Abstract
Cardiac myosin-binding protein C (cMyBP-C) modulates cardiac contractility through putative interactions with the myosin S2 tail and/or the thin filament. The relative contribution of these binding-partner interactions to cMyBP-C modulatory function remains unclear. Hence, we developed a “nanosurfer” assay as a model system to interrogate these cMyBP-C binding-partner interactions. Synthetic thick filaments were generated using recombinant human β-cardiac myosin subfragments (HMM or S1) attached to DNA nanotubes, with 14- or 28-nm spacing, corresponding to the 14.3-nm myosin spacing in native thick filaments. The nanosurfer assay consists of DNA nanotubes added to the in vitro motility assay so that myosins on the motility surface effectively deliver thin filaments to the DNA nanotubes, enhancing thin filament gliding probability on the DNA nanotubes. Thin filament velocities on nanotubes with either 14- or 28-nm myosin spacing were no different. We then characterized the effects of cMyBP-C on thin filament motility by alternating HMM and cMyBP-C N-terminal fragments (C0–C2 or C1–C2) on nanotubes every 14 nm. Both C0–C2 and C1–C2 reduced thin filament velocity four- to sixfold relative to HMM alone. Similar inhibition occurred using the myosin S1 construct, which lacks the myosin S2 region proposed to interact with cMyBP-C, suggesting that the cMyBP-C N terminus must interact with other myosin head domains and/or actin to slow thin filament velocity. Thin filament velocity was unaffected by the C0–C1f fragment, which lacks the majority of the M-domain, supporting the importance of this domain for inhibitory interaction(s). A C0–C2 fragment with phospho-mimetic replacement in the M-domain showed markedly less inhibition of thin filament velocity compared with its phospho-null counterpart, highlighting the modulatory role of M-domain phosphorylation on cMyBP-C function. Therefore, the nanosurfer assay provides a platform to precisely manipulate spatially dependent cMyBP-C binding-partner interactions, shedding light on the molecular regulation of β-cardiac myosin contractility.
Significance
Cardiac myosin-binding protein C (cMyBP-C) is the most frequently mutated protein associated with hypertrophic cardiomyopathy (HCM), a common cause of sudden cardiac death. Despite the importance of cMyBP-C in cardiac contractility, the mechanisms underlying this regulation are unclear due to experimental challenges in studying the complex, transient, weak interactions of cMyBP-C with the contractile proteins of the sarcomere. In this study, we created a nanosurfer synthetic DNA thick filament assay to assess the cMyBP-C interactions with actin and human β-cardiac myosin. We demonstrate actomyosin inhibition by cMyBP-C fragments regardless of recombinant human β-cardiac myosin subfragment (heavy meromyosin [HMM] or S1) and highlight the importance of the cMyBP-C M-domain using cMyBP-C fragments and phosphomimetics.
Introduction
Cardiac myosin-binding protein C (cMyBP-C) is a large, multidomain thick filament-associated sarcomeric protein that modulates cardiac muscle contractility by tuning the speed and efficiency of contraction and relaxation (1, 2, 3). Despite almost 50 years since its discovery (4,5), the mechanisms by which cMyBP-C modulates contractility, including its binding partners and the associated regulatory mechanisms, are not completely understood. The importance of cMyBP-C in regulating contraction was first appreciated in the 1990s when two mutations in MYBPC3, the gene encoding cMyBP-C, were shown to cause hypertrophic cardiomyopathy (HCM), a heritable heart condition characterized by left ventricular hypertrophy and diastolic dysfunction (6,7). Since then, over 350 MYBPC3 mutations have been identified in HCM patients, and it has become the most prominent gene linked to HCM (8). The discovery of these mutations, including nonsense, frameshift, and missense mutations distributed across the protein, has provided substantial motivation toward understanding the role of cMyBP-C in the sarcomere (3). However, despite this increased interest and established importance of cMyBP-C in regulating contraction, the mechanisms by which cMyBP-C regulates contraction have largely remained elusive due to experimental challenges in studying the complex, transient, weak interactions of cMyBP-C in the context of the sarcomere.
Existing studies suggest that cMyBP-C modulates cardiac muscle contractility by sensitizing the thin filament to calcium, while slowing shortening velocity through interactions with actin and/or myosin (9, 10, 11). cMyBP-C exists in two regions of the muscle sarcomere A-band, in seven to nine stripes spaced 43 nm apart. This aligns with the 43-nm helical myosin head repeat on the thick filament (12, 13, 14). cMyBP-C contains 11 subdomains, named sequentially C0–C10, and is anchored to the thick filament backbone at its C terminus by a high-affinity light meromyosin (LMM) myosin-binding site in C10 (15,16). The N terminus extends away from the thick filament and engages in regulatory interactions with myosin and/or actin (10,17,18). The cardiac isoform contains a regulatory cMyBP-C motif or M-domain between C1 and C2 domains, which co-sedimentation assays have established to be the primary interacting region for myosin S2 (19, 20, 21, 22, 23). Studies with actin have found that the M-domain, as well as the C0, C1, and C2 domains, interact with actin (21,24, 25, 26, 27). The N- and C-terminal cMyBP-C interactions to the thin and thick filaments, respectively, may tether the two filaments, thus functioning as a brake on shortening velocity at high calcium concentrations (1,17). Despite this wealth of information, the relative significance of actin vs myosin interactions with distinct N-terminal cMyBP-C domains is still a matter of debate. Additionally, cMyBP-C function itself may be regulated, as the M-domain contains four serines that are thought to be phosphorylated hierarchically to disrupt cMyBP-C interactions with either actin filaments (28, 29, 30) or myosin (31,32). Phosphorylation of the M-domain is considered a regulatory mechanism in response to β-adrenergic stimulation (32).
The complex interactions of cMyBP-C with actin and myosin have made in vitro assessment of cMyBP-C challenging. Some in vitro motility studies have demonstrated inhibition of actomyosin motility by a variety of cMyBP-C N-terminal fragments (30,33). However, mechanistic interpretation of these assays is limited by a lack of control over protein stoichiometry and orientation. Likewise, single-molecule and solution assays do not capture the unique architecture of motor ensembles or cMyBP-C interactions in the context of the sarcomere. To overcome these limitations, we utilized DNA nanotechnology to build a synthetic thick filament that would allow systematic in vitro assessment of cMyBP-C interactions, with control over protein type, stoichiometry, and spacing (Fig. 1 a and S1) (34). We incorporated these synthetic thick filaments in a standard in vitro motility assay to promote thin filament interactions with the decorated DNA nanotubes. Using this nanosurfer assay, we characterized the cMyBP-C domains and interactions that modulate acto-myosin motility.
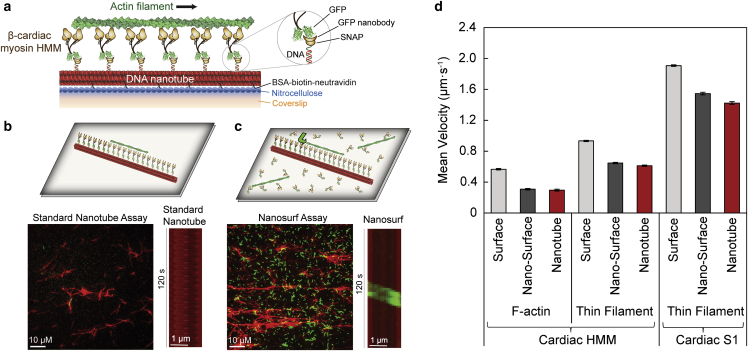
Figure 1.
Optimization of human β-cardiac myosin HMM nanosurfer assay. (a) Schematic of the β-cardiac myosin synthetic thick filament assay with recombinant human β-cardiac myosin HMM (brown) with C-terminal GFP attached to the nanotube (red) via GFP nanobody-SNAP labeled with complementary oligo to the nanotube DNA handle (inset). Nanotubes are attached to the nitrocellulose-coated (blue) coverslip (orange) via BSA-biotin-neutravidin (black Xs). (b) Schematic of standard nanotube assay within a flow cell with motor bound to nanotubes only and nanotubes attached to a coverslip surface blocked with BSA (top). Field of view at 1000× (bottom left) and kymograph (bottom right) for standard nanotube assay (nanotubes in red) with β-cardiac myosin HMM showing the low landing rate of actin filaments (green) on the nanotube surface. (c) Schematic (top) of nanosurfer assay with motor bound to both the nanotubes and the coverslip surface and actin (green) can be observed gliding from the coverslip surface onto the nanotube (green arrow). Field of view at 1000× (bottom left) and kymograph (bottom right) for nanosurfer assay with β-cardiac myosin HMM showing a higher landing rate for actin filaments on the nanotube surface. (d) Velocities of F-actin (left) and regulated thin filaments (middle) with β-cardiac myosin HMM and regulated thin filaments with β-cardiac myosin S1 (right) on a standard in vitro motility surface (light gray), on the coverslip surface in the nanosurfer assay (nano-surface; dark gray), and on the nanotubes in the nanosurfer assay (red). Mean velocities represented as μm·s−1 ± SE. N = 132–390 filaments from three to five independent protein preparations per condition.
