ABSTRACT
Bladder cancer (BCa) is one of the most common cancers in men and is a major threat to the lives and health of older men. Many studies have shown that miR-7, as an important tumor suppressor gene, could directly inhibit some pathways involved in the development of cancer. MiR-7-5p, which was assessed in this study, consists of one arm of miR-7 and acts as a cancer suppressor gene in multiple cancer types. Autophagy, as a common biological process, plays dual roles in the process of cancer. Chemotherapy resistance is a problem in the treatment of BCa. In this study, the data showed that miR-7-5p was obviously down-regulated in BCa tissues and cells compared to their respective controls. In addition, miR-7-5p mimic effectively inhibited migration, invasion and autophagy both in vitro and in vivo. In the mechanistic study, miR-7-5p targeted autophagy-related gene ATG7 to inhibit its expression, which in turn inhibited autophagy. Finally, the migration of BCa cells was inhibited, and chemosensitivity was improved. Overall, our results provide evidence of the role of miR-7-5p as a cancer suppressor gene in BCa and provide new opportunities for the treatment of BCa.
KEYWORDS: MiRNA-7-5p, autophagy, ATG7, BCa, chemoresistance
Graphical Abstract
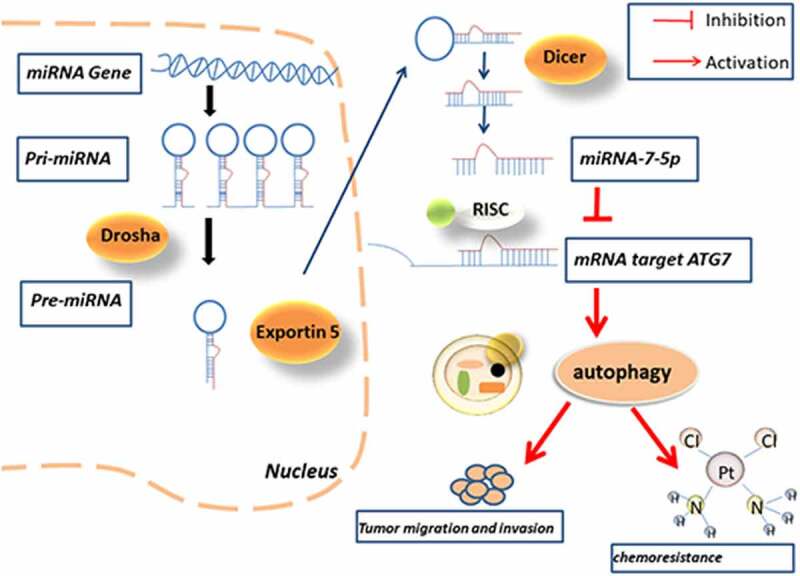
Introduction
BCa is one of the most common urologic cancer types [1]. It is divided into two pathological types: the non-muscle-invasive type and the muscle-invasive type. The latter has a worse prognosis due to its strong invasiveness and tendency to metastasize, and the five-year survival rate is less than 50%[2]. Moreover, metastasis is the major cause of death in BCa patients. The first-line chemotherapy for BCa is based on cisplatin and can improve the five-year overall survival rate [3]. However, cisplatin resistance remains a main barrier to treatment success in these patients. Further research may reveal complex molecular mechanisms regulation the invasion of BCa cells. Therefore, further research on the molecular regulatory mechanism of BCa occurrence and development is helpful to find targeted treatment sites and improve the prognosis of BCa.
MicroRNAs (miRNAs) are single-stranded RNA fragments of 19–25 nucleotides that is highly conserved among species [4]. MiRNA’s primary action site is the 3’- untranslated region (UTR) of the target gene, which relies on the role of the RNA-induced silencing complex (RISC) to inhibit protein translation [5]. It is generally believed that the combination of miRNA with the 3’-UTR of mRNA hinders the translation process but does not affect the stability of mRNA and therefore acts in posttranscriptional regulation [6]. Previous studies have demonstrated that miRNAs play a major role in cell growth, development, and aging. MiRNAs can also act as promoters or inhibitors of many tumors and play a key role in the metastasis of cancers such as BCa, non-small-cell lung cancer (NSCLC) and gastric cancer [6–8]; however, the regulatory mechanisms have yet to be studied in depth.
MiR-7 is a widely studied miRNA and is closely related to the various diseases. In humans, miR-7 is transcribed from miR-7-1, miR-7-2, and miR-7-3, all of which have the same mature miRNA sequence. According to our research, the most famous miRNA in the miR-7 family is miR-7-5p.
Many researchers have found miR-7-5p regulates the expression of various cancer genes in NSCLC, glioblastoma, stomach cancer and other cancers, and revealing the mechanisms regulating miR-7-5p activity may increase our understanding of the causes of various cancers [9–11]. However, few studies of this type have been performed on BCa, so the role of miR-7-5p in the metastasis of BCa and its potential mechanism deserve further exploration.
Autophagy is an evolutionarily conserved process of self-catabolic organelle reuse [12], which is activated in response to starvation, cellular stress and reactive oxygen species and plays a protective role in different conditions while exacerbating the condition [13]. For example, autophagy can maintain a steady state of the heart, and excessive autophagy activity may promote the development of heart failure [14]. Many reports have also confirmed that drug-resistant tumors can keep themselves alive by inducing autophagy [15]. Therefore, autophagy is also considered to be one of the factors inducing chemical resistance. Autophagy is highly regulated in the process of autophagy, and these genes that regulate autophagy are also called autophagy-related genes (ATGs) [16]. The proteins encoded by these autophagy-related genes play an important role in the occurrence and development of autophagy. Autophagy-related gene 7 (ATG7) is one of ATG family which plays a synergistic role in the migration and invasion of BCa. ATG7 is a key protein in the intracellular autophagy response, and previous studies have shown that ATG7 promotes BCa attacks through autophagy by increasing the stability of ARHGDIB mRNA [17]; Zhu et al. revealed that miR-145/FOXO3a/ATG7/autophagy feedback loop could regulate BCa [18]. However, the regulatory role of ATG7 as a miR-7-5p target gene in BCa autophagy and invasion and whether it participates in drug resistance have not been discussed.
