Abstract
Worldwide, there are intensive efforts to identify repurposed drugs as potential therapies against SARS-CoV-2 infection and the associated COVID-19 disease. To date, the anti-inflammatory drug dexamethasone and (to a lesser extent) the RNA-polymerase inhibitor remdesivir have been shown to be effective in reducing mortality and patient time to recovery, respectively, in patients. Here, we report the results of a phenotypic screening campaign within an EU-funded project (H2020-EXSCALATE4COV) aimed at extending the repertoire of anti-COVID therapeutics through repurposing of available compounds and highlighting compounds with new mechanisms of action against viral infection. We screened 8702 molecules from different repurposing libraries, to reveal 110 compounds with an anti-cytopathic IC50 < 20 µM. From this group, 18 with a safety index greater than 2 are also marketed drugs, making them suitable for further study as potential therapies against COVID-19. Our result supports the idea that a systematic approach to repurposing is a valid strategy to accelerate the necessary drug discovery process.
Subject terms: Mechanisms of disease, Viral infection
| Measurement(s) | Cytopathic Effect |
| Technology Type(s) | confocal fluorescence microscopy |
| Factor Type(s) | Cellular toxicity |
| Sample Characteristic - Organism | Chlorocebus sabaeus |
| Sample Characteristic - Environment | continuant |
| Sample Characteristic - Location | Belgium |
Background & Summary
Coronaviruses are enveloped viruses carrying 27 to 31 kb single-stranded positive-sense RNA genomes encoding structural and accessory proteins. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a respiratory infection that was first recorded in December 2019 and declared as a pandemic in March 2020. Since the start of 2020, there has been almost 5.5 M deaths worldwide with at least 307 Mio assessed infection cases1.
Repurposing of known drugs offers a cost and time-effective alternative to the classical antiviral drug discovery approach, which can take many years and involves high costs. Combining high-performance computing and experimental biology is a useful route for drug repurposing efforts in rare, orphan and challenging diseases2 as well as health emergencies3. The high number of repurposing-based clinical trials currently active worldwide are part of an unprecedented effort in fighting SARS-CoV-24. In line with these efforts, EXSCALATE4COV (E4C) is an EU H2020-funded emergency project which uses high-performance computing and screening infrastructures to identify clinical candidates and progress mono- or combination therapies against SARS-CoV-2 viral infections5. Already in the context of the SARS-CoV-2 pandemic, E4C’s supercomputing platform6,7 and high-throughput screening assays have been applied to identify modulators of viral protein function across a panel of targets8–10.
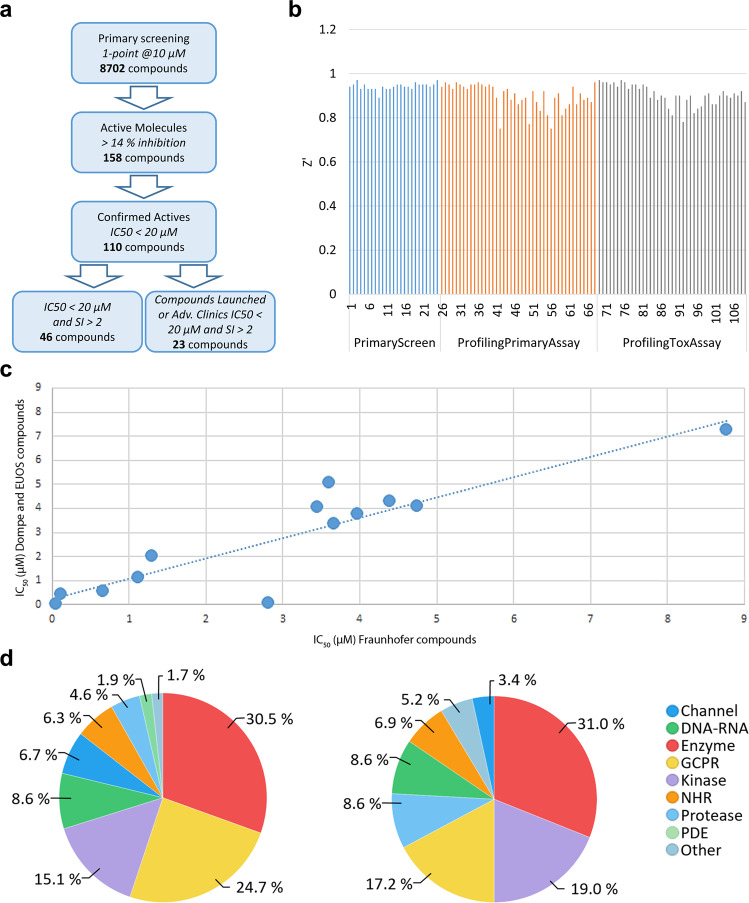
The dataset presented in this study was generated from the phenotypic screening of a large scale repurposing collections totalling ca. 8702 compounds (Fig. 1). The primary efficacy assay readout involved fluorescence imaging measurements of compound induced anti-SARS-CoV-2 cytopathic effects on EGFP expressing African green monkey kidney cells (VeroE6-EGFP)11. Following primary screening, hit profiling and cytotoxicity assessment, 110 compounds had IC50 values below 20 µM and 46 of these have a safety index (ratio of cytotoxic to anti-cytopathic effects) greater than 2 (Workflow shown in Fig. 2). Our results confirmed known, and identified novel repurposed candidates against SARS-CoV-2, including raloxifene (a marketed estrogen receptor antagonist) which, following orthogonal validation studies and independent confirmation by other groups12, has been progressed to clinical investigations against COVID-1913.
Fig. 1.
