Abstract
Plasmacytoid dendritic cells (pDCs) are specialized cells of the immune system that are thought to be the main cellular source of type I interferon alpha (IFNα) in response to viral infections. IFNs are powerful antivirals, whereas defects in their function or induction lead to impaired resistance to virus infections, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19. IFN production needs to be controlled, because sustained IFN production can also have detrimental effects on disease outcome. As such, pDCs are likely important for acute antiviral protection against SARS-CoV-2 infection but could potentially also contribute to chronic IFN levels. Here, we provide a historical overview of pDC biology and summarize existing literature addressing their involvement and importance during viral infections of the airways. Furthermore, we outline recent reports focused on the potential role of pDCs during SARS-CoV-2 infection, as well as the potential for this cellular subset to impact COVID-19 disease outcome.
Keywords: COVID-19, SARS-CoV-2, plasmacytoid dendritic cells, antiviral responses, inflammation
Plasmacytoid dendritic cells (pDCs) produce interferons (IFNs) in response to viral infections, including SARS-CoV-2. IFNs need to be controlled, because sustained levels have detrimental effects on disease outcome. van der Sluis et al. examine the importance of pDCs in protection against SARS-CoV-2, but also how they contribute to chronic inflammation.
Introduction
Over the past century, viral pandemics have detrimentally affected health worldwide. The deadliest of them have been caused by a single member of the Orthomyxoviridae family: influenza A virus (IAV) (Kilbourne, 2006). This changed in late 2019, when a non-influenza pandemic was triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19) (Wu et al., 2020; Zhu et al., 2020). The SARS-CoV-2 pandemic highlighted the vulnerability of the worldwide population to pandemics caused by respiratory viral infections. This has driven efforts to improve our understanding of the mechanisms of viral clearance and the immunopathology that accompanies COVID-19.
Early clinical reports describing SARS-CoV-2 infection and the development of COVID-19 indicated that disease severity was strongly associated with a dysregulated innate immune response, which was characterized by excessive production of pro-inflammatory cytokines and a limited or delayed release of antiviral type I and type III interferons (IFNs) (Del Valle et al., 2020; Lowery et al., 2021). Limited IFN responses may result in insufficient induction of antiviral protection, while the excessive pro-inflammatory cytokines can cause increased cell and tissue damage. During the early stages of the pandemic, many reports suggested that the virus could cause the immune system to go into “overdrive” in some individuals, primarily in the respiratory tract during acute infection, and later systemically (Azkur et al., 2020; Mahmudpour et al., 2020; Mortaz et al., 2020; Nowill and De Campos-Lima, 2020; Sa Ribero et al., 2020; Weatherhead et al., 2020; Zhang et al., 2022). It is still unclear what types of immune cell are responsible for the inflammatory signature observed in severe COVID-19 patients. Multiple studies have reported that plasmacytoid dendritic cells (pDCs), a cell type known to have strong antiviral functions because of production of IFN, declined in numbers during disease development (Liao et al., 2020; Zhou et al., 2020c). We hypothesize that pDCs may play a role in the development of severe COVID-19, as a result of dysfunctionality in their cytokine production. Here, we provide a brief overview of the development of pDCs and their antiviral mechanisms in different virus infection models. We discuss recently published original research and examine whether pDCs constitute an intrinsic part of the host response that may affect COVID-19 disease outcome.
The inflammatory signature of COVID-19
Clinical manifestations of COVID-19 range from asymptomatic to mild or severe (Kim et al., 2020). Mild infections are characterized by symptoms such as fever, cough, malaise, dyspnea, and fatigue, and they can be cleared without medical intervention (Da Rosa Mesquita et al., 2021). A minority of COVID-19 patients develop severe pneumonia, and these patients can develop acute respiratory distress syndrome, which often requires hospitalization and oxygen treatment (Attaway et al., 2021). The likelihood of developing severe COVID-19 increases with age and with pre-existing comorbidities, such as cardiovascular disorder, hypertension, and diabetes (Del Sole et al., 2020; Romero Starke et al., 2020; O’driscoll et al., 2021). Some reports state that disease severity associates with decreased and/or delayed levels of type I and III IFNs, as well as increased levels of pro-inflammatory cytokines, such as TNF-α, interleukin-1b (IL-1b), IL-8, and IL-6, and this reveals a disproportionate immune response following SARS-CoV-2 infection (Del Valle et al., 2020; Galani et al., 2021; Lowery et al., 2021; Bastard et al., 2020; Carvalho et al., 2021; Zhang et al., 2020; Zhou et al., 2020c; Bastard et al., 2021; Pairo-Castineira et al., 2021; Wang et al., 2021). Other reports indicate that prolonged or excessive IFN production may also contribute to severe COVID-19 (Broggi et al., 2020; Carvalho et al., 2021; Lee et al., 2020; Lucas et al., 2020; Major et al., 2020; Sposito et al., 2021), suggesting that the presence of IFN during acute infection is beneficial, whereas sustained IFN production can lead to complications (Wack, 2021). The complexity of IFN in the pathogenesis of SARS-CoV-2 has been reviewed extensively by others (Wong and Perlman, 2022; Zanoni, 2021; Zhang et al., 2022).
Importantly, determining the immune cells involved in the inflammatory response during COVID-19 may assist with the development of therapies that aim to boost (e.g., early IFN) or reduce (e.g., IL-6/IL-8/TNF-α/chronic IFN) the production of pro-inflammatory cytokines. This information might also facilitate the development of targeted screening strategies to identify COVID-19 patients who are more likely to develop severe disease. The key focus of this review is the potential antiviral role of pDC, which is an important, but not sole, producer of IFNs. Furthermore, pDCs can fulfill other immunological functions, such as the activation of cytotoxic T cells and immunosuppressive regulatory T cells (Swiecki and Colonna, 2015), which may play a role for controlling SARS-CoV-2 infection.
The historical angle of pDCs
pDCs are a central part of the innate immune system. After sensing the presence of pathogens, pDCs release high levels of different IFNs, cytokines, and chemokines, and combined with their ability to present antigen, pDCs support shaping the adaptive antiviral immune response (Mathan et al., 2013; Mckenna et al., 2005; Swiecki and Colonna, 2015). As such, pDCs are often referred to as a bridge between the innate and adaptive immune systems. Unfortunately, studying pDCs in diseases such as COVID-19 is a major challenge. Notably, pDCs are an extremely rare cell type, they are difficult to collect from blood or tissue specimens, there are challenges maintaining viable pDCs in culture outside the body, and pDCs have a high degree of phenotypic plasticity making it difficult to characterize them in detail.
pDCs were originally described by multiple groups in the 1980s (Fitzgerald-Bocarsly et al., 1988; Perussia et al., 1985; Ronnblom et al., 1983). They portrayed a cell population with a remarkable phenotype and functionality by (1) extremely low frequency in peripheral blood (less than 0.1%), (2) expression of MHC class I and class II and lack of various lineage markers (i.e., CD3, CD14, CD16, CD19, CD20, CD56), and (3) potent production of IFNα when exposed to viruses. Due to the latter feature, these cells were originally termed “natural interferon producing cells” (NIPCs). Ten years after their discovery, NIPCs were redefined as a subgroup of dendritic cells and, partly because of the morphology, renamed to pDCs (Cella et al., 1999; Siegal et al., 1999).
The characterization of pDCs as DCs has been reassessed within recent years through multi-omics approaches (i.e., single-cell RNA transcriptomics) and multi-parameter flow cytometry. These studies have provided a detailed understanding of pre-pDC and pDC populations in the bone marrow and the periphery. Attempts to identify progenitor cells in the bone marrow have primarily been performed in murine models, although the translatability to human pDC biology remains unclear. The earlier studies in mice suggested that pDCs originate from both myeloid and lymphoid progenitor cell populations, and that various transcription factors determine the developmental path of the pDC lineage from a common precursor DC cell population (Naik et al., 2007; Onai et al., 2007). pDC development seems to be driven by the growth factor FMS-like Tyrosine Kinase 3 Ligand (Flt3L), which activates its receptor FLT3 (CD135) (Brawand et al., 2002; Gilliet et al., 2002), and the transcription factor E2-2 (Cisse et al., 2008). However, a recent paper by Rodrigues et al. (2018) suggested an alternative path of pDC lineage development. Rodrigues et al. (2018) established that murine pDCs do not advance from a myeloid precursor but originate solely from a lymphoid progenitor, defined as an IL-7 receptor-positive (IL-7R+) lymphoid precursor cell, which they named a “pre-pDC.” The development of pDCs from this precursor stem cell is dependent on their capacity to express the transcription factors IFN Regulatory Factor 8 (IRF8), E2-2, and Runt-related transcription factor 2 (Runx2, also known as core-binding factor subunit alpha-1 [CBFα1]) (Rodrigues et al., 2018). Dress et al. (2019) further refined this development path, demonstrating that pre-pDCs are characterized by a Ly6D+IL-7Rα+Flt3+SiglecH+CD2intCD81intCD115− phenotype, and that they originate from a Ly6D+IL-7Rα+ CLP-like progenitor population.
An important distinction between murine and human pDCs is that human pDCs exhibit high expression of the IL-3R alpha chain (CD123), which combined with the IL-3 beta chain subunit (CD131) supports cell survival and the immunological functions of pDC (Grouard et al., 1997; Grzes et al., 2021; Fanning et al., 2006). However, CD123 is not expressed on murine pDCs (Leylek et al., 2019). Murine pDCs express an orthologue called AIC2A (Hara and Miyajima, 1992) with comparable function to human CD123. A thorough description of the human pDC lineage has recently been detailed by Cytlak et al. (2020). These investigators implemented comprehensive immunophenotyping combined with RNA sequencing (RNA-seq) profiling and demonstrated that human pDCs exhibit a high degree of plasticity, but that IRF8 expression is crucial for pDC maturation in bone marrow. This research effort, as well as the efforts of two additional teams of investigators, has generated a comprehensive panel of surface markers (CD123, CD33, CX3CR1, CD2, CD5, CD327, Siglec6, and AXL) that are essential for distinguishing human precursor DCs and DCs from pDCs (See et al., 2017; Villani et al., 2017). Both the See et al. (2017) and Villani et al. (2017) studies highlighted that pDCs are actually unable to produce IL-12. This is contradictory to earlier studies describing IL-12 production as a feature that DCs and pDCs share.
