Abstract
Cellobiose dehydrogenase (CDH) is an extracellular hemoflavoenzyme produced by several wood-degrading fungi. In the presence of a suitable electron acceptor, e.g., 2,6-dichloro-indophenol (DCIP), cytochrome c, or metal ions, CDH oxidizes cellobiose to cellobionolactone. The phytopathogenic fungus Sclerotium rolfsii (teleomorph: Athelia rolfsii) strain CBS 191.62 produces remarkably high levels of CDH activity when grown on a cellulose-containing medium. Of the 7,500 U of extracellular enzyme activity formed per liter, less than 10% can be attributed to the proteolytic product cellobiose:quinone oxidoreductase. As with CDH from wood-rotting fungi, the intact, monomeric enzyme from S. rolfsii contains one heme b and one flavin adenine dinucleotide cofactor per molecule. It has a molecular size of 101 kDa, of which 15% is glycosylation, and a pI value of 4.2. The preferred substrates are cellobiose and cellooligosaccharides; additionally, β-lactose, thiocellobiose, and xylobiose are efficiently oxidized. Cytochrome c (equine) and the azino-di-(3-ethyl-benzthiazolin-6-sulfonic acid) cation radical were the best electron acceptors, while DCIP, 1,4-benzoquinone, phenothiazine dyes such as methylene blue, phenoxazine dyes such as Meldola's blue, and ferricyanide were also excellent acceptors. In addition, electrons can be transferred to oxygen. Limited in vitro proteolysis with papain resulted in the formation of several protein fragments that are active with DCIP but not with cytochrome c. Such a flavin-containing fragment, with a mass of 75 kDa and a pI of 5.1 and lacking the heme domain, was isolated and partially characterized.
The phytopathogenic fungus Sclerotium rolfsii (the anamorph form of the basidiomycete Athelia rolfsii) has been isolated from a wide variety of plants, primarily annuals and herbaceous perennials, but some woody plants are also attacked when they are young (1). S. rolfsii survives on dead plant material in the soil by forming sclerotia which later germinate and attack young plants, causing necrosis by attacking the cell walls. Pectin, hemicellulose, and cellulose are degraded effectively and completely with various enzyme complexes (45, 46). S. rolfsii also produces oxalic acid, which in synergistic action with enzymes causes injury to plant tissue (1).
Cellobiose dehydrogenase (CDH) [EC 1.1.99.18; cellobiose:(acceptor) 1-oxidoreductase] is produced extracellularly by a number of wood- and cellulose-degrading fungi when grown on cellulose. It oxidizes the reducing end of cellobiose and cellooligosaccharides to their corresponding 1,5-lactones, which are subsequently hydrolyzed to the carboxylic acids in aqueous environments. In addition to the cellooligosaccharides, the presumed natural substrates, CDH oxidizes very few other sugars, the most efficient substrates being β-1,4-linked disaccharides with a β-glucose moiety at their reducing end (27). The reduced enzyme is reoxidized by different electron acceptors, such as 2,6-dichloro-indophenol (DCIP), 1,2- or 1,4-benzoquinone and some of their derivatives, cytochrome c (cyt c), metal ions, including Fe(III), and even oxygen, although the latter is only a very poor electron acceptor (27). The natural electron acceptor of the enzyme is presently not known (29, 34, 37, 42, 44).
Cellobiose dehydrogenase has been isolated and characterized from a variety of white-rot fungi (2, 6, 21, 38, 43, 53), soft-rot fungi (13, 15, 20, 50, 52), and one brown-rot fungus (49). Typically, CDH is a monomeric protein consisting of two domains, one containing a hexacoordinate heme b (14) and one containing a noncovalently bound flavin adenine dinucleotide (FAD) (6, 43) or 6-hydroxy FAD (33). These two domains are linked by a protease-sensitive region (17, 35, 39, 52). Limited proteolytic cleavage of CDH leads to an inactive heme peptide and an active FAD domain, which has been termed cellobiose:quinone 1-oxidoreductase (CBQ; EC 1.5.1.1) (24, 54). CBQ carries the catalytic site and reduces cellobiose efficiently. It can be reoxidized by many of the electron acceptors that act on CDH, with the exception of cyt c, which effectively reoxidizes only intact CDH. Frequently, a large percentage of CBQ is found in the culture broth of various CDH-producing organisms.
The in vivo function of CDH is not fully understood. CDH is not an essential component of the lignocellulose-degrading enzyme complex but can enhance both cellulose and lignin degradation (5, 27). CDH also could have a protective function (37) since it can reduce quinones, one of the major antimicrobial systems used by plants.
Most previous work on CDH has been with the enzyme from the white-rot fungus Phanerochaete chrysosporium. As S. rolfsii is a phytopathogen and not a wood-rotting organism, the CDH from this fungus could have different properties and/or in vivo functions. Our objective was to purify CDH from S. rolfsii, to characterize the enzyme, and to compare its properties to those of CDH from other sources. S. rolfsii produces 10- to 50-fold more extracellular CDH activity than P. chrysosporium (4, 23, 46). Since CDH has several attractive properties, e.g., its specificity for β-1,4-linked disaccharides, the increased availability of the S. rolfsii enzyme could enable a range of technological applications, such as biosensors, bioremediation, or biocatalysis. CDH has been used in colorimetric assays (12) and in amperometric biosensors (19) for the detection of lactose. CDH-based biosensors also have been used for the sensitive and selective detection of diphenols, widely distributed toxic pollutants (36). In addition, CDH has a potential role in bioremediation, since it can directly reduce munitions such as 2,4,6-trinitrotoluene and indirectly degrade many more chemicals, including polyacrylate polymers (10). Finally, CDH can be used in biocatalysis for the preparation of organic acids such as lactobionic acid (3).
MATERIALS AND METHODS
Organism and culture conditions.
Stock cultures of S. (Athelia) rolfsii CBS 191.62 (Centraalbureau voor Schimmelcultures, Baarn, The Netherlands) were maintained on glucose-maltose Sabouraud agar plates, which were inoculated with mature sclerotia and incubated at 30°C. For the production of CDH, a medium containing 43 g of α-cellulose (C 8002; Sigma, St. Louis, Mo.) per liter, 80 g of peptone from meat (Merck, Darmstadt, Germany) per liter, 2.5 g of NH4NO3 per liter, 1.5 g of MgSO4 · 7H2O per liter, 1.2 g of KH2PO4 per liter, 0.6 g of KCl per liter, and 0.3 ml of trace element solution (46) per liter was inoculated with several agar plugs (approximately 1-cm2 diameter) taken from 4-day-old agar cultures. Inocula for fermentation cultures were grown in unbaffled 1-liter Erlenmeyer flasks containing 300 ml of medium and were cultivated for 13 days at 30°C with continuous shaking (r = 2.5 cm) at 120 rpm. Fermentations were carried out in a 20-liter stirred-tank fermentor (MBR Bio Reactor, Wetzikon, Switzerland) with a 15-liter working volume, inoculated with 10% (vol/vol) preculture. Fermentation conditions were 30°C, 40% dissolved oxygen tension, and agitation between 200 and 400 rpm. The culture pH, which was initially adjusted to 5.0, was not controlled during the fermentation.
The culture was regularly sampled, samples were clarified by centrifugation, and CDH activity, CBQ activity, and protein concentration (7) were assayed. Total dry matter was determined by filtration through tared filter disks (Schleicher & Schüll ME 25/21, 0.45 μm; Dassel, Germany), dried at 105°C until constant, and weighed.
Enzyme purification procedure.
Mycelia were collected by centrifugation (20 min; 10,000 × g), and the extracellular medium was concentrated using a 50-kDa polysulfone ultrafiltration membrane. Any precipitate was removed by centrifugation (20 min, 10,000 × g), and the clear supernatant was repeatedly dialyzed against water. Aliquots of 500 U of CDH activity were applied to a DEAE Sepharose fast-flow column (2.5 by 8 cm; Amersham-Pharmacia, Uppsala, Sweden), preequilibrated with 50 mM sodium acetate buffer, pH 5.0. The column was eluted with a linear salt gradient (0 to 0.4 M NaCl in the same buffer) in 10 column volumes. Fractions with significant absorptions at both 420 and 450 nm were tested for CDH activity, and those containing CDH activity were pooled.
