Given that IL-1β and Tnfα are thought to be primarily produced by activated macrophages, our data indicate that treatment with the Fc-optimized M81 is unlikely to enhance macrophage-tropic viral infection or trigger the adverse inflammatory signaling that serves as the hallmark of Ab-dependent enhancement (ADE). Fc-effector activities played the predominant role in clearing established infection as all mice treated with ACE2-Fc LALA succumbed to infection. Thus, enhanced Fc-effector functions recruited by M81 ACE2-Fc were critical for viral control during therapy.
In comparison to combined administration (intranasally and intraperitoneally), the single delivery route of M81 (either intranasally at 12.5 mg/kg alone or intraperitoneally at 6.25 mg/kg) was not able to prevent viral neuroinvasion and, as a result, all mice succumbed to infection at a similar rate to control cohorts (fig. S11), although significant reduction in lung viral loads was observed when M81 was delivered intranasally. This suggests that intranasal administration helps to neutralize/inactivate free virus in the mucosal airways, while intraperitoneally delivered ACE2-Fc is necessary for systemic protection to combat viral spread in the whole body.
Engineered ACE2-Fcs potently inhibit the Omicron variant
Given that the Omicron variant (B.1.1.529) has been identified in 171 countries and rapidly outpaced the Delta variant to drive an upsurge of infection globally since January 2022, we sought to examine whether our engineered ACE-Fcs remain effective against this VOC (Fig. 8). Unlike many nAbs that showed markedly reduced efficacy to the Omicron variant (42), M81 displayed higher neutralization potency against PsVB.1.1.529 (IC50 = 0.075 nM) compared to other tested VOCs (Figs. 8, B and C, and 4B), which was, in part, attributed to the extended CLD compared to the shorter variant M86. As expected, M81 and M163 with GASDALIE-Fc retained exceptional ADCP and ADCD activity of RBDB1.1.529-coated beads (Fig. 8D) in contrast to the variants with wt Fc (M55) and LALA-Fc (M58).
Last, we tested the prophylactic efficacy of M58 and M81 in K18-ACE2 mice challenged with an increased titer (1 × 106 FFU) of authentic Omicron virus isolated from patients (Fig. 8, E to G). Consistent with a previous observation (43), the Omicron variant showed substantially lower infectivity and pathogenesis compared to the earlier VOCs, as even untreated K18-hACE2 mice began to recover from 2 dpi. Nonetheless, mice treated with ACE2-Fc underwent rapid recovery by 1 dpi, and the tissue viral load analysis after necropsy on 3 dpi demonstrated a significant viral load reduction in lungs compared to control mice (Fig. 8G). Furthermore, we observed lower viral spread, particularly in the lungs of mice treated with M81 (Fc GASDALIE) compared to its LALA-Fc derivative M58, highlighting the important role of Fc-effector functions in limiting viral replication in infected organs. These data confirm the high efficacy of ACE2-Fc against replication-competent Omicron variant in vivo.
DISCUSSION
ACE2-based therapies have proven effective at countering SARS-CoV-2 infection in humanized organoids (8, 44), animals (15, 45, 46), and a patient with severe COVID-19 (10). These interventions include soluble monomeric ACE2 and dimeric ACE2 Ig-like fusions that have improved avidity and extended serum half-life. As the primary host receptor, recombinant ACE2 derivatives are intrinsically broad neutralizers of ACE2-using coronaviruses that include SARS-CoV-1 and SARS-CoV-2 and those with pandemic potential found in bats. Furthermore, hACE2 decoys have an advantage over Abs elicited by infection or vaccination as they are predisposed to block mutational escape of VOCs that generally emerge with increased affinity to the host receptor. Here, we developed bifunctional ACE2-Fc variants that not only broadly neutralize SARS-CoV-2 VOCs but also engage host innate immune cells through the Fc to eliminate free virions and infected cells. We also showed that both modes of action are required for ACE2-Fc to optimally prevent and control lethal SARS-CoV-2 infection in a K18-hACE2 mouse model.
By using structure-based approaches, we identified three novel ACE2 mutations L79F/M82Y/Q325Y that enhanced RBD binding by up to ~12-fold and improved neutralization potency and breadth against eight SARS-CoV-2 VOCs in PsV assays, including the widespread Delta (B.1.617) and Omicron (B.1.1.529) variant. Structurally, L79F and M82Y (Site-II), together with Y83, orchestrate stacking of F486RBD and stabilize the mobile RBD ridge, while Q325Y (Site-III) forms an additional hydrogen bond to Q506RBD. Given that the ACE2 Site-I helix has already been extensively modified to acquire increased affinity for SARS-CoV-2 RBD (13–16), our additional mutations in Site-II and Site-III could potentially be combined with the validated Site-I mutations to create an ultrapotent ACE2 antiviral. The potential immunogenicity of engineered ACE2, e.g., their propensity to induce adverse T cell activation and autoantibodies that target endogenous ACE2, needs to be examined with caution (15).
Our data suggested that the increased structural flexibility rendered by the CLD dimerization domain in the context of ACE2-Fc likely facilitates S binding and neutralization of SARS-CoV-2 VOCs. However, ACE2-Fc with CLD may become more susceptible to enzymatic cleavage by several SARS-CoV-2 entry–related proteases, e.g., TMPRSS2 and ADAM17, which cleave ACE2 within the CLD region (47). Further studies are needed to test whether the presence of CLD could affect the half-life of ACE2-Fc in sera.
To prevent the potential off-target effects, we introduced H374A/H378A to create a catalytically null ACE2-Fc. Historically, the ACE2-inactive variants were generated by replacing H374 and H378 with Asn (48). We selected Ala over Asn on the basis of the presumption that Ala will introduce minimal perturbation to the overall structure of ACE2 while providing the desired loss of ACE2 activity. Ala introduces no charge to the mutation site and is, therefore, usually a better option for replacing His, a strong, positively charged Zn2+ chelator. Our structural analyses validate our approach as we observed a shift of the catalytic Zn2+ to the substrate binding pocket of ACE2 that not only blocked angiotensin binding but also maintained protein stability through the formation of a new Zn2+ binding site without changing the ACE2-RBD interface.
Our PsV-challenged K18-hACE2 mouse model, adapted from a previously reported protocol (36), provided a safe and inexpensive platform for the dynamic in vivo efficacy assessment of SARS-CoV-2 antivirals, which can be widely used in BSL-2 laboratories. With this preclinical model, the mutational effects elicited by the S protein of SARS-CoV-2 VOCs could be rapidly evaluated, although the viral transduction was limited to the nasal cavity and high PsV titers were required for BLI visualization. In this model, our best-performing variant M81 (5 μg, i.n.), was able to prevent viral transduction as effectively as 25 μg of wtACE2-Fc, M27.
A few ACE2-Fc variants have been developed by others as therapeutic candidates that are capable of viral neutralization, mostly by mechanisms involving direct competition for viral S binding to the host cell surface ACE2 (13–16, 29, 32, 49). However, the bivalent IgG-like ACE2-Fcs not only are capable of avidly interacting with viral antigen but also “profit” from the Fc domain, which can be recognized by effector cells in the host. Our data indicate that ACE2-Fc with “wt” IgG1 Fc only elicited moderate ADCP and ADCD in vitro. No cytotoxic response against S-expressing T lymphoid cells was detected for any of our optimized ACE2-Fc (IgG1 wt) variants. In sharp contrast, potent ADCC, enhanced ADCP, and complement activation were detected when ACE2-Fcs were modified to include the well-known GASDALIE mutations that show increased affinity to FcγRIIIA (50). ACE2740 LFMYQY2HA-Fc GASDALIE (M81) in an IgG1 backbone had ADCP activities comparable to its isoform in an IgG3 backbone (M79), suggesting that, similar to ADCP in HIV-1 Abs, the Fc-dependent phagocytic response is influenced by IgG isotype and hinge length (25, 26).