Methods
DNA nanotube and benzyl-guanine oligonucleotide preparation
Cy5-labeled 10-helix DNA nanotubes composed of 40 single-stranded DNA tiles with 14- or 28-nm spacing between single-stranded protein binding handles were prepared with biotin strands for surface attachment using an annealing protocol as previously described (34). The nanotube DNA handles at 14 or 28 nm are designed to anneal to DNA strands bound to a SNAP protein encoded with the GFP nanobody. The SNAP protein binds benzyl-guanine-treated DNA oligos. To prepare the benzyl-guanine oligo, benzyl-guanine NHS ester (BG-GLA-NHS; New England Biolabs, Ipswich, MA) was covalently linked to C6-amine oligonucleotides (with or without Cy3 modification, C6-amine-Cy3-a′ or C6-amine-a′). This was accomplished by incubating 0.17 mM C6-amine-a′ or C6-amine-b′ with 11.6 mM BG-GLA-NHS in 0.1 M sodium borate, pH 8.5 at 37°C for ∼4 h with rotation. The BG-labeled oligonucleotide was then purified twice through Illustra G-50 micro columns (GE Healthcare, Buckinghamshire, UK) equilibrated in 2 mM Tris, pH 8.5, since the primary amine reacts with any unreacted benzyl-guanine. The final BG-labeled oligonucleotide concentration was determined by measuring Cy3 absorbance (for BG-Cy3-a′ or BG-Cy3-b′) or by estimating the concentration of single-stranded DNA (for BG-a′ or BG-b′) using a NanoDrop spectrophotometer (NanoDrop OneC, Thermo Fisher, Waltham, MA).
GFP Nanobody-SNAP and cMyBP-C-SNAP preparation
The DNA sequence for the GFP nanobody was generated and synthesized by Genewiz (South Plainfield, NJ) as previously described (35). The SNAP-tagged GFP nanobody construct contained from N to C terminus: the GFP nanobody, a flexible (Gly-Ser-Gly)2 linker, the SNAP tag for oligonucleotide labeling, and both FLAG and 6xHis purification tags. N-terminal fragments (C0–C2, C0–C1f, or C1–C2) of the Mus musculus cardiac myosin-binding protein C cDNA were cloned into pBiex-1 and engineered with a C-terminal SNAP-FLAG-6xHis. C0–C2 and C1–C2 phospho-null or phospho-mimetic constructs were created by mutating s273, s282, s302, and s307 to alanines or aspartic acids, respectively. For cMyBP-C constructs containing a linker, either a 10-nm er/k α-helical linker (derived from Sus scrofa’s myosin vi) or a 30-nm linker (derived from the kelch-motif family protein, Trichomonas vaginalis), as previously characterized and described, were inserted between the cMyBP-C C terminus and the N terminus of the SNAP (36, 37, 38). Gly-Ser-Gly repeats were inserted between each element to ensure rotational flexibility. The C0-C2 and C1-C2 fragments contained all structural elements within and including their respective domains, notably including the M-domain between C1 and C2. The C0-C1f fragment contained the C0 and C1 domains with the respective unstructured Pro-Ala-rich linker region, as well as the first 17 amino acids of the M-domain, as described previously (30). The proteins were then transiently transfected (pBiex-1; Escort IV, Sigma-Aldrich, St. Louis, MO) into Sf9 cells, expressed and affinity purified similarly to previously published protocols (35,39). Briefly, cells were lysed at 72 h in lysis buffer: 0.5% IGEPAL, 4 mM MgCl2, 200 mM NaCl, 7% sucrose, 20 mM imidazole (pH 7.5), 0.5 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 μg/ml phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Lysates were centrifuged (176,000 × g, 4°C, 25 min) in a TLA 100.4 rotor (Beckman Coulter, Brea, CA) and bound to anti-FLAG M2 affinity resin (Sigma-Aldrich, St. Louis, MO) at 4°C. The FLAG resin was washed with wash buffer: 20 mM imidazole, 150 mM KCl, 5 mM MgCl2,1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 μg/ml PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin, pH 7.4. To label the SNAP tag, GFP nanobody or cMyBP-C fragments bound to anti-FLAG resin were incubated with excess (>10 μM) BG-oligonucleotide in wash buffer overnight at 4°C with rotation. BG-oligonucleotide-labeled SNAP proteins bound to resin were again washed with wash buffer and eluted using 0.2 mg/ml FLAG peptide (Sigma-Aldrich, St. Louis, MO) overnight at 4°C with rotation. To confirm labeling, proteins were loaded onto a 10% SDS gel followed by Coomassie staining. The protein was stored in 50% glycerol (v/v) at −20°C.
Recombinant human β-cardiac myosin preparation
The human β-cardiac myosin cDNA (AAA51837.1) was purchased from Thermo Fisher Scientific, Waltham, MA. PCR amplification was used to subclone the M2β-HMM fragment containing the first 25 heptads of the N-terminal S2 region (1–1016) into a pshuttle vector (a gift from Dr. Don Winkelmann). A GCN4 leucine zipper was added after the S2 region to promote dimerization, which was followed by an eGFP tag at the C terminus. An N-terminal FLAG tag was also included for purification. Similarly, an M2β-S1 (1–842) construct with a C-terminal eGFP tag and N-terminal FLAG tag was generated and subcloned into a shuttle vector provided by Vector Biolabs, Malvern, PA. Recombinant adenovirus was generated as previously described to express M2β-S1 in C2C12 cells (40). Vector Biolabs produced the initial recombinant adenovirus of M2β-S1-eGFP. High-titer adenovirus was produced by a method developed in the Winkelmann laboratory (41,42) and as previously described (40,43). For expression of β-cardiac myosin eGFP, C2C12 cells (typically 20–30, 145-mm diameter plates) were differentiated and infected with recombinant adenovirus (5 × 108 plaque-forming units [PFU]/ml) diluted into differentiation media as described previously (40). The cells were harvested on day 10 and myosin eGFP was purified by FLAG affinity chromatography as described. The eluted M2β-HMM-eGFP or M2β-S1-eGFP was then ammonium sulfate precipitated and dialyzed into MOPS20 buffer overnight at 4°C. M2β-HMM-eGFP and M2β-S1-eGFP was assessed for purity by Coomassie-stained SDS-polyacrylamide gels and concentrations were determined by eGFP absorbance (ε488 = 55,000 M−1⋅cm−1) or Bradford assay using BSA as a standard.
Actin and reconstituted regulated thin filament preparation
For assays with F-actin, actin was purified from rabbit skeletal muscle using an acetone powder method (44). The actin concentration was determined by absorbance at 290 nm (ε290 = 2.66 × 104 M-1·cm-1). A molar equivalent of tetramethylrhodamine (TRITC) phalloidin (MilliporeSigma, Burlington, MA) was added to stabilize and fluorescently label the F-actin. Calcium-regulated native thin filaments were prepared from mouse ventricular tissue and TRITC labeled as described previously (9).
