Abstract
The nucleic acid contents of individual bacterial cells as determined with three different nucleic acid-specific fluorescent dyes (SYBR I, SYBR II, and SYTO 13) and flow cytometry were compared for different seawater samples. Similar fluorescence patterns were observed, and bacteria with high apparent nucleic acid contents (HNA) could be discriminated from bacteria with low nucleic acid contents (LNA). The best discrimination between HNA and LNA cells was found when cells were stained with SYBR II. Bacteria in different water samples collected from seven freshwater, brackish water, and seawater ecosystems were prelabeled with tritiated leucine and then stained with SYBR II. After labeling and staining, HNA, LNA, and total cells were sorted by flow cytometry, and the specific activity of each cellular category was determined from leucine incorporation rates. The HNA cells were responsible for most of the total bacterial production, and the specific activities of cells in the HNA population varied between samples by a factor of seven. We suggest that nucleic acid content alone can be a better indicator of the fraction of growing cells than total counts and that this approach should be combined with other fluorescent physiological probes to improve detection of the most active cells in aquatic systems.
An important goal in oceanography is to predict the responses of ecosystems to environmental perturbations. Therefore, it is essential to understand what factors control the fluxes of materials and energy within natural ecosystems. It is well recognized that bacteria play an important role in driving these fluxes and that relevant controls may be found in the microbial food webs (1). Over the past 20 years, most of the methods developed by microbial ecologists to evaluate biomass and activity have assumed that all bacterial cells in a community have similar metabolic activities. However, there is some feeling that this assumption is not true (7). The total biomass and bulk activity determined at both population and community levels are regulated by interactions occurring at the cellular level (i.e., virus-bacterium, flagellate-bacterium, bacterium-bacterium interactions). Therefore, which cells are active and which cells are inactive and what the taxonomic identities of the organisms are are central questions today in aquatic microbial ecology. The answers to these questions should be very useful for better estimating the numbers of growing cells and the cell-specific growth rates and for determining taxonomic affiliations. Such information is essential for understanding how phylogenetic components cycle through the active and inactive states. If the fraction of active cells in a natural community is only a small fraction of the total cells, it is important to investigate which factors control activation and inactivation of the cells and to evaluate the specific activities of bacteria in aquatic ecosystems. This could significantly improve descriptions of the bacterial compartment in ecological models.
The question of whether a bacterial cell is active or inactive has been a matter of important debate during the last two decades (3, 11, 23). The controversy is mainly due to some semantic confusion and to the wide diversity of methods that are commonly used to determine the relative proportions of living, active, dormant, inactive, and dead cells (10, 27). There are at least three categories of cells that should be of ecological relevance: the actively growing cells which contribute to production of biomass, the living but inactive cells which do not participate in bacterial production at the time of sampling but have potential activity (often called dormant cells), and the dead and inactive cells that should be considered only organic particles. Although discrimination of these three cellular categories remains unclear (6), the following two methods have been suggested to determine the fractions of actively growing cells in complex assemblages: reduction of a fluorogenic tetrazolium dye, 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), by the electron transport system and determination of the nucleic acid content of cells after staining with a nucleic acid dye.
The CTC method has been used increasingly in recent years to enumerate active cells in aquatic ecosystems (4, 14, 16, 21). Actively respiring cells are generally enumerated by epifluorescence microscopy, but count data can be improved by using flow cytometry (3, 22). Although at times a good correlation has been found between the abundance (or percentage) of CTC-positive cells and bacterial production (5), this method remains controversial, and several authors have recently pointed out several drawbacks of the method (11, 20, 25, 26).
The nucleic acid content of individual cells can be easily analyzed by flow cytometry. This approach has been used in the last few years by aquatic microbiologists to discriminate between different subgroups of bacteria, and at least two subgroups of bacteria were generally found in the aquatic ecosystems studied (6, 15, 17). These groups have slightly different side scatter characteristics, but they differ significantly in fluorescence and, thus, nucleic acid content. Although the different nucleic acid stains used in studies bind to both DNA and RNA and provide similar fluorescence distribution patterns, most of the fluorescence is generally linked to DNA (6). Therefore, the two subgroups are called the high-DNA-content (HDNA) or type II and low-DNA-content (LDNA) or type I groups (6, 15). Different authors have suggested that HDNA cells were more active than LDNA cells (9, 15), and recently, Gasol et al. (6) suggested that the percentage of HDNA cells could be used as a reference for the percentage of actively growing bacteria in marine planktonic environments.
