Abstract
Imaging mass spectrometry has become a mature molecular mapping technology that is used for molecular discovery in many medical and biological systems. While powerful by itself, imaging mass spectrometry can be complemented by the addition of other orthogonal, chemically informative imaging technologies to maximize the information gained from a single experiment and enable deeper understanding of biological processes. Within this review, we describe MALDI, SIMS, and DESI imaging mass spectrometric technologies and how these have been integrated with other analytical modalities such as microscopy, transcriptomics, spectroscopy, and electrochemistry in a field termed multimodal imaging. We explore the future of this field and discuss forthcoming developments that will bring new insights to help unravel the molecular complexities of biological systems, from single cells to functional tissue structures and organs.
Keywords: imaging mass spectrometry, chemical imaging, multimodal analysis, tissue analysis
Graphical Abstract

1. INTRODUCTION
Imaging mass spectrometry (IMS) is a technology that enables the mapping of hundreds to thousands of molecules within biological systems.1–6 The instruments within the field can measure a diverse array of sample types and chemical classes, ranging from low molecular weight metabolites7–10 and signaling molecules11–13 to lipids,14–21 peptides,22–24 and proteins.25–29 A typical IMS experiment involves defining an area of the sample surface to be imaged followed by the desorption and ionization at multiple discrete locations. Each desorbed location, or pixel, is composed of an individual mass spectrum. A molecular image is generated for each signal recorded by plotting the ion intensity of that signal throughout the array of pixels generated from the sample. While there are many types of IMS technologies that have been reported in literature, we highlight here the three most common: matrix-assisted laser desorption/ionization (MALDI),30,31 secondary ion mass spectrometry (SIMS),32–34 and desorption electrospray ionization (DESI).35–38 Additional information on other types of IMS technologies can be found in several recent reviews.1,4,39
Each IMS technology can be coupled to a variety of analytical approaches to enhance the information gained in a single experiment. Although IMS provides rich, chemically informative spectra and mapping capabilities, combining this unique mass specific technology with other analytical approaches can provide additional information for an analyzed sample. Combining two or more imaging modalities is termed multimodal imaging. The accrued advantages include enhanced discrimination between modality specific chemical and instrumental noise from biologically relevant chemical signals, improved sensitivity and specificity of chemical classes not easily analyzed by a single modality alone, and enhanced data mining capabilities. Combining IMS technologies with other analytical approaches provides a more effective means to probe the molecular complexity of biological systems. In this review, we summarize major advancements in the field of multimodal imaging mass spectrometry over the past few years and discuss the exciting future of this field.
2. OVERVIEW OF IMAGING MASS SPECTROMETRY TECHNOLOGIES
2.1. Matrix-Assisted Laser Desorption/Ionization.
MALDI is a common ionization method within the IMS field as it can visualize numerous molecular species over a broad mass range with great molecular diversity. For tissue analysis, sample preparation typically involves sectioning of tissues and thaw mounting these sections onto a glass slide or other target (e.g., conductive surface for high voltage ion sources) and subsequent application of an organic chemical matrix that aids in analyte desorption and ionization.40 Spatial resolution is defined by the ablation area of the laser and the distance between pixels (pitch). A variety of matrices are used for specific molecular classes. Commonly used matrices include 2,5-dihydroxybenzoic acid41 or cyano-4-hydroxycinnamic acid42 for positive ion mode analysis of metabolites, lipids, and peptides. Additionally, 9-aminoacridine43 is typically used for negative ion mode analysis of metabolites, lipids, and proteins, while 1,5-diaminonaphthalene44 is employed for analysis of lipids in both ion polarities. Recently norharmane has been shown to be effective for the analysis of hydrophobic molecules and low molecular weight metabolites.45 Matrix deposition is often performed robotically to best balance reproducibility, analyte extraction, achieve small crystal size, and produce coating homogeneity, although sublimation, sieving, and airbrushing are also used in the field.4,46
As high spatial resolution MALDI IMS capabilities are developed that approach cellular and subcellular resolutions, challenges in sensitivity arise as the number of molecules sampled decreases with smaller pixel sizes. Ultimately, spatial resolution is dependent upon multiple factors such as matrix crystal size, laser focus, and stage motor step precision. Most modern MALDI IMS platforms utilize either a frequency tripled Nd:YAG (355 nm) or nitrogen gas laser (337.7 nm) and can achieve spatial resolutions of 5–20 μm using traditional front-side laser optics.50–52 Improving laser focus beyond 5 μm can be achieved with more advanced setups using lower laser wavelengths (e.g., 213 nm) or changing laser geometries (e.g., transmission-mode), to minimize the effective spot size as described in the next several examples. For instance, Heiles et al. demonstrated a new source that integrates a 213 nm laser allowing for a ~3 μm footprint using front-side laser optics.53 Alternatively, Zavalin and coworkers developed a transmission-mode geometry MALDI source, where the UV-laser is redirected to ablate the sample from the back side. By decoupling the laser and ion optics, higher numerical aperture objectives can be utilized without impeding the ion path, resulting in ~1 μm spot sizes.54 However, a drawback of any high spatial resolution IMS experiment is a reduction in ion abundances; the Caprioli55 and Dreisewerd47 laboratories incorporated a secondary laser perpendicular to the primary ablation plume as a means to enhance ionization of transmission geometry setups. An example of these data sets is featured here (Figure 1A).
Figure 1.

Selected developments within the IMS community. Image of a Vero B cell culture at 1 μm spatial resolution acquired with transition mode MALDI-2 IMS developed by the Dreiswerd lab (A). Panel A is adapted with permission from ref 47. Copyright 2019 Nature Publishing. SIMS image of a coculture of different Pseudomonas aeruginosa strains visualized with different signaling small molecules (B). Panel B is adapted with permission from ref 48 . Copyright 2019 SPIE. Digital Library. Lipid images both positive and negative mode of the mouse uterine tissue using nanoDESI at 10 μm spatial resolution (C). Panel C is adapted with permission from ref 49. Copyright 2019 Nature Publishing.
2.2. Secondary Ion Mass Spectrometry.
SIMS utilizes electrostatically focused primary ions (e.g., Bi+, Cs+, and O−) or clusters (e.g., Au3 +, C60, and Ar) to impact the sample surface, causing a collisional cascade in the top few monolayers of the sample leading to the ejection of secondary ions.2,34 Because of the high energy of the ion beam, analyte ions often undergo significant fragmentation and analysis is typically limited to molecular weight ions <2 kDa.56 The energy of the ion beam plays a critical role in sampling and can be divided into two regimes: static and dynamic SIMS. Static SIMS is defined by low primary ion doses (<1013 cm−2) and beam currents (pA-nA) suitable for surface analysis of elements and molecules. Alternatively, dynamic SIMS has much higher primary ion doses (>1013 cm−2) and beam currents (mA), making it suitable for depth profiling and three-dimensional imaging.57 In general, sample preparation for SIMS analysis is minimal and consists of mounting tissues flatly onto conductive targets and drying prior to introduction into the source vacuum chamber.58,59 Although not required, washing samples to remove salts from tissue prior to analysis can improve ion yield.60,61
The primary advantage of SIMS is its high spatial resolution capabilities because ion beams can be tightly focused using electric fields (1 μm to 30 nm).32 Similar to MALDI, SIMS spatial resolution is defined by the diameter of the ion beam at the surface and the pitch. SIMS has been used for cellular and subcellular analyses62–65 and for the determination of molecular profiles of various disorders, including different cancer types66 and cardiovascular disease.67 It has also been used to monitor signaling between bacterial cocultures and distinct alkyl quinolone messengers between different strains of Pseudomonas aeruginosa (Figure 1B).48 Additionally, SIMS was employed to visualize salt redistribution in brain tissue between healthy and stroke mice.68 While effective for low molecular weight metabolite and lipid analyses, it has not been commonly applied to peptide and protein imaging studies.
2.3. Desorption Electrospray Ionization.
DESI is performed by spraying charged solvent droplets on the surface of the sample where the analytes are desorbed and ionized for subsequent detection by MS.69 The imaging experiment is performed in a continuous raster sampling mode where the target is moved continuously under the DESI spray in a “typwriter-like” motion. Spatial resolution is estimated by the target stage velocity, sampling rate of the mass spectrometer, number of spectra averaged for a single pixel, and distance between adjacent line scans. Although DESI has limited spatial resolution capabilities (~150 μm), it requires minimal to no sample preparation. Tissues sections are mounted onto glass slides and sampling is performed at ambient pressure. Recently, the Laskin lab developed a modified form of this sampling approach, termed nanoDESI, using a liquid microjunction to increase spatial resolution to ~10 μm (Figure 1C).49 While nanoDESI is not discussed in a multimodal imaging context here, we eagerly anticipate the higher spatial resolution of nanoDESI being coupled to other modalities. Moreover, DESI and nanoDESI been used to map metabolite,70–72 lipid,73–76 and drug distributions in a variety of biological systems ranging from plants to diseased mammalian tissue.77–79 Further, additives in DESI solvent can target specific molecules in the case of reactive DESI80 or enhance extraction.70
DESI and nanoDESI IMS are minimally destructive techniques and have been used experimentally in surgical settings to enable intraoperative molecular assessment and aid in real-time intraoperative decisions.36,81,82 Sans et al. molecularly characterized high-grade serous carcinoma, serous borderline ovarian tumors and normal ovarian tissue samples using DESI IMS.83 They identified predictive markers of cancer aggressiveness and built classification models to enable diagnosis and prediction of high-grade serous carcinoma in comparison to normal tissue with a high certainty of ~96%. Similar work from the Eberlin lab has led to the development of a hand-held mass spectrometry device, the MasSpec Pen, which enables in vivo diagnostics during surgery.84–86 This device has been used for classifying ovarian87 and breast cancer88 since it was originally developed. Because it can be operated at atmospheric pressure and requires minimal sample preparation, DESI holds promise for use in clinical and surgical settings.
3. COMBINING MULTIPLE IMAGING MASS SPECTROMETRY TECHNOLOGIES
Because each IMS technology has unique performance characteristics for different molecular classes, there is utility in coupling them together. Technologies such as tandem MS, microextractions, and ion mobility each add additional dimensions to the MS data set, expanding upon the chemical information that can be obtained, either by enabling de novo identification or reducing spectral complexity.
3.1. Spatially Targeted Tandem MS.
Tandem mass spectrometry enables de novo identification of molecules within complex samples.89–91 Tandem MS is generally accomplished by performing isolation within one mass analyzer for subsequent fragmentation and transmittance into another mass analyzer for detection.92 Serially performing these analyses allows for identification of discrete molecules within a biological sample.93,94 In the context of imaging, pixels can be subdivided allowing for a precursor ion scan and subsequent MS/MS scans. This spatially targeted structural information comes at the cost of spatial resolution to accommodate multiple samplings. Additionally, it is often difficult to perform tandem MS analysis within an imaging experiment if the ion of interest is only present within a small number of disperse features or pixels.17 Alternative means of targeting sample regions or locations for identifying key ions include multimodal image-guided surface sampling,95,96 microprobe extraction for offline tandem MS,97–99 and tandem MS on an orthogonal sample.16,30 By using these various strategies, ions that are found in a small number of pixels or structural features can be targeted for analysis.
Tandem MS can be used within an imaging context to identify the differential localization of isomers or isobars within a tissue. Although tandem MS has been reported for several types of IMS technologies,100–102 it is primarily used for SIMS imaging because most ion beams fragment analyte ions during the ionization process.103–107 Any imaging mass spectrometer with MS/MS capabilities can generate fragment ion images108,109 enabling direct visualization and differentiation of isomers and isobars within tissue without prior separation or derivatization. Generally, these experiments are usually limited to surveying a small number of ions within a sample, since the tissue is partially consumed during analysis.
3.2. Multi-Imaging Mass Spectrometry Experiments.
Each of the three types of IMS technologies discussed here provide different molecular coverage and spatial resolution. As a result, investigators have combined some of these to gain further functionality. SIMS and MALDI IMS have been combined to detect multiple chemical classes, such as low molecular weight metabolites and lipids within individual cells95 and tissue.111 Additionally, the combination of MALDI and SIMS has been used to analyze hair to enhance sensitivity and spatial resolution.112,113 Both MALDI and SIMS sources are operated under vacuum using similar tissue preparation protocols. In some cases, SIMS analyses have been shown to be enhanced by the application of a MALDI matrix.114,115 While less common, combining MALDI and DESI together enables analysis of different lipid species116 and comapping of lipids and proteins.117 To our knowledge, there has not yet been a DESI and SIMS multimodal IMS experiment, likely because of the differences in sample preparation. Reaction additives can be employed for DESI analyses for quantitation and derivatization chemistries, bringing additional capabilities to any combination of technologies. Finally, DESI produces multiply charged ions that are often more amenable to tandem MS applications.
