Abstract
The p53 tumor suppressor is a transcription factor with roles in cell development, apoptosis, oncogenesis, aging, and homeostasis in response to stresses and infections. p53 is tightly regulated by the MDM2 E3 ubiquitin ligase. The p53–MDM2 pathway has coevolved, with MDM2 remaining largely conserved, whereas the TP53 gene morphed into various isoforms. Studies on prevertebrate ancestral homologs revealed the transition from an environmentally induced mechanism activating p53 to a tightly regulated system involving cell signaling. The evolution of this mechanism depends on structural changes in the interacting protein motifs. Elephants such as Loxodonta africana constitute ideal models to investigate this coevolution as they are large and long-living as well as having 20 copies of TP53 isoformic sequences expressing a variety of BOX-I MDM2-binding motifs. Collectively, these isoforms would enhance sensitivity to cellular stresses, such as DNA damage, presumably accounting for strong cancer defenses and other adaptations favoring healthy aging. Here we investigate the molecular evolution of the p53–MDM2 system by combining in silico modeling and in vitro assays to explore structural and functional aspects of p53 isoforms retaining the MDM2 interaction, whereas forming distinct pools of cell signaling. The methodology used demonstrates, for the first time that in silico docking simulations can be used to explore functional aspects of elephant p53 isoforms. Our observations elucidate structural and mechanistic aspects of p53 regulation, facilitate understanding of complex cell signaling, and suggest testable hypotheses of p53 evolution referencing Peto’s Paradox.
Keywords: structural variations, model, p53 retrogenes, molecular evolution, Loxodonta africana, lifespan, Peto’s Paradox, intrinsic specificity
Introduction
The p53 family, including p53, p63, and p73, takes up roles in cancer, neurodegeneration, development, inflammation and tissue regeneration, cellular senescence, aging (Chusyd et al. 2021), and stress-induced mechanisms (Anbarasan and Bourdon 2019; Fujita 2019; Beck et al. 2020). P53 family members are suggested to modulate gene expression in both a full-length p53-dependent and -independent manner. Importantly, p53 isoforms with specific activities have been shown to form homo- or hetero-oligomers, providing an additional level of differentiation (Fujita 2019). The p53 transcription factor targets DNA sequences through p53-responsive elements (p53Res) (Bourdon et al. 1997) and coregulates the transactivation of several genes, promoting cell cycle arrest or cellular apoptosis (Lane and Levine 2010), thus coordinating cellular responses (Haronikova et al. 2019). The question arises as of how p53 activity is differentiated in response to the requirements of a cell and, of special interest to humans, how this prevents the development of cancers (Fahraeus and Olivares-Illana 2014; Haronikova et al. 2019; Karakostis and Fahraeus 2019). This important topic can be addressed by tracking the molecular evolution of p53 structures and interaction interfaces.
The p53 gene has evolved from an ancestral p53/p63/p73 gene, which gave rise to three p53-related genes, with distinct roles in mammals (Belyi et al. 2010; Rutkowski et al. 2010; Karakostis et al. 2016; Siau et al. 2016; Hendler et al. 2021). The tp53 transcription factor is a tumor suppressor with key roles in oncogenesis (Mantovani et al. 2019; Salomao et al. 2021). It involves two N-terminal transactivation domains (TAD I and TAD II), a DNA-binding domain (DBD), and a C-terminal oligomerization domain (Wallace et al. 2006; Logotheti et al. 2010). Thirteen isoforms have been shown to be produced through alternative mRNA translation initiation (p53/47) or splicing (full-length p53 or FLp53, p53β, p53γ, Δ40p53α, Δ40p53β, Δ40p53γ, Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, Δ160p53γ) (Bourdon et al. 1997, 2005; Marcel et al. 2010, 2011). The full-length p53 includes the TAD I, which is required for the induction of p53 target genes and proapoptotic factors (p21CDKN1A, Bax, Puma, Noxa of the Bcl-2 family), leading to cell cycle arrest (G1) or apoptosis. The p53/47 isoform at the second in frame AUG (+120), lacking the TAD I but retaining TAD II, is activated during Unfolded Protein Response, following stress to the endoplasmic reticulum, to lead to G2/M arrest (Haronikova et al. 2019). This isoform lacks the BOX-I MDM2-binding epitope (found in the TAD I). Another isoform, the Δ133p53, lacking the first 132 amino acids, corresponding to the TAD I, TAD II, and the first 30 residues of the DBD, is abundant in early passage normal human fibroblasts and decreases in late passage and senescent cells (Fujita 2019). Indeed, these isoforms have various MDM2-binding capacities and are expressed in a stress-dependent fashion, contributing to responses by transactivating downstream pathways (Rozan and El-Deiry 2007; Allen et al. 2014; Chen 2016; Haronikova et al. 2019; Carlsen and El-Deiry 2021). It has been suggested that a quantitatively regulated coexpression of these distinct isoforms is required during cell proliferation (Khoury and Bourdon 2011) and that the expression of different p53 isoforms is induced in response to DNA damage agents or stress to the endoplasmic reticulum (Chen et al. 2009).
In addition, numerous cellular and animal model studies indicate that an unbalance in p53 isoform expression causes cancer, premature aging, and degenerative diseases (Fujita 2019). An abnormal expression of p53 isoforms, or mis-regulation of posttranslational modifications (PTMs) mediating the interactions of p53 pathway, may be induced by mutations leading to carcinogenesis (Khoury and Bourdon 2011; Karakostis et al. 2019; Salomao et al. 2021). Importantly, experiments on mice have shown that extra copies of the p53 gene can be introduced to generate “super p53” animals carrying three or four functional copies of p53, accurately reproducing the behavior of the endogenous gene but with an enhanced response to DNA damage (Garcia-Cao et al. 2002; Moding et al. 2016). As p53 appears to regulate aging and longevity in a context-dependent manner (Feng et al. 2011; Wu and Prives 2018; Fujita 2019), mice with accelerated aging show chronic p53 activation. This suggests that the p53–MDM2 axis leads to elevated p53 activity caused by either early aging or delayed-onset aging (Wu and Prives 2018).
An exciting study discovered that elephants have 20 copies, that is, 40 alleles, of the TP53 genes, compared with the typical number of one copy found (so far) in all other mammals (Sulak et al. 2016). According to the Peto’s Paradox referring to why larger animals with a higher number of cells and cell divisions do not also have a higher cancer incidence (Sulak et al. 2016; Tollis et al. 2017, 2020; Callier 2019), the elephant’s multiple copies are proposed to have evolved in order to defuse enhanced DNA damage response as a way to promote cancer suppression. Despite the large body size and long-life span, elephants exhibit a high resistance to cancer as cancer mortality is estimated to be <5%, as compared with humans reaching up to 25% (Abegglen et al. 2015; Gaughran et al. 2016). During evolution, elephant retrogene p53 sequences have evolved from an ancestral sequence, whereas the initial TP53 retrogene amplification may predate extant elephant species. The closest relative (with sequenced genome) of elephants, the hyrax (Procavia capensis), only contains one copy of TP53 (Abegglen et al. 2015), whereas gene amplification apparently rapidly expanded in Proboscideans in parallel with increased body size and life span (Sulak et al. 2016). It appears that this event may result to an induction of cell death at lower levels of DNA damage, compared with phylogenetically close relatives, like the African rock hyrax, the East African aardvark, and the American southern three-banded armadillo (Callier 2019). However, other long-living large-sized mammals, like the bowhead whale (Balaena mysticetus), have not evolved extra copies of any tumor suppressor gene (Yim et al. 2014; Keane et al. 2015). The variability or truncation of the elephant p53 isoformic sequences (i.e., p53 isoforms of partial length, corresponding to the N′ domains of the canonical p53 full-length sequence), indicates an enhanced functional diversity and the formation of diverse pools of functional p53 proteins. This would increase distinct structural characteristics conferring specialized interacting interfaces for key functional proteins such as MDM2 and downstream activating proteins. In addition to p53 variations, it has been hypothesized that an accumulation of differentially expressed, modified, and activated (by PTMs) p53 pools, collectively or synergistically regulate the response to specific stresses in the cell (Khoury and Bourdon 2011). These PTMs are suggested to have evolved as a result of coevolutionary cues of the p53–MDM2 interactions during the transition from (1) an ancestral early branching invertebrate system (prevertebrate) (Barreira et al. 2021) to (2) a derived mammalian vertebrate system (Karakostis et al. 2016; Karakostis and Fahraeus 2019).
