Abstract
A novel α-amylase (AmyK38) was found in cultures of an alkaliphilic Bacillus isolate designated KSM-K38. Based on the morphological and physiological characteristics and phylogenetic position as determined by 16S ribosomal DNA gene sequencing and DNA-DNA reassociation analysis, it was suggested that the isolate was a new species of the genus Bacillus. The enzyme had an optimal pH of 8.0 to 9.5 and displayed maximum catalytic activity at 55 to 60°C. The apparent molecular mass was approximately 55 kDa, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the isoelectric point was around pH 4.2. This enzyme efficiently hydrolyzed various carbohydrates to yield maltotriose, maltohexaose, maltoheptaose, and, in addition, maltose as major end products after completion of the reaction. The activity was not prevented at all by EDTA and EGTA at concentrations as high as 100 mM. Moreover, AmyK38 was highly resistant to chemical oxidation and maintained more than 80% of its original activity even after incubation for 1 h in the presence of excess H2O2 (1.8 M).
Starch, a main component of our daily diet, is frequently found not only in food residues on dishes but also in food stains on clothes (36). Enzymatic hydrolysis of starch is catalyzed by α-amylase (1,4-α-d-glucan glucanohydrolase; EC 3.2.1.1), β-amylase (1,4-α-d-glucan glucanohydrolase; EC 3.2.1.2), glucoamylase (1,4-α-d-glucan glucanohydrolase; EC 3.2.1.3), α-glucosidase (1,4-α-d-glucan glucanohydrolase; EC 3.2.1.20), and debranching enzymes such as pullulanase (pullulan 6-glucanohydrolase; EC 3.2.1.41) and isoamylase (glycogen 6-glucanohydrolase; EC 3.2.1.68). These amylolytic enzymes, especially α-amylase and pullulanase, are very important, particularly in the food and detergent industries (1, 28). We have found and characterized some unique debranching enzymes, such as a high-alkaline pullulanase (4), an alkali-resistant neopullulanase (16), an alkaline isoamylase (6), and an alkaline amylopullulanase (5) from cultures of alkaliphilic Bacillus strains. These alkaline amylolytic enzymes can be used as effective additives in laundry and automatic dishwashing detergents operating under high alkalinity, as we also reported alkaline cellulases and a highly alkaline protease from alkaliphilic Bacillus strains (21). In particular, the alkaline amylopullulanase is unique in that it hydrolyzes α-1,6 and α-1,4 linkages in various carbohydrates at different active sites (3, 12).
α-Amylases are used widely in technical applications, such as in bread making, production of glucose and/or fructose syrups and fuel ethanol from starches, and desizing of textiles and paper. The demand for α-amylase in laundry and automatic dishwashing detergents has also been growing for several years (36). However, most of the Bacillus α-amylases, such as the enzymes from Bacillus licheniformis (BLA) (29), Bacillus amyloliquefaciens (BAA) (38), and Bacillus stearothermophilus (BSA) (23), are acid or neutral enzymes having pH optima at around 6.5. These neutral enzymes are essentially useless in detergents because the working pH range of detergents is between 8 and 11 (21). Since Horikoshi (13) reported the first alkaline amylase from alkaliphilic Bacillus sp. strain A-40-2 in 1971, many alkaline amylases have been found in cultures of alkaliphilic Bacillus strains (14). Most of the alkaline amylases from these alkaliphilic bacilli reported to date are exo-type amylases, which are not suitable for use in detergents. Generally, α-amylases are metalloenzymes that contain at least one activating and stabilizing Ca2+ ion (37, 39). It is well known that amylases are often inhibited by chelating reagents, such as zeolites, EDTA, and EGTA.
Recently, we found a novel α-amylase (AmyK) from cultures of the alkaline amylopullulanase producer alkaliphilic Bacillus sp. strain KSM-1378 (17) and succeeded in hyperexpressing the amyK gene in Bacillus subtilis cells (20). This enzyme is highly active at alkaline pH compared with BLA, BAA, and BSA. Furthermore, we improved the thermostability of AmyK by deletion of the Arg181-Gly182 residues (18) and substitution of a proline in the enzyme molecule (19). The deletion mutant enzyme also acquired resistance to chelating reagents such as EDTA and EGTA, but the resistance was still lower than that of BLA (unpublished data). In this paper, we report the isolation of a novel α-amylase (AmyK38) from cultures of alkaliphilic Bacillus sp. strain KSM-K38. AmyK38 is an alkaline α-amylase having high resistance to chelating reagents and chemical oxidants. Furthermore, this strain was suggested to be a novel species of Bacillus, as judged by 16S ribosomal DNA (rDNA) gene sequence and DNA-DNA hybridization analysis.
MATERIALS AND METHODS
Organism and culture conditions.