3.3. Microextraction.
Microextraction protocols aim to remove key analytes from bulk material to reduce chemical complexity118 and target specific analytes. Both solid and liquid phase extraction techniques have been coupled to IMS for increased peak capacity and sensitivity. Solid phase microextraction (SPME) comprises a diverse set of solventless techniques that allow for in vivo analysis. An advantage of SPME devices is that most analytes are introduced into the MS system at once. Introducing ions concurrently increases sensitivity and signal-to-noise (S/N) compared to technologies that generate a transient signal.119,120 Furthermore, SPME is used to separate an analyte of interest from bulk material, such as in trace analyte analysis.121
Liquid extractions, such as liquid microjunction (LMJ)96,122and liquid extraction surface analysis (LESA),97,123 also enhance peak capacity and reduce ion suppression. Briefly, extraction solvents are dispensed onto a tissue surface and collected for subsequent liquid chromatography or capillary electrophoresis MS analysis.50 Spatially targeted liquid extractions are advantageous because they can be coupled to different separation techniques, increasing both sensitivity and depth of coverage. For instance, Cahill et al. used an LMJ extraction to image portions of a microfluidic device while it was functioning with future use aimed at biology-based microfluidic devices.124 Typically, this technology utilizes relatively large areas for droplet placement. However, recently microLESA was combined with piezoelectric spotting of trypsin to achieve higher spatial resolution sampling than previously reported.97,125 In addition, microLESA was integrated with autofluorescence microscopy to correlate protein signatures of murine kidney and Staphylococcus aureus abscesses without tissue staining.97 As these techniques continue to improve in spatial resolution and automated platforms become available, combining IMS with spatially targeted microextractions will be adapted to supplement spatial information with deeper molecular coverage and added identification capabilities.
3.4. Ion Mobility.
Ion mobility separations add an orthogonal analytical dimension to IMS that reduces ion interference for improving peak capacity and specificity and aiding in identification of species. Additionally, drift time or collision cross sections (CCS) calculated with the use of standards can be used to help identify isomers and isobars. Ion mobility techniques that have been coupled to IMS include drift tube mass spectrometry (DTIMS),126,127 traveling wave ion mobility spectrometry (TWIMS),128,129 field asymmetric ion mobility spectrometry (FAIMS)130–132 and, more recently, trapped ion mobility spectrometry (TIMS).133–135
A major focus of the ion mobility field is developing higher resolving power platforms, so they can separate isomers and isobars. Park, Fernandez-Lima, and coworkers utilized a buffer gas and ion trapping by electric field gradient for the development of TIMS devices. They report resolving powers of ~200 but up to ~400.136–138 One approach for increasing resolution is by increasing path length and/or number of passes around the mobility device. Notable ion mobility advancements include the development of structures for lossless ion manipulation (SLIM) devices,139,140 as well as the cyclic ion mobility (cIM).141 SLIM devices utilize a separation path length of ~540 m142 for achieving resolving powers over 1800. Using cIM, resolving power of 750 (with 100 passes) has been reported using a reverse sequence peptide pair. Although, sensitivity can be challenging in these devices as ions are lost radially over time. Recently cIM has been integrated with LESA for the improvement of S/N and the number of detected proteins from tissue samples (Figure 2).110
Figure 2.
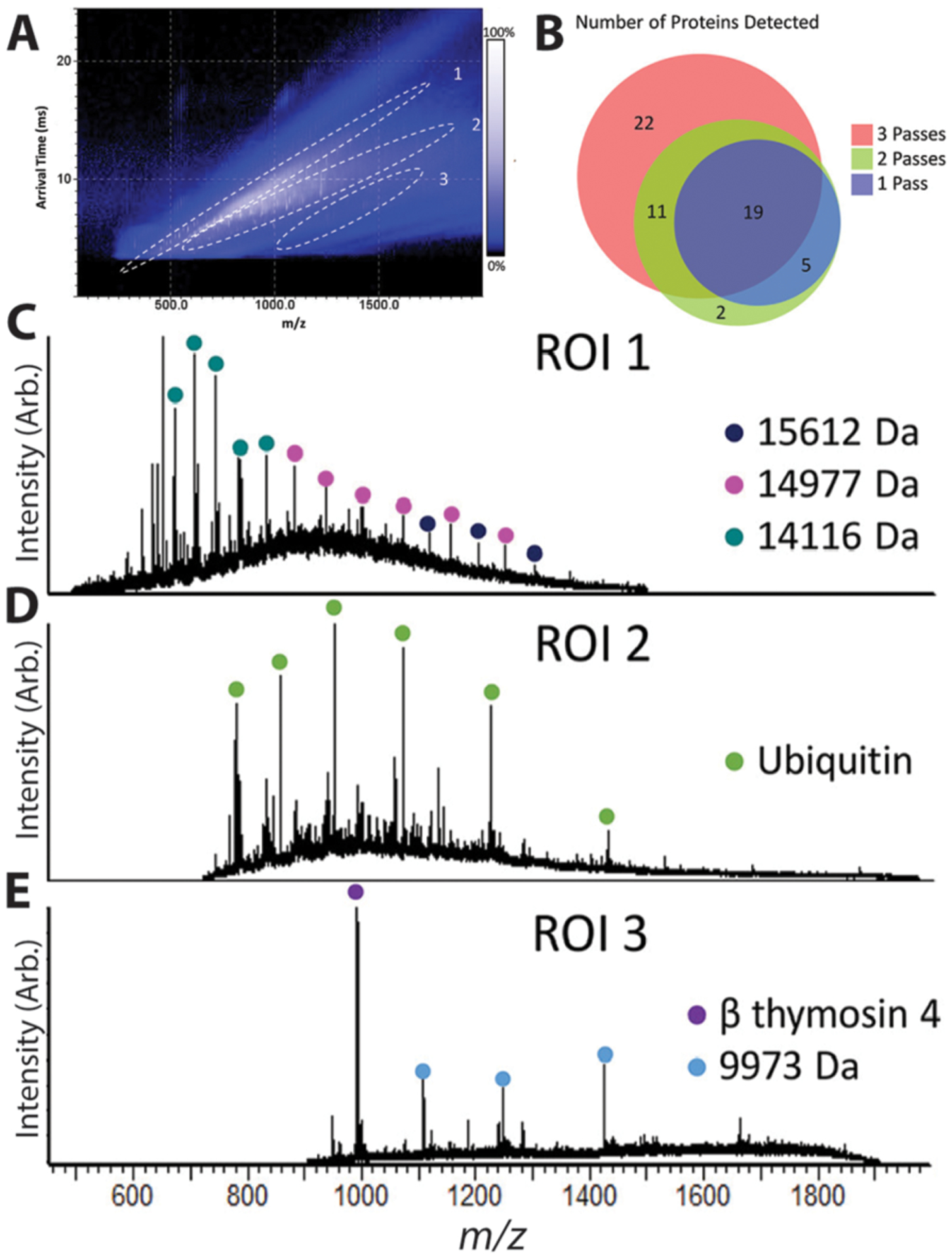
LESA and cIM analysis of a complex murine kidney protein extract. More proteins are detected after each additional cycle of cIM (A). IM heat map generated from a single cycle of cIM (B), where extracted mass spectra from different trend lines contain different protein signatures (C–E). This figure was adapted with permission from ref 110. Copyright 2020 American Chemical Society.
Beyond increasing peak capacity and specificity, trendlines within ion mobility heat maps can be used to identify different molecular species, classes, and subclasses.143 For example, MALDI DTIMS IMS was used successfully to separately image a lipid species ([phosphatidylcholine(34:2)+H]+) from a closely isobaric peptide ion (RPPGFSP).144 Škrášková et al. visualized multiply charged polysialylated gangliosides using DESI TWIMS.116 Recently TIMS has been integrated with MALDI IMS to the separate and map isobaric lipid species directly from tissue.145 Cooper and coworkers introduced a new cylindrical FAIMS device coupled to LESA IMS. This workflow improved the number of detected proteins from what was previously reported as much as 10 times in murine brain, testes, and kidney.146 Clearly, ion mobility has great potential for IMS applications as it provides the ability to overcome the analytical challenges associated with direct sampling of complex biological tissues. Software is continually being developed to more efficiently process highly dimensional ion mobility IMS data.
4. INTEGRATION WITH OTHER TECHNOLOGIES
Non-MS analytical technologies can be integrated with MS technologies to increase chemical coverage. Since each technology has distinctive advantages for different molecular classes, experiments that synergistically incorporate multiple technologies gain unique chemical information neither one can obtain alone. For example, many cell types and functional cell states are uniquely suited for analysis by specific methods, such as transcriptomics or immunostaining, and integrating these can add great utility to the molecular specificity of MS.
4.1. Microscopy.
Microscopy is one of the oldest analytical approaches dating back to the early 17th century and is commonly applied to biological and clinical problems.148–151 Overall, microscopy images are generated by capturing electromagnetic radiation or particle beams as they interact with the sample through reflection, refraction, or diffraction. A series of lenses and objectives focus the light/particles enabling imaging with high magnification.
Brightfield and histological stains, such as hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS), are used extensively in pathology to assess tissue integrity, health, and disease.152–154 Stained tissues have been employed with IMS to connect molecular profiles to histological features of both healthy and diseased tissue, such as cancer155–159 and functional disorders.160,161 Recently, Basu et al. enabled IMS and histology to be rapidly correlated by incorporating matrix precoated slides, templates, and a 10 kHz laser.162 Histological stains are ubiquitous within the scientific and medical field, so multimodal studies that combine IMS and stained microscopy improve interpretation and facilitate collaboration between the technologists and the biologists or physicians. Similarly, simple brightfield images can be used for correlation of IMS signals to specific tissue structures, particularly in cases that have features that are easily distinguished with brightfield microscopy.163,164
Fluorescence microscopy of both endogenous fluorophores165,166 and tagged antibodies167–169 or nucleic acids170,171 have also been fundamental to our understanding of biological systems. For example, Vardi et al. used autofluorescence from chlorophyll and MALDI IMS to study lipid metabolism within algal plaques.172 By targeting key proteins or genes, investigators can parse metabolic pathways and monitor how these change as a function of disease state or demographic. While exceptionally powerful and informative, these technologies are generally limited to studying peptides or proteins since there are few probes available for low molecular weight metabolites and lipids.173,174 IMS has been readily coupled to fluorescence microscopy approaches to tie together cell-type specific immune16,17,175,176 and transcript profiles177–179 to metabolites detected by IMS.180 Immunohistochemistry can be used to histologically classify and contextualize the chemical information obtained by IMS. Moreover, the combination of both IMS and immunohistochemistry produce more rigorous classification schemes than either modality independently. Recently, several laboratories have combined immunohistochemistry and IMS to correlate protein or metabolite signals to specific tissue substructures.181–184 This type of experiment is further extended by recent work where investigators coupled multiplexed immunohistochemistry with MALDI FT-ICR IMS to determine metabolic profiles of cancer cells, incorporating molecular classes.185 The increase in the plexity of the immunofluorescence labels additionally enhances the specificity of chemical profiles created by MS analysis, as these immunofluorescence labels can be correlated to more specific tissue regions or cell types.
Immunofluorescence imaging has been correlated with MALDI IMS176 and fluorescence in situ hybridization (FISH) with SIMS.177 We could not find an example of either being used as part of DESI multimodal studies. This is interesting because there are no experimental considerations preventing IHC or FISH from being combined with all the IMS modalities discussed here. However, technical challenges in coregistering modalities with dramatic differences in spatial resolution may be driving this perceived disparity. Patterson et al. have developed combination experimental and computational pipelines using autofluorescence microscopy to overcome this challenge for multimodal MALDI IMS studies.165,166 It is anticipated that fluorescence microscopy will become an important correlative technology in IMS studies.
Particle-based microscopy, such as scanning electron microscopy (SEM) and transmission electron microscopy (TEM), offer very high spatial resolution because it is not diffraction limited. A series of ion optics are used to focus ions toward a sample. The ions then interact with the sample and the emerging electrons and various energy conversion products (e.g., secondary electrons, X-rays, light) are captured as an image. Because nanoSIMS and electron microscopies have similar sample and operational requirements, they have often been used on the same sample.187 TOF-SIMS has been integrated into a helium ion microscope for 8 nm imaging with the potential for extremely high resolution molecular imaging of biological samples (Figure 3).147 For example, nanoSIMS and TEM have been combined to map dopamine distributions in dense core vesicles, providing key insight into vesicle loading and nanocompartmentalization.188 Another study involves integration of fluorescence microscopy, SIMS, high-energy resolution X-ray photoelectron spectroscopy, and SEM on the same plant root to study bacterial growth and infection on this root.189 The authors combined these four modalities because of the simple and compatible sample preparation involved for each. Beyond registration for enhanced localization, particle-based microscopy is also readily used to assess MALDI matrix application for ensuring matrix coverage and crystal size.190,191
Figure 3.
