Abstract
Today, 2019 Coronavirus (COVID-19) infections are a major health concern worldwide. Therefore, detecting COVID-19 in X-ray images is crucial for diagnosis, evaluation, and treatment. Furthermore, expressing diagnostic uncertainty in a report is a challenging duty but unavoidable task for radiologists. This study proposes a novel CNN (Convolutional Neural Network) model for automatic COVID-19 identification utilizing chest X-ray images. The proposed CNN model is designed to be a reliable diagnostic tool for two-class categorization (COVID and Normal). In addition to the proposed model, different architectures, including the pre-trained MobileNetv2 and ResNet50 models, are evaluated for this COVID-19 dataset (13,824 X-ray images) and our suggested model is compared to these existing COVID-19 detection algorithms in terms of accuracy. Experimental results show that our proposed model identifies patients with COVID-19 disease with 96.71 percent accuracy, 91.89 percent F1-score. Our proposed approach CNN’s experimental results show that it outperforms the most advanced algorithms currently available. This model can assist clinicians in making informed judgments on how to diagnose COVID-19, as well as make test kits more accessible.
Keywords: CNN, COVID-19, Deep Learning, MobileNetv2, ResNet50, X-ray images
1. Introduction
The new type of coronavirus (Covid-19) sickness, which has afflicted the entire planet in the previous year and a half, first appeared in December 2019 in Wuhan, China, and quickly spread throughout the world. It is highly contagious and, in severe cases, can result in acute respiratory distress or multi-organ failure [1], [2], [3], [4].
The World Health Organization designated the disease a “public health emergency of worldwide significance” on January 30, 2020. Reverse transcription polymerase chain reaction (RT-PCR) testing is commonly used to detect COVID-19 [5]. According to WHO, nucleic acid identification in secretory fluid taken from a throat swab by RT–PCR is the most accurate diagnosis of COVID-19 infection. Serial testing may be required to rule out the potential for false negative results, and co-infection with other viruses can impact the accuracy of RT-PCR tests. However, the primary problem with this method is that it suffers from low sensitivity and specificity [6]. In addition, due to the scarcity of RT-PCR test kits in remote rural areas, doctors recommend using medical images for COVID-19 screening [7]. The viral load and virus exposure time are related to RT-PCR false negatives. In other words, the chance of a false negative for RT-PCR is higher when the test is performed too early [8]. These are mistakes that can happen when collecting, transporting, and managing RNA samples. Taking care when collecting throat and nasal swabs can have a big impact on test accuracy. Sample collection is sometimes insufficient, or health personnel do not insert nose swabs far enough into the nose to get a sample with an appropriate virus load. Finding the best sample type at the right moment during an illness can yield the best results with the fewest false negatives [9], [10]. Many countries, however, are unable to conduct widespread testing due to resource restrictions such as testing time (3–4 h per round), basic equipment, experienced people, and reagents [11].
Despite the advantages of CT scan images, similar features are seen between COVID-19 and other lung diseases. Therefore, scanning is not an easy task to carry out. Recently, it has become useful to extract and detect some features from radiological images using machine learning and deep learning techniques [12]. CT can show certain distinctive indications in the lung associated with COVID-19 as a noninvasive imaging method [13], [14]. As a result, these approaches may be useful for early detection and diagnosis of COVID-19. COVID-19 has been identified using volumetric CT images in prior studies [5], [7]. The expense and time involved in adopting the CT technique are the main drawbacks [15]. X-ray imaging has been frequently used for COVID-19 screening in comparison to CT imaging because it requires less imaging time, which is cheaper, and X-ray scanners are generally available even in rural regions [16]. For these reasons, the diagnosis of COVID-19 using X-ray images has been carried out in many studies and presented to the literature [17], [18], [19], [20].
Due to its high feature extraction capability, artificial intelligence utilizing deep learning technology has recently demonstrated tremendous success in medical imaging applications and is thus favoured by researchers [21], [22] are two examples. On pediatric chest radiographs, deep learning has been used to detect and differentiate bacterial and viral pneumonia [23], [24]. Attempts to identify various imaging characteristics of chest CT have also been undertaken in several studies [25], [26].
In general, the CXR approach is one of the first tools for radiologists use to discover chest pathology. Numerous studies have been conducted to identify the COVID-19, according to the COVID-19 scenario [1], [13], [15], [27], [28]. As a result, our research is only focused on the use of X-ray imaging to potentially detect COVID-19 patients. Computer-Aided Diagnosis (CAD) technologies have been enhanced to enable physicians automatically to identify potential disorders of organs on X-ray images, overcoming previous limits [29], [30]. These systems are primarily powered by fast and powerful computer technology (such as CPU and GPU) and used to perform medical vision computing algorithms such as image enhancement, classification, segmentation, and tumor detection [31], [32], [33], [34].
Artificial intelligence approaches such as deep neural networks and machine learning have become the essence of cutting-edge CAD applications in a variety of medical sectors. In recent years, deep learning approaches have been used to automatically analyze multimodal medical images, with promising results for performing radiological tasks [34], [35], [36]. Following a review of recent publications in the literature, a strategy based on a pre-trained ResNet32 model that employs a transfer learning approach to detect COVID-19 disease is proposed. In a sample of 852 CT scan images, the researchers employed tests to identify lung cancer (413 COVID-19 and 439 normal). They were able to achieve training and testing accuracy of up to 96.22 % and 93.01 %, respectively, using the proposed technique in their experimental studies [37]. They designed a strategy based on the DenseNet model that was pre-trained for COVID-19 identification. Their experiments utilizing the COVID-CT dataset, they attained an accuracy of 84.7 % [38]. To extract possibly diseased regions from CT scan images, they recommended using a pre-trained three-dimensional convolutional neural network (CNN). In the experiments, they were able to predict viral pneumonia, COVID-19, and healthy patients with an accuracy of 86.7 % [39].