The elephant’s genetic set-up thus innately confers functional diversity to p53-dependant mechanisms contributing significantly to valuable protein fine tuning. This is in stark contrast to the human set-up where one gene with alternatively activated isoforms is expressed in response to a stress. The existence of multiple p53 forms in elephants, with potentially different activities, provides an ideal model for understanding how p53 activities are executed and regulated. Indeed, substitutions made by retrogene isoforms altering the p53 DBD and its functions, have been investigated and associated with longevity in species lifespan (Bartas et al. 2021). Important for an understanding of P53 diversity in elephants, Sulak et al. (2016) and Haupt and Haupt (2017) investigated the roles of retrogene isoforms (TP53RTGs) forming a “pool of protected p53” thus acting as contributors to an enhanced sensitivity of elephant cells to DNA damage. The two models (the “guardian” or the “decoy” model) explored the potential roles of the TP53RTGs in facilitating the stabilization of the canonical p53, by either participating on the dimerization with canonical p53, or by acting as antagonists, competing for the MDM2 interaction (Sulak et al. 2016; Haupt and Haupt 2017).
Here, we explore the elephants apparently unique system further by focusing on the p53–MDM2 interaction. We consider this interaction important as p53 is primarily regulated by MDM2 and that their interaction depends on the BOX I motif (residing in the TAD I) (Gajjar et al. 2012; Karakostis et al. 2019), which is intrinsically disordered in turn implying greater likelihood of evolutionary changes (Brown et al. 2010). This suggests that the variations identified on the elephant BOX I sequences potentially modify the binding epitope of MDM2 and thus alter the expression levels and activation of p53 (Wasylyk et al. 1999; Naski et al. 2009; Buyukpinarbasili et al. 2016). For our investigation, we focus on the BOX-I sequences of the p53 isoforms in order to test the hypothesis that p53 isoforms induce distinct pools of p53 proteins with variations on the epitopes interacting with MDM2. This has significant implications for functional diversity and the integration of diverse signaling outcomes. Our observations are discussed and compared with previous models, exploring the diverse roles of elephant p53 isoforms (Sulak et al. 2016). We also explore potential translational insights derived from the molecular evolution (interspecies) of the p53–MDM2 axis and the evolved structural modules.
Results
Modeling and experimental results show that elephant p53 isoformic putative proteins exhibit a spectrum of reduced docking capacity to MDM2, strongly indicating a range of an isoform-specific downstream p53 functionality.
Elephant p53 Isoforms Exhibit Diverse Docking Association to MDM2
In this work, we identified p53 sequences in Loxodonta africana, and grouped them based on their MDM2-binding motifs (p53 BOX-I), found at the N′ terminal and within the TAD I. The “XP_010594888.1” (cellular tumor antigen p53 isoform X2 [L. africana]) was assigned as “Type A.” In addition, our findings identified the “XP_003416950” (cellular tumor antigen p53 isoform X1 [L. africana]), long as the canonical p53 sequence and another sequence of 519 aa. It contains an additional 5′ coding sequence upstream of the TAD1 that could have emerged via splicing. This sequence is:
N′MFSINSTLAALVCRTSPPQNPGSLRSLLFHSLSASPLPTGKLLALTCHGDCPALCQKPRGGCWDWEFPFPCAHTGAKSFQLFKSPKPPSWLQLAAGLWRYLVSGLGPCFQGRLHARLRFGGQIALPGAA-C′. Furthermore, a BLAST search has shown a similarity of ∼50% for two other sequences, encoding homologous sequences to p63 (XP_023407389.1 and XP_023407390.1) and p73 (XP_010591460.1) proteins (Lane and Levine 2010).
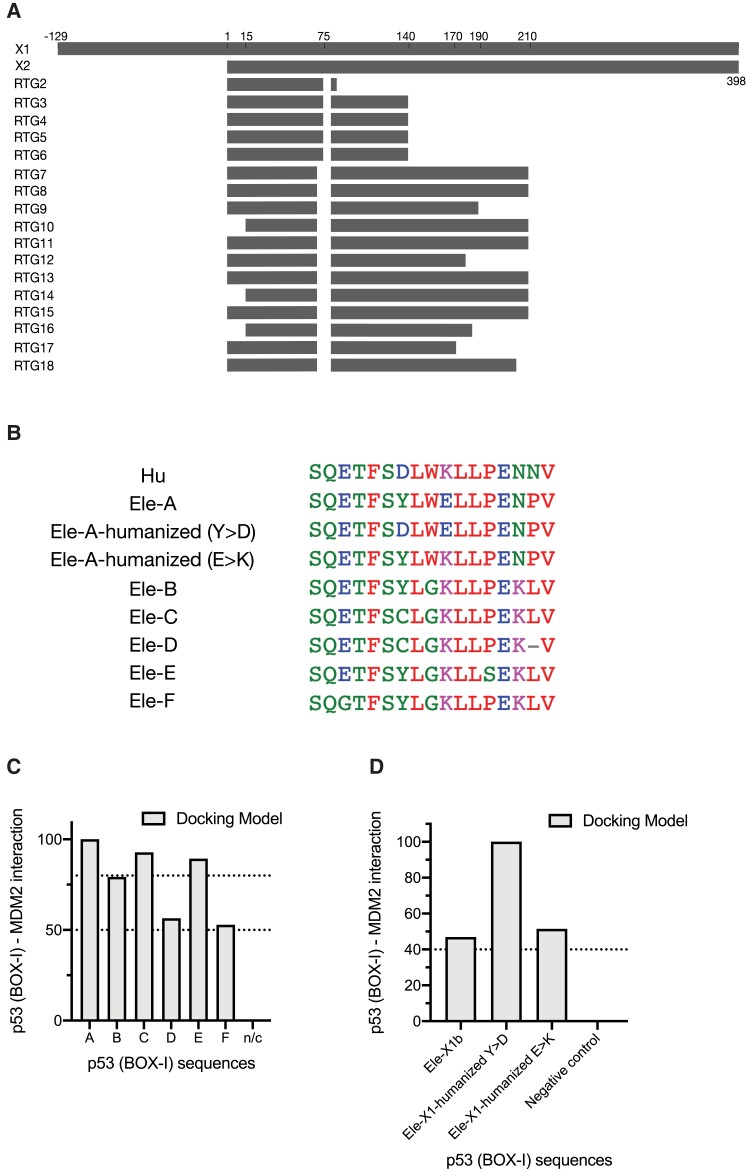
Nineteen sequences were identified and used for the analysis. The retrogene sequences were identified (supplementary tables S1 and S2, Supplementary Material online) and their sequence alignment and phylogenetic relation to the canonical elephant p53 were exposed (supplementary figs. S1 and S2, Supplementary Material online). These alignments were used for the preparation of a graphical illustration representing the putative corresponding topology of each retrogene on the full-length elephant TP53 (fig. 1A). In order to reveal which p53 isoforms might potentially bind MDM2, we first performed a bioinformatic analysis to identify sequence variations on the encoded BOX-I peptides of each isoform. Comparative analysis of the BOX-I sequences from L. africana and the human p53, showed that the BOX-I is conserved in all isoforms (fig. 1B, supplementary figs. S1 and S2 and table S1, Supplementary Material online). However, within these isoforms, the analysis indicated six types of identical sequences, corresponding to BOX-I motifs (supplementary table S1, Supplementary Material online). The generally highly conserved MDM2-interacting BOX-I motif FxxxWxxL is well conserved in only one of the sequences (elephant X1, Type A). The retro gene sequences 2, 5, 6, 7, 8, 9, 11, 13, and 17 (Type B) carry an FxxxGxxL domain. In addition, more variations are present in retrogene 3 (Type C), retrogene 4 (Type D), retrogene 10 (Type E), and retrogenes 12, 14, 15, 16, and 18 (Type F), potentially altering the structures and the binding affinity to MDM2 (fig. 1B, supplementary table S1, Supplementary Material online). Each BOX-I Type was linked to corresponding ENSEMBL references for each p53 sequence, to facilitate a comparative study with previous findings (Sulak et al. 2016) (supplementary table S3, Supplementary Material online). It is noted that ENSLAFG00000025553 (TP53RTG1) and ENSLAFG00000032258 (TP53RTG19) are annotated by ENSEMBL as pseudogenes. For assessing the interaction capacity of each putative protein, the PDB model 1YCR, deriving crystallographic data, was used (Kussie et al. 1996). The 1YCR structure illustrates human MDM2 bound to the p53 TAD. According to this structure, the amino acid sites F, W, and L are positioned to the interphase of MDM2, mediating the interaction. Based on this structure, each type of BOX-I sequence was modeled and the interaction interphases were calculated (fig. 1C). Type A exhibits the highest docking capacity, whereas B, C, and E exhibit a comparatively decreased docking capacity. Types D and F exhibit a poor docking capacity (fig. 1C, supplementary table S1, Supplementary Material online). All the compared sequences carry the FxxxxxxL motif; however, the W to G variation is only present in the sequences exhibiting a decreased docking capacity. These results indicate that the FxxxWxxL motif on the Box-I of p53, increases the binding capacity to MDM2, and variations induce changes on the conformation and the positioning of the BOX-I motif of p53 at the interphase with MDM2.