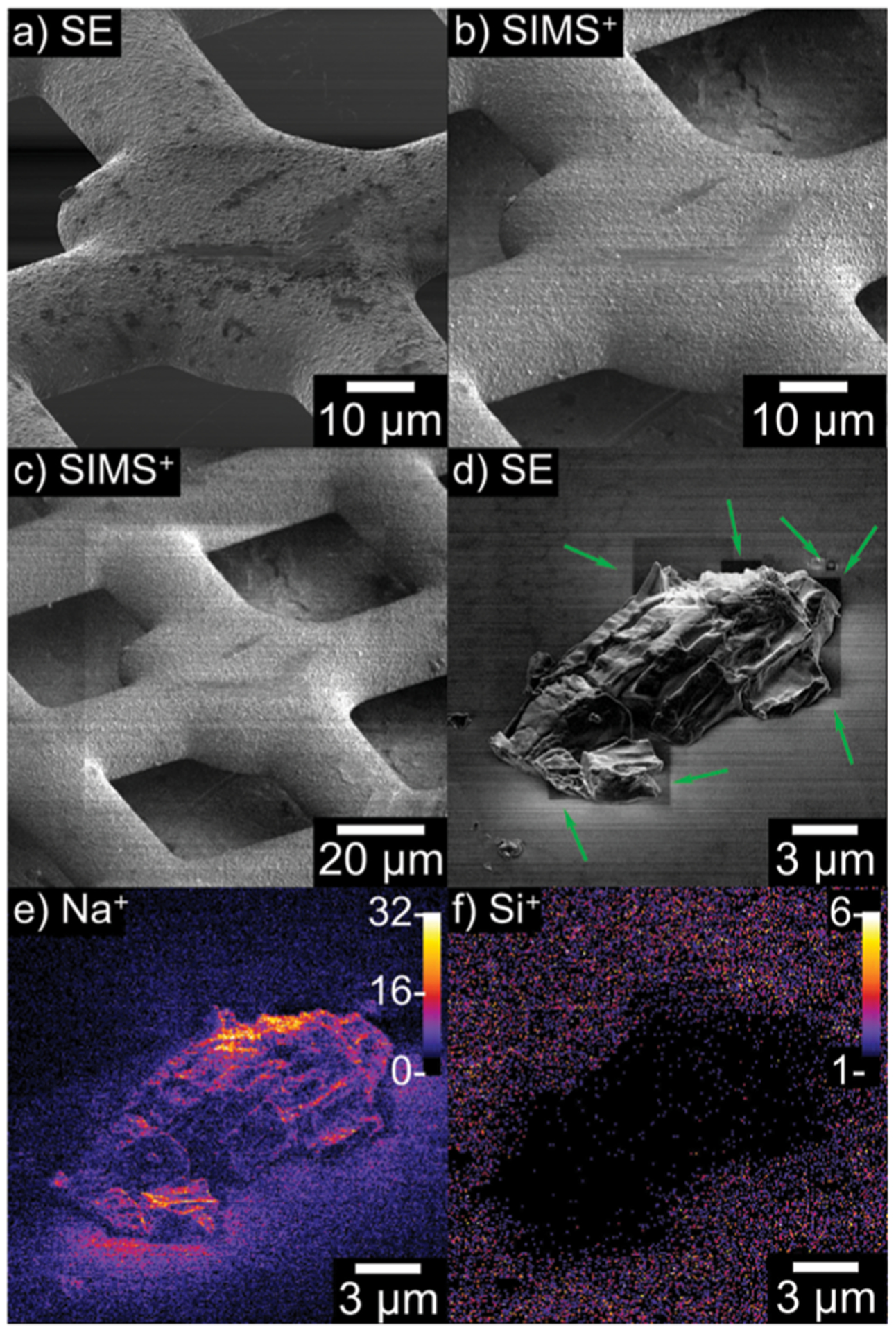
Images of a TEM grid (A–C) and NaCl salt (D–F) crystal generated from the SIMS helium microscope. The SIMS (B, C, E, and F) images are close to the resolution of the microscopy (A and D), demonstrating the power of this technique. This figure was adapted with permission from ref 147. Copyright 2019 Science Direct.
4.2. Spectroscopy.
Spectroscopic imaging includes a suite of optical approaches, such as infrared (IR)192 and Raman, that provide unique spectra of complex chemical mixtures, creating reproducible chemical profiles of different physiological regions and disease states.193 Spectroscopy is used in a wide array of biological studies194–199 because the approaches are generally nondestructive, label-free, and capable of high spatial resolutions (diffraction limited, 250 nm). While each spectroscopic approach activates different, well characterized molecular modes, it is often difficult to correlate spectroscopic signatures of complex mixtures to discrete chemicals. Rather, they provide general information on bond types and functional groups for the entire chemical mixture.200–203 As such, spectroscopy has been coupled to a variety of MS technologies, including IMS, to provide more detailed molecular descriptions of samples.204–206 Because spectroscopic analysis is label-free and nondestructive, both modalities can be performed on the same tissue section.207
Raman and IMS have been correlated to study bacteria,208–210 plants,211 single cells,212 and mammalian organs.213,214 Fourier transform infrared microscopy (FT-IR) has similarly been coupled to IMS for many biological studies.207,215,216 Recently, Rabe et al. developed a method for FT-IR guided MALDI IMS (Figure 4). By coupling these two technologies together, the authors reduced data load and acquisition time by >90%.186 Magnetic resonance imaging has also been used to compliment molecular specificity of IMS with dynamic, whole organ imaging.217–222 While MALDI IMS has primarily been coupled to spectroscopic measurements, we foresee DESI increasingly becoming incorporated with Raman and magnetic resonance imaging. DESI does not have many tissue preparation requirements and can theoretically be directly integrated within either approach. Moreover, Raman and magnetic resonance spectra are not saturated when water is present, unlike IR, which readily is absorbed by water, making them ideal candidates for multimodal DESI experiments.
Figure 4.

MALDI IMS lipid profiles obtained after FT-IR generated segmentation. Murine brain is differentially segmented based on absorbance of 2922 cm−1 based on disease (A) and control mice (B). Using this segmentation, lipid profiles can be generated for different masks for chemical differentiation (C and D). This figure was adapted with permission from ref 186. Copyright 2018 Nature Publishing.
4.3. Transcriptomics.
Transcriptomics technologies measure gene expression through the extraction and amplification of nucleic acids, most generally ribonucleic acids.223–225 Because of the exponential amplification that can occur, this group of technologies is highly sensitive and has been the driving force behind cell typing and classification. There is great interest in coupling IMS with transcriptomics analysis as it would enable simultaneous correlation of gene expression to gene products and biproducts. A majority of the literature combining mass spectrometry and transcriptomics measurements has involved liquid chromatography for bulk proteomics and/or metabolomics.226,227 Toward an imaging context, investigators probed an insect, Carausius morosus, for neuropeptide content with MALDI MS and correlated this to bulk sequencing to uncover peptides with no known homology.228 Although this was not an imaging application, the sample preparation was performed in such a way that this workflow could be adapted to an imaging workflow. As an additional step toward combining IMS with transcriptomics, the Knepper et al. microdissected kidney tubules and performed RNA-seq and proteomics.229 This methodology incorporates spatial information on the microdissected structures, which is an important step toward coupling IMS and transcriptomics directly. Moreover, there has been some exciting work where transcriptomics and proteomics IMS information has been correlated on different bulk samples.230 Work combining two approaches on the same sample provided insight into the link between fatty acids and immunity within breast cancer231 and role of liver X receptors in male reproduction.232 While only a few examples exist, the combination of transcriptomics and IMS will continue to increase in the coming years and has the potential to uncover new connections that span the central dogma of molecular biology.
4.4. Electrochemistry.
Electrochemistry has been vital in the study of electroactive signaling molecules, such as dopamine, epinephrine, serotonin, and histamine within biological systems.233–235 This is partly because it is capable of absolute quantitation of femto- to zeptomole amounts of analyte at μs temporal resolutions. It is highly selective and naturally applicable to the analysis of many low molecular weight metabolites, such as neurotransmitters.236 nanoSIMS/nanoSIMS and electrochemistry are compatible and have been used together often to study nonbiological237 and biological samples alike.236,238,239 Ewing et al. have quantitated L-DOPA concentrations in vesicles and other organelles using nanoSIMS and validated these concentrations with electrochemistry.240 This is particularly important because it enables quantitation of low molecular weight metabolites at high spatial resolutions. Further, Larsson and coworkers quantitated octopamine release during different stimulations using nanoSIMS and an embedded electrode.241 Investigators in the IMS field have also adapted many technologies and developments from electrochemical studies,242 ranging from nanopipettes to electrochemical principals of ion generation. Adopting materials, analyses, or processes from other fields is an effective, time saving process that expedites scientific advancements.
5. FUTURE OF THE FIELD
Mass spectrometry technologies have rapidly advanced with improved sampling, ion transmission, and detector sensitivity, enabling highly sensitive and specific analysis of biological tissues. Under favorable conditions, MS can detect concentrations as low as 10 zeptomoles or approximately 6000 molecules.6 This sensitivity is sufficient for probing most molecules within biological systems and yet there is still much we do not understand. Nevertheless, improvements in the molecular, spatial, temporal, and biological specificity are required to answer remaining questions. IMS is one available tool that provides untargeted, highly multiplexed molecular analysis, but only through the combination with other technologies can we achieve specificity in all these areas mentioned above. Multimodal imaging experiments can significantly improve performance characteristics, including structural identification, throughput, cell type specificity, and dynamic range of MS-based chemical profiles. The present article has summarized the current literature surrounding multimodal IMS and the development of the field to further explore remaining biological questions.
A major task facing multimodal approaches involves efforts to fully integrate the several data sets to enable deeper data mining. This is particularly difficult within an imaging regime because the various technologies can have dramatically different spatial resolutions, data structures, and chemical information. Different computation methods for addressing differences within spatial resolution include various methods of up sampling the data, performing more refined data fusion, as well as other experimental approaches.165,166,243–246 While these are capable of connecting modalities that are similar in resolution, significant experimental and technological capabilities are required to avoid the introduction of artifacts.207 While many imaging modalities are within an order of magnitude of one another, this will influence the capacity to combine lower spatial resolution IMS technologies like DESI with higher resolution molecular imaging approaches, like SEM.
Moreover, technologies provide both overlapping and orthogonal information that is often difficult to correlate. For instance, transcriptomics measures gene expression of different biochemical pathways that result with peptide and metabolomic products. Ideally, a highly expressed gene would indicate the presence of a specific metabolite and IMS would detect this same metabolite in high abundance within the same regions, but this is not always the case. There many transport, degradation, and modification pathways that impede this correlation and create complex data sets that are difficult to interpret. Additional compounding technical factors, such as matrix effects, differences in ionization efficiencies, and limited dynamic range add to this complexity. While transcriptomics is used as an example here, a similar scenario applies to other multimodal approaches, such as protein abundances between IMS and immunofluorescence experiments or lipid analysis by Raman spectroscopy and IMS. While challenging, future multimodal experiments will slowly begin to unravel the complex relationships between the data produced by orthogonal technologies. In connection with this, new machine learning algorithms and approaches will be essential for untangling the abundance of chemical information obtained with multimodal IMS. This will inevitably lead to a more complete picture of biological systems and pathways.
Finally, improving sample preparation and workflows will undoubtedly improve the quality and reproducibility of collected data, particularly as these technologies enter the rigorous domain of medicine and clinical trials. Many technologies cannot be combined “out of the box”, and so there are often trade-offs made to enable multimodal analysis. Although, the gained information from the suboptimal combination of the approaches is often greater than the data of either technique alone. Developing methods that enable multiple imaging modalities to be performed optimally and with minimal spatial compromise will dramatically improve the ability to integrate and discover connections between multimodal data sets. Optimizing the ways and methods behind combining the different approaches is a clear path forward in the field.
In summary, multimodal IMS is a remarkably diverse endeavor that incorporates the best attributes from a variety of scientific disciplines. In the future, multimodal IMS technologies will progressively become more common as the scientific community begins to study more complex biological and medicinal questions. Such studies have the potential to bring together genomic, proteomic, and metabolomic imaging technologies to provide unprecedented insights into biology and medicine.
ACKNOWLEDGMENTS
Support was provided by the NIH Common Fund and National Institute of Diabetes and Digestive and Kidney Diseases (Grant U54DK120058 awarded to J.M.S. and R.M.C.), NIH National Institute of Allergy and Infectious Disease (Grant R01 AI138581 awarded to J.M.S.), the National Science Foundation Major Research Instrument Program (Grant CBET-1828299 awarded to J.M.S. and R.M.C.), and by the NIH National Institute of General Medical Sciences (Grant 2P41GM103391 awarded to R.M.C.). E.K.N. is supported by a National Institute of Environmental Health Sciences training grant (T32ES007028).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jasms.0c00232
The authors declare no competing financial interest.
Contributor Information
Elizabeth K. Neumann, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37205, United States.