To monitor the real-time in vivo efficacy of our lead ACE2-Fc variant, we used the K18-hACE2 mouse model challenged by SARS-CoV-2 nLuc (51). This model is highly susceptible to SARS-CoV-2 infection because of the constitutively high endogenous hACE2, particularly in the brain, posing a major obstacle for soluble ACE2-based antivirals, such as our engineered ACE2-Fc, to achieve full protection (18, 46). However, this model does recapitulate pathological features of SARS-CoV-2 infection such as acute lung injury and acute respiratory disorder syndrome seen in humans (52). As a result, the K18-hACE2 mouse model has been used extensively for evaluating contributions from direct neutralization and Fc-effector activities mediated by nAbs (53) and non-neutralizing Abs (54).
Our in vivo efficacy study demonstrated that the Fc-optimized GASDALIE variant M81 provided a significant benefit when administered prophylactically and therapeutically in K18-hACE2 mice. In both settings, M81 treatments were associated with the marked reduction of virus-induced weight loss, proinflammatory cytokine response, and mortality, particularly in the therapeutic model, when compared to the M58 (LALA-Fc)–treated groups. Presumably, the boosted Fc-effector functions are necessary to efficiently eliminate the viral reservoir when infection is established and considerable numbers of infected cells are present. Given that the human-Fc/mouse-FcγR mismatch may compromise the Fc-effector functionality of ACE-Fcs in K18-hACE2 mice, we expected a better therapeutic outcome in species-matched systems, such as humanized FcγR mice or clinical trials. We did not observe any Fc-related pathogenic or disease-enhancing effects in ACE2-Fc–treated mice. Further studies are required to delineate which Fc-FcγR–mediated pathways confer the improved efficacy of Fc-engineered human IgGs and Fc-fusion molecules considering their immunomodulatory, inflammatory, and cytotoxic activity in other settings.
There is an ongoing debate regarding the benefit/risk of using the Fc-tagged ACE2. While ACE2-Fc generally has increased antiviral activity because of the increased avidity for S antigen, extended serum circulation, and benefits from Fc-effector functions, there is the potential risk of ADE, which should be considered and investigated with caution. There are two major mechanisms by which Abs or Fc-fusion proteins can cause ADE: mechanism 1, by enhancing the viral infection via Fc-mediated interaction with FcγR present on certain effectors cells, e.g., monocyte and macrophage, and mechanism 2, by elevating inflammation and thereby exacerbating immunopathology (55). The common in vitro method used to verify ADE via mechanism 1 is to quantify viral infectivity in the presence of varying concentration of the tested monoclonal antibody (mAb) or Fc-tagged molecule in a host cell that expresses a single FcγR and the viral receptor (56). A few SARS-CoV-2 anti-S mAbs targeting epitopes within the N-terminal domain (57) or RBD (56) have been reported to increase viral infectivity in vitro, but no evidence of increased infection was observed in vivo when mAbs were tested in mice or monkeys (58). Now, there are no available data suggesting that ACE2-Fc can induce ADE in vitro by mechanism 1. Furthermore, our in vivo studies in K18-hACE2 mice demonstrated that treatment with the Fc-enhanced GASDALIE variant (M81) correlated with lower induction of proinflammatory cytokines compared to the Fc-null LALA variant (M58). These data suggest that the GASDALIE ACE2-Fc mutant is unlikely to enhance infection or inflammation via mechanism 2. Notably, a recent study that developed a novel ACE2 dimer fused to a dimerization motif (DDC) and the albumin-binding domain (ABD) instead of the Fc tag (18) showed that multiple-dose administration of this ACE2-DDC-ABD produced promising therapeutic outcomes in the K18-hACE2 mice challenged by a lethal titer of SARS-CoV-2 (2 × 104 PFU, i.n.). Future studies directly comparing ACE2-Fc and its LALA and GASDALIE variants to other non–Fc-tagged ACE2 dimers, such as the ACE2-DDC-ABD, would be needed to better understand the relative contribution of Fc to in vivo effectiveness benefits/risks and whether ACE2-Fc mediates ADE.
Multiple studies confirm the beneficial effect of ACE2 enzymatic activity in anti–SARS-CoV-2 activities of the ACE2–based therapeutics (11, 59). Active sACE2 and ACE2-like enzymes were shown to effectively reduce the vasopressor Ang II in plasma and attenuate acute lung injury caused by SARS-CoV-1 and SARS-CoV-2 by rebalancing renin-angiotensin homeostasis when membrane-bound ACE2 has been stripped and down-regulated by S binding (11, 12, 42). Thus, compared to inactive forms, engineered ACE2 antivirals with intrinsic Ang II–converting activity are expected to be a better therapeutic option, which can not only block viral spread but also modulate the RAS system to prevent lung failure in patients with SARS-CoV-2. Our lead ACE2-Fc variants were prepared as enzymatically inactive variants and lack the angiotensin activity component. The protective effect in K18-hACE2 mice that we observed, therefore, resulted from direct neutralization and the Fc-effector activities of our ACE2-Fc and do not reflect the combined effect of ACE2 as a viral decoy and an enzyme in protection from lung injury. In this regard, a recent study has shown that an enzymatically active ACE2-Fc conferred improved protection as compared to the inactive counterpart when delivered by inhalation to K18-hACE2 mice (59).
To summarize, our data highlight the utility of engineered ACE2-Fcs as broadly effective therapeutics capable of countering SARS-CoV-2 VOCs, including the highly transmissible and immune-evasive Delta and Omicron variants. Our data point toward a crucial role of Fc-effector activities in the antiviral mechanism of ACE2-Fcs. While the engineered ACE2-Fc in wt IgG1 backbone showed moderate Fc functions in vitro, the equivalent Fc-enhancing variants robustly stimulated Fc-effector responses and were capable of better in vivo protection. Overall, our findings fully support the translational relevance of engineered ACE2-Fcs as first-line antivirals for mild to moderate SARS-CoV-2 infection and underlie the importance of Fc-effector functions as an additional antiviral mechanism.
MATERIALS AND METHODS
Structure-based design of ACE2 mutations
Mutation sites within the ACE2 domain were selected on the basis of a structural analysis of the ACE2-RBD interface with the aid of PISA (www.ebi.ac.uk/pdbe/pisa/). Interfaces of two reported high-resolution crystal structures of the complex formed between the wtACE2 and the SARS-CoV-2 RBD and the chimeric RBD composed of the core of SARS-CoV-1 RBD and the receptor binding motif of SARS-CoV-2 [PDB codes: 6M0J (23) and 6VW1 (24), respectively] were analyzed for their BSA and the physiochemical properties of residues at their interfaces. RBD binding–enhancing mutations were selected on the basis of their potential to enhance charge complementarity, the hydrogen bonding network, and/or hydrophobic interactions (e.g., their ability to improve binary packing and reduce steric repulsion, fill empty cavities, or improve aromatic interactions). The enzymatically inactive angiotensin variant was prepared by mutating two zinc-chelating His residues (H374 and H378) in the active site to Ala.
Plasmid construction
To generate the expression plasmids of hACE2 and human IgG1 fusions, the synthetic gene (GenBank, BAJ21180.1; with the original Bam HI site destroyed) encoding the hACE2 PD (residues 1 to 615) and the complete ECD (residues 1 to 740) were fused to the human IgG1 Fc segment (residues D217 to K443) or the codon-optimized human IgG3 Fc (GenBank, AIC59039.1) with full hinge (residues E243 to G520, R509H) or partial hinge (residues P286 to G520, R509H), in which a Bam HI site was inserted between the ACE2 and IgG Fc. The DNA chimera was then cloned into the pACP-tag (m)-2 vector (Addgene, no. 101126) using Nhe I and Not I [New England Biolabs (NEB)] as the restriction sites. All ACE2 mutations were introduced onto the ACE2-IgG backbone by a two-step mutagenesis protocol. Likewise, to generate the engineered Fc variants, gene segments encoding ACE2 PD, ECD, or those with desired ACE2 mutations were fused to the codon-optimized synthetic IgG1 Fc (GenScript) in which GASDALIE or LALA mutations were incorporated. To generate SARS-CoV-2 RBDwt (residues 319 to 541 or residues 319 to 537, for crystallization), RBDB.1.1.7 (residues 329 to 527, N501Y), RBDB.1.351 (residues 329 to 527, K417N/E384K/N501Y), and RBDB1.1.529 (residues 319 to 537; mutations listed in fig. S5B), the respective codon-optimized DNA segments fused with an N-terminal secretion peptide and a C-terminal 6xHis tag were cloned into the pACP-tag (m)-2 vector using either Eco RI/Not I for RBDwt (319 to 541), RBD B.1.1.7, and RBDB.1.351 or Bam HI/Xho I for RBDwt (319 to 537) as restriction enzymes.