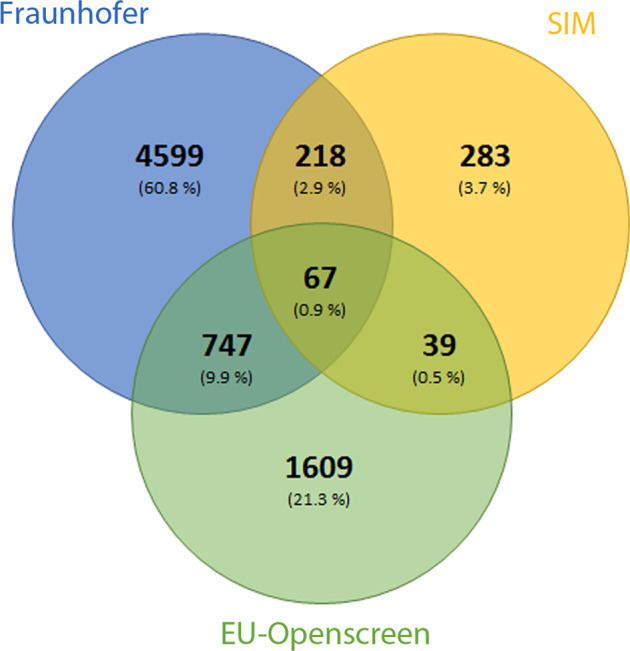
Comparison of the compound collection. Venn diagram showing overlap across the three compound collections in the screened set.
Fig. 2.
High throughput repurposing screening. (a) Workflow and number of compounds at each stage of screening cascade. (b) Z’ factor versus screening order for plates across the 3 experimental phases. All values Z’ > 0.5. (c) Duplicate compound potencies in hit profiling. IC50 values for compounds present in both the Fraunhofer (X axis) and Dompe or EU-OPENSCREEN (Y axis) collections (R2 = 0.81). (d) Distribution of the primary target of screened compounds (lhs) and 110 confirmed hit compounds (rhs). Explanation of keys: “Channel” gathers all cellular channels comprising metal channels and efflux pumps; “DNA-RNA” comprises all cellular DNA/RNA-dependent enzymes; “Enzymes” gathers a large set of metabolic enzymes involved in cellular metabolism/catabolism mechanisms like de-novo syntheses and/or oxidative or proteolytic processing of non-peptidic substrates; “GPCR” comprises G-Protein coupled receptors; “NHR” stands for Nuclear Hormone Receptors; “Proteases” is self-explaining and “Other” categorizes all the cellular proteins classes not previously listed (e.g. glycosylative enzymes, farnesyltransferase and similar).
Methods
Compound screening set
The screened set was composed of three compound libraries from different sources (Fig. 1). Firstly, the Fraunhofer Repurposing Library contains 5632 compounds including 3,400 compounds that have reached clinical use across 600 indications, as well as 1582 preclinical compounds with varying degrees of validation. This library was assembled by an external partner (SPECS; Netherlands) to mirror a set originally established by the Broad Institute. A curated database is available listing the compounds, indications, primary targets (where known) and mechanism of action, as well as analysis tools that can help to determine the mechanism of action and target. Secondly, the EU-OPENSCREEN Probe Library is a collection of 2500 compounds sampled from launched, clinical and preclinical studies aimed to cover multiple therapeutically relevant cellular mechanisms14. Finally, the Dompe’ Farmaceutici S.p.A. proprietary library is a collection of ca 700 candidate drugs which have completed at least phase I clinical trials. All compounds (Table 2) were quality controlled by liquid chromatography/ mass spectrometry (LC/MS) for purity and identity (minimum purity > 90%). The compounds were stored at a concentration of 10 mM in 100% DMSO at –20 °C.
Table 2.
List of files and variables contained46,56. 2a: Metadata information for ChEMBL document report card CHEMBL449556557. 2b: Data file for primary and hit profiling raw data (20201217_primary_PS_HP.xlsx), variables and descriptions. Table 2b contains the information of the Primary Screen sub-table49. A KNIME workflow provided in the code availability section generated the curve fit metrics17. 2c: Data file for primary and hit profiling raw data (20201217_primary_PS_HP.xlsx), variables and descriptions. Table 2c contains the information of the Hit Profiling sub-table49. 2d: Data file for primary and hit profiling raw data (20201217_primary_PS_HP.xlsx), variables and descriptions. Table 2d contains the information of the Hit Profiling Fit Results sub-table49. 2e: Data file for activity information (ACTIVITY.tsv), variables and descriptions49. 2f: Data file for assay information (ASSAY.tsv), variables and descriptions49. 2g: Data file for compound record lists (COMPOUND_RECORD.xlsx), variables and descriptions49. 2h: Data file for Dose_Response_raw_data (ACTIVITY_SUPP.tsv), variables and descriptions49. 2i: Data file for Activity map (ACTIVITY_SUPP.MAP.tsv), variables and descriptions49. 2j: Data file for assay reference text (REFERENCE.tsv), variables and descriptions49.