Fig. 1.
Docking models of the MDM2–p53 N′ terminal (BOX-I) interaction, with calculated GBVI/WSA dG (–kcal/mol) values. (A) Graphical mapping of the topography of each elephant p53 isoformic sequence on the canonical p53. (B) Alignment of the homologous BOX-I sequences from Loxodonta africana and human p53. (C) Docking model of the interaction of MDM2 with different elephant p53 sequences: Type A: X1, Type B: 2, 5, 6, 7, 8, 9, 11, 13, and 17; Type C: retrogene 3; Type D: retrogene 4; Type E: retrogene 10; and Type F: retrogenes 12, 14, 15, 16, and 18. (C) Docking model of the interaction of MDM2 with mutated Type A elephant p53: humanized Y > D or E > K. The humanized elephant p53 sequences exhibit an increased docking capacity, compared with the wt elephant p53 X1 (Type A). To comparatively illustrate the interaction capacity among the Types (C) and the effect of introduced mutations on the full-length (D), the binding affinity values were normalized, setting the highest value at 100. In panel (C), Type A has the highest value and all the measurements were normalized after setting it at 100. In addition, for panel (D), the humanized E > K mutation had the highest value and was set at 100. These results show that variations found in the isoforms, as well as single variants, like the Y > D, strongly effect on the interaction of p53 with MDM2, by modifying the docking interfaces.
In order to explore additional resides modifying the docking capacity, we introduced two mutations, simulating the human BOX-I orthologue, Y to D and E to K. These mutations lead to an increased docking capacity compared with the elephant Type A p53 (fig. 1D, supplementary table S1, Supplementary Material online), indicating that these residues, positioned in between the crucial residues F, W, and L, play a crucial role in the dynamics or the positioning of the BOX-I motif on the MDM2 cleft.
Modeling Six Elephant p53 BOX-I Sequences Interacting with MDM2, Reveals Variable Stereochemistry
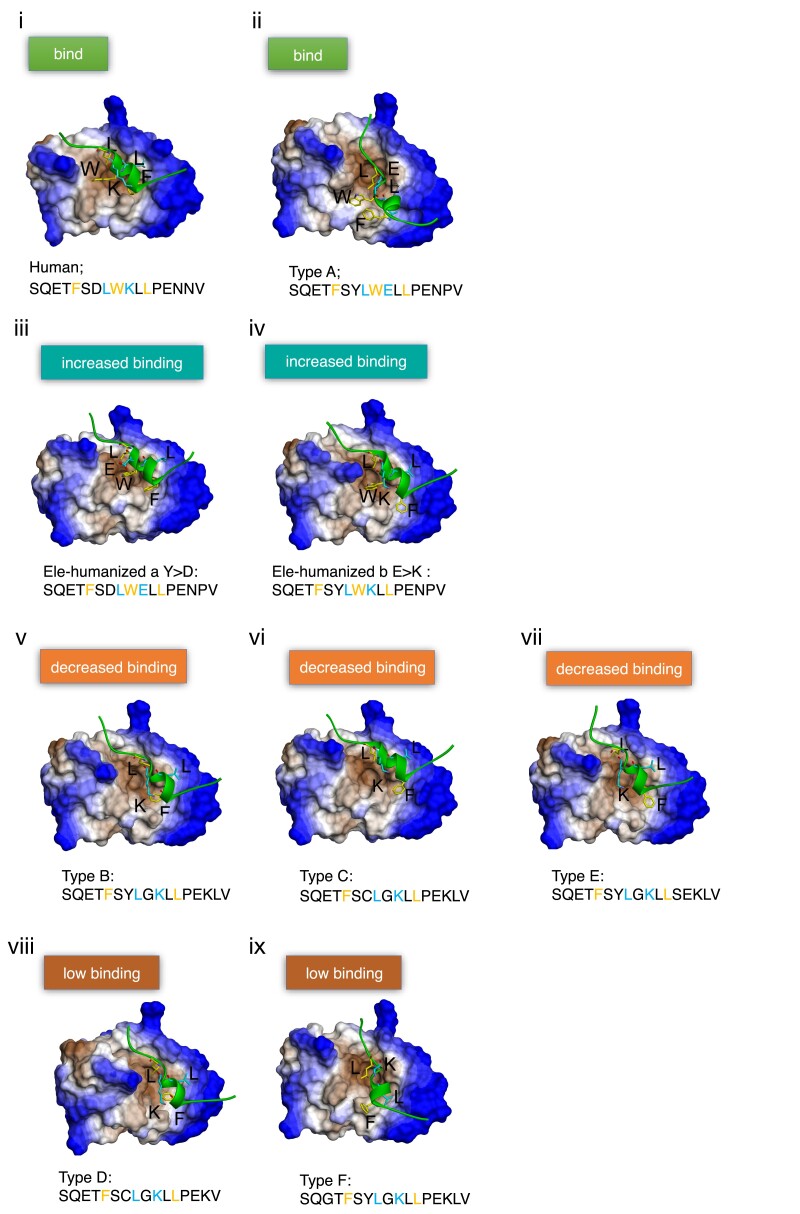
In line with the calculated binding affinity (kcal/mol; Generalized Born/Volume Integral [GBVI]/WSA dG) values of the docking models, the visualization of each model revealed the crucial role of the positioning of the F, W/G, L residues at the MDM2 cleft (fig. 2). Indeed, the human sequence (fig. 2i), as well as the earlier mentioned cross-species mutations of humanized elephant p53 Type A (fig. 2ii and iii), leading to an increased docking capacity. This demonstrates the crucial role of the positioning of the F, W, and L residues extended toward separate directions within the elephant MDM2 cleft. Similarly, the elephant sequence Type A and the above mentioned cross-species mutations Y > D and E > K, show an increased docking efficiency (fig. 2ii and iii).
Fig. 2.
Illustration of the MDM2–p53 peptides docking models, explaining the association of each p53 peptide to MDM2, in human (i) and elephant (iii–ix). The hydrophobicity is visualized in blue for hydrophilic and brown for hydrophobic. Yellow represents the binding pattern FxxxWxxL. Blue letters represent residues that may stabilize the peptide when binding MDM2.
According to the models, sequences of decreased docking capacity due to variations on the BOX-I sequence, exhibit a limited exposure of these residues within the hydrophobic cleft of MDM2. Interestingly, the W > G variation, decreased docking in the isoforms Types B–F (fig. 2v–ix). In addition, the positioning of the Types D and F approaching the MDM2 cleft, was comparatively poor. These results together point toward a model whereby elephant p53 putative proteins may play a fine-tuning regulative role by exhibiting a spectrum of docking capacity on MDM2.