Protein expression and purification
FreeStyle 293F cells (Thermo Fisher Scientific) were grown in FreeStyle 293F medium (Thermo Fisher Scientific) to a density of 1 × 106 cells/ml at 37°C with 8% CO2 with 135-rpm agitation. For production of ACE2-Fc variants, cells were transfected with the corresponding plasmids (100 μg per 108 cells) following the polyethylenimine (PEI) transfection protocol. One week after transfection, cells were pelleted and the supernatant was clarified using a 0.22-μm filter, and protein was purified using protein A resin (Pierce), followed by SEC on a Superose 6 10/300 column (Cytiva) equilibrated with 1× PBS. Monomeric ACE2wt and engineered ACE2LFMYQY2HA plasmids encoding ACE2 (residues 1 to 615) with C-terminal HRV-3C-cleavable 8xHis tag were transfected to FreeStyle 293F cells, and the resulting protein was purified over Ni–nitrilotriacetic acid (NTA) columns (Cytiva). His-tag removal was carried out by overnight HRV-3C (Sigma-Aldrich) digestion at 4°C, and the cleaved protein was then purified on Ni-NTA before being subjected to SEC on Superose 6 10/300 column (Cytiva) equilibrated with PBS.
For recombinant expression of SARS-CoV-2–stabilized spike ectodomain [S-2P (4) and S-6P (60), gifted by J. S. McLellan], RBDwt (residues 319 to 541), RBDB.1.1.7, RBDB.1.351, RBDB.1.1.529, and SARS-CoV RBD (residues 306 to 577; with C-terminal HRV3C-cleavable IgG1 Fc tag and 8xHis tag), plasmids encoding the respective genes were transfected to 293F cells with the same protocol as described above. Supernatants were purified on either Strep-Tactin resin (IBA) for S-2P and S-6P or Ni-NTA columns for SARS-CoV RBD, SARS-CoV-2 RBD, and its variants. S-2P, S-6P, and SARS-CoV RBD were then incubated with HRV3C protease at 4°C overnight, and the mixtures were passed over a Ni-NTA column to remove the protease and cleaved tags. All viral proteins were further purified by SEC on either a Superose 6 10/300 or a HiLoad 16/600 Superdex 200 pg in PBS before being used for indirect ELISA and SPR.
Single-molecule mass photometry
The sample quality and molecular weight of the glycosylated monomeric ACE2615, ACE2-Fcs, and SARS-CoV-2 S-6P were assessed by mass photometry (MP). Purified nontagged ACE2615, ACE2-Fcs, or S-6P was diluted to ~50 nM in PBS, and MP data were acquired and analyzed using a OneMP mass photometer (Refeyn Ltd., Oxford, UK). The estimated molecular weight (75, 230, 270, and 540 kDa for ACE2615, ACE2615-Fc, ACE2740-Fc, and SARS-CoV-2 S-6P, respectively) were used for A280 (absorbance at 280 nm)–based concentration determination (corrected by extinction coefficients).
Enzyme-linked immunosorbent assay
The binding capacity of the purified ACE2-Fcs to various viral antigens was measured by indirect ELISA, as described in (61). Ninety-six–well Nunc Maxisorp plates (Sigma-Aldrich) were coated with SARS-CoV-2 RBDwt (residues 319 to 541) (50 ng), RBDB.1.351 (50 ng), S-2P (75 ng), SB.1.1.7 (75 ng), SB.1.351 (75 ng), SP.1 (75 ng), SB.1.526 (75 ng), and SARS-CoV-1 RBD (50 ng) per well in tris-buffered saline (TBS) at 4°C overnight. Plates were washed with TBS before blocking with TBS + 5% nonfat milk powder and 0.1% Tergitol (blocking buffer) at RT for 2 hours. After washing one time by TBS supplemented with 0.1% Tween 20, serial dilutions of purified ACE2-Fcs (125, 62.5, 20, 10.0, 5.0, 2.5, 0.5, and 0.05 nM) were added and incubated at 4°C overnight. Plates were washed three times and incubated with the goat anti-human IgG Fc secondary Ab conjugated with alkaline phosphatase (AP; Southern Biotech) at a 1:1000 dilution in blocking buffer for 1 hour at RT. Plates were washed three times and developed using the Blue Phos Microwell Phosphatase Substrate System (SeraCare). The reactions were stopped after 5 min of incubation at RT by adding the equivalent volume of the APstop Solution (SeraCare). The plates were then read at 620 nm, and the optical density was recorded by the SpectraMax Plus microplate reader (Molecular Devices). All binding events were measured in triplicate, and each dataset was normalized [OD620 (optical density at 620 nm) at 125 nM as 100%] for cross comparison. GraphPad Prism was used to display the mean and SEM for all groups and used to calculate the area under the curve within the concentration range of 0.05 to 2.5 nM using 5% binding as baseline (Fig. 2D and fig. S2).
Surface plasmon resonance
SPR measurements were carried out as described in (53). All assays were performed on a Biacore 3000 (Cytiva) at RT using 10 mM Hepes (pH 7.5), 150 mM NaCl, and 0.05% Tween 20 as running buffer. For the kinetic measurement of SARS-CoV-2 RBDwt, RBDB.1.1.7, RBDB.1.351, and SARS-CoV-1 RBD binding to ACE2-Fc variants, ~80 to 200 RU (response unit) of ACE2-Fcs were immobilized on a protein A chip (Cytiva), and twofold serial dilutions of the respective viral proteins were then injected as solute analytes with concentrations ranging from 6.25 to 200 nM (SARS-CoV-2 RBDwt and SARS-CoV RBD) or 3.125 to 200 nM (RBDB.1.1.7 and RBDB.1.351). For the kinetic measurement of the nontagged SARS-CoV-2 S-6P binding to ACE2-Fcs, ~60 RU of ACE2-Fcs was loaded on a protein A chip before the serial injection of twofold titrated S-6P (3.125 to 50 nM). To assess monomeric ACE2wt or ACE2LFMYQY2HA binding to SARS-CoV-2 RBD-Fc, ~120 RU of SARS-CoV-2 RBDwt (residues 319 to 591) was immobilized on a protein A chip, and twofold serial dilutions of monomeric ACE2 or the variant were injected with concentrations ranging from 6.25 to 100 nM. For all kinetic assays, the sensor chip was regenerated using 10 mM glycine (pH 2.0) before the next cycle. Sensorgrams were corrected by subtraction of the corresponding blank channel and for the buffer background, and kinetic constants were determined using a 1:1 Langmuir model with the BIAevaluation software (Cytiva), as shown in Fig. 2 (E and F) and fig. S3. The kinetic constants are summarized in table S1. Goodness of fit of the curve was evaluated by the chi square of the fit, with a value below 3 considered acceptable.
Crystallization and structure determination
For crystallographic protein preparation, plasmids encoding ACE2 (residues 1 to 615, LFMYQY2HA, with C-terminal HRV3C-cleavable 8xHis tag) or SARS-CoV-2 RBD (residues 319 to 537) were transfected into Expi293F GnTI cells (Thermo Fisher Scientific) using PEI. The proteins were harvested and purified on Ni-NTA; the C-terminal 8xHis tag on ACE2 was removed by HRV3C digestion as above. The resulting SARS-CoV-2 RBD and the cleaved ACE2 were further purified by gel filtration on Superose 6 10/300 in PBS.