Katerina V. Djambazova, Mass Spectrometry Research Center and Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37235, United States
Richard M. Caprioli, Department of Biochemistry, Mass Spectrometry Research Center, Department of Pharmacology, Department of Chemistry, and Department of Medicine, Vanderbilt University, Nashville, Tennessee 37205, United States
Jeffrey M. Spraggins, Department of Biochemistry, Mass Spectrometry Research Center, and Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37205, United States.
REFERENCES
- (1).Bodzon-Kulakowska A; Suder P Imaging mass spectrometry: Instrumentation, applications, and combination with other visualization techniques. Mass Spectrom. Rev 2016, 35, 147. [DOI] [PubMed] [Google Scholar]
- (2).Xu X; Zhong J; Su Y; Zou Z; He Y; Hou X A brief review on mass/optical spectrometry for imaging analysis of biological samples. Appl. Spectrosc. Rev 2019, 54, 57. [Google Scholar]
- (3).Ho Y-N; Shu L-J; Yang Y-L Imaging mass spectrometry for metabolites: technical progress, multimodal imaging, and biological interactions. WIREs Systems Biology and Medicine 2017, 9, e1387. [DOI] [PubMed] [Google Scholar]
- (4).Buchberger AR; DeLaney K; Johnson J; Li L Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem 2018, 90, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Xu G; Li J Recent advances in mass spectrometry imaging for multiomics application in neurology. J. Comp. Neurol 2019, 527, 2158. [DOI] [PubMed] [Google Scholar]
- (6).Neumann EK; Do TD; Comi TJ; Sweedler JV Exploring the Fundamental Structures of Life: Non-Targeted, Chemical Analysis of Single Cells and Subcellular Structures. Angew. Chem., Int. Ed 2019, 58, 9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).LEón M; Ferreira CR; Eberlin LS; Jarmusch AK; Pirro V; Rodrigues ACB; Favaron PO; Miglino MA; Cooks RG Metabolites and Lipids Associated with Fetal Swine Anatomy via Desorption Electrospray Ionization – Mass Spectrometry Imaging. Sci. Rep 2019, 9, 7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bergman H-M; Lindfors L; Palm F; Kihlberg J; Lanekoff I Metabolite aberrations in early diabetes detected in rat kidney using mass spectrometry imaging. Anal. Bioanal. Chem 2019, 411, 2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Morse N; Jamaspishvili T; Simon D; Patel PG; Ren KYM; Wang J; Oleschuk R; Kaufmann M; Gooding RJ; Berman DM Reliable identification of prostate cancer using mass spectrometry metabolomic imaging in needle core biopsies. Lab. Invest 2019, 99, 1561. [DOI] [PubMed] [Google Scholar]
- (10).Wang M; Dubiak K; Zhang Z; Huber PW; Chen DDY; Dovichi NJ MALDI-imaging of early stage Xenopus laevis embryos. Talanta 2019, 204, 138. [DOI] [PubMed] [Google Scholar]
- (11).Shariatgorji M; Nilsson A; Fridjonsdottir E; Vallianatou T; Källback P; Katan L; Sävmarker J; Mantas I; Zhang X; Bezard E; Svenningsson P; Odell LR; Andrén PE Comprehensive mapping of neurotransmitter networks by MALDI–MS imaging. Nat. Methods 2019, 16, 1021. [DOI] [PubMed] [Google Scholar]
- (12).Watrous JD; Dorrestein PC Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol 2011, 9, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Shank EA; Kolter R Extracellular signaling and multi-cellularity in Bacillus subtilis. Curr. Opin. Microbiol 2011, 14, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Alamri H; Patterson NH; Yang E; Zoroquiain P; Lazaris A; Chaurand P; Metrakos P Mapping the triglyceride distribution in NAFLD human liver by MALDI imaging mass spectrometry reveals molecular differences in micro and macro steatosis. Anal. Bioanal. Chem 2019, 411, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sämfors S; Fletcher JS Lipid Diversity in Cells and Tissue Using Imaging SIMS. Annu. Rev. Anal. Chem 2020, 13, 249. [DOI] [PubMed] [Google Scholar]
- (16).Neumann EK; Comi TJ; Rubakhin SS; Sweedler JV Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical Classification. Angew. Chem., Int. Ed 2019, 58, 5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Neumann EK; Ellis JF; Triplett AE; Rubakhin SS; Sweedler JV Lipid Analysis of 30 000 Individual Rodent Cerebellar Cells Using High-Resolution Mass Spectrometry. Anal. Chem 2019, 91, 7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nemes P; Woods AS; Vertes A Simultaneous Imaging of Small Metabolites and Lipids in Rat Brain Tissues at Atmospheric Pressure by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal. Chem 2010, 82, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Eberlin LS; Dill AL; Golby AJ; Ligon KL; Wiseman JM; Cooks RG; Agar NYR Discrimination of Human Astrocytoma Subtypes by Lipid Analysis Using Desorption Electrospray Ionization Imaging Mass Spectrometry. Angew. Chem., Int. Ed 2010, 49, 5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Angel PM; Spraggins JM; Baldwin HS; Caprioli R Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem 2012, 84, 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Brunelle A; Laprévote O Lipid imaging with cluster time-of-flight secondary ion mass spectrometry. Anal. Bioanal. Chem 2009, 393, 31. [DOI] [PubMed] [Google Scholar]
- (22).Rešetar Maslov D; Svirkova A; Allmaier G; Marchetti-Deschamann M; Kraljevicć Pavelicć S Optimization of MALDI-TOF mass spectrometry imaging for the visualization and comparison of peptide distributions in dry-cured ham muscle fibers. Food Chem 2019, 283, 275. [DOI] [PubMed] [Google Scholar]
- (23).Do TD; Ellis JF; Neumann EK; Comi TJ; Tillmaand EG; Lenhart AE; Rubakhin SS; Sweedler JV Optically Guided Single Cell Mass Spectrometry of Rat Dorsal Root Ganglia to Profile Lipids, Peptides and Proteins. ChemPhysChem 2018, 19, 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Caprioli RM; Farmer TB; Gile J Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem 1997, 69, 4751. [DOI] [PubMed] [Google Scholar]
- (25).Dilillo M; Ait-Belkacem R; Esteve C; Pellegrini D; Nicolardi S; Costa M; Vannini E; Graaf E. L. d.; Caleo M; McDonnell LA Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Sci. Rep 2017, 7, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Garza KY; Feider CL; Klein DR; Rosenberg JA; Brodbelt JS; Eberlin LS Desorption Electrospray Ionization Mass Spectrometry Imaging of Proteins Directly from Biological Tissue Sections. Anal. Chem 2018, 90, 7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Andersson M; Groseclose MR; Deutch AY; Caprioli RM Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat. Methods 2008, 5, 101. [DOI] [PubMed] [Google Scholar]
- (28).Groseclose MR; Andersson M; Hardesty WM; Caprioli RM Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom 2007, 42, 254. [DOI] [PubMed] [Google Scholar]
- (29).Crecelius AC; Cornett DS; Caprioli RM; Williams B; Dawant BM; Bodenheimer B Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J. Am. Soc. Mass Spectrom 2005, 16, 1093. [DOI] [PubMed] [Google Scholar]
- (30).Ryan DJ; Spraggins JM; Caprioli RM Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr. Opin. Chem. Biol 2019, 48, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Angeletti S; Ciccozzi M Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: An updating review. Infect., Genet. Evol 2019, 76, 104063. [DOI] [PubMed] [Google Scholar]
- (32).Eswara S; Pshenova A; Yedra L; Hoang QH; Lovric J; Philipp P; Wirtz T Correlative microscopy combining transmission electron microscopy and secondary ion mass spectrometry: A general review on the state-of-the-art, recent developments, and prospects. Appl. Phys. Rev 2019, 6, 021312. [Google Scholar]
- (33).Gyngard F; Steinhauser ML Biological explorations with nanoscale secondary ion mass spectrometry. J. Anal. At. Spectrom 2019, 34, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gao D; Huang X; Tao Y A critical review of NanoSIMS in analysis of microbial metabolic activities at single-cell level. Crit. Rev. Biotechnol 2016, 36, 884. [DOI] [PubMed] [Google Scholar]
- (35).Zheng Q; Chen H Development and Applications of Liquid Sample Desorption Electrospray Ionization Mass Spectrometry. Annu. Rev. Anal. Chem 2016, 9, 411. [DOI] [PubMed] [Google Scholar]
- (36).Ferreira C; Alfaro CM; Pirro V; Cooks RG Desorption Electrospray Ionization Mass Spectrometry Imaging: Recent Developments and Perspectives. J. Biomol Tech 2019, 30, S46. [Google Scholar]
- (37).Javanshad R; Venter AR Ambient ionization mass spectrometry: real-time, proximal sample processing and ionization. Anal. Methods 2017, 9, 4896. [DOI] [PubMed] [Google Scholar]
- (38).Parrot D; Papazian S; Foil D; Tasdemir D Imaging the Unimaginable: Desorption Electrospray Ionization – Imaging Mass Spectrometry (DESI-IMS) in Natural Product Research. Planta Med 2018, 84, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Shrestha B; Walsh CM; Boyce GR; Nemes P In Advances in MALDI and Laser-Induced Soft Ionization Mass Spectrometry; Springer: 2016; p 149. [Google Scholar]
- (40).Dunham SJB; Neumann EK; Lanni EJ; Ong T-H; Sweedler JV Biomarker Discovery with Mass Spectrometry Imaging and Profiling. In Proteomics for Biological Discovery; John Wiley & Sons: 2019; p 89–123. [Google Scholar]
- (41).Strupat K; Karas M; Hillenkamp F 2,5-Dihydroxybenzoic acid: a new matrix for laser desorption—ionization mass spectrometry. Int. J. Mass Spectrom. Ion Processes 1991, 111, 89. [Google Scholar]
- (42).Beavis RC; Chaudhary T; Chait BT α-Cyano-4-hydroxycinnamic acid as a matrix for matrixassisted laser desorption mass spectromtry. Org. Mass Spectrom 1992, 27, 156. [Google Scholar]
- (43).Vermillion-Salsbury RL; Hercules DM 9-Aminoacridine as a matrix for negative mode matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom 2002, 16, 1575. [Google Scholar]
- (44).Korte AR; Lee YJ MALDI-MS analysis and imaging of small molecule metabolites with 1,5-diaminonaphthalene (DAN). J. Mass Spectrom 2014, 49, 737. [DOI] [PubMed] [Google Scholar]
- (45).Scott AJ; Flinders B; Cappell J; Liang T; Pelc RS; Tran B; Kilgour DPA; Heeren RMA; Goodlett DR; Ernst RK Norharmane matrix enhances detection of endotoxin by MALDI-MS for simultaneous profiling of pathogen, host and vector systems. Pathog. Dis 2016, 74. DOI: 10.1093/femspd/ftw097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Amstalden van Hove ER; Smith DF; Heeren RMA A concise review of mass spectrometry imaging. Journal of Chromatography A 2010, 1217, 3946. [DOI] [PubMed] [Google Scholar]
- (47).Niehaus M; Soltwisch J; Belov ME; Dreisewerd K Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 2019, 16, 925. [DOI] [PubMed] [Google Scholar]
- (48).Cao T; Morales-Soto N; Jia J; Baig N; Dunham SJ; Ellis J; Sweedler J; Shrout J; Bohn P Spatiotemporal dynamics of molecular messaging in bacterial co-cultures studied by multimodal chemical imaging; SPIE, 2019; Vol. 10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yin R; Burnum-Johnson KE; Sun X; Dey SK; Laskin J High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat. Protoc 2019, 14, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Boughton BA; Thinagaran D; Sarabia D; Bacic A; Roessner U Mass spectrometry imaging for plant biology: a review. Phytochem. Rev 2016, 15, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Trim PJ; Snel MF Small molecule MALDI MS imaging: Current technologies and future challenges. Methods 2016, 104, 127. [DOI] [PubMed] [Google Scholar]
- (52).Murray KK; Seneviratne CA; Ghorai S High resolution laser mass spectrometry bioimaging. Methods 2016, 104, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Heiles S; Kompauer M; Müller MA; Spengler B Atmospheric-Pressure MALDI Mass Spectrometry Imaging at 213 nm Laser Wavelength. J. Am. Soc. Mass Spectrom 2020, 31, 326. [DOI] [PubMed] [Google Scholar]