| Metadata name | Metadata content |
|---|---|
| AssayID | CHEMBL4513082 |
| Type | Functional |
| Description | Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 µM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging |
| Format | BAO_0000218 |
| Journal | Tbd |
| Organism | Chlorocebus sabaeus |
| Strain | — |
| Tissue | — |
| Cell Type | Vero C1008 |
| Subcellular Fraction | — |
| Target | CHEMBL4303835 |
| Document | CHEMBL4495565 |
| Cell | CHEMBL4295411 |
| Variable | Unit | Explanation |
|---|---|---|
| Plate Id | dimensionless | Unique Plate identifier. |
| Well Reference | dimensionless | Well Identifier (Composite Row/Column) |
| Compound Id | dimensionless | Compound Identifier |
| CompoundName | dimensionless | Trivial compound name |
| InChi | dimensionless | InChi structural value |
| Concentration [µM] | micromolar | Compound concentration used for primary screen |
| Row | dimensionless | Plate row position identifier |
| Column | dimensionless | Plate column position identifier |
| NumberOfCells | dimensionless | Number of fluorescent cells identified in image analysis (variable identical to variable “ValidObjectCount” in HitProfiling subtable) |
| CellAreaMean | dimensionless | total amount of surface covered by fluorescent cells in image (variable identical to variable “SpotTotalAreaCh2” in HitProfiling subtable) |
| IntensityCell | dimensionless | Total fluorescence intensity measured in image (variable identical to variable “SpotTotalIntenCh2” in HitProfiling subtable) |
| % Confluence | percentage | Percentage effect of “CellAreaMean” relative to positive and negative controls |
| % Inhibition | percentage | Percentage effect of “NumberOfCells” relative to positive and negative controls |
| WellType | dimensionless | WellType identifier (Compound, DMSO CONTROL, REMDESIVIR CONTROL, EMPTY WELL) |
| Variable | Unit | Explanation |
|---|---|---|
| Compound Id | dimensionless | Compound Identifier |
| MotherplateID | dimensionless | Unique identifier of compound storage plate |
| Name | dimensionless | Trivial compound name |
| InChi | dimensionless | InChi structural value |
| Well Reference | dimensionless | Well Identifier (Composite Row/Column) |
| Concentration [µM] | micromolar | Compound concentration used for primary screen |
| Row | dimensionless | Plate row position identifier |
| Column | dimensionless | Plate column position identifier |
| ValidObjectCount_PrimaryAssay | dimensionless | Raw value of number of fluorescent cells identified in image analysis (variable corresponds to variable “NumberOfCells” in Primary screen subtable) |
| SpotTotalAreaCh2_PrimaryAssay | dimensionless | Raw value of total amount of surface covered by fluorescent cells in image (variable corresponds to variable “CellAreaMean” in PrimaryScreen subtable) |
| SpotTotalIntenCh2_PrimaryAssay | dimensionless | Raw value of total fluorescence intensity measured in image (variable corresponds to variable “IntensityCell” in Primary screenSubtable) |
| % Confluence _Primary Assay | percentage | Percentage effect of “CellAreaMean” relative to positive and negative controls in Primary screen |
| % Inhibition_Primary Assay | percentage | Percentage effect of “NumberOfCells” relative to positive and negative controls determined in Primary screen |
| ValidObjectCount_ToxAssay | dimensionless | Raw value of number of fluorescent cells identified in image analysis in cytotoxicity assay |
| SpotTotalAreaCh2_ToxAssay | dimensionless | Raw value of total amount of surface covered by fluorescent cells in image in cytotoxicity assay |
| SpotTotalIntenCh2_ToxAssay | dimensionless | Raw value of total fluorescence intensity measured in image in cytotoxicity assay |
| % Confluence_ToxAssay | percentage | Percentage effect of “SpotTotalAreaCh2_ToxAssay” relative to positive and negative controls |
| % Inhibition_ToxAssay | percentage | Percentage effect of “ValidObjectCount_ToxAssay” relative to positive and negative controls |
| WellType | dimensionless | WellType identifier (COMPOUND, DMSO CONTROL, REMDESIVIR CONTROL, EMPTY) |
| Variable | Unit | Explanation |
|---|---|---|
| Compound Id | dimensionless | Compound Identifier |
| Type | dimensionless | Identifier of result type. It can be “IC50”, “CC50”, “SI”, “BOTTOM”, “TOP”, “SLOPE” |
| Relation | dimensionless | Relation between result type and outcome. It can be “ = ”, “ > ”, “ < ” |
| Value | various | Result value |
| Workflow | dimensionless | Identifier of workflow used to generate result. It can be “KNIME” or “ActivityBase” |
| Unit | dimensionless | Identifier of result unit. It can be “micromolar”, “percent”, “dimensionless” |
| Variable | Unit | Explanation |
|---|---|---|
| ACT_ID | dimensionless | Activity index text collating type of activity measured and progressive index to compound (ACT_INH_001 = primary screen inhibition at fixed concentration for compound 1) |
| CIDX | dimensionless | Compound Identifier |
| AIDX | dimensionless |
Internal code for screening performed: 1_LEY = primary screening (inhibition); 2_LEY = hit profiling (IC50) 3_LEY = cytotoxicity (CC50) 4_LEY = Selectivity Index (CC50/IC50) |
| RIDX | dimensionless | Code for screening. In this case LEY_VERO |
| Type | dimensionless | Description of the data type, could be ‘Inhibition’, ‘Selectivity index’, ‘CC50’ or ‘IC50’ |
| Relation | dimensionless | Relation between data type and outcome. It can be “ = ”, “ > ”, “ < ” |
| Value | dimensionless | Real number |
| Upper value | dimensionless | None given |
| Units | dimensionless | Selectivity index is dimensionless; IC50 or CC50 values are given in microMoles and inhibition is given in %. |
| Text_value | dimensionless | None given |
| Activity_comment | dimensionless | None given |
| CRIDX | dimensionless | Code for screening. In this case LEY_VERO |
| Variable | Unit | Explanation |
|---|---|---|