Validation of methods used to detect and enumerate actively growing cells in the aquatic environment remains difficult and is generally limited to correlations between counts obtained by different methods and the bulk bacterial production determined by incorporation of tritiated leucine or thymidine. One way to determine if the percentage of HDNA bacteria can really be considered a good indicator of the fraction of actively growing cells should be to sort bacteria belonging to the two groups (LDNA and HDNA) after radioactive leucine incorporation and determine the relative specific activity of the cells. Coupling of radioactive labeling followed by cell sorting was developed by Servais et al. (19) and has been used to investigate the relationships among cell size, activity, and genetic diversity (2); Bernard et al. (L. Bernard, P. Lebaron, H. Shäfer, and G. Muyzer, Abstr. Sixth Eur. Mar. Microbiol. Symp., p. 42, 1998) also used flow cytometry and cell sorting to investigate the genetic diversity of actively respiring cells (CTC-positive cells).
In this study, we compared three nucleic acid stains (SYTO 13, SYBR I, and SYBR II) commonly used in aquatic microbiology to determine if they provide similar fluorescence distribution patterns and if at least one of them provides better discrimination between bacterial subgroups with different nucleic acid contents. SYBR II was used to stain bacterial communities from different ecosystems previously labeled with radioactive leucine. Then we sorted the two subgroups of bacterial cells having different nucleic acid contents to determine the specific leucine incorporation rate of each subgroup. The specific activities determined for the different subgroups and at the community level were compared.
MATERIALS AND METHODS
Water samples.
Samples were collected between March and July 2000 along the Mediterranean coast (France) at sites that differed in their physicochemical characteristics. Seawater samples were collected weekly from 15 March to 4 July in the Bay of Banyuls-sur-Mer at the SOLA station (42°29′N, 3°08′E), and additional samples were obtained in the Banyuls-sur-Mer harbor (42°28′N, 3°08′E). Brackish water samples were collected in a coastal lagoon (salinity, 30‰), the Leucate Lagoon (42°49′N, 2°9′E). Samples of freshwater were collected in the Tech River 5 km above the river mouth (42°35′N, 2°58′E). Five liters was collected just below the surface in all of the environments studied except the SOLA station, where water was collected at a depth of 24 m. Samples were processed in the laboratory within 2 h after collection.
Enumeration of total bacterial cells and HNA and LNA cells by flow cytometry.
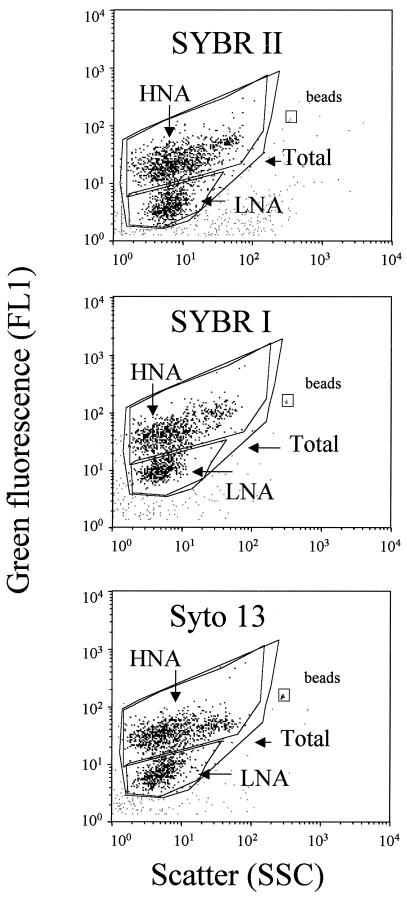
For flow cytometric analyses, three 1-ml subsamples were incubated with three nucleic acid stains, 0.5 μl of SYBR I, 0.5 μl of SYBR II, and 2.5 μM SYTO 13 (Molecular Probes Inc.), for 15 min at room temperature in the dark (13). Counts were obtained with a FacsCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) equipped with an air-cooled argon laser (488 nm, 15 mW). Stained bacteria were discriminated and enumerated by using right angle light scatter (SSC; related to cell size) and green fluorescence measured at 530 ± 30 nm. The volume analyzed and subsequent cell concentration estimate were calculated by weighing a sample before and after a 5-min analysis with the cytometer. Fluorescent beads (diameter, 1.002 μm; Polysciences Europe) were systematically added to each sample analyzed to normalize cell fluorescence emission and light scatter values. A four-log decade was used for all cytograms. In a plot of green fluorescence (FL1) versus red fluorescence (FL3) we were able to distinguish photosynthetic prokaryotes from nonphotosynthetic prokaryotes. The group of cells with high nucleic acid contents (HNA cells) was discriminated from the group of cells with low nucleic acid contents (LNA cells). Each subgroup was delimited on the SSC-versus-FL1 plot by drawing a window (Fig. 1), and cell abundance was determined for each subgroup. The cytometric noise corresponded to particles which could not be assigned to any population, and this noise was sometimes close to the LNA subgroup.
FIG. 1.