In this study, it is aimed to design an artificial intelligence-based COVID-19 diagnosis system. Novelty has been added to the study with a proposed new CNN model, and this model is compared with the most common models available in the literature. On the used dataset, the performance of three different deep learning approaches for classifying lung X-ray images as COVID-19 and normal patients is evaluated and compared. The proposed model consists of fewer convolution layers than the MobileNetv2 and ResNet50 architectures. Therefore, the number of calculated parameters also decreases. In this direction, high performance results are obtained with less processing power and time. The following is an overview of the paper’s structure. The data collection and deep learning models are described in Section 2. The created deep learning application models' parameters and relevant information are provided. Section 3 presents deep learning classifier experimental work and results as well as evaluation metrics such as accuracy, recall, precision, and F1-scores. Section 4 conclusions and suggests areas for future investigation. Finally, discussion is given in Section 5.
2. Material and methods
2.1. Dataset
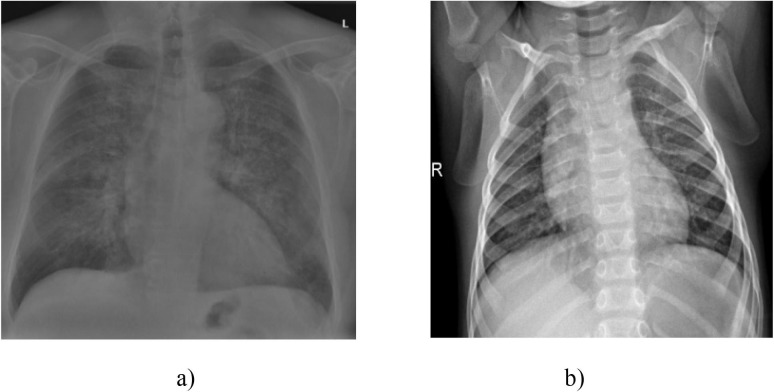
In this study, a open access dataset of X-ray images shared publicly by the Kaggle platform is used to classify COVID-19 patients (https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database) [40]. This data set obtained from X-ray images of Covid-19 consists of two categories: COVID-19 positive and negative (normal); and our dataset includes 13,824 chest X-ray images. A cross-section of the chest X-ray images obtained from this dataset is given in Fig. 1 .
Fig. 1.
X-ray images a) with positive COVID-19 b) normal cases [40].
2.2. Deep learning
Deep learning is a machine learning technique that makes use of many nonlinear information processing layers and is used for feature extraction, pattern analysis and classification in supervised or unsupervised learning [41]. Deep learning, which is a subset of machine learning, is a set of methods based on deep architecture, consisting of artificial neural networks with an increased number of hidden layers and learning a problem-related feature in each layer. In this architecture, the features learned in each layer represent the input data for the next deeper layer. As a result, a structure is established in which the most basic to the most complex quality is learned from the lowest to the highest layer [42], [43].
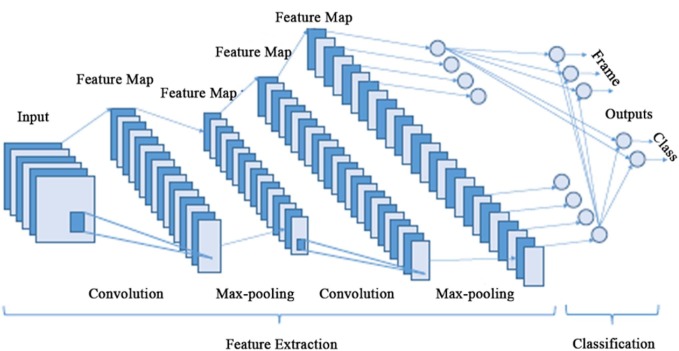
Deep learning methods differ from previous machine learning algorithms in that they require a large amount of data and hardware with significant computational capacity to analyze these data. Most of these algorithms used in image classification applications are based on architectures called Convolutional Neural Networks (CNN), and their structure is shown in Fig. 2 [42].
Fig. 2.
The overall architecture of the CNN.
2.2.1. Convolutional neural network models (CNN)
CNNs have been widely used by researchers in recent years in image and video processing, image classification, object detection and segmentation, which are the widest areas of deep learning applications. They are also used on other signals containing sequential and interrelated data other than the image [44]. CNNs consist of three main layers called the convolution layer, pooling layer and fully connected layer [45]. CNN layers are given below:
a. Input layer
The first layer of the convolutional neural network architecture is the input layer. Image data should be included in the CNN input layer. The size of the data is critical for the model’s effectiveness in this section. The high memory need, the training time, and the test time per image may all rise if the input image size is chosen large. In addition, their chances of success may improve. The input sizes of used models in the application are given in Table 1 .
Table 1.