Standard in vitro motility surface assay
Flow chambers were created using tape placed ∼3 mm apart on a glass slide to adhere to coverslips coated with 0.1% collodion in amyl acetate (Electron Microscopy Sciences, Hatfield, PA). For standard gliding assays with β-cardiac myosin eGFP, myosin was attached to coated coverslips through GFP nanobody with an encoded SNAP protein bound to oligonucleotides annealed to single-stranded DNA nanotube handles (35). Briefly, GFP nanobody SNAP was incubated in flow chambers at ∼0.5–1 μM for 4 min. Excess GFP nanobody SNAP was washed out, and the surface was incubated with assay buffer (AB: 20 mM imidazole, pH 7.5, 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 1 mM DTT) and BSA (AB.BSA: AB + 1 mg/ml BSA) for 2 min. Finally, human β cardiac myosin at ∼0.6–0.8 μM was incubated on the surface for 4 min and washed out with AB.BSA (for experiments with regulated thin filaments, the flow cell was washed with pCa 5 buffer containing an appropriate CaCl2 concentration calculated using Max Chelator from University of California-Davis [UC-Davis]). The final actin imaging solution was added containing TRITC phalloidin-labeled (Invitrogen, Waltham, MA) F-actin (or regulated thin filaments in pCa 5 buffer), 0.1% methylcellulose, 2 mM ATP, 1 mM phosphocreatine, 0.1 mg/ml creatine-phosphokinase, 45 μg/ml catalase, 25 μg/ml glucose oxidase, and 1% glucose. Flow cells were imaged at 1000× magnification on a Nikon (Tokyo, Japan) TiE microscope equipped with a 100× 1.4 NA Plan-Apo oil-immersion objective, 1.5× magnifier, Nikon Perfect Focus System, mercury arc lamp, Evolve EMCCD camera (512 pixel × 512 pixel; Photometrics), and Nikon NIS-Elements software. All in vitro motility assays were performed at room temperature (20°C–23°C).
Standard nanotube assay
The standard nanotube assay, previously reported and characterized (34), was presented for comparison in Fig. 1 b. At room temperature (20°C–23°C), nanotubes were attached to the coverslip surface coated with 0.1% collodion in amyl acetate (Electron Microscopy Sciences, Hatfield, PA) using biotinylated BSA at 0.1 mg/ml in AB (AB: 20 mM imidazole, pH 7.5, 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 1 mM DTT) incubated for 4 min. Excess biotin-BSA was washed out and the surface was incubated with AB.BSA (AB + 1 mg/ml BSA) for 2 min. Next, neutravidin at 0.1 mg/ml in AB.BSA was incubated for 4 min. AB.BSA was added to wash out excess neutravidin. Nanotubes were added at 2–5 nM concentration in AB.BSA.nt (AB.BSA + 5–10 nM random DNA nucleotide mix to reduce non-specific interactions) and incubated for 4 min. Excess nanotubes were washed out of the chamber with AB.BSA.nt. For attachment of β-cardiac myosin eGFP, GFP nanobody SNAP at ∼0.5–1 μM in AB.BSA.nt was incubated in flow chambers for 4 min. Excess GFP nanobody SNAP was washed out, and the surface was incubated with AB.BSA.nt for 2 min. Human β-cardiac myosin eGFP at ∼0.6–0.8 μM was incubated on the surface for 4 min and washed out with AB.BSA.nt. The final actin imaging solution was added containing sheared TRITC phalloidin-labeled (Invitrogen, Waltham, MA) F-actin (sheared to ensure actin filament lengths are shorter than ∼5 μm nanotubes), 0.1% methylcellulose, 2 mM ATP, 1 mM phosphocreatine, 0.1 mg/ml creatine-phosphokinase, 45 μg/ml catalase, 25 μg/ml glucose oxidase, and 1% glucose. For each flow cell, the actin channel was imaged as above at 0.5- to 3-s intervals for 2–3 min. For each actin video, several nanotube images were acquired of the same field of view for overlay.
Nanosurfer assay
Flow chambers were created with nitrocellulose-coated coverslips as described above. At room temperature (20°C–23°C), nanotubes were attached to the coverslip using biotinylated BSA at 0.1 mg/ml in AB (AB: 20 mM imidazole, pH 7.5, 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 1 mM DTT) incubated for 4 min. Excess biotin-BSA was washed out, and the surface was incubated with AB for 2 min. Neutravidin at 0.1 mg/ml in AB was incubated for 4 min. AB was added to wash out excess neutravidin. Nanotubes were added at 2–5 nM concentration in AB.nt (AB + 5–10 nM random DNA nucleotide mix) and incubated for 4 min. Excess nanotubes were washed out of the chamber with AB.nt. For attachment of β-cardiac myosin eGFP, GFP nanobody SNAP at ∼0.5–1 μM in AB.nt was incubated in flow chambers for 4 min. Excess GFP nanobody SNAP was washed out with AB.BSA.nt (AB.nt + 1 mg/ml BSA), and the surface was incubated with AB.BSA.nt for 2 min. For assays with cMyBP-C, N-terminal fragments at ∼0.2–0.6 μM in AB.BSA.nt were incubated for 4 min, to facilitate SNAP-labeled oligo to bind a complementary oligonucleotide on the DNA nanotube. Subsequently, cMyBP-C fragments were then washed out (AB.BSA.nt) before the flow cell was incubated with AB.BSA.nt for 2 min. Finally, human β-cardiac myosin eGFP at ∼0.6–0.8 μM was incubated on the surface for 4 min and washed out with AB.BSA.nt (for experiments with regulated thin filaments, flow cell was washed with pCa 5 buffer containing an appropriate CaCl2 concentration calculated using Max Chelator from UC-Davis). The final actin imaging solution was added (see section “standard nanotube assay”). For each flow cell, the actin and nanotube channels were imaged as described above. Nanotubes with Cy3-labeled oligonucleotides incorporated into the annealing protocol and annealed to protein binding sites were imaged and used as a labeling control for Cy3-labeled cMyBP-C fragments. Nanotubes were imaged and the Cy3 and Cy5 intensities were quantified by manually selecting individual nanotubes using ImageJ and normalizing the Cy3 intensity by the corresponding Cy5 intensity for each detected pixel.
Actin trajectory analysis
Actin trajectories were analyzed using the ImageJ MTrackJ plug-in. Actin movies were corrected for any drift with the TurboReg plug-in. Actin and nanotube channels were merged, and movies were analyzed to identify actin-nanotube gliding events with filaments that move along DNA nanotubes for at least four frames (∼1 s). For the β-cardiac myosin nanosurfer assay, to clearly quantify filaments traveling on the nanotubes, we quantified velocities of the filaments that were first traveling on the coverslip surface, encountered a nanotube, and then turned sharply to glide across the nanotube. We recorded the velocity of the filament while it was on the nanotube surface as the nanotube velocity and the velocity of the filaments traveling on the surrounding coverslip surface in the nanosurfer assay as nano-surface velocity.
Statistical analysis
Data are represented as mean values of pooled filaments ±SEM using n = 82–514 filaments per condition. Experiments were independently conducted at least three times from three to eight independent protein preparations per condition. Statistical analysis was performed using Origin 9 (OriginLab Corporation, Northampton, MA). Statistical significance was calculated for individual experiments using paired Student's t-test. Data were pooled for each condition and paired or unpaired Student's t-tests were conducted to evaluate significance. One-way ANOVA with a Tukey’s posttest was performed to assess significance when evaluating comparisons between multiple conditions (Fig. 3 d, 4 c, and 4 d) with p values ∗p ≤ 0.05 and ∗∗∗p ≤ 0.001.
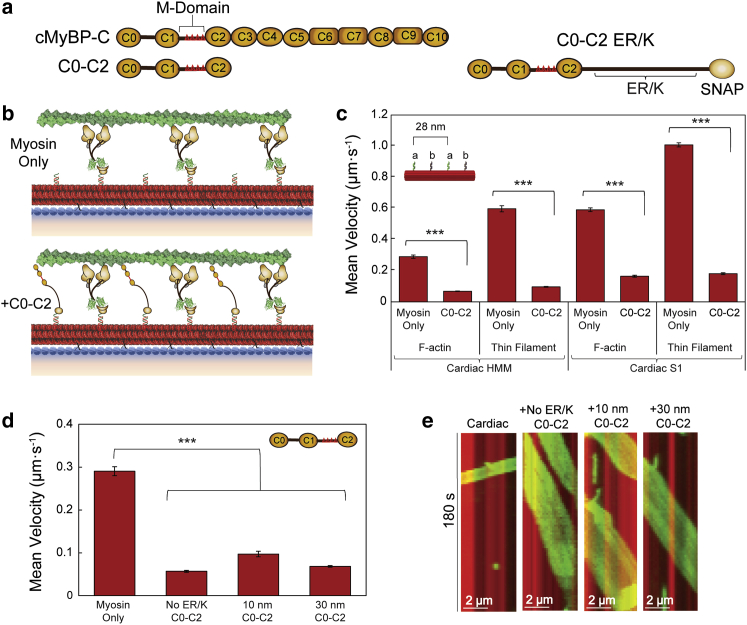
Figure 3.