Flow cytometric analysis of natural seawater collected at the SOLA station. Bacterial cells were stained with SYBR II (a), SYBR I (b) and SYTO 13 (c). HNA and LNA cells are delimited by windows on each cytogram.
Bacterial production.
The [3H]leucine incorporation method (12, 18) was used in this study to estimate bacterial production. Incorporation of [3H]leucine (151 Ci mmol−1; Amersham Corp.) was measured at a concentration of 80 nM (5 nM tritiated leucine and 75 nM nonradioactive leucine); this concentration was shown to saturate leucine incorporation in the different aquatic systems tested (data not shown). Samples were incubated in the presence tritiated leucine for 1 to 2 h in the dark at the in situ temperature. After incubation, cold trichloroacetic acid (TCA) was added (final concentration, 5%), and the samples were filtered through 0.2-μm-pore-size cellulose acetate membrane filters (Sartorius); the filters were rinsed four times with 5 ml of cold 5% TCA. The radioactivity associated with the filters was estimated by liquid scintillation counting, and rates of incorporation into proteins (expressed in picomoles per liter per hour) were calculated.
Leucine incorporation followed by cell sorting by flow cytometry.
In order to estimate the contributions of HNA cells and LNA cells to total bacterial activity, we used a procedure similar to that developed by Servais et al. (19) and used by Bernard et al. (2) to estimate the contributions of different cell size classes to bacterial activity in various aquatic systems. The procedure involves labeling bacteria with [3H]leucine and then sorting different bacterial subpopulations by flow cytometry in order to estimate the contribution of each sorted subpopulation to the activity of the whole bacterial community.
In this study, HNA and LNA cells were sorted after [3H]leucine labeling in order to determine the specific leucine incorporation rates of both subpopulations. In parallel, total bacteria were sorted after similar [3H]leucine labeling to estimate the average specific leucine incorporation rate of the total bacterial community. From a practical point of view, leucine incorporation was performed at a final concentration of 80 nM. Only radioactive leucine (151 Ci mmol−1; Amersham Corp.) was added to maximize the detection limit for labeled bacteria. A 5-ml sample was incubated at the in situ temperature for 2 h in the dark in the presence of [3H]leucine. Incubation was stopped by adding formaldehyde (final concentration, 2%). Labeled bacteria were then stained with SYBR II by using the procedure used to determine total counts.
Labeled and stained bacterial cells were then sorted with the FacsCalibur flow cytometer into three windows defined on a cytogram in which green fluorescence was plotted against SSC (Fig. 1). The first window (Total bacteria) was drawn around the total bacterial population; this window was divided into two windows surrounding the HNA and LNA cells. For all sorting experiments, the salinity of the sterile sheath fluid was adjusted to the salinity of the sample to avoid cell lysis or protein release due to osmotic shock. On the flow cytometer we selected the single-sort mode, in which sorting occurs only if a single target cell is identified. The results give high purity with less emphasis on recovery and the most accurate counts of sorted cells.
Sorted cells were enumerated in this way and collected at the outlet of the flow cytometer directly on a 0.2-μm-pore-size membrane filter (acetate cellulose Sartorius filter). The sorting was stopped when the number of sorted cells was between 300,000 and 500,000 in order to obtain measurable radioactivity on the filter. Ten milliliters of cold 5% TCA was added to the filter in order to precipitate macromolecules and to rinse the membrane. After 10 min, the TCA was eliminated by filtration, and the radioactivity associated with the sorted bacteria was estimated by liquid scintillation counting.
In a previous study, this sorting procedure for the total bacterial population was shown to give results similar to the results obtained by direct measurement of bacterial activity without cell sorting (19). The average specific activity of the total bacterial community (expressed in moles of leucine incorporated per cell per hour) was determined by dividing the radioactivity incorporated by the bacteria in the total-bacteria window by the number of sorted cells in this window. Similarly, the average specific activities of the HNA and LNA cells were calculated by using the radioactivities associated with the cells in the HNA and LNA windows. Duplicate labeling experiments were not performed because of both the high cost and the long time required for these experiments. However, for several samples, a coefficient of variation of 9% was estimated for the specific activity of the total bacterial population determined by radioactive labeling followed by cell sorting after SYBR II staining.
RESULTS
Comparison of nucleic acid stains.
