Abstract
Antimalarial drug resistance is a major threat due to the emerging resistance to all the available drugs in the market. In an approach to develop alternative drugs, a novel class of Pf-DHFR inhibitors was developed using pyrimidine as the core nucleus and substituting the 4- and 6- positions with amines and 4-amino benzoic acid (PABA) to avoid the problem of drug resistance. The resultant compounds 3(a–j) after primary in silico screening and filtering were synthesized using microwave efficiently in high yield and reduced time period compared to conventional synthesis. The antimalarial assay was performed in vitro, against chloroquine-sensitive (3D7) and chloroquine-resistant (Dd2) strains of Plasmodium falciparum using chloroquine as a reference standard. The IC50 values were in the range of 5.26–106.76 µg/ml for 3D7 and in Dd2 the value ranges from 4.71 to 112.98 µg/ml. Compounds 3d, 3e, 3f and 3h showed significant antimalarial activity against both the strains of P. falciparum with no cytotoxicity against fibroblast cell line and 3f was found to be the most potent among them. The hemolysis assay of all the compounds in fresh erythrocytes showed insignificant hemolysis below 5% at a higher dose level. Hence, the present study suggests the possible utility of PABA-substituted pyrimidine scaffold for further development of new Pf-DHFR inhibitors.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-022-03236-w.
Keywords: Pyrimidine, p-Aminobenzoic acid, Pf-DHFR, Microwave organic synthesis, Antimalarial activity, Plasmodium falciparum
Introduction
Dihydrofolate reductase (DHFR) is a well-known enzyme abundant in almost all eukaryotic and prokaryotic cells and has been used as the target for antiprotozoal, antineoplastic, antifungal, antimicrobial and autoimmune disorders (Nzila et al. 2005). The protozoal infection, malaria is one of the most widespread diseases in the world caused by the genus Plasmodium. Plasmodium falciparum and Plasmodium vivax, for which humans are the exclusive mammalian hosts, are the most common species and are responsible for the largest public health burden. However, P. falciparum is the most prevalent malaria parasite in the WHO African Regions. Emerging resistance to the mainstay treatment for Plasmodium falciparum malaria, i.e., artemisinin-based combination (ACTs) treatments and the first-line antimalarials, such as chloroquine and pyrimethamine, there is an urgent need for new alternative capable of overriding the resistance. In 2020, there were an estimated 241 million malaria cases compared to 227 million in 2019 which was associated with disruption to malaria services during the COVID-19 pandemic (World Malaria Report 2021). The clinical efficacy of antimalarial antifolate drugs like pyrimethamine and cycloguanil has been compromised due to multiple mutations occurring in DHFR that lead to drug resistance and are mostly used in combination therapy along with ACTs (Yuthavong et al. 2012). Among the antimalarial drugs currently in clinical use, antifolates have the best defined clinically validated molecular targets such as P. falciparum dihydrofolate reductase (Pf-DHFR) which plays a key role in de novo folate biosynthesis pathway essential for DNA synthesis and the metabolism of certain amino acids in Plasmodium species (Yuthavong et al. 2012). The basis of the anti-infective selectivity of antifolates is that they are potent inhibitors of bacterial and protozoal DHFRs respectively, but are only weak inhibitors of mammalian DHFRs (Schweitzer et al. 1990). The literature indicates that the majority of the DHFR inhibitors currently in use or investigation are folic acid derivatives having di- or tri-amino substitution in nitrogen heterocycles, such as pyrimidine, triazine, pteridines, quinazolines, pyrido-pyrimidines (Hitchings et al. 1948; Agarwal et al. 2005; Morgan et al. 2008; Banjanac et al. 2009; Algul et al. 2011; Sahu et al. 2016). Pyrimidine and triazines particularly with amine linkage at -2, -4 and -6 positions, are known pharmacophores in several structure-based drug design approaches in medicinal chemistry (Sahu et al. 2016; Gogoi et al. 2021). Substituted pyrimidine structures are also common in many marketed drugs, like pyrimethamine and sulfadoxine as antimalarial; trimethoprim, sulfamethazine, sulfadiazine as antibacterial; idoxuridine and trifluridine as antiviral; zidovudine and stavudine as anti-HIV; 5-flourouracil as anticancer etc. (Dansena et al. 2015). 4-amino benzoic acid (PABA) is an essential folate precursor and building block used frequently in various drug design approaches against a wide range of therapeutic activities like antimalarial, antibacterial, antimycobacterial, antifungal etc. (Krátký et al. 2019; Adhikari et al. 2020). Anticancer folate antagonist methotrexate, aminopterin, antimicrobial sulphonamides and sulphones are few examples of drugs containing PABA scaffold in their structure. PABA is one of the important non-protein amino acids utilized by malaria parasite in large quantities to replicate in the mammalian host bloodstream. The combination of bioactive scaffold-like pyrimidine and PABA through the involvement of primary or secondary amine may enhance the folate antagonistic property (Lödige and Hiersch 2015; Vandekerckhove and D’hooghe 2015; Kashyap et al. 2021). Therefore, our aim was to develop a novel class of antimalarials which target the Pf-DHFR enzyme important for nucleic acid synthesis and subsequently inhibit erythrocytic schizont stage of the parasite, thereby preventing invasion of fresh RBCs and lowering of parasite density. These drugs were developed in a very cost-effective way in high yield using microwave organic synthesizer. Since aminopyrimidines have been associated with a wide range of therapeutic activities, the most important reagent for synthesizing the compounds is chloropyrimidine; because of the reactivity of its chlorine atoms toward nucleophiles (Brown 2009). The reactions under conventional heating take many hours to days, microwave irradiation can greatly facilitate the synthesis of various substituted aminopyrimidines through nucleophilic aromatic substitution reaction (Hartung et al. 2006). When two halogens are at meta-positions (for example 4,6-dichloropyrimidine), different amines can be selectively added by purifying the mono-amination intermediates. For the reactions with 4, 6-dichloropyrimidine, the first addition is substantially easier than the addition of the second amino group. A slight excess of dichloropyrimidine should be used for the first amination so that there is no detectable amount of bis-aminated product (Luo et al. 2002; Brown 2009). Hence, new PABA-substituted pyrimidine derivatives were designed and screened in silico to filter out the potential compounds for synthesis. The synthesized compounds were screened for their in vitro antimalarial activity in both chloroquine-sensitive 3D7 as well as chloroquine-resistant Dd2 strains of P. falciparum along with their cytotoxicity studies in human fibroblast cell line.
Results and discussion
In silico drug design
The selected compounds for synthesis were screened using preliminary filters to test Lipinski’s rule of five (Ro5) and predict the likelihood of good oral absorption (Lipinski et al. 1997; Lipinski 2004; Meng et al. 2011). All the titled compounds 3 (a–j) passed the rule of five with zero violation indicating, potential oral bioactivity (Table 1).
Table 1.
“Molinspiration cheminformatics” software predicted molecular properties of the titled compounds 3(a-j)
| Compound code | miLogP | TPSA | n-atoms | MW | n-ON | n-OHNH | n-violation | n-rotb | Volume |
|---|---|---|---|---|---|---|---|---|---|
| 3a | 2.24 | 87.14 | 18 | 244.25 | 6 | 3 | 0 | 4 | 215.51 |
| 3b | 2.61 | 87.14 | 19 | 258.28 | 6 | 3 | 0 | 5 | 232.31 |
| 3c | 3.12 | 87.14 | 20 | 272.31 | 6 | 3 | 0 | 6 | 249.11 |
| 3d | 3.68 | 87.14 | 21 | 286.33 | 6 | 3 | 0 | 7 | 265.91 |
| 3e | 1.78 | 90.38 | 22 | 299.33 | 7 | 3 | 0 | 4 | 268.10 |
| 3f | 2.37 | 81.59 | 23 | 313.36 | 7 | 2 | 0 | 4 | 285.04 |
| 3g | 2.75 | 81.59 | 28 | 327.39 | 7 | 2 | 0 | 5 | 301.84 |
| 3h | 4.07 | 81.59 | 28 | 375.43 | 7 | 2 | 0 | 5 | 339.88 |
| 3i | 2.33 | 87.58 | 22 | 300.32 | 7 | 2 | 0 | 4 | 264.68 |
| 3j | 3.39 | 78.35 | 22 | 298.35 | 6 | 2 | 0 | 4 | 272.49 |
milogP logarithm of partition coefficient of compound between n‐octanol and water; MW molecular weight; n-atoms number of atoms; n‐ON number of hydrogen bond acceptors; n‐OHNH number of hydrogen bonds donors; n-rotb number of rotatable bonds; n-violations number of violations; TPSA topological polar surface area
The predicted toxicity risk using TOPKAT module of Accelrys Discovery studio 3.0 of the compounds showed the entire test compounds 3 (a–j) have no carcinogenicity, skin irritancy and Ames mutagenicity (Table 2).