Ammonium sulfate was added slowly to this pool to give a final concentration of 20% saturation, and the pH was maintained at 5.0 by increasing the buffer concentration to 0.1 M. After removal of the precipitate by centrifugation (15 min, 30,000 × g), approximately 80 U of CDH was applied to a Resource PHE 1-ml column (Amersham-Pharmacia), previously equilibrated with 0.1 M Na-acetate buffer, pH 5.0, containing (NH4)2SO4 (20% saturation) and 0.2 M NaCl (buffer A). CDH was eluted by using a linear gradient of 50 mM Na-acetate buffer, pH 5.0 (0 to 50% in buffer A), in 20 ml.
Standard enzyme assays.
The DCIP assay, measuring both the heme-containing CDH and, when present, the non-heme fragment CBQ, was performed by measuring the time-dependent reduction of 0.3 mM DCIP in 80 mM Na-acetate buffer, pH 4.0, containing 30 mM lactose at an absorption of 520 nm and 25°C. The extinction coefficient for DCIP at 520 nm and pH 4.0 was determined experimentally to be 6.8 mM−1 · cm−1. One unit of enzyme activity was defined as the amount of enzyme that reduces 1 μmol of DCIP per min under the selected assay conditions (pH 4.0, 25°C). Alternatively, CDH activity was specifically determined by following at 550 nm the reduction of 20 μM cyt c in 50 mM Na-succinate buffer, pH 3.5, containing 30 mM lactose. The extinction coefficient was 19.6 mM−1 · cm−1 (11). The pH dependence of both CDH and CBQ activity when using the indicated electron acceptors was determined using the following buffers: 50 mM malic acid (pH 1.7 to 3.0), 50 mM succinic acid (pH 2.8 to 6.4), 50 mM Tris (pH 6.9 to 8.3) for DCIP and cyt c; 50 mM acetate–50 mM morpholineethanesulfonic acid–50 mM Tris (1:1:1) (pH 2.5 to 8.0) for p-benzoquinone and DCIP; and 50 mM Na-acetate (pH 3.0 to 6.0), 50 mM Na-citrate (pH 2.0 to 6.0), 20 mM Tris buffer (pH 6.0 to 9.0) for all others. The spectral properties of DCIP are pH dependent. At 520 nm this dependence is less pronounced than at the more commonly used wavelength of 600 nm. The 6.8 mM−1 · cm−1 extinction coefficient at an absorption of 520 nm was used only for pH values below 6.5. Above pH 6.5, calculations were done with an experimentally determined coefficient of 6.6 mM−1 · cm−1. Activities of β-glucosidase and filter paper cellulase (as an indicator of overall cellulolytic activity) were determined with p-nitrophenyl-β-d-glucopyranoside (Sigma) and filter paper (Whatman No. 1; Maidstone, United Kingdom), respectively, as the substrates as previously described (46).
Steady-state kinetic measurements.
All measurements were performed at 25°C at the pH optimum of the respective electron acceptor. The extinction coefficients used are given in Table 3. All kinetic constants were calculated by nonlinear least-squares regression, fitting the observed data to the Michaelis-Menten equation. Unless otherwise stated, the sugar solutions used for measuring activity and kinetic constants were all prepared in water and left to equilibrate for at least several hours. For the comparison of β- and α-lactose, the sugars were dissolved in buffer (pH 4) immediately before use to avoid extensive mutarotation.
TABLE 3.
Apparent kinetic constants of cellobiose dehydrogenase from S. rolfsii for several electron acceptorsa
| Electron acceptor | Wavelength monitored (nm) | Extinction coefficient (mM−1 · cm−1) | pH optimum | Stoichiometryb | CDH kinetic constants
|
CBQ relative activities | ||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (mM−1 · −1) | ||||||
| DCIP | 520 | 6.8 | 3.2–4.8c | 1 | 15 | 30 | 2,000 | 0.95 |
| 1,4-Benzoquinone | 290 | 2.24 | 3.0–4.2 | 1 | 25 | 30 | 1,200 | n.d. |
| 2,6-Dimethyl-1,4-benzoquinone | 330 | 0.37 | 4.0–5.0 | 1 | 550 | 15 | 27 | 0.3 |
| 2,6-Dimethoxy-1,4-benzoquinone | 395 | 0.58 | 3.5–4.0 | 1 | n.d. | 5e | n.d. | 0.1 |
| 3,5-Di-tert-butyl-1,2-benzoquinone | 420 | 1.4d | 3.0–4.0 | 1 | 53 | 23 | 434 | 0.5 |
| Methylene blue | 620 | 34.3 | 3.1–3.6 | 1 | 4.4 | 18 | 4,100 | 0.04 |
| Methylene green | 654 | 38.1 | 3.4–3.8 | 1 | 4.3 | 18 | 4,190 | 0.15 |
| Meldola's blue | 640 | 5.6 | 3.0–3.8 | 1 | 31 | 24 | 800 | 0.15 |
| ABTS cation radicalf | 436g | 29.3 | 4.0 | 2 | 0.4 | 27 | 67,500 | n.d. |
| Cyt c (horse heart) | 550 | 19.6h | 3.5 | 2 | 0.3 | 34 | 113,000 | 0 |
| K3Fe(CN)6 | 420 | 0.98 | 2.7–3.4 | 2 | 20 | 37 | 1,850 | 1 |
Constants were determined at 25°C using 2 mM cellobiose as the substrate and at the pH optimum of the respective acceptor. Protein was determined using the BCA method. n.d., not determined.
Molecules of electron acceptor reduced per molecule of cellobiose oxidized.
For both CDH and CBQ.
Datum is from Roy et al. (43).
Maximum activity.
The cation radical was prepared by oxidizing ABTS with laccase, the enzyme was removed by ultrafiltration, and the radical was quantified with the help of the extinction coefficient.
Datum is from Niku-Paavola et al. (40).
Datum is from Canevascini et al. (11).
Identification of reaction products.
Putative substrates (50 mM) were incubated with CDH (1.4 U · ml−1), laccase (5.0 U · ml−1; for the regeneration of the electron acceptor), and DCIP (1.5 mM) in 20 mM Na-acetate buffer, pH 4.5, and sparged with oxygen overnight (3) to determine if CDH oxidizes certain sugars. The disappearance of known sugar peaks and the appearance of the acid peaks were monitored by high-performance liquid chromatography (HPLC).
HPLC analyses.
Mono- and disaccharides and their respective carboxylic acids were detected by HPLC on an Aminex HPX-87 C column (7.8 by 300 mm; Bio-Rad, Hercules, Calif.) operated at 85°C, using a refractive index detector and an eluent of 10 mM calcium nitrate at 0.7 ml · min−1. Due to the lack of commercially available standards, only cellobionic, lactobionic, and gluconic acid could be unequivocally identified by comparison with authentic material. The presence of activity was assessed by the disappearance of the sugar substrate and by the appearance of a peak with a much higher retention time, which is typical for carboxylic acids.
Protein measurements.
Protein concentrations were determined using either the Bradford dye-binding assay (Coomassie blue; Bio-Rad) or the bicinchoninic acid (BCA) assay (Sigma) according to the protocols of the manufacturers and using bovine serum albumin as the standard.
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on an Amersham-Pharmacia Multiphor II system using precast ExcelGel SDS gradient gels (8–18). Native PAGE was carried out on an Amersham-Pharmacia Phast System using precast PhastGel gradient gels (8–25) with both high- and low-molecular-weight standards (Amersham-Pharmacia). Proteins were visualized by staining with Coomassie blue. All procedures were according to the manufacturer's recommendations.
