Abstract
Zika virus (ZIKV) causes significant human diseases without specific therapy. Previously we found erythrosin B, an FDA-approved food additive, inhibited viral NS2B−NS3 interactions, leading to inhibition of ZIKV infection in cell culture. In this study, we performed pharmacokinetic and in vivo studies to demonstrate the efficacy of erythrosin B against ZIKV in 3D mini-brain organoid and mouse models. Our results showed that erythrosin B is very effective in abolishing ZIKV replication in the 3D organoid model. Although pharmacokinetics studies indicated that erythrosin B had a low absorption profile, mice challenged by a lethal dose of ZIKV showed a significantly improved survival rate upon oral administration of erythrosin B, compared to vehicle control. Limited structure−activity relationship studies indicated that most analogs of erythrosin B with modifications on the xanthene ring led to loss or reduction of inhibitory activities towards viral NS2B−NS3 interactions, protease activity and antiviral efficacy. In contrast, introducing chlorine substitutions on the isobenzofuran ring led to slightly increased activities, suggesting that the isobenzofuran ring is well tolerated for modifications. Cytotoxicity studies indicated that all derivatives are nontoxic to human cells. Overall, our studies demonstrated erythrosin B is an effective antiviral against ZIKV both in vitro and in vivo.
KEY WORDS: Flavivirus, Zika virus, Dengue virus, Antiviral, Protease inhibitor, Erythrosin B
Abbreviations: aa, amino acid; AUC, area under he curve; DENV, dengue virus; DMSO, dimethyl sulfoxide; dpi, day post infection; EB, erythrosin B; FDA, US Food and Drug Administration; FRET, fluorescence resonance energy transfer; ip, intraperitoneal; NS, non-structural protein; ORF, open reading frame; PFU, plaque-forming unit; PK, pharmacokinetic; PP, polyprotein precursor; SAR, structure−activity relationship; SLC, split luciferase complementation; UTR, untranslated region; WHO, World Health Organization; ZIKV, Zika virus
Graphical abstract
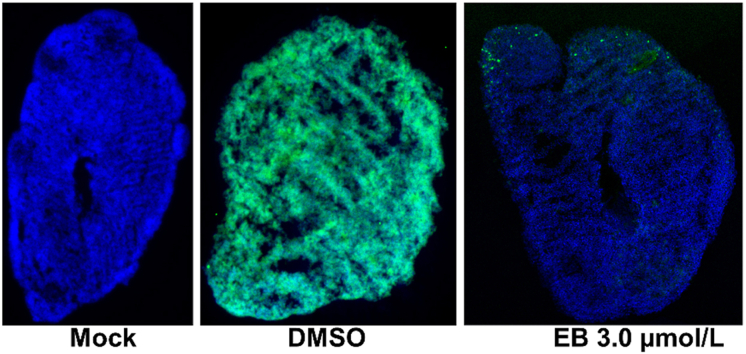
Erythrosin B (EB), an FDA-approved food dye, can protect 3D mini-brain organoid from infection of Zika virus. This figure shows the immunofluorescence imaging of slices of organoid infected with mock solution or ZIKV PRVABC59 (green) treated with EB or DMSO control. Blue, DAPI.
1. Introduction
Zika virus (ZIKV) is a member of the genus Flavivirus. ZIKV and many other flaviviruses such as dengue virus (DENV) are significant human pathogens1. ZIKV outbreaks have occurred worldwide, leading to devastating diseases, including central nervous system malformations such as Guillain-Barré syndrome, microcephaly and congenital Zika syndrome2, 3, 4, 5, 6. DENVs cause life-threatening diseases such as dengue shock syndrome and dengue hemorrhagic fever, now together termed as severe dengue by WHO7. DENV infection threatens 3.9 billion people, leading to 22,000 deaths per year worldwide. A DENV vaccine was recently approved in a few countries8. However, it is not effective for young children9, and may pose an increased risk for naïve children10. Although there are effective vaccines for a few flaviviruses, safe and effective vaccines for many flaviviruses including ZIKV have not been developed or approved by US Food and Drug Administration (FDA).
The flaviviral genome is composed of 5ʹ-untranslated region (UTR), a single open reading frame (ORF), and 3ʹ-UTR. A polyprotein precursor (PP) encoded by the ORF will be co- and post-translationally processed by viral and cellular proteases into individual functional proteins11. NS3 is a multi-functional viral protein with activities of a serine protease, an RNA triphosphatase, a nucleoside triphosphatase, and a helicase12, 13, 14. The viral NS3 protease is located at the N-terminal 184 amino acids (aa) of viral NS3 protein and requires a hydrophobic core of about 40 aa in length within viral NS2B protein as an essential cofactor15, 16, 17. The NS2B binding site on the NS3 protease is highly conserved and essential (Supporting Information Fig. S1)18, 19, 20, 21, 22. Crystal structures of the viral proteases and their complexes with inhibitors have been determined15,22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, including the most recent ZIKV NS2B−NS3 protease31, 32, 33, 34. NS2B binding is required for NS3 function.
In recent studies, we identified several clinically-used and FDA-approved compounds as inhibitors against viral NS2B−NS3 protease complex17,35, 36, 37, 38, 39, including erythrosin B (EB), an FDA-approved food additive. EB functions as an inhibitor to abolish the essential binding of co-factor NS2B to the viral protease NS3. Docking and mutagenesis studies confirmed that EB bound in a pocket formed by residues Y23, I25, Q28, F46, and L58 of the DENV3 NS3 protease36. It is noted that except Q28 which is neither conserved nor essential, all other 4 residues are highly conserved and essential for binding of EB to NS3 (Fig. S1)36. Consistently, in vitro, EB was found as a broad-spectrum inhibitor against multiple flaviviruses with an appreciable therapeutic window. EB was also found to inhibit virus replication in multiple cell types including lung carcinoma cells, human placental epithelial cells, and human neural progenitor and stem cells.
In this study, we explored the in vivo antiviral efficacy of EB using 3D organoid and mouse models. We found that EB significantly reduced ZIKV infection on a 3D-mini forebrain organoid model derived from pluripotent neural stem cells. The survival rate of mice infected by a lethal dose of ZIKV was significantly improved by EB treatment, compared to that by vehicle control. To improve the potency and reduce potential side effects, we performed a limited scale of structure−activity relationship (SAR) study. We found that modification of I on the xanthene ring led to the loss of antiviral activity, whereas introducing chlorine substitutions on the isobenzofuran ring maintained the same level of potency in inhibition of NS2B−NS3 interaction, protease activity, and antiviral efficacy, indicating that the isobenzofuran ring is tolerated for modifications.