The organism used was Bacillus sp. strain KSM-K38, which was originally isolated from a soil sample collected in Tochigi, Japan. The soil sample (0.5 g) was suspended in 5 ml of sterilized water and then heated at 80°C for 15 min. A sample (0.1 ml) of the suspension was spread on blue starch agar and incubated at 30°C for 2 days. The blue starch agar was composed of 0.5% (wt/vol) starch azure (Sigma, St. Louis, Mo.), 1.5% (wt/vol) tryptone (Difco Laboratories, Detroit, Mich.), 0.5% (wt/vol) Soytone (Difco), 0.5% (wt/vol) NaCl, 0.5% (wt/vol) Na2CO3 (separately autoclaved), and 1.5% (wt/vol) agar (pH 10). Colonies that had formed a clear zone around their margins were picked up as α-amylase producers. The isolates were inoculated individually into 5-ml aliquots of an alkaline liquid medium in 50-ml test tubes and cultured, with shaking, at 30°C for 2 days. The alkaline liquid medium contained 1% (wt/vol) soluble starch (Wako Pure Chemical, Kyoto, Japan), 1.5% (wt/vol) tryptone (Difco), 0.5% (wt/vol) Soytone (Difco), 0.5% (wt/vol) NaCl, and 0.5% (wt/vol) Na2CO3 (separately autoclaved; pH 10). Among the isolates obtained in this way, Bacillus sp. strain KSM-K38 was found to produce a novel alkaline α-amylase that is highly resistant to chelating reagents and chemical oxidants. It was propagated at 30°C for 3 days, with shaking, in 100-ml aliquots of the alkaline liquid medium placed in 500-ml flasks. After removal of cells by centrifugation (12,000 × g for 30 min) at 5°C, the supernatant was used as the starting material for purification of AmyK38. Taxonomic characteristics of this strain were examined according to the methods of Gordon et al. (11) and Nielsen et al. (27). This strain has been deposited as a patent strain (FERM BP-6946) in the National Institute of Bioscience and Human Technology Agency of Japan.
16S rDNA gene sequencing.
16S rDNA fragments from Bacillus sp. strain KSM-K38 were analyzed using PCR-direct sequencing, as described by Shima et al. (31). 16S rDNA sequences were aligned using the CLUSTAL X multiple-alignment program (34), and nucleotide substitution rates (Knuc values) were calculated. Sites involving gaps were excluded from all analyses. A phylogenetic tree was inferred by the neighbor-joining method (30) in the CLUSTAL X program version 1.64b. The similarity values of the sequences were calculated using the GENETYX-MAC program version 9.0 (SDC Software Development, Tokyo, Japan).
DNA base composition and DNA-DNA hybridization.
Genomic DNA was prepared according to the method of Marmur (24). The G+C content of the DNA was determined by high-performance liquid chromatography (HPLC) of the derived deoxyribonucleosides as described by Tamaoka and Komagata (33). Levels of DNA relatedness were determined by the method of Ezaki et al. (10) using photobiotin-labeled DNA probes and microplates. Bacillus agaradhaerens (DSM 8721T) and Bacillus clarkii (DSM 8720T) were used as reference strains for the DNA-DNA hybridization test.
Purification of AmyK38.
Enzyme purification was done below 5°C. The centrifugal supernatant of the culture broth was treated with ammonium sulfate, and the fraction that precipitated at 80% saturation was collected. Precipitates formed were dissolved in a small volume of 10 mM Tris-HCl buffer (pH 7.0), and the solution was dialyzed overnight against 250 volumes of the same buffer. The retentate was then applied to a column of DEAE-Toyopearl 650 M (5 by 18 cm; Tosoh, Tokyo, Japan) that had been equilibrated with 10 mM Tris-HCl buffer (pH 7.0). The column was initially washed with 1.7 liters of 0.3 M NaCl in the same buffer, and proteins were eluted with a 3.0-liter linear gradient of 0.3 to 1.0 M NaCl in the same buffer, at a flow rate of 8.6 ml min−1. The active fractions were combined and concentrated by ultrafiltration (PM-10, 10,000-Mr cutoff; Millipore, Bedford, Mass.). The concentrate obtained was put on a column of Toyopearl HW-55F (1.5 by 94 cm; Tosoh) that had been equilibrated with 10 mM Tris-HCl buffer (pH 7.0) containing 0.2 M NaCl. Proteins were eluted with the equilibration buffer, at a flow rate of 0.21 ml min−1. Protein in column effluents was routinely monitored by measuring the absorbance at 280 nm. The active fractions were combined and dialyzed overnight against 10 mM glycine-NaOH buffer (pH 10). The resultant retentate was used exclusively for further experiments as the final preparation of purified AmyK38. For comparison, we also purified a commercially available thermostable α-amylase from B. licheniformis (BLA) (Termamyl; Novo Nordisk, Bagsvaerd, Denmark) to homogeneity by the method reported previously (17).
Enzyme assay.
α-Amylase activity was routinely measured at 50°C in a 1-ml reaction mixture that contained 0.5 ml of a 1.0% (wt/vol) solution of soluble starch (from potato; Sigma) in 50 mM glycine-NaOH buffer (pH 10) and 0.1 ml of a suitably diluted solution of enzyme. The reducing sugar formed was measured by the dinitrosalicylic acid procedure (25). One unit of enzymatic activity was defined as the amount of protein that produced 1 μmol of reducing sugar as glucose per min under the conditions of the assay. Maltooligosaccharides in the G3 to G7 range were purchased from Hayashibara Biochemical (Kurashiki, Japan) and Funakoshi (Tokyo, Japan). Other polysaccharides used as substrates were the products of Sigma. Protein was determined using a protein assay kit (Bio-Rad, Richmond, Calif.) with bovine serum albumin as standard protein.
Oxidative stability test.
AmyK38 and BLA (2.0 U ml−1) were each incubated with up to 1.8 M H2O2 at pH 10 in 50 mM glycine-NaOH buffer and at 30°C for up to 60 min. Samples (0.2 ml) were taken at the indicated intervals and immediately added to a solution of catalase (200 μg ml−1) to quench remaining H2O2. The solution (0.1 ml) was used for the measurement of the residual activity under the standard conditions of the assay.