C0–C2 inhibits β-cardiac HMM and S1 nanotube motility. (a) Schematic of cMyBP-C domains C0–C10 with the M-domain (red triangles) containing four phosphorylatable serines in the linker region between the C1 and C2 domains and the N-terminal fragment used, C0–C2. C0–C2 schematic (right) shown with the encoded ER/K linker and SNAP tag. (b) Schematics of synthetic thick filaments with β-cardiac myosin HMM bound to oligo a′ at 28-nm intervals (myosin only, top) and interdigitated C0–C2 containing M-domain, bound to oligo b′ (bottom). (c) Velocities of F-actin and regulated thin filaments on nanotubes decorated with β-cardiac myosin HMM (left) or β-cardiac myosin S1 (right) bound to oligo a′ alone versus myosin + C0–C2 containing a 30-nm ER/K bound to oligo b′ in the pattern shown (inset). (d) Mean velocities of F-actin filaments on nanotubes decorated with β-cardiac myosin HMM only versus β-cardiac HMM nanotubes with interdigitated C0–C2 (inset) containing no ER/K helix, a 10-nm ER/K helix, or a 30-nm ER/K helix. (e) Kymographs of F-actin filaments (green) traveling on a nanotube (red) with β-cardiac myosin HMM only versus cardiac nanotubes with interdigitating C0–C2 containing no ER/K helix, a 10-nm ER/K helix, or a 30-nm ER/K helix. Mean velocities represented as μm·s−1 ± SE. N = 82–514 filaments from three to eight independent protein preparations per condition. Significance calculated using Student's t-test or one-way ANOVA where appropriate. Significance is denoted as ∗∗∗p ≤ 0.001.
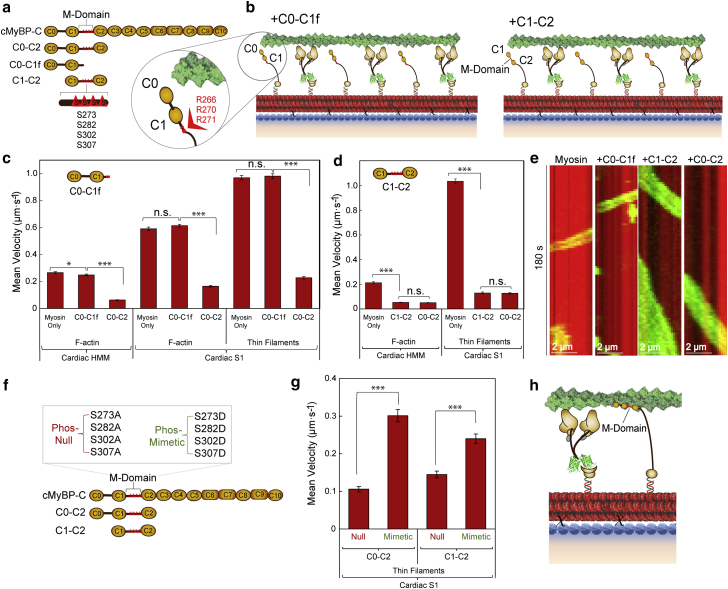
Figure 4.
cMyBP-C M-domain essential for inhibition of β-cardiac myosin HMM and S1 nanotube motility. (a) Schematic of cMyBP-C domains C0–C10 containing the M-domain (red triangles) in the linker region between the C1 and C2 domains and the N-terminal fragments used, including C0–C2, C0–C1f, and C1–C2. The four phosphorylatable serines in the M-domain are represented by red triangles (S273, S282, S302, S307). (b) Schematics of synthetic thick filaments with β-cardiac myosin HMM bound to oligo a′ at 28-nm intervals and interdigitated C0–C1f (left) or C1–C2 (right) containing the entire M-domain, bound to oligo b′. C0–C1f (inset) contains the first 17 amino acids of the M-domain including several arginine residues (R266, R270, R271) oriented away from the actin filament. (c) Velocities of F-actin on nanotubes decorated with β-cardiac myosin HMM (left) and either F-actin (middle) or regulated thin filaments (right) on nanotubes decorated with β-cardiac myosin S1. β-Cardiac nanotubes were either labeled with the motor alone or interdigitated with C0–C1f or C0–C2 containing 30-nm ER/K helices. (d) Velocities of F-actin on nanotubes decorated with β-cardiac myosin HMM (left) and regulated thin filaments on nanotubes decorated with β-cardiac myosin S1 (right) versus β-cardiac nanotubes with interdigitated C1–C2 or C0–C2 containing 30-nm ER/K helices. (e) Kymographs of F-actin filaments (green) traveling on a nanotube (red) with β-cardiac myosin HMM alone versus cardiac nanotubes with interdigitating C0–C1f, C1–C2, or C0–C2. (f) Schematic of cMyBP-C domains C0–C10 with the M-domain (red triangles) containing four phosphorylatable serines in the linker region between the C1 and C2 domains mutated to alanines (phospho-null) or aspartic acids (phospho-mimetic) within the C0–C2 and C1–C2 N-terminal fragments. (g) Mean nanotube velocities of regulated thin filaments on nanotubes decorated with β-cardiac myosin S1 and interdigitating phospho-null or phospho-mimetic C0–C2 (left) or C1–C2 (right) containing 30-nm ER/K helices. (h) Schematic of β-cardiac myosin (brown) synthetic thick filament with cMyBP-C N-terminal fragment (yellow) bound to actin (green), functioning as a tether between thick and thin filaments and reducing velocity. Mean velocities represented as μm·s−1 ± SE. N = 90–410 filaments from three to four independent protein preparations per condition. Significance was calculated using Student's t-test. Significance is denoted as ∗p ≤ 0.05, ∗∗∗p ≤ 0.001.
Results
Nanosurfer motility assay
We previously highlighted the use of synthetic nanotube thick filaments as a tool to characterize the behavior of ensembles of myosin motor proteins (34). These synthetic filaments are derived from DNA nanotubes with myosins spaced 14 or 28 nm apart. The 14-nm spacing matches that of the layers (i.e., crowns) of myosin heads projecting from vertebrate thick filament, where the heads are arranged in a three-start helix (45). However, the linear repeat of myosin heads along the thick filament surface is 43 nm, which would be the spacing between myosin head interactions with the thin filament in the sarcomere.
Our previous study primarily utilized two different highly processive unconventional myosins, V and VI (34). We reported an initial characterization of nanotube motility with recombinant β-cardiac myosin S1, demonstrating proof of concept of this assay for muscle myosins. However, the broader application of this assay to cardiac myosin was limited by the infrequent actin gliding events per camera field of view, consistent with the low duty ratio of cardiac myosins (Fig. 1 b). Therefore, in this study we developed the nanosurfer assay, with myosin present on both the nanotube and the surrounding coverslip surface (Fig. 1 c). The presence of myosin on the coverslip surface recruits actin filaments to the surface and greatly increases the probability that a motile actin filament will encounter a nanotube. Using the nanosurfer assay, we observed about a two-orders-of-magnitude increase in motile events along DNA nanotubes during the ∼2-min data acquisition time (Fig. 1 c). Bi-directional motility of actin filaments was observed along DNA nanotubes, consistent with actin filaments gliding in opposing directions, which is determined by the filament polarity when interacting with nanotubes (Video S1). To distinguish motility surface filaments from those gliding on the DNA nanotube (i.e., nanotube motility), analysis of nanotube motility events was limited to actin filaments that travel across the coverslip surface and then turn sharply to glide across the nanotube (Fig. 1 c schematic, green arrow). However, the diameter of actin filaments and individual DNA nanotubes (∼10 nm) are significantly lower than the optical resolution limit of our standard epi-fluorescence microscope (∼200 nm). Hence, our assay cannot distinguish between motile events driven by myosins on the nanotube and those on the surface gliding parallel within ∼100 nm of the nanotube. To control for this possibility, we compared motility on nanotubes labeled with two distinct oligos (a and b), only one of which is complementary to the oligo on the eGFP nanobody (a′) that is used to attach myosin to the nanotube surface (Fig. S2 a). Assays under otherwise identical conditions were quantified for both the number of events per field of view where filaments turn and glide over the nanotube (Fig. S2 b), and the fraction of all apparent actin filament encounters with nanotubes, including crossover events, that result in sharp turn-and-glide events (Fig. S2 c). DNA nanotubes patterned with myosin (a-a′) exhibit ∼sevenfold higher number of motile events per field of view compared with control nanotubes patterned with a non-complementary oligo (b-a′). Further, any actin filament encountering a DNA nanotube patterned with myosin (a-a′) exhibited an equal probability of either crossing over the nanotube or turn and glide along the nanotube (fraction of gliding events, ∼0.5; Fig. S2 c). In contrast, ∼90% of actin filaments encountering a DNA nanotube without myosin (b-a′) cross over the nanotube and only occasionally turn and glide (Fig. S2 c), suggesting that the control nanotubes (b-a′) are not a physical barrier that forces the actin filaments gliding on the surface to turn and appear as if gliding on the nanotube surface. Taken together, these observations demonstrate that the turn-and-glide events observed along DNA nanotubes are driven primarily by myosins patterned on the DNA nanotube.