Three blue nucleic acid stains which are commonly used (SYBR I, SYBR II, and SYTO 13) were compared to determine if they provided the same counts of total, HNA, and LNA cells and to determine which of them resulted in better discrimination of LNA and HNA cells. The mean fluorescence and scatter values for each category of cells and the mean ratios determined for HNA and LNA cells are shown in Table 1. For each category of cells, the mean scatter values did not vary significantly for the different dyes, and the highest ratio of HNA cells to LNA cells was obtained with SYBR II. In contrast, the mean intensity of fluorescence varied greatly for the different stains, and the same trend was observed for each category of cells. The fluorescence of SYBR I-stained cells was greater than that of SYTO 13- or SYBR II-stained cells. However, the highest ratio of fluorescence of HNA cells to fluorescence of LNA cells was observed with SYBR II-stained cells. Therefore, although SYBR I-stained cells were more fluorescent and better discriminated from the background fluorescence (noise) than cells stained with the other dyes, better discrimination between HNA and LNA cells was obtained with SYBR II-stained cells. These differences between dyes are shown at least in part in Fig. 1; for this sample, the ratios of the mean fluorescence values for HNA and LNA cells were 6.3, 4.4, and 5.2 for subsamples stained with SYBR II, SYBR I, and SYTO 13, respectively. The correlation coefficients determined for total, HNA, and LNA cell counts are shown in Table 2. Total and HNA cell counts were highly correlated for the three dyes, but the correlation for the LNA cell counts was not as good. This was due to the low signal/noise ratio generally observed when natural communities are stained (Fig. 2). On the basis of these results, SYBR II was chosen for cell sorting experiments because the first objective of this study was to discriminate HNA cells from LNA cells. We preferred using the terms HNA and LNA rather than HDNA and LDNA because the fluorescence of SYBR II-stained bacteria depends on both DNA and RNA.
TABLE 1.
Scatter and fluoresence values for total, HNA, and LNA cells as determined after staining with different nucleic acid dyes (SYTO 13, SYBR I, and SYBR II)a
| Nucleic acid stain | Mean scatter
|
Mean fluorescence
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total cell value | HNA cell value | LNA cell value | Ratio of HNA cell value to LNA cell value | Total cell value | HNA cell value | LNA cell value | Ratio of HNA cell value to LNA cell value | |
| SYTO 13 | 0.024 ± 0.006 | 0.025 ± 0.008 | 0.016 ± 0.003 | 1.61 ± 0.46 | 0.184 ± 0.034 | 0.262 ± 0.048 | 0.056 ± 0.011 | 4.75 ± 0.84 |
| SYBR I | 0.022 ± 0.005 | 0.024 ± 0.007 | 0.015 ± 0.003 | 1.63 ± 0.43 | 0.252 ± 0.025 | 0.329 ± 0.033 | 0.079 ± 0.014 | 4.32 ± 0.96 |
| SYBR II | 0.024 ± 0.007 | 0.026 ± 0.008 | 0.016 ± 0.003 | 1.72 ± 0.45 | 0.154 ± 0.025 | 0.210 ± 0.026 | 0.043 ± 0.010 | 4.88 ± 1.36 |
Means ± standard deviations were determined for samples collected at different times at the SOLA station (n = 16). The scatter and fluorescence values were normalized with fluorescent beads.
TABLE 2.
Correlation coefficients for total, HNA, and LNA cell counts obtained with the different dyes for samples taken at different times at the SOLA station (n = 16)
| Cells | Correlation coefficients
|
||
|---|---|---|---|
| SYTO 13 vs SYBR I | SYTO 13 vs SYBR II | SYBR I vs SYBR II | |
| Total | 0.97 | 0.96 | 0.97 |
| HNA | 0.82 | 0.79 | 0.87 |
| LNA | 0.47 | 0.48 | 0.56 |
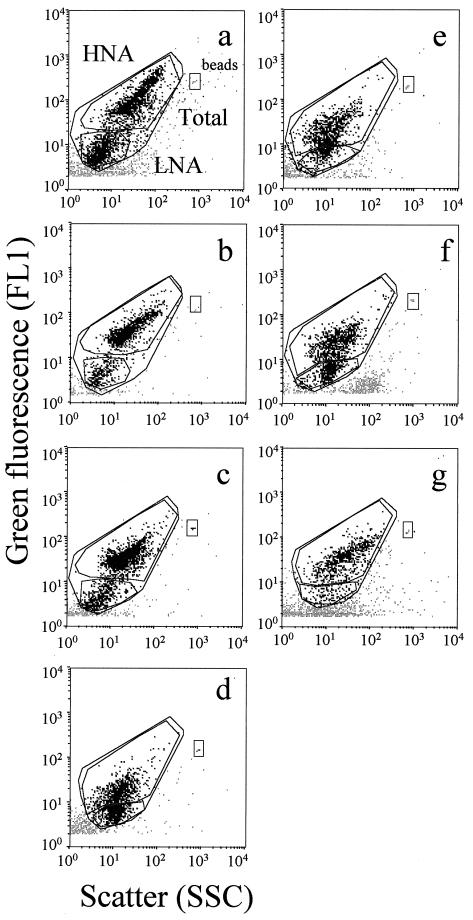
FIG. 2.