In this study, qPCR data showed a significant decrease in the expression level of miR-7-5p in BCa tissues and cell lines. Immunohistochemistry (IHC) and Western blotting demonstrated that ATG7 expression is significantly higher in BCa tissues and cell lines than in respective control. The results further reveal that the in vitro miR-7-5p inhibits the protein translation of ATG7, does not affect the mRNA transcription of ATG7, and inhibits the occurrence of autophagy, thus inhibiting the migration and invasion of BCa. In addition, forced expression of ATG7 was found to attenuate the increase in cisplatin sensitivity induce by up regulated of miR-7-5p. We also found that miR-7-5p in a T24T xenograft transplantation model could inhibit tumor progression.
Materials and methods
Tissues samples
A total of 47 BCa tissues from patients undergoing total cystectomy at the First Affiliated Hospital of Wannan Medical College. All tissues were identified as tumor or normal tissues by the pathology department, stored in a liquid nitrogen tank. This study was approved by the Research Ethics Committee of the First Affiliated Hospital of Wannan Medical College, and all patients signed a written informed consent form.
Cell culture
The human metastatic BCa cell line T24T was kindly provided by Dr. Li (Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China). Human bladder epithelial cells (SV-HUC-1) and BCa cells (EJ) were obtained from FuHeng BioLogy (China). T24T and EJ cells were cultured with RPMI 1640 (Gibco), and SV-HUC-1 cells were cultured with F-12 K (Gibco) medium at a temperature of 37°C with 5% CO2.
Chemical reagents
Bafilomycin A1 (Baf A1) (HY-100558) and cisplatin (HY-17394) were purchased from MCE (China). BBN was purchased from Merck (Darmstadt, Germany).
MiRNA transfection
The kits and transfection reagents used to knockdown and overexpress miR-7-5p were obtained from RiboBio. The cell density reached 30–50% before transfection. The transfection process included dilution of mimic and inhibitor, preparation of mixed solution, addition of transfection reagent, and finally culture in a 5% CO2 incubator at 37°C. RNA and protein were extracted 24 h or 48 h later to verify the transfection effect.
Quantitative reverse-transcription PCR (qRT-PCR)
To detect the expression level of miR-7-5p, total RNA was extracted, and the First-Strand cDNA Synthesis Kit (TIANGEN) and RT Reagent Kit were used for stem loop detection. cDNA was generated from miRNA according to the instructions, and the U6 level was used as an internal control.
Cell migration and invasion assay
For the migration experiment, 1 × 104 cells were suspended in serum-free RPMI 1640 in the upper chamber without matrix glue, and 500 μl fresh normal RPMI 1640 was added to the lower chamber. In the invasion experiment, the upper chamber was precoated with matrix glue, and 3 × 104 cells suspended in serum-free RPMI 1640 were added. After 24 h of incubation, the invasive cells were fixed with 4% methanol for 30 minutes and stained with 0.1% crystal violet for 30 minutes. After that, a cotton swab was used to remove cells from the upper chamber. Images were acquired with a light microscope (100X).
Wound healing assay
T24T and EJ cells transfected with mimic NC, miR-7-5p, anti-NC and anti-miR-7-5p were plated one day before the experiment, and the fusion rate was close to 100% after overnight incubation. On the second day, the cell layer was scratched along a vertical line with a 200-µl pipette tip perpendicular to the cell plane. After wounding was completed, the cells were washed 2–3 times with PBS, the no adherent cells were washed away, and the medium was replaced with fresh serum-free medium. The cells were then placed in a 37°C 5% CO2 incubator for culture. The cells were observed under a microscope at 0 h and 24 h, the width of the scratches was measured, and the cells were photographed. ImageJ was used to calculate the wound healing rate.
Western blot analysis
Cells were collected and lysed with RIPA buffer, and protein quantification was performed using the BCA method. Antibodies against human LC3B (Proteintech Group, Rosemont, IL, USA 1:2500), SQSTM1/p62 (Proteintech Group, Rosemont, IL, USA1:8000), Beclin1 (Proteintech Group, Rosemont, IL, USA1:8000), β-actin (Proteintech Group Rosemont IL USA1:5000), and ATG7 (Proteintech Group, Rosemont, IL, USA1:2000) were added to antibody dilution buffer and incubated with samples at 4°C overnight. β-Actin was used as a control for the examined proteins.
Luciferase reporter assay
Transfection was performed when the cell density reached 60% to 80% in a six-well plate. After transfection for the corresponding time, we discarded the culture medium, washed the cells with PBS once, completely covered the cells with cell lysis solution, fully mixed the sample, and added cell lysis solution for approximately 30 min. Cell lysate was taken and added to the plate. Each condition was set up in triplicate wells. The luciferase kit was purchased from GenePharma (Shanghai, China).
IHC
IHC is used to make enzyme-labeled antibodies by connecting enzymes to antibodies through covalent bonds. Based on the specific catalytic effect of enzymes on substrates, colored insoluble products or particles with certain electron densities are generated, and various antigen components on the cell surface and inside cells can be visualized under an ordinary microscope or an electron microscope. First, the tissue was dewaxed and rehydrated; antigen repair, antibody incubation, dehydration and sealing were performed, and then the results were observed.