The input image size of used models.
| Number | Network | Input Image Size |
|---|---|---|
| 1 | CNN_Model (Proposed) | 224 × 224x3 |
| 1 | MobileNetv2 | 224 × 224x3 |
| 2 | ResNet50 | 224 × 224x3 |
b. Convolutional layer
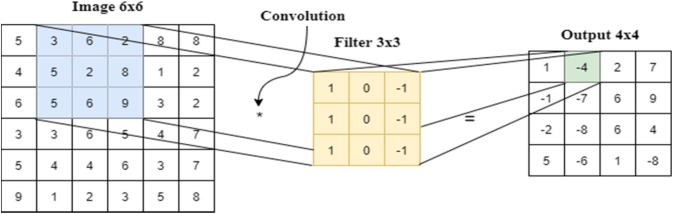
This layer can be defined as feature extraction filters (kernels) on images [3]. Each filter represents a particular matrix that performs the convolution operation on the input image. An example of convolutional layer is given in Fig. 3 . The proposed model, which includes 5 convolution layers (detailed in Section 3.3) are the Sequential model of Keras.
Fig. 3.
Outline of convolutional layer.
c. The pooling layer
One of the most common ways in deep learning architectures can be to include pooling layers between convolution layers. By decreasing parameters and computational effort in the network topology, the pooling layer reduces overfitting. This layer’s aim is to reduce the size of the feature maps in order to deal with the image’s complexity. The proposed model in our study, which includes max pooling layers, is the Sequential model of Keras.
d. Activation layer
A non-linear layer (or activation layer) is added immediately after each convolution layer. This layer’s objective is to add nonlinearity to a system that primarily computes linear operations throughout its convolutional levels. The function in the activation layer processes the systems’ input and decides the value that the system will generate in response to it. ReLu activation function which is given in Eq. (1) is used in each layer of the proposed model and softmax function is used in the last layer.
| (1) |
e. Fully Connected layer
In the CNN models, each neuron in one layer is connected to a neuron in another layer through fully connected layers. In principle, fully connected layers act as multi-layer perceptrons (MLPs). The only difference is that the data to feed the fully connected layer is in the form created by the convolutional layers. In this study, the number of classes (COVID-19, and Normal) is 2. For this reason, the output value of the fully connected layer 2 of our model is given.
f. Dropout
To prevent the network from memorizing, the dropout layer is utilized. The model is capable of memorizing training data and severe learning. The learning ability of the network is lost if it engages in an excessive learning process. Some network nodes are disabled at random throughout the dropout process [46]. In this study, 2 dropout layers are used to prevent over-learning, and the area of the first layer is determined as 108x108x128 and the area of the other dropout layer is 128.
2.2.2. Convolutional neural network models
In order to compare the study’s outcomes, some of the most well-known CNN models utilized in previous experiments are presented in this section. On the ImageNet dataset, these architectures are deep learning models with pre-trained weights. Prediction, feature extraction, and classification can all be done with these models. A pre-trained image classification model can be utilized as a starting point for learning a new task because it has learned to extract powerful and useful features from photos. In the image classification part, pre-trained ResNet50 and MobileNetv2 architectures are used and analyzed. MobileNetV2 is a neural network design that achieves outstanding results when it comes to balancing resource constraints and recognition accuracy. MobileNetV2 is a neural network design that achieves outstanding results when it comes to balancing resource constraints and recognition accuracy. It is also one of the most significant benefits of being useable on mobile devices and embedded systems. Deep CNN designs face a number of challenges, including network optimization, vanishing gradient concerns, and distortion issues. The ResNet50 design uses residual blocks to address all of these issues in the training process, including as saturation and accuracy loss [47]. In our study, these models are chosen because they are frequently used in deep learning applications [48], [49]. Pre-trained networks and some features are given in Table 2 .
Table 2.
Pre-trained networks and some features.
| Number | Network | Depth | Parameter (million) | Input Image Size |
|---|---|---|---|---|
| 1 | MobileNetv2 | 53 | 3.5 | 224 × 224 |
| 2 | ResNet50 | 50 | 25.6 | 224 × 224 |
Table 3 shows the parameter which is explained below:
Table 3.
Parameters.
| Predicted |
|||
|---|---|---|---|
| Negative | Positive | ||
| Actual Cases | Negative | True Negatives (tn) | True Positives (tp) |
| Positive | False Negatives (fn) | False Positives (fp) | |
True negative: which is the proportion of COVID-19 negative cases which are classified correctly?
Calculated as: tn/tn + fp.
False Positive: which is the proportion of COVID-19 negative cases which are classified incorrectly as positive? Calculated as: fp/tn + fp.
False Negative: which is the proportion of COVID-19 positive cases which are classified incorrectly as negative?
Calculated as: fn/fn + tp.
True Positive: proportion of COVID-19 positive cases that are correctly classified.
Calculated as: tp/fn + tp.
The loss functions are a function that map to a real number to represent the error rate of the model as well as its performance. In short, it calculates how much the prediction made by the model deviates from the true value. In this case, the loss of a good model should be close to 0. The loss function should fit the model and the problem. In this study, the Binary Cross Entropy loss function, which is used in binary classification tasks, is used. The mathematical calculation of the function is given in the Eq. (2) [50] .
| (2) |
In the equation, N represents the number of samples of the data, yi represents the true value of the ith data, and ŷi represents the estimated value of the ith data. Loss functions enable the model to learn by reducing the error in the estimation with the help of optimization algorithms. In this study, Adam optimization algorithm, one of the stochastic gradient descent algorithms, is used as the optimization algorithm. Performance comparisons between classifications are evaluated according to various parameters given below.