| AIDX | dimensionless |
Internal code for screening performed: 1_LEY = primary screening (inhibition); 2_LEY = hit profiling (IC50) 3_LEY = cytotoxicity (CC50) 4_LEY = Selectivity Index (CC50/IC50) |
| RIDX | dimensionless | Code for screening. In this case LEY_VERO |
| ASSAY DESCRIPTION | dimensionless | Free text describing the content of AIDX |
| ASSAY_TYPE | dimensionless | Text code for assay type F = functional, T = toxicology |
| ASSAY_TEST_TYPE | dimensionless | Text code for assay conditions: in this case = in vitro |
| ASSAY_ORGANISM | dimensionless | Organism source according to BAO ontology: in this case = “Chlorocebus sabaeus” for green monkey |
| ASSAY_STRAIN | dimensionless | None given |
| ASSAY_TAX_ID | dimensionless | Taxonomy ID: in this case = 60711 |
| ASSAY_SOURCE | dimensionless | None given |
| ASSAY_TISSUE | dimensionless | None given |
| ASSAY_CELL_TYPE | dimensionless | Cell type name: in this case = VERO-E6 |
| ASSAY_SUBCELLULAR_FRACTION | dimensionless | None given |
| TARGET_TYPE | dimensionless | Description of target: In this case = “Organism” for primary screen and for IC50 or = “Cell Line” for cytotoxicity in absence of virus |
| TARGET_NAME | dimensionless | Target name: In this case = “Severe acute respiratory syndrome coronavirus 2” or = VERO-E6 |
| TARGET_ACCESSION | dimensionless | None given |
| TARGET_ORGANISM | dimensionless | None given |
| Variable | Unit | Explanation |
|---|---|---|
| CIDX | dimensionless | Compound Identifier |
| RIDX | dimensionless | Code for screening. In this case LEY_VERO |
| COMPOUND_NAME | dimensionless | Compound Trivial Name |
| COMPOUND_KEY | dimensionless | Compound Trivial name or IUPAC |
| Variable | Unit | Eplanation |
|---|---|---|
| SAM_ID | dimensionless | Sample index text collating type of activity measured and progressive index to compound (SAM_CC50_001 = measurements target (IC50 or CC50) for compound 1) |
| TYPE | dimensionless | Text describing collating type of value type (concentrations or readouts) and type of experiment |
| RELATION | dimensionless | Relation between data type and outcome. It can be “ = ”, “ > ”, “ < ” |
| VALUE | dimensionless | Real number |
| UNIT | dimensionless | Readout values are given in µM and inhibition values are given in %. |
| REG_ID | dimensionless | Text collating type of experiment with index of molecules and index of concentrations (all in triplicates) |
| Variable | Unit | Explanation |
|---|---|---|
| ACT_ID | dimensionless | Activity index text collating type of activity measured and progressive index to compound (ACT_INH_001 = primary screen inhibition at fixed concentration for compound 1) (see above ACTIVITY.XLSX) |
| SAM_ID | dimensionless | Sample index text collating type of activity measured and progressive index to compound (SAM_CC50_001 = measurements target (IC50 or CC50) for compound 1) |
| Variable | Unit | Explanation |
|---|---|---|
| RIDX | dimensionless | Code for screening. In this case LEY_VERO |
| PUBMED_ID | dimensionless | Future PUBMED_ID (link for accepted paper) |
| JOURNAL_NAME | dimensionless | Journal Name where paper is or will be published |
| YEAR | dimensionless | Year of publication |
| VOLUME | dimensionless | Journal Volume where paper is or will be published |
| ISSUE | dimensionless | Journal Issue where paper is or will be published |
| FIRST_PAGE | dimensionless | Integer for first page where paper is or will be published |
| LAST_PAGE | dimensionless | Integer for last page where paper is or will be published |
| REF_TYPE | dimensionless | Reference type: Dataset |
| TITLE | dimensionless | Cytopathic SARS-CoV-2 screening on VERO-E6 cells in a large-scale repurposing effort |
| DOI | dimensionless | To be given by Editor/Publisher |
| PATENT_ID | dimensionless | None given |
| ABSTRACT | dimensionless | Text of paper abstract |
| AUTHORS | dimensionless | Authors’ list |
Cell and virus culture
The African green monkey kidney cell line (Vero E6) was previously engineered to constitutively express GFP15. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% v/v foetal calf serum (FCS; Biowest), 0.075% Sodium Bicarbonate (7.5% solution, Gibco) and 1x Pen-strep (Gibco) and kept under 5% CO2 on 37 °C. Assay medium contained 2% FCS. SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 recovered from a nasopharyngeal swab taken from an asymptomatic patient returning from Wuhan, China in the beginning of February 2020 was sequenced on a MinION platform (Oxford Nanopore). After serial passaging on Huh7 and Vero E6 cells, infectious content of the virus stock was determined by titration on Vero E6 cells using the Spearman-Kärber method. All virus-related work was carried out in certified, high-containment biosafety level-3 facilities in the Rega Institute at KU Leuven.
Primary anti-cytopathicity assay development: 384-well plates
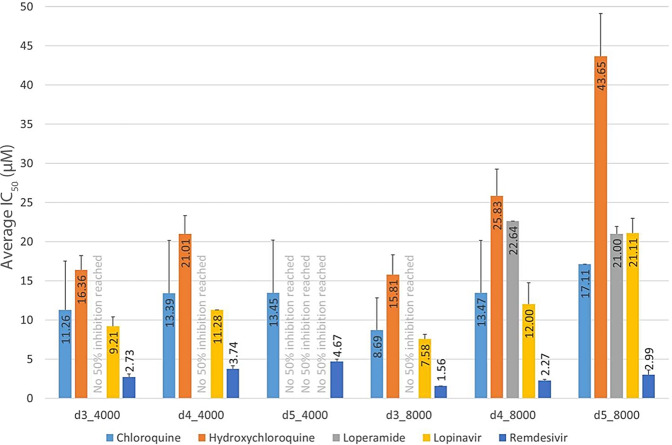
The primary assay protocol was optimised with respect to cell seeding, viral MOI and pharmacological response. Cells were seeded at 4000 and 8000 cells/well in 384-well plates. The following day, cells were incubated with the control compounds and the virus at different MOI (0.01 and 0.001). A panel of control compounds was tested to evaluate the pharmacological performance of the assay. Compounds tested were: chloroquine, hydroxychloroquine, and loperamide at a starting concentration of 50 µM; lopinavir and remdevisir at a starting concentration of 100 µM. A dose response curve was achieved with serial dilution at seven different concentration points following a half-log dilution schema. Remdevisir was the most active reference compound under all experimental conditions16, (Fig. 3) and was selected as the positive control used at 20 µM final concentration in the primary assay. Positive control remdesivir showed IC50 comparable to literature10 as depicted in Fig. 4.