Table 2.
TOPKAT module of Accelrys Discovery studio 3.0-predicted toxicity risks of the titled compounds 3(a-j)
| Compound code | Mouse female NTP carcinogen |
Mouse male NTP carcinogen |
Rat male NTP carcinogen |
Ames mutagen | Skin irritancy |
|---|---|---|---|---|---|
| 3a | − | − | − | − | − |
| 3b | − | − | − | − | − |
| 3c | − | − | − | − | − |
| 3d | − | − | − | − | − |
| 3e | − | − | − | − | − |
| 3f | − | − | − | − | − |
| 3g | − | − | − | − | − |
| 3h | − | − | − | − | − |
| 3i | − | − | − | − | − |
| 3j | − | − | − | − | − |
The negative symbol ( −) indicates absence of any toxicity risk, such as carcinogenicity, Ames mutagenicity and skin irritancy
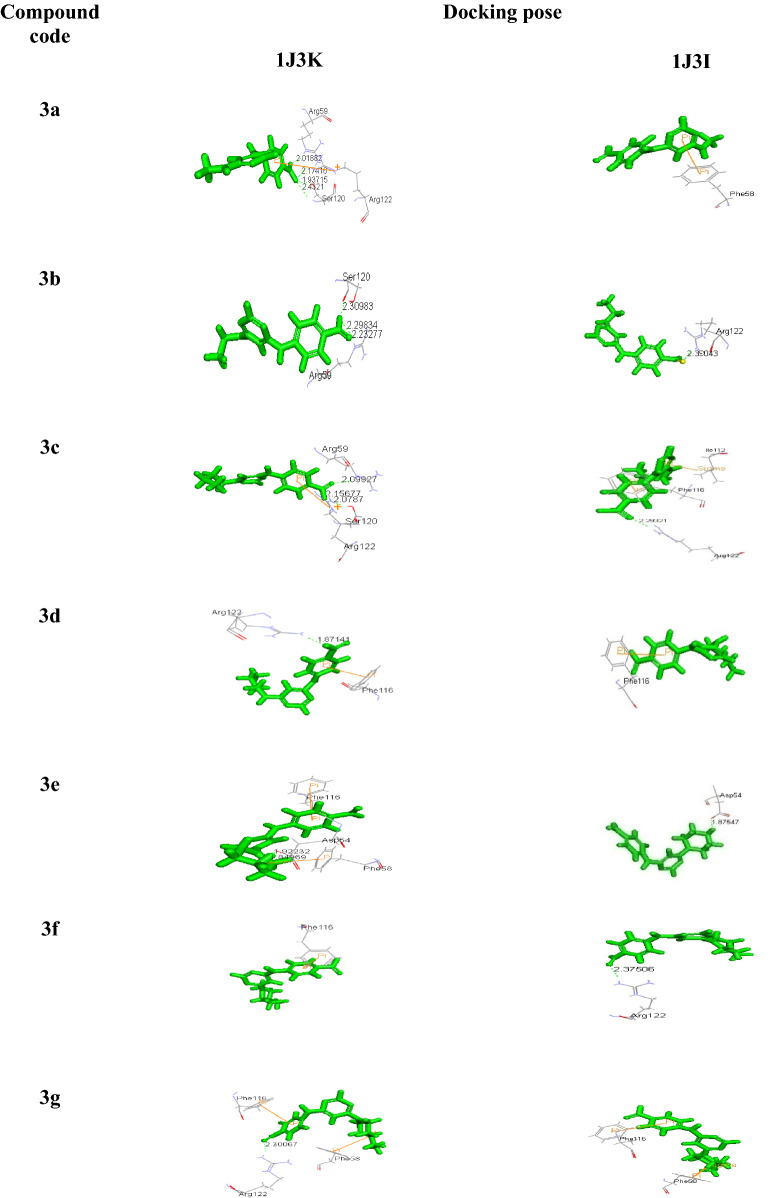
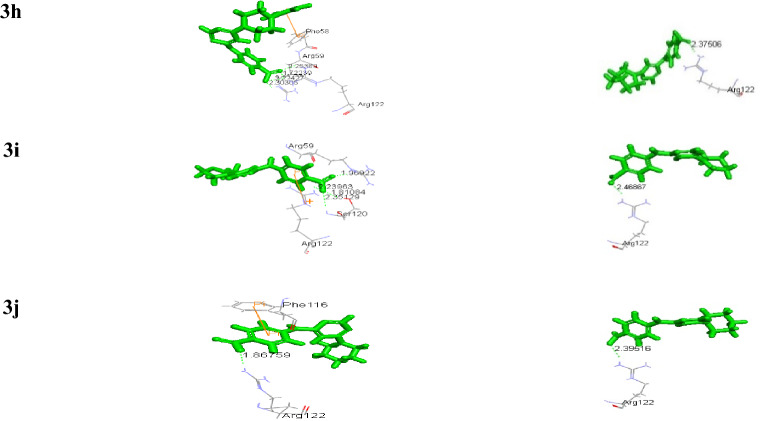
Molecular docking study
Docking studies using Accelrys Discovery Studio 3.0 of all the compounds with both the wild (1J3I.pdb) and quadruple mutant (1J3K.pdb) Plasmodium falciparum–dihydrofolate reductase–thymidylate synthase complex (Pf–DHFR–TS), exhibited significant binding affinities as evident from the binding energies in comparison to the reference compounds (WR99210), which is the originally bound ligand with 1J3K with binding energy (−144.15 kcal/mol) (Yuvaniyama et al. 2003). The binding energies of all the titled compounds were in the range of −292.47 to −407.25 (Kcal/mol) for 1J3K and −242.34 to −331.03 (Kcal/mol) for 1J3I (Table 3). Docking pose of the titled compounds revealed the presence of different types of interaction including a hydrogen-bonding pattern in the active site of both 1J3I and 1J3K along with the formation of cation–pi, pi–pi, and pi–sigma interactions with the amino acids. Compounds 3d, 3e, 3f and 3h were found to be active antimalarials which showed pi–pi and pi–cation interaction on the active site residues Phe116, Phe58, whereas Arg122 and Asp54 formed a hydrogen bond with amino pyrimidine moiety due to the COOH group of phenyl ring linked to pyrimidine ring. Also, it has been observed that among the selected compounds, the binding energies of all the four mentioned compounds were very low (−319.09, −407.25, −375.25 and −292.47) for 1J3K and (−243.73, −276.28, −324.71 and −324.70) for 1J3I, making them the most stable compounds. All the other Compounds had also exhibited higher negative binding energy compared to reference compound WR99210 with H-bonded, pi–pi and pi–cation interaction into the active site however, the antimalarial activity being mild.
Table 3.
Docking interactions and binding energy of the compounds 3(a-j) with 1J3K and 1J3I
| Compound code | Docking interaction | ( −) Binding energy (Kcal/mol) | ||
|---|---|---|---|---|
| 1J3K | 1J3I | 1J3K | 1J3I | |
| 3a | Arg59, Ser120, Arg122 | Phe58 | 293.41 | 263.49 |
| 3b | Arg59, Ser120 | Arg122 | 311.44 | 253.69 |
| 3c | Arg59, Ser120, Arg122 | Ile112, Phe116, Arg122 | 306.94 | 242.81 |
| 3d | Phe116, Arg122 | Phe116 | 319.09 | 243.73 |
| 3e | Asp54, Phe58, Phe116 | Asp54 | 407.25 | 276.28 |
| 3f | Phe116 | Arg122 | 375.25 | 324.71 |
| 3g | Phe58, Phe116, Arg122 | Phe58, Phe116 | 370.02 | 331.03 |
| 3h | Phe58, Arg59, Arg122 | Arg122 | 292.47 | 324.70 |
| 3i | Arg59, Ser120, Arg122 | Arg122 | 298.40 | 242.34 |
| 3j | Phe116, Arg122 | Arg122 | 304.80 | 244.85 |
Most of the compounds had shown strong charge mediated H-bond with Arg122 in the dock poses which is the important interaction site of mutant 1J3K, thus making them a potent inhibitor of Pf-DHFR (Yuthavong et al. 2012) (Table 4).
Table 4.