2. Results and discussion
2.1. Chemistry
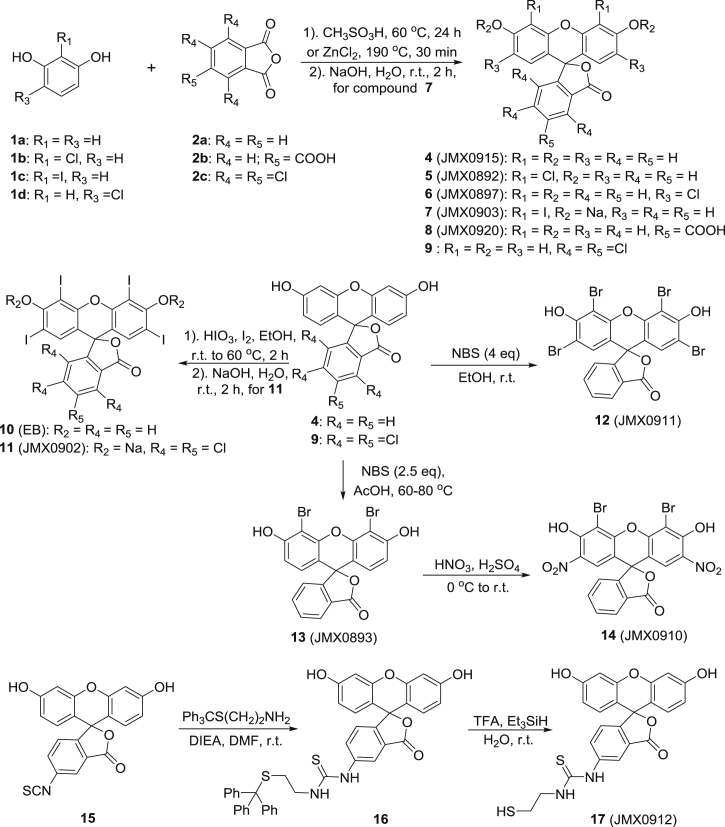
The general synthetic routes of erythrosin B and its derivatives were summarized in Scheme 1. Coupling of substituted resorcinols 1a−d with various phthalic anhydrides 2a−c afforded fluorescein derivatives 4–9. Iodination of 4 and 9 produced derivatives 10 and 11, respectively. Bromination of 4 with NBS (4.0 or 2.5 eq) gave tetrabromo- and dibromo-derivatives 12 and 13, respectively. Further nitration of 13 yielded compound 14. Commercially available isothiocyanate 15 was coupled with trityl-protected cysteamine to give the intermediate 16, which was then converted into the final product 17 via acidic deprotection. The detailed synthetic procedures of these derivatives followed the reported protocols in literatures42, 43, 44, 45. The structures and purities of all synthesized compounds were evaluated by 1H and 13C NMR and HPLC analysis, respectively. All compounds are >95% pure.
Scheme 1.
The general synthetic routes of erythrosin B (EB) and its derivatives.
2.2. Erythrosin B protects 3D mini-forebrain organoid from ZIKV infection
Previously, we found that EB inhibited ZIKV infection in neural progenitor and placenta cells relevant to ZIKV pathogenesis using 2D cell culture. Here we further investigated whether EB could protect against ZIKV-associated neurological damage using a 3D mini-forebrain organoid model, as we described previously35,37.
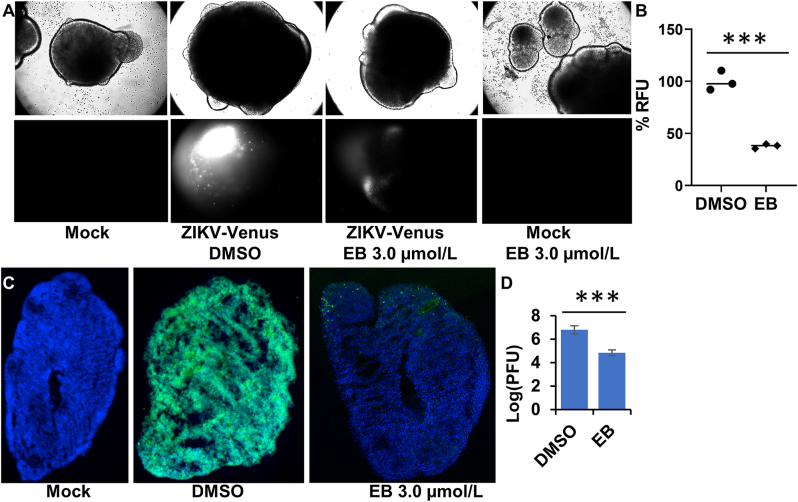
Following published protocol35,37, we generated the 3D mini-forebrain organoid from induced pluripotent stem cells. We first evaluated if EB could damage the 3D organoid. Our results showed that EB treatment did not lead to any changes in morphology of the 3D organoids (Fig. 1A, upper panel), suggesting that EB at 3.0 μmol/L is not toxic to the organoid. In contrast, temoporfin, an inhibitor blocking NS2B−NS3 interactions we identified previously17, showed significant toxicity to the 3D organoids (Supporting Information Fig. S2), even though it only showed moderate cellular cytotoxicity17. As shown, upon temoporfin treatment, the organoids fell apart and lost clear organoid boundary, indicating toxicity. Next, we used a full-length infectious ZIKV clone expressing Venus fluorescent protein (ZIKV-Venus)35,37 to evaluate the antiviral efficacy of EB in the organoid model. Our results showed that organoids treated with DMSO showed significant fluorescence, indicating successful ZIKV infection (Fig. 1A, lower panel, and Fig. S2). In contrast, EB-treated organoids showed greatly reduced fluorescence [Fig. 1A (lower panel) and B, and Fig. S2], suggesting less viral infection and replication. Our results suggest that ZIKV infection in the 3D organoid is significantly reduced by EB treatment.
Figure 1.