The path from the discovery of pDCs to what we know today highlights the challenges the field has met when studying these cells during steady-state conditions and disease. However, technological advancements have clarified that pDCs have a unique lineage/specific developmental path, immunological function (e.g., induction of specific cytokines and IFNs), and phenotypic profile (cellular surface markers) that set them aside from other known DC subsets. More importantly, advances in our fundamental understanding of pDC biology have made it possible to explore the potential role of pDCs in emerging viral infections, such as SARS-CoV-2.
pDCs are sentinels of the innate immune system
A central feature of pDCs is their ability to sense pathogens and mediate a rapid and strong IFN response. pDCs sense a broad range of danger/pattern-associated molecular patterns (DAMPs or PAMPs) through a variety of pattern recognition receptors (PRRs). pDCs can also mediate direct cell killing in a natural killer cell-like manner (Chaperot et al., 2006; Hardy et al., 2007) and facilitate the generation of adaptive immune responses through the process of antigen uptake and presentation to T cells (Oberkampf et al., 2018; Van Beek et al., 2020; Tel et al., 2010, 2013a, 2013b). In other words, pDCs possess great immunological flexibility and functionality (Karrich et al., 2014).
pDCs are often defined as the most powerful NIPC. The meaning of this label was addressed by Ito et al. (2006), who compared the immunological responses of highly purified human CD11c+ myloid (mDCs) versus pDCs. The investigators demonstrated that over the first 12 h following viral sensing, pDCs dedicate approximately 60% of their transcriptional activity to the production of type I IFNα isoforms. Compared with mDCs, pDCs produced at least 150 times more type I IFNα. Another relevant feature of pDCs is the endogenous high expression levels of the endosomal Toll-like receptors (TLRs) 7 and 9. This confers pDCs with the potential to rapidly respond to endosomal uptake of nucleic acids, including both single/double-stranded RNA and DNA (Ito et al., 2006; Bruni et al., 2015; Bao and Liu, 2013). Importantly, this enables pDCs to sense a very broad range of viruses, including cytomegalovirus (CMV), influenza, hepatitis C virus (HCV), HIV, rotavirus, and various coronaviruses (Deal et al., 2013; Thomas et al., 2014; Yun et al., 2021). On activation via either TLR7 or TLR9, pDCs produce IFNα through a dedicated pathway that involves MyD88-IRAK1/4 and TRAF6 signaling cascades and eventually induces phosphorylation of the transcription factor IRF7 (Liu, 2005). Constitutive high expression of IRF7 in pDCs also facilitates their strong and rapid IFNα production. Following IFNα production by pDCs, paracrine and autocrine activation of the type I IFN receptor (IFNAR) drives a positive-feedback loop and maximizes the delayed but full activation status of the pDCs (Abbas et al., 2020; Kim et al., 2014). In some situations, pDCs may also produce type I IFNβ, IFNω, and type III IFNλ1 in addition to IFNα, as demonstrated by exposure to HSV-1, influenza, and SARS-CoV-2 (Ito et al., 2006; Onodi et al., 2021; Severa et al., 2021).
pDCs can engulf extracellular material through endocytosis and cellular and viral material through phagocytosis (Bruni et al., 2015; Tel et al., 2010) and perform “self” sensing via intracellular autophagy (Frenz et al., 2014; Lee et al., 2007). These processes facilitate activation of cytosolic sensors, for example, the double-stranded RNA sensor retinoic acid-inducible gene I (RIG-I) (Yoneyama and Fujita, 2008). Although some studies suggest that pDCs have low RIG-I expression (Kato et al., 2005; Kumagai et al., 2007), others suggest that on TLR7/9 activation RIG-I expression increases rapidly in an IFNα-independent manner (Szabo et al., 2014) and enables cytosolic RNA sensing in pDCs. There are also reports describing the capacity of pDCs to sense cytosolic DNA (Deb et al., 2020; Paijo et al., 2016) by constitutive expression of the DNA sensors cyclic GMP-AMP synthase (cGAS) and Stimulator of IFN genes (STING). However, the levels of these DNA sensors in pDCs are not upregulated by IFN stimulation (Paijo et al., 2016), as has been noted to occur in other cell types (Ma et al., 2015). Nevertheless, exposure of pDCs to double-stranded DNA, or the specific STING ligand 2′3′-cGAMP, drives pDCs to generate high concentrations of IFNα in an IRF3-dependent manner, which is distinct from TLR-mediated sensing, where IFNα production is regulated by IRF7 (Deb et al., 2020; Paijo et al., 2016; Bode et al., 2016; Laustsen et al., 2018).
There are opposing views as to whether pDCs express cell surface TLRs. Stimulation of pDCs with lipopolysaccharides (LPS) does not induce activation, suggesting non-functional or absent TLR4 (Piccioli et al., 2009). It has been shown that pDCs isolated from peripheral blood can express TLR1/2 and respond to lipoprotein ligands by both secreting IFNα and increasing expression of activation markers (Raieli et al., 2019). This is highly relevant for viral sensing, because TLR2 senses glycoproteins from multiple viruses when combined in a heterodimer with TLR1 or TLR6 (Bieback et al., 2002; Boehme et al., 2006; Leoni et al., 2012; Zhu et al., 2007; Zheng et al., 2021b). In summary, pDCs are equipped with various means to sense pathogen danger signals from both the intracellular and the extracellular environment, as well as the capacity to initiate antiviral responses.
pDCs and the viral synapse
In addition to their ability to sense and respond to immunostimulatory PAMPs, some data suggest that pDCs can respond directly to virus-infected cells (Donaghy et al., 2009; Yun et al., 2021). Both Assil et al. (2019) and Yun et al. (2021) demonstrated pDC sensing of virally infected cells occurs through cell-to-cell contact. The pDC and virally infected cell form a virus-sensing synapse that is composed of integrin complexes, such as lymphocyte function-associated antigen 1 (LFA-1) and intracellular adhesion molecule 1 (ICAM-1). Synapse involving cells infected with both RNA (Assil et al., 2019) and DNA viruses (Yun et al., 2021) initiated pDC signaling. Signaling is facilitated by the transfer of virus-derived PAMPs across the synapse and their endocytosis by pDCs. The endocytosed PAMPs subsequently activate TLR7/9 and induce an antiviral response. Interestingly, following viral sensing, pDCs diversify into three distinct populations based on the expression of PD-L1 and CD80 (Alculumbre et al., 2018). These populations include P1 (PD-L1highCD80low; IFNα producing), P2 (PD-L1highCD80high), and P3 (PD-L1lowCD80high; antigen presentation). For example, recognition of cell-free SARS-CoV-2 skews the pDC population toward an IFN-producing P1 population, as compared with what has been observed for influenza-stimulated pDCs (Onodi et al., 2021). Furthermore, pDCs sensing virus-infected cells will push more cells toward the P1 population, as compared with pDCs sensing free virions, and this correlates with a stronger type I IFN signal (Yun et al., 2021; Alculumbre et al., 2018). These observations indicate that only a proportion of pDCs are equipped to produce IFNα on viral sensing. This contention has also been supported by studies using droplet-based microfluidic approaches, where only a limited fraction of pDCs seems to be capable of secreting IFNα. Importantly, this initial IFNα response then primes additional pDCs, via an autocrine feedback mechanism, ultimately enlarging the population of IFNα-producing pDCs (Van Eyndhoven et al., 2021; Wimmers et al., 2018). In summary, pDCs have evolved mechanisms to sense and respond to both cell-free and cell-associated viruses via a virus-sensing synapse.
pDCs and the generation of antiviral immunity
Various attempts have been made to determine the in vivo importance of pDCs for the generation of immunity to viral infections. Many of these studies have implemented antibodies targeting the cell surface markers Gr-1 (also known as Ly-6G/Ly-6C) and bone marrow stromal antigen 2 (BST-2, also known as pDC antigen-1 [PDCA-1]) to achieve in vivo depletion of murine pDCs. Importantly, these markers are also expressed by additional cell types, making it difficult to attribute any observed reductions in antiviral to the elimination of pDCs (Asselin-Paturel et al., 2001, 2003; Blasius et al., 2006; Krug et al., 2003). More conclusive evidence has since been presented in a study by Wang et al. (2006), who demonstrated the recruitment of pDCs to the lungs during the early stages of infection with respiratory syncytial virus (RSV). Here, depletion of pDCs by a pDC-specific antibody cocktail led to an increase in viral replication and decreased resistance to infection (Wang et al., 2006). Further evidence of a role for pDCs in promoting the generation of antiviral immunity has been provided by data from the Colonna laboratory, which used a diphtheria toxin (DT)-based model to specifically ablate pDCs (Swiecki et al., 2010). In this model, pDCs were demonstrated to be important for the induction of NK cell activation, as well as the expansion of virus-specific cytotoxic T cells in two separate virus infection models (Swiecki et al., 2010). Other investigators have found that the induction of CD4 and CD8 T cells is diminished during lymphocytic choriomeningitis virus (LCMV) infection in a pDC-deficient mouse strain. Interestingly, in this model, the induction of CD8 T cells, but not CD4 T cells, could be rescued by administration of exogenous type I IFN (Cervantes-Barragan et al., 2012).
Although pDCs appear to facilitate the generation of antiviral immunity, in most animal models this role appears to be redundant. For example, IFN from pDCs is important for resistance to infection with Newcastle disease virus (NDV) in a murine model only when alveolar macrophages, which are the main IFN producers in this model, are eliminated (Kumagai et al., 2007). Likewise, ablation of pDCs resulted in loss of antiviral immunity to Ross River virus infection only in mitochondrial antiviral-signaling protein (MAVS)-deficient mice and not in wild-type mice (Haist et al., 2021). Infection models of coronavirus and herpes simplex virus (HSV) are, however, important exceptions. Ablation of pDCs with antibodies against the surface marker PDCA-1 reduces resistance to infection with the mouse hepatitis virus (MHV) coronavirus (Cervantes-Barragan et al., 2007). Furthermore, ablation of murine pDCs using the DT-based model mildly increased susceptibility to infection with HSV-2, although the pDC protective effect is not mediated through inhibition of viral replication at the site of infection (Swiecki et al., 2013). In support of pDCs playing a non-redundant role in HSV infection, ablation of pDCs leads to increased viral loads in an HSV-1 intravenous infection model (Takagi et al., 2011).
Besides their role in T cell stimulation and the induction of IFN-stimulated genes (ISGs), multiple examples highlight a complex antiviral role of pDCs during early viral infections. However, future studies are warranted to confirm this and to reach a better understanding of the direct and indirect effects of pDC viral sensing and activation on the establishment of antiviral immunity.
pDCs and inflammation during virus infection
Besides the release of substantial amounts of IFNα, pDCs also release a series of pro-inflammatory cytokines that are important for the recruitment of antiviral effector cells to the site of infection. This has been elegantly shown in a mouse model of SARS-CoV infection, where pDCs are recruited to the lungs after intranasal challenge with SARS-CoV. pDC recruitment is followed by an increase in chemokine secretion and a subsequent infiltration of various immune cells to the site of infection (Chen et al., 2010). Whether the inflammation induced by pDCs is protective or pathogenic is difficult to assess, and this is likely to be different depending on the virus and tissue. However, in the case of RSV, depletion of murine pDCs increased inflammatory responses and pathology, suggesting that pDC-induced inflammation is mainly protective (Wang et al., 2006). Evidence of pDCs as early promoters of inflammation during virus infection is rare, but extensive research with bacterial infection models and with models of non-infectious diseases suggests that pDCs are important inducers of inflammation. For example, pDC ablation reduces in vivo inflammatory responses after infection with Listeria monocytogenes and following systemic stimulation with TLR agonists (Takagi et al., 2011). This is at least partially due to pDCs inhibiting anti-inflammatory T regulatory cells (Tregs). The notion that pDCs can suppress Tregs is supported by research showing pDCs subdue the recruitment of a specialized Treg subset into adipose tissue in obese mice, which promotes the inflammatory phenotype of metabolic diseases such as diabetes (Li et al., 2021a). Contrary to the early recruitment of pDCs into the lungs during SARS-CoV infection in mice, the recruitment of pDCs into the central nervous system (CNS) in experimental autoimmune encephalitis (EAE), a common mouse model of multiple sclerosis, seems to be delayed and follows T cell recruitment and not vice versa (Gao et al., 2009). This indicates that pDCs are not always the first immune cell to infiltrate inflamed tissue.