Docking poses of the pyrimidine derivatives of 4-amino benzoic acid in mutant type Pf-DHFR-TS (1J3K) and (1J3I) protein
Pharmacokinetics
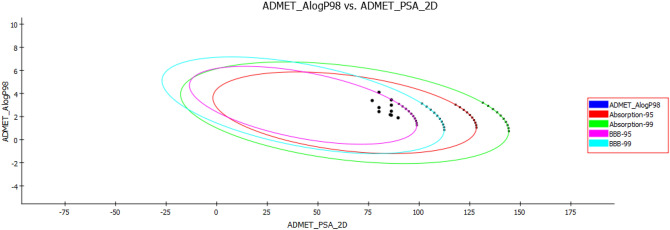
The pharmacokinetics of the compounds under study was explored by (ADME) absorption, distribution, metabolism, excretion prediction of the best active compounds after in silico and docking studies. The in-built ADMET model of Accelrys Discovery Studio 3.0 was used for the prediction of human intestinal absorption. The well-absorbed compounds were plotted in the confidence ellipses using the descriptors AlogP98 and 2D polar surface area (PSA_2D). It was observed that most of the selected compounds were within 95% and 99% ellipse region and are well-absorbed compounds (Fig. 1) (Adhikari et al. 2020).
Fig. 1.
ADME prediction of the compounds 3(a-j) for human intestinal absorption
Chemistry
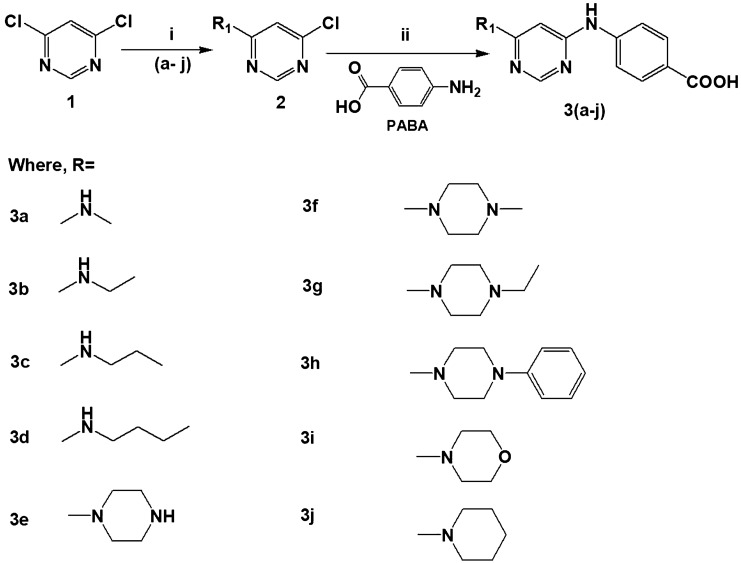
Scheme 1 depicts the synthesis of 4,6-diaminopyrimidine derivatives. First, the synthesis of 4-amino-6-chloropyrimidine 2 (a-j) was obtained by the reaction of 4,6-dichloro pyrimidine (1) with different primary and secondary amines (a-j), in presence of a base triethylamine using microwave organic synthesis at 30–35 °C temperature, 1 bar pressure, 15 W power for 15 min. The intermediates 2 (a-j) were then washed using various solvents like water and ethanol to remove unreacted products if any and then dried. Whereas, the synthesis of final compounds 3 (a-j) was achieved by the reaction of mono-substituted pyrimidine 2 (a–j) with an equimolar amount of 4-aminobenzoic acid and stirred in 10 ml closed vessel in the microwave at 160 °C, 15 bar pressure and 50 W power for 30–40 min. All the synthesized compounds 3 (a-j) were then recrystallized, properly dried and characterized through various physicochemical properties like solubility, physical state, color, melting point and determination of Rf value by TLC using methanol: ethyl acetate: chloroform (0.5:0.5:1) as solvent system. Various spectroscopic techniques Fourier-transform infrared spectroscopy (FTIR), mass spectrometry (MS), nuclear magnetic resonance spectroscopy (NMR) 1H and 13C NMR were used for the confirmation of the synthesized compounds. Characteristic FTIR peaks for functional groups were observed in the region 3400–3200 cm−1 due to secondary N–H stretching, 3000–2500 cm−1 due to OH group present in COOH of the phenyl ring, 1650–1700 cm−1 due to C = O and 1600–1150 cm−1 due to C–C, C–N stretching. 1H-NMR of the compounds showed proton signals in terms of chemical shift (δ) value in between 4.3–5.2 ppm due to NH linked to pyrimidine and in between 9–11 ppm due to acidic proton of COOH at the para position. 13C-NMR showed the appearance of carbon signal at about 166–170 ppm for carboxyl and at about 150–160 ppm for pyrimidine nucleus. The mass spectrometry showed an ion as (M + H) + for each compound.
Scheme 1.
Reagents and conditions: (i) primary and secondary amines (a-j), 2-propanol, 30–35 °C, 1 bar, 15 W, 15 min. (iii) p-amino benzoic acid (PABA), Dimethyl sulfoxide (DMSO), 160 °C, 15 bars, 50 W, 30–40 min
In vitro antimalarial screening
The results of antimalarial activity were shown in Table 5 which depicts the parasite inhibitory activity of all the synthesized compounds against P. falciparum strains 3D7 (chloroquine-sensitive) and Dd2 (chloroquine-resistant). The compounds 3a to 3j displayed an IC50 value in the range of 5.26–106.76 µg/ml for 3D7 and for Dd2 the IC50 value range from 4.71–112.98 µg/ml. Out of all the ten synthesized compounds four compounds 3d, 3e, 3f and 3h containing n-butyl amine, piperazine, 4-methylpiperazine and 4-phenylpiperazine showed comparatively better activity against both chloroquine-sensitive and -resistant strain (3D7 and Dd2). The compound 3f substituted with 4-methyl piperazine was found to be the most active antimalarial among all the compounds having IC50 4.71 µg/ml in CQ resistant strain (Dd2) and IC50 5.26 µg/ml in CQ sensitive strain (3D7) with resistance index 0.89. Similarly, the piperazine substituted compound 3e showed good antimalarial activity with IC50 7.06 µg/ml (Dd2) and 5.82 µg/ml (3D7). The results were partially in correlation with the research work of Das et al., where piperazine substituted s-triazine moiety provided good antimalarial activity (Das et al. 2020). Whereas, the replacement with 4-phenylpiperazine i.e., compound 3h showed moderate antimalarial activity with IC50 8.95 µg/ml (Dd2) and 7.46 µg/ml (3D7). Substitution with 4-ethylpiperazine however reduced the inhibitory activity considerably. The compound with butylamine substitution (3d) showed mild to moderate activity in Dd2 and 3D7 having IC50 11.36 and 6.18 µg/ml respectively. The decrease in chain length of amines, such as propylamine (3c), ethylamine (3b) and methylamine (3a) respectively, reduced the inhibition of parasite with negligible antimalarial activity. The results were found to be in line with the previously published research involving PABA-substituted s-triazine derivatives (Adhikari et al. 2020).
Table 5.
In vitro antimalarial activity of title compounds 3(a-j) on Plasmodium falciparum chloroquine-sensitive (3D7) and chloroquine-resistant (Dd2) strains and Resistance index (RI)
| Compounds | 3D7 | DD2 | RI | ||||
|---|---|---|---|---|---|---|---|
| 5 µg/ml (%-inhibition) | 50 µg/ml (%-inhibition) | IC50 µg/ml | 5 µg/ml (%-inhibition) | 50 µg/ml (%-inhibition) | IC50 µg/ml | ||
| 3a | 0 | 23 | 106.76 ± 0.57 | 0 | 21 | 112.98 ± 0.51 | 1.05 |
| 3b | 0 | 27 | 92.90 ± 0.22 | 0 | 18.5 | 105.23 ± 0.95 | 1.13 |
| 3c | 7.5 | 35 | 34.02 ± 0.98 | 13.5 | 57 | 30.35 ± 1.12 | 0.89 |
| 3d | 40.5 | 72 | 6.18 ± 0.01 | 22.5 | 59 | 11.36 ± 1.16 | 1.84 |
| 3e | 43 | 86 | 5.82 ± 0.007 | 37.5 | 88 | 7.06 ± 0.32 | 1.21 |
| 3f | 47.5 | 88 | 5.26 ± 0.004 | 53.5 | 90 | 4.71 ± 0.21 | 0.89 |
| 3 g | 4 | 27 | 64.58 ± 2.94 | 7.5 | 57.5 | 39.43 ± 0.07 | 0.61 |
| 3 h | 33.5 | 71 | 7.46 ± 0.01 | 28 | 81 | 8.95 ± 1.46 | 0.59 |
| 3i | 2 | 31 | 69.68 ± 0.34 | 6 | 40.5 | 61.72 ± 0.94 | 0.88 |
| 3j | 4 | 33 | 63.30 ± 0.27 | 13.5 | 43 | 60.86 ± 1.13 | 0.96 |
| CQ | – | – | 0.7 | – | – | 1.2 | |
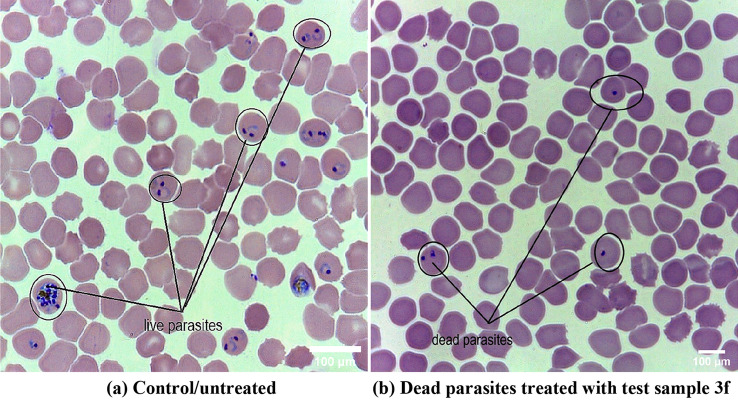
The Fig. 2 represents the morphology of live and dead parasites after antimalarial screening. Image (a) represents the live ring and schizonts stage P. falciparum treated with 0.01% DMSO and image (b) represents the dead parasites of P.falciparum treated with test compound (3f) after 40 h of incubation. Both images had been taken in 100× magnification with light microscope (Olympus).