Isoelectric focusing in the range of pH 2.5 to 5 was performed on the Phast System using precast, dry gels (PhastGel dry IEF; Amersham-Pharmacia) rehydrated with carrier ampholytes (7.5 parts Pharmalyte, pH 2.5 to 5, and 2.5 parts Ampholine, pH 3.5 to 5; Amersham-Pharmacia). The low-pI marker protein kit (pH 2.8 to 6.5; Amersham-Pharmacia) was used to determine pI values. Either protein bands were stained with silver following the instruction manual of the manufacturer or the CBQ and CDH bands were visualized by activity staining (43).
MALDI-MS.
The MALDI-MS (matrix-assisted laser desorption ionization-mass spectrometry) measurements were carried out with a DYNAMO linear time-of-flight spectrometer (Thermo BioAnalysis, Hemel Hempstead, United Kingdom) operated with delayed extraction off and deflector on. CDH was mixed with an equal volume of matrix solution (20 g of sinapinic acid per liter in a 7.3:1 [vol/vol] mixture of acetonitrile and 1.0 g of trifluoroacetic acid per liter in water). The sample (1 μl; corresponding to approximately 0.1 pmol of protein) was placed on a probe, air dried, and analyzed. The mass axis was externally calibrated with bovine serum albumin (66.441 kDa).
Determination of the prosthetic groups.
Heme was identified and its stoichiometry was estimated by conversion to the pyridine hemochrome (22). This procedure consisted of adding 1 volume of dry pyridine to the buffered enzyme solution (pH 7), vortexing for a minute, and then adding NaOH to a final concentration of 0.2 M. After centrifugation (10 min, 10,000 × g) to remove any precipitate, spectra of both the reduced and oxidized forms of the pyridine hemochrome were recorded. Reduction was performed with sodium dithionite, and oxidation was performed with K3[Fe(CN)6]. The localization of the α-peak of the reduced form and the extinction coefficient (reduced-oxidized spectrum) at this wavelength were compared to published data (55). FAD was removed from the enzyme, and its identity was compared with that of commercial FAD (F-6625; Sigma) using thin-layer chromatography and spectroscopic methods (6, 38).
Partial proteolysis of CDH.
Partial proteolysis of CDH was performed as previously reported (11, 52). CDH was incubated with papain (Sigma) for about 2 h, and the cyt c activity was monitored. The resulting protein fragments were separated on a Mono Q column (1 ml; Amersham-Pharmacia) pre-equilibrated with 20 mM Tris buffer, pH 7.5, and eluted with a 1 M NaCl gradient.
Antigenic and genetic comparison.
The polyclonal antibodies against CDH from P. chrysosporium that were used in this study were the same as those previously reported (35). Immunohybridization experiments were performed according to the dot blot test as described in the instruction manual for the picoBlue Immunoscreening Kit (Stratagene, La Jolla, Calif.).
Purified DNA from S. rolfsii was digested with EcoRI, KpnI, SalI, and SacI (Stratagene), and the resulting fragments were separated and blotted according to standard procedures (8). The probe used for hybridization was a 32P-labeled 1.2-kb DNA fragment of the C-terminal domain of P. chrysosporium CDH (35) or the complete 2.8-kb DNA of Sporotrichum thermophile CDH (52).
Carbohydrate content.
The carbohydrate content of purified CDH was estimated by the phenol-sulfuric acid method using mannose as the standard (16).
Reaction of CDH with oxygen.
To determine if oxygen can act as an electron acceptor of CDH, 16 μM CDH was incubated with 300 μM cellobiose in sodium succinate buffer at pH 4.5 and the absorbance was monitored at 564 nm, a wavelength that is isosbestic for the flavin. Formation of H2O2 in the CDH reaction was estimated by a peroxidase assay as previously described (6), using horseradish peroxidase and guaiacol.
Chemicals.
All chemicals were of the highest grade of purity available. 2,2′-Azino-di-(3-ethyl-benzthiazolin-6-sulfonic acid) (ABTS), DCIP, cyt c from horse heart, lactobionic acid, α-lactose, cellobiose, the cellooligosaccharides, lactitol and cellobiitol, and recombinant molecular weight standards for SDS-PAGE were purchased from Sigma. Ca-cellobionate was from ICN Pharmaceuticals (Costa Mesa, Calif.), and thiocellobiose was from Toronto Research Chemicals (Toronto, Canada). β-Lactose, the substituted and unsubstituted quinones, and the redox dyes were purchased from Aldrich (Steinheim, Germany). Stock solutions were prepared as follows: 2,6-dimethoxy-1,4-benzoquinone (15 mM) was dissolved in dimethyl sulfoxide (DMSO); 2,6-dimethyl-1,4-benzoquinone (50 mM) was dissolved in DMSO-ethanol (1:1); 3,5-di-tert-butyl-1,2-benzoquinone (1 mM) was dissolved in ethanol-DMSO-water (5:1:4). Mannobiose, mannotriose, and galactobiose (β-1,3 and β-1,4) were obtained from Megazyme (Bray, Ireland). Allolactose was provided by Sergio Riva (CNR, Milan, Italy). The lactobiono-1,5-lactone was prepared by dissolving lactobionic acid in 2-methoxyethanol and then heating and distilling after addition of toluene (56). The lactone was freeze-dried under vacuum. The laccase was a partially purified enzyme preparation from Trametes pubescens (3).
RESULTS
Production and purification of CDH.
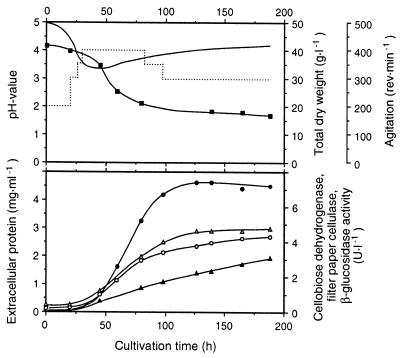
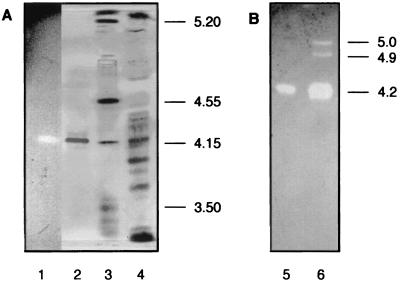
Strain CBS 191.62 was previously identified as an excellent producer of CDH activity (46). Pure cellulosic substrates such as α-cellulose or microcrystalline Avicel were best for CDH production. A 15-liter laboratory fermentation of S. rolfsii grown on α-cellulose as the main carbon source yielded 7,400 U of cyt c activity per liter (measuring only the flavoheme enzyme CDH) and 7,500 U of DCIP activity per liter in the extracellular medium after 130 h of cultivation (Fig. 1). Only a very small fraction (approximately 7%) of the DCIP-reducing activity could be attributed to the flavin-only fragment CBQ, and only minor bands corresponding to CBQ were visible following separation of the proteins by isoelectric focusing and activity staining with DCIP (Fig. 2B, lane 6).
FIG. 1.
Time course of a laboratory cultivation of S. rolfsii CBS 191.62 in a 20-liter stirred-tank reactor (working volume, 15 liters) using a medium based on α-cellulose (42.6 g · liter−1). The temperature was controlled at 30°C, and the pH, initially adjusted to 5.0, was allowed to float. Aeration was automatically varied from 0.1 to 1.0 volume of air per fluid volume per min to maintain a partial O2 pressure of 40% of air saturation. Solid line, pH value; dotted line, agitation. Symbols: ■, total dry matter; ○, extracellular protein; ●, CDH activity (DCIP assay); ▴, β-glucosidase activity; ▵, filter paper cellulase activity.
FIG. 2.
Isoelectric focusing of the crude culture filtrate from S. rolfsii and the purified CDH preparation. (A) Lane 1 shows purified CDH (activity stained as for panel B). On the silver-stained gel, lane 2 represents purified CDH, lane 3 represents pI standards (Amersham-Pharmacia), and lane 4 represents the culture supernatant. (B) Gel incubated with DCIP and lactose (activity staining). Lane 5 shows purified CDH and lane 6 shows the supernatant.