(A) ZIKV organoid infected with ZIKV-Venus. The 3D organoids were infected with PBS (Mock), or ZIKV untreated (DMSO), or ZIKV treated with EB (3.0 μmol/L), or Mock treated with EB (3.0 μmol/L). Upper panel, bright field image of intact organoids. Lower panel, Venus fluorescence image (excitation 515 nm, emission 528 nm) of the intact 3D organoids. (B) Relative mean fluorescence intensity of ZIKV-Venus infected organoids treated with DMSO or EB (3.0 μmol/L). n = 3. ∗∗∗P < 0.001. (C) Immunofluorescence assay (IFA) imaging of slices of organoid infected with ZIKV PRVABC59. The 3D organoids were infected with PBS (Mock), or ZIKV untreated (DMSO), or ZIKV treated with EB. IFA using anti-E 4G2 antibody (green); blue, DAPI. (D) ZIKV production from the 3D organoids at 5 dpi. Culture supernatants were collected, and virus production was quantified by PFU assay. n = 3. Error bars in panel D represent the standard deviations at each concentration. ∗∗∗P < 0.001. Student t-test was used for data in panels B and D.
We next used immunostaining to evaluate viral antigen expression in the 3D organoid. Using the 4G2 antibody which recognizes flaviviral envelope protein, we showed that DMSO-treated organoid is fully susceptible to ZIKV infection, showing viral antigen expression throughout the organoid (Fig. 1B). In contrast, only very limited viral antigen expression was found for EB-treated organoids.
We next quantified virus production using plaque-forming unit assay. Our results showed that EB treatment reduced ZIKV production by the 3D organoids up to 2-log order, compared to the DMSO control. Collectively, our results demonstrate that EB treatment can prevent ZIKV infection in human cortical tissue.
2.3. Pharmacokinetic (PK) analysis of EB
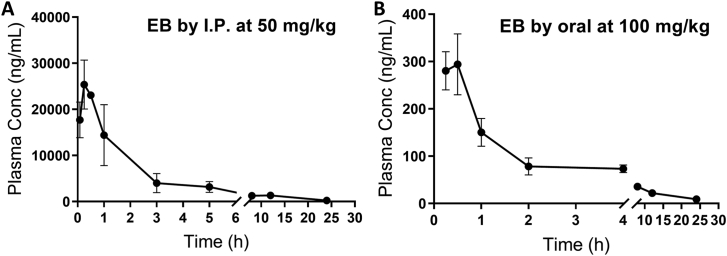
To study the PK properties, EB was first administered to female B6 mice at 50 mg/kg by intraperitoneal (ip) injection (Fig. 2A, Table 1). EB was absorbed rapidly in mice and reached the plasma peak concentration in 15 min. The clearance of EB is relatively slower with a t1/2 about 2 h. To examine the oral bioavailability of EB, the compound was administered to female B6 mice at 100 mg/kg by oral gavage (Fig. 2B). Compared to that with ip administration, the absorption of EB was also very rapid. It reached the plasma peak concentration in about 20 min. The clearance is also relatively slow with t1/2 of about 3 h. However, the Cmax and AUC were both much lower in the oral administrated group than those in the ip one, indicating that EB has relatively low absorption via the oral route.
Figure 2.
Pharmacokinetic study of EB. Adult female B6 mice were given a single dose of EB at (A) 50 mg/kg (ip) or (B) 100 mg/kg (oral). Plasma was obtained at various times after dosing. EB was extracted from the plasma and then analyzed by LC−MS/MS as described in the Experimental Section. n = 3.
Table 1.
Pharmacokinetic parameters of erythrosine B in female B6 mice after either ip or oral administration. Values represent means ± SD (n = 3).
| Parameter | Unit | ip |
Oral |
|---|---|---|---|
| 50 mg/kg | 100 mg/kg | ||
| Cmax | ng/mL | 26,800 ± 2884 | 325 ± 12.0 |
| Tmax | h | 0.3 ± 0.1 | 0.4 ± 0.1 |
| t1/2 | h | 2.0 ± 0.1 | 3.0 ± 0.4 |
| AUC 0−∞ | ng/mL∙h | 54,233 ± 14,449 | 860 ± 43 |
| Vz/F_obs | L/kg | 2.8 ± 0.6 | 504 ± 61 |
| CL/F_obs | L/kg/h | 1.0 ± 0.2 | 116 ± 6 |
2.4. In vivo antiviral efficacy
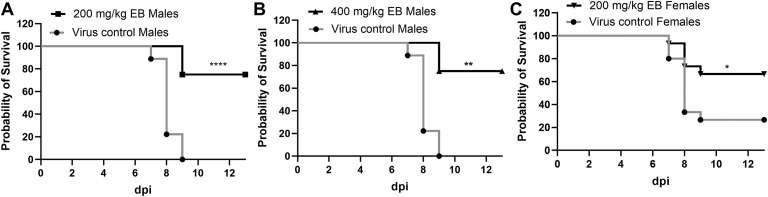
We next performed an in vivo antiviral efficacy study using an IfnαβR−/− ZIKV mouse model as we described previously17. Because we observed differences in susceptibility to ZIKV challenge between male and female mice, we evaluated male and female mice separately. Our results showed that male mice are more susceptible to ZIKV challenge than females. 1.7 × 104 PFU ZIKV PRVABC59 led to 100% moribundity of male mice. In contrast, 1.7 × 105 PFU ZIKV only resulted in a 75% moribundity of female mice (Fig. 3). Compared to the vehicle control group with 100% lethality, male mice treated with 200 mg/kg EB daily for 7 days survived 75% (Fig. 3A). Increasing dosage to 400 mg/kg did not further improve the survival rate (Fig. 3B). For female mice, treatment with 200 mg/kg daily for 7 days significantly improved the survival rate to 67.7%, compared to the 25% survival rate in the vehicle control group (Fig. 3C). Overall, the results indicated that EB treatment significantly improved mouse survival rate upon a lethal challenge of ZIKV.
Figure 3.
In vivo antiviral activity of EB against ZIKV. (A) and (B) Survival percentage for four-week-old A129 male mice infected with ZIKV PRVABC59 (1.7 × 104 PFU) and treated with EB 200 mg/kg (n = 12) (A) or EB 400 mg/kg (n = 4) (B) or vehicle (n = 9) via oral gavage. (C) Survival percentage of female mice infected with ZIKV (1.7 × 105 PFU) and treated with EB 200 mg/kg (n = 15) or vehicle (n = 15). All survival curves were compared using the Log-rank test. ∗∗∗∗P < 0.0001; ∗∗P < 0.005; ∗P < 0.05.
2.5. Screening EB derivatives
We next performed a small scale of SAR study (Table 2). We re-synthesized EB and generated 10 EB derivatives and evaluated their potency of inhibition towards NS2B−NS3 interactions, protease activity, anti-Zika virus, and cell viability (Table 2).
Table 2.
Effects of EB derivatives on NS2B–NS3 interaction (IC50-SLC), protease (IC50-pro), anti-ZIKV (EC50) and cell viability (CC50). All in micromolar scale.