Flow cytometric analysis of water samples collected in the Tech River, including samples Tech 1 (a), Tech 2 (b), and Tech 3 (c); in the Leucate Lagoon (d); at the SOLA station, including samples SOLA 1 (e) and SOLA 2 (f); and in the Banyuls-sur-Mer harbor (g). HNA and LNA cells are delimited by windows on each cytogram.
Cell sorting experiments.
Seven ecosystems, including freshwater, brackish water, and marine water ecosystems, were sampled for the cell sorting experiments. After leucine incorporation, samples were fixed and stained with SYBR II. Then the total, HNA, and LNA cells were sorted, and the radioactivity in each cell fraction was determined. All of the results are shown in Table 3, whereas the cytograms corresponding to the samples are shown in Fig. 2.
TABLE 3.
Total counts and percentages of HNA and LNA cells in the samples analyzed in the cell sorting experiments
| Sample | Total count (108 cells liter−1) | % of HNA cells | % of LNA cells | Sp act (10−23 mol of leucine liter−1 h−1)a
|
Ratio of HNA cell sp act to LNA cell sp act | Production (pmol liter−1 h−1)b
|
% of total production
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total cells | HNA cells | LNA cells | Total cells | HNA cells | LNA cells | HNA cells | LNA cells | |||||
| Tech 1 | 1.61 | 53.4 | 46.6 | 51 | 75 | 6 | 12.5 | 82 | 64.5 | 3.5 | 78.6 | 4.2 |
| Tech 2 | 5.29 | 73.5 | 26.5 | 288 | 408 | 5.5 | 74.2 | 1,523 | 1,588 | 4.8 | 104 | 0.3 |
| Tech 3 | 2.5 | 72.0 | 28.0 | 170 | 233 | 13 | 17.9 | 425 | 420 | 6.5 | 98.8 | 1.5 |
| Leucate | 6.02 | 39.9 | 60.1 | 62 | 143 | NDc | ND | 373 | 343 | ND | 91.9 | ND |
| Harbor | 1.02 | 81.4 | 18.6 | 421 | 464 | 29 | 16 | 429 | 385 | 5 | 89.7 | 1.2 |
| SOLA 1 | 0.9 | 51.1 | 48.9 | 48 | 68 | 25 | 2.7 | 43.2 | 31.3 | 11 | 72.4 | 25.4 |
| SOLA 2 | 1.48 | 48.6 | 51.4 | 40 | 59 | 15.6 | 3.8 | 59 | 42.7 | 11.6 | 72.4 | 19.7 |
Specific activity was determined after cell sorting.
Determined by multiplying the specific activity by the count.
ND, not determined.
Two bacterial subgroups could be discriminated for all natural communities, but the fluorescence and scatter parameters varied greatly between samples (Fig. 2). The best discrimination was obtained for freshwater samples. In brackish water and seawater samples, discrimination between the two subgroups was more subjective.
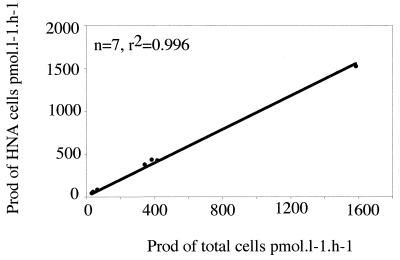
The percent contribution of HNA cells to the total cell counts varied from 39.9 to 81.4% (Table 3). The specific activity of HNA cells was always higher than those of total and LNA cells (Table 3). The highest specific activities of the total community and of the HNA bacteria were in two samples taken in the harbor of Banyuls-sur-Mer and in the Tech River (Table 3). The specific activity of the LNA cells was generally very low compared with the specific activity of the HNA cells. The ratio of the specific activities of the HNA and LNA cells was lowest for the SOLA station samples (2.7 and 3.8). In these samples, the contribution of the LNA cells was not insignificant (20 to 25% of the total bacterial community). The bacterial communities in these samples also were the less active bacterial communities. In other samples, the contribution of LNA cells to the total production was low and never represented more than 4.2% of the total production. In contrast, the contribution of HNA cells to production was very high, and the average contribution of these cells to the total activity was sometimes close to 100%. The lowest contributions were found for the less active communities (SOLA station samples). Some contribution values were slightly higher than 100% because the relative contributions of the HNA, LNA, and total cells were determined with different subsamples. The correlation between total bacterial production and HNA cell production was very good (Fig. 3).
FIG. 3.
Correlation between HNA cell activity and total bacterial production (Prod) determined from seven samples.