Electrophoretic analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done essentially as described by Laemmli (22) on slab gels (10% [wt/vol] acrylamide, 70 by 50 mm, 2.0-mm thickness), and the gels were stained for protein with Coomassie brilliant blue R-250. The molecular mass was estimated by SDS-PAGE (10% [wt/vol] acrylamide gel) with low-range molecular mass standards (Bio-Rad), which included phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa). Isoelectric points (pIs) of proteins were measured using a Multiphore II gel electrofocusing system, a polyacrylamide gel plate, and a Broad pI calibration kit (Pharmacia Fine Chemica AB, Uppsala, Sweden), which included amyloglucosidase (pI 3.50); methyl red (pI 3.75); soybean trypsin inhibitor (pI 4.55); β-lactoglobulin A (pI 5.20); bovine carbonic anhydrase b (pI 5.85); human carbonic anhydrase b (pI 6.55); horse myoglobin, acidic band (pI 6.85); horse myoglobin, basic band (pI 7.35); lentil lectin, acidic band (pI 8.15); lentil lectin, middle band (pI 8.45); lentil lectin, basic band (pI 8.65); and trypsinogen (pI 9.30).
Chromatographic analysis of the products of hydrolysis of carbohydrates.
The hydrolysis products of the purified AmyK38 were measured by HPLC. The purified enzyme was incubated at 30°C with soluble starch as substrate in 10 mM potassium phosphate buffer (pH 8.0). Samples were removed at intervals and heated immediately in boiling water for 5 min to terminate the reaction. The products were analyzed by HPLC with a carbohydrate column (4.6 by 250 mm; Waters, Milford, Mass.) with acetonitrile-water (70:30, vol/vol) as eluent at a flow rate of 1.4 ml min−1, and they were measured with data analysis software, 805 Data Station (Waters), using authentic maltooligosaccharides.
Analysis of anomeric configuration.
The anomeric configuration of products of soluble starch hydrolyzed by the purified AmyK38 was determined by measuring the optical rotation of the hydrolysate (15). A reaction mixture (1 ml), consisting of 1% (wt/vol) soluble starch (from potato; Sigma) in 10 mM potassium phosphate buffer (pH 8.0) and enzyme (4.7 U ml−1), was placed in a cuvette with a 5.0-cm light path. The change in optical rotation of the mixture was monitored at room temperature in a highly sensitive SEPA-200 polarimeter (Horiba, Tokyo, Japan) by using the sodium line (589 nm). The mutarotation of the hydrolysate was observed by adding 2 drops of 28% ammonium solution after the optical rotation had become approximately constant.
Analysis of calcium in the AmyK38 molecule.
The enzyme samples were dialyzed overnight against 10 mM glycine-NaOH buffer (pH 10) at 5°C. The resultant retentate and dialysate portions were hydrolyzed with nitric acid and hydrogen peroxide. Calcium concentrations of both hydrolysates were measured by atomic absorption at 393.36 nm using an inductively coupled plasma emission spectral system (SPS1200VR; Seiko Electron, Tokyo, Japan). Milli-Q water (Millipore) was used to make the buffer and the standard solution of calcium.
Sequencing of amino-terminal regions of protein.
The enzyme sample was blotted on a polyvinylidene difluoride membrane (Prosorb; Applied Biosystems, Foster City, Calif.), which had been wetted with methanol. The N-terminal amino acid sequence of the protein was determined directly by a protein sequencer (model 476A; Applied Biosystems).
Nucleotide sequence accession number.
The 16S rDNA sequence data of KSM-K38 have been submitted to the DDBJ, GenBank, and EMBL data banks with the accession no. AB044748. An extensive search of the scientific literature (PubMed; http://www.ncbi.nlm.nih.gov/PubMed/) and databases (nr-aa, PIR, and Swiss-Prot) was performed to collect the 16S rDNA sequences of Bacillus strains using the BLAST2 program (2). Sequences incorporated in the present study are under the following accession numbers: B. agaradhaerens DSM 8721T, X76445; B. clarkii DSM 8720T, X76444; Bacillus alcalophilus DSM 485T, X76436; Bacillus pseudofirmus DSM 8715T, X76439; Bacillus pseudalcaliphilus DSM 8725T, X76449; Bacillus halodurans ATCC 27557T, AB021187; Bacillus halodenitrificans ATCC 49067T, AB021186; Bacillus horikoshii DSM 8719T, X76443; Bacillus halmapalus DSM 8723T, X76447; and Bacillus niacini IFO 15566T, AB021194.
RESULTS AND DISCUSSION
Taxonomic characteristics of strain KSM-K38.
The physiological and biochemical characteristics of strain KSM-K38 were identified. The organism was capable of growing over a pH range from 9 to 11, but no growth was observed at pH 7. The range of temperature for growth was between 15 and 40°C with optimal growth around 30°C. It was a strict aerobe that was spore forming (cylindrical, central, or subterminal endospores), gram positive, motile, rod shaped (1.0 to 1.2 by 1.8 to 3.8 μm), and peritrichous. It was positive for production of catalase and oxidase and hydrolysis of starch, casein, gelatin, Tween 40, and Tween 60 and was negative for formation of indole and H2S, production of urease, deamination of phenylalanine, and growth in 15% (wt/vol) NaCl. The organism was able to grow on d-glucose, d-galactose, d-fructose, d-mannose, d-xylose, d-ribose, l-arabinose, d-mannitol, glycerol, sucrose, lactose, maltose, melibiose, trehalose, and d-raffinose but not on inositol, d-sorbitol, esculin, and rhamnose. Thus, this isolate is an obligately alkaliphilic Bacillus strain. The doubling time was 40 min when the organism was grown at 30°C in the soluble starch medium described above.