Biochemical Data Confirm the Structural Interface Models
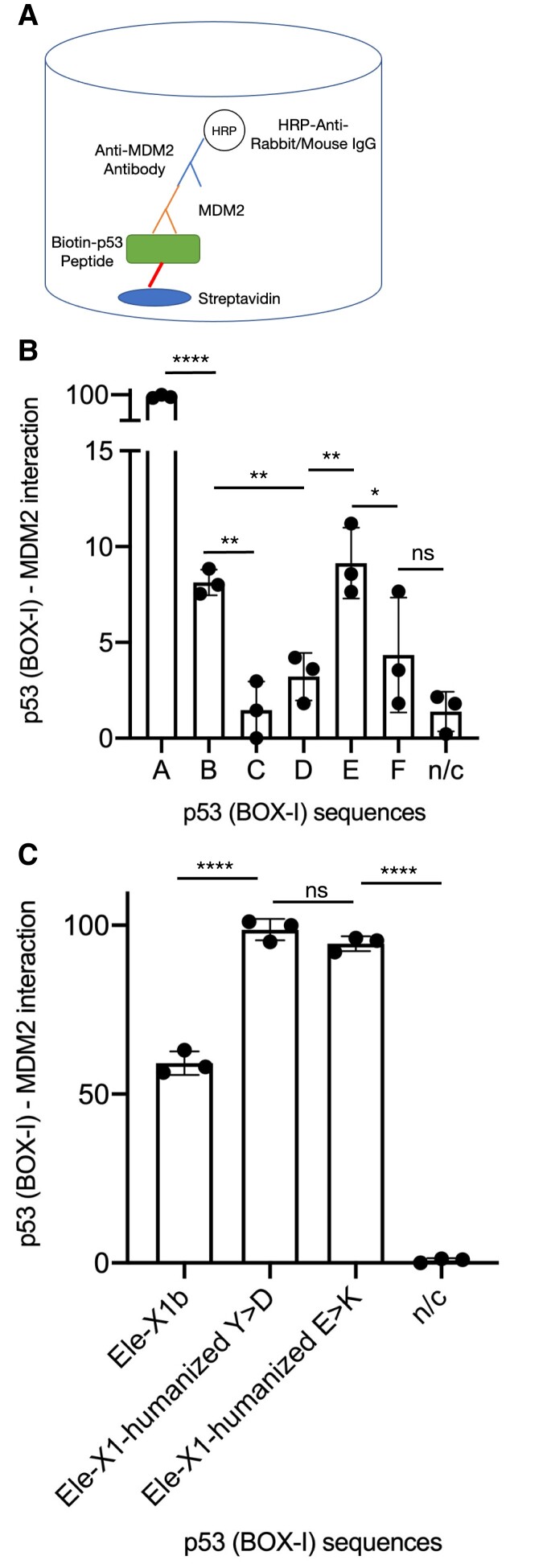
Our docking model simulations were further tested in vitro by Sandwich ELISA, using recombinant human MDM2 protein (fig. 3A). The experimental results are in line with the bioinformatic predictive models, showing an increased binding of the elephant p53, Type A, followed by the Types B and E, whereas the Types D and F showed decreased binding (fig. 3B). These results indicate that MDM2 structure is well conserved in human and elephant, and that the cross-species proteins retain their binding relationships. However, the elephant p53 Type C did not interact with recombinant human MDM2. In order to uncover why this result is not in line with the bioinformatic docking model, we performed another docking calculation in order to compare the docking of this isoform to human and elephant MDM2 proteins (supplementary fig. 3, Supplementary Material online). It emerged that the Type C isoform (Y > C in the elephant and D > C compared with the human) (fig. 1B) shows a high docking efficiency to elephant MDM2, but a decreased one to human MDM2.
Fig. 3.
Interaction of MDM2–p53 (protein–peptide) carried out by Sandwich ELISA. These results are in line with the docking models. The statistical significance was calculated by t-test (two-tailed) and the P-values are indicated as follows: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***), and P ≤ 0.0001 (****). Unpaired test results are indicated on the graph. Nonsignificant measurements are indicated by “ns” and negative controls as: “n/c.” The y-axis is the normalized (%) values derived from the ELISA measurements, which indicate the interaction of MDM2 with each of the p53 peptides. The y-axis represents the “p53 (BOX-I)–MDM2 interaction” and the bars show the standard deviation of the measurements. The y-axis in (B) is divided in two segments, for illustrative purposes. (A) Scheme illustrating the set-up of the sandwich ELISA. Biotinylated p53 peptides are fixed on streptavidin-coated plates. An anti-MDM2 antibody tagged with HRP and a secondary anti-IgG were used. (B) Elephant (Loxodonta africana) p53 peptides Types: A–F exhibit a variable capacity to bind MDM2. (C) The interaction of MDM2 with humanized Type A elephant p53: Y > D or E > K, is increased compared with the wt Type A. Experiments were tested in three technical replicates (see supplementary table S4, Supplementary Material online).
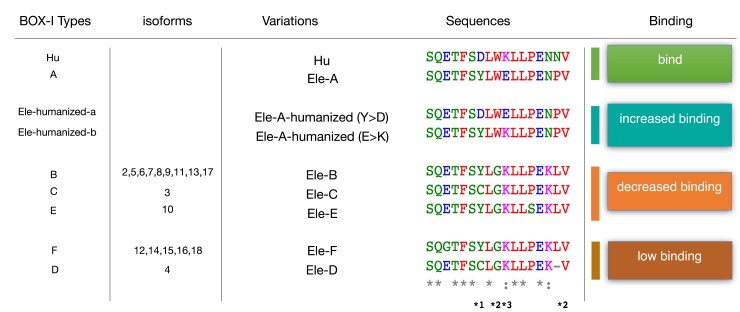
This observation is in line with figure 1C (docking model using elephant MDM2) and figure 3B (ELISA using human MDM2). In addition, the mutations Y to D and E to K, simulating the human orthologues, exhibited an increased binding capacity, in line with the models (figs. 1D and 3C). These results collectively demonstrate that peptides encoded by human p53 and elephant isoforms carrying the conserved residue corresponding W23 in human p53 have an increased MDM2-binding affinity, whereas peptides carrying the W > G variation show decreased binding activity with MDM2 (fig. 4). Indeed, the W > G and P > L/-variations prevent binding, whereas the Y > D or the E > K variations increase binding.
Fig. 4.
Summary of human and elephant p53 sequences, grouping the different BOX-I domains into six types (A–F) illustrating their capacity to interact/dock with MDM2. *1: The Y > D increases binding; *2: W > G and P > L/- prevent binding; *3: The E > K increases binding, depending on additional variations.
We conclude that a group of the p53 isoforms putatively expressed in the elephant L. africana retain the capacity to find to MDM2, thus forming distinct pools of variable (gradually decreasing) binding affinity. Isoforms with the W residue are expected to be regulated by MDM2 via well-described pathways, signaling p53 for degradation under normal conditions. However, an underlying mechanism, combining activities synergistically deriving from these isoforms, may have evolved to orchestrate and fine-tune a selective activation of specific tumor suppressors in response to comorbiditing stress signals (Abegglen et al. 2015) (fig. 4).
Discussion
The p53–MDM2 axis has been thoroughly studied in humans and mice, as well as in a range of other species. It is well-established that during normal conditions, the MDM2 E3 ubiquitin ligase binds the p53 protein via the BOX-I motif and catalyses the poly-ubiquitination of p53, targeting it for degradation via the 26S proteasomal pathway. Several studies have shown the interaction interfaces of p53–MDM2 (Chene 2003; Moll and Petrenko 2003; Pant et al. 2013) and have directly linked the prevention of this interaction to the stabilization or/and activation of p53. In addition, it is worth-noting that the abrogation of the p53–MDM2 (protein–protein) interaction constitutes a druggable target in anticancer therapies (e.g., Nutlin 3a), aiming to stabilize p53. Moreover, preventing the capacity of p53 isoforms to bind MDM2, directly promotes the stabilization, activation, and downstream signaling functions of p53 (Gajjar et al. 2012; Nguyen et al. 2018; Haronikova et al. 2019).