Representative video depicting a nanosurfer assay with F-actin filaments (green) traveling on nanotubes (red) labeled with β-cardiac myosin HMM spaced 14 nm apart. Two actin filaments are seen traveling in opposite directions on the nanotube in the center.
In the nanosurfer assay, the velocity of an F-actin filament traveling along a nanotube, with an ensemble of human β-cardiac myosin HMM spaced at 14-nm intervals (Fig. 1 d: nanotube; 0.30 ± 0.01 μm·s−1; n = 132 pooled filaments from four independent protein preparations), is similar to the velocity of an F-actin filament traveling across the human β-cardiac myosin HMM-coated coverslip surface (Fig. 1 d: nano-surface; 0.31 ± 0.01 μm·s−1; n = 139). However, these velocities were significantly lower than those obtained, using matched β-cardiac myosin preparations, using a standard in vitro motility surface gliding assay (Fig. 1 d: surface; 0.57 ± 0.01 μm·s−1; n = 390 filaments). Hence, to account for any surface effects on actin gliding speeds, all future comparisons are made using matched nanosurfer assays with equivalent assay conditions. These trends were also observed with fully activated (pCa 5), calcium-regulated, native thin filaments with either β-cardiac myosin HMM and or β-cardiac S1 (data from three independent protein preparations of each subfragment) (Fig. 1 d). Notably, ∼twofold higher speeds were observed with S1 compared with HMM fragments. Adhikari et al. previously reported twofold higher actin-activated ATPase activity for human cardiac S1 compared with HMM (46), while it is still unclear how this difference in ATPase activity affect the actin gliding velocity in the motility assay. With β-cardiac myosin HMM, thin filaments had an in vitro motility surface velocity of 0.93 ± 0.01 μm·s−1 (n = 300 filaments), a nano-surface velocity of 0.65 ± 0.01 μm·s−1 (n = 206 filaments), and a nanotube velocity of 0.61 ± 0.01 μm·s−1 (n = 206 filaments). The increased velocity for thin filaments compared with F-actin (Fig. 1 d), regardless of the myosin subfragment, has been described previously (47). With β-cardiac myosin S1, the regulated thin filaments had an in vitro motility surface velocity of 1.91 ± 0.01 μm·s−1 (n = 359 filaments), a nano-surface velocity of 1.55 ± 0.02 μm·s−1 (n = 270 filaments), and a nanotube velocity of 1.42 ± 0.02 μm·s−1 (n = 270 filaments) (Fig. 1 d). In contrast, the average velocity of F-actin filaments in a standard in vitro motility surface assay with β-cardiac myosin S1 was 1.24 ± 0.01 μm·s−1 (n = 360 filaments), once again demonstrating slower velocities associated with F-actin compared with activated thin filaments. Taken together, these data provide baseline motility conditions for the nanosurfer assay and establish the nanosurfer assay as a robust method to study β-cardiac myosin ensembles.
Motor spacing does not affect β-cardiac HMM motility
Next, we examined the impact of β-cardiac myosin motor spacing on actin gliding speeds. In our previous study with myosin V, myosin VI, and β-cardiac myosin S1, in a standard nanotube assay, we found no significant differences in nanotube ensemble velocity when myosin was spaced at 14 nm compared with 28 nm (34). Using the nanosurfer assay with human β-cardiac myosin HMM or S1 spaced at either 14-nm or 28-nm intervals (Figs. 2 a and b), the velocities of either F-actin or regulated thin filaments traveling on nanotubes were quantified (Fig. 2 c). With HMM, we found no significant difference between F-actin velocities (data from five independent protein preparations) at 14 nm (0.30 ± 0.01 μm·s−1; n = 162 filaments) versus 28 nm (0.32 ± 0.01 μm·s−1; n = 143 filaments), as visually represented in the kymographs in Fig. 2 d.
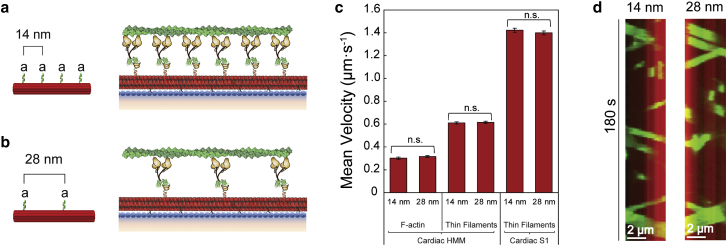
Figure 2.
Motor spacing does not affect β-cardiac HMM motility. (a and b) Schematic of the synthetic thick filament with nanotube motor spacing of 14 nm (a) or 28 nm (b). (c) Velocities of F-actin (left) and regulated thin filaments (middle) on nanotubes decorated with β-cardiac myosin HMM or regulated thin filaments on nanotubes decorated with β-cardiac myosin S1 (right) at 14- or 28-nm spacing in the nanosurfer assay. (d) Kymographs of F-actin filaments (green) traveling on a nanotube (red) with β-cardiac myosin HMM at 14-nm (left) or 28-nm (right) spacing. Mean velocities represented as μm·s−1 ± SE. N = 143–270 filaments from three to five independent protein preparations per condition. Comparisons were performed using Student's t-test. n.s., not significant
Similarly, there was no significant difference in regulated thin filament velocities at pCa 5 with human β-cardiac myosin HMM or S1 at the different spacing (data from three independent protein preparations of each subfragment). With the HMM spaced at 14 nm versus 28 nm, regulated thin filament velocities were 0.61 ± 0.01 μm·s−1 (n = 206 filaments) and 0.62 ± 0.01 μm·s−1 (n = 175 filaments), respectively. With S1 spaced at 14 nm versus 28 nm, regulated thin filament velocities were 1.42 ± 0.02 μm·s−1 (n = 270 filaments) and 1.40 ± 0.02 μm·s−1 (n = 240 filaments), respectively. These data indicate that 28-nm spaced myosins on nanotubes can be used with interdigitated cMyBP-C so that any effects of cMyBP-C on actin filament velocities cannot be attributed simply to myosin density/spacing on the nanotube.
C0–C2 inhibits β-cardiac HMM and S1 nanotube motility
We used the nanosurfer assay as a tool to investigate the role of cMyBP-C putative interactions with actin and myosin in modulating thin filament velocity. The cMyBP-C protein contains 11 domains, C0–C10, with the N-terminal domains, C0–C2, thought to be most critical for its regulatory interactions with actin and myosin (Fig. 3 a) (10,17,18). Further, the M-domain linker between the C1 and C2 domains contains four phosphorylatable serines that, once phosphorylated in response to adrenergic stimuli, may modulate cMyBP-C’s impact on cardiac contractility (28, 29, 30, 31, 32). Hence, the C0–C2 N-terminal fragment was used to simulate the influence of the full-length cMyBP-C. Human β-cardiac myosin spaced at 28-nm intervals (Fig. 3 b, myosin only) on the nanotube was interdigitated with C0–C2, also at 28-nm intervals (Fig. 3 b,+C0–C2), containing either no ER/K α-helical linker, a 10-nm ER/K α-helical linker, or a longer 30-nm ER/K α-helical linker to potentially facilitate cMyBP-C interactions with both actin and myosin (Fig. S1 b and c). The surface around the nanotubes was blocked with BSA prior to the introduction of C0–C2 (see section “methods”). Hence, C0–C2 fragments should avidly bind to the nanotubes via their DNA handles. However, a ∼30% reduction in surface actin gliding velocities were observed, consistent with residual non-specific binding to the BSA-coated nano-surface (30) (Fig. S3). Nonetheless, when C0–C2 with a 30-nm ER/K linker was present on a nanotube decorated with β-cardiac myosin HMM (data from eight independent protein preparations), ensemble velocity of F-actin was reduced fourfold from 0.29 ± 0.01 μm·s−1 (n = 211 filaments) to 0.07 ± 0.002 μm·s−1 (n = 389 filaments) (Fig. 3 c; Video S2). Likewise, with the presence of C0–C2, regulated thin filament velocity at pCa 5 on β-cardiac myosin HMM nanotubes (data from three independent protein preparations) was reduced sixfold from 0.60 ± 0.02 μm·s−1 (n = 171 filaments) to 0.10 ± 0.003 μm·s−1 (n = 223 filaments). These effects are comparable with a previously reported ∼fivefold decrease in actin gliding speeds in a standard in vitro motility assay with chicken skeletal muscle myosin and cMyBP-C adsorbed at high concentrations (0.4 μM) (30).