Accuracy: This ratio represents the percentage of samples classified as true. It is given in Eq. (3) is a measure of how well the learning model is [51], [52];
| (3) |
True Positive Rate (Recall, (TPR)): It gives how many of the actually correct ones are marked correct in the prediction [51] and is given in Eq. (4).
| (4) |
Precision: It gives how much of what is marked as correct in the prediction section is actually correct. It is given in Eq. (5) [51].
| (5) |
F1-Score (F1-Score): It is the harmonic mean of the precision and recall. which is shown in Eq. (6). It can be used as a comparison metric between two models. For example, if we consider that a model has high precision and a very low recall value, and the situation is the opposite in the second model, it would be more accurate to choose the F1-score in the comparison to be made between these two models.
| (6) |
3. Experimental results
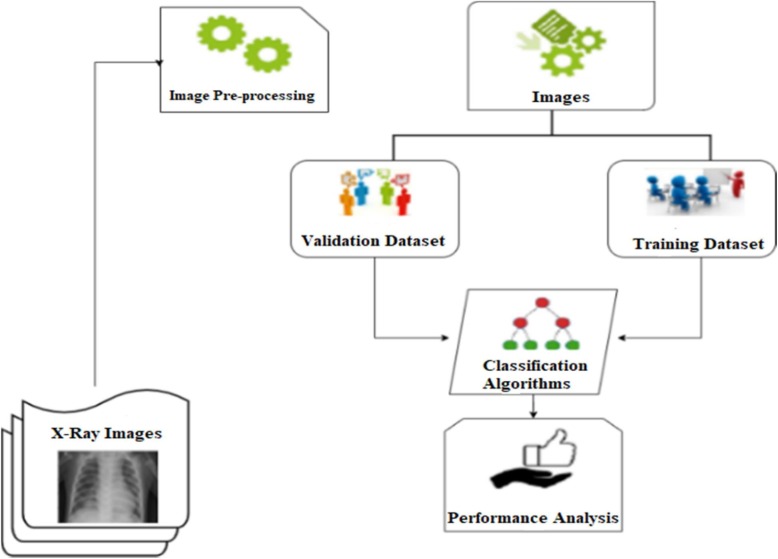
In this part, the models trained with the training data are then tested (evaluated) with the test data. Models are compared according to accuracy and F1-score performance metrics. The model is trained using computational resources provided by Google Colaboratory. The block diagram of whole study is illustrated in Fig. 4 .
Fig. 4.
Block diagram of the designed system.
3.1. Pre-processing
All X-ray pictures were gathered into a single dataset and scaled to a standardized size of 224x224 pixels so that they could be used in the deep learning pipeline. Image labeling is then used to indicate whether each image in the dataset has positive COVID-19.
3.2. Datasets and their application to the model
CNN models create parameters based on inputs of a fixed size. To avoid losing features in the images, the X-ray images in the dataset are scaled to 224x224 after reading. The data set is randomly divided into 80 % training and 20 % testing for testing. The number of images used as training and testing sets is given Table 4 . The dataset can be accessed via the given link (https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database).
Table 4.
The number of images used as training and testing sets.
| Name | COVID-19 cases | Normal cases |
|---|---|---|
| Train | 2919 | 8140 |
| Test | 707 | 2058 |
3.3. Network architecture
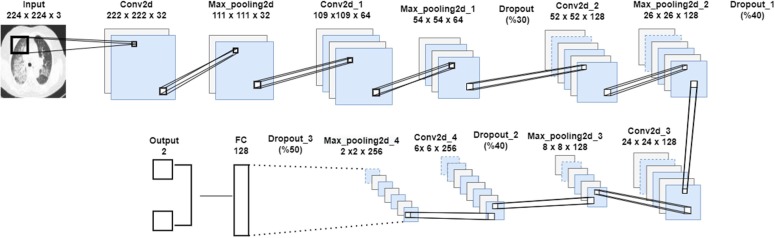
In this study, a CNN model is proposed for the diagnosis of COVID-19 from lung radiography X-ray images. The architecture of the model is given in Fig. 5 . The proposed model, which includes convolution and max pooling layers, is the Sequential model of Keras. The CNN model, which consists of sequential convolution and pooling layers, is trained with the X-ray images dataset and the results are observed. Pre-trained and proven MobileNetv2, ResNet50 models with the proposed model dataset are trained with the same dataset with 80 % training and 20 % testing. The proposed CNN model details are given in Table 5 .
Fig. 5.
A summary on our proposed CNN model.
Table 5.
The proposed CNN model details.
| Layer (type) | Output Shape | Parameter |
|---|---|---|
| Conv2D | 222 × 222 × 32 | 896 |
| MaxPooling2D | 111 × 111 × 32 | 0 |
| Conv2D | 109 × 109 × 64 | 18,496 |
| MaxPooling2D | 54 × 54 × 64 | 0 |
| Dropout | 54 × 54 × 64 | 0 |
| Conv2D | 52 × 52 × 128 | 73,856 |
| MaxPooling2D | 26 × 26 × 128 | 0 |
| Dropout | 26 × 26 × 128 | 0 |
| Conv2D | 24 × 24 × 128 | 147,584 |
| MaxPooling2D | 8 × 8 × 128 | 0 |
| Dropout | 8 × 8 × 128 | 0 |
| Conv2D | 6 × 6 × 256 | 295,168 |
| MaxPooling2D | 2 × 2 × 256 | 0 |
| Flatten | 1024 | 0 |
| Dense | 128 | 131,200 |
| Dropout | 128 | 0 |
| Dense | 2 | 258 |
| Total Params: 667.458 Trainable Params: 667.458 Non-trainable Params: 0 | ||
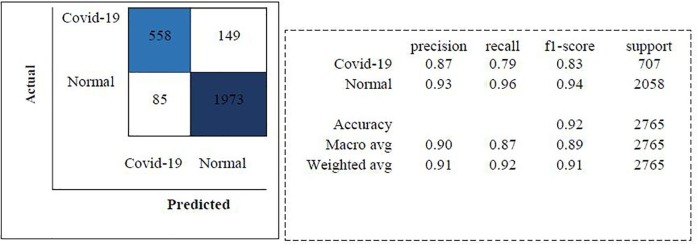
The trainings are carried out in 30 batch sizes and using the Adam optimization algorithm. The learning rate is determined as 1e-4. ReLu is used as activation function in each layer of the proposed model and softmax function is used in the last layer. The models trained with the training data are then tested with the test data. Models are compared according to accuracy and F1-score performance metrics. As a result of the test of the proposed model, an accuracy rate of 96.71 % is obtained. The classification report and confusion matrix of the models are given in Fig. 6, Fig. 7, Fig. 8 , respectively.