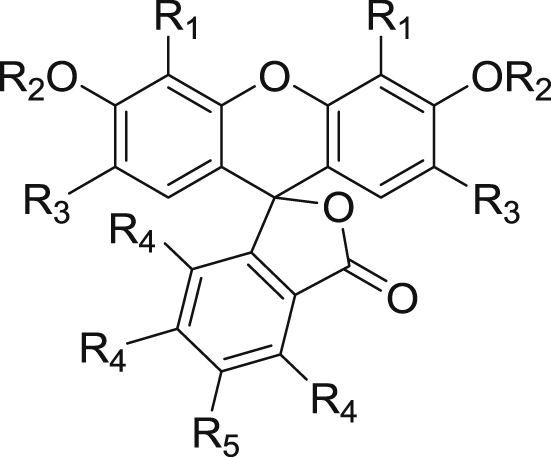
| Compd. | R1 | R2 | R3 | R4 | R5 | IC50-SLC | IC50-pro | EC50 | CC50 |
|---|---|---|---|---|---|---|---|---|---|
| EB | I | H | I | H | H | 0.42 | 7.9 | 0.62 | >200 |
| JMX0892 | Cl | H | H | H | H | 50 | >60 | >15 | >200 |
| JMX0893 | Br | H | H | H | H | 16 | >60 | >15 | >200 |
| JMX0897 | H | H | Cl | H | H | 48 | 23 | >15 | >200 |
| JMX0902 (Acid Red 94) | I | Na | I | Cl | Cl | 0.43 | 2.6 | 0.30 | >200 |
| JMX0903 | I | Na | H | H | H | 15 | 60 | >15 | >200 |
| JMX0910 | Br | H | NO2 | H | H | 13 | 47 | >15 | >200 |
| JMX0911 | Br | H | Br | H | H | 3.4 | 45 | >15 | >200 |
| JMX0912 (Fluorescein-SH) | H | H | H | H | 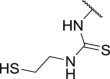 |
>100 | 88 | >15 | >200 |
| JMX0915 | H | H | H | H | H | >100 | >100 | >15 | >200 |
| JMX0920 | H | H | H | H | COOH | 53 | >100 | >15 | >200 |
Previously we demonstrated that EB inhibited flavivirus replication by inhibiting the binding of co-factor NS2B to viral NS3 protease, leading to inhibition of the viral NS2B−NS3 protease activity36. Therefore, we used previously-developed split luciferase complementation (SLC) assay and protease assay using a fluorescence resonance energy transfer (FRET) substrate to quantify the effects of these EB derivatives17,35, 36, 37.
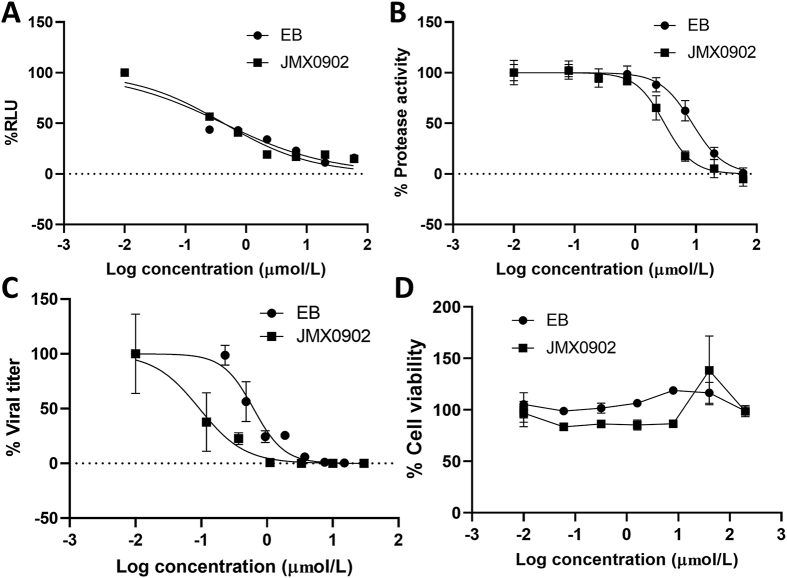
Using the NS2B−NS3 SLC assay as we described previously17,35, 36, 37, we determined the effects of these modifications on inhibition of the NS2B−NS3 interactions. Apparently, removal of I at both R1 and R3 positions of the fluorescein xanthyl part (compounds JMX0915, JMX0920, JMX0912) led to a drastic reduction in activity to inhibit NS2B−NS3 interactions (IC50-SLC > 50 μmol/L). Inserting Cl at either R1 or R3 position (JMX0892 and JMX0897) also abolished the inhibitory activity against the NS2B−NS3 interaction (IC50-SLC ∼50 μmol/L). Removing I at R3 position (JMX0903) or replacing I with Br at R1 positions (JMX0893, JMX0910, JMX0911) moderately impacted the inhibitory activity against the NS2B−NS3 interactions. Introducing Cl at both R4 and R5 positions (JMX0902) retained the same level of potency with an IC50-SLC of 0.43 μmol/L (Fig. 4A).
Figure 4.
Inhibition of NS2B−NS3 interaction by the SLC assay (A), NS2B−NS3 protease activity (B), anti-Zika virus activity (C) by EB and JMX0902 and cytotoxicity of EB and JMX0902 (D). (A) Dose-dependent inhibition of SLC upon binding of NLuc-NS2B49−66 to GST-CLuc-NS3. n = 3. (B) Dose−response inhibitions of the His-NS2B/His-MBP-NS3 protease activity. n = 3. (C) Dose-dependent inhibition of ZIKV infectivity. n = 3. (D) Cell viability assay. A549 cells were incubated with various concentrations of compounds and then assayed for viability at 48 h post incubation. n = 3. Error bars in all panels represent the standard deviations at each concentration. The DMSO control was set as 100%.
Next, we evaluated their inhibitory effect on the NS2B−NS3 protease activity, using His-tag NS2B and MBP-tagged NS3 proteins with a FRET substrate Abz-RRRR↓SAG-nTyr substrate, as described previously17,35, 36, 37. Similarly, all derivatives, except JMX0902, greatly reduced their potency in inhibition of the viral protease activity (Table 2). Compound JMX0902 showed slightly improved inhibitory activity against the NS2B−NS3 protease activity (IC50-pro) (Fig. 4B).