Representative video depicting a nanosurfer assay with F-actin filaments (green) traveling on nanotubes (red) labeled with interdigitated β-cardiac myosin HMM and C0–C2 (+10 nm ER/K) spaced 14 nm apart. Actin filaments can be seen moving slower on the C0–C2-decorated β-cardiac HMM nanotubes (nanotube velocity) before accelerating onto the surrounding motility surface (nano-surface) coated with β-cardiac myosin HMM.
We found no significant impact of the ER/K linker or linker length on the inhibitory capacity of C0–C2, as all C0–C2 ER/K constructs inhibited F-actin-nanotube motility ∼fourfold (Fig. 3 d; data from n = 82–389 filaments pooled from three to eight independent protein preparations), as visually represented in the kymographs in Fig. 3 e. Together, these studies demonstrate the robust use of the nanosurfer assay to recapitulate the effects of cMyBP-C on actomyosin motility.
To distinguish between cMyBP-C effects being imparted through its interaction with actin and/or myosin, we utilized nanotubes decorated with β-cardiac myosin S1, which lacks the S2 domain that has been implicated as the primary cMyBP-C interacting region in myosin (19, 20, 21, 22, 23). β-Cardiac S1 and HMM displayed similar velocity inhibition with the C0–C2 fragment (Fig. 3 c). Specifically, the presence of C0–C2 with β-cardiac S1 (data from three independent protein preparations) reduced F-actin velocity almost fourfold (0.59 ± 0.01 μm·s−1 to 0.16 ± 0.01 μm·s−1; n = 180 and 267, respectively) and regulated thin filament velocity almost sixfold (1.01 ± 0.01 μm·s−1 to 0.18 ± 0.01 μm·s−1; n = 450 and 514, respectively). These data suggest that the presence of C0–C2 on both S1 and myosin patterned nanotubes does not require C0–C2 interactions with myosin S2 to slow filament velocity. The potential that this slowing results in part from C0–C2 interactions with actin is supported by the capacity of nanotubes with only C0–C2 to recruit actin filaments, which is not the case for unlabeled nanotubes that do not recruit actin filaments (Fig. S4 a). Specifically, when the C0–C2 fragment was attached to the nanotubes without motor, it pulled more F-actin filaments (237.0 filaments per field of view ±37 standard deviation; Fig. S4 b) onto nanotubes than motor in the absence of C0–C2 (146.7 filaments per field of view ±22 standard deviation; Fig. S4 c). The presence of both C0–C2 and β-cardiac myosin HMM on nanotubes yielded greater actin recruitment on nanotubes than either of them alone (326.7 filaments per field of view ±116 standard deviations; Fig. S4 d). Robust actin recruitment to nanotubes was still seen by C0–C2, even when the surface was blocked in the standard nanotube assay (Fig. S4 e)
cMyBP-C M-domain phosphorylation and structurally dependent inhibition of β-cardiac myosin HMM and S1 nanotube motility
To investigate the role of individual cMyBP-C domains in the inhibition of actomyosin motility, we examined alternative N-terminal fragments, C0–C1f and C1–C2, in our nanosurfer assay (Fig. 4 a). The C0–C1f fragment contains the C0 and C1 domains, as well as the first 17 amino acids of the M-domain (Fig. 4 a). This segment of the M-domain contains several arginine residues proposed to be essential for cMyBP-C interaction(s) resulting in actomyosin inhibition (Fig. 4 b, left, inset) (30,48,49). However, it lacks the four phosphorylatable serines found in the M-domain (Fig. 4 a) (30,50). The C1–C2 fragment contains the C1 and C2 domains, as well as the entire M-domain linker region between C1 and C2. Using the nanosurfer assay, human β-cardiac myosin spaced at 28-nm intervals on the nanotube was interdigitated with either C0–C1f (Fig. 4 b, left), C1–C2 (Fig. 4 b, right), or C0–C2, each spaced at 28 nm. The presence of C0–C1f did not significantly affect F-actin or thin filament motility driven by either β-cardiac myosin HMM or S1 (Fig. 4 c; data from n = 100–274 filaments pooled from three to four independent protein preparations). In contrast to C0–C1f, the C1–C2 fragment was as inhibitory of F-actin and thin filament motility as C0–C2 (Fig. 4 d). Specifically, both C1–C2 and C0–C2 reduced F-actin velocity on β-cardiac myosin HMM nanotubes ∼fourfold, and reduced thin filament velocity on β-cardiac myosin S1 nanotubes ∼eightfold (Fig. 4 d and e; data from n = 90–270 filaments pooled from three to four independent protein preparations). These data are consistent with similar inhibitory effects observed with the addition of C1–C2 or C0–C2 to a standard in vitro motility assay (30,33).
The C1–C2 fragment contains the entire M-domain, including the phosphorylatable serines (S273, S282, S302, S307) that regulate cardiac contractility. Hence, we generated phospho-null (Ser to Ala) and phospho-mimetic (Ser to Asp) C0–C2 and C1–C2 N-terminal fragments (Fig. 4 f), as previously reported for the C0–C3 fragment (1,50). Mass spectrometry suggests that our insect-cell-expressed N-terminal fragments are not phosphorylated (data not shown). The phospho-null C0–C2 and C1–C2 fragments inhibited thin filament velocity over β-cardiac myosin S1 nanotubes to a similar extent to unmodified fragments (Fig. 4 g; data from n = 240–270 filaments pooled from three independent protein preparations). However, the phospho-mimetic C0–C2 and C1–C2 mutants exhibited less inhibition (Fig. 4 g), as previously reported for the identical phospho-mimetic substitutions in the C0–C3 fragment used in the in vitro motility assay (1,50). These results highlight the importance of the phospho-serines in regulating cMyBP-C mediated inhibition of β-cardiac myosin function.
Discussion
In this study, we utilized DNA nanotechnology to create a synthetic myosin thick filament with incorporated cMyBP-C. This synthetic thick filament controls the stoichiometry and spatial configuration of protein-protein interactions to provide mechanistic insights into cMyBP-C function. Using synthetic thick filaments with an assortment of cMyBP-C N-terminal fragments, we characterized the effects of cMyBP-C domains on the velocity of either F-actin or regulated thin filaments. Our findings suggest that the binding of cMyBP-C to actin contributes at least in part to the inhibitory effect of cMyBP-C on actomyosin motility over nanotubes (Fig. 4 h) and that the cMyBP-C M-domain is crucial to this inhibition. Further, phospho-mimetic cMyBP-C fragments, displaying diminished inhibition of thin filament velocity on nanotubes, reproduce the established modulatory role of M-domain phosphorylation on cMyBP-C function (1,28,29,51). Hence, while the synthetic thick filament does not capture the inherent three-dimensional nature of the cardiac muscle sarcomere and the strain-dependent activation of myosin (52), our study demonstrates a robust approach to investigate subtle changes in contractility through genetically inherited mutations and/or physiological regulation through post-translational modifications.
In vitro motility assays with native thick filaments from mouse cardiac tissue maintain the myosin to cMyBP-C stoichiometry and spatial relationships (1,26). These were critical to establish that cMyBP-C mediates thin filament slowing and calcium sensitization within the C-zone. However, such native thick filament assays are not suitable for molecular structure-function studies with systematic perturbations to the myosin or cMyBP-C, due to the high cost of animal model design. Our nanosurfer assay fills this need by defining the stoichiometry and spatial relations between myosin and cMyBP-C and the relative ease in expression of human β-cardiac myosin and cMyBP-C fragments that decorate the nanotube surface.