Fig. 6.
Classification Report and Confusion Matrix for the proposed CNN model.
Fig. 7.
Classification Report and Confusion Matrix for the MobileNetv2 model.
Fig. 8.
Classification Report and Confusion Matrix for the ResNet50 model.
Although the MobileNetv2 architecture is generally designed for low-computing systems such as mobile devices, it can be used like any other architecture. With the model designed using the MobileNetv2 architecture, a test accuracy rate of 95.73 % is achieved. The ResNet50 architecture uses residual blocks to pass output from a previous layer to the next. This helps alleviate the vanishing gradient problem. With the model designed using the ResNet50 architecture, a test accuracy rate of 91.54 % is achieved.
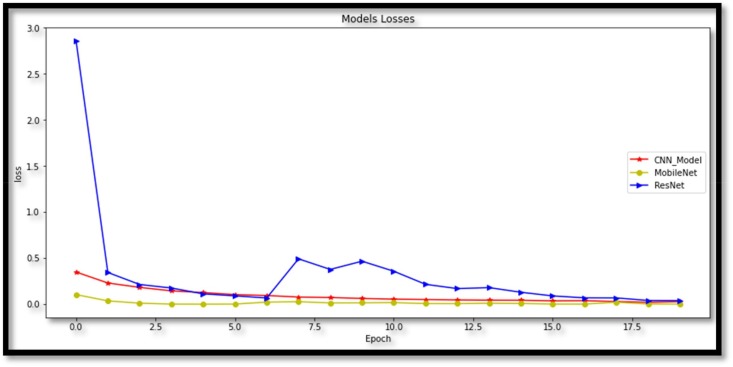
The goal of this study is to detect patients with covid-19 and classify them as covid-19 possitive or covid-19 negative by using X-ray images. These classes are COVID-19 and normal (non-COVID-19). In addition, the loss function is extremely important to understand the status of predictions in training. The difference between the estimated and actual output is measured in losses (i.e. the label). Accuracy and loss are supposed to be inversely proportional based on the definitions in these graphs: low loss levels are expected for high accuracy values. The epoch is a network designer-defined parameter. The number of epochs chosen ensures that the loss is minimal and that it does not worsen in subsequent epochs. As a result, the accuracy value attained is the highest and does not increase in subsequent epochs, indicating that the network has reached stability and that future epochs will not improve performance. Because both models find stability with a number of epochs less or equal to 20, the number of epochs is set to 20 in Fig. 9 .
Fig. 9.
Loss graph of models.
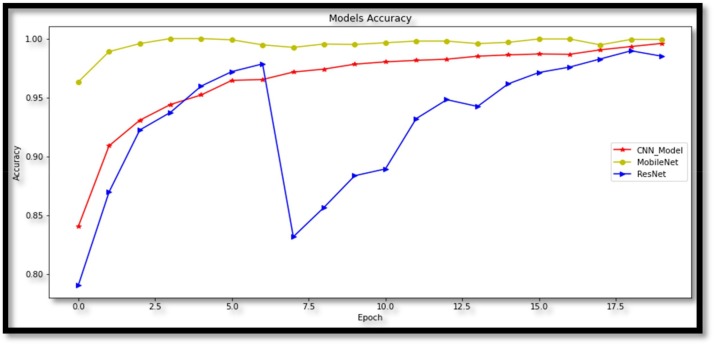
In addition to the proposed Convolutional Neural Network model, the training is carried out using two pre-trained network architectures, MobileNetv2 and ResNet50 models. These three models are evaluated by comparing each other according to their performance. The running performance of the proposed deep learning frameworks is evaluated on the basis of accuracy and F1-score. While the proposed model reaches 99.08 % training accuracy, other models; MobileNetv2 and ResNet50 achieved training accuracy of 99.93 % to 98.50 %, respectively. In Fig. 10 , the accuracy graph of the models over 20 epochs is given.
Fig. 10.
Accuracy graph of models.
The difference in the number of data between the two classes in the training data may affect the model performance. The class with more numbers can suppress the other class. In the training data used in the study, the normal class consists of 8140 and the COVID-19 class consists of 2919 samples. In this case, it is not appropriate to only take accuracy as a performance metric. Comparisons are made by taking into account the F1-score performance metric, which is the harmonic mean of precision and recall values, apart from accuracy. The models and performance metrics used in this study are given in Table 6 .
Table 6.