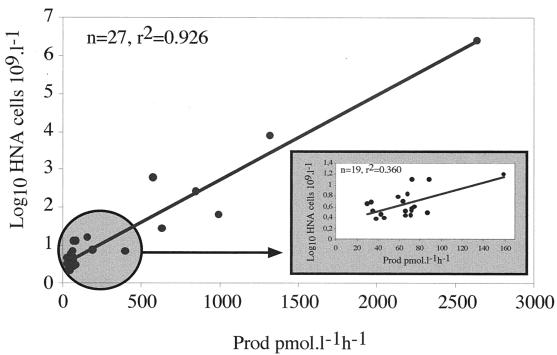
Although it was not possible to apply the cell sorting procedure to more samples, the correlation between HNA cell counts and total production was determined for a large number of samples. These samples included the temporal samples obtained at the SOLA station to investigate the potential use of HNA cell counts as an indicator of community activity (Fig. 4). The two parameters were highly correlated, but this correlation was due in part to the wide range of production values. When values were determined only for samples collected weekly at the SOLA station (n = 19), the correlation was not as good, and the relationship between HNA cell counts and bulk activity was far from clear.
FIG. 4.
Correlation between HNA cell counts and total bacterial production (Prod) determined from all samples. (Inset) Correlation determined only from SOLA station samples (data in the circle).
DISCUSSION
Comparison of nucleic acid dyes.
The different methods used to determine total cell counts were compared to determine if at least one of the three dyes commonly used provides better discrimination of HNA cells. The comparison was performed with samples collected weekly at the SOLA station. The total cell counts correlated well, and the results obtained by all of the methods are comparable. This result is in good agreement with the results of other comparative studies (6, 13, 17), suggesting that all these dyes can be used to determine total cell counts. However, the greater mean fluorescence of total, HNA, and LNA cells stained with SYBR I was not found in a previous study (13). This may be partially explained by the low level of bacterial production recorded at the SOLA sampling station since less active cells have a lower RNA content and SYBR I has a higher quantum yield when it binds to DNA than when it binds to RNA, whereas SYTO 13 and SYBR II have a lower affinity for DNA and a higher affinity for RNA (13). These differences have little effect on detection of HNA cells, and the good correlations obtained for HNA cell counts suggest that all of the dyes can be used to enumerate HNA cells. Inversely, the correlations for LNA cell counts were not so good. This can be explained by the lower fluorescence of LNA cells stained with SYTO 13 and SYBR II, which results in more interactions between cells and noise and, consequently, less-accurate counts. Obviously, this interaction has an insignificant effect on the total cell counts but is more apparent when LNA cells are counted. Therefore, SYBR I should be preferred when accurate quantification of both LNA and HNA cells is desired. However, SYBR II was preferred to the other dyes for cell sorting experiments for two reasons. First, this dye provides the highest fluorescence and scatter ratios for HNA and LNA cells and the best discrimination of the two subgroups. Second, assuming that SYBR II stains RNA with a higher quantum yield than other dyes (14) and that LNA cells are nongrowing cells (6), this dye should provide better discrimination of the two subgroups in the most productive ecosystems by increasing the fluorescence of actively growing cells (HNA cells) with no increase in the fluorescence of LNA cells.
Activities of HNA and LNA cells.
The high correlation between the activity of HNA sorted cells and the total cell activity suggested that HNA cells were responsible for a very large fraction of the bulk activity. The calculated contribution of HNA cells to the bulk activity was sometimes as great as 100% of the total bacterial production. This confirms the results of Gasol et al. (6) which suggested that HNA cells are the dynamic members of natural communities. Furthermore, HNA cells represented significant fractions of the total communities and accounted for at least 40%, and sometimes up to 80% of the total cells. This does not necessarily mean that all HNA cells are active members of the community nor that all HNA cells have high cell-specific rates of production, but it clearly demonstrates that bacterial cells in natural waters are not equally active and that an important fraction of natural communities, at least the fraction consisting of LNA cells, is inactive or dead. It also confirms previous hypotheses based on the use of specific tests to determine the fraction of inactive or dead cells (8, 11, 28). The small but significant contribution of LNA cells to the bulk activity found for the two samples collected at the SOLA station was more likely due to poor discrimination between the two subgroups (Fig. 2) and probably due to inclusion of some active cells in the LNA cell subgroup. Poor discrimination between the two subgroups was always observed when the differences between the specific activities of LNA and HNA cells were the lowest, as was the case for the SOLA station samples (Table 3). Similar poor discrimination was reported by Gasol and del Giorgio (7), and this suggests that it may sometimes be difficult to obtain accurate HNA cell counts when the two subgroups are not clearly separated. In this case, the reproducibility of HNA cell count data may have been low for different samples for a given operator and for different operators for a given sample. Objective separation between the two subgroups should be improved by developing appropriate software.