Despite their pro-inflammatory properties in a series of inflammation models, and despite their ability to release mainly T helper 1 (Th1)-associated cytokines, there is rather limited evidence to support the hypothesis that pDCs are strong drivers of excessive pathogenic inflammation during viral infections. To the contrary, pDC function seems to be mostly directed at producing antiviral IFNs, which promote Th1 differentiation and protective antiviral immunity.
The role of pDCs in SARS-CoV-2
Because SARS-CoV-2 has a positive sense single-stranded RNA genome, PRRs triggered by RNA, including TLR7 and RIG-I, dominate the early response. As highlighted previously, TLR7 and RIG-I are functional pathways within pDCs. Importantly non-RNA PRRs, like TLR2, may also contribute to the early response against the virus (discussed in detail below). As such, pDCs have the potential for strong antiviral sensing of SARS-CoV-2. In the following sections, we will review the current understanding of the role of pDCs during SARS-CoV-2 infection (Figure 1 ).
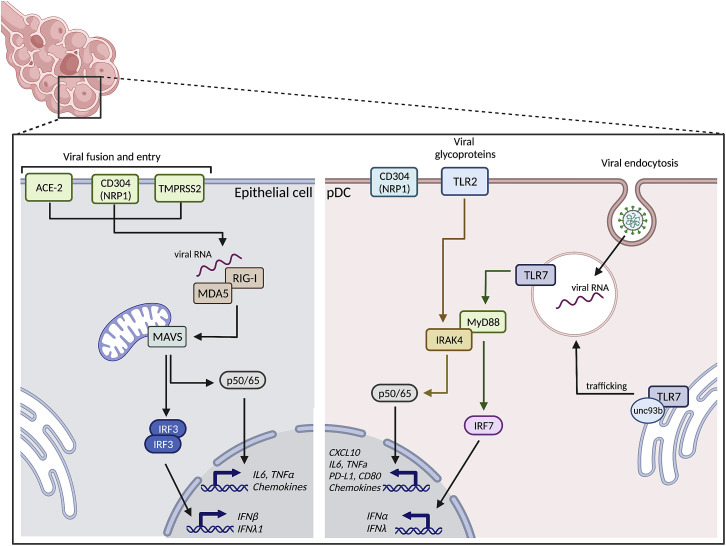
Figure 1.
Sensing of SARS-CoV-2
SARS-CoV-2 genomic RNA can be sensed by different RNA-driven pattern recognition receptors. Which RNA sensor is activated is determined by the mechanisms of viral entry: membrane fusion (indicated for the epithelial cell on the left) or viral particle uptake (indicated for the pDC on the right). Fusion of the SARS-CoV-2 membrane with the cellular membrane of epithelial cells requires the cellular receptors ACE2, TMRPSS2, and in some cases, NRP1/CD304. This allows the viral RNA to be detected by cytosolic RNA sensors RIG-I/MDA5, which will activate transcription factors IRF3 and nuclear factor κB (NF-κB), leading to production of type I IFNα, type III IFNλ1, and various pro-inflammatory cytokines and chemokines. SARS-CoV-2 utilizes several mechanisms to counter sensing via the RIG-I/MDA5 pathway and subsequent IFN production, which is reviewed in detail elsewhere (Min et al., 2021). Uptake of viral particles, for example, by endocytosis, does not require the receptors that mediate fusion of the viral and cellular membranes. Instead, the virus is engulfed into a cellular vesicle, where it triggers TLR7, starting a signaling cascade that includes MyD88 and IRAK4, ultimately activating the transcription factors IRF7 and NF-κB that will facilitate production of type I IFNα, type III IFNλ1, CXCL10, and other cytokines and chemokines. The interaction of SARS-CoV-2 with NRP1/CD304 can inhibit IFNα production by pDCs, but the molecular mechanisms are currently unknown. pDCs can also sense SARS-CoV-2’s glycosylated envelope protein via TLR2, and this leads specifically to IL-6 production in pDCs. Figure was created with BioRender.com.
SARS-CoV-2 enters cells via two mechanisms: membrane fusion and viral particle uptake. Depending of the mode of entry, different innate immune PRRs could be triggered. Fusion of the viral and cellular membranes requires the cellular receptors angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), and in some cases, neuropilin 1 (NRP1) (Wu et al., 2020; Zhou et al., 2020a; Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Hoffmann et al., 2020). Following fusion, the viral genome is released into the cytoplasm of the host cell. As reported for epithelial cells, viral genome release can trigger RIG-I and MDA5 (Sampaio et al., 2021; Yin et al., 2021; Thorne et al., 2021; Yamada et al., 2021). However, some studies suggest that the virus antagonizes intracellular RNA PRRs. This is mediated by Nsp5 (Liu et al., 2021) and other viral proteins (Oh and Shin, 2021). Importantly, it has been shown that lung epithelial cells in culture can sense SARS-CoV-2 and produce type I IFNβ and type III IFNλ1, but this occurred only after the initiation of virus replication (Wahl et al., 2021; Yin et al., 2021; Hatton et al., 2021; Vanderheiden et al., 2020). This might indicate that multiple rounds of viral replication are needed before innate antiviral defense mechanisms are initiated. The relevance of this in vitro observation to the antiviral environment following viral exposure in vivo remains unclear.
In general, viruses are unable to easily infect and replicate in macrophages and DCs. This is also the case for SARS-CoV-2, and several studies have now shown that these innate immune cells express little to no ACE2 and TMPRSS2 (Abassi et al., 2020; Dalskov et al., 2020; Lu et al., 2021; Onodi et al., 2021; Severa et al., 2021; Van Der Sluis et al., 2022). However, DCs and macrophages are specialized in the uptake of foreign material, which usually leads to the triggering of PRRs located in their endosomal compartments. Myeloid cells can take up SARS-CoV-2 via several C-type lectins, including Dendritic Cell-Specific ICAM-3 (DC-SIGN), C-type lectin domain family 10 member A (CLEC10A), and Tweety family member 2 (TTYH2) (Lu et al., 2021). However, there are conflicting reports regarding the ability of myeloid cells to elicit the production of pro-inflammatory and antiviral cytokines in response to SARS-CoV-2 (Niles et al., 2021; Zheng et al., 2021a). Alveolar macrophages seem incapable of sensing SARS-CoV-2 (Dalskov et al., 2020). It has also been suggested that lung epithelial cells are needed for macrophages to produce antiviral cytokines after paracrine IFNβ feedback (Thorne et al., 2021). Others suggested that human lymphatic endothelial cells are needed for myeloid DCs to produce antiviral cytokines, possibly by sensing double-stranded RNA intermediates via TLR3 (Sposito et al., 2021). However, it has been hypothesized that myeloid cells play a major role in the overall pulmonary inflammatory response by contributing to local cell damage and innate sensing via the cGAS-STING pathway (Domizio et al., 2022) or via FcγR-mediated uptake (Junqueira et al., 2022).
pDCs can clearly sense SARS-CoV-2 and produce different pro-inflammatory cytokines and IFNs (Asano et al., 2021; Onodi et al., 2021; Severa et al., 2021; Van Der Sluis et al., 2022), although they are not permissive to virus infection because they lack expression of ACE2 and TMPRSS2 (Onodi et al., 2021; Severa et al., 2021; Van Der Sluis et al., 2022). As such, viral uptake by pDCs is likely mediated via endocytosis, although this has not been formally demonstrated. By using pDCs from patients carrying deleterious mutations in specific genes or genetically edited stem cell-generated pDCs, we and others have identified several critical factors required for pDCs to sense SARS-CoV-2 (Figure 1). Importantly, TLR7 allows pDCs to detect SARS-CoV-2 RNA (Asano et al., 2021; Fallerini et al., 2021; Van Der Sluis et al., 2022). Viral detection through TLR7 requires UNC93B1, possibly because of its role in controlling TLR7 trafficking (Onodi et al., 2021) followed by MyD88 and IRAK4 activation (Onodi et al., 2021; Van Der Sluis et al., 2022). Next, IRF7 is activated, which is needed for IFNα production by pDCs. However, SARS-CoV-2 sensing via TLR7 also induces production of CXCL10 and other cytokines (Mantovani et al., 2022; Onodi et al., 2021; Van Der Sluis et al., 2022). The RIG-I pathway that is important for SARS-CoV-2 sensing in epithelial cells seems not to play a role in the pDCs (Van Der Sluis et al., 2022). In contrast, TLR2, which has been reported to sense the SARS-CoV-2 envelope protein (Zheng et al., 2021b), seems to induce strong IL-6 production independent of TLR7 sensing (Van Der Sluis et al., 2022).
SARS-CoV-2 antagonizes IFN production by pDCs
Innate sensing of SARS-CoV-2 should result in the production of IFNs and the protection of permissive cells from infection. Indeed, SARS-CoV-2 induces IFN and ISG expression in the upper and lower respiratory tract in humans (Cheemarla et al., 2021; Delorey et al., 2021; Desai et al., 2020; Lieberman et al., 2020; Ziegler et al., 2021), and IFN production is associated with protection against SARS-CoV-2 (Bastard et al., 2020, 2021; Pairo-Castineira et al., 2021; Wang et al., 2021). In line with this, viral replication studies have shown that SARS-CoV-2 is highly sensitive to IFN-induced antiviral mechanisms both in vitro and in vivo. For example, rhinovirus-induced IFN protects human epithelial organoids from SARS-CoV-2 infection (Cheemarla et al., 2021). Pharmacological provision of IFN either by direct nasal IFN administration (Bessiere et al., 2021; Hatton et al., 2021) or indirect induction by innate immune agonists targeting the STING pathway (Li et al., 2021b) can also protect mice against primary infection. However, IFN administration offers less protection when administered after primary infection, as demonstrated in clinical trials that investigated the potential of subcutaneous injection of type III IFNλ1 (Feld et al., 2021; Jagannathan et al., 2021) and type I IFNβ (Ader et al., 2021; Consortium et al., 2021). Importantly, type I IFNβ can be correlated to a more severe disease outcome when administered to patients who required high-flow oxygen at time of hospitalization (Kalil et al., 2021). This indicates that clinical administration of IFN needs to be timed carefully in relation to disease development (Anjum et al., 2021; Sodeifian et al., 2022). Interestingly, inhaled nebulized IFNβ enhances the odds of survival in a randomized, double-blind, placebo-controlled, phase 2 pilot trial (Monk et al., 2021). Follow-up studies and similar trials with nebulized type I IFNα2b are in progress (ClinicalTrials.gov: NCT04988217, NCT05381363, and NCT04469491); the latter has given promising results in an uncontrolled exploratory study (Zhou et al., 2020b).
Multiple antiviral evasion strategies are employed by SARS-CoV-2, and these have been extensively reviewed elsewhere (Lowery et al., 2021; Kasuga et al., 2021; Kim and Shin, 2021). However, it is important to note that SARS-CoV-2 seems to employ a particular unusual evasion strategy to avoid the antiviral functions of pDCs. NRP1, also known as CD304, is a phenotypic marker of pDCs, and its engagement by anti-CD304 antibodies reduces pDC IFN production (Fanning et al., 2006; Grage-Griebenow et al., 2007; Van Der Sluis et al., 2022). Because SARS-CoV-2 also binds to CD304 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020), the virus can exploit this pathway and dampen IFN production to promote viral replication. We recently showed that removal of CD304 from pDCs, using CRISPR-Cas9, enhanced IFN production by pDCs exposed to SARS-CoV-2 (Van Der Sluis et al., 2022). The exact mechanism behind this phenomenon is still unclear. However, our data indicate a link between viral spike proteins binding to CD304 and intracellular signaling that impairs IFN production.