Accuracy comparision our proposed model vs existing models.
| Number | Architecture of model | Training accuracy (%) | Test accuracy (%) | F1 score (%) |
|---|---|---|---|---|
| 1 | CNN_Model (Proposed) | 99.08 | 96.71 | 97 |
| 2 | MobileNetv2 | 99.93 | 95.73 | 96 |
| 3 | ResNet50 | 98.50 | 91.54 | 91 |
With the proposed CNN model, 96.71 % test accuracy and 97 % F1-score have been achieved, and its usability in the diagnosis of COVID-19 has been proven. In addition, as a result of testing with Convolutional Neural Network architectures that have proven themselves in ImageNet, a test accuracy rate of at least 91.54 % is obtained. As can be seen from the results, CNNs provide higher accuracy and speed in the diagnosis of COVID-19 with X-ray images compared to other medical tests. The performances of pretrained models are compared to those of other proposed models in Table 6. In convolutional neural networks, some methods have been developed to avoid overfitting and underfitting the model. The overfitting is not occured in our study due to the use of dropout and max pooling layers. Comparisons of with other methods are given in Table 7 .
Table 7.
Comparisons of with other studies
| Class | Data Size | Reference | Methods | Result (Accuracy) |
|---|---|---|---|---|
| COVID-19 and non COVID-19 or pneumonia | 413 COVID-19 439 Normal or Pneumonia (CT images) |
[37] | ResNet50 | 93.01 % |
| COVID-19 Pneumonia |
108 COVID-19 86 Pneumonia Totally 1020 images (CT images) |
[53] | AlexNet | 78.92 % |
| COVID-19 Normal, Pneumonia |
358 COVID-19 (+) 5538 COVID-19 (-) 8066 Healthy (Chest X-ray) |
[54] | COVID-NET | 93.3% |
| COVID-19, Normal, Pneumonia |
127 COVID-19 (+) 127 COVID-19 (-) (Chest X-ray) 127 Pneumonia |
[55] | ResNet50 + SVM | 95.33 % |
| COVID-19 Normal Pneumonia |
225 COVID-19 1583 Normal 4292 Pneumonia (X-ray images) |
[56] | CNN | 98.50 % |
| COVID-19 Normal |
142 COVID-19 142 Normal (X-ray images) |
[57] | nCOVnet | 88.1 % |
| COVID-19 Normal, Pneumonia |
285 COVID-19 285 Normal 285 Pneumonia (X-ray images) |
[58] | CNN | 94.03 % |
Different types of viral pneumonia have similar CXR scan images, making it difficult for radiologist to distinguish COVID-19 from other viral pneumonia. This limitation can result in misdiagnosis, as well as non-COVID-19 viral pneumonia being misdiagnosed as COVID-19 pneumonia [49], [59]. Another disadvantage of this study is that the networks are trained on individual slices (images), rather than using all available samples for each subject.
4. Discussion
The findings show that deep learning with CNNs can have a considerable impact on the automatic detection and extraction of important information from X-ray images, which is relevant to the diagnosis of the COVID-19. Various attempts have been made to develop a reliable diagnostic model using deep learning techniques and many studies have been recommended in the literature. Also, due to the early stage of the disease, the number of labeled data points available is limited and most previous methods are evaluated using limited data. Extensive experiments are performed on a relatively large dataset, taking into account several factors, to determine the best-performing model for automated COVID-19 screening.
Our currently proposed work consists of three different models used to perform the aforementioned classification tasks. For classification tasks, we have considered MobileNetv2, ResNet50, and the proposed CNN models. We have achieved different levels of accuracy as shown in Table 6. Test accuracy of 96.71 % and F1 score of percent are attained using the proposed CNN model, and its applicability in the diagnosis of COVID-19 is demonstrated. When the accuracy rate obtained in our study is compared to studies published in the literature and cited in the paper, it is clear that it is high.
The proposed work has several restrictions as well. COVID-19 and other viral pneumonias exhibit similar CXR scan pictures, making it difficult for radiologist to discriminate between them. This limitation can lead to misdiagnosis, as well as the misdiagnosis of non-COVID-19 viral pneumonia as COVID-19 pneumonia. Another flaw in this study is that the networks are trained on individual slices (images) rather than all of the samples available for each person. Our proposed study does not consider the same patient data to classify COVID-19 positive X-ray images as severe, moderate, and mild. Severity classification would be more useful if the same patient data could be obtained that would show disease progression and thus classify images into three classes. In addition, X-ray machines require less maintenance in terms of their reagents compared to RT-PCR and hence the operating cost is relatively low. In further studies, other current deep learning-based approaches will be investigated, and it is planned to work on popular deep learning techniques, hybrid methods and more data.
More publicly available chest image datasets can be collected and produced for future usage. By adding more chest X-ray samples to the training dataset, we can achieve higher accuracy with the model architecture we used before. In the future, an architecture can be prepared that will perform the successful grouping of chest X-rays in aforementioned classes in one go. The performance of deep learning models cannot be enhanced without the availability of high-quality data. Other research areas include data construction and annotation, as well as metadata information. In the future, our AI-model will help large-scale investigations aimed at determining the presence of “invisible cases,” such as research aimed at determining how many people without noticeable symptoms could be infected by the virus.