Transmission electron microscopy (TEM)
TEM is an important means to study the fine structure of organisms. In the ultrastructure, the autophagosome is a bilayer membrane enclosing cytoplasmic components such as mitochondria and segmented endoplasmic reticulum. Autophagosomes can be observed well by electron microscopy.
CCK-8 assay
CCK-8 assays were used to detect the growth vitality of T24T and EJ cells (Cellorlab Laboratories, Shanghai). Cell suspensions (100 μL/well) were inoculated into 96-well plates. The plates were placed in an incubator for preculture for 24 h (at 37°C and 5% CO2). After adding 10 μL CCK-8 solution to each well (to avoid the formation of bubbles in wells, this might affect the OD reading), the plates were placed in the incubator for 1 ~ 4 h. The absorbance at 450 nm was measured with an enzyme plate instrument.
In vivo model
T24T cells were stably transfected with lentiviruses to overexpress miR-7-5p or miR-NC. Then, approximately 5.0 × 106 cells in PBS were injected into the right subcutaneous area of mice (BALB/C, 4 weeks old, n = 3 per group). The location and size of the tumors in the mice were observed weekly. Four weeks after injection, the mice were sacrificed to obtain the tumors, and then samples were removed for continuous sectioning and immunohistochemical detection. The study was approved by the local Ethics Committee of The Wannan Medical College.
Statistical analysis
All our data were analyzed using GraphPad Prism Software Version 7.0a (GraphPad, San Diego, CA) and SigmaPlot v10.0 (Systat Software Inc). The results were evaluated by Student’s t-test, with P < 0.05 considered to indicate significance.
Results
1. MiR-7-5p is expressed at relatively low levels in BCa tissues and cells
To explore the role of miR-7-5p in BCa, a database (http://gepia2.cancer-pku.cn/#index) was used to predict the expression of miR-7 in human BCa tissues, and it was found that it was significantly lower in cancerous tissues than in adjacent tissue. In the 47 patients’ BCa tissues we collected, the expression level of miR-7-5p in cancerous tissues was consistent with that predicted for miR-7 (Figure 1(a, b)). Second, we selected three cell lines: human ureteral epithelial cells (SV-HUC-1), human metastatic BCa cells (T24T) and human BCa cells (EJ). The RT-PCR results showed a significant decrease in the expression of miR-7-5p in T24T and EJ cells compared to SV-HUC-1 cells (Figure 1(c)). These data suggested that miR-7-5p may play a role as a cancer suppressor gene in BCa. Then, T24T and EJ cells were transfected with mimic NC, miR-7-5p mimic (miR-7-5p), anti-miR-7-5p or anti NC, and qPCR was applied to determine mimic transfection efficiency. Since the inhibitor binds with miRNA to inhibit the function of miRNA, it does not alter the generation of miRNA, so the expression level of miRNA-7-5p was not detected by PCR (Figure 1(d)).
Figure 1.
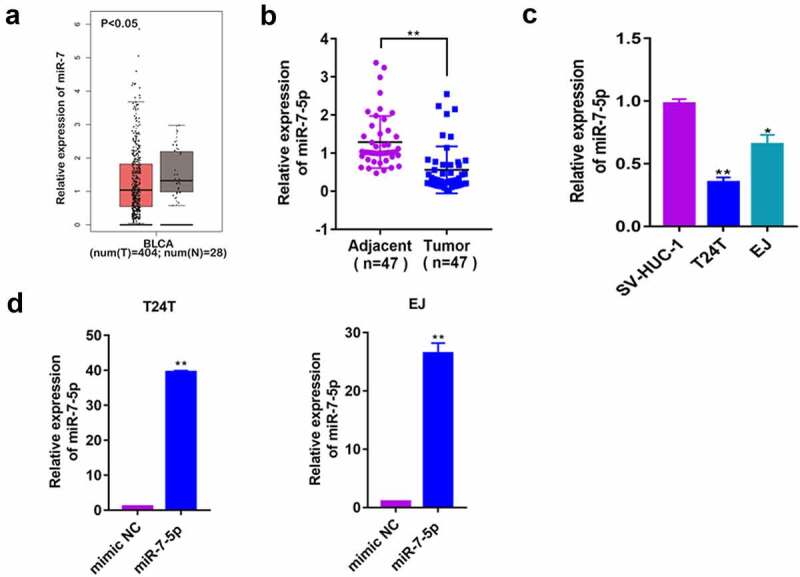
Baseline assessment of miR-7-5p levels in tissues and cells and construction of overexpression and inhibition models. (a) The database predicted the expression of miR-7 in BCa tissues. (b) The expression of miR-7-5p in 47 BCa tissues and adjacent tissues. (c) The expression of miR-7-5p in the SV-HUC-1, T24T, and EJ cells. (d) MiR-7-5p mimic was applied to T24T and EJ cell lines. (*p < 0.05, ** p < 0.01).
2. MiR-7-5p inhibits the migration and invasion of T24T and EJ cells
To verify whether miR-7-5p could inhibit the migration and invasion of BCa cells, the effect of miR-7-5p on BCa cells was observed experimentally. Transwell experiments showed that overexpression of miR-7-5p inhibited the ability of T24T and EJ cells to migrate and invade compared to that in the NC group, while inhibition of miR-7-5p increased the number of T24T and EJ cells passing through the chamber (Figure 2(a, b)). The histogram showed the suppression rate and would healing rate (Figure 2(c, f)). Subsequent wound healing experiments in miR-7-5p-overexpressing or miR-7-5p inhibitor-treated T24T and EJ cells showed the same results. Compared with the mimic NC group, the miR-7-5p group had a reduced would healing rate, while the anti-miR-7-5p group had an accelerated would healing rate compared with that in the anti-NC group (Figure 2(d, e)).