- (54).Zavalin A; Yang J; Hayden K; Vestal M; Caprioli RM Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal. Bioanal. Chem 2015, 407, 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Spivey EC; McMillen JC; Ryan DJ; Spraggins JM; Caprioli RM Combining MALDI-2 and transmission geometry laser optics to achieve high sensitivity for ultra-high spatial resolution surface analysis. J. Mass Spectrom 2019, 54, 366. [DOI] [PubMed] [Google Scholar]
- (56).Wogelred L; Höök F; Agnarsson B; Sjövall P Toward multiplexed quantification of biomolecules on surfaces using time-of-flight secondary ion mass spectrometry. Biointerphases 2018, 13, 03B413. [DOI] [PubMed] [Google Scholar]
- (57).Sodhi RNS Time-of-flight secondary ion mass spectrometry (TOF-SIMS):—versatility in chemical and imaging surface analysis. Analyst 2004, 129, 483. [DOI] [PubMed] [Google Scholar]
- (58).Yoon S; Lee TG Biological tissue sample preparation for time-of-flight secondary ion mass spectrometry (ToF–SIMS) imaging. Nano Convergence 2018, 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Schaepe K; Jungnickel H; Heinrich T; Tentschert J; Luch A; Unger WES In Characterization of Nanoparticles; Hodoroaba V-D, Unger WES, Shard AG, Eds.; Elsevier: 2020; p 481. [Google Scholar]
- (60).Gilmore IS; Heiles S; Pieterse CL Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging. Annu. Rev. Anal. Chem 2019, 12, 201. [DOI] [PubMed] [Google Scholar]
- (61).Nunez J; Renslow R; Cliff JB; Anderton CR NanoSIMS for biological applications: Current practices and analyses. Biointerphases 2018, 13, 03B301. [DOI] [PubMed] [Google Scholar]
- (62).Tian H; Six DA; Krucker T; Leeds JA; Winograd N Subcellular Chemical Imaging of Antibiotics in Single Bacteria Using C60-Secondary Ion Mass Spectrometry. Anal. Chem 2017, 89, 5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Massonnet P; Heeren RMA A concise tutorial review of TOF-SIMS based molecular and cellular imaging. J. Anal. At. Spectrom 2019, 34, 2217. [Google Scholar]
- (64).Yin L; Zhang Z; Liu Y; Gao Y; Gu J Recent advances in single-cell analysis by mass spectrometry. Analyst 2019, 144, 824. [DOI] [PubMed] [Google Scholar]
- (65).Li H-W; Hua X; Long Y-T Graphene quantum dots enhanced ToF-SIMS for single-cell imaging. Anal. Bioanal. Chem 2019, 411, 4025. [DOI] [PubMed] [Google Scholar]
- (66).Gularyan SK; Gulin AA; Anufrieva KS; Shender V; Shakhparonov MI; Bastola S; Antipova NV; Kovalenko TF; Rubtsov YP; Latyshev YA; Potapov AA; Pavlyukov MS Investigation of inter- and intra-tumoral heterogeneity of glioblastoma using TOF-SIMS. Mol. Cell. Proteomics 2020, 19, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Mezger STP; Mingels AMA; Bekers O; Cillero-Pastor B; Heeren RMA Trends in mass spectrometry imaging for cardiovascular diseases. Anal. Bioanal. Chem 2019, 411, 3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mulder IA; Ogrinc Potočnik N; Broos LAM; Prop A; Wermer MJH; Heeren RMA; van den Maagdenberg AMJM Distinguishing core from penumbra by lipid profiles using Mass Spectrometry Imaging in a transgenic mouse model of ischemic stroke. Sci. Rep 2019, 9, 1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wiseman JM; Ifa DR; Venter A; Cooks RG Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat. Protoc 2008, 3, 517. [DOI] [PubMed] [Google Scholar]
- (70).Wang X; Hou Y; Hou Z; Xiong W; Huang G Mass Spectrometry Imaging of Brain Cholesterol and Metabolites with Trifluoroacetic Acid-Enhanced Desorption Electrospray Ionization. Anal. Chem 2019, 91, 2719. [DOI] [PubMed] [Google Scholar]
- (71).Huang L; Mao X; Sun C; Luo Z; Song X; Li X; Zhang R; Lv Y; Chen J; He J; Abliz Z A graphical data processing pipeline for mass spectrometry imaging-based spatially resolved metabolomics on tumor heterogeneity. Anal. Chim. Acta 2019, 1077, 183. [DOI] [PubMed] [Google Scholar]
- (72).Martin R; Midey A; Olivos H; Shrestha B; Claude E Spatial Mapping of Lipids and Neurotransmitters in Rat Brain Section Using DESI Ion Mobility Mass Spectrometry. J. Biomol Tech 2019, 30, S38. [Google Scholar]
- (73).Nguyen SN; Kyle JE; Dautel SE; Sontag R; Luders T; Corley R; Ansong C; Carson J; Laskin J Lipid Coverage in Nanospray Desorption Electrospray Ionization Mass Spectrometry Imaging of Mouse Lung Tissues. Anal. Chem 2019, 91, 11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Nagai K; Uranbileg B; Chen Z; Fujioka A; Yamazaki T; Matsumoto Y; Tsukamoto H; Ikeda H; Yatomi Y; Chiba H; Hui S-P; Nakazawa T; Saito R; Koshiba S; Aoki J; Saigusa D; Tomioka Y Identification of novel biomarkers of hepatocellular carcinoma by high-definition mass spectrometry: Ultrahigh-performance liquid chromatography quadrupole time-of-flight mass spectrometry and desorption electrospray ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom 2020, 34, e8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Liu C; Qi K; Yao L; Xiong Y; Zhang X; Zang J; Tian C; Xu M; Yang J; Lin Z; Lv Y; Xiong W; Pan Y Imaging of Polar and Nonpolar Species Using Compact Desorption Electrospray Ionization/Postphotoionization Mass Spectrometry. Anal. Chem 2019, 91, 6616. [DOI] [PubMed] [Google Scholar]
- (76).Unsihuay D; Qiu J; Swaroop S; Nagornov KO; Kozhinov AN; Tsybin YO; Kuang S; Laskin J Imaging of triglycerides in tissues using nanospray desorption electrospray ionization (Nano-DESI) mass spectrometry. Int. J. Mass Spectrom 2020, 448, 116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Feider CL; Woody S; Ledet S; Zhang J; Sebastian K; Breen MT; Eberlin LS Molecular Imaging of Endometriosis Tissues using Desorption Electrospray Ionization Mass Spectrometry. Sci. Rep 2019, 9, 15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Banerjee S; Wong AC-Y; Yan X; Wu B; Zhao H; Tibshirani RJ; Zare RN; Brooks JD Early detection of unilateral ureteral obstruction by desorption electrospray ionization mass spectrometry. Sci. Rep 2019, 9, 11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Tamura K; Horikawa M; Sato S; Miyake H; Setou M Discovery of lipid biomarkers correlated with disease progression in clear cell renal cell carcinoma using desorption electrospray ionization imaging mass spectrometry. Oncotarget 2019, 10, 1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Zheng Q; Ruan X; Tian Y; Hu J; Wan N; Lu W; Xu X; Wang G; Hao H; Ye H Ligand–protein target screening from cell matrices using reactive desorption electrospray ionization-mass spectrometry via a native-denatured exchange approach. Analyst 2019, 144, 512. [DOI] [PubMed] [Google Scholar]
- (81).Alfaro CM; Pirro V; Keating MF; Hattab EM; Cooks RG; Cohen-Gadol AA J. Neurosurg 2020, 132, 180. [DOI] [PubMed] [Google Scholar]
- (82).DeHoog RJ; Zhang J; Alore E; Lin JQ; Yu W; Woody S; Almendariz C; Lin M; Engelsman AF; Sidhu SB; Tibshirani R; Suliburk J; Eberlin LS Preoperative metabolic classification of thyroid nodules using mass spectrometry imaging of fine-needle aspiration biopsies. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Sans M; Gharpure K; Tibshirani R; Zhang J; Liang L; Liu J; Young JH; Dood RL; Sood AK; Eberlin LS Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging. Cancer Res 2017, 77, 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Zhang J; Rector J; Lin JQ; Young JH; Sans M; Katta N; Giese N; Yu W; Nagi C; Suliburk J; Liu J; Bensussan A; DeHoog RJ; Garza KY; Ludolph B; Sorace AG; Syed A; Zahedivash A; Milner TE; Eberlin LS Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med 2017, 9, eaan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Feider CL; Krieger A; DeHoog RJ; Eberlin LS Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem 2019, 91, 4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Hall OM; Peer CJ; Figg WD Tissue preservation with mass spectroscopic analysis: Implications for cancer diagnostics. Cancer Biol. Ther 2018, 19, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Sans M; Zhang J; Lin JQ; Feider CL; Giese N; Breen MT; Sebastian K; Liu J; Sood AK; Eberlin LS Performance of the MasSpec Pen for Rapid Diagnosis of Ovarian Cancer. Clin. Chem 2019, 65, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).St John ER; Balog J; McKenzie JS; Rossi M; Covington A; Muirhead L; Bodai Z; Rosini F; Speller AV; Shousha S Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: towards an intelligent knife for breast cancer surgery. Breast Cancer Res 2017, 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Tran NH; Zhang X; Xin L; Shan B; Li M De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Kumar D; Yadav AK; Dash D In Proteome Bioinformatics; Springer: 2017; p 17. [Google Scholar]
- (91).Jadid SE; Touahni R; Moussa A In Proceedings of the New Challenges in Data Sciences: Acts of the Second Conference of the Moroccan Classification Society; Association for Computing Machinery: Kenitra, Morocco, 2019; Article 4. [Google Scholar]
- (92).El-Aneed A; Cohen A; Banoub J Mass Spectrometry, Review of the Basics: Electrospray, MALDI, and Commonly Used Mass Analyzers. Appl. Spectrosc. Rev 2009, 44, 210. [Google Scholar]
- (93).Korte AR; Yagnik GB; Feenstra AD; Lee YJ Multiplex MALDI-MS imaging of plant metabolites using a hybrid MS system. Methods Mol. Biol 2015, 1203, 49. [DOI] [PubMed] [Google Scholar]
- (94).Ellis SR; Paine MRL; Eijkel GB; Pauling JK; Husen P; Jervelund MW; Hermansson M; Ejsing CS; Heeren RMA Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat. Methods 2018, 15, 515. [DOI] [PubMed] [Google Scholar]
- (95).Comi TJ; Neumann EK; Do TD; Sweedler JV microMS: A Python Platform for Image-Guided Mass Spectrometry Profiling. J. Am. Soc. Mass Spectrom 2017, 28, 1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Comi TJ; Makurath MA; Philip MC; Rubakhin SS; Sweedler JV MALDI MS Guided Liquid Microjunction Extraction for Capillary Electrophoresis–Electrospray Ionization MS Analysis of Single Pancreatic Islet Cells. Anal. Chem 2017, 89, 7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Ryan DJ; Patterson NH; Putnam NE; Wilde AD; Weiss A; Perry WJ; Cassat JE; Skaar EP; Caprioli RM; Spraggins JM MicroLESA: Integrating Autofluorescence Microscopy, In Situ Micro-Digestions, and Liquid Extraction Surface Analysis for High Spatial Resolution Targeted Proteomic Studies. Anal. Chem 2019, 91, 7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Ryan DJ; Nei D; Prentice BM; Rose KL; Caprioli RM; Spraggins JM Protein identification in imaging mass spectrometry through spatially targeted liquid micro-extractions. Rapid Commun. Mass Spectrom 2018, 32, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Delcourt V; Franck J; Quanico J; Gimeno J-P; Wisztorski M; Raffo-Romero A; Kobeissy F; Roucou X; Salzet M; Fournier I Spatially-Resolved Top-down Proteomics Bridged to MALDI MS Imaging Reveals the Molecular Physiome of Brain Regions. Mol. Cell. Proteomics 2018, 17, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Kompauer M; Heiles S; Spengler B Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 2017, 14, 90. [DOI] [PubMed] [Google Scholar]
- (101).Prentice BM; McMillen JC; Caprioli RM Multiple TOF/TOF events in a single laser shot for multiplexed lipid identifications in MALDI imaging mass spectrometry. Int. J. Mass Spectrom 2019, 437, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Pasilis SP; Kertesz V; Van Berkel GJ; Schulz M; Schorcht S HPTLC/DESI-MS imaging of tryptic protein digests separated in two dimensions. J. Mass Spectrom 2008, 43, 1627. [DOI] [PubMed] [Google Scholar]
- (103).Fisher GL Structural Assessment of (Sub-)Monolayer Coatings in Device Processing at High Spatial Resolving Power by TOF-SIMS Tandem MS Imaging. Microsc. Microanal 2019, 25, 732. [Google Scholar]