5. Conclusions
COVID-19 has had a major negative impact on our life, ranging from public health to the global economy. Using chest X-ray images from COVID-19 patients as both normal and COVID-19 positive, we describe in this work a deep learning-based method for automatically identifying COVID-19 disease. The most popular models in the literature (MobileNetv2 and ResNet50) are examined with the novel CNN model that has been developed for the study. Three alternative deep learning algorithms are tested for accuracy in this study in order to effectively identify and classify COVID-19 patients. The proposed CNN model has a test accuracy of 96.71 percent and an F1 score of 97 percent, and its applicability in the diagnosis of COVID-19 has been demonstrated. There are fewer convolution layers in the proposed CNN model than in the MobileNetv2 and ResNet50 designs. As a result, the number of calculated parameters is reduced. As a result, high-performance results can be achieved while using minimal computing power. We expect that using a computer-assisted diagnostic tool to diagnose COVID-19 cases will considerably increase the speed and accuracy of diagnosis. Furthermore, the proposed CNN model can be utilized to extract sensitive information from X-ray images in order to identify patients. The development of a larger dataset for the evaluation of classification algorithms is another contribution to the article. In terms of accuracy, the proposed model has been demonstrated to outperform a number of existing strategies. As can be observed from the results, the proposed CNN model outperforms previous medical tests in terms of accuracy and speed in diagnosing COVID-19 using X-ray images. This proposed model can be employed as an alternative or helper system to RT-PCR in remote places where identification kits and specialist physicians are scarce. Our future goal is to get around hardware restrictions that prevent us from using larger image sets to train our proposed model and compare its performance with a larger number of existing methods. One of the limitations of this research is the difficulty of finding a labelled COVID-19 pneumonia dataset. Using more images in the training section is thought to assist us improve the model’s performance in the future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L.i. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q.i., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q.i., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G.e., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B.o., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox LeAnne, Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., Hu X., Chen J., Liu B.o. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleve G., Rizzotti L., Dellaglio F., Torriani S. Development of reverse transcription (RT)-PCR and real-time RT-PCR assays for rapid detection and quantification of viable yeasts and molds contaminating yogurts and pasteurized food products. Appl. Environ. Microbiol. 2003;69(7):4116–4122. doi: 10.1128/AEM.69.7.4116-4122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahreini F., Najafi R., Amini R., Khazaei S., Bashirian S. Reducing false negative PCR test for COVID-19. Int. J. Mater. Child Health AIDS. 2020;9(3):408. doi: 10.21106/ijma.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Internal Med. 2020;173(4):262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S., Agarwal R., Rehan M.A., Pathak S., Agarwal P., Gupta Y., Consul S., Gupta N., Ritika, Goenka R., Rajwade A., Gopalkrishnan M. A compressed sensing approach to pooled RT-PCR testing for COVID-19 detection. IEEE Open J. Signal Process. 2021;2:248–264. doi: 10.1109/OJSP.2021.3075913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.E. Özbay, F.A. Özbay. Derin Öğrenme ve Sınıflandırma Yaklaşımları ile BT görüntülerinden Covid-19 Tespiti. Dicle Üniversitesi Mühendislik Fakültesi Mühendislik Dergisi, 12(2), 211-219.
- 13.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroft L.J., van der Velden L., Girón I.H., Roelofs J.J., de Roos A., Geleijns J. Added value of ultra–low-dose computed tomography, dose equivalent to chest x-ray radiography, for diagnosing chest pathology. J. Thoracic Imaging. 2019;34(3):179. doi: 10.1097/RTI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process. Control. 2021;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 18.J. Zhang, Y. Xie, Y. Li, C. Shen, Y. Xia. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338, 27. 2020.
- 19.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142:110495. doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Basu, S. Mitra, N. Saha. Deep learning for screening covid-19 using chest x-ray images. In 2020 IEEE Symposium Series on Computational Intelligence (SSCI) (pp. 2521-2527). IEEE. 2020.
- 21.Xia C., Li X., Wang X., Kong B., Chen Y., Yin Y., Wu X. October). A multi-modality network for cardiomyopathy death risk prediction with CMR images and clinical information. Springer; Cham: 2019. pp. 577–585. [Google Scholar]
- 22.Kong B., Wang X., Bai J., Lu Y.i., Gao F., Cao K., Xia J., Song Q.i., Yin Y. Learning tree-structured representation for 3D coronary artery segmentation. Comput. Med. Imaging Graph. 2020;80:101688. doi: 10.1016/j.compmedimag.2019.101688. [DOI] [PubMed] [Google Scholar]
- 23.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., McKeown A., Yang G.e., Wu X., Yan F., Dong J., Prasadha M.K., Pei J., Ting M.Y.L., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V.A.N., Wen C., Zhang E.D., Zhang C.L., Li O., Wang X., Singer M.A., Sun X., Xu J., Tafreshi A., Lewis M.A., Xia H., Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Rajaraman S., Candemir S., Kim I., Thoma G., Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl. Sci. 2018;8(10):1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depeursinge A., Chin A.S., Leung A.N., Terrone D., Bristow M., Rosen G., Rubin D.L. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution CT. Invest. Radiol. 2015;50(4):261. doi: 10.1097/RLI.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthimopoulos M., Christodoulidis S., Ebner L., Christe A., Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 27.Ng M.Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Kuo M.D. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol.: Cardiothoracic Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Liu F., Li J., Zhang T., Wang D., Lan W. Clinical and CT imaging features of the COVID-19 pneumonia: focus on pregnant women and children. J. Infect. 2020;80(5):e7–e13. doi: 10.1016/j.jinf.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merk D.R., Karar M.E., Chalopin C., Holzhey D., Falk V., Mohr F.W., Burgert O. Image-guided transapical aortic valve implantation sensorless tracking of stenotic valve landmarks in live fluoroscopic images. Innovations. 2011;6(4):231–236. doi: 10.1097/IMI.0b013e31822c6a77. [DOI] [PubMed] [Google Scholar]