Nineteen TP53RTGs have been accumulated primarily by segmental duplication and drift, following an initial retrotransposition of the p53 gene. Studies by Sulak and collaborators indicated that most of these RTGs are under the control of the RTE-type non-LTR retrotransposon (RTE1_LA) promoter. Using RNA-Seq data, the authors showed that TP53 and TP53RTG12 genes were transcribed in African and Asian elephant samples (Sulak et al. 2016). This work largely supports previous studies (Cortez et al. 2014; Reddy et al. 2015); whereas the TP53RTG3 and TP53RTG18 transcripts were considerably less abundant (Sulak et al. 2016). Though TP53RTG12 is annotated as a pseudogene at the Ensemble database, an elephant-specific protein band of the expected size for TP53RTG19 (22.3 kDa) was identified by Sulak et al., as well as a band at the size of TP53RTG12 protein (19.6 kDa). Further, these authors employed treatment with low doses of different genotoxic agents (mitomycin c, doxorubicin) or with UV-C in order to induce an up-regulation of TP53 signaling and apoptosis (Cas3/7 activity) aiming to study whether elephant cells exhibit an enhanced TP53 response in response. However, these experiments did not indicate the activation of an expanded TP53 gene repertoire (Sulak et al. 2016). Correlating with previous findings (Karakostis et al. 2016), the abrogation of the MDM2–p53 PPI by nutlin 3a lead to the stabilization of p53, with cells were resistant to apoptosis. Indeed, nutlin 3a prevents p53 from MDM2-mediated degradation. However, the activation of p53 further requires PTMs (e.g., phosphorylation by ATM, acetylation by p300), mediated in response to stress signals (Meek and Anderson 2009; Haronikova et al. 2019).
Previously, Karakostis et al. (2019) described a mechanism activating p53 (S15 phosphorylation) in response to DNA damage (doxorubicin) that requires the MDM2–p53 mRNA interaction. Indeed, it appears that MDM2 and p53 have coevolved since the early Metazoan with the BOX-I encoding sequence evolving two MDM2 interactions, one RNA for controlling synthesis that takes place in prevertebrates and one protein for regulating degradation that occurs in vertebrates. The MDM2–p53 interaction also evolved to impose allosteric regulation of both proteins as they gained complex interactomes to increase and their functional repertoires. Hence, the p53–MDM2 interactions not only serve to control p53 expression but also as ligands to control their respective conformations and functions. In mammals that carry a single p53 gene, the MDM2–p53 interaction is controlled by alternatively spliced or translated isoforms. In elephants, in contrast, the retrogenes serve to differentiate the function and regulation of p53 into specific isoforms that contribute to an enhanced sensitivity to cellular stresses, such as DNA damage. Indeed, isoforms derived by gene duplication followed by subfunctionalization, have been hypothesized to allow an intrinsic specificity, by partitioning ancestral interacting partners along each lineage (Wheeler and Harms 2021). Elephant species show variations in the TP53 sequences and copy numbers, with the smallest species expressing a higher number (Tollis et al. 2021). Such variations may be epigenetically introduced (mutations) or be endogenously expressed via alternative splicing mechanisms and synthesis of several retrogenes. Hints concerning the coevolution of the p53–MDM2 interaction reflected by variations on the sequences expressing the binding interphases, are crucial for the appropriate regulation of the P53–MDM2 pathway, constituting the basis for the evolution of specific activation mechanisms of p53 regulation in response to diver stresses. Conceptually, this interpretation is embedded in the hypothesis that an accumulation of differentially expressed and structurally modified p53 pools (either isoformic or modified by PTMs), collectively and synergistically regulates the response to specific stresses in the cell (Khoury and Bourdon 2011). To address this paradigm, Sulak et al. (2016) developed a model for TP53RTG function to explore whether the retrogenes act as “decoys” or “guardians” of the canonical p53 with the view to understand how several TP53RTGs contribute to an enhanced sensitivity of elephant cells to DNA damage. Indeed, the proposed “guardian” model, is supported by the interaction between TP53 and the retrogress TP53RTG12, but not between TP53RTG12 and MDM2. According to this model, Sulak et al. suggested that TP53RTG proteins dimerize with canonical TP53 to block the formation of TP53 tetramers, thus preventing the MDM2-dependant negative regulation of the canonical TP53 protein. The alternative “decoy” model, describing TP53RTGs acting as decoys for the MDM2 interaction, was also tested and discussed by the authors (Sulak et al. 2016). Although confirming the dynamics of the oligomerization states will be challenging, both hypotheses (models) provide a testable approach to address TP53RTGs functions.
Here, we aim to advance previous findings and add to the discussions, by focusing on the interaction between MDM2 and each p53 BOX-I motif constituting the MDM2-binding epitope. Our research provides novel experimental and in silico evidence that reveal how elephant p53 isoforms may have modified BOX-I motifs to exhibit reduced binding capacity to MDM2 (Kubbutat et al. 1997; Klein and Vassilev 2004; Coffill et al. 2016). Alternations in the coding sequences of p53 BOX-I isoforms induce structural variations in the elephant p53, as shown by modeling and experimental (ELISA) studies, constituting a range of putative functional p53 proteins with different activities toward various stress inducers. Our findings support previous co-IP evidence, which show that the Type B BOX-I isoforms (corresponding to the TP53RTG12) prevent the interaction of p53 with MDM2 (Sulak et al. 2016) (figs. 1 and 3). Additionally, these findings also show how other BOX-I Types (corresponding to TP53RTGs) also limit the interaction with MDM2, constitutively contributing in a range of MDM2-binding p53 molecules with variable binding efficiencies (decreased), thus preventing MDM2-dependant control.
Our observations do not exclude the “guardian model” proposed by Sulak et al (2016). Indeed, our results add by suggesting a range of district and/or contributing activities of these isoforms, potentially dynamically regulating the activation of p53 toward various stresses. We note that even though these p53 isoforms are truncated compared with the canonical full-length p53, they retain the TAD and parts of the DBD regulating downstream targets (fig. 1A). After all, the canonical p53 regulates the expression of more than 3,000 genes and the interactions between p53 isoforms are considered to be direct via oligomerization or indirect, via promoter-dependent oligomerization (Anbarasan and Bourdon 2019).
Structural Variations on the p53–MDM2 Interface in Different Species
Despite intensive academic or commercial (biomedical) studies over the last 40 years (Marcel et al. 2018; Duffy et al. 2020; Levine 2020; Yang et al. 2021), we lack a clear picture of how p53 may be regulated, how it functions and how cells differentiate p53 activity to best suit intracellular and extracellular changes to their environment (Salomao et al. 2021). It is still very much an open question how the human p53, which is expressed from a single gene can have so many different activities and how these are all regulated. Insights into the molecular evolution of the ancestral p53/p63/p73 gene to the mammalian p53 family of genes and comparative studies on the MDM2–p53 pathway, are beginning to unravel the biological roles of p53 homologs in different species. Mechanistic and structural studies on interacting partners can be approached by employing different model systems. In our current study, we introduced the p53 isoforms from elephant as an interesting model system for structural studies of the p53–MDM2 interplay. From the analysis so far, as reflected by the high number of p53 isoformic sequences (both the high copy number and the existence of several diverse or/and partial p53 sequences), it emerges that the elephant may employ a sequence-specific strategy to respond to p53-mediated regulation, depending on a stress. It remains unclear whether the p53 PTM palette (including phosphorylation, acetylation, methylation, ubiquitination, sumoylation, etc.), is conserved in the elephant as it is for example in the human, although the documented functional diversity of elephant p53 sequences escaping MDM2-binding and protein degradation, points toward a mechanism whereby p53 employs isoformic sequences of variable BOX-I motifs and partial DBDs. These complexes (1) form dimers with the canonical p53 protein, preventing its degradation via MDM2 (guardian model, suggested by Sulak et al 2016); or/and (2) independently contribute to p53 mechanisms potentially substituting the function of PTMs-mediated regulation, described in human (e.g., during the DNA damage response). The physiological significance and the function (1) of extra copies of the gene and (2) of the expression of various isoforms, are highly relevant for further studies of the paradigm. For example, in connection to human or other systems expressing a single p53 gene, the acquisition of variations during molecular evolution, integrated on a single p53 gene, may elegantly explain the employment of PTMs that can tangle differential expression and signal-specific responses, thus compensating or substituting the need of several isoforms or copy number variations. Importantly, genetically engineered mouse models indicate that p53 may also regulate longevity and aging, in a context-specific manner (Donehower 2009). Additionally, single nucleotide variations such as the mouse P72, were shown to better retain self-renewal functions of stem/progenitor cells compared with R72 mice (Zhao et al. 2018). Overexpression of an N-terminal truncated p53 that starts at the second AUG and lacks the first 40 amino acids (Δ40p53) results in premature aging in mice and altered stem cell pluripotency (Ota et al. 2017; Levandowski et al. 2021). Surprisingly, this phenotype is dependent on the presence of full-length p53. In addition, a truncated carboxy-terminal p53 fragment leads to activated p53 and mice expressing this truncated version along with p53wt (p53+/m) exhibited enhanced resistance to spontaneous tumors compared with wild-type (p53+/+) mice, but an early onset of aging (Tyner et al. 2002). Recurrently in species, increased TP53 expression comes with a cost, whereas the elephants, expressing 20 copies and 19 truncated isoforms of p53, potentially may potentially compensate it (Sulak et al. 2016), even though some of these result from retrotransposition.