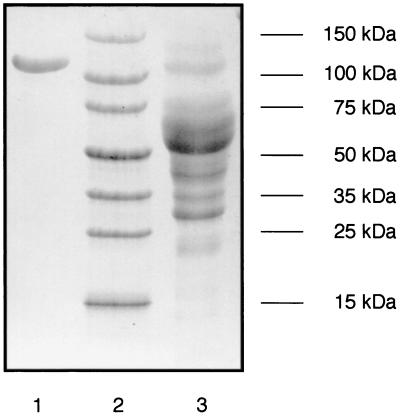
No CDH activity was lost during harvest, removal of the mycelia by centrifugation, and initial concentration and desalting of the medium prior to separation on DEAE Sepharose. Both purification steps, i.e., anion exchange and hydrophobic interaction chromatography, gave single activity peaks. This two-step procedure (Table 1) yielded a reddish-brown protein that was apparently homogenous as judged by SDS-PAGE (Fig. 3). The purified CDH had a specific activity of 18 U · mg−1 for cyt c reduction and using the BCA protein assay (66 U · mg−1 when using the Bradford protein assay). The molecular mass was 101 kDa as determined by MALDI-MS, 110 kDa as estimated by SDS-PAGE (Fig. 3), and 126 kDa according to native PAGE (data not shown). The isoelectric point was 4.2 as judged by isoelectric focusing and comparison to standard proteins (Fig. 2A). Activity staining of the culture supernatant after isoelectric focusing highlighted the major CDH band at pH 4.2 and showed two additional minor bands at pHs 4.9 and 5.0, probably representing small amounts of CBQ (Fig. 2B). Due to the low activity of these DCIP-reducing enzymes, no attempts were made to isolate them. The purified CDH preparation had two additional minor bands with a difference in pI of ±0.05 compared to that of the main protein band. These isoenzymes were not removed by the described purification procedure. They comprise less than 5% of the total activity and presumably represent differences in glycosylation. The N terminus of the purified enzyme was blocked and could not be sequenced.
TABLE 1.
Purification of cellobiose dehydrogenase from S. rolfsii
| Purification stage | Sp act (U · mg−1) witha:
|
A420/A280 | Yield (%) | Purification (fold) | |
|---|---|---|---|---|---|
| DCIPb | cyt cc | ||||
| Supernatant | 2.7 | 2.6 | 0.08 | 100 | 1 |
| DEAE Sepharose | 26 | 28 | 0.22 | 80 | 11 |
| Resource PHE | 63 | 66 | 0.51 | 55 | 25 |
Protein was determined using the Bradford protein assay. Using the BCA protein assay, the following results were obtained for pure CDH: DCIP activity, 16 U · mg−1; cyt c activity, 18 U · mg−1.
Activity was measured using DCIP and cellobiose at pH 4.0; this assay detects both CBQ and CDH.
Activity was measured using cyt c and cellobiose at pH 3.5; this assay detects only CDH.
FIG. 3.
SDS-PAGE of the supernatant from S. rolfsii (lane 3) and the purified CDH preparation (lane 1). Lane 2 shows recombinant molecular size markers (Sigma).
Cofactors of CDH.
The heme cofactor was identified as protoheme IX (heme b) and estimated as approximately 1 heme per CDH molecule. FAD was estimated as 1 nucleotide per molecule by spectrophotometric analysis, and its identity was confirmed with thin-layer chromatography.
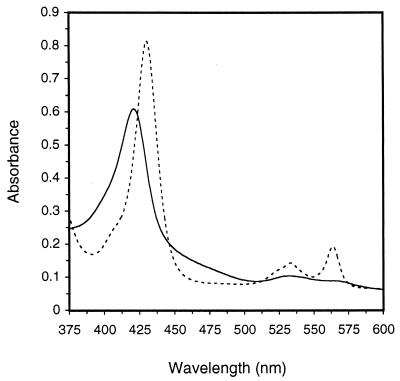
The major peak of the oxidized spectrum (Fig. 4) at an absorption of 421 nm can be attributed to the heme cofactor, whereas the broad absorbance shoulder between 450 and 500 nm can be mainly attributed to the FAD group. The extinction coefficients for the oxidized state of the enzyme at 421, 460, 531, and 563 nm were 105, 24, 12, and 9 mM−1 · cm−1, respectively. Upon reduction of CDH at pH 4, strong peaks appeared at 429, 533, and 564 nm (Fig. 4), representing the Soret, β-, and α-peaks of a typical heme protein. Absorption between 450 and 500 nm decreased drastically, presumably representing the reduced form of FAD. The extinction coefficients for 429, 460, 533, and 564 nm were 140, 10, 17, and 28 mM−1 · cm−1, respectively, for the reduced enzyme. All values were calculated using the BCA protein assay and a molecular size of 101 kDa for the enzyme.
FIG. 4.
Spectra of the oxidized (——) and reduced states (––––) of CDH. Reduction was performed with cellobiose.
CBQ.
Papain degradation of CDH resulted in the formation of several protein fragments that were active with DCIP but not with cyt c. The main fraction was isolated by anion exchange chromatography to apparent homogeneity as judged by SDS-PAGE (data not shown). This enzyme preparation, termed CBQ, was used for all subsequent characterizations. The molecular mass of this protein was estimated to be 75 kDa by SDS-PAGE, and the pI was determined to be 5.1 by isoelectric focusing. The extinction coefficients for absorption at 420 and 450 nm for this fragment in the oxidized form were 6.5 and 8.5 mM−1 · cm−1, respectively. The specific activity was 39 U · mg−1 (using the DCIP activity and BCA protein assays).
Carbohydrate content.
The heme-containing, intact CDH strongly bound to a concanavalin A column and was eluted with 0.4 M methyl-d-mannopyranoside but could not be eluted with glucose, mannose, or methyl-d-glucopyranoside. These results suggest the protein is highly glycosylated with mannose-rich oligosaccharide units. The carbohydrate content was estimated to be 15% using the phenol-sulfuric acid method with mannose as the standard.
Antigenic and genetic comparisons.
Polyclonal rabbit antibodies against P. chrysosporium CDH showed strong cross-reaction with S. rolfsii CDH. Despite this antigenic similarity between the two CDH proteins, no hybridization was detectable on a genetic level. Neither the radiolabeled 1.2-kb gene fragment of the flavin-containing C terminus of P. chrysosporium nor the complete 2.8-kb S. thermophile CDH gene hybridized with the genomic DNA of S. rolfsii in Southern blots (data not shown).
Catalytic properties, electron donors.
The kinetic constants determined for the oxidation of cellobiose and cellooligosaccharides were between 0.1 and 0.6 mM for the Michaelis constant (Km) and 24 to 27 s−1 for the kinetic constant kcat when using DCIP as the electron acceptor at 25°C (Table 2). The catalytic efficiencies, kcat/Km, indicate that these are the preferred and presumably in vivo substrates of S. rolfsii CDH. A second group of substrates, including β-lactose, thiocellobiose, and xylobiose, had somewhat higher Km values (2.0 to 5.4 mM) but had kcat values similar to those determined for the cellodextrins (26 to 27 s−1). A third group, including maltose and glucose, had very high apparent Km values and low catalytic activities, yet the C-1-oxidized product of glucose, i.e., gluconic acid, was unequivocally identified by HPLC using an authentic standard, indicating that these oxidation reactions occur. In a similar manner, the reaction products of the CDH-catalyzed oxidation of cellobiose and lactose, i.e., cellobionic acid and lactobionic acid, were confirmed. Although the reaction product could not be unequivocally identified for maltose oxidation, HPLC analysis showed the disappearance of the substrate and the formation of an acidic product during prolonged incubation with CDH.
TABLE 2.