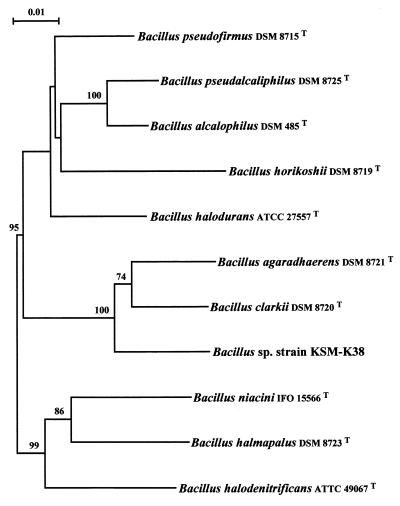
For further characterization of strain KSM-K38, we constructed a phylogenetic tree based on comparison of the 16S rDNA gene sequence of this strain and those of 10 type strains of Bacillus spp., as shown in Fig. 1. Similarity values were as low as 90.5 to 95.5% compared with these Bacillus strains. The sequence of strain KSM-K38 had the closest match (95.5% homology) with that from B. agaradhaerens. The next highest similarity was with B. clarkii (94.8% homology). The DNA-DNA hybridization of strain KSM-K38 with B. clarkii and B. agaradhaerens revealed a low association (less than 23%), as shown in Table 1. Moreover, the G+C contents of the DNA of strain KSM-K38, B. clarkii, and B. agaradhaerens were 46.2, 42.0, and 38.2 mol%, respectively. On the basis of the results of the phenotypic characteristics, the G+C content of genomic DNA, the 16S rDNA similarity, and the level of DNA-DNA hybridization, Bacillus sp. strain KSM-K38 is not closely related to any of the strains of Bacillus compared. Thus, we suggest that the isolate is a new species of the genus Bacillus.
FIG. 1.
Unrooted phylogenetic tree based on the 16S rDNA sequences of strain KSM-K38 and representative Bacillus strains. The searched sequences having similarity with less than 90.5% identity were omitted from the figure. The numbers at internal nodes are the percentages of bootstrap values derived from 1,000 samples in which the group to the right of the node was monophyletic. Bootstrap probability values less than 50% were omitted from the figure. Bar = 0.01 Knuc unit, representing 0.01 inferred substitutions per nucleotide position. T, type strain (see Materials and Methods).
TABLE 1.
Levels of relatedness between Bacillus sp. strain KSM-K38 and related Bacillus strains
| Strain | G+C content (mol%) | % DNA-DNA hybridization with strain:
|
||
|---|---|---|---|---|
| Bacillus sp. strain KSM-K38 | Bacillus agaradhaerens DSM 8721T | Bacillus clarkii DSM 8720T | ||
| Bacillus sp. strain KSM-K38 | 46.2 | 100 | 10 | 6 |
| Bacillus agaradhaerens DSM 8721T | 38.2 | 22 | 100 | NTa |
| Bacillus clarkii DSM 8720T | 42.0 | 22 | NTa | 100 |
NT, not tested.
Purification and physicochemical properties of AmyK38.
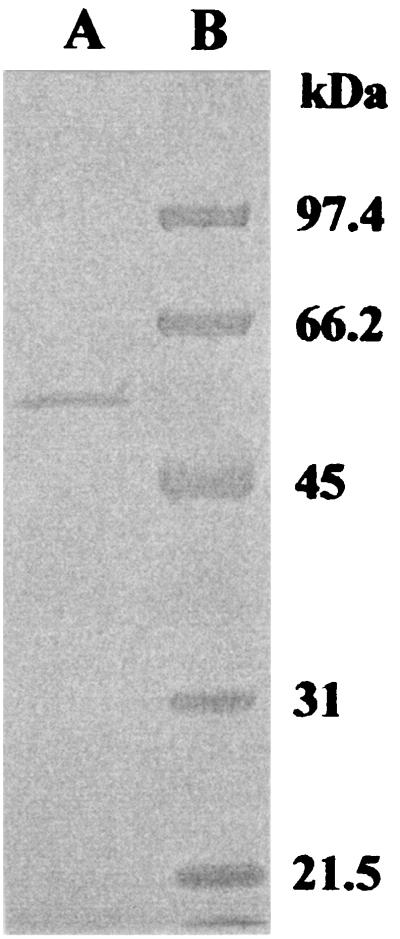
AmyK38 was purified to homogeneity from cultures of Bacillus sp. strain KSM-K38 by precipitation with ammonium sulfate and the subsequent two-step column chromatographic procedures, as summarized in Table 2. Approximately 817-fold purification to a specific activity as high as 4,221 U mg of protein−1 was obtained for the α-amylase activity when measured at 50°C and at pH 10 in 50 mM glycine-NaOH buffer. The protein was homogeneous, as judged by SDS-PAGE, as shown in Fig. 2. The subunit molecular mass of the purified enzyme was approximately 55 kDa by SDS-PAGE, a value that is similar to the molecular masses of BLA, BAA, BSA, and AmyK. The pI value was around pH 4.2. The N-terminal amino acid sequence was determined to be Asp-Gly-Leu-Asn-Gly-Thr-Met-Met-Gln-Tyr-Tyr-Glu-Trp. A comparison of the N-terminal amino acid sequence of the purified enzyme with those of other α-amylases, such as AmyK, BLA, BAA, and BSA, revealed high homology. The strong homology appeared after the fourth amino acid, Asn.
TABLE 2.