Jellett et al. (9) suggested that the percent contribution of HDNA cells to the bacterial community could be used as an active cell index. This was congruent with the results previously reported by Li et al. (15) showing that HDNA cells grew faster than LDNA cells. This idea was further investigated by Gasol et al. (6), and these authors suggested that the percentage of HDNA cells could be used as a reference for the percentage of actively growing bacteria in marine planktonic environments. Their results were based on different correlations obtained for counts of LDNA, HDNA, and total bacteria during microcosm experiments. In this study, we directly measured the activity of targeted cells, and we confirmed that most of the bulk activity resulted from HNA cells.
Significance of the HNA and LNA subgroups.
The significance of HNA and LNA cells remains questionable from a physiological point of view. The high nucleic acid content of HNA cells can be explained in different ways. If we assume that HNA cells are active cells, they may include virus-infected cells in which viruses can multiply, cells containing plasmids, growing cells with a significant RNA content, and/or cells with multiple copies of the genome. Inversely, Gasol et al. (6) hypothesized that LNA cells may include an important fraction of dead cells because most of these cells are not responsive to environmental changes; assuming that active bacteria were included in the LNA fraction, then the separation between LNA and HNA cells should not exist. We fully agree with this hypothesis, but if we assume that LNA cells are inactive and nonviable cells with damaged membranes having more or less degraded DNA, then HNA cells should be intact cells containing at least a single genome and should obviously include cells with a wide range of activities from inactive to rapidly growing. This hypothesis was supported by the fact that the fluorescence intensity of some isolated marine bacteria in late stationary phase containing one genome was always within the range of intensities found for the HNA subgroup (data not shown). The heterogeneity of individual cell-specific activities in the HNA subgroup was also suggested by the poor correlation found between HNA cell counts and total production at the SOLA station. This poor correlation may be explained by the low activity at this oligotrophic station and, more likely, by the fact that the activity of HNA cells may be heterogeneous and, thus, HNA cells may include a wide gradient of activity from inactive to very active. For all samples, HNA cells were always distributed in a wide range of scatter values since the ratios of the width of the peak to the height of the peak on scatter histograms (as given by the flow cytometer software and called coefficients of variation) ranged from 80 to 110. As SSC is clearly related to cell size (6, 24), this finding means that the cell volumes of HNA cells vary within a wide range of sizes. Therefore, if we consider the relationship which was recently demonstrated between cell size and activity (2, 19), the data suggest that the activity of HNA cells varies greatly within the HNA subgroup. This is congruent with the idea developed by Gasol et al. (6) that “bacterial single cell activity in a given aquatic assemblage varies continuously from high to low metabolism.”
Conclusion.
From this study, we concluded that the HNA subgroup is responsible for the most important part of the bulk activity and that the activity in this subgroup is heterogeneous. We suggest that nucleic acid content alone can be used as a better indicator of the fraction of growing cells than total cell counts and that this approach should be combined with other physiological parameters to improve detection of the most active cells in aquatic systems. To our knowledge, this is the first study which demonstrated by a direct approach that a fraction of bacteria is responsible for the bulk activity and that significant fractions of natural assemblages are composed of inactive cells. Although there is strong evidence that the nucleic acid content of individual cells is not enough to discriminate between the less active cells and the more active cells, additional techniques based on multiparametric approaches should be developed to combine the staining procedure used to stain nucleic acids with other fluorescent physiological probes in order to provide more accurate evaluation of cell-specific activities. Our results address new questions, such as how and at what rate LNA cells are produced and how long they persist in the environment. Another question involves the diversity of species in both fractions. Do some species occur in both fractions, and if so, what are the consequences of this heterogeneity in terms of diversity and function?
ACKNOWLEDGMENTS
The FACSCalibur cytometer was funded by the Région Languedoc-Roussillon (France), the Centre National de la Recherche Scientifique (CNRS-INSU-SDV, France), and EU Eloise Project PL95049. This work was funded in part by Rhône-Poulenc (France) and CNRS.
At the time of this study, Pierre Servais was employed by the Centre National de la Recherche Scientifique (CNRS-INSU, France).