Fig. 3.
Control compound profiling. Five control compounds were tested: chloroquine (light-blue); hydroxychloroquine (orange bars); loperamide (grey); lopinavir (yellow) and remdesivir (blue). The IC50 was measured and reported on the y-axis at different days of incubation. The days of incubation (d) and the number of cells seeded (4000 or 8000) were reported on the x-axis. Data are reported as the mean.
Fig. 4.
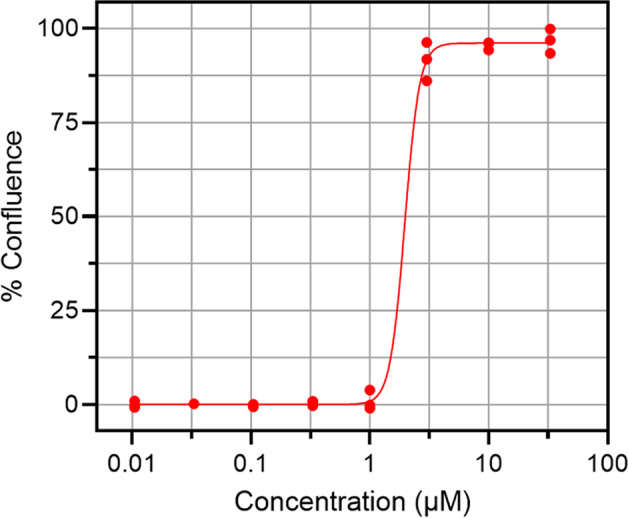
Remdesivir curve fitting example. Performance of positive control. %Confluence versus compound concentration for Remdesivir, IC50 = 1.7 µM, in accord with literature10.
Hit identification and compound profiling
To measure inhibition of the SARS-CoV-2 cytopathic effect, 384-well imaging plates (Greiner #781092) were spotted with test compounds and controls (16 positive and 16 negative control wells per plate) using an acoustic dispenser (Echo, Labcyte) to yield 10 µM final test compound concentration at 0.1% vol/vol DMSO. For concentration response studies, eight concentrations of a semi-log dilution series from 33 µM to 10 nM at 0.33% DMSO were used. One day before infection (Day -1), test plates were equilibrated to room temperature and 30 µL of Vero E6 EGFP cells were added at 8,000 cells/well. On the day of infection (Day 0), plates were transported to the CAPS-IT robotic system for the addition of virus (MOI = 0.001) using a liquid handler (EVO 100, Tecan) to a final volume of 60 µl, and incubated at 37 °C, 5% CO2 for 4 days. Plates were then imaged on an Arrayscan XTI, (Thermofisher). Parallel assessments of the underlying cytotoxicities of the compounds were performed as described above in dose response studies, but without virus infection and using sodium-selenite (20 µM final) as the intra-plate positive cytotoxicity control.
Image acquisition and analysis
At day four post-infection, the GFP signal was captured using wide field fluorescence imaging by exciting at 485–20 nm and emitting with the BGRFRN filter set. A 5 X objective captured 80% of the well bottom area in the 384 plate. The optimal exposure time was determined based on fluorescence intensity and was set as 0.023 seconds. A 2 × 2 binning was used and autofocus plane count was reduced to increase image acquisition speed. An image analysis protocol was developed in-house by using the SpotDetector bioapplication (Cellomics, Thermofisher). After background reduction on the raw image files, a fixed fluorescent intensity threshold was determined for the identification of fluorescent cells and their morphologoical parameters were then determined. The two most relevant extracted parameters describing the anticytopathic effects of compounds cells were: i) the number of fluorescent cells in each well (NumberOfCells); and ii) the area of each well covered by fluorescence cells (CellAreaMean).
Test compound results were normalised relative to the corresponding intra-plate controls. For cytopathicity experiments the positive control (100% inhibition of virus induced cytopathicity) were 16 remdesivir (20 µM) containing wells per 384 well plate in column 24. For cytotoxity experiments the positive control (100% cytotoxicity) were 16 sodium-selenite (20 µM) containing wells in column 24. The negative controls (0% effect) for both cytotoxicity and anti-cytopathic experiments were 16 wells with DMSO (0.1% vol/vol) in column 23. The normalised value of “CellAreaMean” was termed “% Confluence” whilst the normalised value of “NumberOfCells” was termed “% Inhibition”. Although, it might be expected that number of cells in each well would scale linearly with the area of the well covered by the cells, compound exposure can also induce changes in cell morphology and dimensions due to poly-pharmacological effects unrelated to any anti-viral properties. Therefore, whilst both parameters are of interest and are made available for re-analysis in the raw data sets provided, the parameter “% Confluence” was used for the purposes of reporting compound effects in ChEMBL and in subsequent compound selection and prioritisation.
Large scale data analysis for Primary and Hit Profiling studies was performed in two ways. The first method used commercial software (ActivityBase, IDBS, Version 8.0.5.4) in a procedure which was aligned with in-house data management policies. Hit profiling dose response data (% Confluence versus compound concentration) were fitted to 4-parameter logistic functions to give the IC50 for the anti-cytopathic effect or the CC50 for the cytoxicity effects in the absence of virus. Assay quality was assessed using the Z′ factor calculation (Fig. 2B) with Z’ factor > 0.5 as the threshold for assay acceptance17. The cytotoxity index was defined as CC50/IC50. A higher CI value indicates a wider window between the anti-cytopathic effects and possible underlying in-vitro toxicity. Individuals seeking to reanalyse the data may not have access to the ActivityBase software, therefore, a second method was established in the KNIME environment to calculate IC50 values from the raw data. The workflow is deposited in github repository and replicates the ActivityBase analysis18.