Purification of amylase produced by Bacillus sp. strain KSM-K38
| Purification step | Total protein (mg) | Total act (U) | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture filtrate | 243 | 1,254 | 5 | 100 | 1.0 |
| 80% ammonium sulfate precipitation | 120 | 789 | 7 | 63 | 1.3 |
| DEAE-Toyopearl chromatography | 0.20 | 569 | 2,917 | 45 | 565.0 |
| Toyopearl HW-55F chromatography | 0.04 | 168 | 4,221 | 13 | 817.6 |
FIG. 2.
SDS-PAGE of the purified enzyme from Bacillus sp. strain KSM-K38. The purified enzyme (0.28 μg) was visualized by Coomassie brilliant blue staining for protein (lane A). Lane B shows molecular mass markers (calibration in kilodaltons).
Substrate specificity.
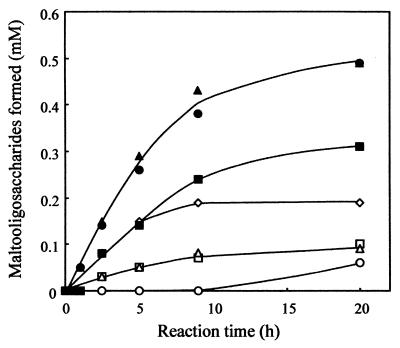
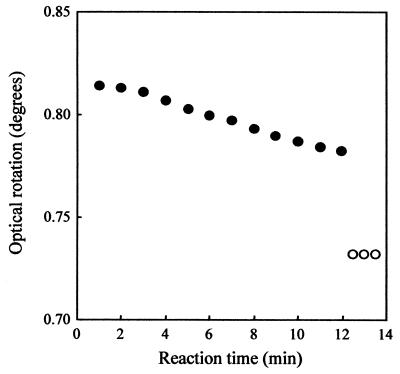
The purified enzyme was examined for its ability to hydrolyze various carbohydrates under the standard conditions of the assay, as shown in Table 3. Of the substrates tested, soluble starch was hydrolyzed by the enzyme, and amylopectin, glycogen, amylose, and dextrin were also hydrolyzed to a lesser extent. No reaction was observed with dextran; pullulan; or α-, β-, and γ-cyclodextrins. The product pattern of the purified enzyme with soluble starch (0.5% [wt/vol]) as substrate was examined by HPLC. The major products were maltotriose (G3) and maltohexaose (G6), with intermediate products being maltose (G2) and maltoheptaose (G7), as shown in Fig. 3. The typical molar ratios of products at equilibrium reached after 20 h were as follows: G7, 0.19 mM; G6, 0.49 mM; maltopentaose (G5), 0.10 mM; maltotetraose (G4), 0.09 mM; G3, 0.49 mM; G2, 0.31 mM; glucose (G1), 0.06 mM. This hydrolysis pattern was consistent with those of endo-type amylases. The anomeric configuration of the products was determined by measurement of optical rotation of the hydrolysate, as shown in Fig. 4. An abrupt downward shift of optical rotation occurred upon addition of ammonia solution after a 12-min incubation. This indicates that the hydrolysis products have an α-anomeric configuration, and hence, the enzyme is classified as an α-amylase.
TABLE 3.
Substrate specificity of the purified enzyme
| Substratea | Relative activity (%)b |
|---|---|
| Soluble starch (potato) | 100 |
| Starch (sweet potato) | 100 |
| Starch (corn) | 105 |
| Amylose (corn) | 65 |
| Amylopectin (potato) | 74 |
| Glycogen (bovine muscle) | 58 |
| Pullulan | 1 |
| Dextran | 0 |
| Dextrin (corn) | 27 |
| α-Cyclodextrin | 0 |
| β-Cyclodextrin | 0 |
| γ-Cyclodextrin | 0 |
Added at 0.5% (wt/vol).
The activity was measured at 50°C and at pH 10.0 in 50 mM glycine-NaOH buffer. The values shown are the percentages of the activity obtained using soluble starch as substrate, which is taken as 100%.
FIG. 3.
Analysis of products of hydrolysis of soluble starch by HPLC. The reaction (0.3 U ml−1) was done at 30°C and at pH 8.0 in 10 mM potassium phosphate buffer. Samples were taken at the indicated intervals and boiled for 5 min to terminate the reaction. The products were analyzed by HPLC, as described in Materials and Methods. Symbols used: ○, G1; ▪, G2; ▴, G3; ▵, G4; □, G5; ●, G6; ⋄, G7.
FIG. 4.
Optical rotation of the action of the purified enzyme with soluble starch. The symbols indicate the optical rotations before (●) and after (○) addition of alkali to the digests, as described in Materials and Methods.
Effect of pH on activity and stability.
The effect of pH on the activity of AmyK38 was examined with soluble starch as the substrate in 50 mM buffers (acetate, pH 3.5 to 6.0; potassium phosphate, pH 6.0 to 8.0; glycine-NaOH, pH 9.0 to 10.5; carbonate, pH 10.0 to 12.0). The purified enzyme showed catalytic activity from pH 5.5 to 10.5 and was an alkaline enzyme, having a pH optimum of 8.0 to 9.5 in the buffers. More than 50% of the maximum activity was detectable between pH 6.5 and 10. At pH 9, the specific activity of AmyK38 is approximately fivefold greater than that of BLA (17). Ca2+ ion (1 mM) inhibited the AmyK38 activity by 25 to 30% over a range from pH 8.0 to 10 (data not shown). To examine the pH stability of the purified enzyme, the enzyme (2.0 U ml−1) was preincubated at the indicated pH in 10 mM Britton-Robinson buffer and at 40°C for 30 min, and then samples (0.1 ml) were used to measure the residual activity under the standard conditions of the assay. The enzyme was very stable, with more than 80% of the original activity detected over the wide range of pHs from 6 to 11.