Apparent kinetic constants of cellobiose dehydrogenase from S. rolfsii for several electron donorsa
| Substrate | Kinetic constants
|
||
|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | |
| Cellobiose | 0.12 | 27 | 225 |
| Thiocellobiose | 2.3 | 27 | 11.7 |
| Cellotriose | 0.49 | 26 | 53.1 |
| Cellotetraose | 0.60 | 24 | 40.0 |
| Cellopentaose | 0.54 | 24 | 44.4 |
| Lactose (equilibrated) | 2.4 | 26 | 10.8 |
| β-Lactose (70 to 75%) | 2.0 | 26 | 13.0 |
| α-Lactose | >54 | <17 | <0.32 |
| Xylobiose | 5.4 | 27 | 5.0 |
| Maltose | 240 | 0.8 | 3.3 × 10−3 |
| Glucose | 1,250 | 1.5 | 1.2 × 10−3 |
Kinetics were measured using the standard DCIP assay at pH 4.0 and 25°C; protein was determined using the BCA method.
When the temperature was increased to 45°C, both the Km and kcat values of cellobiose and lactose increased to give slightly lower catalytic efficiencies than at 25°C. The Km and kcat values determined at this higher temperature for cellobiose were 0.28 mM and 45 s−1, respectively, giving a kcat/Km ratio of 164 mM−1 · s−1, while the corresponding values found for lactose (equilibrated solution) were 4.4 mM, 42 s−1, and 9.6 mM−1 · s−1, respectively.
CDH did not oxidize the monosaccharides d-galactose, d-mannose, or d-xylose, the oligosaccharides gentiobiose (Glc–β-1,6-Glc), sophorose (Glc–β-1,2-Glc), melibiose (Gal–α-1,6-Glc), allolactose (Gal–β-1,6-Glc), galactobiose (Gal–β-1,4-Gal and Gal–β-1,3-Gal), mannobiose (Man–β-1,4-Man), or mannotriose (Man–β-1,4-Man-β-1,4-Man), or the sugar alcohols lactitol and cellobiitol.
Catalytic properties, electron acceptors.
The specificity of CDH for its electron donor is relatively high, but it can transfer electrons to a wide range of different substrates (Table 3). Cyt c could be reduced only by the intact flavoheme protein CDH, while all other acceptors studied could also be reduced by CBQ. Both cyt c and the ABTS radical had remarkably low Km values and the highest catalytic efficiencies. DCIP (a benzoquinone imine), unsubstituted 1,4-benzoquinone, the phenothiazine dyes methylene blue and methylene green, Meldola's blue (a substituted phenoxazine), and ferricyanide have Km values that are two orders of magnitude higher than that for cyt c but are still excellent substrates. Substitution of 1,4-benzoquinone with two methyl or methoxy groups resulted in significantly reduced activities, which is evident from the catalytic constant and/or the catalytic efficiency. No activity was found with 20 mM pyrrolo-quinolinoquinone.
Reaction with oxygen.
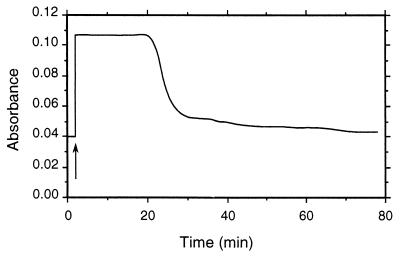
Addition of cellobiose to CDH in aerated buffer led to an immediate increase in absorbance at 564 nm (reduction of the enzyme). After depletion of cellobiose, the reduced CDH was slowly but completely reoxidized to its native form (Fig. 5). Samples taken after 30 min (cellobiose being depleted and the enzyme largely reoxidized) were tested for hydrogen peroxide, and 0.7 μmol of this reduced oxygen species was found for each μmol of cellobiose oxidized.
FIG. 5.
The reaction of CDH with cellobiose and oxygen (air) was monitored at 564 nm. Addition of cellobiose (marked by an arrow) led to the immediate reduction of the heme cofactor of the enzyme. After depletion of cellobiose (after approximately 20 min), the enzyme was slowly but completely reoxidized.
Inhibition of CDH.
No inhibition was detected for cellobiose and lactose for concentrations of up to 5 and 200 mM, respectively. A weak inhibition of CDH by its primary product, lactobiono-1,5-lactone, was found, while the acid, derived by hydrolysis of the lactone, is practically noninhibitory. The inhibition constants determined for the oxidation of lactose are listed in Table 4. An aqueous equilibrium mixture of lactobionic acid at 25°C contains 84% acid and 16% lactone (18). Hydrolysis of the lactone in aqueous solutions is rather slow (KH, 2.14 × 10−4 s−1 at pH 4 [18]), but fungi often produce lactonases that accelerate this process and in this way could relieve inhibition.
TABLE 4.
Kinetic constants for competitive inhibition of lactose oxidation by its primary product, lactobiono-1,5-lactone, and by lactobionic acid or glucono-1,5-lactonea
| Inhibitor | Kinetic constants
|
|||
|---|---|---|---|---|
| Km (mM) | Ki (mM) | Km/Ki | kcat (s−1) | |
| Lactobionic acid, equilibrated | 2.9 | 510 | 0.006 | 24 |
| Ca-lactobionate | 2.4 | 1,600 | 0.002 | 25 |
| Lactobiono-1,5-lactone | 2.6 | 31 | 0.08 | 25 |
| Glucono-1,5-lactone | 2.3 | 105 | 0.02 | 25 |
The electron acceptor used was DCIP.
pH and temperature dependence of activity and stability.
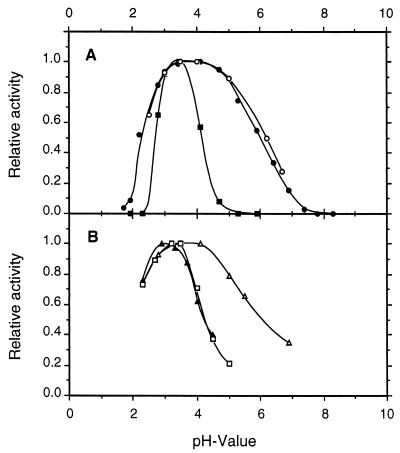
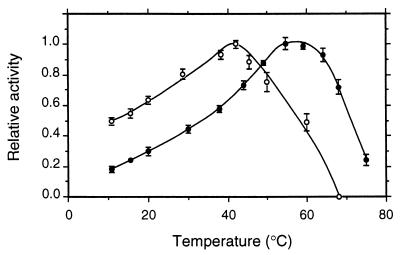
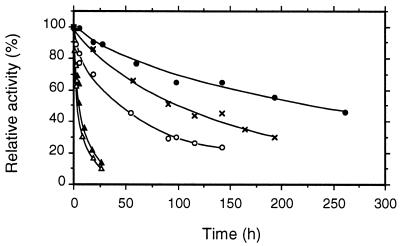
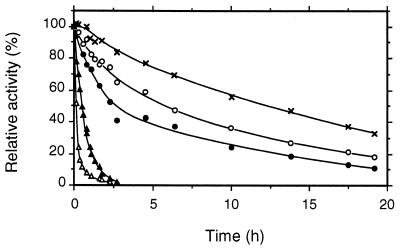
pH optima for CDH and CBQ were between pHs 3 and 5 and depended upon the electron acceptor (Fig. 6; pH optima for several additional cosubstrates are given in Table 3). Most quinone acceptors have broad activity maxima around pH 4, whereas the phenothiazines (e.g., methylene blue) or phenoxazines (Meldola's blue) and ferricyanide and cyt c have more narrow optima around pH 3 to 3.5. The temperature optimum also depended on the electron acceptor. For DCIP this was 55°C, whereas cyt c activity was lost more quickly, with the apparent optimum being 42°C (Fig. 7). The corresponding activation energies, calculated from the Arrhenius plot, were 30 and 17 kJ · mol−1 for DCIP and cyt c, respectively. At 35°C CDH stability was highest at pH 5.7, with a half-life of activity of 225 h (Fig. 8). CDH was especially sensitive to pH values below 3, losing 50% of its initial activity in approximately 10 min at pH 2.2. When the temperature was increased to 46°C, the stability optimum shifted to pH 5.0, with a half-life of 12 h (Fig. 9). Again, CDH was rapidly inactivated at low pH values (t1/2 [pH 2.2] = 4 min).