In the current study, we make use of a naturally evolved system, the Elephant’s genome, endogenously expressing a variety of truncated and variable p53 sequences. As such, the Elephant p53 sequences constitute an ideal model for exploring the multifaceted functions of p53. Even though elephant p53 retrogenes have not been shown to directly function as transcription factors, they might enhance DNA damage sensitivity and induction of apoptosis by regulating the p53 signaling pathway (Sulak et al. 2016). Since many of the encoded proteins (i.e., the p53 retrogene 12) retain the MDM2 interaction motif at the TAD and dimerization sites at the DBD, they might act in favor of p53 activation via binding competition to the MDM2 complex or by dimerization with the canonical p53 (Kubbutat et al. 1998; Sulak et al. 2016). In correlation with this, our results show that retrogene 12 along with retrogenes 14, 15, 16, and 18, form the group “Type F,” which shows poor binding to MDM2 (fig. 4) strongly indicating that these putative proteins escape MDM2-dependant regulation. In addition, our in silico analysis and the experimental ELISA findings both show that all the peptides carrying the W > G variation exhibit decreased binding activity with MDM2 (figs. 1 and 3). In fact, the alignment of the elephant BOX-I sequences of the canonical p53 and the TP53RTGs, illustrates at least two vital substitutions which are conserved in all the TP53RTGs (W > G and N > K) (fig. 4). This indicates that these sequences derive from a common initial duplication. This is in line with Sulak et al. (2016), providing evidence confirming that these isoforms result from a single retrotransposition event followed by repeated rounds of segmental duplication of chromosomal loci containing the TP53RTG genes.
Thus collectively these variations contribute to the formation of altered p53 BOX-I structures, thus resulting to a range of interaction capacities for these isoforms. These results are in line with previous co-IP data, which show that the TP53RTG12 isoform (BOX-I, Type B), coimmunoprecipitated with p53 but not with MDM2, suggests that these isoforms dimerize with p53, whereas preventing the interaction with MDM2 (requiring tetramerization of p53), and thus act as guardians against MDM2-dependant p53 degradation. These findings, like the guardian model proposed by Sulak et al. (2016), support our thesis that p53 peptides exhibit a spectrum of reduced p53–MDM2 interaction implicated in p53 activity. Moreover, considering that these isoforms carry the TADs and partial sequences of the DBD (fig. 1A), it is likely that their reduced binding capacity to MDM2 allows these isoforms/retrogenes to evade degradation via specific processes, that is, the activation of specific pools of downstream genes independently from the canonical FL p53. Additional PTMs identified in human, including acetylation of C-terminal lysines (K370, K372, K373, K381, K382, and K386) by p300/CBP, which boosts p53 binding to its target gene loci to activate downstream pathways, like cell cycle arrest, senescence or apoptosis, remain to be addressed in the elephant p53 isoforms (Liu et al. 2019; Chen et al. 2020). Much remains to be done to decipher the physiological significance of these findings; for now, Peto’s Paradox has opened Pandora’s Box of important questions revolving around the all-round function of p53.
Implications in Clinical Translation Research
It is argued that the p53–MDM2 axis constitutes a druggable target for cancer therapeutics (Hientz et al. 2017; Sharma et al. 2019; Karagiannakos et al. 2022). We assert that addressing the coevolution of the p53–MDM2 interaction may refine predictive studies aiming to identify and characterize cancer biomarkers for translation research. In respect to diagnostics, immune biomarkers from serum, for example, the cytokines (tumor necrosis factor-alpha, interferon-gamma, and interleukins), which are known to induce p53 up-regulation have also been documented to be indicative of immune responses to various pathologies in the elephant (Edwards et al. 2020). Additionally, identification and characterization of the functional implications of such variants and of their physiological significance to onco-immunology and aging, constitute an unforeseen opportunity for personalized medicine (Abegglen et al. 2015). Here, we show that the elephant BOX-I Type F isoform, has an E to G variation at residue 17. We note that variations on E17 in human p53 are cancer mutants, frequently associated with female genital cancers (Petitjean et al. 2007; Ganguly and Chen 2015). The K24N mutation found in colorectal cancers, is thought to implicate ubiquitination by TRAF6, thereby facilitating p53 acetylation by p300 (Lai et al. 2022). Isothermal titration calorimetry measurements demonstrated that this structural variation did not prevent the binding affinity for p53TAD–MDM2 (Zhan et al. 2013). However, the K24N human mutation was shown by computational and NMR secondary chemical shift analysis, to reduce the helicity in residues 18–27, from an average of ∼10–5% by disrupting the Asp21–Lys24 salt bridge, which stabilizes the local partial helices. In parallel, it also reduced the probability of contact formation between residues 21 and 24, by ∼50% thus increasing the flexibility of the p53 TAD and affecting several molecular interactions of p53 as well as posttranslational regulation (Ganguly and Chen 2015).
Other alternations on the BOX-I motif documented here via the comparative interspecies analysis, have also been suggested to be implicated in cancer cell lines with inactivating TP53 point mutations (i.e., P27, Sonkin 2015 and N29, Thirion et al. 2002). In addition, the cross-species mutations Y > D and E > K, showed an increased docking efficiency. Interestingly, the glutamic acid (E), asparagine (N), tryptophan (W), and tyrosine (Y) residues are highly conserved in disordered proteins as they play crucial role in forming protein–protein interfaces (Brown et al. 2010). As such, it emerges that cross-species studies, like our pilot study presented here, on genetic variations of p53 can lead to several novel insights into altered structural and functional outcomes of p53 action (Wang et al. 2007; Lu et al. 2009; Belyi et al. 2010; Joerger et al. 2014; Coffill et al. 2016; Bartas et al. 2019; Biscotti et al. 2019; Fischer 2019). Derived discoveries will in turn lead to novel, testable hypotheses concerning clinical aspects of p53 with links to the development of drug-resistance or cancer progression (Somarelli, Gardner, et al. 2020; Salomao et al. 2021). Accumulating evidence present how comparative genomic studies aiming to shed light on complex human traits with several translational aspects (Sulak et al. 2016; Tollis et al. 2019; Somarelli, Boddy, et al. 2020; Somarelli, Gardner, et al. 2020; Farre et al. 2021; Stakyte et al. 2021) indicate the structural basis of the interfaces, which can be used for the development of highly specific antibodies to target neoantigens and cancer mutations that are difficult to target in conventional ways (Hsiue et al. 2021).
Insights into structural modifications and activation toward signaling mechanisms, illuminate the factors that prevent or promote carcinogenesis. Studies on the p53 isoforms (Zhang et al. 2019; Levandowski et al. 2021) will help to illuminate how their respective activities are regulated in response to cellular damages and pathogen infections (Breton et al. 2021; Mehta et al. 2021). This will not only result in a better understanding of p53 roles and regulation in elephants and in its role in protecting elephants from cancers as well as probably other “inflictions”, such as parasites (Lynsdale et al. 2017), but it also facilitates the development of new therapeutic strategies for human, based on structural molecular data.