Figure 2.
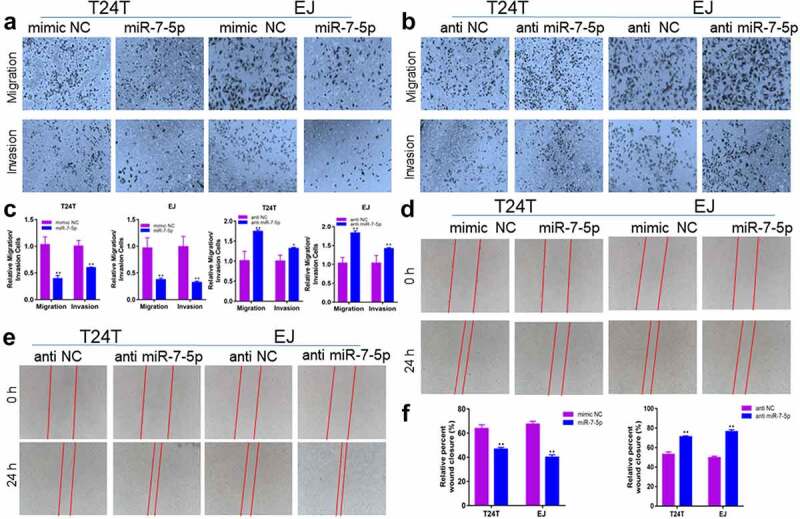
MiR-7-5p suppresses the migration and invasion of BCa cells. (a) Compared with the mimic NC group, the miR-7-5p group showed inhibition of the migration and invasion of BCa cells. (b) Compared with the anti-NC group. The anti-miR-7-5p group showed increased migration and invasion compared with the anti-NC group. (c) The histograms represent the migration and invasion rates of the indicated cells. (d, e) Wound healing assays also verified that miR-7-5p could inhibit the migration of BCa cells. (f) The statistical results of the scratch wound cell migration assay. Red lines represent the scratch area 24 h after the scratch. (*p < 0.05, ** p < 0.01).
3. MiR-7-5p regulates autophagy in T24T and EJ cells
Previous studies have demonstrated that high levels of migration and invasion in BCa cells are caused by high levels of autophagy. Therefore, we further explored whether miR-7-5p regulates autophagy levels in BCa cells. Western blot assays showed that miR-7-5p inhibited the accumulation of LC3BI to LC3BII compared to that in the NC group (mimic NC), reducing the protein levels of Beclin 1 but increasing the protein expression of P62 (Figure 3(a)). Inhibiting miR-7-5p (with anti-miR-7-5p) expression promoted the conversion of LC3BI to LC3BII, which increased the protein level of Beclin 1 and reduced the protein level of P62 compared to that in the NC group (anti-NC) (Figure 3(b)). The TEM results showed that in EJ cells, the number of autophagolysosomes (ASS) was significantly inhibited after the mimic miR-7-5p, while the anti-miR-7-5p promoted the production of ASS (Figure 3(c)). Figure 3(d) showed a histogram of ASS number in the treatment group versus the control group. These results demonstrated that miR-7-5p could reduce autophagy levels in BCa cells.
Figure 3.
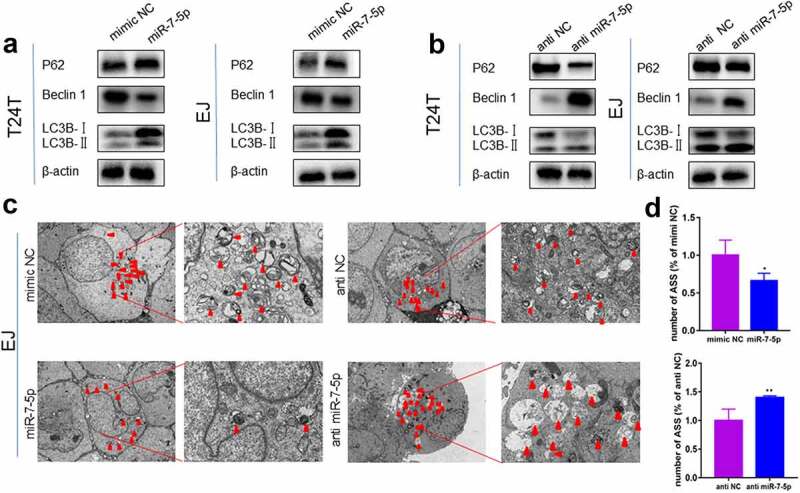
MiR-7-5p inhibits the level of autophagy, and autophagy marker expression changes correspondingly. (a, b) Western blot data reveled that after miR-7-5p mimic treatment of T24T and EJ cell lines, the expression level of the autophagy-related marker Beclin 1 decreased, the expression level of P62 increased, and the ratio of LC3BI to LC3BII decreased. The opposite result was obtained after anti-miR-7-5p treatment. (c, d) The TEM results revealed that the formation of autophagosomes in EJ cells was inhibited by miR-7-5p. The histogram shows a statistical diagram of APLs. The red arrows represent ASS.