The purified nontagged ACE2615(LFMYQY2HA) was mixed with excess RBD (molar ratio of 1:5) and incubated on ice for 2 hours. The mixture was then deglycosylated by Endo Hf (NEB) in 1× PBS at RT overnight. Endo Hf was removed by repeated loading onto amylose resin (NEB), and the crude ACE2-RBD mixture was further purified on a HiLoad 16/600 Superdex 200, which was preequilibrated in 10 mM tris (pH 8.0) and 100 mM ammonium acetate. The complex fractions were pooled and concentrated to ~7.5 mg/ml for crystallization. Crystallization trials were performed using the vapor diffusion hanging drop method with a 1:1.5 ratio of protein-to-well solution. Rod-shaped crystals were obtained in 0.2 M ammonium sulfate, 0.1 M MES (pH 6.5), and 20% (w/v) PEG-8000 after ~3 weeks of incubation at 21°C. Crystals were snap-frozen in the crystallization condition supplemented with 20% 2-methyl-2,4-pentanediol (MPD). X-ray diffraction data were collected at the SSRL beamline 9-2 and were processed with HKL-3000 (62). The structure was solved by molecular replacement in PHASER from the CCP4 suite (63) using 6M0J (23) and 1R4L (30) as independent searching models for the RBD and ACE2 moiety, respectively. Iterative cycles of model building and refinement were done in Coot and Phenix (64). Data collection and refinement statistics are shown in table S2.
ACE2 enzyme activity assay
Angiotensin-converting activity was determined using the fluorometirc ACE2 assay kit (BioVision). Briefly, the wtACE2-Fcs (M27 and M31) and H374A/H378A bearing M33 and M81 were diluted in assay buffer to the final concentrations of 1.56, 3.13, 6.25, 12.5, 25, and 50 nM, and the reactions were set up with/without ACE2 inhibitors as described in the manufacturer protocol. The time course measurements [excitation/emission (Ex/Em) = 320/420 nm] were performed in the EnSpire multimode plate reader (PerkinElmer). The initial linear regions in fig. S4D were used to calculate the slopes d(RFU)/d(t) in given ACE2-Fc concentrations shown in Fig. 3G. To measure its kinetic activity for the ACE2 native substrate Ang II as described in (65), 1 μl of diluted ACE2-Fc (3.13 to 50 nM) were added to the 9-μl reaction premix containing 1 μl of Ang II (AnaSpec; 10 mM) and 8 μl of developer/enzyme mix from the Phenylalanine Detection Kit (BioAssay Systems). The resulting kinetic curves were showed in fig. S4 (H and I).
SARS-CoV-2 pseudo-virus production
PsVs for in vitro neutralization assays, live cell imaging, and in vivo efficacy studies were produced using the SARS-CoV-2 S–pseudo-typed lentiviral kit (NR-52948, BEI Resources) as described in (37). The resulting PsV lentiviral particles with SARS-CoV-2 Swt expressed on the surface contained the reporter genes of synthetic firefly luciferase (Luc2) and synthetic Zoanthus sp. ZsGreen1. To generate PsV pseudo-typed with spikes of different SARS-CoV-2 VOCs, spike–pseudo-typing vector plasmids, including D614G (NR-53765, BEI Resources); P.1 [gift from R. Petrovich, etc., from the National Institute of Environmental Health Sciences (NIEHS)]; B.1.1.7, B1.351, B.1.429, P.1, B.1.526, and B.1.617.2 (InvivoGen); and B.1.1.529 (Sino Biological) were used in lieu of the Swt plasmid. Sixteen to 24 hours after seeding, 293T cells (Thermo Fisher Scientific) were cotransfected with the respective spike plasmid or vesicular stomatitis virus glycoprotein (positive control), lentiviral backbone, and three helper plasmids encoding Gag, Tat1b, and Rev1b (BEI Resources). At 72 hours after transfection, the supernatant was harvested and clarified by 0.45-μm filters. To determine viral titers, hACE2-expressing 293T cells [gift from A. Malloy, Uniformed Services University of the Health Science (USUHS)] were infected with serial PsV dilutions. Forty-eight to 60 hours after infection, the luciferase signal was detected by the Bright-Glo Luciferase Assay System (Promega) for titer estimations (37). PsVs were concentrated by the homemade fourfold lentivirus concentrator (protocol of MD Anderson) and stored at 4°C for short-term use or −20°C for longer storage.
In vitro neutralization assay
For in vitro neutralization assays, 50 μl of serial dilutions of Synagis, monomeric ACE2 (M14), and selected ACE2-Fcs (M27, M31, M81, and M86) (final concentration, 0.005 to 50 ng/μl) were preincubated with 50 μl of SARS-CoV-2 spike PsV (~106 RLU/ml) of WA-1/2020 isolate or eight VOCs in 96-well plates at 37°C for 1 hour. Subsequently, hACE2-expressing 293T cells (1.25 × 104 cells per well) in 50-μl culture medium were added and incubated at 37°C for 48 hours. Microscopic live-cell imaging for ZsGreen was performed using the All-in-One Fluorescence Microscope BZ-X (Keyence) (Fig. 4C and figs. S6 and S7), and the luciferase signal was further measured by the Bright-Glo Luciferase Assay System (Promega). Data analysis and normalization followed the protocol as described previously (66).
ADCC assay
The assay was carried out as previously described (38). Briefly, for evaluation of anti–SARS-CoV-2 ADCC activity, parental CEM.NKr CCR5+ cells were mixed at a 1:1 ratio with CEM.NKr-Spike cells. These cells were stained by AquaVivid (Thermo Fisher Scientific) for viability assessment and by a cell proliferation dye eFluor670 (Thermo Fisher Scientific) and subsequently used as target cells. Overnight rested peripheral blood mononuclear cells were stained with another cellular marker eFluor450 (Thermo Fisher Scientific) and used as effector cells. Stained effector and target cells were mixed at a 10:1 ratio in 96-well V-bottom plates. Titrated concentrations (0.5 to 20 μg/ml) of ACE2-Fc variants were added to the appropriate wells. The plates were subsequently centrifuged for 1 min at 300g and incubated at 37°C, 5% CO2 for 5 hours before being fixed in a 2% PBS-formaldehyde solution. Because CEM.NKr-Spike cells express green fluorescent protein (GFP), ADCC activity was calculated using the following formula: [(% of GFP+ cells in Targets + Effectors) − (% of GFP+ cells in Targets + Effectors + Ab)]/(% of GFP+ cells in Targets) × 100 by gating on transduced live target cells. All samples were acquired on an LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo v10 (Tree Star).
ADCP assay
ADCP assays were carried out as previously described (27). Briefly, streptavidin-coated 1-μm fluorescent microspheres were coated with biotinylated SARS-CoV-2 S-6P, RBDwt, or RBDB.1.1.529 (residues 319 to 541) overnight at 4°C. Following washing, the beads were incubated with purified ACE2-Fcs at varying concentrations (0.02 to 5 μg/ml) for 3 hours at 37°C and were analyzed in duplicate. For monocyte ADCP, THP-1 cells were used as effector cells. Cells were added to the bead/Ab mixture and incubated overnight to allow phagocytosis. Samples were then fixed and analyzed via flow cytometry to define the fraction and fluorescent intensity of cells that phagocytosed one or more beads.
To estimate ADCP efficiency for cellular elimination, CEM.NKr-Spike cells were used as target cells and were labeled with a proliferation dye eFluor450. THP-1 cells were used as effector cells and were stained with another proliferation dye eFluor670. Stained target and effector cells were mixed at a 5:1 ratio in 96-well plates. Titrated concentrations (0.78 to 50 μg/ml) of ACE2-Fc variants were added to the appropriate wells. After an overnight incubation at 37°C and 5% CO2, cells were fixed with a 2% PBS-formaldehyde solution. ADCP was determined by flow cytometry, gating on THP-1 cells that were triple positive for GFP, eFluor450, and eFluor670 cellular dyes. All samples were acquired on an LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo v10 (Tree Star). The resulting ADCP curves were shown in fig. S9.