Fig. 2.
In vitro acute toxicity screening against Plasmodium falciparum malaria parasites
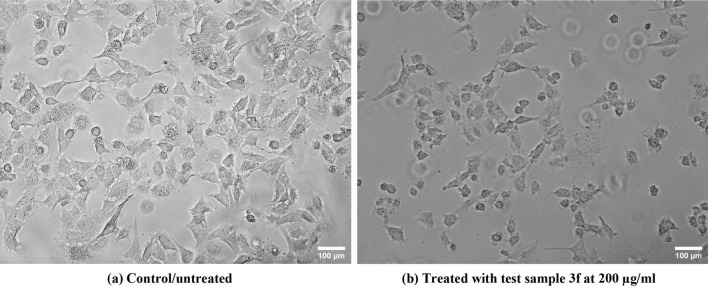
Meanwhile, the compounds that showed significant in vitro antimalarial activity (3d, 3e, 3f and 3h) were tested for in vitro cytotoxicity in human fibroblast cell line by MTT assay and were found to be non-toxic at the highest dose concentration of 200 µg/ml (Fig. 3). The cytotoxicity data were represented in IC50 values which were found to be 99.00 µg/ml for compound 3d, 97.65 µg/ml for compound 3e, 97.08 µg/ml for compound 3f and 99.20 µg/ml for compound 3h respectively.
Fig. 3.
In vitro cytotoxicity test on human fibroblast cells. Represents (a) control (b) 200 µg/ml dosage concentration of the test compound (3f). Both images had been taken in 40× magnification of the phase contrast microscope (Magnus INVI)
Structure–activity relationship (SAR) study
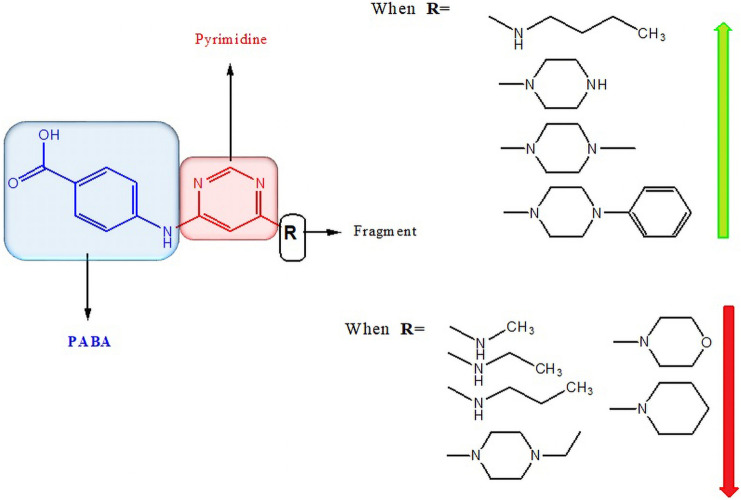
A structure–activity relationship (SAR) study was generated from the in vitro antimalarial screening results (Fig. 4) and it had been observed that compounds containing 4-methylpiperazine, piperazine, 4-phenyl piperazine and n-butyl amine showed better activity than compounds having methylamine, ethylamine, propylamine, 4-ethyl piperazine, morpholine, piperidine and found to be partially in agreement with Maurya et al. (2019). The increase in chain length in case of aliphatic primary amines is directly proportional to enhanced activity, for example, compound 3d with butylamine (Adhikari et al. 2020). However, secondary cyclic amines showed better activity than primary aliphatic amines. Since most of the active compounds contain 4-methylpiperazine, piperazine and 4-phenyl piperazine, indicate that increase in the chain length at para position of piperazine ring decreases activity; for instance, compound 4g with 4-ethylpiperazine and involvement of an aromatic group at para position enhances the activity. Also, the incorporation of morpholine and piperidine reduces the activity drastically.
Fig. 4.
Structure–activity relationship of PABA Substituted pyrimidine derivatives
Effect on fresh erythrocytes (hemolysis assay)
The effect of the synthesized compounds on fresh erythrocytes showed minimal percentage hemolysis below 5% after 3 h incubation period at 37 °C (Table 6). The fresh erythrocytes were treated with test compounds at different concentrations (50, 100, 250, 500 μg/mL) and compared with the standard control saponin 0.05% which showed 100% hemolysis (Sinha et al. 2019).
Table 6.
%-Hemolysis on normal erythrocytes with effect from the compounds 3 (a-j)
| Compounds | 100 µg/ml (Absorbance) | %-Hemolysis in 100 µg/ml | 50 µg/ml (Absorbance) | %-Hemolysis in 50 µg/ml |
|---|---|---|---|---|
| 3a | 0.201 ± 0.051 | 1.260 | 0.179 ± 0.076 | 0.468 |
| 3b | 0.207 ± 0.039 | 1.476 | 0.191 ± 0.087 | 0.900 |
| 3c | 0.259 ± 0.023 | 3.350 | 0.185 ± 0.0007 | 0.684 |
| 3d | 0.192 ± 0.006 | 0.9365 | 0.183 ± 0.008 | 0.612 |
| 3e | 0.245 ± 0.006 | 2.845 | 0.211 ± 0.000 | 1.621 |
| 3f | 0.247 ± 0.025 | 2.917 | 0.207 ± 0.031 | 1.476 |
| 3g | 0.204 ± 0.009 | 1.368 | 0.197 ± 0.019 | 1.116 |
| 3h | 0.218 ± 0.025 | 1.873 | 0.182 ± 0.025 | 0.576 |
| 3i | 0.222 ± 0.047 | 2.017 | 0.176 ± 0.002 | 0.360 |
| 3j | 0.227 ± 0.053 | 2.197 | 0.183 ± 0.0007 | 0.612 |
| CQ | 0..233 ± 0.003 | 2.413 | 0.198 ± 0.001 | 1.15 |
| Blank | 0.166 ± 0.033 | 0 | 0.166 ± 0.033 | 0 |
| Saponin | 2.776 ± 0.066 | 100 | 2.776 ± 0.066 | 100 |
Experimental section
In silico screening
The in silico study involves designing a virtual library of compounds by substituting the two chlorine atoms of the 4,6-dichloropyrimidine ring using 4-aminobenzoic acid as one of the common substituents and primary or secondary amines as other substituents. The designed library of all the compounds was preliminarily screened through “Molinspiration chem. informatics” for their oral bioavailability by checking various physicochemical properties (Table1). Subsequently, prediction of ADMET, toxicity risks and docking of the designed compounds in the binding pocket of the target protein with validation of the docking protocol was done by root mean square deviation (RMSD) calculation using “Accelrys Discovery Studio3.0.” The in-built ADMET model of Accelrys Discovery Studio 3.0 uses the descriptors AlogP98 and 2D polar surface area (PSA_2D) providing a plot of confidence ellipse indicating human intestinal absorption. TOPKAT module of Accelrys Discovery Studio 3.0 provides toxicity risks, such as carcinogenicity, mutagenicity, and irritancy, etc., evaluating the molecular structure of the compounds (Table 2). The toxicity models were based on the technical reports conducted by National Cancer Institute and National Toxicology Program (NTP) utilizing hybrid mice and inbred rats for determining rodent carcinogenicity (Egan et al. 2000; Venkataramana et al. 2011).