4. MiR-7-5p and miR-7-5p-mediated autophagy is essential for human BCa invasion
To explore the relationship between BCa invasion and autophagy, T24T and EJ cell lines were treated with n-butyl-n-(4-hydroxybutyl-nitrosamine) (BBN), a genotoxic bladder carcinogen, and Bafilomycin A1(Baf A1), a late-stage inhibitor of autophagy. Cell migration and invasion were observed 24 h later. The results showed that BBN significantly promoted the ability of T24T and EJ cells to migrate and invade compared to that in the normal group, while Baf A1 inhibited the ability of T24T and EJ cells to migrate and invade (Figure 4(a, b)). Similarly, our results showed that miR-7-5p significantly reduced the number of migrating and invading T24T and EJ cells, while the addition of BBN treatment with miR-7-5p reduced the low migration and invasive force caused by the miR-7-5p (Figure 4(d, e)). The histogram showed migrating and invading cells relative to the control group (Figure 4(c, f)). These results indicated that miR-7-5p-mediated autophagy was important for BCa migration and invasion.
Figure 4.
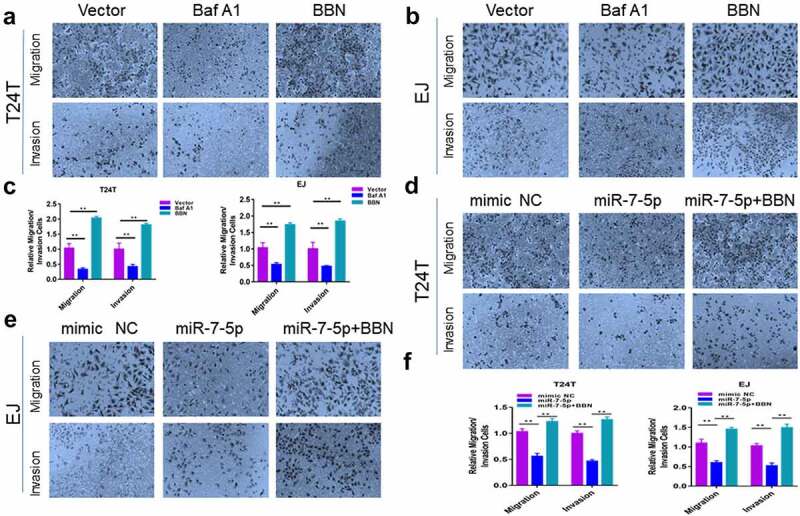
MiR-7-5p mediates autophagy inhibition, migration and invasion. (a, b) Transwell assay results demonstrated that after 24 h of treatment with BBN and Baf A1 in T24T and EJ cell lines, respectively, there were significantly fewer cells passing through the chamber in the Baf A1 treatment group compared to the control group, and the number of cells passing through the chamber increased significantly after BBN treatment. (d, e) In both cell lines, there was an obvious decrease in the number of cells passing through the chamber after miR-7-5p compared to mimic NC treatment, indicating a decrease in cell migration and invasion. However, when BBN was added after miR-7-5p treatment, the number of cells passing through the chamber was increased compared to that with miR-7-5p alone. (c, f) Cell count results from the Transwell invasion assay. The bar chart represents the average of three independent experiments.
5. ATG7 is a target gene of miR-7-5p
To explore how miR-7-5p regulated the level of autophagy in BCa, the TargetScan tool was used to explore potential targets of miR-7-5p. ATG7 was predicted as a potential target for miR-7-5p. Subsequently, we proved whether the predicted results were correct. The experiment found that after miR-7-5p treatment, the expression level of ATG7 protein in T24T and EJ cells was downregulated (Figure 5(a)), and the expression level of mRNA was not significantly changed (Figure 5(b)), indicating that ATG7 may be targeted by miR-7-5p. To further confirm whether ATG7 is a direct target for miR-7-5p, we constructed a mutant 3’-UTR for the full-length wild-type (WT) ATG7 and ATG7 vector (Figure 5(c)), and a double luciferase reporter gene was used. The data showed that the luciferase activity in ATG7-WT 293 T cells decreased significantly. After transfecting ATG7-MUT vectors (containing mutations in the seed region), the inhibition of miR-7-5p was attenuated. It was shown that miR-7-5p significantly reduced the WT but not the MUT luciferase signal, confirming its binding activity.
Figure 5.
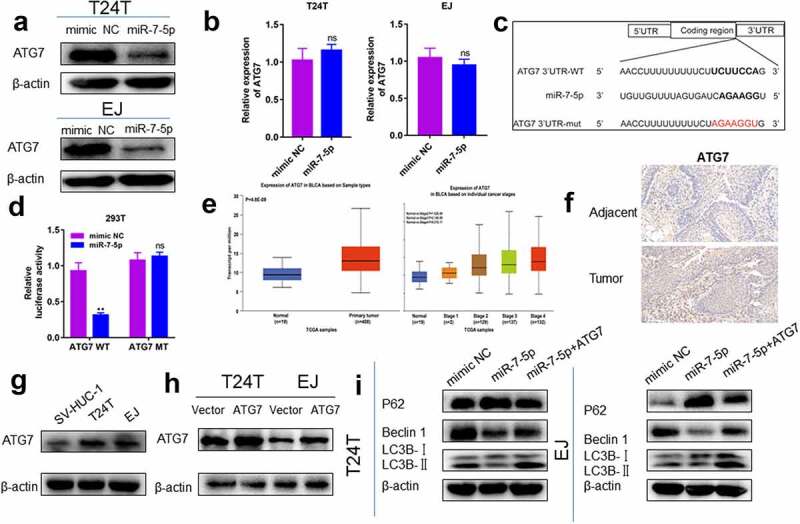
MiR-7-5p targets ATG7 to inhibit autophagy. (a) Western blot experiments showed a significant decrease in ATG7 protein expression after miR-7-5p transfection compared to mimic NC transfection. (b) The PCR results showed no change in mRNA levels at ATG7 after miR-7-5p treatment of T24T and EJ cells for 48 h. (c) MiR-7-5p could bind to the 3’-UTR of ATG7. The red areas are mutated nucleotides. (d) In 293 T cells, the WT or MUT ATG7 3’-UTR luciferase reporter gene was transfected along with miR-7-5p mimic or mimic NC, and the cells were lysed 24 h later to assess the relative luciferase activity. (e) The database predicted that ATG7 was expressed at obviously higher levels in 404 BCa tissues than in 28 adjacent tissues. The expression of ATG7 in bladder urothelial carcinoma (BLCA) based on individual cancer stages. (f) ATG7 immunohistochemical staining of BCa and paracancer tissues. (g) The Western blot experiment data reveled that ATG7 expression was obviously higher in T24T and EJ cells than in SV-HUC-1 cells. (h, i)The expression of ATG7 was increased by instant transfection of plasmids, and the Western blot experiment showed that the expression of miR-7-5p inhibited the occurrence of autophagy, but the miR-7-5p co-expression responded to the occurrence of autophagy.