Using plaque reduction assay, we demonstrated that except JMX0902, all other derivatives did not show appreciable antiviral efficacy towards Zika virus (EC50 > 15 μmol/L), which is consistent with their low protease inhibitory activity. In contrast, our data showed that JMX0902 was slightly more potent than EB in inhibition of Zika virus replication, with an EC50 of 0.3 μmol/L (Fig. 4C), which is in agreement with its better inhibitory activity to the NS2B−NS3 protease than that of EB.
Finally, we measured the cell cytotoxicity of these compounds towards A549 cells. Our data showed that all derivatives are well tolerated by A549 cells, with CC50 > 200 μmol/L (Fig. 4D).
3. Conclusions
Flaviviruses represent significant human pathogens. During the last two decades, frequent outbreaks of flaviviruses such as West nile virus, DENV, and most recently ZIKV, occurred worldwide. Unfortunately, no specific therapy exists to treat flavivirus infections. A recent study shows that orthosteric and allosteric inhibitors against viral NS2B−NS3 protease provide a promise for therapeutic development17,35, 36, 37,40.
Previously, we identified EB as a potent inhibitor abolishing flavivirus NS2B−NS3 interactions, resulting in inhibition of the viral protease activity and viral replication in 2D cell culture36. In this study, we investigated the antiviral efficacy of EB using 3D organoid and in vivo mouse models. We showed that the food dye EB can effectively impair the viral replication in a 3D mini-brain organoid model and greatly improved the survival rate of mice challenged by a lethal dose of Zika virus. Our SAR studies indicated that iodine substitutions at R1 and R3 positions of the xanthene ring are essential for EB's biological activities, and chlorine substitutions are allowed on the isobenzofuran ring of EB. Overall, our results showed that EB is the most effective compound among those analogs to combat Zika infections.
4. Experimental
4.1. Compounds
4.1.1. General chemistry information
Materials and solvents in reagent grade were purchased from commercial vendors and directly used without further purification. The reference erythrosin B was also procured from Sigma−Aldrich. All reactions were performed in dry glassware with magnetic stirring under a nitrogen atmosphere. The Silica gel 60 with a particle size of 0.063–0.200 mm (70–230 mesh, flash) was used for preparative column chromatography. The F254 plates with Silica gel 60 (Merck, Darmstadt) were used for analytical thin layer chromatography. UV (254 nm) was used to visualize the developed chromatograms. A Brucker-300 (1H NMR, 300 MHz; 13C NMR, 75 MHz) spectrometer was used to record NMR spectra. TMS was used as an internal reference to record 1H and 13C NMR spectra, with chemical shifts in ppm and J values in Hz. A Shimadzu analytical HPLC system (model: CBM-20A LC-20AD SPD-20A UV/Vis) was used to determine purities of final compounds, as we described previously41.
4.1.1.1. 3′,6′-Dihydroxy-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (4, JMX0915)
Red solid. HPLC purity 95.5% (tR = 14.72 min). 1H NMR (300 MHz, DMSO-d6) δ 10.09 (s, 2H), 7.99 (d, J = 7.5, 1H), 7.79 (td, J = 7.5, 1.3 Hz, 1H), 7.71 (td, J = 7.5, 1.1 Hz, 1H), 7.26 (d, J = 7.6 Hz, 1H), 6.68 (t, J = 1.4 Hz, 2H), 6.55 (d, J = 1.4 Hz, 4H). 13C NMR (75 MHz, DMSO-d6) δ 168.7, 159.5, 152.4, 151.9, 135.5, 130.0, 129.0, 126.2, 124.6, 124.0, 112.6, 109.6, 102.2.
4.1.1.2. 4′,5′-Dichloro-3′,6′-dihydroxy-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (5, JMX0892)
Yellow solid. HPLC purity 98.0% (tR = 14.59 min). 1H NMR (300 MHz, CD3OD) δ 8.03–7.96 (m, 1H), 7.77 (td, J = 7.5, 1.3 Hz, 1H), 7.69 (td, J = 7.4, 1.1 Hz, 1H), 7.24–7.16 (m, 1H), 6.68 (d, J = 2.2 Hz, 2H), 6.61–6.48 (m, 4H). 13C NMR (75 MHz, CD3OD) δ 171.6, 161.3, 154.1, 136.5, 131.0, 130.1, 128.3, 125.8, 125.4, 113.6, 111.4, 103.6.
4.1.1.3. 2′,7′-Dichloro-3′,6′-dihydroxy-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (6, JMX0897)
Orange solid. HPLC purity 99.2% (tR = 16.48 min). 1H NMR (300 MHz, DMSO-d6) δ 11.08 (s, 2H), 8.01 (d, J = 7.4 Hz, 1H), 7.82 (t, J = 7.4 Hz, 1H), 7.74 (t, J = 7.3 Hz, 1H), 7.33 (d, J = 7.5 Hz, 1H), 6.91 (s, 2H), 6.65 (s, 2H). 13C NMR (75 MHz, DMSO-d6) δ 168.3, 155.2, 151.5, 150.1, 135.9, 130.5, 128.2, 125.9, 125.1, 124.0, 116.3, 110.5, 103.8.
4.1.1.4. Sodium 4′,5′-diiodo-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthene]-3′,6′-bis(olate) (7, JMX0903)
Red solid. HPLC purity 95.0% (tR = 16.68 min). 1H NMR (300 MHz, DMSO-d6) δ 8.10–8.01 (m, 1H), 7.53–7.39 (m, 2H), 7.12–7.03 (m, 1H), 6.58 (d, J = 9.3 Hz, 2H), 6.16 (d, J = 9.3 Hz, 2H). 13C NMR (75 MHz, DMSO-d6) δ 176.2, 169.6, 157.3, 155.4, 140.6, 133.7, 130.1, 129.8, 128.8, 128.1, 127.9, 120.2, 110.0, 79.3.
4.1.1.5. 3′,6′-Dihydroxy-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthene]-5-carboxylic acid (8, JMX0920)
Yellow solid. HPLC purity 99.1% (tR = 13.52 min). 1H NMR (300 MHz, DMSO-d6) δ 13.53 (s, 1H), 10.17 (s, 2H), 8.40 (s, 1H), 8.29 (dd, J = 8.0, 1.5 Hz, 1H), 7.39 (d, J = 8.0, 1H), 6.70 (d, J = 2.2 Hz, 2H), 6.61 (d, J = 8.6 Hz, 2H), 6.55 (dd, J = 8.7, 2.3 Hz, 2H). 13C NMR (75 MHz, DMSO-d6) δ 167.8, 166.0, 159.7, 156.1, 151.8, 136.1, 132.9, 129.2, 126.8, 125.5, 124.6, 112.7, 108.9, 102.3.