Effect of temperature on activity and stability.
The activity of AmyK38 was measured at various temperatures at pH 10 in 50 mM glycine-NaOH buffer. The alkaline enzyme showed catalytic activity from 20 to 80°C, and the optimal temperature was around 55 to 60°C. To examine the temperature stability of the enzyme, the time course of the thermal inactivation of the enzyme was monitored at pH 10 in 50 mM glycine-NaOH buffer. The enzyme retained full activity after 60 min of incubation at 30°C but lost 80% of the original activity after 30 min of incubation at 50°C in the absence of Ca2+ ions. This divalent cation (1 mM) did not protect it from the thermal inactivation of the enzyme at all. BLA was very stable under the same conditions (data not shown).
Effects of metal ions and laundry surfactants.
AmyK38 was incubated with various metal ions (1 mM each) for 30 min at 30°C and at pH 10 in 50 mM glycine-NaOH buffer, and the residual activity was measured. Mn2+ ions inhibited the activity by 20%. Other metal ions, including Al3+, Fe3+, Ca2+, Co2+, Hg2+, Ag+, Cu2+, Ni2+, Fe2+, Mg2+, Zn2+, Ba2+, Be2+, Pd2+, Sr2+, Na+ (50 mM), and K+ (50 mM), were without effects on the activity. The enzyme was quite stable to incubation at 40°C for 1 h with various surfactants (0.1% [wt/vol] each), such as SDS, polyoxyethylene alkyl sulfate, polyoxyethylene alkyl ether, sodium α-sulfonated fatty acid ester, and alkyl glucoside. Linear alkylbenzene sulfonate and alkyl sulfate slightly inhibited the activity. These properties, together with the above results, of AmyK38 fulfill the essential requirements for enzymes that can be used as effective additives in laundry and automatic dishwashing detergents.
Effects of chemical oxidants and chelating reagents.
Inactivation by chemical oxidation has been reported previously for an α-amylase from B. subtilis (26), as in the cases of an alkaline protease (subtilisin) (32) and various proteins and peptides (8). The oxidative stability of AmyK38 was then examined by measuring the residual activities after incubation with 0.6 M H2O2 at 30°C and at pH 10, with BLA, which is the most thermostable natural α-amylase reported so far (29, 35), as control. AmyK38 retained full activity even over the course of 1 h, but the enzymatic activity of BLA rapidly decreased (half-life [t1/2] = ∼3 min) in the presence of excess H2O2. Moreover, AmyK38 maintained more than 80% of its original activity even after a 1-h incubation with 1.8 M H2O2 (data not shown). These results indicate that AmyK38 is strongly resistant to chemical oxidation.
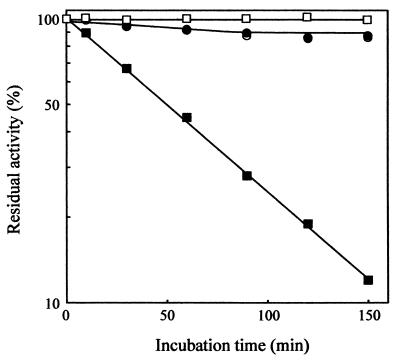
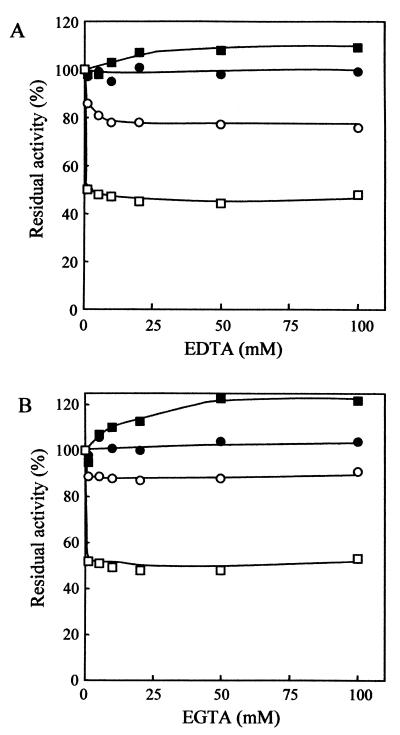
Effects of chelating reagents on the activity of AmyK38 were examined with BLA as control. AmyK38 and BLA were incubated with 1 mM EDTA at pH 10 in 50 mM glycine-NaOH buffer and at 40°C for up to 150 min. As shown in Fig. 5, both enzymes were stable in the absence of EDTA at least up to 150 min. In the presence of 1 mM EDTA, AmyK38 retained full activity even after incubation for 150 min, but BLA lost 88% of its original activity. Both enzymes were incubated with EDTA and EGTA at concentrations up to 100 mM in 50 mM glycine-NaOH buffer (pH 10) at 30 or 45°C for 30 min. As shown in Fig. 6A, AmyK38 retained full activity in the presence of EDTA at concentrations as high as 100 mM at both 30 and 45°C. When incubated at 45°C, the activity was rather activated by EDTA. In contrast, the activity of BLA was reduced by 1 mM EDTA to 80 and 50% of the original activity after incubation at 30 and 45°C, respectively. Similar results were observed with EGTA (Fig. 6B). Analysis of atomic absorption spectra showed that the calcium content of the EDTA-treated AmyK38 is almost zero, while that of the untreated enzyme varies from 0.05 to 1 mol/mol of protein possibly due to the weak binding affinity of this cation for the enzyme molecule. The high resistance of AmyK38 to chelating reagents is very interesting in that calcium is essential for the manifestation of activity and the maintenance of structural rigidity of α-amylases reported to date.