Then, we further detected the expression of ATG7 in BCa tissues and cell lines. First, the expression of ATG7 in BCa tissue was predicted in the database (http://ualcan.path.uab.edu/analysis.html), and it was found that the expression of ATG7 in cancerous tissues was obviously higher than that in adjacent tissues. Moreover, the expression of ATG7 increased persistently from stage 1 to stage 4 (Figure 5(e)). Second, IHC results also revealed that the expression level of ATG7 in tumor tissue was higher than that in adjacent tissue (Figure 5(f)). Western blot data showed that the protein level of ATG7 in BCa cells was obviously higher than that in SV-HUC-1 cells (Figure 5(g)). The relationship between ATG7 and miR-7-5p was further verified.
We further verified that miR-7-5p regulated the level of autophagy in BCa cells by targeting ATG7. T24T and EJ cells were transfected with ATG7 and vector (Figure 5(h)). T24T and EJ cells overexpressing ATG7 were cultured with or without miR-7-5p, and the expression level of autophagy-related proteins was measured by Western blot assay. After overexpression of ATG7, the inhibitory effect of miR-7-5p on autophagy levels of T24T and EJ cells was partially weakened (Figure 5(i)).
6. MiR-7-5p elevates cisplatin chemosensitivity by targeting ATG7 in T24T and EJ cells
Cisplatin, as one of the basic chemotherapy drugs, has a very high curative effect in BCa, but cisplatin resistance can occur. To understand whether miR-7-5p and ATG7 are related to the sensitivity to cisplatin, T24T and EJ cells transfected with mimic NC and miR-7-5pwere treated with different concentrations of cisplatin. CCK-8 assays data showed that miR-7-5p increased the chemosensitivity of the two cell lines to cisplatin (Figure 6(a)). When these cells were also treated with BBN, We found that the addition of BBN could reverse the increase of miR-7-5p dependent cisplatin sensitivity (Figure 6(b)). Finally, miR-7-5p-transfected cells were also forced to overexpress ATG7, and the results were similar to those after BBN treatment (Figure 6(c)). Taken together, these findings demonstrate that miR-7-5p increases the sensitivity of BCa cells to cisplatin-induced cell death by reducing autophagy levels.
Figure 6.
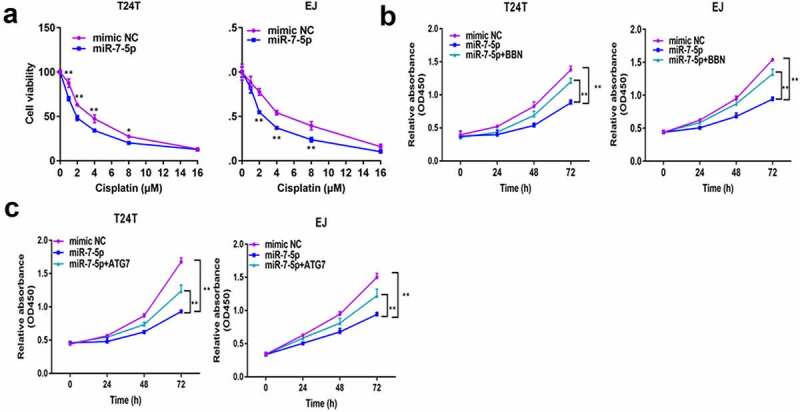
MiR-7-5p increases BCa cell cisplatin sensitivity via ATG7 inhibition. (a) T24T and EJ cells were transfected with mimic NC and miR-7-5p and then cultured with cisplatin at different concentrations for 48 h to detect the cell viability with the CCK-8 assay. (b) T24T and EJ cells were transfected with NC or miR-7-5p mimic and cultured with cisplatin[T24T (4 µM), EJ (5 µM)]; then the cells were treated with or without BBN. The indicated cisplatin concentration before CCK-8 evaluation [T24T (4 µM), EJ (5 µM)]. (c) Other conditions remained unchanged, but BBN was applied to cells with simultaneous expression of miR-7-5p and forced expression of ATG7.
7. MiR-7-5p inhibits the growth and metastasis of T24T xenografts
MiR-7-5p inhibits the progress of BCa in vivo
To further explore the effects of miR-7-5p in vivo, we injected T24T cells transfected with miR-7-5p and miR-NC into the right skin of two groups of nude mice (four-week-old females, n = 3 per group). The results showed a significant decrease in tumor volume and weight in mice in the miR-7-5p group compared to those in the miR-NC group (Figure 7(a, b)). In addition, the IHC results showed that the expression of ATG7, Ki-67, and N-cadherin was inhibited by miR-7-5p, but the expression of P62 was promoted (Figure 7(c)). Overall, our results indicate that miR-7-5p inhibits tumor growth, migration, and invasion in vivo.