Fate of pDCs during COVID-19: The pDC desert phenomenon
In theory, pDCs are ideal candidates to sense and protect against a respiratory viral infection such as SARS-CoV-2. However, pDCs could also potentially contribute to lung pathology through excessive or prolonged production of type I IFN. Type I IFN-induced pathology has been shown in the context of SARS-CoV-2 (Broggi et al., 2020; Carvalho et al., 2021; Lucas et al., 2020; Major et al., 2020; Sposito et al., 2021; Domizio et al., 2022) and other viral infections like influenza (Davidson et al., 2015; Lee and Ashkar, 2018; Mcnab et al., 2015). The detrimental effects can include tissue damage (Broggi et al., 2020; Major et al., 2020) and a state of IFN desensitization (Sandler et al., 2014). As described earlier, data from murine models indicate that pDC subsets mainly provide protection during viral infections, including infections with coronaviruses (Cervantes-Barragan et al., 2012; Chen et al., 2010; Swiecki et al., 2010; Wang et al., 2006). Speculations regarding the role of pDCs during SARS-CoV-2 are primarily based on correlations between cell numbers and disease severity, as well as studies that explore the functionality of pDCs isolated from COVID-19 patients ex vivo. For the purpose of this review, we have summarized articles with information pertaining to pDCs and COVID-19. This information is provided in alphabetical order in Table 1 . A plurality of articles report on the frequency of pDCs in blood, from various disease stages, determined by flow cytometry or their unique genetic signature in, for example, RNA sequence analysis. Of the 23 articles that quantified pDC frequencies or cell counts in peripheral blood during COVID-19, 19 observed a significant reduction in circulating pDCs in infected individuals (Arunachalam et al., 2020; Benard et al., 2021; Hadjadj et al., 2020; Kreutmair et al., 2021; Kulkarni-Munje et al., 2021; Kvedaraite et al., 2021; Laing et al., 2020; Li et al., 2021c; Matic et al., 2020; Perez-Gomez et al., 2021; Peruzzi et al., 2020; Rebillard et al., 2021; Severa et al., 2021; Van Der Sluis et al., 2022; Vono et al., 2021; Wilk et al., 2020; Winheim et al., 2021; Zingaropoli et al., 2021; Sanchez-Cerrillo et al., 2020). Three studies observed a similar trend that was not statistically significant (Schulte-Schrepping et al., 2020; Xu et al., 2020; Zhou et al., 2020c), and one study observed no change (Shi et al., 2021). When grouped by severity of disease, 10 studies observed that pDC numbers declined more in patients with severe disease than with mild/moderate disease (Benard et al., 2021; Kreutmair et al., 2021; Kulkarni-Munje et al., 2021; Kvedaraite et al., 2021; Laing et al., 2020; Peruzzi et al., 2020; Severa et al., 2021; Van Der Sluis et al., 2022; Winheim et al., 2021; Zingaropoli et al., 2021), whereas three studies did not observe this trend (Hadjadj et al., 2020; Rebillard et al., 2021; Wilk et al., 2020). In general, this information strongly suggests that pDC numbers decline in peripheral blood during SARS-CoV-2 infection, and that this is augmented during severe disease, resulting in a phenomenon we call “the pDC desert” (Figure 2 ). The mechanisms underlying the induction of the “pDC desert” are currently unclear. However, “the pDC desert” has also been reported during infection with other RNA viruses, such as HIV and HCV (Kanto et al., 2004; Van Der Sluis et al., 2022; Ulsenheimer et al., 2005). The “pDC desert” can also persist throughout the chronic stage of infections with hepatitis B virus (HBV), HCV, or HIV. As such, it will be interesting to evaluate the pDC population in people suffering from long COVID.
Table 1.
Fate of pDCs during COVID-19
| Article | Description of COVID-19 disease stage and other study participants | Sample collection | Observations regarding pDCs |
|---|---|---|---|
| Arunachalam et al. (2020) |
|
blood was collected |
|
| Bénard et al. (2021) |
|
blood collection at symptom onset (≤24 h) and 1–7 days thereafter, or after recovery BALF was collected from non-COVID-19 individuals with other pulmonary diseases |
|
| Hadjadj et al. (2020) |
|
blood samples from 8–12 days after symptom onset were analyzed |
|
| Kreutmair et al. (2021) |
|
first blood sample (t1) was collected at 0–2 days after hospital admission, t2 after 3–5 days, t3 after 6–9 days, t4 after 11–15 days, and t5 after 19–96 days |
|
| Kulkarni-Munje et al. (2021) |
|
blood was collected at time of hospitalization |
|
| Kvedaraite et al. (2021) |
|
blood was collected at a mean of 12.9 (range 6–19) days for moderate and 14.6 (5–24) days for severe cases after symptom onset |
|
| Laing et al. (2020) |
|
blood collection within 24 h of study recruitment and at days 3 and 9 thereafter |
|
| Li et al. (2021c) |
|
blood was collected at 27–47 days after symptom onset or after positive PCR result; all patients were in the recovery stage and without symptoms at time of sample collection |
|
| Liao et al. (2020) |
|
BALF was collected |
|
| Matic et al. (2020) |
|
blood was collected the first day of hospitalization |
|
| Perez-Gomez et al. (2021) |
|
blood was collected at a median of 3 (IQR 2–23) days after hospitalization and 14 (9–31) days after symptom onset for 7-month follow-up, blood was collected at a median of 201 (181–221) days after hospitalization and 208 (189–230) days after symptom onset |
|
| Peruzzi et al. (2020) |
|
blood was collected |
|
| Rebillard et al. (2021) |
|
first blood sample was collected at a median of 12 (range 3–23) days, and the follow-up sample 11.5 (3–23) days after symptom onset, and 6.5 (range 2–23) and 6 (2–23) days after positive PCR test result |
|
| Saichi et al. (2021) |
|
blood was collected at time of hospital admission (approximately 10–12 days after symptom onset) and 4 days thereafter |
|
| Sanchez-Cerillo et al. (2020) |
|
blood was collected from all patients BALF sample was collected from 23 of the 24 critical patients |
|
| Schulte-Schrepping et al. (2020) |
|
blood was collected |
|
| Severa et al. (2021) |
|
blood was collected |
|
| Shi et al. (2021) |
|
blood was collected |
|
| Van Der Sluis et al. (2022) |
|
blood was collected at time of hospitalization (0–4 days after symptom onset, n = 22; 5–8 days, n = 39; 9–12 days, n = 39; and ≥13 days, n = 16) and 5 days thereafter |
|
| Vono et al. (2021) |
|
blood was collected at 5 visits: v1, 0–5 days after symptom onset; v2, 6–14 days; v3, 15–22 days; v4, 23–35 days; v5, 36–81 days |
|
| Wilk et al. (2020) |
|
blood was collected ≤48 h after hospital admission and ventilation status included in some of the analyses; from 1 patient, 2 samples were included, of which the first was without ventilation and the second with ventilation |
|
| Winheim et al. (2021) |
|
blood collection within 3 weeks of positive PCR test result and at various time points thereafter |
|
| Xu et al. (2020) |
|
blood was collected from all cases BALF was collected from 4 mild and 10 severe cases |
|
| Zhou et al., 2020c |
|
blood was collected at a median of 9 and 15 days after symptom onset for mild and severe cases, respectively blood was collected at a median of 13 and 30 days after symptom onset for APs and CPs, respectively |
|
| Zingaropoli et al. (2021) |
|
blood was collected on hospital admission |
|
| Zulu et al. (2021) |
|
blood was collected |
|
Articles are listed in alphabetical order.
AP, acute patient; ARDS, acute respiratory distress syndrome; BALF, bronchoalveolar lavage fluid; BMI, body mass index; CP, convalescent patient; CRP, C-reactive protein; HC, healthy control subject; IQR, interquartile range; LRTI, lower respiratory tract infection; WHO, World Health Organization; PBMC, Peripheral Blood Mononuclear Cells; mTORC, mammalian target of rapamycin complex 1; cDC; classical DC.
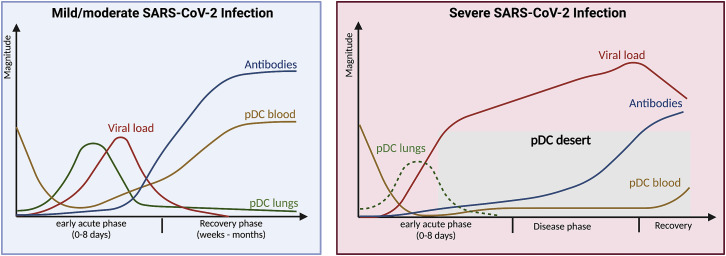
Figure 2.
Fate of pDCs during COVID-19 and the pDC desert phenomenon
During a mild/moderate infection with SARS-CoV-2 (blue panel), the early acute phase of viral infection is characterized by the decline of pDCs from peripheral blood (orange/yellow line), as they migrate to the lungs (green line) to aid in the respiratory tract’s antiviral mechanisms to inhibit virus replication and reduce viral loads (red line). After mounting adaptive immunity and antibody production (blue line), the virus will be cleared and the recovery phase begins. This includes disappearance of pDCs from the lungs, by unknown mechanisms, and increased peripheral numbers facilitated by replenishment from the bone marrow. With a severe SARS-CoV-2 infection (red panel), pDC numbers decline in peripheral blood to lower levels, as compared with a mild/moderate infection. At the same time, pDCs seem to be absent from the lungs (green dotted line). During this time of severe disease, pDC numbers remain low, akin to a “pDC desert” (gray rectangle), until the recovery phase begins and peripheral pDC numbers increase. Figure was created with BioRender.com.
If the pDCs are no longer in peripheral blood, where do they go? One possibility is that pDCs migrate to the lungs to contribute to the antiviral defense. The presence of pDCs in bronchoalveolar lavage fluid (BALF) of COVID-19 patients has been investigated. Here, pDC frequency was noted to increase in the lungs during mild COVID-19 (Liao et al., 2020), and this has also been observed during non-COVID-19 pulmonary disease (Benard et al., 2021). In contrast with these observations, pDC frequency in the lungs drastically declines during severe COVID-19 (Liao et al., 2020; Sanchez-Cerrillo et al., 2020). Collectively, this suggests that early pDC infiltration into the lungs is beneficial during SARS-CoV-2 infection.
Most investigators have reported that pDC numbers increase during COVID-19 recovery (i.e., between 7 and 42 days after symptom onset) (Benard et al., 2021; Laing et al., 2020; Li et al., 2021c; Perez-Gomez et al., 2021; Shi et al., 2021; Van Der Sluis et al., 2022; Winheim et al., 2021; Zhou et al., 2020c). Some studies have even reported that pDC numbers are restored to levels similar to uninfected controls (Laing et al., 2020; Li et al., 2021c; Shi et al., 2021; Winheim et al., 2021), with the exception of one study that reported that pDC numbers remained reduced for up to 7 months postinfection (Perez-Gomez et al., 2021). The reasons for these different observations are currently unclear.