FIG. 5.
Effect of EDTA on the activities of AmyK38 and BLA. AmyK38 (●) and BLA (▪) (each at 2.0 U ml−1) were each incubated with 1 mM EDTA at pH 10 in 50 mM glycine-NaOH buffer and at 40°C for up to 150 min. As control, the former (○) and the latter (□) enzymes were also incubated under the same conditions without EDTA. Samples (0.1 ml) were taken after the indicated times, and then the residual activity in the sample was immediately measured under the standard conditions of the enzyme assay. The values shown are percentages of the respective original activities, which are taken as 100%.
FIG. 6.
Effects of graded concentrations of EDTA (A) or EGTA (B) on the activity of AmyK38 and BLA. (A) Both enzymes (each at 2.0 U ml−1) were incubated with the indicated concentrations of EDTA at pH 10 in 50 mM glycine-NaOH buffer at both 30°C (AmyK38, ●; BLA, ○) and 45°C (AmyK38, ▪; BLA, □) for 30 min. Samples (0.1 ml) were taken after incubation, and then the residual activities in the samples were immediately measured under the standard conditions of the enzyme assay. The values shown are percentages of the respective original activities, which are taken as 100%. (B) AmyK38 and BLA (each at 2.0 U ml−1) were each treated with EGTA under the same conditions described above.
The primary goals for an optimally performing detergent α-amylase are high activity and stability in the temperature range from 40 to 60°C under alkaline pH conditions (7). Our alkaline α-amylase, AmyK38, characteristically shows high resistance to chemical oxidants and chelating reagents. Inactivation by chemical oxidation of an enzyme occurs mainly by oxidation of a methionine residue to its sulfoxide derivative. The oxidative inactivation hampers the industrial production and applications of enzymes and proteins. It is one of the most serious problems in the detergent industry because laundry and automatic dishwashing detergent formulations often contain bleach (1). To solve such problems, the oxidative stability of enzymes, subtilisins for example (9), has been improved by replacing oxidizable methionine with nonoxidizable amino acids using site-directed mutagenesis. However, we often encounter the reduction of catalytic activities of the improved mutant enzymes, including α-amylases. AmyK38 is highly resistant to chemical oxidation and exhibits high catalytic activity at alkaline pH compared with commercially available, neutral α-amylases such as BLA, BAA, and BSA. Moreover, our enzyme is very stable to incubation with chelating reagents, which are indispensable ingredients in detergent formulations. Therefore, our α-amylase is a high-performing enzyme even in detergent formulations. We are now cloning the gene for and crystallizing the promising AmyK38 to analyze the tertiary structure and clarify the molecular mechanism of the high resistance to chelating reagents and oxidative stability of the enzyme.
ACKNOWLEDGMENT
We thank Y. Matsui for measuring atomic absorption spectra of the enzyme.
REFERENCES
- 1.Aehle W. Development of new amylases. In: van Ee J H, Misset O, Baas E J, editors. Enzymes in detergency. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 213–229. [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ara K, Igarashi K, Hagihara H, Sawada K, Kobayashi T, Ito S. Separation of functional domains for the α-1,4 and α-1,6 hydrolytic activities of a Bacillus amylopullulanase by limited proteolysis with papain. Biosci Biotechnol Biochem. 1996;60:634–639. doi: 10.1271/bbb.60.634. [DOI] [PubMed] [Google Scholar]
- 4.Ara K, Igarashi K, Saeki K, Kawai S, Ito S. Purification and some properties of an alkaline pullulanase from alkalophilic Bacillus sp. KSM-1876. Biosci Biotechnol Biochem. 1992;56:62–65. [Google Scholar]
- 5.Ara K, Igarashi K, Saeki K, Takaiwa M, Uemura T, Hagihara H, Kawai S, Ito S. Purification and characterization of an alkaline amylopullulanase with both α-1,4 and α-1,6 hydrolytic activity from alkalophilic Bacillus sp. KSM-1378. Biochim Biophys Acta. 1995;1243:315–324. doi: 10.1016/0304-4165(94)00148-q. [DOI] [PubMed] [Google Scholar]
- 6.Ara K, Saeki K, Ito S. Purification and characterization of an alkaline isoamylase from an alkalophilic strain of Bacillus. J Gen Microbiol. 1993;139:781–786. [Google Scholar]
- 7.Bisgaard-Frantzen H, Svendsen A, Norman B, Pedersen S, Kjærulff S, Outtrup H, Borchert T V. Development of industrially important α-amylases. J Appl Glycosci. 1999;46:199–206. [Google Scholar]
- 8.Brot N, Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 9.Estell D A, Graycar T P, Wells J A. Engineering an enzyme by site-directed mutagenesis to be resistant to chemical oxidation. J Biol Chem. 1985;260:6518–6521. [PubMed] [Google Scholar]
- 10.Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol. 1989;39:224–229. [Google Scholar]
- 11.Gordon R E, Haynes W C, Pang C H. The genus Bacillus. U.S. Washington, D.C.: Department of Agriculture; 1973. [Google Scholar]