Molecular docking studies
Molecular docking analysis was used to screen the active antimalarial compounds after preliminary screening, which was then docked at the binding sites of the wild and mutant Pf-DHFR complex. The X-ray crystallographic structures of the protein were obtained from Protein Data Bank (PDB ID. 1J3I and 1J3K).
Ligand preparation and optimization
A library of pyrimidine analogs was designed using ChemDrawultra-8.0. The “Prepare ligand” protocol of Discovery Studio 3.0 was used to prepare the ligands which verify and optimize the structures by removing duplicate structures, generating isomers and tautomer’s, standardizing the charges of common groups and calculates the ions and ionization of the ligand’s functional groups, energy minimization of all the ligands were done by applying CHARMm force field to construct stable structures.
Protein preparation
The protein was refined in the protein workspace of Accelrys Discovery Studio 3.0 which involves removing water molecules, the chains B, C, D and co-crystallized ligand WR99210. However, the cofactor NADPH was retained and the chain A bound to the prototype WR99210 was used for docking study. This refined protein was then simulated by applying CHARMm forcefield and finally; the binding site of the receptor was defined from receptor cavities as a sphere (28.00, 5.89121, 59.83, and 16.10) around the active site of chain A.
Docking
The docking was performed using CDOCKER docking protocol where the most stable conformer was docked into the active site of the selected protein. The protocol was validated by RMSD calculation between the five docked poses of WR99210 and the ligand's X-ray docking pose. The orientation of WR99210 was taken as reference and the RMSD value was less than 2 Å (Sahu et al. 2019).
Binding energy calculation
For the top five final poses, the average binding energy of each complex was calculated and the top-scoring (highest negative binding energy) compound was selected for the generation of docking pose.
Chemistry
The chemicals and solvents used for synthesizing the selected compounds were of analytical grade and used without further purification. The synthesis was performed using microwave-assisted organic synthesis in CEM Discover Microwave Synthesizer (CEM, Discover System Model No. 908010) and the progress of the reactions was monitored by thin-layer chromatography. The synthesized compounds were analyzed by various physicochemical characterizations, such as appearance, melting point and spectral analyses, such as UV, FTIR, mass spectrometry, 1H NMR and 13C NMR. The melting point of all the final compounds 3(a-j) was determined by the BUCHI Melting Point M560 apparatus at 10 °C temperature gradients. UV/Visible Spectrophotometer (UV3200; Lab India Analytical) was used to obtain the (λ max) wavelength of maximum absorption. Bruker ALPHA-T FTIR Spectrometer was used to record the FTIR spectra for determining different functional groups. Waters ZQ-4000 Mass Spectrometer was used to record the mass spectra (MS) of the synthesized compounds using electrospray ionizer as ionization method. The nuclear magnetic resonance (NMR) spectra (1H NMR and 13C NMR) of the synthesized compounds were recorded using DMSO-d6 as solvent by Bruker Avance II 400 MHz FT-NMR Spectrometer. The multiplicity of 1H NMR signal is indicated as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet) together with Coupling constant (J) value quoted in Hz.
The general synthetic procedure for the synthesis of the titled compounds
Synthesis of the intermediates 2(a-j) and the final compounds were done by nucleophilic substitution reaction depicted in Scheme 1.
Synthetic procedure for 4-amino-6-chloro pyrimidine 2(a-j)
The first step of the reaction scheme involves the substitution of the chlorine at the fourth position of the molecule under microwave irradiation. 0.004 mol of the first reactant 4,6-dichloropyrimidine was dissolved in 6 ml 2-propanol followed by the addition of 0.003 mol of various primary and secondary amines. The reaction mixture was stirred in a 10 ml closed vessel in a microwave synthesizer at 30 °C, 1 bar pressure, 15-W power for 15 min. The product was dried and recrystallized with a suitable solvent to remove impurities. The recrystallized product obtained was dried, weighed and confirmed by melting point, TLC and FTIR analysis.
Synthetic procedure for final compounds 3(a-j)
The final step of the reaction scheme involves the synthesis of the proposed 4,6-disubstituted-pyrimidine derivatives using microwave irradiation. For that reaction, 0.002 mol of the first intermediate was dissolved in 3 ml DMSO followed by the addition of double molar 4-aminobenzoic acid in 2 ml DMSO solution. The reaction was stirred in 10 ml closed microwave vessel at 160 °C, 10 bar pressure and 50-W power for 30 min. The final product was recrystallized, dried and evaluated for physicochemical parameters together with different spectral analyses.
Characterization and spectral data
4-(6-(methylamino)pyrimidin-4-ylamino)benzoic acid (3a): Yield 61.76%; Orange solid; mp 96–98 °C; Rf 0.48; λmax 285 nm; FTIR (cm−1): 3388 (N–H stretch, secondary), 3075 (C–H stretch), 2921 (OH stretch, COOH), 1685 (C=O stretch), 1593 (N–H bend), 1523 (C–C stretch, aromatic), 1425 (C=N stretch, aromatic), 1380 (C–H bend), 1314 (C–N stretch, aromatic), 1256 (C–O stretch) 1080 (C–N stretch, aliphatic); 1H NMR (400 MHz, DMSO-d6) δ(ppm): 2.19 (s, 3H, CH3), 5.26 (s, 2H, 2NH) 6.51 (s, 1H, Ar–H), 6.45 (s, 1H, Ar–H), 7.63–7.58 (d, J = 20 Hz, 2H, Ar–H) 8.23–8.15 (d, J = 32 Hz, 2H, Ar–H), 9.56 (s, 1H, COOH); 13C NMR (400 MHz, DMSO-d6) δ(ppm): 165.43, 163.97, 163.43, 158.97, 156.60, 131.22, 112.65, 103.54, 99.07, 27.06; ; MS (m/z) calculated for C12H12N4O2: 244.10. Found (M+H)+: 245.10.
4-(6-(ethylamino)pyrimidin-4-ylamino)benzoic acid (3b): Yield 65.17%; Yellow solid; mp 78–80 °C; Rf 0.46; λmax 245 nm; FTIR (cm−1): 3432, 3224 (N–H stretch, secondary), 2977, 2917 (C–H stretch, aliphatic), 2522 (OH stretch, COOH), 1697 (C=O stretch), 1591 (N–H bend), 1515 (C–C stretch, aromatic), 1436 (C=N stretch, aromatic), 1406 (C–H bend), 1310 (C–N stretch, aromatic), 1254 (C–O stretch), 1077 (C–N stretch, aliphatic); 1H NMR (400 MHz, DMSO-d6) δ(ppm): 1.09–1.05 (t, J = 16 Hz, 3H, CH3), 3.28–2.48 (q, 2H, CH2), 5.26 (s, 2H, 2NH), 6.45 (s, 1H, Ar–H), 6.57–6.55 (d, J = 8 Hz, 2H, Ar–H), 7.60–7.58 (d, J = 8 Hz, 2H, Ar–H), 8.21 (s, 1H, Ar–H), 10.12 (s, 1H, COOH); 13C NMR (400 MHz, DMSO-d6) δ(ppm): 167.53, 165.44, 162.84, 158.46, 153.02, 131.23, 116.79, 112.65, 103.48, 34.98, 14.59; MS (m/z) calculated for C13H14N4O2: 258.11. Found (M+H)+: 259.15.
4-(6-(propylamino)pyrimidin-4-ylamino)benzoic acid (3c): Yield 60.11%; Brown solid; mp 129–131 °C; Rf 0.59; λmax 298 nm; FTIR (cm−1): 3389 (N–H stretch, secondary), 2921 d(C–H stretch, aromatic), 1685 (C=O stretch), 1592 (N–H bend), 1527 (C–C stretch, aromatic), 1412 (C–H bend), 1317 (C–N stretch, aromatic), 1260 (C–O stretch), 1084 (C–N stretch, aliphatic); 1H NMR (400 MHz, DMSO-d6) δ(ppm): 0.68–0.61 (t, J = 28 Hz, 3H, CH3), 1.25–1.06 (m, 2H, CH2), 2.79–2.68 (t, J = 44 Hz, 2H, CH2), 5.04 (s, 2H, 2NH), 5.84 (s, 1H,Ar–H), 6.31–6.30 (d, J = 4 Hz, 2H, Ar–H), 7.669–7.662 (d, J = 2.8 Hz, 2H, Ar–H), 8.15 (s, 1H, Ar–H), 10.13 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ(ppm): 168.13, 167.82, 166.87, 152.51, 151.48, 131.17, 119.86, 116.93, 112.69, 45.44, 19.37, 13.48; MS (m/z) calculated for C14H16N4O2 : 272.13. Found (M+H)+: 273.13.