- (104).Angerer TB; Velickovic D; Nicora CD; Kyle JE; Graham DJ; Anderton C; Gamble LJ Exploiting the Semidestructive Nature of Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry Imaging for Simultaneous Localization and Confident Lipid Annotations. Anal. Chem 2019, 91, 15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Bruinen AL; Fisher GL; Heeren RM In Imaging Mass Spectrometry; Springer: 2017; p 165. [Google Scholar]
- (106).Ogrinc Potočnik N; Fisher GL; Prop A; Heeren RMA Sequencing and Identification of Endogenous Neuropeptides with Matrix-Enhanced Secondary Ion Mass Spectrometry Tandem Mass Spectrometry. Anal. Chem 2017, 89, 8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Fu T; Houël E; Amusant N; Touboul D; Genta-Jouve G; Della-Negra S; Fisher GL; Brunelle A; Duplais C Biosynthetic investigation of γ-lactones in Sextonia rubra wood using in situ TOF-SIMS MS/MS imaging to localize and characterize biosynthetic intermediates. Sci. Rep 2019, 9, 1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Sugiura Y; Zaima N; Setou M; Ito S; Yao I Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem 2012, 403, 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Takeo E; Sugiura Y; Uemura T; Nishimoto K; Yasuda M; Sugiyama E; Ohtsuki S; Higashi T; Nishikawa T; Suematsu M; Fukusaki E; Shimma S Tandem Mass Spectrometry Imaging Reveals Distinct Accumulation Patterns of Steroid Structural Isomers in Human Adrenal Glands. Anal. Chem 2019, 91, 8918. [DOI] [PubMed] [Google Scholar]
- (110).Sisley EK; Ujma J; Palmer M; Giles K; Fernandez-Lima FA; Cooper HJ LESA Cyclic Ion Mobility Mass Spectrometry of Intact Proteins from Thin Tissue Sections. Anal. Chem 2020, 92, 6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Desbenoit N; Walch A; Spengler B; Brunelle A; Römpp A Correlative mass spectrometry imaging, applying time-of-flight secondary ion mass spectrometry and atmospheric pressure matrix-assisted laser desorption/ionization to a single tissue section. Rapid Commun. Mass Spectrom 2018, 32, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Erne R; Bernard L; Steuer AE; Baumgartner MR; Kraemer T Hair Analysis: Contamination versus Incorporation from the Circulatory System—Investigations on Single Hair Samples Using Time-of-Flight Secondary Ion Mass Spectrometry and Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chem 2019, 91, 4132. [DOI] [PubMed] [Google Scholar]
- (113).Lanni EJ; Masyuko RN; Driscoll CM; Aerts JT; Shrout JD; Bohn PW; Sweedler JV MALDI-guided SIMS: Multiscale Imaging of Metabolites in Bacterial Biofilms. Anal. Chem 2014, 86, 9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Dowlatshahi Pour M; Malmberg P; Ewing A An investigation on the mechanism of sublimed DHB matrix on molecular ion yields in SIMS imaging of brain tissue. Anal. Bioanal. Chem 2016, 408, 3071. [DOI] [PubMed] [Google Scholar]
- (115).Cai L; Sheng L; Xia M; Li Z; Zhang S; Zhang X; Chen H Graphene Oxide as a Novel Evenly Continuous Phase Matrix for TOF-SIMS. J. Am. Soc. Mass Spectrom 2017, 28, 399. [DOI] [PubMed] [Google Scholar]
- (116).Škrášková K; Claude E; Jones EA; Towers M; Ellis SR; Heeren RMA Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods 2016, 104, 69. [DOI] [PubMed] [Google Scholar]
- (117).Eberlin LS; Liu X; Ferreira CR; Santagata S; Agar NYR; Cooks RG Desorption Electrospray Ionization then MALDI Mass Spectrometry Imaging of Lipid and Protein Distributions in Single Tissue Sections. Anal. Chem 2011, 83, 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Prosen H; Zupančič-Kralj L Solid-phase microextraction. TrAC, Trends Anal. Chem 1999, 18, 272. [Google Scholar]
- (119).Gómez-Ríos GA; Mirabelli MF Solid Phase Microextraction-mass spectrometry: Metanoia. TrAC, Trends Anal. Chem 2019, 112, 201. [Google Scholar]
- (120).Psillakis E; Kalogerakis N Developments in liquid-phase microextraction. TrAC, Trends Anal. Chem 2003, 22, 565. [Google Scholar]
- (121).Wang C-H; Su H; Chou J-H; Lin J-Y; Huang M-Z; Lee C-W; Shiea J Multiple solid phase microextraction combined with ambient mass spectrometry for rapid and sensitive detection of trace chemical compounds in aqueous solution. Anal. Chim. Acta 2020, 1107, 101. [DOI] [PubMed] [Google Scholar]
- (122).Van Berkel GJ; Kertesz V; Koeplinger KA; Vavrek M; Kong A-NT Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J. Mass Spectrom 2008, 43, 500. [DOI] [PubMed] [Google Scholar]
- (123).Eikel D; Vavrek M; Smith S; Bason C; Yeh S; Korfmacher WA; Henion JD Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Commun. Mass Spectrom 2011, 25, 3587. [DOI] [PubMed] [Google Scholar]
- (124).Cahill JF; Khalid M; Retterer ST; Walton CL; Kertesz V In Situ Chemical Monitoring and Imaging of Contents within Microfluidic Devices Having a Porous Membrane Wall Using Liquid Microjunction Surface Sampling Probe Mass Spectrometry. J. Am. Soc. Mass Spectrom 2020, 31, 832. [DOI] [PubMed] [Google Scholar]
- (125).Quanico J; Franck J; Cardon T; Leblanc E; Wisztorski M; Salzet M; Fournier I NanoLC-MS coupling of liquid microjunction microextraction for on-tissue proteomic analysis. Biochim. Biophys. Acta, Proteins Proteomics 2017, 1865, 891. [DOI] [PubMed] [Google Scholar]
- (126).May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA Conformational Ordering of Biomolecules in the Gas Phase: Nitrogen Collision Cross Sections Measured on a Prototype High Resolution Drift Tube Ion Mobility-Mass Spectrometer. Anal. Chem 2014, 86, 2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Valentine SJ; Koeniger SL; Clemmer DE A Split-Field Drift Tube for Separation and Efficient Fragmentation of Biomolecular Ions. Anal. Chem 2003, 75, 6202. [DOI] [PubMed] [Google Scholar]
- (128).Zhong Y; Hyung S-J; Ruotolo BT Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst 2011, 136, 3534. [DOI] [PubMed] [Google Scholar]
- (129).Shvartsburg AA; Smith RD Fundamentals of Traveling Wave Ion Mobility Spectrometry. Anal. Chem 2008, 80, 9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Purves RW; Guevremont R Electrospray Ionization High-Field Asymmetric Waveform Ion Mobility Spectrometry–Mass Spectrometry. Anal. Chem 1999, 71, 2346. [DOI] [PubMed] [Google Scholar]
- (131).Kolakowski BM; Mester Z Review of applications of high-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential mobility spectrometry (DMS). Analyst 2007, 132, 842. [DOI] [PubMed] [Google Scholar]
- (132).Guevremont R High-field asymmetric waveform ion mobility spectrometry: A new tool for mass spectrometry. Journal of Chromatography A 2004, 1058, 3. [PubMed] [Google Scholar]
- (133).Fernandez-Lima F; Kaplan DA; Suetering J; Park MA Gas-phase separation using a trapped ion mobility spectrometer. Int. J. Ion Mobility Spectrom 2011, 14, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Michelmann K; Silveira JA; Ridgeway ME; Park MA Fundamentals of Trapped Ion Mobility Spectrometry. J. Am. Soc. Mass Spectrom 2015, 26, 14. [DOI] [PubMed] [Google Scholar]
- (135).Groessl M; Graf S; Knochenmuss R High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst 2015, 140, 6904. [DOI] [PubMed] [Google Scholar]
- (136).Ridgeway ME; Lubeck M; Jordens J; Mann M; Park MA Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom 2018, 425, 22. [Google Scholar]
- (137).Fernandez-Lima F; Kaplan D; Park M Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev. Sci. Instrum 2011, 82, 126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Adams KJ; Montero D; Aga D; Fernandez-Lima F Isomer separation of polybrominated diphenyl ether metabolites using nanoESI-TIMS-MS. Int. J. Ion Mobility Spectrom 2016, 19, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Ibrahim YM; Hamid AM; Deng L; Garimella SV; Webb IK; Baker ES; Smith RD New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst 2017, 142, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Deng L; Ibrahim YM; Baker ES; Aly NA; Hamid AM; Zhang X; Zheng X; Garimella SVB; Webb IK; Prost SA; Sandoval JA; Norheim RV; Anderson GA; Tolmachev AV; Smith RD Ion Mobility Separations of Isomers based upon Long Path Length Structures for Lossless Ion Manipulations Combined with Mass Spectrometry. ChemistrySelect 2016, 1, 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem 2019, 91, 8564. [DOI] [PubMed] [Google Scholar]
- (142).Deng L; Webb IK; Garimella SV; Hamid AM; Zheng X; Norheim RV; Prost SA; Anderson GA; Sandoval JA; Baker ES Serpentine ultralong path with extended routing (SUPER) high resolution traveling wave ion mobility-MS using structures for lossless ion manipulations. Anal. Chem 2017, 89, 4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Woods AS; Ugarov M; Egan T; Koomen J; Gillig KJ; Fuhrer K; Gonin M; Schultz JA Lipid/Peptide/Nucleotide Separation with MALDI-Ion Mobility-TOF MS. Anal. Chem 2004, 76, 2187. [DOI] [PubMed] [Google Scholar]
- (144).McLean JA; Ridenour WB; Caprioli RM Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom 2007, 42, 1099. [DOI] [PubMed] [Google Scholar]
- (145).Spraggins JM; Djambazova KV; Rivera ES; Migas LG; Neumann EK; Fuetterer A; Suetering J; Goedecke N; Ly A; Van de Plas R; Caprioli RM High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (timsTOF) Mass Spectrometry. Anal. Chem 2019, 91, 14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Griffiths RL; Hughes JW; Abbatiello SE; Belford MW; Styles IB; Cooper HJ Comprehensive LESA Mass Spectrometry Imaging of Intact Proteins by Integration of Cylindrical FAIMS. Anal. Chem 2020, 92, 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Klingner N; Heller R; Hlawacek G; Facsko S; von Borany J Time-of-flight secondary ion mass spectrometry in the helium ion microscope. Ultramicroscopy 2019, 198, 10. [DOI] [PubMed] [Google Scholar]
- (148).Araki T The history of optical microscope. Mechanical Engineering Reviews 2017, 4, 16. [Google Scholar]
- (149).van Helvoort T; Sankaran N How Seeing Became Knowing: The Role of the Electron Microscope in Shaping the Modern Definition of Viruses. Journal of the History of Biology 2019, 52, 125. [DOI] [PubMed] [Google Scholar]
- (150).Murphy RF Building cell models and simulations from microscope images. Methods 2016, 96, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Sahl SJ; Hell SW; Jakobs S Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol 2017, 18, 685. [DOI] [PubMed] [Google Scholar]
- (152).Kumarasinghe MP; Bourke MJ; Brown I; Draganov PV; McLeod D; Streutker C; Raftopoulos S; Ushiku T; Lauwers GY Pathological assessment of endoscopic resections of the gastrointestinal tract: a comprehensive clinicopathologic review. Mod. Pathol 2020, 33, 986. [DOI] [PubMed] [Google Scholar]
- (153).Deschaine MA; Lehman JS The interface reaction pattern in the skin: an integrated review of clinical and pathological features. Hum. Pathol 2019, 91, 86. [DOI] [PubMed] [Google Scholar]
- (154).Picard C; Orbach D; Carton M; Brugieres L; Renaudin K; Aubert S; Berrebi D; Galmiche L; Dujardin F; Leblond P; Thomas-Teinturier C; Dijoud F Revisiting the role of the pathological grading in pediatric adrenal cortical tumors: results from a national cohort study with pathological review. Mod. Pathol 2019, 32, 546. [DOI] [PubMed] [Google Scholar]
- (155).Sun C; Liu W; Mu Y; Wang X 1,1′-binaphthyl-2,2′-diamine as a novel MALDI matrix to enhance the in situ imaging of metabolic heterogeneity in lung cancer. Talanta 2020, 209, 120557. [DOI] [PubMed] [Google Scholar]
- (156).Wang Y; Hinz S; Uckermann O; Hönscheid P; von Schönfels W; Burmeister G; Hendricks A; Ackerman JM; Baretton GB; Hampe J; Brosch M; Schafmayer C; Shevchenko A; Zeissig S Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2020, 1865, 158579. [DOI] [PubMed] [Google Scholar]