- 12.Hatada Y, Igarashi K, Ozaki K, Ara K, Hitomi J, Kobayashi T, Kawai S, Watabe T, Ito S. Amino acid sequence and molecular structure of an alkaline amylopullulanase from Bacillus that hydrolyzes α-1,4 and α-1,6 linkages in polysaccharides at different active sites. J Biol Chem. 1996;271:24075–24083. doi: 10.1074/jbc.271.39.24075. [DOI] [PubMed] [Google Scholar]
- 13.Horikoshi K. Production of alkaline enzymes by alkalophilic microorganisms. II. Alkaline amylase produced by Bacillus no. A-40-2. Agric Biol Chem. 1971;35:1783–1791. [Google Scholar]
- 14.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyun H H, Zeikus J G. General biochemical characterization of the thermostable extracellular β-amylase from Clostridium thermosulfurogenes. Appl Environ Microbiol. 1985;49:1162–1167. doi: 10.1128/aem.49.5.1162-1167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi K, Ara K, Saeki K, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene that encodes a neopullulanase from an alkalophilic Bacillus. Biosci Biotechnol Biochem. 1992;56:514–516. doi: 10.1271/bbb.56.514. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi K, Hatada Y, Hagihara H, Saeki K, Takaiwa M, Uemura T, Ara K, Ozaki K, Kawai S, Kobayashi T, Ito S. Enzymatic properties of a novel liquefying α-amylase from an alkaliphilic Bacillus isolate and entire nucleotide and amino acid sequences. Appl Environ Microbiol. 1998;64:3282–3289. doi: 10.1128/aem.64.9.3282-3289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igarashi K, Hatada Y, Ikawa K, Araki H, Ozawa T, Kobayashi T, Ozaki K, Ito S. Improved thermostability of a Bacillus α-amylase by deletion of an arginine-glycine residue is caused by enhanced calcium binding. Biochem Biophys Res Commun. 1998;248:372–377. doi: 10.1006/bbrc.1998.8970. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi K, Ozawa T, Ikawa-Kitayama K, Hayashi Y, Araki H, Endo K, Hagihara H, Ozaki K, Kawai S, Ito S. Thermostabilization by proline substitution in an alkaline, liquefying α-amylase from Bacillus sp. strain KSM-1378. Biosci Biotechnol Biochem. 1999;63:1535–1540. doi: 10.1271/bbb.63.1535. [DOI] [PubMed] [Google Scholar]
- 20.Ikawa K, Araki H, Tsujino Y, Hayashi Y, Igarashi K, Hatada Y, Hagihara H, Ozawa T, Ozaki K, Kobayashi T, Ito S. Hyperexpression of the gene for a Bacillus α-amylase in Bacillus subtilis cells: enzymatic properties and crystallization of the recombinant enzyme. Biosci Biotechnol Biochem. 1998;62:1720–1725. doi: 10.1271/bbb.62.1720. [DOI] [PubMed] [Google Scholar]
- 21.Ito S, Kobayashi T, Ara K, Ozaki K, Kawai S, Hatada Y. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles. 1998;2:185–190. doi: 10.1007/s007920050059. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Manning G B, Campbell L L. Thermostable α-amylase of Bacillus stearothermophilus. J Biol Chem. 1961;236:2952–2957. [PubMed] [Google Scholar]
- 24.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Miller G L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 26.Mitchell E D, Jr, Loring R, Carraway K L. Oxidative inactivation of B. subtilis α-amylase by chloroperoxidase. Proc Okla Acad Sci. 1981;61:43–48. [Google Scholar]
- 27.Nielsen P, Fritze D, Priest F G. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology. 1995;141:1745–1761. [Google Scholar]
- 28.Norman B E. A novel debranching enzyme for application in the glucose syrup industry. Starch/Stärke. 1982;34:340–346. [Google Scholar]
- 29.Saito N. A thermophilic extracellular α-amylase from Bacillus licheniformis. Arch Biochem Biophys. 1973;155:290–298. doi: 10.1016/0003-9861(73)90117-3. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Shima S, Yanagi M, Saiki H. The phylogenetic position of Hydrogenobacter acidophilus based on 16S rRNA sequence analysis. FEMS Microbiol Lett. 1994;119:119–122. doi: 10.1111/j.1574-6968.1994.tb06877.x. [DOI] [PubMed] [Google Scholar]
- 32.Stauffer C E, Etson D. The effect on subtilisin activity of oxidizing a methionine residue. J Biol Chem. 1969;244:5333–5338. [PubMed] [Google Scholar]
- 33.Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomazic S J, Klibanov A M. Why is one Bacillus α-amylase more resistant against irreversible thermoinactivation than another? J Biol Chem. 1988;263:3092–3096. [PubMed] [Google Scholar]
- 36.UpaDek H, Kottwitz B. Application of amylases in detergents. In: van Ee J H, Misset O, Baas E J, editors. Enzymes in detergency. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 203–212. [Google Scholar]
- 37.Vallee B L, Stein E A, Sumerwell W N, Fischer E H. Metal content of α-amylases of various origins. J Biol Chem. 1959;234:2901–2905. [PubMed] [Google Scholar]
- 38.Welker N E, Campbell L L. Unrelatedness of Bacillus amyloliquefaciens and Bacillus subtilis. J Bacteriol. 1967;94:1124–1130. doi: 10.1128/jb.94.4.1124-1130.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto T The Amylase Research Society of Japan, editors. Handbook of amylases and related enzymes. Oxford, England: Pergamon Press; 1988. Bacterial α-amylase (liquefying and saccharifying types) of Bacillus subtilis and related bacteria; pp. 40–45. [Google Scholar]