FIG. 6.
pH profiles for CDH and CBQ from S. rolfsii. CDH was assayed with DCIP (●), cyt c (■), methylene blue (□), ferricyanide (▴), and p-benzoquinone (▵); CBQ was assayed with DCIP (○). Buffers used were as described in the Materials and Methods section; the extinction coefficients are given in Table 3. The total assay time was 3 min. Mean values are shown for duplicate experiments; the standard deviations did not exceed 5%.
FIG. 7.
Temperature dependence of CDH activity measured with cellobiose and DCIP (●) or cyt c (○). The total assay time was 3 min. Data shown are the mean values from two independent replicates ± standard deviations.
FIG. 8.
pH stability of CDH at 35°C. CDH (0.2 mg) was incubated in an appropriate buffer, and the activity was monitored by DCIP assay. The different buffers used were 50 mM Na-citrate for pHs 3.2 to 5.7 and 20 mM Tris for pH 7.6. Mean values are shown for duplicate experiments; the standard deviations did not exceed 5%. Symbols: pH 3.2 (▴), pH 4.0 (○), pH 5.0 (×), pH 5.7 (●), and pH 7.6 (▵).
FIG. 9.
pH stability of CDH at 46°C. Conditions and symbols are the same as those for Fig. 8.
DISCUSSION
The true in vivo function of cellobiose dehydrogenase is not known. It has been speculated that the enzyme is involved in cellulose and/or lignin degradation (27). Whereas S. rolfsii produces a complete cellulase system (46), strain CBS 191.62 formed no ligninase or laccase activity under the growth conditions we used, suggesting a possible involvement of CDH in cellulose degradation. As a plant pathogen, S. rolfsii preferentially degrades young plant material containing an abundance of cellulose but very little lignin. The role of CDH in cellulose degradation could be to relieve the product inhibition of the cellulases (cellobiohydrolases and endoglucanases) by removing cellobiose (32). Alternatively, CDH could be directly involved in cellulose degradation via a mechanism based on hydroxyl radicals. Such a mechanism, based on the CDH-catalyzed reduction of Fe(III), has been suggested for the brown-rot fungus Coniophora puteana (31).
An extracellular, cellobiose-oxidizing enzyme was reported from S. rolfsii CPC 142 and was described as a monomeric protein of 64 kDa with a pI of 5.18 (47). The enzyme oxidized cellobiose, cellooligosaccharides, and lactose, and could be reoxidized by DCIP and also by cyt c, although the catalytic efficiency with the latter electron acceptor was several orders of magnitude lower. The absorption spectra of the enzyme indicated the presence of neither a heme nor an FAD group. Since the catalytic properties of the enzyme were similar to those of the first described cellobiose dehydrogenases (13, 15)—at that time a term commonly used for the flavin-only enzyme—it also was termed cellobiose dehydrogenase, and it was speculated that it might possibly carry a different cofactor.
A heme protein has characteristic peaks in its spectrum and because of its strong absorption can be easily recognized even at low concentrations in both the oxidized and the reduced form. The FAD group can be missed more easily, especially if the enzyme is present only at low concentrations or is in its reduced form (25). Since all hitherto characterized cellobiose dehydrogenases, including the one described in this work, contain a flavin prosthetic group, and since we found no indication of a flavin-devoid cellobiose dehydrogenase during our studies, we think it likely that the first described cellobiose-oxidizing enzyme from S. rolfsii was a flavin-containing CBQ species. Some important characteristics of the enzyme, including the molecular weight, the pI value, and its substrate specificity, correspond to the properties we describe in this paper for CBQ, the flavoprotein fragment obtained by in vitro proteolytic cleavage with papain. Some distinct differences, however, are obvious, e.g., the reaction with cyt c or the pH optimum with DCIP.
CDH is produced by S. rolfsii CBS 191.62 on cellulose-based media during the exponential phase of growth and in parallel with cellulolytic and hemicellulolytic enzymes (46), again suggesting a role in cellulose degradation. CDH production reaches levels of up to 7,400 U per liter (410 mg · liter−1) of fermentation medium or 39 mg per g of extracellular protein. Yields of CDH from P. chrysosporium were 140 U · liter−1 (14 mg · liter−1) under optimized conditions (4) and 800 U · liter−1 (52 mg · liter−1) in response to the addition of calf serum to the medium (23). Reported yields for other organisms range from 50 to 270 U · liter−1 (43). This level of CDH produced by S. rolfsii should stimulate studies on potential technological applications of CDH, e.g., for biosensors (36) or biocatalysis (3). Recently, interest has increased in the biodegradation of persistent chemicals by CDH, e.g., the depolymerization of an insoluble polyacrylate polymer (9). Here CDH is acting indirectly by reducing both ferric iron and molecular oxygen, thereby generating the hydroxyl radical which is responsible for this depolymerization. The high reduction potential and the nonspecific reaction of the hydroxyl radical should facilitate the oxidation of a wide range of different chemicals (10).
The purification protocol reported in this study is easy and efficient and results in high yields of apparently homogenous CDH. The absorbance ratio A420/A280 determined for S. rolfsii CDH was 0.51, which is in accordance with those reported for pure CDH from Trametes versicolor (43) or from P. chrysosporium (6, 25). There is agreement that a good carbohydrate substrate of CDH is composed of at least two sugar residues and that these are β-1,4 linked. Cellobiose, cellooligosaccharides, and lactose are the common denominators with respect to substrate specificity for all CDH enzymes described to date. In studying CDH from P. chrysosporium, it was suggested that both the orientation of the C-3 and C-4 hydroxyl groups and the presence of a C-6 hydroxyl group at the reducing end of a β-1,4-linked disaccharide substrate are essential for binding, whereas the orientation of the C-2 hydroxyl group is of less importance (28). In this respect, S. rolfsii CDH is clearly different from the P. chrysosporium enzyme. Mannobiose, a good substrate of the latter enzyme, is not oxidized by S. rolfsii CDH, while xylobiose is an excellent substrate for S. rolfsii CDH, with a kcat value comparable to that of the presumed in vivo substrate cellobiose, but is not oxidized at all by the P. chrysosporium enzyme. The Humicola insolens CDH, which is active with xylobiose, has an extremely high Km value and a very low catalytic efficiency (kcat/Km) compared to cellobiose (50). The ability of the S. rolfsii CDH to efficiently oxidize xylobiose might enable the use of this enzyme in a coupled assay for the measurement of xylanase kinetics in which xylose would not interfere (50, 51).
The reductive activity of CDH from S. rolfsii is quite similar to that of enzymes from other sources (6, 43, 50, 52). Based on the catalytic efficiencies determined, cyt c and the ABTS radical are the favored electron acceptors. It has been postulated that CDH reacts with oxygen to produce superoxide radicals (38). These anions can reduce cyt c, leading to an indirect reduction of cyt c by CDH. The addition of superoxide dismutase did not reduce cyt c activity in a photometric assay of S. rolfsii CDH, hence we conclude that cyt c reduction is not mediated by the superoxide radical, as has been reported by others (6). CDH can be reoxidized by oxygen, albeit slowly, which is in agreement with previous reports (30, 34, 48). However, a substantial amount of H2O2 produced in this reaction has not been accounted for. Possibly, the hydrogen peroxide formed is degraded in the presence of trace amounts of iron according to Fenton's reaction (41).
A preliminary comparison of CDH with the flavin-only fragment CBQ showed that the substrate specificity for the electron acceptor and the pH optimum are very similar (Table 5). The main difference is that CBQ does not react with cyt c. The catalytic activities of CBQ measured with DCIP and cellobiose are higher than that for CDH, indicating that the FAD domain efficiently reacts with cellobiose and quinones regardless of the presence of the heme domain. CBQ also reduces ferricyanide efficiently (Table 3), showing that the heme is not essential for one-electron reductions. Unlike the S. rolfsii CBQ, the FAD fragment of P. chrysosporium CDH had significantly reduced activity with Fe(CN)63− compared to that of the intact enzyme (26), whereas the heme-devoid CDH 6.4 from T. versicolor was not reoxidized by ferricyanide ions (43).