- 30.Messerli M., Kluckert T., Knitel M., Rengier F., Warschkow R., Alkadhi H., Leschka S., Wildermuth S., Bauer R.W. Computer-aided detection (CAD) of solid pulmonary nodules in chest x-ray equivalent ultralow dose chest CT-first in-vivo results at dose levels of 0.13 mSv. Eur. J. Radiol. 2016;85(12):2217–2224. doi: 10.1016/j.ejrad.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Tian X., Wang J., Du D., Li S., Han C., Zhu G., Tan Y., Ma S., Chen H., Lei M. Medical imaging and diagnosis of subpatellar vertebrae based on improved Laplacian image enhancement algorithm. Comput. Methods Programs Biomed. 2020;187:105082. doi: 10.1016/j.cmpb.2019.105082. [DOI] [PubMed] [Google Scholar]
- 32.He T., Hu J., Song Y., Guo J., Yi Z. Multi-task learning for the segmentation of organs at risk with label dependence. Med. Image Analysis. 2020;61:101666. doi: 10.1016/j.media.2020.101666. [DOI] [PubMed] [Google Scholar]
- 33.Hannan R., Free M., Arora V., Harle R., Harvie P. Accuracy of computer navigation in total knee arthroplasty: a prospective computed tomography-based study. Med. Eng. Phys. 2020;79:52–59. doi: 10.1016/j.medengphy.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Chen L., Bentley P., Mori K., Misawa K., Fujiwara M., Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Analysis. 2019;58:101539. doi: 10.1016/j.media.2019.101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao F., Yoon H., Wu T., Chu X. A feature transfer enabled multi-task deep learning model on medical imaging. Expert Syst. Appl. 2020;143:112957. [Google Scholar]
- 36.Kim M., Yan C., Yang D., Wang Q., Ma J., Wu G. Biomedical information technology. Academic Press; 2020. Deep learning in biomedical image analysis; pp. 239–263. [Google Scholar]
- 37.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S. Deep transfer learning based classification model for COVID-19 disease. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. Zhao, Y. Zhang, X. He, P. Xie. Covid-ct-dataset: a ct scan dataset about covid-19. arXiv preprint arXiv:2003.13865, 490. 2020.
- 39.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., Lang G., Li Y., Zhao H., Liu J., Xu K., Ruan L., Sheng J., Qiu Y., Wu W., Liang T., Li L. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database.
- 41.Deng L., Yu D. Deep learning: methods and applications. Found. Trends Signal Process. 2014;7(3–4):197–387. [Google Scholar]
- 42.G. Işık, H. Artuner. Recognition of radio signals with deep learning Neural Networks. In 2016 24th Signal Processing and Communication Application Conference (SIU) (pp. 837-840). IEEE. 2016.
- 43.Rawat W., Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017;29(9):2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 44.L. Bottou, C. Cortes, J.S. Denker, H. Drucker, I. Guyon, L.D. Jackel, ... & V. Vapnik. Comparison of classifier methods: a case study in handwritten digit recognition. In Proceedings of the 12th IAPR International Conference on Pattern Recognition, Vol. 3-Conference C: Signal Processing (Cat. No. 94CH3440-5) (Vol. 2, pp. 77-82). IEEE. 1994.
- 45.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep Learning. [Google Scholar]
- 46.Suh M.H., Zangwill L.M., Manalastas P.I.C., Belghith A., Yarmohammadi A., Medeiros F.A., Diniz-Filho A., Saunders L.J., Weinreb R.N. Deep retinal layer microvasculature dropout detected by the optical coherence tomography angiography in glaucoma. Ophthalmology. 2016;123(12):2509–2518. doi: 10.1016/j.ophtha.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theckedath D., Sedamkar R.R. Detecting affect states using VGG16, ResNet50 and SE-ResNet50 networks. SN Comput. Sci. 2020;1(2):1–7. [Google Scholar]
- 48.S. Hossain, R. Rahman, M.S. Ahmed, M.S. Islam. Pneumonia Detection by Analyzing Xray Images Using MobileNET, ResNET Architecture and Long Short Term Memory. In 2020 30th International Conference on Computer Theory and Applications (ICCTA) (pp. 60-64). IEEE. 2020.
- 49.Ibrahim A.U., Ozsoz M., Serte S., Al-Turjman F., Yakoi P.S. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cogn. Comput. 2021:1–13. doi: 10.1007/s12559-020-09787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.X. Mao, Q. Li, H. Xie, R.Y. Lau, Z. Wang. (2016). Multi-class generative adversarial networks with the L2 loss function. arXiv preprint arXiv:1611.04076, 5, 00102.
- 51.Burkov A. The Hundred-Page Machine Learning Book. 2019;Vol. 1:3–5. [Google Scholar]
- 52.M.E. Sahin, H. Ulutas, Esra, Yuce. A deep learning approach for detecting pneumonia in chest X-rays. Avrupa Bilim ve Teknoloji Dergisi, (28), 562-567.
- 53.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. Wang, A. Wong, COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images, 2020 arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 55.P.K. Sethy, S.K. Behera, Detection of Coronavirus Disease (COVID-19) Based on Deep Features, 2020.
- 56.Sekeroglu B., Ozsahin I. Detection of COVID-19 from Chest X-Ray images using convolutional neural networks. SLAS Technol. Transl. Life Sci. Innov. 2020;25:553–565. doi: 10.1177/2472630320958376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. Ahammed, M.S. Satu, M.Z. Abedin, M.A. Rahaman, S.M.S. Islam. Early Detection of Coronavirus Cases Using Chest X-ray Images Employing Machine Learning and Deep Learning Approaches. medRxiv 2020.
- 59.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]