The nanosurfer assay also overcomes a key limitation of our previously reported standard nanotube assay (34). The standard nanotube assay is well suited to study high-duty-ratio motors, which readily recruit actin filaments from solution and display abundant motility. In contrast, the use of low-duty-ratio β-cardiac myosin nanotubes is limited by infrequent engagement of actin filaments from the surrounding solution (Fig. 1 b). To overcome this challenge, we seeded β-cardiac myosin subfragments at a sufficient density on the surface to which the nanotubes were attached (see section “methods”) so that the coverslip surface myosin recruited actin filaments from solution, which then encountered the neighboring nanotubes, substantially enhancing the frequency of nanotube motile events (Fig. 1 c). The actin gliding speeds on nanotubes and the surrounding surface are indistinguishable; however, these speeds are significantly reduced compared with that observed in the standard in vitro motility assay (Fig. 1 d). These data suggest that the surface conditions and/or the mode of myosin attachment likely contribute to the lower speeds in the nanosurfer assay. Specifically, the myosin attachment strategy (see section “methods”) may reduce the efficient transfer of myosin displacements to the actin filament, leading to slower velocities. Hence, we focused on motility comparisons between conditions only within the nanosurfer assay. In addition, similar flexibility by the addition of a variable length ER/K α-helical linker to the cMyBP-C fragment ensured that cMyBP-C on the nanotube was spatially free to interact with β-cardiac myosin and/or actin filaments (Fig. S1 b and c). However, the ER/K α-helical linker was not essential for the observed cMyBP-C effects on actin filament motility, as constructs lacking the ER/K α-helical linker demonstrated equivalent inhibition. Overall, the multiple rotational elements in the myosin and cMyBP-C attachment, combined with myosin’s access to numerous sites along the actin filament (Fig. S2 c), yields significant conformational freedom without limiting protein-protein interactions.
We observed that slowing of F-actin or fully activated thin filament velocity by the C0–C2 N- terminal fragment was structurally localized to the C1–C2 domains, with the M-domain linker between C1 and C2 being a critical component (Fig. 4 c and d). The importance of the M-domain is emphasized by phospho-mimetic replacement of four serines within the M-domain that reduce the observed slowing of thin filament velocity, as reported previously (1,50). To further characterize this regulatory domain, we utilized the C0–C1f fragment, which contains only the first 17 amino acids of the M-domain but lacks the four phosphorylatable serines (30,50). Interestingly, a previous study showed inhibition of thin filament velocity by C0–C1f in a standard in vitro motility assay, and it was proposed that several C-terminal arginine residues in this fragment may be particularly important for this inhibition (Fig. 4 b, inset) (30,48). The inability of the C0–C1f to slow thin filament velocity in our nanosurfer assay (Fig. 4 c), despite the presence of several C-terminal arginine residues (Fig. 4 b, inset), further supports the importance of the entire M-domain. A potential source for this discrepancy is the weaker binding of C0–C1f to actin so that its potential inhibitory effects manifest only when high concentrations of this fragment are used in the standard in vitro motility assay (30).
In vitro, both actin and myosin binding have been described for the cMyBP-C N terminus (17, 18, 19,21,22,24, 25, 26, 27). Whether one or both of these binding-partner interactions is responsible for the modulatory capacity of cMyBP-C is still a matter of debate. The myosin S2 domain was the first structural element identified in cMyBP-C binding (19). Using the β-cardiac S1 subfragment, which is devoid of the S2 segment, substantial slowing of actin gliding speeds in the presence of the C0–C2 and C1–C2 fragments on the nanotubes remained (Fig. 4 d), as reported previously using the S1 subfragment and C1–C2 in the in vitro motility assay (53). Therefore, myosin S2 binding by the cMyBP-C N terminus is not an absolute requirement for cMyBP-C modulation of actomyosin interactions. However, the nanosurfer assay as described here cannot rule out cMyBP-C N terminus binding to myosin via the myosin regulatory light chain (22) or to a yet defined region of the motor domain (32) as contributing to the inhibition of actin filament velocity observed on the nanotubes. However, the fact that nanotubes patterned only with C0–C2 recruit actin filaments to the nanotube surface does support the literature evidence (1,9,17,18,24, 25, 26, 27, 28, 29), suggesting that the cMyBP-C inhibitory effect in vitro may in part be due to its interaction with actin. In fact, recent in vivo super-resolution imaging data suggest that the cMyBP-C N terminus in mouse myocardium resides predominantly near the thin filament regardless of the muscle’s active state and that only when activated would the myosin head domain be in close enough proximity to interact with the cMyBP-C terminus (18). At least through actin binding, the slowing of thin filament velocity may arise, in part, due to drag forces imposed by cMyBP-C binding and tethering the thin filament to the nanotube (54), thereby slowing actomyosin kinetics (55,56). In addition, cMyBP-C may be competing with myosin for binding sites on actin, thus reducing the number of force-generating motors, as suggested previously (25,57,58).
Conclusion
In conclusion, we have established the DNA synthetic thick filament nanosurfer assay as a robust tool for the characterization of β-cardiac myosin and cMyBP-C interactions. We have recapitulated actomyosin inhibition by cMyBP-C N-terminal fragments on recombinant human β-cardiac myosin DNA nanotubes, emphasizing the importance of the M-domain and its phosphorylation in regulating actomyosin motility. Future studies using the nanosurfer assay can further our understanding of the molecular mechanisms underlying contractile regulation by cMyBP-C, and, more importantly, under loaded conditions where the heart normally operates.
Author contributions
A.M.T., A.R., D.M.W., C.M.Y., and S.S. designed research. A.M.T., W.T., D.V.R., D.V., A.R., and S.B.P. performed research. C.M.Y. and D.M.W. provided key reagents. A.M.T., D.V., A.R., D.M.W., C.M.Y., and S.S. analyzed data. The manuscript was written by A.M.T., D.M.W., C.M.Y., and S.S. and approved by all authors.
Acknowledgments
The authors would like to thank Ruth Sommese for technical expertise and Michael Ritt for helpful discussions and technical assistance. Research was supported by the NIH (1R35GM126940 to S.S.; 1R01HL150953 to D.W., C.Y., and S.S.; 1F30HL146089-01 to A.T.) and supported in part by a generous gift to D.W. from Arnold and Mariel Goron.
Declaration of interests
The authors declare no competing interests.
Editor: David Sept
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.05.013.