Additional unresolved issues are the temporal association between severe COVID-19 and lower pDC numbers, potential defects in pDC migration, or pDCs that are more prone to undergoing cell death on SARS-CoV-2 infection. Interestingly, one study reported that pDCs from severe cases displayed increased pro-apoptotic pathways (Saichi et al., 2021). Another study reported that pDCs from mild COVID-19 cases express higher levels of the Fas receptor CD95, as compared with pDCs from uninfected people, and Fas expression increased with disease severity (Kreutmair et al., 2021). It is not known if other cells, such as lung epithelial cells, upregulate the ligand for Fas during COVID-19 and induce pDC apoptosis.
Not only are pDC numbers negatively affected during COVID-19, their function is also altered. This indicates pDC exhaustion or anergy. Ex vivo stimulation of pDCs isolated from severe COVID-19 cases results in reduced IFNα and TNF-α production (Arunachalam et al., 2020). Single-cell RNA-seq has shown that pDCs from severe cases are enriched for inflammatory signaling pathways, such as TNF-α, IL-6, p53, and mTORC, whereas IFNα-related and MYC-related signaling pathways are reduced (Saichi et al., 2021). This suggests that pDCs remaining during severe disease are more likely to produce pro-inflammatory cytokines and less likely to produce antiviral IFNs. This response pattern potentially contributes to the development of severe COVID-19. One study has reported that pDCs from older (i.e., ≥60 years) COVID-19 patients more frequently express both TNF-α and IL-6 after stimulation with various TLR ligands, as compared with pDCs from younger (i.e., <60 years) COVID-19 patients (Zulu et al., 2021). Interestingly, increased TNF-α/IL-6 and decreased IFN production correlated positively with body mass index (BMI). As such, pDCs in older and obese SARS-CoV-2-infected individuals appear to be more likely to produce pro-inflammatory cytokines than younger and leaner individuals. This is consistent with known risk factors for severe COVID-19. In some cases, functional changes in pDCs have been noted to persist for up to 27–47 days postinfection, even after symptoms have disappeared (Li et al., 2021c).
An important question pertaining to pDC migration is: are pDCs a second line of immune cell sensors? Because the entry route of SARS-CoV-2 is through the respiratory tract, multiple immune cells might initiate antiviral detection and release factors facilitating pDC infiltration (Lommatzsch et al., 2007). Benard et al. (2021) suggested that increased IL-3 levels in the lungs could be the main driver for pDC recruitment. They supported this contention using murine models, showing that pDCs are recruited to the lungs via an IL-3- and CXCL12-dependent mechanism. The authors also found that IL-3 was associated with COVID-19 survival (Benard et al., 2021). Furthermore, the IL-3 receptor, CD123, has been shown by others to be expressed at higher levels during the acute phase of COVID-19 (Shi et al., 2021). Moreover, IL-3 and pDC numbers decline with age (Benard et al., 2021; Li et al., 2021c; Garbe et al., 2012), which is one of the risk factors for severe COVID-19. However, it is also possible that low numbers of pDCs reside in the lungs under steady-state conditions prior to infection, and they facilitate a feedback response that supports pDC migration from peripheral blood to the lungs. Future studies on human pDC migration may be able to answer this question.
Following viral sensing through TLR7, pDCs can become functionally exhausted (Macal et al., 2018). In mice infected with LCMV, pDCs rapidly produce type I IFN. However, during chronic infection, TLR7-mediated type I IFN signaling limits de novo pDC generation in the bone marrow and supports the self-renewal of exhausted pDCs in the periphery. The lack of pDC replenishment and their loss of function caused by exhaustion are linked to downregulation of the E2-2 transcription factor. It has been hypothesized that these mechanisms may explain the decreased pDC numbers in the periphery during chronic infection with viruses such as HBV, HCV, and HIV. Thus, it may be informative to assess pDC exhaustion in severe COVID-19.
Sex-related hormones can also account for differences in pDC function, and pDCs obtained from healthy female donors produce more type I IFN in response to TLR7 stimulation than pDCs obtained from men (Griesbeck et al., 2015; Guery, 2021; Webb et al., 2018). This appears to be beneficial during the acute phase of viral infection but can have detrimental effects when the infections persists (Meier et al., 2009) or may increase the likelihood of developing IFN-mediated diseases (Webb et al., 2018). Similarly, sex-related differences have been described in the context of COVID-19, where mortality is reported to be higher among males than females (O’driscoll et al., 2021; Gebhard et al., 2020). Although pDCs may provide some additional protection via increased IFN production during acute infection, other factors between males and females are also likely to contribute, such as immunological differences in non-classical monocyte and T cell activation (Takahashi et al., 2020) or behavioral differences (Galasso et al., 2020). This suggests that it may be beneficial to include sex- and gender-sensitive analysis in the ongoing therapeutic treatment studies of COVID-19 (Gebhard et al., 2020).
The role of IFN during COVID-19 is complex and extensively reviewed by others (Wong and Perlman, 2022; Zanoni, 2021; Zhang et al., 2022). Although pDCs are important producers of IFN, they also fulfill other immunological functions, such as the activation of cytotoxic T cells and anti-inflammatory regulatory T cells (Swiecki and Colonna, 2015). Unfortunately, these functions have not yet been studied in the context of COVID-19. Current literature indicates that on infection with SARS-CoV-2, pDCs migrate from peripheral blood into the respiratory tract and contribute to the suppression of SARS-CoV-2 replication via their IFN production. However, it is unclear whether and how pDCs could contribute to the sustained detrimental IFN production that accompanies severe disease. We know that pDC numbers decline drastically in severe disease, and the pDCs that remain seem to be impaired when it comes to IFN production (see Table 1). Interestingly, the remaining pDCs appear to be more prone to produce other inflammatory cytokines, such as TNF-α and IL-6, and it is possible that such effect could contribute to a limited extent to COVID-19 severity via non-IFN pro-inflammatory cytokines (see Table 1).
Concluding remarks
Despite pDCs having pro-inflammatory properties, there is limited evidence that pDCs drive pathogenicity during COVID-19. Instead, pDCs appear to play a beneficial role, likely through their ability to produce high amounts of type I IFNs that restrict SARS-CoV-2 spread during the acute phase of infection. Future studies should determine whether the low frequency of pDCs in the respiratory tract, as observed in some cases of severe COVID-19, contributes to COVID-19 pathogenicity.
Acknowledgments
We apologize to the individuals whose work was not cited in this article. We thank Prof. Martin Tolstrup (Aarhus University, Denmark) for critical reading of the manuscript. This project has received funding from the Danish Independent Research Fund (4183-00275B) and the Lundbeck Foundation (R238-2016-2708).
Declaration of interests
M.R.J. is a founder and shareholder of UNIKUM Therapeutics. The remaining authors declare no competing interests.
References
- Abassi Z., Knaney Y., Karram T., Heyman S.N. The lung macrophage in SARS-CoV-2 infection: a friend or a foe? Front. Immunol. 2020;11:1312. doi: 10.3389/fimmu.2020.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas A., Vu Manh T.P., Valente M., Collinet N., Attaf N., Dong C., Naciri K., Chelbi R., Brelurut G., Cervera-Marzal I., et al. The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection. Nat. Immunol. 2020;21:983–997. doi: 10.1038/s41590-020-0731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ader F., Peiffer-Smadja N., Poissy J., Bouscambert-Duchamp M., Belhadi D., Diallo A., Delmas C., Saillard J., Dechanet A., Mercier N., Dupont A., et al. An open-label randomized controlled trial of the effect of lopinavir/ritonavir, lopinavir/ritonavir plus IFN-beta-1a and hydroxychloroquine in hospitalized patients with COVID-19. Clin. Microbiol. Infect. 2021;27:1826–1837. doi: 10.1016/j.cmi.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alculumbre S.G., Saint-André V., Di Domizio J., Vargas P., Sirven P., Bost P., Maurin M., Maiuri P., Wery M., Roman M.S., et al. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol. 2018;19:63–75. doi: 10.1038/s41590-017-0012-z. [DOI] [PubMed] [Google Scholar]
- Anjum F.R., Anam S., Abbas G., Mahmood M.S., Rahman S.U., Goraya M.U., Abdullah R.M., Luqman M., Ali A., Akram M.K., Chaudhry T.H. Type I IFNs: a blessing in disguise or partner in crime in MERS-CoV-SARS-CoV-and SARS-CoV-2-induced pathology and potential use of type I IFNs in synergism with IFN-gamma as a novel antiviral approach against COVID-19. Viral Immunol. 2021;34:321–329. doi: 10.1089/vim.2020.0085. [DOI] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021;6:eabl4348. doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Paturel C., Boonstra A., Dalod M., Durand I., Yessaad N., Dezutter-Dambuyant C., Vicari A., O'garra A., Biron C., Brière F., Trinchieri G. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 2001;2:1144–1150. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- Asselin-Paturel C., Brizard G., Pin J.J., Brière F., Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol. 2003;171:6466–6477. doi: 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- Assil S., Coléon S., Dong C., Décembre E., Sherry L., Allatif O., Webster B., Dreux M. Plasmacytoid dendritic cells and infected cells form an interferogenic synapse required for antiviral responses. Cell Host Microbe. 2019;25:730–745.e6. doi: 10.1016/j.chom.2019.03.005. [DOI] [PubMed] [Google Scholar]
- Attaway A.H., Scheraga R.G., Bhimraj A., Biehl M., Hatipoğlu U. Severe covid-19 pneumonia: pathogenesis and clinical management. BMJ. 2021;372:n436. doi: 10.1136/bmj.n436. [DOI] [PubMed] [Google Scholar]
- Azkur A.K., Akdis M., Azkur D., Sokolowska M., Van De Veen W., Brüggen M.C., O'mahony L., Gao Y., Nadeau K., Akdis C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M., Liu Y.J. Regulation of TLR7/9 signaling in plasmacytoid dendritic cells. Protein Cell. 2013;4:40–52. doi: 10.1007/s13238-012-2104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Orlova E., Sozaeva L., Lévy R., James A., Schmitt M.M., Ochoa S., Kareva M., Rodina Y., Gervais A., Le Voyer T., et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 2021;218:e20210554. doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénard A., Jacobsen A., Brunner M., Krautz C., Klösch B., Swierzy I., Naschberger E., Podolska M.J., Kouhestani D., David P., et al. Interleukin-3 is a predictive marker for severity and outcome during SARS-CoV-2 infections. Nat. Commun. 2021;12:1112. doi: 10.1038/s41467-021-21310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessiere P., Wasniewski M., Picard-Meyer E., Servat A., Figueroa T., Foret-Lucas C., Coggon A., Lesellier S., Boué F., Cebron N., et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog. 2021;17:e1009427. doi: 10.1371/journal.ppat.1009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieback K., Lien E., Klagge I.M., Avota E., Schneider-Schaulies J., Duprex W.P., Wagner H., Kirschning C.J., Ter Meulen V., Schneider-Schaulies S. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 2002;76:8729–8736. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Giurisato E., Cella M., Schreiber R.D., Shaw A.S., Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- Bode C., Fox M., Tewary P., Steinhagen A., Ellerkmann R.K., Klinman D., Baumgarten G., Hornung V., Steinhagen F. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur. J. Immunol. 2016;46:1615–1621. doi: 10.1002/eji.201546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme K.W., Guerrero M., Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 2006;177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- Brawand P., Fitzpatrick D.R., Greenfield B.W., Brasel K., Maliszewski C.R., De Smedt T. Murine plasmacytoid pre-dendritic cells generated from Flt3 ligand-supplemented bone marrow cultures are immature APCs. J. Immunol. 2002;169:6711–6719. doi: 10.4049/jimmunol.169.12.6711. [DOI] [PubMed] [Google Scholar]
- Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni D., Chazal M., Sinigaglia L., Chauveau L., Schwartz O., Desprès P., Jouvenet N. Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons. Sci. Signal. 2015;8:ra25. doi: 10.1126/scisignal.aaa1552. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Van Der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Jarrossay D., Facchetti F., Alebardi O., Nakajima H., Lanzavecchia A., Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Lewis K.L., Firner S., Thiel V., Hugues S., Reith W., Ludewig B., Reizis B. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci. USA. 2012;109:3012–3017. doi: 10.1073/pnas.1117359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaperot L., Blum A., Manches O., Lui G., Angel J., Molens J.P., Plumas J. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J. Immunol. 2006;176:248–255. doi: 10.4049/jimmunol.176.1.248. [DOI] [PubMed] [Google Scholar]
- Cheemarla N.R., Watkins T.A., Mihaylova V.T., Wang B., Zhao D., Wang G., Landry M.L., Foxman E.F. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med. 2021;218:e20210583. doi: 10.1084/jem.20210583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., Den Hollander N.S., Kant S.G., et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, W. H. O. S. T. Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., et al. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytlak U., Resteu A., Pagan S., Green K., Milne P., Maisuria S., Mcdonald D., Hulme G., Filby A., Carpenter B., et al. Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. 2020;53:353–370.e8. doi: 10.1016/j.immuni.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Rosa Mesquita R., Francelino Silva Junior L.C., Santos Santana F.M., Farias De Oliveira T., Campos Alcântara R., Monteiro Arnozo G., Rodrigues Da Silva Filho E., Galdino Dos Santos A.G., Oliveira Da Cunha E.J., Salgueiro De Aquino S.H., Freire De Souza C.D. Clinical manifestations of COVID-19 in the general population: systematic review. Wien Klin. Wochenschr. 2021;133:377–382. doi: 10.1007/s00508-020-01760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalskov L., Møhlenberg M., Thyrsted J., Blay-Cadanet J., Poulsen E.T., Folkersen B.H., Skaarup S.H., Olagnier D., Reinert L., Enghild J.J., et al. SARS-CoV-2 evades immune detection in alveolar macrophages. EMBO Rep. 2020;21:e51252. doi: 10.15252/embr.202051252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S., Maini M.K., Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 2015;35:252–264. doi: 10.1089/jir.2014.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal E.M., Lahl K., Narváez C.F., Butcher E.C., Greenberg H.B. Plasmacytoid dendritic cells promote rotavirus-induced human and murine B cell responses. J. Clin. Invest. 2013;123:2464–2474. doi: 10.1172/JCI60945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb P., Dai J., Singh S., Kalyoussef E., Fitzgerald-Bocarsly P. Triggering of the cGAS-STING pathway in human plasmacytoid dendritic cells inhibits TLR9-mediated IFN production. J. Immunol. 2020;205:223–236. doi: 10.4049/jimmunol.1800933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sole F., Farcomeni A., Loffredo L., Carnevale R., Menichelli D., Vicario T., Pignatelli P., Pastori D. Features of severe COVID-19: a systematic review and meta-analysis. Eur. J. Clin. Invest. 2020;50:e13378. doi: 10.1111/eci.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., Abbondanza D., Fleming S.J., Subramanian A., Montoro D.T., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N., Neyaz A., Szabolcs A., Shih A.R., Chen J.H., Thapar V., Nieman L.T., Solovyov A., Mehta A., Lieb D.J., Kulkarni A.S., et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat. Commun. 2020;11:6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domizio J.D., Gulen M.F., Saidoune F., Thacker V.V., Yatim A., Sharma K., Nass T., Guenova E., Schaller M., Conrad C., et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghy H., Bosnjak L., Harman A.N., Marsden V., Tyring S.K., Meng T.C., Cunningham A.L. Role for plasmacytoid dendritic cells in the immune control of recurrent human herpes simplex virus infection. J. Virol. 2009;83:1952–1961. doi: 10.1128/JVI.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dress R.J., Dutertre C.A., Giladi A., Schlitzer A., Low I., Shadan N.B., Tay A., Lum J., Kairi M.F.B.M., Hwang Y.Y., et al. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 2019;20:852–864. doi: 10.1038/s41590-019-0420-3. [DOI] [PubMed] [Google Scholar]
- Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., Paciosi F., Schiaroli E., Baldassarri M., Fava F., Palmieri M., et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife. 2021;10:e67569. doi: 10.7554/eLife.67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S.L., George T.C., Feng D., Feldman S.B., Megjugorac N.J., Izaguirre A.G., Fitzgerald-Bocarsly P. Receptor cross-linking on human plasmacytoid dendritic cells leads to the regulation of IFN-alpha production. J. Immunol. 2006;177:5829–5839. doi: 10.4049/jimmunol.177.9.5829. [DOI] [PubMed] [Google Scholar]
- Feld J.J., Kandel C., Biondi M.J., Kozak R.A., Zahoor M.A., Lemieux C., Borgia S.M., Boggild A.K., Powis J., Mccready J., et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir. Med. 2021;9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P., Feldman M., Mendelsohn M., Curl S., Lopez C. Human mononuclear cells which produce interferon-alpha during NK(HSV-FS) assays are HLA-DR positive cells distinct from cytolytic natural killer effectors. J. Leukoc. Biol. 1988;43:323–334. doi: 10.1002/jlb.43.4.323. [DOI] [PubMed] [Google Scholar]
- Frenz T., Graalmann L., Detje C.N., Döring M., Grabski E., Scheu S., Kalinke U. Independent of plasmacytoid dendritic cell (pDC) infection, pDC triggered by virus-infected cells mount enhanced type I IFN responses of different composition as opposed to pDC stimulated with free virus. J. Immunol. 2014;193:2496–2503. doi: 10.4049/jimmunol.1400215. [DOI] [PubMed] [Google Scholar]
- Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Galasso V., Pons V., Profeta P., Becher M., Brouard S., Foucault M. Gender differences in COVID-19 attitudes and behavior: panel evidence from eight countries. Proc. Natl. Acad. Sci. USA. 2020;117:27285–27291. doi: 10.1073/pnas.2012520117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Majchrzak-Kita B., Fish E.N., Gommerman J.L. Dynamic accumulation of plasmacytoid dendritic cells in lymph nodes is regulated by interferon-beta. Blood. 2009;114:2623–2631. doi: 10.1182/blood-2008-10-183301. [DOI] [PubMed] [Google Scholar]
- Garbe K., Bratke K., Wagner S., Virchow J.C., Lommatzsch M. Plasmacytoid dendritic cells and their Toll-like receptor 9 expression selectively decrease with age. Hum. Immunol. 2012;73:493–497. doi: 10.1016/j.humimm.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Gebhard C., Regitz-Zagrosek V., Neuhauser H.K., Morgan R., Klein S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex Differ. 2020;11:29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M., Boonstra A., Paturel C., Antonenko S., Xu X.L., Trinchieri G., O'garra A., Liu Y.J. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grage-Griebenow E., Löseke S., Kauth M., Gehlhar K., Zawatzky R., Bufe A. Anti-BDCA-4 (neuropilin-1) antibody can suppress virus-induced IFN-alpha production of plasmacytoid dendritic cells. Immunol. Cell Biol. 2007;85:383–390. doi: 10.1038/sj.icb.7100048. [DOI] [PubMed] [Google Scholar]
- Griesbeck M., Ziegler S., Laffont S., Smith N., Chauveau L., Tomezsko P., Sharei A., Kourjian G., Porichis F., Hart M., et al. Sex differences in plasmacytoid dendritic cell levels of IRF5 drive higher IFN-alpha production in women. J. Immunol. 2015;195:5327–5336. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouard G., Rissoan M.C., Filgueira L., Durand I., Banchereau J., Liu Y.J. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzes K.M., Sanin D.E., Kabat A.M., Stanczak M.A., Edwards-Hicks J., Matsushita M., Hackl A., Hässler F., Knoke K., Zahalka S., et al. Plasmacytoid dendritic cell activation is dependent on coordinated expression of distinct amino acid transporters. Immunity. 2021;54:2514–2530.e7. doi: 10.1016/j.immuni.2021.10.009. [DOI] [PubMed] [Google Scholar]
- Guery J.C. Sex differences in primary HIV infection: revisiting the role of TLR7-driven type 1 IFN production by plasmacytoid dendritic cells in women. Front. Immunol. 2021;12:729233. doi: 10.3389/fimmu.2021.729233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haist K.C., Carpentier K.S., Davenport B.J., Morrison T.E. Plasmacytoid dendritic cells mediate control of Ross River virus infection via a type I interferon-dependent, MAVS-independent mechanism. J. Virol. 2021;95:e01538-20. doi: 10.1128/JVI.01538-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Miyajima A. Two distinct functional high affinity receptors for mouse interleukin-3 (IL-3) EMBO J. 1992;11:1875–1884. doi: 10.1002/j.1460-2075.1992.tb05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy A.W., Graham D.R., Shearer G.M., Herbeuval J.P. HIV turns plasmacytoid dendritic cells (pDC) into TRAIL-expressing killer pDC and down-regulates HIV coreceptors by Toll-like receptor 7-induced IFN-alpha. Proc. Natl. Acad. Sci. USA. 2007;104:17453–17458. doi: 10.1073/pnas.0707244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton C.F., Botting R.A., Dueñas M.E., Haq I.J., Verdon B., Thompson B.J., Spegarova J.S., Gothe F., Stephenson E., Gardner A.I., et al. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat. Commun. 2021;12:7092. doi: 10.1038/s41467-021-27318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Kanzler H., Duramad O., Cao W., Liu Y.J. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood. 2006;107:2423–2431. doi: 10.1182/blood-2005-07-2709. [DOI] [PubMed] [Google Scholar]
- Jagannathan P., Andrews J.R., Bonilla H., Hedlin H., Jacobson K.B., Balasubramanian V., Purington N., Kamble S., De Vries C.R., Quintero O., Feng K., et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira C., Crespo Â., Ranjbar S., De Lacerda L.B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M.R., Ho F., et al. FcgammaR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–584. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil A.C., Mehta A.K., Patterson T.F., Erdmann N., Gomez C.A., Jain M.K., Wolfe C.R., Ruiz-Palacios G.M., Kline S., Regalado Pineda J., et al. Efficacy of interferon beta-1a plus remdesivir compared with remdesivir alone in hospitalised adults with COVID-19: a double-bind, randomised, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2021;9:1365–1376. doi: 10.1016/S2213-2600(21)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanto T., Inoue M., Miyatake H., Sato A., Sakakibara M., Yakushijin T., Oki C., Itose I., Hiramatsu N., Takehara T., et al. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J. Infect. Dis. 2004;190:1919–1926. doi: 10.1086/425425. [DOI] [PubMed] [Google Scholar]