TABLE 5.
A comparison of apparent kinetic constants for CDH and CBQ from S. rolfsii
| Substrate or electron acceptor | Kinetic constants fora:
|
|||||
|---|---|---|---|---|---|---|
| CDH
|
CBQ
|
|||||
| Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | |
| Cellobiose | 0.12 | 27 | 225 | 0.090 | 48 | 530 |
| DCIP | 0.015 | 30 | 2,000 | 0.018 | 49 | 2,700 |
Kinetics were measured using cellobiose and DCIP at pH 4.0 and 25°C; protein was determined using the BCA method.
In conclusion, the flavoheme enzyme CDH isolated from the phytopathogenic fungus S. rolfsii shows a number of similarities to the enzymes described to date from wood-degrading fungi, e.g., P. chrysosporium or S. thermophile. In contrast, different digests of S. rolfsii DNA did not hybridize with CDH genes from P. chrysosporium or S. thermophile, which suggests low sequence similarity between these enzymes. The relatively high level of CDH production by the wild-type strain of S. rolfsii should facilitate studies on potential technological applications of this enzyme.
ACKNOWLEDGMENTS
We thank Claudia Großwindhager and Alois Sachslehner (Universität für Bodenkultur) for performance of the fermentation work, Christiane Galhaup (Universität für Bodenkultur) for the T. pubescens laccase, Fritz Altmann (Universität für Bodenkultur) for making the MALDI-MS measurements, Sergio Riva for his gift of allolactose, and Klaus D. Kulbe (Universität für Bodenkultur) for his encouragement and interest in our work. We also thank Christian Obinger (Universität für Bodenkultur) and Mike Gold (Oregon Graduate Institute of Technology) for valuable discussions on heme-containing enzymes and CDH.
This work was supported by the Austrian Research Foundation (FWF P14537-MOB) and the European Commission (FAIR CT 96-1048).
REFERENCES
- 1.Aycock R. Symposium on Sclerotium rolfsii. Phytopathology. 1961;51:107–128. [Google Scholar]
- 2.Ayers A R, Ayers S B, Eriksson K-E. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur J Biochem. 1978;90:171–181. doi: 10.1111/j.1432-1033.1978.tb12588.x. [DOI] [PubMed] [Google Scholar]
- 3.Baminger U, Ludwig R, Galhaup C, Leitner C, Kulbe K D, Haltrich D. Continuous enzymatic regeneration of redox mediators used in biotransformation reactions employing flavoproteins. J Mol Catal B Enzym. 2001;11:541–550. [Google Scholar]
- 4.Bao W, Lymar E, Renganathan V. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 1994;42:642–646. [Google Scholar]
- 5.Bao W, Renganathan V. Cellobiose oxidase of Phanerochaete chrysosporium enhances crystalline cellulose degradation by cellulases. FEBS Lett. 1992;302:77–80. doi: 10.1016/0014-5793(92)80289-s. [DOI] [PubMed] [Google Scholar]
- 6.Bao W, Usha S N, Renganathan V. Purification and characterization of cellobiose dehydrogenase, a novel extracellular hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys. 1993;300:705–713. doi: 10.1006/abbi.1993.1098. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brown T. Analysis of DNA sequences by blotting and hybridisation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley; 1994. pp. 4.9.1–4.9.4. [Google Scholar]
- 9.Cameron M D, Aust S D. Degradation of chemicals by reactive radicals produced by cellobiose dehydrogenase from Phanerochaete chrysosporium. Arch Biochem Biophys. 1999;367:115–121. doi: 10.1006/abbi.1999.1257. [DOI] [PubMed] [Google Scholar]
- 10.Cameron M D, Timofeevski S, Aust S D. Enzymology of Phanerochaete chrysosporium with respect to the degradation of recalcitrant compounds and xenobiotics. Appl Microbiol Biotechnol. 2000;54:751–758. doi: 10.1007/s002530000459. [DOI] [PubMed] [Google Scholar]
- 11.Canevascini G, Borer P, Dreyer J-L. Cellobiose dehydrogenases of Sporotrichum (Chrysosporium) thermophile. Eur J Biochem. 1991;198:43–52. doi: 10.1111/j.1432-1033.1991.tb15984.x. [DOI] [PubMed] [Google Scholar]
- 12.Canevascini G, Etienne K, Meyer H. A direct enzymatic lactose assay using cellobiose-(lactose-) dehydrogenase from Sporotrichum thermophile. Z Lebensm Unters Forsch. 1982;175:125–129. [Google Scholar]
- 13.Coudray M-R, Canevascini G, Meier H. Characterization of a cellobiose dehydrogenase in the cellulolytic fungus Sporotrichum (Chrysosporium) thermophile. Biochem J. 1982;203:277–284. doi: 10.1042/bj2030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox M C, Rogers M S, Cheesman M, Jones G D, Thomson A J, Wilson M T, Moore G R. Spectroscopic identification of the haem ligands of cellobiose oxidase. FEBS Lett. 1992;307:233–236. doi: 10.1016/0014-5793(92)80774-b. [DOI] [PubMed] [Google Scholar]
- 15.Dekker R F H. Induction and characterization of a cellobiose dehydrogenase produced by a species of Monilia. J Gen Microbiol. 1980;120:309–316. [Google Scholar]
- 16.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 17.Dumonceaux T J, Bartholomew K A, Charles T C, Moukha S M, Archibald F S. Cloning and sequencing of a gene encoding cellobiose dehydrogenase from Trametes versicolor. Gene. 1998;210:211–219. doi: 10.1016/s0378-1119(98)00084-5. [DOI] [PubMed] [Google Scholar]
- 18.Dutta S K, Mukherjee S K. Study of the lactone-acid-salt equilibria and the hydrolysis kinetics for lactobiono-δ-lactone. Ind J Chem. 1971;9:229–232. [Google Scholar]
- 19.Elmgren M, Lindquist S-E, Henriksson G. Cellobiose oxidase crosslinked to a redox polymer matrix at an electrode surface—a new biosensor. J Electroanal Chem. 1992;341:257–273. [Google Scholar]
- 20.Fähnrich P, Irrgang K. Conversion of cellulose to sugars and cellobionic acid by the extracellular enzyme system of Chaetomium cellulolyticum. Biotechnol Lett. 1982;4:775–780. [Google Scholar]
- 21.Fang J, Liu W, Gao P J. Cellobiose dehydrogenase from Schizophyllum commune: purification and study of some catalytic, inactivation, and cellulose-binding properties. Arch Biochem Biophys. 1998;353:37–46. doi: 10.1006/abbi.1998.0602. [DOI] [PubMed] [Google Scholar]
- 22.Fuhrhop J-H, Smith K M. Laboratory methods in porphyrin and metalloporphyrin research. Amsterdam, The Netherlands: Elsevier Scientific Publishing Company; 1975. [Google Scholar]
- 23.Habu N, Igarashi K, Samejima M, Pettersson B, Eriksson K-E L. Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol Appl Biochem. 1997;26:97–102. [PubMed] [Google Scholar]
- 24.Habu N, Samejima M, Dean J F D, Eriksson K-E L. Release of FAD domain from cellobiose oxidase by proteases from cellulolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1993;327:161–164. doi: 10.1016/0014-5793(93)80162-n. [DOI] [PubMed] [Google Scholar]
- 25.Henriksson G. Structure, function and applications of cellobiose dehydrogenase from Phanerochaete chrysosporium. Ph.D. thesis. Uppsala, Sweden: Uppsala University; 1995. [Google Scholar]
- 26.Henriksson G, Johansson G, Pettersson G. Is cellobiose oxidase from Phanerochaete chrysosporium a one-electron reductase? Biochim Biophys Acta. 1993;1144:184–190. doi: 10.1016/0005-2728(93)90171-b. [DOI] [PubMed] [Google Scholar]