Conclusion and Perspective
The elephant has naturally evolved several p53 isoforms of variable lengths and sequences. It thus constitutes an ideal model for studying the functions of p53 variants in this natural “experiment.” With the discovery that elephants have 20 copies of p53 and strong evidence that elephants only rarely get cancer, a whole new front of research has opened up for studies of p53 (Sulak et al. 2016; Vazquez et al. 2018). In this context, the elephant constitutes an ideal model to comprehend how molecular evolution may lead to the expression of multiple p53 isoforms with district RNA and protein structures and functions as opposed to human where p53 incorporates multiple functions under a tightly regulated signaling mechanism (employing PTMs).
In this work, we employed bioinformatic modeling analysis to identify the homology of the BOX-I motifs of p53 isoforms from the elephant L. africana and calculate their docking capacities (binding) to the main p53 regulator; MDM2. We also used in vitro synthesized peptides and recombinant versions of human MDM2, to experimentally confirm the interactions in vitro, employing sandwich ELISA. These results suggest that mutations altering the structures of the interacting partners (MDM2–p53 interface) may have dramatic functional consequences for cell signaling mechanisms, constituting potentially critical therapeutic or diagnostic targets (Karagiannakos et al. 2022). Our findings support previous models pointing toward contributing roles of p53 retrogenes in the regulation of p53 and increased sensitivity to DNA damage responses (Sulak et al. 2016). Furthermore, we show that the modified BOX-I sequences of these isoforms induce structural variations that prevent the interaction with MDM2, thus evading MDM2-dependant degradation. Moreover, their truncated DBDs indicate a selective transactivation of downstream targets.
We envision that our observations and conclusions form a solid basis for further experiments aiming to address how each isoform contributes to the activation of p53, as a result of structural variability. The methodology presented here for the first time demonstrates how in silico docking simulations can be used to explore functional aspects of these p53 isoforms and sets the basis for perspective studies aiming to explore the dynamics of the interactions with MDM2 under stress-inducing conditions. Studies using elephant cell lines or tissues to measure the expression levels of the isoforms (qPCR) and knock-down assays (siRNAs) targeting combinations of the p53 isoforms, are highly anticipated to probe functional and physiological aspects, potentially contributing to the dissection/resolution of the Peto’s Paradox. In a broader view, such studies may also address the mechanisms whereby tumor suppressor genes and duplications regulate cellular senescence to drive lifespan and body mass (Farre et al. 2021; Tejada-Martinez et al. 2022).
Materials and Methods
In silico Analysis and Phylogenetic Characterization for the Determination of p53 Sequences in L. africana
Homologous sequences of human p53 and MDM2 were identified by searches in NCBI (http://www.nlm.nih.gov/). Nineteen isoformic sequences were identified and used for the analysis. The sequences are the following: X1 (XP_010594888.1) and X2 (XP_003416950.2) sharing an identical BOX-I sequence; isoform 1 (KF715855.1); isoform 2 (KF715856.1); isoform 3 (KF715857.1); isoform 4 (KF715858.1); isoform 5 (KF715859.1); isoform 6 (KF715860.1); isoform 7 (KF715861.1); isoform 8 (KF715862.1); isoform 9 (KF715863.1); isoform 10 (KF715864.1); isoform 11 (KF715865.1); isoform 12 (KF715866.1); isoform 13 (KF715867.1); isoform 14 (KF715868.1); isoform 15 (KF715869.1); isoform 16 (KF715870.1); isoform 17 (KF715871.1); and isoform 18 (KF715872.1). The human p53 sequence BAC16799.1, was used for the comparative analysis. Sequences were analyzed in silico with the ClustalOmega software (Sievers et al. 2011). The BOX-1 sequences (MDM2-binding motif) of the elephant p53 sequences were analyzed. Sequence alignment, comparisons, and phylogeny were generated using the ClustalOmega software for Multiple Sequence Alignment (EMBL, EBI) (Sievers et al. 2011) and the tree is a Neighbour-joining tree. Sequences were grouped based on their variations into six types and a graphical representation is shown (fig. 1A, supplementary figs. S1 and S2 and table S1, Supplementary Material online). A phylogenetic tree of the BOX-I sequences indicates the phylogeny among the elephant p53 types (supplementary fig. S2, Supplementary Material online). This information may be of interest for exploring the molecular evolution of the BOX-I motif, its coevolution with MDM2, and the putative duplication events that lead to the formation of several BOX-I isoforms in the elephant. The corresponding ENSEMBL id numbers of each BOX-I Type are mentioned on supplementary table S3, Supplementary Material online.
Modeling L. africana p53–MDM2 interaction
The homology modeling approach was applied for constructing the MDM2 and p53 protein structures. The 1YCR crystal structure, illustrates MDM2 bound to the p53 TAD, modeling the sequences >1YCR_2|Chain B|P53| SQETFSDLWKLLPEN and >1YCR_1|Chain A|MDM2|Homo sapiens (9606) (Kussie et al. 1996). The modeled sequence of human MDM2 is the following:
ETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVV. 1YCR_1 was used as a template to build the L. africana MDM2 protein structure (Kussie et al. 1996; Berman et al. 2000). The sequence of the elephant MDM2 is the following: ETLVRPKPLLLKLLKSVGAQKDTYTMKEVIFYLGQYIMAKRLYDEKQQHMVYCSNDLLGDLFGEPSFSVKDHRKIYTMIYRNLVV. Homology modeling was performed using the Molecular Operating Environment (MOE; Chemical Computing Group Inc., Montreal, QC, Canada) package (Vilar et al. 2008) along with the CHARMM27 forcefield (Brooks et al. 2009).
The modeled MDM2 structure was further analyzed with energy minimization within the MOE package (Berman et al. 2000). To build the tertiary structures of the p53 peptides, the human p53 from pdb id.: 1YCR (Kussie et al. 1996) was used as a template, which was further optimized by applying CHARMM27 forcefield (Brooks et al. 2009). These different MDM2 and p53 protein structures were considered for the protein–peptide docking protocol, to calculate their binding patterns. In order to achieve different conformations of p53 peptides with MDM2 protein, the rigid body docking protocol described in MOE (Berman et al. 2000) was implemented. The CHARMM27 forcefield (Vilar et al. 2008) was used to rank the binding score of individual p53 peptides with MDM2 protein, using the GBVI/WSA dG (kcal/mol) scoring function (Labute 2008; Padariya et al. 2021). Furthermore, considering molecular mechanics a refinement of individual clusters protein–peptide complex was performed and best 100 poses were analyzed. The BIOVIA Discovery Studio visualizer (Dassault Systèmes, BIOVIA Corp., San Diego, CA, USA) package was used to prepare representation of the MDM2–p53 complexes.
Peptide Synthesis and Protein–Protein Sandwich ELISA
Each of the six types elephant BOX-I peptide was synthesized in vitro with a conjugated Biotin-N-terminal tag, followed by SGSG spacer, then the amino acid sequence terminating with a C-terminal amide, and obtained from Mimotopes (Australia). Recombinant human glutathione S-transferase (GST)-tagged MDM2 protein was used for the MDM2–p53, protein–peptide sandwich ELISA, following the protocol published previously (Picksley et al. 1994). Peptides from Mimotopes were resuspended in DMSO to 1 mg/ml and then diluted into Elisa Buffer (used throughout the entire experiment which is PBS plus 0.1% Tween-20 and 3% BSA). White, high protein binding 96-well ELISA plates (Costar) were coated with streptavidin (1 µg/100 µl in H2O) by incubation overnight at 37 °C. Wells were washed 3× with PBS + Tween-20 (0.1%) and then blocked with 100 µl of ELISA buffer at room temperature for 60 min. Peptides were added at 100 ng/well in 100 ul of ELISA buffer to allow capture of the biotinylated peptide into the well at room temperature for 60 min. After washing three times with PBS + Tween-20 (0.1%), MDM2 was diluted to 100 ng per well in 100 µl of ELISA buffer. After incubation at room temperature for 60 min, wells were washed three times with PBS + Tween-20 (0.1%). The MDM2 antibody was diluted 1:1000 in ELISA buffer and 100 µl was added per well and after incubation at room temperature for 60 min, wells were washed three times with PBS + Tween-20 (0.1%). The secondary HRP-anti-IgG antibody (DAKO, P260 for mouse and P217 for rabbit) was diluted 1:1000 in ELISA buffer and 100 µl was added per well and after incubation at room temperature for 60 min, wells were washed three times with PBS + Tween-20 (0.1%), and enhanced chemiluminescence solution was added followed by reading the plate with a reader (Perkin Elmer). GST-tagged human MDM2 (purchased from Dundee University protein production facilities, DU43570) (fig. 3A). The human p53 peptide Biotin-SGSG-SQETFSDLWKLLPENNV was used as a positive control. Each sample was tested in triplicates and the Prism software was used for the preparation of the graphs. The values were normalized to the highest value (i.e., 100% was set for the largest mean in each dataset and 0% for the smallest mean). The statistical significance was calculated by t-test (two-tailed) comparing the values of each pair of Type (BOX-I) interaction with MDM2 and the P-values are indicated in the graph using the asterisk symbol scoring system: P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***), and P ≤ 0.0001 (****). Nonsignificant comparisons are indicated by “ns” and negative controls as: “n/c.” Both paired and unpaired t-tests were performed (supplementary table S4, Supplementary Material online). Scores derived from the unpaired t-test are indicated (fig. 3B and C).