- Karrich J.J., Jachimowski L.C.M., Uittenbogaart C.H., Blom B. The plasmacytoid dendritic cell as the Swiss army knife of the immune system: molecular regulation of its multifaceted functions. J. Immunol. 2014;193:5772–5778. doi: 10.4049/jimmunol.1401541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga Y., Zhu B., Jang K.J., Yoo J.S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021;53:723–736. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kilbourne E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.U., Kim M.J., Ra S.H., Lee J., Bae S., Jung J., Kim S.H. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin. Microbiol. Infect. 2020;26:948–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kaiser V., Beier E., Bechheim M., Guenthner-Biller M., Ablasser A., Berger M., Endres S., Hartmann G., Hornung V. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. Eur. J. Immunol. 2014;44:807–818. doi: 10.1002/eji.201343806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Shin E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021;53:750–760. doi: 10.1038/s12276-021-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutmair S., Unger S., Núñez N.G., Ingelfinger F., Alberti C., De Feo D., Krishnarajah S., Kauffmann M., Friebel E., et al. Distinct immunological signatures discriminate severe COVID-19 from non-SARS-CoV-2-driven critical pneumonia. Immunity. 2021;54:1578–1593.e5. doi: 10.1016/j.immuni.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A., Veeraswamy R., Pekosz A., Kanagawa O., Unanue E.R., Colonna M., Cella M. Interferon-producing cells fail to induce proliferation of naive T cells but can promote expansion and T helper 1 differentiation of antigen-experienced unpolarized T cells. J. Exp. Med. 2003;197:899–906. doi: 10.1084/jem.20021091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni-Munje A., Palkar S., Shrivastava S., Lalwani S., Mishra A.C., Arankalle V.A. Disease-duration based comparison of subsets of immune cells in SARS CoV-2 infected patients presenting with mild or severe symptoms identifies prognostic markers for severity. Immun. Inflamm. Dis. 2021;9:419–434. doi: 10.1002/iid3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Takeuchi O., Kato H., Kumar H., Matsui K., Morii E., Aozasa K., Kawai T., Akira S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Kvedaraite E., Hertwig L., Sinha I., Ponzetta A., Hed Myrberg I., Lourda M., Dzidic M., Akber M., Klingström J., Folkesson E., et al. Major alterations in the mononuclear phagocyte landscape associated with COVID-19 severity. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2018587118. e2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., Mckenzie D.R., Hayday T.S., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Laustsen A., Bak R.O., Krapp C., Kjær L., Egedahl J.H., Petersen C.C., Pillai S., Tang H.Q., Uldbjerg N., et al. Interferon priming is essential for human CD34+ cell-derived plasmacytoid dendritic cell maturation and function. Nat. Commun. 2018;9:3525. doi: 10.1038/s41467-018-05816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J., Ashkar A.A. The dual nature of type I and type II interferons. Front. Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni V., Gianni T., Salvioli S., Campadelli-Fiume G. Herpes simplex virus glycoproteins gH/gL and gB bind Toll-like receptor 2, and soluble gH/gL is sufficient to activate NF-kappaB. J. Virol. 2012;86:6555–6562. doi: 10.1128/JVI.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leylek R., Alcántara-Hernández M., Lanzar Z., Lüdtke A., Perez O.A., Reizis B., Idoyaga J. Integrated cross-species analysis identifies a conserved transitional dendritic cell population. Cell Rep. 2019;29:3736–3750.e8. doi: 10.1016/j.celrep.2019.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wang G., Sivasami P., Ramirez R.N., Zhang Y., Benoist C., Mathis D. Interferon-alpha-producing plasmacytoid dendritic cells drive the loss of adipose tissue regulatory T cells during obesity. Cell Metab. 2021;33:1610–1623.e5. doi: 10.1016/j.cmet.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ferretti M., Ying B., Descamps H., Lee E., Dittmar M., Lee J.S., Whig K., Kamalia B., Dohnalová L., Uhr G., Zarkoob H., et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021;6:eabi9007. doi: 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Garg M., Jia T., Liao Q., Yuan L., Li M., Wu Z., Wu W., Bi Y., George N., Papatheodorou I., et al. Single-cell analysis reveals the immune characteristics of myeloid cells and memory T cells in recovered COVID-19 patients with different severities. Front. Immunol. 2021;12:781432. doi: 10.3389/fimmu.2021.781432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lieberman N.A.P., Peddu V., Xie H., Shrestha L., Huang M.L., Mears M.C., Cajimat M.N., Bente D.A., Shi P.Y., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18:e3000849. doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Liu Y., Qin C., Rao Y., Ngo C., Feng J.J., Zhao J., Zhang S., Wang T.Y., Carriere J., Savas A.C., Zarinfar M., et al. SARS-CoV-2 Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. mBio. 2021;12:e0233521. doi: 10.1128/mBio.02335-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommatzsch M., Bratke K., Bier A., Julius P., Kuepper M., Luttmann W., Virchow J.C. Airway dendritic cell phenotypes in inflammatory diseases of the human lung. Eur. Respir. J. 2007;30:878–886. doi: 10.1183/09031936.00036307. [DOI] [PubMed] [Google Scholar]
- Lowery S.A., Sariol A., Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: implications for COVID-19. Cell Host Microbe. 2021;29:1052–1062. doi: 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., Ran X., Damani-Yokota P., Tang H., Karakousi T., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304–1319.e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Li B., Yu Y., Iyer S.S., Sun M., Cheng G. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep. 2015;16:202–212. doi: 10.15252/embr.201439366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Jo Y., Dallari S., Chang A.Y., Dai J., Swaminathan S., Wehrens E.J., Fitzgerald-Bocarsly P., Zúñiga E.I. Self-renewal and toll-like receptor signaling sustain exhausted plasmacytoid dendritic cells during chronic viral infection. Immunity. 2018;48:730–744.e5. doi: 10.1016/j.immuni.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133:155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major J., Crotta S., Llorian M., Mccabe T.M., Gad H.H., Priestnall S.L., Hartmann R., Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani S., Daga S., Fallerini C., Baldassarri M., Benetti E., Picchiotti N., Fava F., Gallì A., Zibellini S., Bruttini M., et al. Rare variants in Toll-like receptor 7 results in functional impairment and downregulation of cytokine-mediated signaling in COVID-19 patients. Genes Immun. 2022;23:51–56. doi: 10.1038/s41435-021-00157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan T.S.M.M., Figdor C.G., Buschow S.I. Human plasmacytoid dendritic cells: from molecules to intercellular communication network. Front. Immunol. 2013;4:372. doi: 10.3389/fimmu.2013.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic S., Popovic S., Djurdjevic P., Todorovic D., Djordjevic N., Mijailovic Z., Sazdanovic P., Milovanovic D., Ruzic Zecevic D., Petrovic M., et al. SARS-CoV-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PLoS One. 2020;15:e0241097. doi: 10.1371/journal.pone.0241097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna K., Beignon A.S., Bhardwaj N. Plasmacytoid dendritic cells: linking innate and adaptive immunity. J. Virol. 2005;79:17–27. doi: 10.1128/JVI.79.1.17-27.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnab F., Mayer-Barber K., Sher A., Wack A., O'garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D., et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y.Q., Huang M., Sun X., Deng F., Wang H., Ning Y.J. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput. Struct. Biotechnol. J. 2021;19:4217–4225. doi: 10.1016/j.csbj.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.P., et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E., Tabarsi P., Varahram M., Folkerts G., Adcock I.M. The immune response and immunopathology of COVID-19. Front. Immunol. 2020;11:2037. doi: 10.3389/fimmu.2020.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O'keeffe M., Bahlo M., et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Niles M.A., Gogesch P., Kronhart S., Ortega Iannazzo S., Kochs G., Waibler Z., Anzaghe M. Macrophages and dendritic cells are not the major source of pro-inflammatory cytokines upon SARS-CoV-2 infection. Front. Immunol. 2021;12:647824. doi: 10.3389/fimmu.2021.647824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowill A.E., De Campos-Lima P.O. Immune response resetting as a novel strategy to overcome SARS-CoV-2-induced cytokine storm. J. Immunol. 2020;205:2566–2575. doi: 10.4049/jimmunol.2000892. [DOI] [PubMed] [Google Scholar]
- Oberkampf M., Guillerey C., Mouriès J., Rosenbaum P., Fayolle C., Bobard A., Savina A., Ogier-Denis E., Enninga J., et al. Mitochondrial reactive oxygen species regulate the induction of CD8(+) T cells by plasmacytoid dendritic cells. Nat. Commun. 2018;9:2241. doi: 10.1038/s41467-018-04686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Cauchemez S., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- Oh S.J., Shin O.S. SARS-CoV-2 nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. Cells. 2021;10:530. doi: 10.3390/cells10030530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Onodi F., Bonnet-Madin L., Meertens L., Karpf L., Poirot J., Zhang S.Y., Picard C., Puel A., Jouanguy E., et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J. Exp. Med. 2021;218:e20201387. doi: 10.1084/jem.20201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijo J., Döring M., Spanier J., Grabski E., Nooruzzaman M., Schmidt T., Witte G., Messerle M., Hornung V., Kaever V., Kalinke U. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. 2016;12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pérez-Gómez A., Vitallé J., Gasca-Capote C., Gutierrez-Valencia A., Trujillo-Rodriguez M., Serna-Gallego A., Muñoz-Muela E., Jiménez-Leon M.d.L.R., Rafii-El-Idrissi Benhnia M., Rivas-Jeremias I., et al. Dendritic cell deficiencies persist seven months after SARS-CoV-2 infection. Cell. Mol. Immunol. 2021;18:2128–2139. doi: 10.1038/s41423-021-00728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perussia B., Fanning V., Trinchieri G. A leukocyte subset bearing HLA-DR antigens is responsible for in vitro alpha interferon production in response to viruses. Nat. Immun. Cell Growth Regul. 1985;4:120–137. [PubMed] [Google Scholar]
- Peruzzi B., Bencini S., Capone M., Mazzoni A., Maggi L., Salvati L., Vanni A., Orazzini C., Nozzoli C., Morettini A., et al. Quantitative and qualitative alterations of circulating myeloid cells and plasmacytoid DC in SARS-CoV-2 infection. Immunology. 2020;161:345–353. doi: 10.1111/imm.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccioli D., Sammicheli C., Tavarini S., Nuti S., Frigimelica E., Manetti A.G.O., Nuccitelli A., Aprea S., Valentini S., et al. Human plasmacytoid dendritic cells are unresponsive to bacterial stimulation and require a novel type of cooperation with myeloid dendritic cells for maturation. Blood. 2009;113:4232–4239. doi: 10.1182/blood-2008-10-186890. [DOI] [PubMed] [Google Scholar]