Data Records
The analysed data and raw data have been made available for download and reuse (Table 1). An overview of all analysed Primary Screening and Hit Profiling data in the ChEMBL repository can be found in the document report card CHEMBL4495565, whilst individual data sets are available for viewing and download from four assay report cards (Table 1). Raw data files for Primary and Hit Profiling have also been made available by the ChEMBL administrators on a FTP server and through the Figshare platform. The variables present in the tables of ChEMBL entries and the raw data files are defined in Table 2.
Table 1.
Description and location of data records.
| Description | Reference |
|---|---|
| ChEMBL Document Report Card for the complete study | Data available48. |
| Single concentration primary screen for anti-cytopathic effect of compounds (Confluence, %) in ChEMBL DB | Data available49. |
| Hit profiling results for compound anti-cytopathic effect (IC50) in ChEMBL DB | Data available48. |
| Hit profiling results for compound cytotoxic effect (CC50) in ChEMBL DB | Data available50. |
| Derived cytotoxity index results (CI) in ChEMBL DB | Data available51. |
| Data from the primary screen and hit profiling deposited on CHEMBL FTP server | All screening results in separate files52. |
| Figshare record with figures, tables and Primary data sets available for download | Data can be found at53–55 |
Technical Validation
Screening Assay
The primary screen resulted in 158 hits (% confluence > 14%) (CHEMBL4495565 – Fig. 2a). At all phases (Primary, Profiling and Cytotoxicity) the Z’ exceeded > 0.5, indicating acceptable assay quality (Fig. 2b). Sources of compounds from the three different libraries used in the screening campaign are shown in Table 3. Primary hit compounds were cherry-picked and progressed to dose response profiling and cytotoxicity assessment. In profiling studies, identical compounds present in different libraries (Fig. 1) showed consistent potency (Fig. 2c,R2 = 0.81), suggesting good assay reproducibility with respect to compound origin. Some 110 compounds gave IC50 values below 20 µM and were classified as confirmed hits. From the 110 compounds, 66 have a SI > 2 (Supp. Table S1 and Fig. 2a) and of this group 18 compounds are either marketed drugs or in advanced clinical trials (Fig. 2a). (CHEMBL449565). Fig. 2d shows the distribution of primary therapeutic targets annotated for the screened collection (lhs) and the 110 confirmed hits (rhs). No changes in the targets of the confirmed hits relative to those of the screened compound set were observed.
Table 3.
Source of compounds from the three different libraries used in the screening campaign.
Assay properties and influence on hit compound identification
SARS-CoV infects and replicates more efficiently in some cell types, such as VeroE6, FRhK4, Caco2, LLCMK2, compared to Calu3 and Hek293T cells, while there is a very low efficiency of replication in U251 and MDCK cells under the same multiplicity of infection. Therefore, cell model selection in SARS-COV-2 phenotypic screening studies is important, and recent reports have shown differences in compound potencies depending which cell line has been used in the primary assay19–27. The cell line used in this study, Vero E-6 kidney epithelial cells from the African green monkey28, has been extensively used for SARS-CoV-like virus studies29–33. In those models, cell viability and virus titre were usually verified after 3–5 days post infection (p.i.)27,34 and in our assay the incubation of compounds for a period of 4 days p.i. resulted in a robust readout. Nevertheless, it has already been reported that for the same active drug, infection with different virus MOI may result in variable safety index values35, suggesting that cytotoxicity analyses in all studies could be somewhat limited. It should also be noted that the VeroE6-EGFP cell line used for this study is not sensitive to ACE inhibitors or some SARS-CoV active drugs such as ribavirin and glycyrrhizin36–39, which are the subject of ongoing clinical trials We cannot exclude, therefore, that alternative experimental setups may lead to partially different hit populations, as others have observed19. Nevertheless, the GFP reporter line presents an opportunity to perform fast, automated and homogeneous high-throughput screening, with a high signal-to-noise ratio and low variability and is fit-for-purpose for hit identification studies.
Within the confirmed compounds with IC50 < 20 µM, some 70% modulate the intracellular signalling pathways. Notable groups are the inhibitors of the Growth Factor Receptors (PDGFR) (like masitinib or tandutinib), Dihydrofolate Reductase (DHFR) (trimetrexate) and Estrogen Receptor Modulator (clomiphene and raloxifene). In addition, many show common protein targets such as the Phosphatidylinositol 3-Kinase (PI3K) (VPS34-IN-1) or mTOR protein (VE-822) two key elements of pro-survival signalling. Finally, where a therapeutic indication was annotated, the majority of the compounds were associated with cancer and anti-infective (antifungal and anti-malaria) therapy. These observations suggest that drugs associated with cell survival and growth may be an optimal choice for antiviral therapies for SARS-CoV-2, if adequate safety and exposure/efficacy can be achieved. Comparing the hit population in this dataset with reported studies show five compounds (amodiaquine, ciclesonide, eltrombopag, loperamide, niclosamide) that are also reported by Jeon et al., who screened 3000 FDA approved drugs against Vero cells15. Similarly, two (amodiaquine, chlorpromazine) overlap with a set of 20 inhibitors identified by Weston29, also in Vero cells. The common compounds have an antifungal, antimalarial or anticancer activity, suggesting again that drugs for these indications may contain the most promising antiviral compounds. Among compounds selected for further studies, raloxifene was prioritised as it has been found active in an independent phenotypic assays of SARS CoV-2 viral infections in VERO-E6 cells40 and against coronavirus OCR43 in LLC-MK2 cells41. This compound can inhibit RNA replication42 and related estrogenic receptor modulators have been found to be active against in-vitro viral infections43. Interestingly, there have been evidence from other datasets44 that several SERMS, either agonist or antagonists, are active at SARS-CoV 2 viral entry level and we are also exploring this and other possible mechanisms of action for this class of compounds.