- (157).Andersen MK; Krossa S; Høiem TS; Buchholz R; Claes BSR; Balluff B; Ellis SR; Richardsen E; Bertilsson H; Heeren RMA; Bathen TF; Karst U; Giskeødegård GF; Tessem M-B Simultaneous Detection of Zinc and Its Pathway Metabolites Using MALDI MS Imaging of Prostate Tissue. Anal. Chem 2020, 92, 3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Marsilio S; Newman SJ; Estep JS; Giaretta PR; Lidbury JA; Warry E; Flory A; Morley PS; Smoot K; Seeley EH; Powell MJ; Suchodolski JS; Steiner JM Differentiation of lymphocytic-plasmacytic enteropathy and small cell lymphoma in cats using histology-guided mass spectrometry. J. Vet. Intern. Med 2020, 34, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Cordeiro FB; Jarmusch AK; León M; Ferreira CR; Pirro V; Eberlin LS; Hallett J; Miglino MA; Cooks RG Mammalian ovarian lipid distributions by desorption electrospray ionization–mass spectrometry (DESI-MS) imaging. Anal. Bioanal. Chem 2020, 412, 1251. [DOI] [PubMed] [Google Scholar]
- (160).Zhang G; Zhang J; DeHoog RJ; Pennathur S; Anderton CR; Venkatachalam MA; Alexandrov T; Eberlin LS; Sharma K DESI-MSI and METASPACE indicates lipid abnormalities and altered mitochondrial membrane components in diabetic renal proximal tubules. Metabolomics 2020, 16, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Severiano DLR; Oliveira-Lima OC; Vasconcelos GA; Lemes Marques B; Almeida de Carvalho G; Freitas EMM; Xavier CH; Gomez MV; Pinheiro ACO; Gomez RS; Vaz BG; Pinto MCX Cerebral Lipid Dynamics in Chronic Cerebral Hypoperfusion Model by DESI-MS Imaging. Neuroscience 2020, 426, 1. [DOI] [PubMed] [Google Scholar]
- (162).Basu SS; Regan MS; Randall EC; Abdelmoula WM; Clark AR; Gimenez-Cassina Lopez B; Cornett DS; Haase A; Santagata S; Agar NYR Rapid MALDI mass spectrometry imaging for surgical pathology. npj Precision Oncology 2019, 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Sun C; Zhang M; Dong H; Liu W; Guo L; Wang X A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. J. Pharm. Biomed. Anal 2020, 179, 113014. [DOI] [PubMed] [Google Scholar]
- (164).Sun C; Liu W; Ma S; Zhang M; Geng Y; Wang X Development of a high-coverage matrix-assisted laser desorption/ionization mass spectrometry imaging method for visualizing the spatial dynamics of functional metabolites in Salvia miltiorrhiza Bge. Journal of Chromatography A 2020, 1614, 460704. [DOI] [PubMed] [Google Scholar]
- (165).Patterson NH; Tuck M; Lewis A; Kaushansky A; Norris JL; Van de Plas R; Caprioli RM Next Generation Histology-Directed Imaging Mass Spectrometry Driven by Autofluorescence Microscopy. Anal. Chem 2018, 90, 12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Patterson NH; Tuck M; Van de Plas R; Caprioli RM Advanced Registration and Analysis of MALDI Imaging Mass Spectrometry Measurements through Autofluorescence Microscopy. Anal. Chem 2018, 90, 12395. [DOI] [PubMed] [Google Scholar]
- (167).Goltsev Y; Samusik N; Kennedy-Darling J; Bhate S; Hale M; Vazquez G; Black S; Nolan GP Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Eng J; Thibault G; Luoh S-W; Gray JW; Chang YH; Chin K In Biomarkers for Immunotherapy of Cancer: Methods and Protocols; Thurin M, Cesano A, Marincola FM, Eds.; Springer: New York, NY, 2020; p 521. [Google Scholar]
- (169).Rashid R; Gaglia G; Chen Y-A; Lin J-R; Du Z; Maliga Z; Schapiro D; Yapp C; Muhlich J; Sokolov A; Sorger P; Santagata S Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer. Sci. Data 2019, 6, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Lin T; Jin H; Gong L; Yu R; Sun S; Yang L; Zhang P; Han P; Cheng J; Liu L; Wei Q Surveillance of non-muscle invasive bladder cancer using fluorescence in situ hybridization: Protocol for a systematic review and meta-analysis. Medicine (Philadelphia, PA, U. S.) 2019, 98, e14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Miedema J; Andea AA Through the looking glass and what you find there: making sense of comparative genomic hybridization and fluorescence in situ hybridization for melanoma diagnosis. Mod. Pathol 2020, 33, 1318. [DOI] [PubMed] [Google Scholar]
- (172).Schleyer G; Shahaf N; Ziv C; Dong Y; Meoded RA; Helfrich EJN; Schatz D; Rosenwasser S; Rogachev I; Aharoni A; Piel J; Vardi A In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids. Nature Microbiology 2019, 4, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Solier C; Langen H Antibody-based proteomics and biomarker research—Current status and limitations. Proteomics 2014, 14, 774. [DOI] [PubMed] [Google Scholar]
- (174).Budnik B; Levy E; Harmange G; Slavov N SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 2018, 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Kriegsmann K; Longuespée R; Hundemer M; Zgorzelski C; Casadonte R; Schwamborn K; Weichert W; Schirmacher P; Harms A; Kazdal D; Leichsenring J; Stenzinger A; Warth A; Fresnais M; Kriegsmann J; Kriegsmann M Combined Immunohistochemistry after Mass Spectrometry Imaging for Superior Spatial Information. Proteomics: Clin. Appl 2019, 13, 1800035. [DOI] [PubMed] [Google Scholar]
- (176).Kaya I; Michno W; Brinet D; Iacone Y; Zanni G; Blennow K; Zetterberg H; Hanrieder J Histology-Compatible MALDI Mass Spectrometry Based Imaging of Neuronal Lipids for Subsequent Immunofluorescent Staining. Anal. Chem 2017, 89, 4685. [DOI] [PubMed] [Google Scholar]
- (177).Tominski C; Lösekann-Behrens T; Ruecker A; Hagemann N; Kleindienst S; Mueller CW; Höschen C; Kögel-Knabner I; Kappler A; Behrens S Insights into Carbon Metabolism Provided by Fluorescence In Situ Hybridization-Secondary Ion Mass Spectrometry Imaging of an Autotrophic, Nitrate-Reducing, Fe(II)-Oxidizing Enrichment Culture. Appl. Environ. Microbiol 2018, 84, e02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Huber K; Kunzke T; Buck A; Langer R; Luber B; Feuchtinger A; Walch A Multimodal analysis of formalin-fixed and paraffin-embedded tissue by MALDI imaging and fluorescence in situ hybridization for combined genetic and metabolic analysis. Lab. Invest 2019, 99, 1535. [DOI] [PubMed] [Google Scholar]
- (179).Dekas AE; Orphan VJ In Methods in Enzymology; Klotz MG, Ed.; Academic Press: 2011; Vol. 486, p 281. [DOI] [PubMed] [Google Scholar]
- (180).Jones MA; Cho SH; Patterson NH; Van de Plas R; Spraggins JM; Boothby MR; Caprioli RM Discovering New Lipidomic Features Using Cell Type Specific Fluorophore Expression to Provide Spatial and Biological Specificity in a Multimodal Workflow with MALDI Imaging Mass Spectrometry. Anal. Chem 2020, 92, 7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Vanickova L; Guran R; Kollár S; Emri G; Krizkova S; Do T; Heger Z; Zitka O; Adam V Mass spectrometric imaging of cysteine rich proteins in human skin. Int. J. Biol. Macromol 2019, 125, 270. [DOI] [PubMed] [Google Scholar]
- (182).Zhang Z; Sugiura Y; Mune T; Nishiyama M; Terada Y; Mukai K; Nishimoto K Immunohistochemistry for aldosterone synthase CYP11B2 and matrix-assisted laser desorption ionization imaging mass spectrometry for in-situ aldosterone detection. Curr. Opin. Nephrol. Hypertens 2019, 28, 105. [DOI] [PubMed] [Google Scholar]
- (183).Prentice BM; Hart NJ; Phillips N; Haliyur R; Judd A; Armandala R; Spraggins JM; Lowe CL; Boyd KL; Stein RW; Wright CV; Norris JL; Powers AC; Brissova M; Caprioli RM Imaging mass spectrometry enables molecular profiling of mouse and human pancreatic tissue. Diabetologia 2019, 62, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Bien T; Perl M; Machmüller AC; Nitsche U; Conrad A; Johannes L; Müthing J; Soltwisch J; Janssen K-P; Dreisewerd K MALDI-2 Mass Spectrometry and Immunohistochemistry Imaging of Gb3Cer, Gb4Cer, and Further Glycosphingolipids in Human Colorectal Cancer Tissue. Anal. Chem 2020, 92, 7096. [DOI] [PubMed] [Google Scholar]
- (185).Prade VM; Kunzke T; Feuchtinger A; Rohm M; Luber B; Lordick F; Buck A; Walch A De novo discovery of metabolic heterogeneity with immunophenotype-guided imaging mass spectrometry. Mol. Metab 2020, 36, 100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Rabe J-H; Sammour, D. A; Schulz S; Munteanu B; Ott M; Ochs K; Hohenberger P; Marx A; Platten M; Opitz CA; Ory DS; Hopf C Fourier Transform Infrared Microscopy Enables Guidance of Automated Mass Spectrometry Imaging to Predefined Tissue Morphologies. Sci. Rep 2018, 8, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Jiang H; Kilburn MR; Decelle J; Musat N NanoSIMS chemical imaging combined with correlative microscopy for biological sample analysis. Curr. Opin. Biotechnol 2016, 41, 130. [DOI] [PubMed] [Google Scholar]
- (188).Lovrić J; Dunevall J; Larsson A; Ren L; Andersson S; Meibom A; Malmberg P; Kurczy ME; Ewing AG Nano Secondary Ion Mass Spectrometry Imaging of Dopamine Distribution Across Nanometer Vesicles. ACS Nano 2017, 11, 3446. [DOI] [PubMed] [Google Scholar]
- (189).Liu W; Huang L; Komorek R; Handakumbura PP; Zhou Y; Hu D; Engelhard MH; Jiang H; Yu X-Y; Jansson C; Zhu Z Correlative surface imaging reveals chemical signatures for bacterial hotspots on plant roots. Analyst 2020, 145, 393. [DOI] [PubMed] [Google Scholar]
- (190).Strnad Š; Pražienková V; Sýkora D; Cvačka J; Maletínská L; Popelová A; Vrkoslav V The use of 1,5-diaminonaphthalene for matrix-assisted laser desorption/ionization mass spectrometry imaging of brain in neurodegenerative disorders. Talanta 2019, 201, 364. [DOI] [PubMed] [Google Scholar]
- (191).Yang H; Su R; Wishnok JS; Liu N; Chen C; Liu S; Tannenbaum SR Magnetic silica nanoparticles for use in matrix-assisted laser desorption ionization mass spectrometry of labile biomolecules such as oligosaccharides, amino acids, peptides and nucleosides. Microchim. Acta 2019, 186, 104. [DOI] [PubMed] [Google Scholar]
- (192).Baker MJ; Trevisan J; Bassan P; Bhargava R; Butler HJ; Dorling KM; Fielden PR; Fogarty SW; Fullwood NJ; Heys KA; Hughes C; Lasch P; Martin-Hirsch PL; Obinaju B; Sockalingum GD; Sulé-Suso J; Strong RJ; Walsh MJ; Wood BR; Gardner P; Martin FL Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc 2014, 9, 1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Masyuko R; Lanni EJ; Sweedler JV; Bohn PW Correlated imaging – a grand challenge in chemical analysis. Analyst 2013, 138, 1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Rinia HA; Burger KNJ; Bonn M; Müller M Quantitative Label-Free Imaging of Lipid Composition and Packing of Individual Cellular Lipid Droplets Using Multiplex CARS Microscopy. Biophys. J 2008, 95, 4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Le TT; Duren HM; Slipchenko MN; Hu C-D; Cheng J-X Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J. Lipid Res 2010, 51, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Li J; Condello S; Thomes-Pepin J; Ma X; Xia Y; Hurley TD; Matei D; Cheng J-X Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Tipping WJ; Lee M; Serrels A; Brunton VG; Hulme AN Stimulated Raman scattering microscopy: an emerging tool for drug discovery. Chem. Soc. Rev 2016, 45, 2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Meister K; Niesel J; Schatzschneider U; Metzler-Nolte N; Schmidt DA; Havenith M Label-Free Imaging of Metal–Carbonyl Complexes in Live Cells by Raman Microspectroscopy. Angew. Chem., Int. Ed 2010, 49, 3310. [DOI] [PubMed] [Google Scholar]
- (199).Yamakoshi H; Dodo K; Palonpon A; Ando J; Fujita K; Kawata S; Sodeoka M Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J. Am. Chem. Soc 2012, 134, 20681. [DOI] [PubMed] [Google Scholar]
- (200).Lazaro-Pacheco D; Shaaban AM; Rehman S; Rehman I Raman spectroscopy of breast cancer. Appl. Spectrosc. Rev 2020, 1, 439. [Google Scholar]
- (201).Ralbovsky NM; Lednev IK Raman spectroscopy and chemometrics: A potential universal method for diagnosing cancer. Spectrochim. Acta, Part A 2019, 219, 463. [DOI] [PubMed] [Google Scholar]