Figure 7.
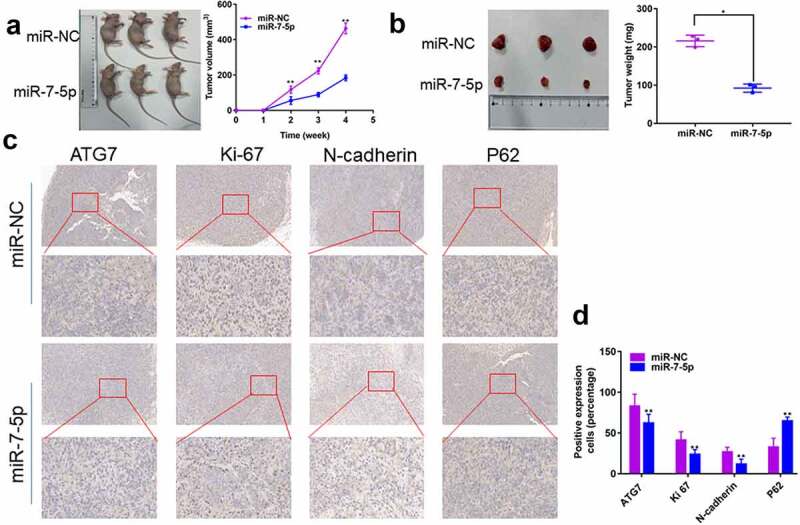
MiR-7-5p inhibits the development of tumors in vivo. (a, b) T24T cells (5 × 106) stably transduced and expressing miR-7-5p or miR-NC were implanted subcutaneously into nude BALB/c mice (four weeks old, female), and tumor size and volume were evaluated weekly. After 28 days, tumors were collected; the tumors were lower in weight and volume in miR-7-5p mimic mice than in mimic NC mice. (c) IHC staining showed that miR-7-5p mimic transfection resulted in decreased expression of ATG7, Ki-67 and N-cadherin and increased expression of P62 in tumors. The data are shown as the mean ± SEM, **P < 0.01 (Student’s t-test).
Discussion
MiR-7-5p is expressed at lower levels in BCa tissues than in adjacent tissues, suggesting that miR-7-5p may act as an anticancer gene. Here, we found that an increase in miR-7-5p expression inhibited the migration, invasion, as well as autophagy of BCa cells, suggesting that this molecule could be a new therapeutic target for BCa. In addition, our experimental data also showed that miR-7-5p reduced autophagy levels inhibited the invasion of BCa migration and promoted cisplatin sensitivity by reducing ATG7 expression.
The role of autophagy in cancer is currently controversial. On the one hand, a study found that autophagy promotes the development of diseases [19] and that autophagy inhibits the malignant biological behavior of diseases [20]. Previous studies have found that LncRNA MEG3/ miR-7-5p /EGFR axis mediated cardiac autophagy [21] and that miR-7-5p targeted RAF1 and inhanced crizotinib-induced cytotoxicity and autophagic flux in lymphoma cells [22]. These studies may suggest that miR-7-5p has different effects on autophagy in different diseases. However, it is not clear whether miR-7-5p affects the autophagy level of BCa.
ATG7 is an autophagy-related gene and is also involved in the progress of many cancers. Previous studies have reported that CD44s and ATG7 were involved in BCa cells invasion and lung metastasis [23]. ATG7andmiR-138-5pregulated the self-renewal and invasion of lung cancer stem-like cells [24]. ATG7 targeted the ETS2/miRNA196b/FOXO1/p27 axis in BCa cells [25]. These results indicated that ATG7 affects the level of autophagy and is also related to the migration and invasion ability of BCa. In addition, in previous studies, miR-7-5p targeted NOVA2 DNA methylation suppressed the metastasis of NSCLC [9]; miR-7-5p regulated gastric cancer stem cell invasion [11]; and lncRNA LINC00240 suppressed invasion and migration in NSCLC by sponging miR-7-5p [26]. These data show that miR-7-5p also has different effects on the migration and invasion in different cancers, but the effects in BCa are unclear. Then, we used a database to predict that miR-7-5p could target ATG7, suggesting that miR-7-5p might inhibit the autophagy of BCa cells by downregulating ATG7, thus inhibiting migration and invasion. Our experimental results proved that miR-7-5p could inhibit the migration and invasion of BCa. However, inhibition of miR-7-5p expression showed the opposite result, and a luciferase assay proved that miR-7-5p could target ATG7, further verifying our hypothesis.
Finally, the current treatments for BCa mainly include surgery, radiotherapy, chemotherapy and biotherapy [27]. However, the survival rate of patients with metastatic BCa is poor, and chemotherapy resistance is a common cause of BCa recurrence and treatment failure. In previous studies, we found that miRNAs may act as regulators of increased chemical sensitivity in cancer therapy. For example, miR-27b-3p and ATG10 regulated chemoresistance in colorectal cancer [28]. The miR-146a-5p/TRAF6/NF-kB p65 axis regulated pancreatic cancer chemoresistance [29]. Therefore, miRNA is worthy of further research in the context of drug resistance mechanisms. Our study found that miR-7-5p can increase the sensitivity of BCa cells to cisplatin, and this effect is caused by a reduction in the level of autophagy. We confirmed this result by adding BBN to cells with and without ATG7 overexpression. Therefore, intervention with miR-7-5p expression in BCa may play a role in BCa treatment.