ADCD assay
As described in (27), the selected ACE2-Fc variants at varied concentrations (0.02 to 5 μg/ml) were incubated with multiplex assay microspheres coated with SARS-CoV-2 RBDwt, RBDB1.1.529, or S-6P for 2 hours at RT. Lyophilized guinea pig complement was resuspended according to the manufacturer’s instructions (Cedarlane), and 2 μl per well was added in veronal buffer with 0.1% gelatin (Boston BioProducts). After washing, the mixtures of ACE2-Fc/microspheres were incubated with guinea pig complement serum at RT with shaking for 1 hour. Samples were washed, sonicated, and incubated with goat anti–guinea pig C3 Ab conjugated with biotin (Immunology Consultants Laboratory) at RT for 1 hour followed by incubation with streptavidin R–phycoerythrin (PE; Agilent Technologie) at RT for 30 min. After a final wash and sonication, samples were resuspended in Luminex sheath fluid, and complement deposition was determined on a MAGPIX (Luminex Corp.) instrument to define the median fluorescence intensity of PE from two independent replicates. Assays performed without ACE2-Fc and without complement serum were used as negative controls.
In vivo efficacy of ACE2-Fcs in K18-hACE2 mice challenged with SARS-CoV-2 PsVs
K18-hACE2 transgenic mice were purchased from the Jackson Laboratory. All mice were maintained under a specific pathogen–free condition at the National Institute of Health Animal Facility. All animal experiments were performed according to Institute of Laboratory Animal Resources guidelines, and the protocol (LM-113-B) was approved by the National Cancer Institute Animal Care and Use Committee.
For in vivo PsV-based inhibition assays, 6- to 8-week-old K18-hACE2 mice were intranasally treated with Synagis (control IgG; 25 μg), M27, or M81 (5 or 25 μg) 1 hour before challenge by SARS-CoV-2 PsVD614G or PsVB.1.617.2 (~108 RLU, i.n.). Dynamic luciferase signal was acquired 4, 8, and 12 dpi by the IVIS Spectrum In Vivo Imaging System (PerkinElmer) 10 min after intranasal delivery of 200 μg of d-luciferin (LUCK, GoldBio). Tissues (nasal cavity, trachea, and lung) were collected 13 dpi and stored in −80°C before processing.
For PK studies, C57BL/6J mice were intravenously injected with 100 μg (5 mg/kg) of two engineered ACE2-Fc M81 or M86 or intraperitoneally with 375 μg (12.5 mg/kg) of M55, M58, M81, and M163. Before and after injection, serum samples were collected at different time points (10 min, 1 hour, 6 hours, 24 hours, and 48 hours for intravenous injection and 30 min, 2 hours, 6 hours, 10 hours, 24 hours, and 48 hours for intraperitoneal injection), and the ACE2-Fc serum concentration was estimated by indirect ELISA in which SARS-CoV-2 RBDwt (200 ng per well) was used as the capturing molecule, and goat anti-human IgG conjugated with AP (1:1000 dilution; Southern Biotech) was used as secondary Ab as described above. The pharmacokinetic profiles were summarized in fig. S8 (A and C). The alanine transaminase (ALT) and aspartate transaminase (AST) concentrations in sera before and 48 hours after ACE2-Fc intravenous injection were assessed using commercial ALT and AST assay kits (Catachem) and monitored at 340 nm for 15 min with a microplate reader (BioAssay Systems).
For the quantitative real-time polymerase chain reaction (PCR) of tissues from PsV-challenged mice, the tissues were lysed in TRIzol reagent (Invitrogen), and total RNA was isolated using by phenol/chloroform extraction. Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the qSript cDNA SuperMix (Quantabio). The primer sequences used in this study are listed in table S3. The relative level of each mRNA was calculated as fold change compared with control groups after normalizing with Gapdh.
In vivo efficacy of ACE2-Fcs in K18-hACE2 mice challenged with SARS-CoV-2 nLuc and Omicron VOC (B.1.1.529)
All experiments were approved by the Institutional Animal Care and Use Committees (IACUCs) of and Institutional Biosafety Committee of Yale University (IBSCYU). All the animals were housed under specific pathogen–free conditions in the facilities provided and supported by Yale Animal Resources Center (YARC). hACE2 transgenic B6 mice (heterozygous) were obtained from the Jackson Laboratory. Six- to 8-week-old male and female mice were used for all the experiments. The heterozygous mice were crossed and genotyped to select heterozygous mice for experiments by using the primer sets recommended by the Jackson Laboratory.
For in vivo efficacy studies, 6- to 8-week-old male and female mice were challenged intranasally with 1 × 105 FFU of SARS-CoV-2 nLuc WA/2020 (obtained from C. B. Wilen, Yale University) or 1 × 106 FFU of Omicron VOC (B.1.1.529; sequence confirmed and isolated a patient in Yale New Haven Hospital and obtained from C. B. Wilen) in 25- to 30-μl volume under anesthesia (0.5 to 5% isoflurane) delivered using a precision Dräger vaporizer with an oxygen flow rate of 1 liter/min. For prophylaxis, purified ACE2-Fc proteins were administered intranasally at 12.5 mg/kg or 6.25 mg/kg for intraperitoneal injection, 5 hours before infection of K18-hACE2 mice. For therapy, the same amounts (intranasally, intraperitoneally, or both) were administered at 1 dpi. The starting body weight was set to 100%. For survival experiments, mice were monitored every 6 to 12 hours starting 6 days after virus administration. Lethargic and moribund mice or those that lost more than 20% of their body weight were euthanized and considered to have succumbed to infection for Kaplan-Meier survival plots.
BLI of SARS-CoV-2 infection
All standard operating procedures and protocols for IVIS imaging of SARS-CoV-2–infected animals under ABSL-3 conditions were approved by IACUC, IBSCYU, and YARC. All the imaging was carried out using the IVIS Spectrum (PerkinElmer) in an XIC-3 animal isolation chamber (PerkinElmer) that provided biological isolation of anesthetized mice or individual organs during the imaging procedure. All mice were anesthetized via isoflurane inhalation (3 to 5% isoflurane, oxygen flow rate of 1.5 liter/min) before and during BLI using the XGI-8 Gas Anesthesia System. Before imaging, 100 μl of nanoluciferase substrate, furimazine (NanoGlo, Promega, Madison, WI), diluted 1:40 in endotoxin-free PBS was retro-orbitally administered to mice under anesthesia. The mice were then placed into XIC-3 animal isolation chamber (PerkinElmer) presaturated with isoflurane and oxygen mix. The mice were imaged in both dorsal and ventral position on the indicated dpi. The intention of imaging in dorsal position was to visualize virus neuroinvasion. Hence, dorsal view in the figures show only the head of the animal to demonstrate the signal emitted from the brain. The animals were then imaged again after euthanasia and necropsy by supplementing additional 200 μl of the substrate on to the exposed intact organs. Infected areas were identified by carrying out whole-body imaging after necropsy and were isolated, washed in PBS to remove residual blood, and placed onto a clear plastic plate. Additional droplets of furimazine in PBS (1:40) were added to organs and soaked in substrate for 1 to 2 min before BLI.
Images were acquired and analyzed with Living Image v4.7.3 in vivo software package (PerkinElmer Inc.). Image acquisition exposures were set to auto, with imaging parameter preferences set in order of exposure time, binning, and f/stop, respectively. Images were acquired with luminescent f/stop of 2, with photographic f/stop of 8, and with binning set to medium. Comparative images were compiled and batch-processed using the image browser with collective luminescent scales. Photon flux was measured as luminescent radiance (p/s/cm2/sr). Luminescent signals were regarded as background when the minimum threshold setting resulted in displayed radiance above nontissue-containing or known uninfected regions.
Measurement of viral burden
Indicated organs (nasal cavity, brain, and lungs) from infected or uninfected mice were collected, weighed, and homogenized in 1 ml of serum-free RPMI containing penicillin-streptomycin and 1.5 mm of zirconium beads with a BeadBug 6 homogenizer (Benchmark Scientific, T Equipment Inc.). Viral titers were measured using two highly correlative methods. First, the total RNA was extracted from homogenized tissues using an RNeasy plus mini kit (QIAGEN) and was reverse-transcribed with an iScript advanced cDNA kit (Bio-Rad) followed by a SYBR Green real-time PCR assay for determining copies of SARS-CoV-2 N gene RNA using primers that are listed in table S3.