4-(6-(butylamino)pyrimidin-4-ylamino)benzoic acid (3d): Yield 69.91%; Off white solid; mp 223–225 °C; Rf 0.72 λmax 240 nm; FTIR (cm−1): 3282 (N–H stretch, secondary), 3061 (C–H stretch, aromatic), 2957 (C–H stretch, aliphatic), 2868 (OH stretch, COOH), 1639 (C=O stretch), 1591 (N–H bend), 1510 (C–C stretch, aromatic), 1461 (C=N stretch, aromatic), 1366 (C–H bend, aliphatic), 1309 (C–N stretch, aromatic), 1229 (C–O stretch), 1165 (C–N aliphatic stretch); 1H NMR (400 MHz, DMSO-d6) δ(ppm): 0.64–0.61 (t, J = 12 Hz, 3H, CH3), 1.10–0.93 (m, 2H, CH2), 1.27–1.24 (t, J = 12 Hz, 2H, CH2), 2.98–1.97 (m, 2H, CH2), 5.13 (s, 2H, 2NH), 5.88 (s, 1H, Ar–H), 6.34–6.31 (d, J = 12 Hz, 2H, Ar–H), 7.66–7.64 (d, J = 8 Hz, 2H, Ar–H), 8.10 (s, 1H, Ar–H), 10.15 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ(ppm): 167.51, 166.87, 152.82, 151.48, 142.91, 131.13, 119.86, 116.93, 112.69, 45.44, 30.18, 19.37, 13.48; MS (m/z) calculated for C15H18N4O2 : 286.14. Found (M+H)+: 287.05.
4-(6-(piperazin-1-yl)pyrimidin-4-ylamino)benzoic acid (3e): Yield 74%; Yellow solid; mp 172–174 °C; Rf 0.50; λmax 278 nm; FTIR (cm−1): 3390 (N–H stretch, secondary), 2912 (C–H stretch, aliphatic), 2525 (OH stretch, COOH), 1670 (C=O stretch), 1596 (N–H bend), 1517 (C–C stretch, aromatic), 1424 (C=N stretch, aromatic), 1381 (C–H bend, aliphatic), 1173 (C–N stretch, aliphatic); 1H NMR (400 MHz, DMSO-d6) δ(ppm): 2.17 (s, 1H, NH); 2.81–2.68 (q, 4H, CH2); 3.77–3.74 (t, J = 12 Hz, 4H, CH2); 4.34 (s, 1H, NH), 4.82 (s, 1H, Ar–H); 6.37–6.29 (d, J = 32 Hz, 2H, Ar–H); 7.15 (s, 1H, Ar–H); 8.98–8.96 (d, J = 8 Hz, 2H, Ar–H); 9.81 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ(ppm): 167.97, 167.42, 160.47, 152.18, 131.93, 124.70, 116.84, 101.17, 57.79, 53.58; MS (m/z) calculated for C15H17N5O2 : 299.14. Found (M+H)+: 300.15.
4-(6-(4-methylpiperazin-1-yl)pyrimidin-4-ylamino)benzoic acid (3f): Yield 77.93%; Yellow solid; mp 220–222 °C; Rf 0.66; λmax 283 nm; FTIR (cm−1): 3361 (N–H stretch, secondary), 2991 (C–H stretch, aliphatic), 2533 (OH stretch, COOH), 1666 (C=O stretch), 1600 (N–H bend), 1516 (C–C stretch, aromatic), 1436 (C=N stretch, aromatic), 1379 (C–H bend, aliphatic), 1245 (C–O stretch), 1177 (C–N stretch, aliphatic); 1H NMR (400 mHz) DMSO-d6, δ(ppm): 2.29 (s, 3H, N-CH3); 2.51–2.42 (t, J = 36 Hz, 4H, CH2); 4.67–4.63 (t, J = 16Hz, 4H, CH2); 4.81 (s, 1H, NH), 6.49 (s, 1H, Ar–H); 6.56–6.54 (d, J = 8 Hz, 2H, Ar–H); 7.33 (s, 1H, Ar–H) 8.36–8.31 (d, J = 20 Hz, 2H, Ar–H); 9.95 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 167.59, 167.47, 160.42, 152.20, 131.10, 124.70, 116.84, 101.66, 57.91, 53.24, 40.06; MS (m/z) calculated for C16H19N5O2 : 313.15. Found (M+H)+: 314.19.
4-(6-(4-ethylpiperazin-1-yl)pyrimidin-4-ylamino)benzoic acid (3g): Yield 68.25%; Brown solid; mp 259–261 °C; Rf 0.65 λmax 267 nm; FTIR (cm−1): 3387 (N–H stretch, secondary), 2923 (C–H stretch, aliphatic), 2524 (OH stretch, COOH), 1673 (C=O stretch), 1597 (N–H bend), 1518 (C–C stretch, aromatic), 1413 (C=N stretch, aromatic), 1383 (C–H bend, aliphatic), 1230 (C–O stretch) 1168 (C–N stretch, aliphatic); 1H NMR (400 mHz) DMSO-d6, δ(ppm): 1.99–1.87 (t, J = 48 Hz, 3H, CH3); 2.19–2.05 (q, 2H, CH2); 2.55–2.48 (t, J = 28 Hz, 4H, CH2); 2.81–2.76 (t, J = 20 Hz, 4H, CH2); 4.65 (s, 1H, NH), 5.37 (s, 1H, Ar–H); 6.65–6.63 (d, J = 8 Hz, 2H, Ar–H); 6.89 (s, 1H, Ar–H) 7.17–7.15 (d, J = 8 Hz, 2H, Ar-H); 9.02 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 167.08, 161.93, 157.84, 150.57, 131.00, 119.05, 114.89, 101.68, 55.23, 51.14, 49.71, 14.31; MS (m/z) calculated for C17H21N5O2 : 327.17. Found (M+H)+: 328.07.
4-(6-(4-phenylpiperazin-1-yl)pyrimidin-4-ylamino)benzoic acid (3h): Yield 66.87%; Yellow solid; mp 230–232 °C; Rf 0.53; λmax 258 nm; FTIR (cm−1): 3342 (N–H stretch, secondary), 3074 (C–H stretch, aromatic), 2828 (OH stretch, COOH), 1670 (C=O stretch), 1578 (N–H bend), 1522 (C–C stretch, aromatic), 1414 (C=N stretch, aromatic), 1384 (C–H bend, aliphatic), 1336 (C–N stretch, aromatic), 1228 (C–O stretch), 1157 (C–N aliphatic stretch); 1H NMR (400 mHz) DMSO-d6, δ(ppm): 2.55–2.48 (t, J = 28 Hz, 4H, CH2); 3.65–3.47 (t, J = 28 Hz, 4H, CH2), 4.63 (s, 1H, NH), 5.29 (s, 1H, Ar–H), 6.65 (s, 1H, Ar–H), 6.79–6.74 (t, J = 20Hz, 2H, Ar–H), 7.08–7.03 (t, J = 20Hz, 2H, Ar–H), 7.17–7.09 (t, J = 32 Hz, 1H, Ar–H), 7.63-7.53 (d, J = 40Hz, 2H, Ar–H), 7.81–7.73 (d, J = 32Hz, 2H, Ar–H), 9.96 (s, 1H, COOH); 13C NMR (100 MHz, DMSO-d6) δ(ppm): 167.93, 167.75, 159.82, 157.82, 152.16, 131.17, 129.15, 121.45, 117.01, 116.79, 113.20, 85.47, 57.75, 55.23; MS (m/z) calculated for C21H21N5O2 : 375.17. Found (M+H)+: 376.18.
4-(6-morpholinopyrimidin-4-ylamino)benzoic acid (3i): Yield 81.96%; Yellow solid; mp 153–155 °C; Rf 0.46; λmax 265 nm; FTIR (cm−1): 3379 (N–H stretch, secondary), 3230 (OH stretch, COOH), 2911 (C–H stretch), 1665 (C=O stretch), 1597 (N–H bend), 1511 (C–C stretch, aromatic), 1435 (C=N stretch, aromatic), 1377 (C–H bend, aliphatic), 1242 (C–O stretch), 1172 (C–N stretch, aliphatic); 1H NMR (400 mHz) DMSO-d6, δ(ppm): 2.92–2.68 (t, J = 96 Hz, 4H, N-CH2), 4.24–4.13 (t, J = 44 Hz, 4H, CH2), 5.31(s, 1H, NH), 5.83 (s, 1H, Ar–H), 6.02 (s 1H, Ar–H), 6.91-6.86 (d, J = 20 Hz, 2H, Ar–H), 8.30–8.24 (d, J = 24 Hz, 2H, Ar–H), 9.78 (s, 1H, COOH); 13C NMR (400 MHz) DMSO-d6, δ(ppm): 167.60, 167.18, 157.79, 152.16, 131.17, 121.32, 117.82, 116.82, 65.76, 44.01; MS (m/z) calculated for C15H16N4O3: 300.12. Found (M+H)+: 301.03.