Conclusion
Overall, our results provide new insights into the role of miR-7-5p in BCa migration, invasion, autophagy, and chemoresistance by regulating ATG7, thereby affecting tumor progression, and may provide new ideas for the treatment of BCa.
Acknowledgements
Chong Wang and Zhao Tang contributed equally to this work. Conception and design: Chong Wang, Ze Zhang,Tiantian Liu,Jingwei Zhang, Yawei Li and Houbao Huang.
Funding Statement
The National Natural Science Foundation of China (81802559); Natural Science Foundation of Anhui Province (1908085MH285); Anhui university provincial natural science research Foundation (KY2018A0260); Research Fund of Wannan Medical College for Middle-aged and Youth (WK2020F32);Funding of ‘Climbing Peak’ Training Program for Innovative Technology team of Yijishan Hospital, Wannan Medical College (KDF2019015); Funding of ‘Peak’ Training Program for Scientific Research of Yijishan Hospital, Wannan Medical College (KGF2019J09).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics Approval and Consent to Participate
This study was approved by the ethics committee of The First Affiliated Hospital of Wannan Medical College. Written informed consent was obtained from all subjects.
References
- [1].Kaufman DS, Shipley WU, Feldman AS.. Bladder cancer. Lancet. 2009;374:239–249. [DOI] [PubMed] [Google Scholar]
- [2].Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108. [DOI] [PubMed] [Google Scholar]
- [3].Williams SB, Shan Y, Jazzar U, et al. Comparing survival outcomes and costs associated with radical cystectomy and trimodal therapy for older adults with muscle-invasive bladder cancer. JAMA Surg. 2018;153:881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim VN, Han J, Siomi MC.. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology. 2009;10:126–139. [DOI] [PubMed] [Google Scholar]
- [5].Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. [DOI] [PubMed] [Google Scholar]
- [6].Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin HF, Wang J-F, Shao M, et al. Down-regulation of miR-7 in gastric cancer is associated with elevated LDH-A expression and chemoresistance to cisplatin. Front Cell Dev Biol. 2020;8:555937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cai H, Yang X, Gao Y, et al. Exosomal microRNA-9-3p secreted from BMSCs downregulates ESM1 to suppress the development of bladder cancer. Mol Ther Nucleic Acids. 2019;18:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiao H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gajda E, Godlewska M, Mariak Z, et al. Combinatory treatment with miR-7-5p and drug-loaded cubosomes effectively impairs cancer cells. Int J Mol Sci. 2020;21:5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xin L, Liu L, Liu C, et al. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J Cell Physiol. 2020;235:2643–2654. [DOI] [PubMed] [Google Scholar]
- [12].Lapierre LR, Kumsta C, Sandri M, et al. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johansson I, Monsen VT, Pettersen K, et al. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy. 2015;11:1636–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cao DJ, Hill JA. Titrating autophagy in cardiac plasticity. Autophagy. 2011;7:1078–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nguyen HG, Yang JC, Kung H-J, et al. Targeting autophagy overcomes Enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33:4521–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature Reviews Immunology. 2013;13:722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhu J, Tian Z, Li Y, et al. ATG7 promotes bladder cancer invasion via autophagy-mediated increased ARHGDIB mRNA stability. Adv Sci (Weinh). 2019;6:1801927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhu J, Li Y, Luo Y, et al. A feedback loop formed by ATG7/Autophagy, FOXO3a/miR-145 and PD-L1 regulates stem-like properties and invasion in human bladder cancer. Cancers (Basel). 2019;11:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guo C, Li X, Xie J, et al. Long noncoding RNA SNHG1 activates autophagy and promotes cell invasion in bladder cancer. Front Oncol. 2021;11:660551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [20].Mortensen M, Soilleux EJ, Djordjevic G, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cao Y, Wen J, Li Y, et al. Uric acid and sphingomyelin enhance autophagy in iPS cell-originated cardiomyocytes through lncRNA MEG3/miR-7-5p/EGFR axis. Artificial Cells, Nanomedicine, and Biotechnology. 2019;47:3774–3785. [DOI] [PubMed] [Google Scholar]
- [22].Sorrentino D, Frentzel J, Mitou G, et al. High levels of miR-7-5p potentiate crizotinib-induced cytokilling and autophagic flux by targeting RAF1 in NPM-ALK positive lymphoma cells. Cancers (Basel). 2020;12:2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu J, Huang G, Hua X, et al. CD44s is a crucial ATG7 downstream regulator for stem-like property, invasion, and lung metastasis of human bladder cancer (BC) cells. Oncogene. 2019;38:3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhou Q, Cui F, Lei C, et al. ATG7-mediated autophagy involves in miR-138-5p regulated self-renewal and invasion of lung cancer stem-like cells derived from A549 cells. Anticancer Drugs. 2021;32:376–385. [DOI] [PubMed] [Google Scholar]
- [25].Zhu J, Li Y, Tian Z, et al. ATG7 overexpression is crucial for tumorigenic growth of bladder cancer in vitro and in vivo by targeting the ETS2/miRNA196b/FOXO1/p27 axis. Molecular Therapy - Nucleic Acids. 2017;7:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ku GW, Kang Y, Yu S-L, et al. LncRNA LINC00240 suppresses invasion and migration in non-small cell lung cancer by sponging miR-7-5p. BMC Cancer. 2021;21:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chua MMJ, Lee M, Dominguez I. Cancer-type dependent expression of CK2 transcripts. PloS one. 2017;12:e0188854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sun W, Li J, Zhou L, et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics. 2020;10:1981–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meng Q, Liang C, Hua J, et al. A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: functional validation and clinical significance. Theranostics. 2020;10:3967–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]