Usage Notes
We suggest that other similar screening data sets are available from the Covid-19 portal45, the NCATS databases46 and a newly developed collection called “The COVID-19 Drug and Gene Set Library”47.
Supplementary information
Acknowledgements
This work has been supported by the project “EXaSCale smArt pLatform Against paThogEns for CoronaVirus – Exscalate4CoV or E4C”4, which is funded through EU’s H2020-SC1-PHE-CORONAVIRUS-2020 emergency call, (grant N. 101003551). It collects 18 European public Institutions and a private company (Dompe’ Farmaceutici SpA) in the leading role and brings together high-end computational facilities, drug discovery and BSL-3 high-throughput screening resources. FAIRification of data for community use was supported by funding from the European Union’s Horizon 2020 programme of EOSC-LIFE, under grant agreement number 824087. Part of this research work was performed using the ‘Caps-It’ research infrastructure (project ZW13–02) that was financially supported by the Hercules Foundation and Rega Foundation, KU Leuven. We thank Andrew Leach, Anna Gaulton, Emma Manners and James Blackshaw of the European Bioinformatics Institute (Hinxton) for preparing FAIR data sets. We thank Yulia Gerhardt and Peter Maas of SPECS and Joshua Bitker (ex-Broad) for input into the selection and quality control of the Fraunhofer compound library.
Author contributions
A.Z., P.G., P.L., D.I., A.B. conceived the study. P.L., M.K., O.K. and L.V. carried out the tissue culture work, cytotoxicity assays, analysed the data and prepared the initial manuscript. A.Z., J.R., D.I. and P.G. analysed the data and prepared the initial manuscript. J.C. conceived the study, carried out the tissue culture work and virus experiments. C.C. conceived the study and supervised the work. S.W. carried out the tissue culture work, virus experiments and cytotoxicity assays. O.K. carried out the tissue culture work, cytotoxicity assays, analysed the data. M.K. carried out the tissue culture work, cytotoxicity assays and analysed the data. M.W. carried out all the plate preparations and logistics. All authors contributed to the text of the manuscript, read, and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Code availability
We developed a more comprehensive open-source KNIME workflow for the curve fitting calculations17.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01532-x.
References
- 1.SARS-CoV-2 infection cases https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases (2022).
- 2.Ballard C, et al. Drug repositioning and repurposing for Alzheimer disease. Nat Rev Neurol. 2020;16(12):661–673. doi: 10.1038/s41582-020-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low ZY, Farouk IA, Lal SK. Drug Repositioning: New Approaches and Future Prospects for Life-Debilitating Diseases and the COVID-19 Pandemic Outbreak. Viruses. 2020;12(9):1058. doi: 10.3390/v12091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID-19 Studies from the World Health Organisation Database https://clinicaltrials.gov/ct2/who_table (2022).
- 5.Exscalate4CoV Homepage www.exscalate4cov.eu (2022).
- 6.Beccari AR, Cavazzoni C, Beato C, Costantino G. LiGen: a high performance workflow for chemistry driven de novo design. J Chem Inf Model. 2013;53(6):1518–1527. doi: 10.1021/ci400078g. [DOI] [PubMed] [Google Scholar]
- 7.Talarico C, et al. Combining Molecular Dynamics and Docking Simulations to Develop Targeted Protocols for Performing Optimized Virtual Screening Campaigns on The hTRPM8 Channel. Int J Mol Sci. 2020;21(7):2265. doi: 10.3390/ijms21072265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grottesi A, et al. Computational Studies of SARS-CoV-2 3CLpro: Insights from MD Simulations. Int J Mol Sci. 2020;21(15):5346. doi: 10.3390/ijms21155346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gervasoni S, et al. A Comprehensive Mapping of the Druggable Cavities within the SARS-CoV-2 Therapeutically Relevant Proteins by Combining Pocket and Docking Searches as Implemented in Pockets 2.0. Int J Mol Sci. 2020;21(14):5152. doi: 10.3390/ijms21145152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannalire R, et al. SARS-CoV-2 Entry Inhibitors: Small Molecules and Peptides Targeting Virus or Host Cells. Int J Mol Sci. 2020;21(16):5707. doi: 10.3390/ijms21165707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu H, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. Korean researchers find Raloxifene effective in treating COVID-19 https://www.youtube.com/watch?v = cv2zhgtjTJM (2021).
- 13.Italy okays trial of osteoporosis drug to treat COVID-19 https://www.reuters.com/article/us-health-coronavirus-italy-drug-idUSKBN27C136 (2021).
- 14.Probe compound sets https://www.probes-drugs.org/compoundsets (2022).
- 15.Ellinger B, et al. A phenotypic screening approach to identify anticancer compounds derived from marine fungi [published correction appears in Assay Drug Dev Technol. 2014 May;12(4):247] Assay Drug Dev Technol. 2014;12:162–175. doi: 10.1089/adt.2013.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iversen PW, Eastwood BJ, Sittampalam GS, Cox KL. A comparison of assay performance measures in screening assays: signal window, Z’ factor, and assay variability ratio. J Biomol Screen. 2006;11(3):247–252. doi: 10.1177/1087057105285610. [DOI] [PubMed] [Google Scholar]
- 18.Fraunhofer-ITMP / KNIME workflow (https://github.com/Fraunhofer-ITMP/KNIME/releases/tag/v1.0) (2022).