4.1.1.6. Sodium 4,5,6,7-tetrachloro-2′,4′,5′,7′-tetraiodo-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthene]-3′,6′-bis(olate) (11, JMX0902)
Dark red solid. HPLC purity 95.1% (tR = 21.08 min). 13C NMR (75 MHz, DMSO-d6) δ 171.6, 164.4, 157.3, 145.4, 143.4, 136.9, 132.6, 130.3, 128.3, 127.7, 127.2, 110.7, 96.4, 75.3.
4.1.1.7. 2′,4′,5′,7′-tetrabromo-3′,6′-dihydroxy-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (12, JMX0911)
Dark red solid. HPLC purity 95.6% (tR = 12.78 min). 1H NMR (300 MHz, DMSO-d6) δ 8.14–8.08 (m, 1H), 7.61–7.49 (m, 2H), 7.19–7.15 (m, 1H), 6.99 (s, 2H). 13C NMR (75 MHz, DMSO-d6) δ 168.9, 168.1, 157.0, 153.0, 140.4, 132.8, 130.4, 130.1, 128.8, 128.7, 128.6, 117.7, 109.7, 98.9.
4.1.1.8. 4′,5′-dibromo-3′,6′-dihydroxy-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (13, JMX0893)
Yellow solid. HPLC purity 99.3% (tR = 16.37 min). 1H NMR (300 MHz, CD3OD) δ 8.04–7.97 (m, 1H), 7.77 (td, J = 7.4, 1.4 Hz, 1H), 7.69 (td, J = 7.4, 1.1 Hz, 1H), 7.27–7.21 (m, 1H), 6.69 (d, J = 8.8 Hz, 2H), 6.59 (d, J = 8.8 Hz, 2H). 13C NMR (75 MHz, CD3OD) δ 171.0, 158.5, 150.8, 136.7, 131.3, 128.3, 128.0, 126.1, 125.4, 113.6, 112.7, 99.4.
4.1.1.9. 4′,5′-Ddibromo-3′,6′-dihydroxy-2′,7′-dinitro-3H-spiro[isobenzofuran-1,9′-xanthen]-3-one (14, JMX0910)
Yellow solid. HPLC purity 97.0% (tR = 18.44 min). 1H NMR (300 MHz, DMSO-d6) δ 8.07 (d, J = 7.1 Hz, 1H), 7.84 (td, J = 7.4, 1.4 Hz, 1H), 7.76 (td, J = 7.4, 1.2 Hz, 1H), 7.48 (d, J = 7.4 Hz, 1H), 7.14 (s, 2H). 13C NMR (75 MHz, DMSO-d6) δ 168.3, 164.3, 153.3, 137.6, 134.4, 130.9, 129.9, 127.9, 127.3, 125.7, 105.4.
4.1.1.10. 1-(3′,6′-Dihydroxy-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthen]-5-yl)-3-(2-mercaptoethyl)thiourea (17, JMX0912)
Yellow solid. HPLC purity 95.0% (tR = 14.88 min). 1H NMR (300 MHz, DMSO-d6) δ 10.05 (s, 1H), 8.83 (s, 2H), 8.29–8.20 (m, 2H), 7.75 (d, J = 8.2 Hz, 1H), 7.19 (d, J = 8.3 Hz, 1H), 6.68 (d, J = 2.2 Hz, 2H), 6.64–6.54 (m, 4H), 3.68 (q, J = 6.5 Hz, 2H), 2.73 (q, J = 7.4 Hz, 2H), 2.45 (t, J = 8.1 Hz, 1H). 13C NMR (75 MHz, DMSO-d6) δ 180.6, 168.5, 159.6, 158.6, 158.1, 152.0, 147.2, 141.2, 129.7, 129.0, 126.6, 124.1, 117.1, 113.2, 112.67, 109.8, 102.3, 46.9, 22.8.
4.2. Split luciferase complementation (SLC) assay
The SLC assay was performed using purified proteins as previously described36,39,46. All experiments described here and below were performed in triplicate, unless specified. The software suite GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA) was used to fit dose-dependent titration points using non-linear regression function to determine all inhibitory IC50/EC50/CC50 values.
4.3. Protease inhibition assay
Protease inhibition assay was carried out with the His-tagged NS2B (50 nmol/L), DENV2 NS3-MBP fusion protein (50 nmol/L), and a FRET substrate Abz-RRRRSAG-nTyr (NeoScientific, Cambridge, MA, USA) as we described previously17,35, 36, 37.
4.4. Cytotoxicity assay
A WST-8 cell proliferation assay (Enzo Life Science, Farmingdale, NY, USA) was used to determine cytotoxicity for EB and derivatives, as described previously17,35, 36, 37,47,48.
4.5. Plaque forming unit (PFU) assays
The effect of erythrosin B on ZIKV PRVABC59 was determined by a PFU viral plaque reduction assay using A549 cells [American Type Culture Collection (ATCC), Manassa, VA, USA] as described previously17,35, 36, 37,47,48. No mycoplasma contamination was detected in all cells.
4.6. Culture of 3D mini-brain organoid
Generation of the 3D mini-brain organoid mimicking forebrain was cultured as described previously35,37.
4.7. Immunofluorescence assay
The 3D mini-brain organoids at Day 20 were treated with DMSO or erythrosin B (3.0 μmol/L) as described previously35,37. At Day 21, the organoids were treated with DMSO or EB (3.0 μmol/L) and infected with ZIKV PRVABC59 at MOI of 1 or mock. MOI was estimated by estimation of cell numbers based on organoid volume. At 5 days post infection (dpi), ZIKV titer in culture supernatant was evaluated by the PFU assay. At 7 dpi, the 3D organoids were fixed and sectioned for immunohistochemistry staining, as described previously35,37. An Olympus DP71 fluorescence imaging system was used to record fluorescence images. Quantification of relative fluorescence unit (RFU) was done using the ImageJ suite49. Quantification of virus yield was carried out using the PFU assay as described above. Student t-test was used to calculate statistical significance.
4.8. Pharmacokinetics of EB
C57BL/6 (B6) mice (female, 20–22 g) were obtained from breeding stocks maintained at the University of Arizona. All mice were housed under conditions of controlled temperature (22 °C) with on-off light cycle, with food and water provided ad libitum. EB was given to mice by intraperitoneal injection (ip) at 50 mg/kg (in 50% PEG containing 10% ethanol and 10% DMSO) or by oral gavage at 100 mg/kg (in 10% Solutol HS 15 containing 10% DMSO). The University of Arizona Animal Care and Use Committee approved all animal studies.