We used nanoluciferase activity as an efficient surrogate for a plaque assay. Dilutions from infected cell homogenates were applied on a Vero E6 monolayer. Twenty-four hours after infection, infected Vero E6 cells were washed with PBS, lysed with passive lysis buffer, and transferred into a 96-well solid white plate (Costar Inc.), and nanoluciferase activity was measured using a Tristar multiwell luminometer (Berthold Technology) for 2.5 s by adding 20 μl of Nano-Glo substrate in nanoluciferase assay buffer (Promega Inc.). An uninfected monolayer of Vero E6 cells treated identically served as a control for determining background and obtaining normalized relative light units. The data were processed and plotted using GraphPad Prism 8 v8.4.3.
Analyses of signature inflammatory cytokines mRNA expression
Brain, lung, and nose samples were collected from mice at the time of necropsy. Total RNA was extracted using an RNeasy plus mini kit (QIAGEN) and was reverse-transcribed with the iScript advanced cDNA kit (Bio-Rad) followed by a SYBR Green Real-time PCR assay for determining the relative expression of selected inflammatory cytokines, i.e., Il6, Ccl2, Cxcl10, and Ifng, using primers listed in table S3. The reaction plate was analyzed using the CFX96 touch real-time PCR detection system. The relative cytokine mRNA levels were calculated with the formula ΔCt(target gene) = Ct(target gene) − Ct(Gapdh). The fold increase was determined using the 2−ΔΔCt method comparing treated mice to uninfected controls.
Quantification and statistical analysis
Data were analyzed and plotted using GraphPad Prism software (La Jolla). Statistical significance for pairwise comparisons was derived by applying nonparametric Mann-Whitney test (two-tailed). To obtain statistical significance for survival curves, grouped data were compared by log-rank (Mantel-Cox) test. To assess statistical significance for grouped data, we used two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests. P values lower than 0.05 were considered statistically significant. P values were indicated as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank J. S. McLellan from University of Texas, Austin for sharing the expression plasmids of SARS-CoV-2 S-2P, S-6P, SARS-CoV-1 RBD-Fc, and monomeric ACE2. We thank BEI Resources for sharing the SARS-CoV-2–related reagents, including pseudo-typed lentiviral kits (NR-53816 and NR-53817), recombinant SB.1.1.7 (NR-55311), SB.1.351 (NR-55311), SB.1526 (NR-55438), SP.1 (NR-55307), etc. We thank K. Pratt and P. Vir from the Department of Medicine, USUHS for providing laboratory resources and technical support to this project. We thank A. Malloy and Z. Lu from the Department of Pediatric, USUHS for sharing the hACE2-expressing HEK293T cell line. We thank R. Petrovich and N. P. Martin from NIEHS for sharing the SP.1 plasmid for PsV production. The views expressed in this manuscript are those of the authors and do not reflect the official policy or position of the Uniformed Services University, the U.S. Army, the Department of Defense, the National Institutes of Health, Department of Health and Human Services, or the U.S. government, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Funding: This work was supported by USU, Bethesda, MD intramural funds to M.P.; in part by funding to F.J.G. from the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research; and in part by NIH grant R01AI163395 to W.M. This work was also supported by Canadian Institutes of Health Research (CIHR) operating Pandemic and Health Emergencies Research grant #177958 and an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) #41027 to A.F. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. G.B.-B. is a recipient of the FRQS Ph.D. fellowship, and S.P.A. is a recipient of the CIHR Ph.D. fellowship. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences.
Author contributions: Y.C., L.S., I.U., P.D.U., F.J.G., and M.P. conceptualized this study, designed the experiments, analyzed data, generated figures, and wrote the manuscript. Y.C. and M.P. designed the ACE2-Fc variants. Y.C., S.G., R.S., D.W., and D.N.N. produced, purified, and characterized the proteins. Y.C. and S.M. performed SPR kinetics. Y.C. and W.D.T. solved and analyzed the crystal structure. L.S. and S.D. generated PsVs and performed neutralization assays. L.S. performed live-cell imaging. L.S., Y.L., and Y.C. designed, optimized, and carried out in vivo inhibition studies on PsV-challenged mice. G.B.-B., S.P.A., A.P.H., and L.M. performed in vitro ADCC, ADCP, and ADCD assays. I.U. and P.D.U. designed, optimized, and performed the in vivo efficacy studies on SARS-CoV-2 nLuc infected mice. I.U., P.D.U., and L.S. analyzed the mouse tissues and quantified viral loads and cytokines. R.S., W.D.T., P.D.U., A.F., G.B.-B., S.P.A., D.N.N., M.E.A., and F.J.G. critically reviewed, edited, and commented on the manuscript. M.P., F.J.G., W.M., A.F., M.E.A., and G.P. funded the work. Every author has read, edited, and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S11
References
REFERENCES AND NOTES
- 1.Vlasova A. N., Diaz A., Damtie D., Xiu L., Toh T.-H., Lee J. S.-Y., Saif L. J., Gray G. C., Novel canine coronavirus isolated from a hospitalized patient with pneumonia in East Malaysia. Clin. Infect. Dis. 74, 446–454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdulla Z. A., Al-Bashir S. M., Al-Salih N. S., Aldamen A. A., Abdulazeez M. Z., A summary of the SARS-CoV-2 vaccines and technologies available or under development. Pathogens 10, 788 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey W. T., Carabelli A. M., Jackson B., Gupta R. K., Thomson E. C., Harrison E. M., Ludden C., Reeve R., Rambaut A.; COVID-19 GENOMICS UK (COG-UK) Consortium, Peacock S. J., Robertson D. L., SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A. C., Tortorici M. A., Snijder J., Xiong X., Bosch B.-J., Rey F. A., Veesler D., Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 114, 11157–11162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q., Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tikellis C., Thomas M. C., Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. 2012, 256294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R. A., Stahl M., Leopoldi A., Garreta E., Del Pozo C. H., Prosper F., Romero J. P., Wirnsberger G., Zhang H., Slutsky A. S., Conder R., Montserrat N., Mirazimi A., Penninger J. M., Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batlle D., Wysocki J., Satchell K., Soluble angiotensin-converting enzyme 2: A potential approach for coronavirus infection therapy? Clin. Sci. (Lond.) 134, 543–545 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Zoufaly A., Poglitsch M., Aberle J. H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C., Neuhold S., Haider D., Stiasny K., Bergthaler A., Puchhammer-Stoeckl E., Mirazimi A., Montserrat N., Zhang H., Slutsky A. S., Penninger J. M., Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 8, 1154–1158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi T., Hoshizaki M., Minato T., Nirasawa S., Asaka M. N., Niiyama M., Imai M., Uda A., Chan J. F.-W., Takahashi S., An J., Saku A., Nukiwa R., Utsumi D., Kiso M., Yasuhara A., Poon V. K.-M., Chan C. C.-S., Fujino Y., Motoyama S., Nagata S., Penninger J. M., Kamada H., Yuen K.-Y., Kamitani W., Maeda K., Kawaoka Y., Yasutomi Y., Imai Y., Kuba K., ACE2-like carboxypeptidase B38-CAP protects from SARS-CoV-2-induced lung injury. Nat. Commun. 12, 6791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A. S., Liu D., Qin C., Jiang C., Penninger J. M., A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M. A., Rettko N. J., Zha S., Yamin R., Kao K., Rosenberg O. S., Ravetch J. V., Wiita A. P., Leung K. K., Lim S. A., Zhou X. X., Hobman T. C., Kortemme T., Wells J. A., Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 28046–28055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan K. K., Dorosky D., Sharma P., Abbasi S. A., Dye J. M., Kranz D. M., Herbert A. S., Procko E., Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi Y., Suzuki T., Arimori T., Ikemura N., Mihara E., Kirita Y., Ohgitani E., Mazda O., Motooka D., Nakamura S., Sakai Y., Itoh Y., Sugihara F., Matsuura Y., Matoba S., Okamoto T., Takagi J., Hoshino A., Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat. Commun. 