4-(6-(piperidin-1-yl)pyrimidin-4-ylamino)benzoic acid (3j): Yield: 63.25%; Yellow solid; mp 174–176 °C; Rf 0.51; λmax 290 nm; FTIR (cm-1): 3326 (N–H stretch, secondary), 2932 (C–H stretch), 2856 (OH stretch, COOH), 1658 (C=O stretch), 1576 (N–H bend), 1507 (C–C stretch, aromatic), 1439 (C=N stretch, aromatic), 1375 (C–H bend, aliphatic), 1240 (C–O stretch) 1176 (C–N stretch, aliphatic); 1H NMR (400 mHz) DMSO-d6, δ (ppm): 2.67–1.44 (m, 6H, 6CH2); 3.54–3.40 (t, J = 56 Hz, 4H, CH2); 4.237 (s, 1H, NH); 6.00 (s, 1H, Ar–H); 6.81 (s, 1H, Ar–H), 7.79–7.77 (d, J = 8 Hz, 2H, Ar–H); 8.23–8.19 (d, J = 16 Hz, 2H, Ar–H); 9.44 (1H, s, COOH); 13C NMR (400 MHz) DMSO-d6, δ (ppm): 167.09, 161.62, 157.84, 145.13, 131.07, 120.40, 117.62, 101.17, 44.63, 25.40, 24.96; MS (m/z) calculated for C16H18N4O2: 298.14. Found (M+H)+: 299.19.
In vitro antimalarial activity
Giemsa-stained slide method is used for in vitro antimalarial evaluation of the test compounds. It is a low-cost alternative for testing a small number of compounds and is classically referred to as the Minimum Inhibitory Concentration (MIC), the method which is suitable for distinguishing both susceptible and resistant isolates. Parasites are incubated with the test compound and then parasitemia of control and treated groups are compared by counting Giemsa-stained parasites by light microscopy.
Parasite preparation
The CQ sensitive 3D7 and CQ resistant Dd2 strains of P. falciparum were thawed by bringing cryopreserved parasite strains to room temperature from liquid nitrogen or (-80°C). 12% NaCl solution (1/5th of original volume) and 1.6% NaCl solution were added gradually with 1 ml increment and centrifuged at 2000 rpm for 5 min. The supernatant was discarded and sufficient fresh medium was added to wash 2–3 times to remove lysed parasitized RBCs. The strains were then routinely maintained as continuous culture in malariology division of ICMR-RMRC-NE Region Dibrugarh, following Trager and Jensen method with slight modification. A 5% hematocrit of the parasites were maintained in RPMI-1640 medium supplemented with 25 mM HEPES, 0.2% d-glucose, 0.1% gentamycin sulfate, 0.1% amphotericin B, 0.5% Albumax-II, 0.01% hypoxanthine and 1.77 mM NaHCO3 at a pH of 7.2. The red blood cells used were O+ and the culture flask was incubated at 37 °C and 5% CO2 level in CO2 incubator with regular parasitemia assessment and medium change. The culture medium was prepared as an incomplete medium weekly (without sodium bicarbonate) and stored at 4 °C. Sodium bicarbonate was added freshly after warming to 37 °C before use in culture (Radfar et al. 2009).
Synchronization of parasites
The P. falciparum malaria parasite is usually asynchronous during in vitro growth with all asexual stages of the parasite present. The generation of cultures containing highly synchronized ring-stage parasites is necessary for in vitro antimalarial assay. So, the asynchronous parasites of P. falciparum were synchronized after obtaining a 2–3% parasitemia having sufficient rings with 5% d-sorbitol treatment. This causes differential osmotic lysis of the trophozoites, schizonts and retaining only the ring-stage parasites. In a 15 ml falcon tube, the culture stock was centrifuged at 3000 rpm for 10 min followed by discarding the supernatant. Then three times 5% d-sorbitol to the original volume of the parasite was added and incubated at 37 °C for 20–25 min. The culture was then centrifuged at 1200 rpm for 10 min and the supernatant was discarded. Finally, the culture was washed three times at 2500 rpm for 3 min each with sufficient RPMI medium and 50% HCT was prepared to assess parasitemia by taking a thin smear (Trager and Jensen 1976, 1977, 1997; Trager and Jenson 1978).
In vitro antimalarial assay
The synthesized compounds were screened against CQ sensitive 3D7 and CQ resistant Dd2 strains of P. falciparum adopting the micro assay methods of Rieckmann et al. (1978) with minor modification. The test compounds were dissolved in 1:200 dimethyl sulfoxide to obtain a stock solution of 5000 μg/ml. For preliminary screening, two different dosages 5 and 50 μg/ml were used by further dilution of the stock with incomplete media (without NaHCO3). For carrying out the assay, an initial ring-stage parasitemia of ~1% at 3% hematocrit in the total volume of 200 μl of medium RPMI-1640 is uniformly maintained in a 96-well flat bottom microtiter plate. The test compound in 20 μl volume was added at the required concentration (5 and 50 μg/ml) in duplicate wells and the negative control (without test) wells were made with only complete medium and 0.5% DMSO. The reference drug chloroquine was run concurrently at its predetermined IC50 dosage as the positive control (Basco et al. 1994; Bhat et al. 2013). The test plate was then incubated at 37 °C and 5% CO2 environment. After 36–40 h incubation, the blood smears from each well were prepared, fixed with methanol and stained with 10% Giemsa stain to observe under microscope. Based on the parasite morphology, the parasites were scored as dead or alive and %-dead rings along with trophozoites and schizonts were determined by counting a total of 100 asexual parasites. The compounds were further processed to determine IC50 values with multiple lower dosages using GraphPad Prism software. The following formula was used to determine the percentage inhibition.
Resistance index (RI)
The degree of resistance was determined by comparing the activity of the synthesized pyrimidine derivatives of 4-aminobenzoic acid on the chloroquine-sensitive and chloroquine-resistant strains of P. falciparum using the following formula (Sinha et al. 2019).
Hemolysis assay
Hemolytic effect on normal erythrocytes was examined for all the synthesized compounds by incubating uninfected erythrocytes with test compounds in phosphate-buffered saline (PBS). Briefly, fresh erythrocytes were washed using PBS thrice by centrifuging for 5 min at 1600 rpm. The final supernatant was discarded and the remaining pellet was re-suspended in PBS to obtain 2% hematocrit. Then a total of 100 μl volume of the re-suspended pellet was maintained in triplicate using a 96-well sterile culture plate with the test compounds at the different desired concentrations. PBS alone (for baseline values) and 0.05% saponin in PBS (for 100% hemolysis) were employed as negative and positive controls respectively. The test plate was then incubated at 37 °C for 3 h followed by centrifuging the test samples at 1500 rpm for 5 min. The supernatant was then monitored spectrophotometrically using iMark Microplate Reader by taking absorbance at 415 nm for determination of the hemolytic activity which was quantified in terms of hemoglobin release (Sinha et al. 2019). The %-hemolysis was measured using the following formula:
Toxicity studies
The synthesized compounds (3d, 3e, 3f and 3h) of pyrimidine derivatives were examined for the in vitro cytotoxicity screening against human fibroblast cell line by MTT assay method. The fibroblast cells were kept under starvation for 1 h followed by treatment with different concentrations of the test sample (0, 100, 150, 200 µg) for 48 h in clear 96-well microplate at 37 °C and 5% CO2 incubator. The test drugs containing media were removed, washed with PBS and 10% MTT reagent in 200 μl total volume of media was added followed by 3 h incubation. The culture medium was removed and 100 μl of DMSO was added. The plate was then gently shaken to solubilize the formed formazan and the absorbance was measured using a microplate reader at a wavelength of 570 nm. The percentage growth inhibition was calculated by subtracting the background and the blank, and concentration of test drug inhibiting 50% cell growth (IC50) was generated from the dose–response curve for the cell line (Sharma et al. 2021).