Heparinized capillary tubes were used to collect blood samples through the tail vein at various time as indicated in the figure legends after dosing. Plasma was prepared and stored at −30 °C until analysis. Plasma sample (5 μL) was mixed with 30 μL methanol and 10 μL of internal standard (IS; cholic acid-d4 1 μg/mL), then diluted with 760 μL water before loaded onto an ISOLUTE® C18 SPE Columns (1 mL/100 mg, Biotage, Salem, NH, USA). After washed with 1 mL water, the analytes were eluted from the cartridges with 1 mL methanol, dried with nitrogen, and reconstituted in 100 μL methanol prior to LC–MS/MS analysis.
LC–MS/MS was used to detect EB. The LC–MS system is composed of a Sciex Qtrap6500+ Mass Spectrometer (AB SCIEX, Framingham, MA, USA) and an Agilent 1290 UPLC system (Agilent Technologies, Santa Clara, CA, USA). An ACQUITY C18 column (2.1 mm × 100 mm, 1.7 μm, Waters) was used to separate analytes at temperature of 35 °C, with mobile phase A containing 10 mmol/lammonium acetate in water and mobile phase B containing 10 mmol/L ammonium acetate in acetonitrile. Elution was programed as follows: 10% B (0–0.5 min), 10% B→90% B (0.5–0.6 min), 90% B (0.6–3 min), 90% B→10% B (3–3.1 min), 10% B (3.1–5.5 min), with a flow rate of 0.2 mL/min. The MS was operated in the negative ion mode, using electrospray ionization. The ion spray temperature and voltage were set at 500 °C and 4500 V, respectively. Ion source gas 1, 2 and curtain gas were set at 25, 25, 20 psi, respectively. EB and the internal standard (IS, cholic acid-d4) were detected using Multiple Reaction Monitoring (MRM), with a dwell time of 150 msec per transition, at m/z 834.5/662.8 and 411.0/411.0, respectively. For quantitative analysis of erythrosine B, standards (5–1000 ng/mL in 10 μL methanol), along with 10 μL IS (at 1 μg/mL in methanol), were added to 5 μL of blank mouse plasma to construct the calibration curve.
PK solver (Microsoft, Redmond, WA, USA) was used to calculate the pharmacokinetic parameters by assuming a noncompartmental model.
4.9. In vivo protection efficacy
As we described previously17, a ZIKV survival animal model was used to evaluate the in vivo antiviral activity of EB, which was approve by the Wadsworth Center IACUC and Institutional Biosafety Committees.
1.7 × 104 or 1.7 × 105 PFU of the PRVABC59 strain was administered to a group of four-week-old A129 male (M) and female (F) mice by subcutaneous injection, respectively. Then, the infected mice were treated with EB at 200 mg/kg [n = 12 (M); n = 15 (F)] or 400 mg/kg [n = 4 (M)] of body weight or with vehicle control [n = 9 (M), or n = 15 (F)] every day through oral gavage for 7 consecutive days post-infection (dpi). Mice were examined daily for signs of illness and mortality for 14 days. The Log-rank test was used to compare survival curves.
Acknowledgments
This study was partially supported by grants AI131669, AI140726, and AI141178 from the National Institute of Allergy and Infectious Diseases (NIAID, USA), the National Institutes of Health (Hongmin Li and Jia Zhou). Additionally, Jia Zhou is partly supported by the John D. Stobo, M.D. Distinguished Chair Endowment Fund at UTMB. Hongmin Li is additionally supported by NIH grants AI133219, AI134568, AI140406, and AI140491, USA, and by the R. Ken and Donna Coit Endowed Chair fund in Drug Discovery.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.10.017.
Author contributions
Zhong Li, Jimin Xu, David Butler, Yongcheng Song, Qing-Yu Zhang, Jia Zhou and Hongmin Li conceived the concept of the study. Zhong Li, Jimin Xu, Yuekun Lang, Xiangmeng Wu, Saiyang Hu, Subodh Kumar Samrat, and Anil M. Tharappel performed the experiments. Zhong Li, Jimin Xu, Xiangmeng Wu, Yongcheng Song, Qing-Yu Zhang, Jia Zhou and Hongmin Li wrote the manuscript.
Conflicts of interest
The authors declare that they have no potential conflicts of interest involving the contents of this article.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Brecher M., Zhang J., Li H. The flavivirus protease as a target for drug discovery. Virol Sin. 2013;28:326–336. doi: 10.1007/s12250-013-3390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvet G., Aguiar R.S., Melo A.S., Sampaio S.A., de Filippis I., Fabri A., et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 3.Thomas D.L., Sharp T.M., Torres J., Armstrong P.A., Munoz-Jordan J., Ryff K.R., et al. Local transmission of Zika virus—Puerto Rico, November 23, 2015−January 28, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:154–158. doi: 10.15585/mmwr.mm6506e2. [DOI] [PubMed] [Google Scholar]
- 4.Martines R.B., Bhatnagar J., Keating M.K., Silva-Flannery L., Muehlenbachs A., Gary J., et al. Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses —Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:159–160. doi: 10.15585/mmwr.mm6506e1. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Morales A.J. Zika and microcephaly in Latin America: an emerging threat for pregnant travelers?. Trav Med Infect Dis. 2016;14:5–6. doi: 10.1016/j.tmaid.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 7.WHO Dengue and severe dengue. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue Available from:
- 8.Hadinegoro S.R., Arredondo-Garcia J.L., Capeding M.R., Deseda C., Chotpitayasunondh T., Dietze R., et al. Efficacy and long-term safety of a Dengue vaccine in regions of endemic disease. N Engl J Med. 2015;373:1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 9.Aguiar M., Stollenwerk N., Halstead S.B. The impact of the newly licensed Dengue vaccine in endemic countries. PLoS Neglected Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson N.M., Rodriguez-Barraquer I., Dorigatti I., Mier Y.T.-R.L., Laydon D.J., Cummings D.A. Benefits and risks of the Sanofi-Pasteur dengue vaccine: modeling optimal deployment. Science. 2016;353:1033–1036. doi: 10.1126/science.aaf9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 12.Warrener P., Tamura J.K., Collett M.S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 14.Bartelma G., Padmanabhan R. Expression, purification, and characterization of the RNA 5'-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002;299:122. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- 15.Erbel P., Schiering N., D'Arcy A., Renatus M., Kroemer M., Lim S., et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 16.Brecher M., Li Z., Liu B., Zhang J., Koetzner C.A., Alifarag A., et al. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B−NS3 protease. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Brecher M., Zhang J., Sakamuru S., Liu B., Huang R., et al. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B−NS3 interaction. Cell Res. 2017;27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prusis P., Lapins M., Yahorava S., Petrovska R., Niyomrattanakit P., Katzenmeier G., et al. Proteochemometrics analysis of substrate interactions with dengue virus NS3 proteases. Bioorg Med Chem. 2008;16:9369–9377. doi: 10.1016/j.bmc.2008.08.081. [DOI] [PubMed] [Google Scholar]