- 27.Henriksson G, Johansson G, Pettersson G. A critical review of cellobiose dehydrogenases. J Biotechnol. 2000;78:93–113. doi: 10.1016/s0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- 28.Henriksson G, Sild V, Szabo I J, Pettersson G, Johansson G. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim Biophys Acta. 1998;1383:48–54. doi: 10.1016/s0167-4838(97)00180-5. [DOI] [PubMed] [Google Scholar]
- 29.Henriksson G, Zhang L, Li J, Ljungquist P, Reitberger T, Pettersson G, Johansson G. Is cellobiose dehydrogenase from Phanerochaete chrysosporium a lignin degrading enzyme? Biochim Biophys Acta. 2000;1480:83–91. doi: 10.1016/s0167-4838(00)00096-0. [DOI] [PubMed] [Google Scholar]
- 30.Hyde S M, Wood P M. Kinetic and antigenic similarities for cellobiose dehydrogenase from the brown rot fungus Coniophora puteana and the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett. 1996;145:439–444. [Google Scholar]
- 31.Hyde S M, Wood P M. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe(III) reduction by cellobiose dehydrogenase and Fe(II) oxidation at a distance from the hyphae. Microbiology. 1997;143:259–266. doi: 10.1099/00221287-143-1-259. [DOI] [PubMed] [Google Scholar]
- 32.Igarashi K, Samejima M, Eriksson K-E L. Cellobiose dehydrogenase enhances Phanerochaete chrysosporium cellobiohydrolase I activity by relieving product inhibition. Eur J Biochem. 1998;253:101–106. doi: 10.1046/j.1432-1327.1998.2530101.x. [DOI] [PubMed] [Google Scholar]
- 33.Igarashi K, Verhagen M F, Samejima M, Schülein M, Eriksson K-E L, Nishino T. Cellobiose dehydrogenase from the fungi Phanerochaete chrysosporium and Humicola insolens. A flavohemoprotein from Humicola insolens contains 6-hydroxy-FAD as the dominant active cofactor. J Biol Chem. 1999;274:3338–3344. doi: 10.1074/jbc.274.6.3338. [DOI] [PubMed] [Google Scholar]
- 34.Kremer S M, Wood P M. Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an iron(III) reductase: kinetic comparison with neutrophil NADPH oxidase and yeast flavocytochrome b2. Eur J Biochem. 1992;205:133–138. doi: 10.1111/j.1432-1033.1992.tb16760.x. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Nagalla S R, Renganathan V. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:1329–1335. doi: 10.1128/aem.62.4.1329-1335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindgren A, Stoica L, Ruzgas T, Ciucu A, Gorton L. Development of a cellobiose dehydrogenase modified electrode for amperometric detection of diphenols. Analyst. 1999;124:527–532. [Google Scholar]
- 37.Morpeth F. Cellobiose oxidoreductases. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Boca Raton, Fla: CRC Press; 1991. pp. 337–348. [Google Scholar]
- 38.Morpeth F F. Some properties of cellobiose-oxidase from the white-rot fungus Sporotrichum pulverulentum. Biochem J. 1985;228:557–564. doi: 10.1042/bj2280557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moukha S M, Dumonceaux T J, Record E, Archibald F S. Cloning and analysis of Pycnoporus cinnabarinus cellobiose dehydrogenase. Gene. 1999;234:23–33. doi: 10.1016/s0378-1119(99)00189-4. [DOI] [PubMed] [Google Scholar]
- 40.Niku-Paavola M L, Raaska L, Itävaara M. Detection of white-rot fungi by a non-toxic stain. Mycol Res. 1990;94:27–31. [Google Scholar]
- 41.Nutt A, Salumets A, Henriksson G, Sild V, Johansson G. Conversion of O2 species by cellobiose dehydrogenase (cellobiose oxidase) and glucose oxidase—a comparison. Biotechnol Lett. 1997;19:379–383. [Google Scholar]
- 42.Renganathan V, Bao W. Cellobiose dehydrogenase. ACS Symp Ser. 1994;566:179–187. [Google Scholar]
- 43.Roy B P, Dumonceaux T, Koukoulas A A, Archibald F S. Purification and characterization of cellobiose dehydrogenases from the white rot fungus Trametes versicolor. Appl Environ Microbiol. 1996;62:4417–4427. doi: 10.1128/aem.62.12.4417-4427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy B P, Paice M G, Archibald F S, Misra S K, Misiak L E. Creation of metal-complexing agents, reduction of manganese dioxide, and promotion of manganese peroxidase-mediated Mn(III) production by cellobiose:quinone oxidoreductase from Trametes versicolor. J Biol Chem. 1994;269:19745–19750. [PubMed] [Google Scholar]
- 45.Sachslehner A, Haltrich D, Gübitz G, Nidetzky B, Kulbe K D. Efficient production of mannan-degrading enzymes by the basidiomycete Sclerotium rolfsii. Appl Biochem Biotechnol. 1998;70–71:939–953. doi: 10.1007/BF02920205. [DOI] [PubMed] [Google Scholar]
- 46.Sachslehner A, Haltrich D, Nidetzky B, Kulbe K D. Production of hemicellulose- and cellulose-degrading enzymes by various strains of Sclerotium rolfsii. Appl Biochem Biotechnol. 1997;63–65:189–201. doi: 10.1007/BF02920424. [DOI] [PubMed] [Google Scholar]
- 47.Sadana J C, Patil R V. The purification and properties of cellobiose dehydrogenase from Sclerotium rolfsii and its role in cellulolysis. J Gen Microbiol. 1985;131:1917–1923. [Google Scholar]
- 48.Samejima M, Eriksson K-E. A comparison of the catalytic properties of cellobiose:quinone oxidoreductase and cellobiose oxidase from Phanerochaete chrysosporium. Eur J Biochem. 1992;207:103–107. doi: 10.1111/j.1432-1033.1992.tb17026.x. [DOI] [PubMed] [Google Scholar]
- 49.Schmidhalter D R, Canevascini G. Isolation and characterization of the cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana (Schum ex Fr.) Karst. Arch Biochem Biophys. 1993;300:559–563. doi: 10.1006/abbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 50.Schou C, Christensen M H, Schülein M. Characterization of a cellobiose dehydrogenase from Humicola insolens. Biochem J. 1998;330:565–571. doi: 10.1042/bj3300565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schou C, Rasmussen G, Kaltoft M-B, Henrissat B, Schülein M. Stereochemistry, specificity and kinetics of the hydrolysis of reduced cellodextrins by nine cellulases. Eur J Biochem. 1993;217:947–953. doi: 10.1111/j.1432-1033.1993.tb18325.x. [DOI] [PubMed] [Google Scholar]
- 52.Subramaniam S S, Nagalla S R, Renganathan V. Cloning and characterization of a thermostable cellobiose dehydrogenase from Sporotrichum thermophile. Arch Biochem Biophys. 1999;365:223–230. doi: 10.1006/abbi.1999.1152. [DOI] [PubMed] [Google Scholar]
- 53.Temp U, Eggert C. Novel interaction between laccase and cellobiose dehydrogenase during pigment synthesis in the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1999;65:389–395. doi: 10.1128/aem.65.2.389-395.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood J D, Wood P M. Evidence that cellobiose:quinone oxidoreductase from Phanerochaete chrysosporium is a breakdown product of cellobiose oxidase. Biochim Biophys Acta. 1992;1119:90–96. doi: 10.1016/0167-4838(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 55.Yamanaka T. Iron. In: Otsuka S, Yamanaka T, editors. Metalloproteins. Amsterdam, The Netherlands: Elsevier; 1988. pp. 95–265. [Google Scholar]
- 56.Zhang Z, Fukunaga K, Sugimura Y, Nakao K, Shimizu T. Synthesis of glycolipids: dialkyl N-[N-(4-lactonamidobutyl) succinamoyl]-l-glutamates. Carbohydr Res. 1996;290:225–232. doi: 10.1016/0008-6215(96)00140-1. [DOI] [PubMed] [Google Scholar]