12, 3802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan K. K., Tan T. J. C., Narayanan K. K., Procko E., An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci. Adv. 7, eabf1738 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havranek B., Chan K. K., Wu A., Procko E., Islam S. M., Computationally designed ACE2 decoy receptor binds SARS-CoV-2 spike (S) protein with tight nanomolar affinity. J. Chem. Inf. Model. 61, 4656–4669 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Hassler L., Wysocki J., Gelarden I., Sharma I., Tomatsidou A., Ye M., Gula H., Nicoleascu V., Randall G., Pshenychnyi S., Khurram N., Kanwar Y., Missiakas D., Henkin J., Yeldandi A., Batlle D., A novel soluble ACE2 protein provides lung and kidney protection in mice susceptible to lethal SARS-CoV-2 infection. J. Am. Soc. Nephrol. ASN.2021091209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao T., Lu J., Zhang J., Johnson R. I., McKay L. G. A., Storm N., Lavine C. L., Peng H., Cai Y., Rits-Volloch S., Lu S., Quinlan B. D., Farzan M., Seaman M. S., Griffiths A., Chen B., A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat. Struct. Mol. Biol. 28, 202–209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller A., Leach A., Thomas J., McAndrew C., Bentley E., Mattiuzzo G., John L., Mirazimi A., Harris G., Gamage N., Carr S., Ali H., van Montfort R., Rabbitts T., A super-potent tetramerized ACE2 protein displays enhanced neutralization of SARS-CoV-2 virus infection. Sci. Rep. 11, 10617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao L., Goreshnik I., Coventry B., Case J. B., Miller L., Kozodoy L., Chen R. E., Carter L., Walls A. C., Park Y.-J., Strauch E.-M., Stewart L., Diamond M. S., Veesler D., Baker D., De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inal J. M., Decoy ACE2-expressing extracellular vesicles that competitively bind SARS-CoV-2 as a possible COVID-19 therapy. Clin. Sci. (Lond.) 134, 1301–1304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu T. H., Crowley A. R., Backes I., Chang C., Tay M., Broge T., Tuyishime M., Ferrari G., Seaman M. S., Richardson S. I., Tomaras G. D., Alter G., Leib D., Ackerman M. E., Hinge length contributes to the phagocytic activity of HIV-specific IgG1 and IgG3 antibodies. PLOS Pathog. 16, e1008083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson S. I., Lambson B. E., Crowley A. R., Bashirova A., Scheepers C., Garrett N., Abdool Karim S., Mkhize N. N., Carrington M., Ackerman M. E., Moore P. L., Morris L., IgG3 enhances neutralization potency and Fc effector function of an HIV V2-specific broadly neutralizing antibody. PLOS Pathog. 15, e1008064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler S. E., Crowley A. R., Natarajan H., Xu S., Weiner J. A., Bobak C. A., Mattox D. E., Lee J., Wieland-Alter W., Connor R. I., Wright P. F., Ackerman M. E., Distinct features and functions of systemic and mucosal humoral immunity among SARS-CoV-2 convalescent individuals. Front. Immunol. 11, 618685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barros E. P., Casalino L., Gaieb Z., Dommer A. C., Wang Y., Fallon L., Raguette L., Belfon K., Simmerling C., Amaro R. E., The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys. J. 120, 1072–1084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leist S. R., Dinnon K. H. III, Schäfer A., Tse L. V., Okuda K., Hou Y. J., West A., Edwards C. E., Sanders W., Fritch E. J., Gully K. L., Scobey T., Brown A. J., Sheahan T. P., Moorman N. J., Boucher R. C., Gralinski L. E., Montgomery S. A., Baric R. S., A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towler P., Staker B., Prasad S. G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N. A., Patane M. A., Pantoliano M. W., ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 279, 17996–18007 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S. L., Marino T., Fang W. H., Russo N., Himo F., Peptide hydrolysis by the binuclear zinc enzyme aminopeptidase from Aeromonas proteolytica: A density functional theory study. J. Phys. Chem. B 112, 2494–2500 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S., Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 11, 2070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benton D. J., Wrobel A. G., Roustan C., Borg A., Xu P., Martin S. R., Rosenthal P. B., Skehel J. J., Gamblin S. J., The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 118, e2022586118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozono S., Zhang Y., Ode H., Sano K., Tan T. S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K., SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 12, 848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu L., Wittrock K. N., Clabaugh G. C., Srivastava V., Cho M. W., A structural landscape of neutralizing antibodies against SARS-CoV-2 receptor binding domain. Front. Immunol. 12, 647934 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng S. H., Lam B., Kung Y. J., Lin J., Liu L., Tsai Y. C., Ferrall L., Roden R. B. S., Wu T. C., Hung C. F., A novel pseudovirus-based mouse model of SARS-CoV-2 infection to test COVID-19 interventions. J. Biomed. Sci. 28, 34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford K. H. D., Eguia R., Dingens A. S., Loes A. N., Malone K. D., Wolf C. R., Chu H. Y., Tortorici M. A., Veesler D., Murphy M., Pettie D., King N. P., Balazs A. B., Bloom J. D., Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaudoin-Bussieres G., Richard J., Prevost J., Goyette G., Finzi A., A new flow cytometry assay to measure antibody-dependent cellular cytotoxicity against SARS-CoV-2 Spike-expressing cells. STAR Protoc. 2, 100851 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischinger S., Fallon J. K., Michell A. R., Broge T., Suscovich T. J., Streeck H., Alter G., A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo M., Kim H. S., Tong R. K., Bainbridge T. W., Vernes J.-M., Zhang Y., Lin Y. L., Chung S., Dennis M. S., Zuchero Y. J. Y., Watts R. J., Couch J. A., Meng Y. G., Atwal J. K., Brezski R. J., Spiess C., Ernst J. A., Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J. Biol. Chem. 292, 3900–3908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackerman M. E., Moldt B., Wyatt R. T., Dugast A.-S., McAndrew E., Tsoukas S., Jost S., Berger C. T., Sciaranghella G., Liu Q., Irvine D. J., Burton D. R., Alter G., A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng S. M. S., Mok C. K. P., Leung Y. W. Y., Ng S. S., Chan K. C. K., Ko F. W., Chen C., Yiu K., Lam B. H. S., Lau E. H. Y., Chan K. K. P., Luk L. L. H., Li J. K. C., Tsang L. C. H., Poon L. L. M., Hui D. S. C., Peiris M., Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 28, 486–489 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuai H., Chan J. F.-W., Hu B., Chai Y., Yuen T. T.-T., Yin F., Huang X., Yoon C., Hu J.-C., Liu H., Shi J., Liu Y., Zhu T., Zhang J., Hou Y., Wang Y., Lu L., Cai J.-P., Zhang A. J., Zhou J., Yuan S., Brindley M. A., Zhang B.-Z., Huang J.-D., To K. K.-W., Yuen K.-Y., Chu H., Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Wysocki J., Ye M., Hassler L., Gupta A. K., Wang Y., Nicoleascu V., Randall G., Wertheim J. A., Batlle D., A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS-CoV-2 infection in human kidney organoids. J. Am. Soc. Nephrol. 32, 795–803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Zeng E., Zhang L., Wang W., Jin Y., Sun J., Huang S., Yin W., Dai J., Zhuang Z., Chen Z., Sun J., Zhu A., Li F., Cao W., Li X., Shi Y., Gan M., Zhang S., Wei P., Huang J., Zhong N., Zhong G., Zhao J., Wang Y., Shao W., Zhao J., Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 7, 65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., Dutta S., Xiong S., Chan M., Chan K. K., Fan T. M., Bailey K. L., Lindeblad M., Cooper L. M., Rong L., Gugliuzza A. F., Shukla D., Procko E., Rehman J., Malik A. B., Engineered ACE2 decoy mitigates lung injury and death induced by SARS-CoV-2 variants. Nat. Chem. Biol. 18, 342–351 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zipeto D., Palmeira J. D. F., Arganaraz G. A., Arganaraz E. R., ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front. Immunol. 11, 576745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore M. J., Dorfman T., Li W., Wong S. K., Li Y., Kuhn J. H., Coderre J., Vasilieva N., Han Z., Greenough T. C., Farzan M., Choe H., Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78, 10628–10635 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.N. Iwanaga, L. Cooper, L. Rong, B. Beddingfield, J. Crabtree, R. A. Tripp, X. Qin, J. K. Kolls, Novel ACE2-IgG1 fusions with improved in vitro and in vivo activity against SARS-CoV2. bioRxiv 2020.06.15.152157 [Preprint]. 24 July 2020. https://doi.org/10.1101/2020.06.15.152157.