- 19.Huang R, et al. Biological activity-based modeling identifies antiviral leads against SARS-CoV-2. Nat Biotechnol. 2021;39:747–753. doi: 10.1038/s41587-021-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CZ, et al. Drug Repurposing Screen for Compounds Inhibiting the Cytopathic Effect of SARS-CoV-2. Frontiers in Pharmacology. 2021;11:2005. doi: 10.3389/fphar.2020.592737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiser, K. et al. Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2 Preprint at https://www.biorxiv.org/content/10.1101/2020.04.21.054387v1 (2020)
- 22.Jeon S, et al. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob Agents Chemother. 2020;64(7):e00819–20. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riva, L. et al. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.16.044016v1 (2020)
- 24.Touret F, et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep. 2020;10(1):13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mark Dittmar,M., etal. Drug repurposing screens reveal FDA approved drugs active against SARS-Cov-2. Preprint at https://www.biorxiv.org/content/10.1101/2020.06.19.161042v1 (2020) 9 [DOI] [PMC free article] [PubMed]
- 26.Bocci G, et al. Virtual and In Vitro Antiviral Screening Revive Therapeutic Drugs for COVID-19. ACS Pharmacol Transl Sci. 2020;3(6):1278–1292. doi: 10.1021/acsptsci.0c00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai W, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammerman, N.C., Beier-Sexton, M., Azad, A.F., Growth and maintenance of Vero cell lines. Curr Protoc Microbiol. 2008;11: A.4E.1-A.4E.7. 10.1002/9780471729259.mca04es11 [DOI] [PMC free article] [PubMed]
- 29.Kistner O, et al. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine. 1998;16(9-10):960–968. doi: 10.1016/s0264-410x(97)00301-0. [DOI] [PubMed] [Google Scholar]
- 30.Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12(1):128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett PN, Portsmouth D, Ehrlich HJ. Vero cell culture-derived pandemic influenza vaccines: preclinical and clinical development. Expert Rev Vaccines. 2013;12(4):395–413. doi: 10.1586/erv.13.21. [DOI] [PubMed] [Google Scholar]
- 32.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogando NS, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivens T, et al. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome-associated coronavirus. J Virol Methods. 2005;129(1):56–63. doi: 10.1016/j.jviromet.2005.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weston S, et al. Broad Anti-coronavirus Activity of Food and Drug Administration-Approved Drugs against SARS-CoV-2 In Vitro and SARS-CoV In Vivo. J Virol. 2020;94(21):e01218–20. doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So LK, et al. Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet. 2003;361(9369):1615–1617. doi: 10.1016/s0140-6736(03)13265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J., Jr Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326(4):905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cinatl J, et al. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/s0140-6736(03)13615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi L, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78(20):11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan S, et al. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacological Research. 2020;159:104960. doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, et al. Identification of Potent and Safe Antiviral Therapeutic Candidates Against SARS-CoV-2. Front Immunol. 2020;11:586572. doi: 10.3389/fimmu.2020.586572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzikov M, et al. (2020) Identification of inhibitors of SARS-CoV-2 3CL-Pro enzymatic activity using a small molecule repurposing screen. ACS Pharmacol. Transl. Sci. 2021;4:1096–1110. doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda M, et al. Raloxifene inhibits hepatitis C virus infection and replication. FEBS Open Bio. 2012;2:279–283. doi: 10.1016/j.fob.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spike-ACE2 protein-protein interaction https://opendata.ncats.nih.gov/covid19/assay?aid=1 (2022).
- 45.The Covid-19 portal https://www.covid19dataportal.org/ (2022).
- 46.NCATS databases https://ncats.nih.gov/expertise/covid19-open-data-portal (2022).
- 47.The COVID-19 Drug and Gene Set Library https://maayanlab.cloud/covid19/ (2022). [DOI] [PMC free article] [PubMed]
- 48.2022. Hit profiling results IC50. ChEMBL. CHEMBL4513083
- 49.2022. Single concentration primary. ChEMBL. CHEMBL4513082
- 50.2022. Hit profiling results (CC50) ChEMBL. CHEMBL4513084
- 51.2022. Cytotoxicity index results. ChEMBL. CHEMBL4513085
- 52.Result files of the screening campaign https://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLNTD/set24_fraunhofer_sarscov2_vero/ (2022).
- 53.Zaliani A. 2022. Dose Response Results. figshare. [DOI]
- 54.Zaliani A. 2022. Primary Screening. figshare. [DOI]
- 55.Zaliani A. 2022. Molecule library and Results summary. figshare. [DOI]
- 56.General data depository https://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLNTD/set24_fraunhofer_sarscov2_vero/ (2022).
- 57.2022. Cytopathic SARS-Cov2 screening on VERO-E6 cells in a large repurposing effort. ChEMBL. CHEMBL4495565 [DOI] [PMC free article] [PubMed]
- 58.Fraunhofer Repurposing Library https://s3.amazonaws.com/data.clue.io/repurposing/downloads/repurposing_drugs_20180907.txt (2022).
- 59.EU-OPENSCREEN Bioactive set https://www.probes-drugs.org/compounds/standardized#compoundset=353@AND (2022).
- 60.DOMPE_SIM collection https://mediate.exscalate4cov.eu/downloads/Dompe_SIM_Collection.txt (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2022. Hit profiling results IC50. ChEMBL. CHEMBL4513083
- 2022. Single concentration primary. ChEMBL. CHEMBL4513082
- 2022. Hit profiling results (CC50) ChEMBL. CHEMBL4513084
- 2022. Cytotoxicity index results. ChEMBL. CHEMBL4513085
- Zaliani A. 2022. Dose Response Results. figshare. [DOI]
- Zaliani A. 2022. Primary Screening. figshare. [DOI]
- Zaliani A. 2022. Molecule library and Results summary. figshare. [DOI]
- 2022. Cytopathic SARS-Cov2 screening on VERO-E6 cells in a large repurposing effort. ChEMBL. CHEMBL4495565 [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
We developed a more comprehensive open-source KNIME workflow for the curve fitting calculations17.