- (202).Wiercigroch E; Szafraniec E; Czamara K; Pacia MZ; Majzner K; Kochan K; Kaczor A; Baranska M; Malek K Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta, Part A 2017, 185, 317. [DOI] [PubMed] [Google Scholar]
- (203).Gautam R; Vanga S; Ariese F; Umapathy S Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ. Techniques and Instrumentation 2015, 2, 8. [Google Scholar]
- (204).Lasch P; Noda I Two-Dimensional Correlation Spectroscopy for Multimodal Analysis of FT-IR, Raman, and MALDI-TOF MS Hyperspectral Images with Hamster Brain Tissue. Anal. Chem 2017, 89, 5008. [DOI] [PubMed] [Google Scholar]
- (205).Tai T; Karácsony O; Bocharova V; Van Berkel GJ; Kertesz V Topographical and Chemical Imaging of a Phase Separated Polymer Using a Combined Atomic Force Microscopy/Infrared Spectroscopy/Mass Spectrometry Platform. Anal. Chem 2016, 88, 2864. [DOI] [PubMed] [Google Scholar]
- (206).Balbekova A; Lohninger H; van Tilborg GAF; Dijkhuizen RM; Bonta M; Limbeck A; Lendl B; Al-Saad KA; Ali M; Celikic M; Ofner J Fourier Transform Infrared (FT-IR) and Laser Ablation Inductively Coupled Plasma–Mass Spectrometry (LA-ICP-MS) Imaging of Cerebral Ischemia: Combined Analysis of Rat Brain Thin Cuts Toward Improved Tissue Classification. Appl. Spectrosc 2018, 72, 241. [DOI] [PubMed] [Google Scholar]
- (207).Neumann EK; Comi TJ; Spegazzini N; Mitchell JW; Rubakhin SS; Gillette MU; Bhargava R; Sweedler JV Multimodal Chemical Analysis of the Brain by High Mass Resolution Mass Spectrometry and Infrared Spectroscopic Imaging. Anal. Chem 2018, 90, 11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Chisanga M; Linton D; Muhamadali H; Ellis DI; Kimber RL; Mironov A; Goodacre R Rapid differentiation of Campylobacter jejuni cell wall mutants using Raman spectroscopy, SERS and mass spectrometry combined with chemometrics. Analyst 2020, 145, 1236. [DOI] [PubMed] [Google Scholar]
- (209).Dunham SJB; Ellis JF; Baig NF; Morales-Soto N; Cao T; Shrout JD; Bohn PW; Sweedler JV Quantitative SIMS Imaging of Agar-Based Microbial Communities. Anal. Chem 2018, 90, 5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Morales-Soto N; Dunham SJB; Baig NF; Ellis JF; Madukoma CS; Bohn PW; Sweedler JV; Shrout JD Spatially dependent alkyl quinolone signaling responses to antibiotics in Pseudomonas aeruginosa swarms. J. Biol. Chem 2018, 293, 9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Burkhow SJ; Stephens NM; Mei Y; Dueñas ME; Freppon DJ; Ding G; Smith SC; Lee Y-J; Nikolau BJ; Whitham SA; Smith EA Characterizing virus-induced gene silencing at the cellular level with in situ multimodal imaging. Plant Methods 2018, 14, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Ali A; Abouleila Y; Shimizu Y; Hiyama E; Watanabe TM; Yanagida T; Germond A Single-Cell Screening of Tamoxifen Abundance and Effect Using Mass Spectrometry and Raman-Spectroscopy. Anal. Chem 2019, 91, 2710. [DOI] [PubMed] [Google Scholar]
- (213).Ryabchykov O; Popp J; Bocklitz T Fusion of MALDI Spectrometric Imaging and Raman Spectroscopic Data for the Analysis of Biological Samples. Front. Chem 2018, 6. DOI: 10.3389/fchem.2018.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Bergholt MS; Serio A; McKenzie JS; Boyd A; Soares RF; Tillner J; Chiappini C; Wu V; Dannhorn A; Takats Z; Williams A; Stevens MM Correlated Heterospectral Lipidomics for Biomolecular Profiling of Remyelination in Multiple Sclerosis. ACS Cent. Sci 2018, 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Bakry R; Rainer M; Huck CW; Bonn GK Protein profiling for cancer biomarker discovery using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and infrared imaging: A review. Anal. Chim. Acta 2011, 690, 26. [DOI] [PubMed] [Google Scholar]
- (216).Petit VW; Réfrégiers M; Guettier C; Jamme F; Sebanayakam K; Brunelle A; Laprévote O; Dumas P; Le Naour F Multimodal Spectroscopy Combining Time-of-Flight-Secondary Ion Mass Spectrometry, Synchrotron-FT-IR, and Synchrotron-UV Microspectroscopies on the Same Tissue Section. Anal. Chem 2010, 82, 3963. [DOI] [PubMed] [Google Scholar]
- (217).Lohöfer F; Hoffmann L; Buchholz R; Huber K; Glinzer A; Kosanke K; Feuchtinger A; Aichler M; Feuerecker B; Kaissis G; Rummeny EJ; Höltke C; Faber C; Schilling F; Botnar RM; Walch AK; Karst U; Wildgruber M Molecular imaging of myocardial infarction with Gadofluorine P – A combined magnetic resonance and mass spectrometry imaging approach. Heliyon 2018, 4, e00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Lohöfer F; Buchholz R; Glinzer A; Huber K; Haas H; Kaissis G; Feuchtinger A; Aichler M; Sporns PB; Höltke C; Stölting M; Schilling F; Botnar RM; Kimm MA; Faber C; Walch AK; Zernecke A; Karst U; Wildgruber M Mass Spectrometry Imaging of atherosclerosis-affine Gadofluorine following Magnetic Resonance Imaging. Sci. Rep 2020, 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Srimany A; George C; Naik HR; Pinto DG; Chandrakumar N; Pradeep T Developmental patterning and segregation of alkaloids in areca nut (seed of Areca catechu) revealed by magnetic resonance and mass spectrometry imaging. Phytochemistry 2016, 125, 35. [DOI] [PubMed] [Google Scholar]
- (220).Verbeeck N; Spraggins JM; Murphy MJM; Wang H.-d.; Deutch AY; Caprioli RM; Van de Plas R Connecting imaging mass spectrometry and magnetic resonance imaging-based anatomical atlases for automated anatomical interpretation and differential analysis. Biochim. Biophys. Acta, Proteins Proteomics 2017, 1865, 967. [DOI] [PubMed] [Google Scholar]
- (221).Marshall DD; Powers R Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc 2017, 100, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Cassat JE; Moore JL; Wilson KJ; Stark Z; Prentice BM; Van de Plas R; Perry WJ; Zhang Y; Virostko J; Colvin DC; Rose KL; Judd AM; Reyzer ML; Spraggins JM; Grunenwald CM; Gore JC; Caprioli RM; Skaar EP Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl. Med 2018, 10, eaan6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Wang Z; Gerstein M; Snyder M RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet 2009, 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Horgan RP; Kenny LC Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. Obstetrician & Gynaecologist 2011, 13, 189. [Google Scholar]
- (225).Kulkarni A; Anderson AG; Merullo DP; Konopka G Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr. Opin. Biotechnol 2019, 58, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Wang Z; Shi H; Yu S; Zhou W; Li J; Liu S; Deng M; Ma J; Wei Y; Zheng Y; Liu Y Comprehensive transcriptomics, proteomics, and metabolomics analyses of the mechanisms regulating tiller production in low-tillering wheat. Theor. Appl. Genet 2019, 132, 2181. [DOI] [PubMed] [Google Scholar]
- (227).Angelidis I; Simon LM; Fernandez IE; Strunz M; Mayr CH; Greiffo FR; Tsitsiridis G; Ansari M; Graf E; Strom T-M; Nagendran M; Desai T; Eickelberg O; Mann M; Theis FJ; Schiller HB An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun 2019, 10, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Liessem S; Ragionieri L; Neupert S; Büschges A; Predel R Transcriptomic and Neuropeptidomic Analysis of the Stick Insect, Carausius morosus. J. Proteome Res 2018, 17, 2192. [DOI] [PubMed] [Google Scholar]
- (229).Sung C-C; Chen L; Limbutara K; Jung HJ; Gilmer GG; Yang C-R; Lin S-H; Khositseth S; Chou C-L; Knepper MA RNA-Seq and protein mass spectrometry in microdissected kidney tubules reveal signaling processes initiating lithium-induced nephrogenic diabetes insipidus. Kidney Int 2019, 96, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Noble KV; Reyzer ML; Barth JL; McDonald H; Tuck M; Schey KL; Krug EL; Lang H Use of Proteomic Imaging Coupled With Transcriptomic Analysis to Identify Biomolecules Responsive to Cochlear Injury. Front. Mol. Neurosci 2018, 11. DOI: 10.3389/fnmol.2018.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Kawashima M; Tokiwa M; Nishimura T; Kawata Y; Sugimoto M; Kataoka TR; Sakurai T; Iwaisako K; Suzuki E; Hagiwara M; Harris AL; Toi M High-resolution imaging mass spectrometry combined with transcriptomic analysis identified a link between fatty acid composition of phosphatidylinositols and the immune checkpoint pathway at the primary tumour site of breast cancer. Br. J. Cancer 2020, 122, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Jarvis S; Gethings L; Gadeleta R; Claude E; Winston R; Williamson C; Bevan C In Society for Endocrinology BES 2018; BioScientifica: 2018; Vol. 59. [Google Scholar]
- (233).Dunevall J; Majdi S; Larsson A; Ewing A Vesicle impact electrochemical cytometry compared to amperometric exocytosis measurements. Current Opinion in Electrochemistry 2017, 5, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Chen C-C; Cang C; Fenske S; Butz E; Chao Y-K; Biel M; Ren D; Wahl-Schott C; Grimm C Patch-clamp technique to characterize ion channels in enlarged individual endolysosomes. Nat. Protoc 2017, 12, 1639. [DOI] [PubMed] [Google Scholar]
- (235).Leyrer-Jackson JM; Olive MF; Gipson CD In Glutamate Receptors: Methods and Protocols; Burger C, Velardo MJ, Eds.; Springer: New York, NY, 2019; p 107. [Google Scholar]
- (236).Phan NTN; Li X; Ewing AG Measuring synaptic vesicles using cellular electrochemistry and nanoscale molecular imaging. Nature Reviews Chemistry 2017, 1, 0048. [Google Scholar]
- (237).Li K; Liu J; Grovenor CRM; Moore KL NanoSIMS Imaging and Analysis in Materials Science. Annu. Rev. Anal. Chem 2020, 13, 273. [DOI] [PubMed] [Google Scholar]
- (238).Ranjbari E; Majdi S; Ewing A Analytical Techniques: Shedding Light upon Nanometer-Sized Secretory Vesicles. Trends in Chemistry 2019, 1, 440. [Google Scholar]
- (239).Chadwick GL; Jiménez Otero F; Gralnick JA; Bond DR; Orphan VJ NanoSIMS imaging reveals metabolic stratification within current-producing biofilms. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Thomen A; Najafinobar N; Penen F; Kay E; Upadhyay PP; Li X; Phan NTN; Malmberg P; Klarqvist M; Andersson S; Kurczy ME; Ewing AG Subcellular Mass Spectrometry Imaging and Absolute Quantitative Analysis across Organelles. ACS Nano 2020, 14, 4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Larsson A; Majdi S; Oleinick A; Svir I; Dunevall J; Amatore C; Ewing AG Intracellular Electrochemical Nano-measurements Reveal that Exocytosis of Molecules at Living Neurons is Subquantal and Complex. Angew. Chem., Int. Ed 2020, 59, 6711. [DOI] [PubMed] [Google Scholar]
- (242).Baker LA; Jagdale GS On the intersection of electrochemistry and mass spectrometry. Current Opinion in Electrochemistry 2019, 13, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Lin L-E; Chen C-L; Huang Y-C; Chung H-H; Lin C-W; Chen K-C; Peng Y-J; Ding S-T; Wang M-Y; Shen T-L; Hsu C-C Precision biomarker discovery powered by microscopy image fusion-assisted high spatial resolution ambient ionization mass spectrometry imaging. Anal. Chim. Acta 2020, 1100, 75. [DOI] [PubMed] [Google Scholar]
- (244).Van de Plas R; Yang J; Spraggins J; Caprioli RM Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods 2015, 12, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Lin L-E; Chen C-L; Huang Y-C; Chung H-H; Lin C-W; Chen K-C; Peng Y-J; Ding S-T; Wang M-Y; Shen T-L; Hsu C-C High Spatial Resolution Ambient Ionization Mass Spectrometry Imaging Using Microscopy Image Fusion Determines Tumor Margins. bioRxiv 2019, 657494. [DOI] [PubMed] [Google Scholar]
- (246).Vollnhals F; Audinot J-N; Wirtz T; Mercier-Bonin M; Fourquaux I; Schroeppel B; Kraushaar U; Lev-Ram V; Ellisman MH; Eswara S Correlative Microscopy Combining Secondary Ion Mass Spectrometry and Electron Microscopy: Comparison of Intensity–Hue–Saturation and Laplacian Pyramid Methods for Image Fusion. Anal. Chem 2017, 89, 10702. [DOI] [PubMed] [Google Scholar]