- 19.Chambers T.J., Grakoui A., Rice C.M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falgout B., Pethel M., Zhang Y.M., Lai C.J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lescar J., Luo D., Xu T., Sampath A., Lim S.P., Canard B., et al. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antivir Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Noble C.G., Seh C.C., Chao A.T., Shi P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J Virol. 2012;86:438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aleshin A., Shiryaev S., Strongin A., Liddington R. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assenberg R., Mastrangelo E., Walter T.S., Verma A., Milani M., Owens R.J., et al. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandramouli S., Joseph J.S., Daudenarde S., Gatchalian J., Cornillez-Ty C., Kuhn P. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol. 2010;84:3059–3067. doi: 10.1128/JVI.02044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammamy M.Z., Haase C., Hammami M., Hilgenfeld R., Steinmetzer T. Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B−NS3 protease. ChemMedChem. 2013;8:231–241. doi: 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- 27.Luo D., Wei N., Doan D.N., Paradkar P.N., Chong Y., Davidson A.D., et al. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo D., Xu T., Hunke C., Gruber G., Vasudevan S.G., Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo D., Xu T., Watson R.P., Scherer-Becker D., Sampath A., Jahnke W., et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robin G., Chappell K., Stoermer M.J., Hu S.H., Young P.R., Fairlie D.P., et al. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J Mol Biol. 2009;385:1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 31.Lei J., Hansen G., Nitsche C., Klein C.D., Zhang L., Hilgenfeld R. Crystal structure of Zika virus NS2B−NS3 protease in complex with a boronate inhibitor. Science. 2016;353:503–505. doi: 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Li Y., Loh Y.R., Phoo W.W., Hung A.W., Kang C., et al. Crystal structure of unlinked NS2B−NS3 protease from Zika virus. Science. 2016;354:1597–1600. doi: 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]
- 33.Phoo W.W., Li Y., Zhang Z., Lee M.Y., Loh Y.R., Tan Y.B., et al. Structure of the NS2B−NS3 protease from Zika virus after self-cleavage. Nat Commun. 2016;7:13410. doi: 10.1038/ncomms13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang C., Keller T.H., Luo D. Zika virus protease: an antiviral drug target. Trends Microbiol. 2017;25:797–808. doi: 10.1016/j.tim.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Li Z., Lang Y., Sakamuru S., Samrat S., Trudeau N., Kuo L., et al. Methylene blue is a potent and broad-spectrum inhibitor against Zika virus in vitro and in vivo. Emerg Microbes Infect. 2020;9:2404–2416. doi: 10.1080/22221751.2020.1838954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., Sakamuru S., Huang R., Brecher M., Koetzner C.A., Zhang J., et al. Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B−NS3 protease. Antivir Res. 2018;150:217–225. doi: 10.1016/j.antiviral.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Xu J., Lang Y., Fan X., Kuo L., D'Brant L., et al. JMX0207, a niclosamide derivative with improved pharmacokinetics, suppresses Zika virus infection both in vitro and in vivo. ACS Infect Dis. 2020;6:2616–2628. doi: 10.1021/acsinfecdis.0c00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Zhang J., Li H. In: Viral proteases and their inhibitors. Gupta S.P., edtor, editors. Elsevier; San Diego, CA: 2017. Flavivirus NS2B/NS3 Protease: structure, function, and inhibition; pp. 163–188. [Google Scholar]
- 39.Lang Y., Li Z., Li H. Analysis of protein−protein interactions by split luciferase complementation assay. Curr Protoc Toxicol. 2019;82:e90. doi: 10.1002/cptx.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Y., Huo T., Lin Y.L., Nie S., Wu F., Hua Y., et al. Discovery, X-ray crystallography and antiviral activity of allosteric inhibitors of Flavivirus NS2B−NS3 protease. J Am Chem Soc. 2019;141:6832–6836. doi: 10.1021/jacs.9b02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudra J.S., Ding Y., Neelakantan H., Ding C., Appavu R., Stutz S., et al. Suppression of cocaine-evoked hyperactivity by self-adjuvanting and multivalent peptide nanofiber vaccines. ACS Chem Neurosci. 2016;7:546–552. doi: 10.1021/acschemneuro.5b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benkovics G., Afonso D., Darcsi A., Beni S., Conoci S., Fenyvesi E., et al. Novel beta-cyclodextrin-eosin conjugates. Beilstein J Org Chem. 2017;13:543–551. doi: 10.3762/bjoc.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomorski A., Adamczyk J., Bishop A.C., Krezel A. Probing the target-specific inhibition of sensitized protein tyrosine phosphatases with biarsenical probes. Org Biomol Chem. 2015;13:1395–1403. doi: 10.1039/c4ob02256d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietrancosta N., Kessler A., Favre-Besse F.C., Triballeau N., Quentin T., Giros B., et al. Rose bengal analogs and vesicular glutamate transporters (VGLUTs) Bioorg Med Chem. 2010;18:6922–6933. doi: 10.1016/j.bmc.2010.06.069. [DOI] [PubMed] [Google Scholar]
- 45.Park S., Foote P.K., Krist D.T., Rice S.E., Statsyuk A.V. UbMES and UbFluor: novel probes for ring-between-ring (RBR) E3 ubiquitin ligase PARKIN. J Biol Chem. 2017;292:16539–16553. doi: 10.1074/jbc.M116.773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z., Brecher M., Zhang J., Sakamuru S., Liu B., Huang R., et al. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B−NS3 interaction. Cell Res. 2017;27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H., Liu L., Jones S.A., Banavali N., Kass J., Li Z., et al. Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antivir Res. 2013;97:232–239. doi: 10.1016/j.antiviral.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H., Zhou B., Brecher M., Banavali N., Jones S.A., Li Z., et al. S-Adenosyl-homocysteine is a weakly bound inhibitor for a flaviviral methyltransferase. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.