Conclusion
The titled 4-aminobenzoic acid-substituted pyrimidine derivatives were developed as a novel class of potential antimalarial agents. In vitro results have outlined the significance of compound 3d, 3e, 3f and 3h as promising antimalarial agents. The compound 3f substituted with methyl piperazine and PABA has been found to be the most potent with no cytotoxicity having high binding energy and H-bond interaction with Arg122. The reported compounds were designed as folic acid analogs using 1J3I and 1J3K protein targeting Pf-DHFR enzyme and the significant in vitro antimalarial activity results revealed their probable mechanism of action might be through Pf-DHFR inhibition.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Adhikari N, Kashyap A, Shakya A, et al. Microwave assisted synthesis, docking and antimalarial evaluation of hybrid PABA-substituted 1, 3, 5-triazine derivatives. J Heterocycl Chem. 2020;57:2389–2399. doi: 10.1002/jhet.3955. [DOI] [Google Scholar]
- Agarwal A, Srivastava K, Puri SK, Chauhan PM. Synthesis of 2, 4, 6-trisubstituted pyrimidines as antimalarial agents. Bioorg Med Chem. 2005;13:4645–4650. doi: 10.1016/j.bmc.2005.04.061. [DOI] [PubMed] [Google Scholar]
- Algul O, Paulsen JL, Anderson AC. 2, 4-Diamino-5-(2′-arylpropargyl) pyrimidine derivatives as new nonclassical antifolates for human dihydrofolate reductase inhibition. J Mol Graph Model. 2011;29:608–613. doi: 10.1016/j.jmgm.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjanac M, Tatic I, Ivezic Z et al (2009) Pyrimido-pyrimidines: a novel class of dihydrofolate reductase inhibitors. Food Technol Biotechnol 47
- Basco LK, Ramiliarisoa O, Le Bras J. In vitro activity of pyrimethamine, cycloguanil, and other antimalarial drugs against African isolates and clones of Plasmodium falciparum. Am J Trop Med Hyg. 1994;50:193–199. doi: 10.4269/ajtmh.1994.50.193. [DOI] [PubMed] [Google Scholar]
- Bhat HR, Singh UP, Gahtori P, et al. 4-Aminoquinoline-1, 3, 5-triazine: design, synthesis, in vitro antimalarial activity and docking studies. New J Chem. 2013;37:2654–2662. doi: 10.1039/c3nj00317e. [DOI] [Google Scholar]
- Brown DJ. The pyrimidines. New York: John Wiley & Sons; 2009. [Google Scholar]
- Dansena H, Dhongade HJ, Chandrakar K. Pharmacological potentials of pyrimidine derivative: a review. Asian J Pharm Clin Res. 2015;8:171–177. [Google Scholar]
- Das Arpita, Ghosh Surajit K., Bhat Hans Raj, Kalita Junmoni, Kashyap Ankita, Adhikari Nayana. Docking, Synthesis and Antimalarial Evaluation of Hybrid Phenyl Thiazole 1,3,5-Triazine Derivatives. Current Bioactive Compounds. 2020;16(5):639–653. doi: 10.2174/1573407215666190308154139. [DOI] [Google Scholar]
- Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- Gogoi P, Shakya A, Ghosh SK, et al. In silico study, synthesis, and evaluation of the antimalarial activity of hybrid dimethoxy pyrazole 1,3,5-triazine derivatives. J Biochem Mol Toxicol. 2021;35:e22682. doi: 10.1002/jbt.22682. [DOI] [PubMed] [Google Scholar]
- Hartung CG, Backes AC, Felber B, et al. Efficient microwave-assisted synthesis of highly functionalized pyrimidine derivatives. Tetrahedron. 2006;62:10055–10064. doi: 10.1016/j.tet.2006.08.065. [DOI] [Google Scholar]
- Hitchings GH, Elion GB, Vander Werff H, Falco EA. Pyrimidine derivatives as antagonists of pteroylglutamic acid. J Biol Chem. 1948;174:765–766. doi: 10.1016/S0021-9258(18)57361-0. [DOI] [PubMed] [Google Scholar]
- Kashyap A, Choudhury AAK, Saha A, et al. Microwave-assisted synthesis of hybrid PABA-1,3,5-triazine derivatives as an antimalarial agent. J Biochem Mol Toxicol. 2021;35:e22860. doi: 10.1002/jbt.22860. [DOI] [PubMed] [Google Scholar]
- Krátký M, Konečná K, Janoušek J, et al. 4-Aminobenzoic acid derivatives: converting folate precursor to antimicrobial and cytotoxic agents. Biomolecules. 2019;10:E9. doi: 10.3390/biom10010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Lödige M, Hiersch L. Design and synthesis of novel hybrid molecules against malaria. Int J Med Chem. 2015;2015:e458319. doi: 10.1155/2015/458319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Chen L, Poindexter GS. Microwave-assisted synthesis of aminopyrimidines. Tetrahedron Lett. 2002;43:5739–5742. doi: 10.1016/S0040-4039(02)01190-5. [DOI] [Google Scholar]
- Maurya SS, Bahuguna A, Khan SI, et al. N-Substituted aminoquinoline-pyrimidine hybrids: synthesis, in vitro antimalarial activity evaluation and docking studies. Eur J Med Chem. 2019;162:277–289. doi: 10.1016/j.ejmech.2018.11.021. [DOI] [PubMed] [Google Scholar]
- Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J, Haritakul R, Keller PA. Antimalarial activity of 2, 4-diaminopyrimidines. Lett Drug Des Discov. 2008;5:277–280. doi: 10.2174/157018008784619843. [DOI] [Google Scholar]
- Nzila A, Ward SA, Marsh K, et al. Comparative folate metabolism in humans and malaria parasites (part I): pointers for malaria treatment from cancer chemotherapy. Trends Parasitol. 2005;21:292–298. doi: 10.1016/j.pt.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radfar A, Méndez D, Moneriz C, et al. Synchronous culture of Plasmodium falciparum at high parasitemia levels. Nat Protoc. 2009;4:1899. doi: 10.1038/nprot.2009.198. [DOI] [PubMed] [Google Scholar]
- Rieckmann KH, Campbell GH, Sax LJ, Mrema JE. Drug sensitivity of Plasmodium falciparum. An in vitro microtechnique. Lancet Lond Engl. 1978;1:22–23. doi: 10.1016/s0140-6736(78)90365-3. [DOI] [PubMed] [Google Scholar]
- Sahu S, Ghosh SK, Kalita J, et al. Design, synthesis and antimalarial screening of some hybrid 4-aminoquinoline-triazine derivatives against pf-DHFR-TS. Exp Parasitol. 2016;163:38–45. doi: 10.1016/j.exppara.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Sahu S, Ghosh SK, Gahtori P, et al. In silico ADMET study, docking, synthesis and antimalarial evaluation of thiazole-1, 3, 5-triazine derivatives as Pf-DHFR inhibitor. Pharmacol Rep. 2019;71:762–767. doi: 10.1016/j.pharep.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Schweitzer BI, Dicker AP, Bertino JR. Dihydrofolate reductase as a therapeutic target. FASEB J. 1990;4:2441–2452. doi: 10.1096/fasebj.4.8.2185970. [DOI] [PubMed] [Google Scholar]
- Sharma MG, Pandya J, Patel DM, et al. One-pot assembly for synthesis of 1,4-dihydropyridine scaffold and their biological applications. Polycycl Aromat Compd. 2021;41:1495–1505. doi: 10.1080/10406638.2019.1686401. [DOI] [Google Scholar]
- Sinha S, Batovska DI, Medhi B, et al. In vitro anti-malarial efficacy of chalcones: cytotoxicity profile, mechanism of action and their effect on erythrocytes. Malar J. 2019;18:1–11. doi: 10.1186/s12936-019-3060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Cultivation of erythrocytic stages. Bull World Health Organ. 1977;55:361. [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Continuous culture of Plasmodium falciparum: its impact on malaria research. Int J Parasitol. 1997;27:989–1006. doi: 10.1016/S0020-7519(97)00080-5. [DOI] [PubMed] [Google Scholar]
- Trager W, Jenson JB. Cultivation of malarial parasites. Nature. 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove S, D’hooghe M. Quinoline-based antimalarial hybrid compounds. Bioorg Med Chem. 2015;23:5098–5119. doi: 10.1016/j.bmc.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Venkataramana CHS, Sravani KR, Singh SS, Madhavan V. In silico ADME and toxcity studies of some novel indole derivatives. J Appl Pharm Sci. 2011;1:159. [Google Scholar]
- World Malaria Report 2021. https://www.who.int/publications-detail-redirect/9789240040496. Accessed 1 Mar 2022
- Yuthavong Y, Tarnchompoo B, Vilaivan T, et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci. 2012;109:16823–16828. doi: 10.1073/pnas.1204556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Mol Biol. 2003;10:357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.