- 50.Saunders K. O., Conceptual approaches to modulating antibody effector functions and circulation half-life. Front. Immunol. 10, 1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oladunni F. S., Park J.-G., Pino P. A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., Chiem K., Headley C., Dwivedi V., Parodi L. M., Alfson K. J., Staples H. M., Schami A., Garcia J. I., Whigham A., Platt R. N. II, Gazi M., Martinez J., Chuba C., Earley S., Rodriguez O. H., Mdaki S. D., Kavelish K. N., Escalona R., Hallam C. R. A., Christie C., Patterson J. L., Anderson T. J. C., Carrion R. Jr., Dick E. J. Jr., Hall-Ursone S., Schlesinger L. S., Alvarez X., Kaushal D., Giavedoni L. D., Turner J., Martinez-Sobrido L., Torrelles J. B., Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 11, 6122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCray P. B. Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H. P., Halabi C., Sigmund C. D., Meyerholz D. K., Kirby P., Look D. C., Perlman S., Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ullah I., Prévost J., Ladinsky M. S., Stone H., Lu M., Anand S. P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., Gasser R., Fink C., Chen Y., Tauzin A., Goyette G., Bourassa C., Medjahed H., Mack M., Chung K., Wilen C. B., Dekaban G. A., Dikeakos J. D., Bruce E. A., Kaufmann D. E., Stamatatos L., McGuire A. T., Richard J., Pazgier M., Bjorkman P. J., Mothes W., Finzi A., Kumar P., Uchil P. D., Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity 54, 2143–2158.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beaudoin-Bussieres G., Chen Y., Ullah I., Prévost J., Tolbert W. D., Symmes K., Ding S., Benlarbi M., Gong S. Y., Tauzin A., Gasser R., Chatterjee D., Vézina D., Goyette G., Richard J., Zhou F., Stamatatos L., McGuire A. T., Charest H., Roger M., Pozharski E., Kumar P., Mothes W., Uchil P. D., Pazgier M., Finzi A., A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 38, 110368 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee W. S., Wheatley A. K., Kent S. J., DeKosky B. J., Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 5, 1185–1191 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Wang S., Wang J., Yu X., Jiang W., Chen S., Wang R., Wang M., Jiao S., Yang Y., Wang W., Chen H., Chen B., Gu C., Liu C., Wang A., Wang M., Li G., Guo C., Liu D., Zhang J., Zhang M., Wang L., Gui X., Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcγRIIB and virus-antibody complex with bivalent interaction. Commun. Biol. 5, 262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y., Soh W. T., Kishikawa J.-I., Hirose M., Nakayama E. E., Li S., Sasai M., Suzuki T., Tada A., Arakawa A., Matsuoka S., Akamatsu K., Matsuda M., Ono C., Torii S., Kishida K., Jin H., Nakai W., Arase N., Nakagawa A., Matsumoto M., Nakazaki Y., Shindo Y., Kohyama M., Tomii K., Ohmura K., Ohshima S., Okamoto T., Yamamoto M., Nakagami H., Matsuura Y., Nakagawa A., Kato T., Okada M., Standley D. M., Shioda T., Arase H., An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell 184, 3452–3466.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D., Edwards R. J., Manne K., Martinez D. R., Schäfer A., Alam S. M., Wiehe K., Lu X., Parks R., Sutherland L. L., Oguin T. H. III, Danal C. M., Perez L. G., Mansouri K., Gobeil S. M. C., Janowska K., Stalls V., Kopp M., Cai F., Lee E., Foulger A., Hernandez G. E., Sanzone A., Tilahun K., Jiang C., Tse L. V., Bock K. W., Minai M., Nagata B. M., Cronin K., Gee-Lai V., Deyton M., Barr M., Holle T. V., Macintyre A. N., Stover E., Feldman J., Hauser B. M., Caradonna T. M., Scobey T. D., Rountree W., Wang Y., Moody M. A., Cain D. W., De Marco C. T., Denny T. N., Woods C. W., Petzold E. W., Schmidt A. G., Teng I.-T., Zhou T., Kwong P. D., Mascola J. R., Graham B. S., Moore I. N., Seder R., Andersen H., Lewis M. G., Montefiori D. C., Sempowski G. D., Baric R. S., Acharya P., Haynes B. F., Saunders K. O., In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 184, 4203–4219.e32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. Zhang, K. K. Narayanan, L. Cooper, K. K. Chan, C. A. Devlin, A. Aguhob, K. Shirley, L. Rong, J. Rehman, A. B. Malik, E. Procko, An engineered ACE2 decoy receptor can be administered by inhalation and potently targets the BA.1 and BA.2 omicron variants of SARS-CoV-2. bioRxiv 2022.03.28.486075 [Preprint]. 28 March 2022. 10.1101/2022.03.28.486075. [DOI] [PMC free article] [PubMed]
- 60.Hsieh C.-L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H.-C., Javanmardi K., le K. C., Wrapp D., Lee A. G., Liu Y., Chou C.-W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherburn R., Tolbert W. D., Gottumukkala S., Hederman A. P., Beaudoin-Bussières G., Stanfield-Oakley S., Tuyishime M., Ferrari G., Finzi A., Ackerman M. E., Pazgier M., Incorporating the cluster A and V1V2 targets into a minimal structural unit of the HIV-1 envelope to elicit a cross-clade response with potent Fc-effector functions. Vaccines (Basel) 9, 975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu P., Wysocki J., Serfozo P., Ye M., Souma T., Batlle D., Jin J., A fluorometric method of measuring carboxypeptidase activities for angiotensin II and apelin-13. Sci. Rep. 7, 45473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrara F., Temperton N., Pseudotype neutralization assays: From laboratory bench to data analysis. Methods Protoc. 1, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mou H., Quinlan B. D., Peng H., Liu G., Guo Y., Peng S., Zhang L., Davis-Gardner M. E., Gardner M. R., Crynen G., DeVaux L. B., Voo Z. X., Bailey C. C., Alpert M. D., Rader C., Gack M. U., Choe H., Farzan M., Mutations derived from horseshoe bat ACE2 orthologs enhance ACE2-Fc neutralization of SARS-CoV-2. PLOS Pathog. 17, e1009501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss M. S., Global indicators of X-ray data quality. J. Appl. Cryst. 34, 130–135 (2001). [Google Scholar]
- 69.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popov A. N., Bourenkov G. P., Choice of data-collection parameters based on statistic modelling. Acta Crystallogr. D Biol. Crystallogr. 59, 1145–1153 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Brunger A. T., Free R value: Cross-validation in crystallography. Methods Enzymol. 277, 366–396 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Y. Liu, J. Liu, K. S. Plante, J. A. Plante, X. Xie, X. Zhang, Z. Ku, Z. An, D. Scharton, C. Schindewolf, V. D. Menachery, P.-Y. Shi, S. C. Weaver, The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv 2021.03.08.434499 [Preprint]. 9 March 2021. 10.1101/2021.03.08.434499. [DOI]
- 74.Zalevsky J., Chamberlain A. K., Horton H. M., Karki S., Leung I. W. L., Sproule T. J., Lazar G. A., Roopenian D. C., Desjarlais J. R., Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S11
References