Supporting material
References
- 1.Previs M.J., Previs S.B., et al. Warshaw D.M. Molecular mechanics of cardiac myosin binding protein-C in native thick filaments. Science. 2012;337:1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moss R.L., Fitzsimons D.P., Ralphe J.C. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 2015;116:183–192. doi: 10.1161/circresaha.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris S.P., Lyons R.G., Bezold K.L. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 2011;108:751–764. doi: 10.1161/circresaha.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starr R., Offer G. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Lett. 1971;15:40–44. doi: 10.1016/0014-5793(71)80075-3. [DOI] [PubMed] [Google Scholar]
- 5.Offer G., Moos C., Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. J. Mol. Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 6.Bonne G., Carrier L., et al. Schwartz K. Cardiac myosin binding protein–C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 7.Watkins H., Conner D., et al. Seidman C.E. Mutations in the cardiac myosin binding protein–C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 8.Carrier L., Mearini G., et al. Cuello F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 2015;573:188–197. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mun J.Y., Previs M.J., et al. Craig R. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heling L.W.H.J., Geeves M.A., Kad N.M. MyBP-C: one protein to govern them all. J. Muscle Res. Cell Motil. 2020;41:91–101. doi: 10.1007/s10974-019-09567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris S.P. Making waves: a proposed new role for myosin-binding protein C in regulating oscillatory contractions in vertebrate striated muscle. J. Gen. Physiol. 2021;153:e202012729. doi: 10.1085/jgp.202012729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flashman E., Redwood C., et al. Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004;94:1279–1289. doi: 10.1161/01.res.0000127175.21818.c2. [DOI] [PubMed] [Google Scholar]
- 13.Luther P.K., Bennett P.M., et al. Moss R.L. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J. Mol. Biol. 2008;384:60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K., Harris S.P., et al. Craig R. Orientation of myosin binding protein C in the cardiac muscle sarcomere determined by domain-specific immuno-EM. J. Mol. Biol. 2015;427:274–286. doi: 10.1016/j.jmb.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okagaki T., Weber F.E., et al. Reinach F.C. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alyonycheva T.N., Mikawa T., et al. Fischman D.A. Isoform-specific interaction of the myosin-binding proteins (MyBPs) with skeletal and cardiac myosin is a property of the C-terminal immunoglobulin domain. J. Biol. Chem. 1997;272:20866–20872. doi: 10.1074/jbc.272.33.20866. [DOI] [PubMed] [Google Scholar]
- 17.Luther P.K., Winkler H., et al. Liu J. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahmanseresht S., Lee K.H., et al. Previs M.J. The N terminus of myosin-binding protein C extends toward actin filaments in intact cardiac muscle. J. Gen. Physiol. 2021;153:e202012726. doi: 10.1085/jgp.202012726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruen M., Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C 1 1Edited by J. Karn. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 20.Howarth J.W., Ramisetti S., et al. Rosevear P.R. Structural insight into unique cardiac myosin-binding protein-C motif. J. Biol. Chem. 2012;287:8254–8262. doi: 10.1074/jbc.m111.309591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris S.P., Rostkova E., et al. Moss R.L. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ. Res. 2004;95:930–936. doi: 10.1161/01.res.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- 22.Ratti J., Rostkova E., et al. Pfuhl M. Structure and interactions of myosin-binding protein C domain C0. J. Biol. Chem. 2011;286:12650–12658. doi: 10.1074/jbc.m110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfuhl M., Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J. Muscle Res. Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 24.Kensler R.W., Shaffer J.F., Harris S.P. Binding of the N-terminal fragment C0–C2 of cardiac MyBP-C to cardiac F-actin. J. Struct. Biol. 2011;174:44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris S.P., Belknap B., et al. Galkin V.E. C0 and C1 N-terminal Ig domains of myosin binding protein C exert different effects on thin filament activation. Proc. Natl. Acad. Sci. Unit. States Am. 2016;110:125a–1563. doi: 10.1016/j.bpj.2015.11.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Previs M.J., Prosser B.L., Warshaw D.M., et al. Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risi C., Patra M., et al. Galkin V.E. Interaction of the C2 ig-like domain of cardiac myosin binding protein-C with F-actin. J. Mol. Biol. 2021;433:167178. doi: 10.1016/j.jmb.2021.167178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Previs M.J., Mun J.Y., et al. Craig R. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:3239–3244. doi: 10.1073/pnas.1522236113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer J.F., Kensler R.W., Harris S.P. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 2009;284:12318–12327. doi: 10.1074/jbc.m808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weith A., Sadayappan S., et al. Warshaw D.M. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruen M., Prinz H., Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 32.Nag S., Trivedi D.V., et al. Spudich J.A. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 2017;24:525–533. doi: 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razumova M.V., Shaffer J.F., et al. Harris S.P. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay. J. Biol. Chem. 2006;281:35846–35854. doi: 10.1074/jbc.m606949200. [DOI] [PubMed] [Google Scholar]
- 34.Hariadi R.F., Sommese R.F., et al. Sivaramakrishnan S. Mechanical coordination in motor ensembles revealed using engineered artificial myosin filaments. Nat. Nanotechnol. 2015;10:696–700. doi: 10.1038/nnano.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommese R.F., Hariadi R.F., et al. Sivaramakrishnan S. Patterning protein complexes on DNA nanostructures using a GFP nanobody. Protein Sci. 2016;25:2089–2094. doi: 10.1002/pro.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivaramakrishnan S., Spudich J.A. Systematic control of protein interaction using a modular ER/K α-helix linker. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20467–20472. doi: 10.1073/pnas.1116066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson C.J., Sivaramakrishnan S. Harnessing the unique structural properties of isolated α-helices. J. Biol. Chem. 2014;289:25460–25467. doi: 10.1074/jbc.r114.583906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik R.U., Dysthe M., et al. Sivaramakrishnan S. ER/K linked GPCR-G protein fusions systematically modulate second messenger response in cells. Sci. Rep. 2017;7:7749. doi: 10.1038/s41598-017-08029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rai A., Vang D., et al. Sivaramakrishnan S. Dynamic multimerization of Dab2-Myosin VI complexes regulates cargo processivity while minimizing cortical actin reorganization. J. Biol. Chem. 2021;296:100232. doi: 10.1074/jbc.ra120.012703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swenson A.M., Tang W., et al. Yengo C.M. Omecamtiv mecarbil enhances the duty ratio of human β-cardiac myosin resulting in increased calcium sensitivity and slowed force development in cardiac muscle. J. Biol. Chem. 2017;292:3768–3778. doi: 10.1074/jbc.m116.748780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., White H.D., et al. Forgacs E. Omecamtiv Mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 2015;54:1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q., Moncman C.L., Winkelmann D.A. Mutations in the motor domain modulate myosin activity and myofibril organization. J. Cell Sci. 2003;116:4227–4238. doi: 10.1242/jcs.00709. [DOI] [PubMed] [Google Scholar]
- 43.Winkelmann D.A., Forgacs E., Stock A.M., et al. Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat. Commun. 2015;6 doi: 10.1038/ncomms8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardee J.D., Spudich J.A. Purification of muscle actin. Methods Enzymol. 1982;85 Pt B(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 45.Squire J.M. Muscle myosin filaments: cores, crowns and couplings. Biophys. Rev. 2009;1:149–160. doi: 10.1007/s12551-009-0017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adhikari A.S., Trivedi D.V., et al. Ruppel K.M. β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 2019;10:2685. doi: 10.1038/s41467-019-10555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraser I.D.C., Marston S.B. In vitro motility analysis of actin-tropomyosin regulation by troponin and calcium. J. Biol. Chem. 1995;270:7836–7841. doi: 10.1074/jbc.270.14.7836. [DOI] [PubMed] [Google Scholar]
- 48.Bhuiyan M.S., Gulick J., et al. Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C’s interactions. J. Mol. Cell. Cardiol. 2012;53:838–847. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bezold K.L., Shaffer J.F., et al. Harris S.P. A gain-of-function mutation in the M-domain of cardiac myosin-binding protein-C increases binding to actin. J. Biol. Chem. 2013;288:21496–21505. doi: 10.1074/jbc.m113.474346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weith A.E., Previs M.J., et al. Warshaw D.M. The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. J. Muscle Res. Cell Motil. 2012;33:449–459. doi: 10.1007/s10974-012-9312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barefield D., Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J. Mol. Cell. Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fusi L., Brunello E., et al. Irving M. Thick filament mechano-sensing is a calcium-independent regulatory mechanism in skeletal muscle. Nat. Commun. 2016;7:13281. doi: 10.1038/ncomms13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaffer J.F., Razumova M.V., et al. Harris S.P. Myosin S2 is not required for effects of myosin binding protein-C on motility. FEBS Lett. 2007;581:1501–1504. doi: 10.1016/j.febslet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Colson B.A. What a drag!: skeletal myosin binding protein-C affects sarcomeric shortening. J. Gen. Physiol. 2019;151:614–618. doi: 10.1085/jgp.201912326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenberg M.J., Shuman H., Ostap E. Inherent force-dependent properties of β-cardiac myosin contribute to the force-velocity relationship of cardiac muscle. Biophys. J. 2014;107:L41–L44. doi: 10.1016/j.bpj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung J., Nag S., et al. Spudich J.A. Harmonic force spectroscopy measures load-dependent kinetics of individual human β-cardiac myosin molecules. Nat. Commun. 2015;6:7931–7939. doi: 10.1038/ncomms8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walcott S., Docken S., Harris S.P. Effects of cardiac myosin binding protein-C on actin motility are explained with a drag-activation-competition model. Biophys. J. 2015;108:339a–13. doi: 10.1016/j.bpj.2014.11.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inchingolo A.V., Previs S.B., et al. Kad N.M. Revealing the mechanism of how cardiac myosin-binding protein C N-terminal fragments sensitize thin filaments for myosin binding. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:6828–6835. doi: 10.1073/pnas.1816480116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative video depicting a nanosurfer assay with F-actin filaments (green) traveling on nanotubes (red) labeled with β-cardiac myosin HMM spaced 14 nm apart. Two actin filaments are seen traveling in opposite directions on the nanotube in the center.
Representative video depicting a nanosurfer assay with F-actin filaments (green) traveling on nanotubes (red) labeled with interdigitated β-cardiac myosin HMM and C0–C2 (+10 nm ER/K) spaced 14 nm apart. Actin filaments can be seen moving slower on the C0–C2-decorated β-cardiac HMM nanotubes (nanotube velocity) before accelerating onto the surrounding motility surface (nano-surface) coated with β-cardiac myosin HMM.