- Raieli S., Trichot C., Korniotis S., Pattarini L., Soumelis V. TLR1/2 orchestrate human plasmacytoid predendritic cell response to gram+ bacteria. PLoS Biol. 2019;17:e3000209. doi: 10.1371/journal.pbio.3000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébillard R.M., Charabati M., Grasmuck C., Filali-Mouhim A., Tastet O., Brassard N., Daigneault A., Bourbonnière L., Anand S.P., Balthazard R., et al. Identification of SARS-CoV-2-specific immune alterations in acutely ill patients. J. Clin. Invest. 2021;131:e145853. doi: 10.1172/JCI145853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues P.F., Alberti-Servera L., Eremin A., Grajales-Reyes G.E., Ivanek R., Tussiwand R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018;19:711–722. doi: 10.1038/s41590-018-0136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Starke K., Petereit-Haack G., Schubert M., Kämpf D., Schliebner A., Hegewald J., Seidler A. The age-related risk of severe outcomes due to COVID-19 infection: a rapid review, meta-analysis, and meta-regression. Int. J. Environ. Res. Public Health. 2020;17:5974. doi: 10.3390/ijerph17165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnblom L., Ramstedt U., Alm G.V. Properties of human natural interferon-producing cells stimulated by tumor cell lines. Eur. J. Immunol. 1983;13:471–476. doi: 10.1002/eji.1830130608. [DOI] [PubMed] [Google Scholar]
- Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saichi M., Ladjemi M.Z., Korniotis S., Rousseau C., Ait Hamou Z., Massenet-Regad L., Amblard E., Noel F., Marie Y., et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat. Cell Biol. 2021;23:538–551. doi: 10.1038/s41556-021-00681-2. [DOI] [PubMed] [Google Scholar]
- Sampaio N.G., Chauveau L., Hertzog J., Bridgeman A., Fowler G., Moonen J.P., Dupont M., Russell R.A., Noerenberg M., Rehwinkel J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021;11:13638. doi: 10.1038/s41598-021-92940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cerrillo I., Landete P., Aldave B., Sanchez-Alonso S., Sanchez-Azofra A., Marcos-Jimenez A., Avalos E., Alcaraz-Serna A., De Los Santos I., Mateu-Albero T., et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J. Clin. Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler N.G., Bosinger S.E., Estes J.D., Zhu R.T.R., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., Del Prete G.Q., et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Gunther P., Mcgovern N., Irac S.E., Gunawan M., Beyer M., Handler K., et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:eaag3009. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severa M., Diotti R.A., Etna M.P., Rizzo F., Fiore S., Ricci D., Iannetta M., Sinigaglia A., Lodi A., Mancini N., Criscuolo E., et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PLoS Pathog. 2021;17:e1009878. doi: 10.1371/journal.ppat.1009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Liu X., Cao Q., Ma P., Le W., Xie L., Ye J., Wen W., Tang H., Su W., Zheng Y., Liu Y. High-dimensional single-cell analysis reveals the immune characteristics of COVID-19. Am. J. Physiol. Lung Cell Mol. Physiol. 2021;320:L84–L98. doi: 10.1152/ajplung.00355.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Sodeifian F., Nikfarjam M., Kian N., Mohamed K., Rezaei N. The role of type I interferon in the treatment of COVID-19. J. Med. Virol. 2022;94:63–81. doi: 10.1002/jmv.27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito B., Broggi A., Pandolfi L., Crotta S., Clementi N., Ferrarese R., Sisti S., Criscuolo E., Spreafico R., Long J.M., et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184:4953–4968.e16. doi: 10.1016/j.cell.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Gilfillan S., Colonna M. Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 2013;9:e1003728. doi: 10.1371/journal.ppat.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A., Magyarics Z., Pazmandi K., Gopcsa L., Rajnavolgyi E., Bacsi A. TLR ligands upregulate RIG-I expression in human plasmacytoid dendritic cells in a type I IFN-independent manner. Immunol. Cell Biol. 2014;92:671–678. doi: 10.1038/icb.2014.38. [DOI] [PubMed] [Google Scholar]
- Takagi H., Fukaya T., Eizumi K., Sato Y., Sato K., Shibazaki A., Otsuka H., Hijikata A., Watanabe T., et al. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity. 2011;35:958–971. doi: 10.1016/j.immuni.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tel J., Aarntzen E.H.J.G., Baba T., Schreibelt G., Schulte B.M., Benitez-Ribas D., Boerman O.C., Croockewit S., Oyen W.J.G., et al. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res. 2013;73:1063–1075. doi: 10.1158/0008-5472.CAN-12-2583. [DOI] [PubMed] [Google Scholar]
- Tel J., Lambeck A.J.A., Cruz L.J., Tacken P.J., de Vries I.J.M., Figdor C.G. Human plasmacytoid dendritic cells phagocytose, process, and present exogenous particulate antigen. J. Immunol. 2010;184:4276–4283. doi: 10.4049/jimmunol.0903286. [DOI] [PubMed] [Google Scholar]
- Tel J., Schreibelt G., Sittig S.P., Mathan T.S.M., Buschow S.I., Cruz L.J., Lambeck A.J.A., Figdor C.G., de Vries I.J.M. Human plasmacytoid dendritic cells efficiently cross-present exogenous Ags to CD8+ T cells despite lower Ag uptake than myeloid dendritic cell subsets. Blood. 2013;121:459–467. doi: 10.1182/blood-2012-06-435644. [DOI] [PubMed] [Google Scholar]
- Thomas J.M., Pos Z., Reinboth J., Wang R.Y., Wang E., Frank G.M., Lusso P., Trinchieri G., Alter H.J., Marincola F.M., Thomas E. Differential responses of plasmacytoid dendritic cells to influenza virus and distinct viral pathogens. J. Virol. 2014;88:10758–10766. doi: 10.1128/JVI.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L.G., Reuschl A.K., Zuliani-Alvarez L., Whelan M.V.X., Turner J., Noursadeghi M., Jolly C., Towers G.J. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021;40:e107826. doi: 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsenheimer A., Gerlach J.T., Jung M.C., Gruener N., Wächtler M., Backmund M., Santantonio T., Schraut W., Heeg M.H.J., et al. Plasmacytoid dendritic cells in acute and chronic hepatitis C virus infection. Hepatology. 2005;41:643–651. doi: 10.1002/hep.20592. [DOI] [PubMed] [Google Scholar]
- Van Beek J.J.P., Flórez-Grau G., Gorris M.A.J., Mathan T.S.M., Schreibelt G., Bol K.F., Textor J., De Vries I.J.M. Human pDCs are superior to cDC2s in attracting cytolytic lymphocytes in melanoma patients receiving DC vaccination. Cell Rep. 2020;30:1027–1038.e4. doi: 10.1016/j.celrep.2019.12.096. [DOI] [PubMed] [Google Scholar]
- Van Der Sluis R.M., Cham L.B., Gris-Oliver A., Gammelgaard K.R., Pedersen J.G., Idorn M., Ahmadov U., Hernandez S.S., Cémalovic E., Godsk S.H., et al. TLR2 and TLR7 mediate distinct immunopathological and antiviral plasmacytoid dendritic cell responses to SARS-CoV-2 infection. EMBO J. 2022;41:e109622. doi: 10.15252/embj.2021109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyndhoven L.C., Chouri E., Subedi N., Tel J. Phenotypical diversification of early IFNalpha-producing human plasmacytoid dendritic cells using droplet-based microfluidics. Front. Immunol. 2021;12:672729. doi: 10.3389/fimmu.2021.672729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94:e00985-20. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vono M., Huttner A., Lemeille S., Martinez-Murillo P., Meyer B., Baggio S., Sharma S., Thiriard A., Marchant A., et al. Robust innate responses to SARS-CoV-2 in children resolve faster than in adults without compromising adaptive immunity. Cell Rep. 2021;37:109773. doi: 10.1016/j.celrep.2021.109773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wack A. Monocyte and dendritic cell defects in COVID-19. Nat. Cell Biol. 2021;23:445–447. doi: 10.1038/s41556-021-00685-y. [DOI] [PubMed] [Google Scholar]
- Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.Y., Mao T., Klein J., Dai Y., Huck J.D., Jaycox J.R., Liu F., Zhou T., Israelow B., Wong P., Coppi A., et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–288. doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- Wang H., Peters N., Schwarze J. Plasmacytoid dendritic cells limit viral replication, pulmonary inflammation, and airway hyperresponsiveness in respiratory syncytial virus infection. J. Immunol. 2006;177:6263–6270. doi: 10.4049/jimmunol.177.9.6263. [DOI] [PubMed] [Google Scholar]
- Weatherhead J.E., Clark E., Vogel T.P., Atmar R.L., Kulkarni P.A. Inflammatory syndromes associated with SARS-CoV-2 infection: dysregulation of the immune response across the age spectrum. J. Clin. Invest. 2020;130:6194–6197. doi: 10.1172/JCI145301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb K., Peckham H., Radziszewska A., Menon M., Oliveri P., Simpson F., Deakin C.T., Lee S., Ciurtin C., et al. Sex and pubertal differences in the type 1 interferon pathway associate with both X chromosome number and serum sex hormone concentration. Front. Immunol. 2018;9:3167. doi: 10.3389/fimmu.2018.03167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., Mckechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers F., Subedi N., Van Buuringen N., Heister D., Vivié J., Beeren-Reinieren I., Woestenenk R., Dolstra H., Piruska A., et al. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018;9:3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winheim E., Rinke L., Lutz K., Reischer A., Leutbecher A., Wolfram L., Rausch L., Kranich J., Wratil P.R., et al. Impaired function and delayed regeneration of dendritic cells in COVID-19. PLoS Pathog. 2021;17:e1009742. doi: 10.1371/journal.ppat.1009742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.Y.R., Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy? Nat. Rev. Immunol. 2022;22:47–56. doi: 10.1038/s41577-021-00656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y., Liu L., Zhang S., Zhang Z. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Yun T.J., Igarashi S., Zhao H., Perez O.A., Pereira M.R., Zorn E., Shen Y., Goodrum F., Rahman A., Sims P.A., Farber D.L., Reizis B. Human plasmacytoid dendritic cells mount a distinct antiviral response to virus-infected cells. Sci. Immunol. 2021;6:eabc7302. doi: 10.1126/sciimmunol.abc7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I. Interfering with SARS-CoV-2: are interferons friends or foes in COVID-19? Curr. Opin. Virol. 2021;50:119–127. doi: 10.1016/j.coviro.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., COVID Human Genetic Effort. Cobat A., Casanova J.L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603:587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wang Y., Li K., Meyerholz D.K., Allamargot C., Perlman S. Severe acute respiratory syndrome coronavirus 2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 2021;223:785–795. doi: 10.1093/infdis/jiaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., Jonsson C.B., Kanneganti T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chen V., Shannon C.P., Wei X.S., Xiang X., Wang X., Wang Z.H., Tebbutt S.J., Kollmann T.R., Fish E.N. Interferon-alpha2b treatment for COVID-19. Front. Immunol. 2020;11:615275. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.W., Wong Y.C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.Y., Lau T.T.K., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Martinez J., Huang X., Yang Y. Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood. 2007;109:619–625. doi: 10.1182/blood-2006-06-027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Miao V.N., Owings A.H., Navia A.W., Tang Y., Bromley J.D., Lotfy P., Sloan M., Laird H., Williams H.B., et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell. 2021;184:4713–4733.e22. doi: 10.1016/j.cell.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingaropoli M.A., Nijhawan P., Carraro A., Pasculli P., Zuccalà P., Perri V., Marocco R., Kertusha B., Siccardi G., Del Borgo C., et al. Increased sCD163 and sCD14 plasmatic levels and depletion of peripheral blood pro-inflammatory monocytes, myeloid and plasmacytoid dendritic cells in patients with severe COVID-19 pneumonia. Front. Immunol. 2021;12:627548. doi: 10.3389/fimmu.2021.627548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulu M.Z., Sureshchandra S., Pinski A.N., Doratt B., Shen W., Messaoudi I. Obesity correlates with pronounced aberrant innate immune responses in hospitalized aged COVID-19 patients. Front. Immunol. 2021;12:760288. doi: 10.3389/fimmu.2021.760288. [DOI] [PMC free article] [PubMed] [Google Scholar]