REFERENCES
- 1.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 2.Bernard L, Courties C, Servais P, Troussellier M, Petit M, Lebaron P. Relationships between bacterial cell-size, productivity and genetic diversity in aquatic environments using cell sorting and flow cytometry. Microb Ecol. 2000;40:148–158. doi: 10.1007/s002480000046. [DOI] [PubMed] [Google Scholar]
- 3.Choi J W, Sherr E B, Sherr B F. Dead or alive? A large fraction of ET-inactive marine bacterioplankton cells, as assessed by reduction of CTC, can become ETS-active cells with incubation and substrate addition. Aquat Microb Ecol. 1999;16:27–35. [Google Scholar]
- 4.del Giorgio P A, Scarborough G. Increase in the proportion of metabolically active bacteria along gradients of enrichment in freshwater and marine plankton: implication for estimates of bacterial growth and production rates. J Plankton Res. 1995;17:1905–1924. [Google Scholar]
- 5.del Giorgio P A, Prairie Y T, Bird D F. Coupling between rates of bacterial production and the abundance of metabolically active bacteria in lakes, enumerated using CTC reduction and flow cytometry. Microb Ecol. 1997;34:144–154. doi: 10.1007/s002489900044. [DOI] [PubMed] [Google Scholar]
- 6.Gasol J M, Zweifel U L, Peters F, Furhman J A, Hagström Å. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol. 1999;65:4475–4483. doi: 10.1128/aem.65.10.4475-4483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasol J M, del Giorgio P A. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar. 2000;64:197–224. [Google Scholar]
- 8.Gasol J P, del Giorgio P A, Massana R, Duarte C M. Active versus inactive bacteria: size-dependence in a coastal marine plankton community. Mar Ecol Prog Ser. 1995;128:91–97. [Google Scholar]
- 9.Jellett J F, Li W K W, Dickie P M, Boraie A, Keplay P E. Metabolic activity of bacterioplankton communities assessed by flow cytometry and single carbon substrate utilization. Mar Ecol Prog Ser. 1996;136:213–225. [Google Scholar]
- 10.Joux F, Lebaron P. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2000;2:1523–1535. doi: 10.1016/s1286-4579(00)01307-1. [DOI] [PubMed] [Google Scholar]
- 11.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchman D, K'Nees F, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebaron P, Parthuisot N, Catala P. Comparison of blue nucleic acid dyes for the flow cytometric enumeration of bacteria in aquatic systems. Appl Environ Microbiol. 1998;64:1724–1730. doi: 10.1128/aem.64.5.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebaron P, Servais P, Troussellier M, Courties C, Vives-Rego J, Muyzer G, Bernard L, Guindulain T, Schäfer H, Pukall R, Stackebrandt E. Changes in bacterial community structure in seawater mesocosms differing in their nutrient status. Aquat Microb Ecol. 1999;19:255–267. [Google Scholar]
- 15.Li W K W, Jelett J F, Dickie P M. DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol Oceanogr. 1995;40:1485–1495. [Google Scholar]
- 16.Lovejoy C, Legendre L, Klein B, Tremblay J E, Ingram R G, Therriault J C. Bacterial activity during early winter mixing (Gulf of St. Lawrence, Canada) Aquat Microb Ecol. 1996;10:1–13. [Google Scholar]
- 17.Marie D, Partenski F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Servais P. Bacterial production measured by 3H-thymidine and 3H-leucine in various aquatic ecosystems. Arch Hydrobiol Beih Ergebn Limnol. 1992;37:73–81. [Google Scholar]
- 19.Servais P, Courties C, Lebaron P, Troussellier M. Coupling bacterial activity measurements with cell sorting by flow cytometry. Microb Ecol. 1999;38:180–189. doi: 10.1007/s002489900160. [DOI] [PubMed] [Google Scholar]
- 20.Servais, P., H. Agogué, C. Courties, F. Joux, and P. Lebaron. Are the actively respiring cells (CTC+) those responsible for bacterial production in aquatic environments? FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 21.Sherr B F, del Giorgio P A, Sherr E B. Estimating the abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat Microb Ecol. 1999;18:117–131. [Google Scholar]
- 22.Sieracki M E, Cucci T L, Nicinski J. Flow cytometric analysis of 5-cyano-2,3-ditolyl tetrazolium chloride activity of marine bacterioplankton in dilution cultures. Appl Environ Microbiol. 1999;65:2409–2417. doi: 10.1128/aem.65.6.2409-2417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson L H. A case for bacterial dormancy in aquatic systems. Microb Ecol. 1978;4:127–133. doi: 10.1007/BF02014283. [DOI] [PubMed] [Google Scholar]
- 24.Troussellier M, Courties C, Lebaron P, Servais P. Flow cytometric discrimination of bacterial populations in seawater based on SYTO 13 staining of nucleic acids. FEMS Microbiol Ecol. 1999;29:319–330. [Google Scholar]
- 25.Ullrich S, Karrasch B, Hoppe H, Jeskulke K, Mehrens M. Toxic effects on bacterial metabolism of the redox dye 5-cyano-2,3-ditolyltetrazolium chloride. Appl Environ Microbiol. 1996;62:4587–4593. doi: 10.1128/aem.62.12.4587-4593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullrich S, Karrash B, Hoppe H G. Is the CTC dye technique an adequate approach for estimating active bacterial cells? Aquat Microb Ecol. 1999;17:207–209. [Google Scholar]
- 27.Vives-Rego J, Lebaron P, Nebe-Von Caron G. Current and future applications of flow cytometry in aquatic microbiology. FEMS Microbiol Rev. 2000;24:429–448. doi: 10.1111/j.1574-6976.2000.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 28.Zweifel U L, Hagström Å. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]