Supplementary material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Inserm, European Regional Development Fund (ENOCH, CZ.02.1.01/0.0/0.0/16_019/0000868), MH CZ—DRO (MMCI, 00209805), Cancerforskningsfonden Norr, Cancerfonden (160598), Vetenskapsradet. The APC was funded by the International Centre for Cancer Vaccine Science, University of Gdansk (Fundacja na rzecz Nauki Polskiej: MAB/3/2017). U.K. is supported by the grant: 2020/36/C/NZ2/00108, from The National Science Centre (Narodowe Centrum Nauki, Krakow, Poland). The International Centre for Cancer Vaccine Science project is carried out within the International Research Agendas program of the Foundation for Polish Science cofinanced by the European Union under the European Regional Development Fund. Authors would also like to thank the PL-Grid Infrastructure, Poland for providing their hardware and software resources.
Contributor Information
Monikaben Padariya, International Centre for Cancer Vaccine Science, University of Gdansk, ul. Kładki 24, Gdansk, Poland.
Mia-Lyn Jooste, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh, UK.
Ted Hupp, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh, UK.
Robin Fåhraeus, International Centre for Cancer Vaccine Science, University of Gdansk, ul. Kładki 24, Gdansk, Poland; Inserm UMRS1131, Institut de Génétique Moléculaire, Université Paris 7, Hôpital St. Louis, Paris, France; Research Centre for Applied Molecular Oncology (RECAMO), Masaryk Memorial Cancer Institute, Brno, Czech Republic; Department of Medical Biosciences, Umeå University, Umeå, Sweden.
Borek Vojtesek, Research Centre for Applied Molecular Oncology (RECAMO), Masaryk Memorial Cancer Institute, Brno, Czech Republic.
Fritz Vollrath, Department of Zoology, Zoology Research and Administration Building, University of Oxford, Oxford, UK; Save the Elephants Marula Manor, Marula Lane, Karen P.O. Box 54667, Nairobi, Kenya.
Umesh Kalathiya, International Centre for Cancer Vaccine Science, University of Gdansk, ul. Kładki 24, Gdansk, Poland.
Konstantinos Karakostis, Inserm UMRS1131, Institut de Génétique Moléculaire, Université Paris 7, Hôpital St. Louis, Paris, France; Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona, Bellaterra (Barcelona), Spain.
Author contributions
M.P., M.-L.J., T.H., U.K. conducted the experiments; T.H., R.F., B.V., F.V., K.K. conceptualized the study, designed the experiments, and wrote the paper.
Data availability
The article complies with the MBE's policy on the availability of data and materials.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314:1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. 2014. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 3:e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbarasan T, Bourdon JC. 2019. The emerging landscape of P53 isoforms in physiology, cancer and degenerative diseases. Int J Mol Sci. 20:6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreira SN, Nguyen AD, Fredriksen MT, Wolfsberg TG, Moreland RT, Baxevanis AD. 2021. Aniprotdb: a collection of consistently generated metazoan proteomes for comparative genomics studies. Mol Biol Evol. 38:4628–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartas M, Brazda V, Cerven J, Pecinka P. 2019. Characterization of P53 family homologs in evolutionary remote branches of Holozoa. Int J Mol Sci. 21:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartas M, Brazda V, Volna A, Cerven J, Pecinka P, Zawacka-Pankau JE. 2021. The changes in the P53 protein across the animal kingdom point to its involvement in longevity. Int J Mol Sci. 22:8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Turnquist C, Horikawa I, Harris C. 2020. Targeting cellular senescence in cancer and aging: roles of P53 and its isoforms. Carcinogenesis 41:1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ. 2010. The origins and evolution of the P53 family of genes. Cold Spring Harb Perspect Biol. 2:a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscotti MA, Barucca M, Carducci F, Forconi M, Canapa A. 2019. The p53 gene family in vertebrates: evolutionary considerations. J Exp Zool B Mol Dev Evol. 332:171–178. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. 1997. Further characterisation of the P53 responsive element—identification of new candidate genes for trans-activation by P53. Oncogene 14:85–94. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. 2005. P53 isoforms can regulate P53 transcriptional activity. Genes Dev. 19:2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton Y, Barat C, Tremblay MJ. 2021. The balance between p53 isoforms modulates the efficiency of HIV-1 infection in macrophages. J Virol. 95:e0118821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. 2009. CHARMM: the biomolecular simulation program. J Comput Chem. 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Daughdrill GW. 2010. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 27:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyukpinarbasili N, Gucin Z, Ersoy YE, Ilbak A, Kadioglu H, Muslumanoglu M. 2016. P53 expression and relationship with MDM2 amplification in breast carcinomas. Ann Diagn Pathol. 21:29–34. [DOI] [PubMed] [Google Scholar]
- Callier V. 2019. Core concept: solving Peto's paradox to better understand cancer. Proc Natl Acad Sci U S A. 116:1825–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen L, El-Deiry WS. 2021. Differential p53-mediated cellular responses to DNA-damaging therapeutic agents. Int J Mol Sci. 22:11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. 2016. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 6:a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu S, Tao Y. 2020. Regulating tumor suppressor genes: post-translational modifications. Signal Transduct Target Ther. 5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, Peng J. 2009. P53 isoform delta113p53 is a p53 target gene that antagonizes P53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 23:278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P. 2003. Inhibiting the p53–MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 3:102–109. [DOI] [PubMed] [Google Scholar]
- Chusyd DE, Ackermans NL, Austad SN, Hof PR, Mielke MM, Sherwood CC, Allison DB. 2021. Aging: what we can learn from elephants. Front Aging. 2:726714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffill CR, Lee AP, Siau JW, Chee SM, Joseph TL, Tan YS, Madhumalar A, Tay BH, Brenner S, Verma CS, et al. 2016. The p53–Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes Dev. 30:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493. [DOI] [PubMed] [Google Scholar]
- Donehower LA. 2009. Using mice to examine P53 functions in cancer, aging, and longevity. Cold Spring Harb Perspect Biol. 1:a001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Synnott NC, O'Grady S, Crown J. 2020. Targeting p53 for the treatment of cancer. Semin Cancer Biol. 79:58–67. [DOI] [PubMed] [Google Scholar]
- Edwards KL, Miller MA, Siegal-Willott J, Brown JL. 2020. Serum health biomarkers in African and Asian elephants: value ranges and clinical values indicative of the immune response. Animals (Basel) 10:1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus R, Olivares-Illana V. 2014. MDM2's social network. Oncogene 33:4365–4376. [DOI] [PubMed] [Google Scholar]
- Farre X, Molina R, Barteri F, Timmers P, Joshi PK, Oliva B, Acosta S, Esteve-Altava B, Navarro A, Muntane G. 2021. Comparative analysis of mammal genomes unveils key genomic variability for human life span. Mol Biol Evol. 38:4948–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Lin M, Wu R. 2011. The regulation of aging and longevity: a new and complex role of p53. Genes Cancer 2:443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. 2019. Conservation and divergence of the p53 gene regulatory network between mice and humans. Oncogene 38:4095–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K. 2019. P53 isoforms in cellular senescence- and ageing-associated biological and physiological functions. Int J Mol Sci. 20:6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, Fahraeus R. 2012. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for P53 activation following DNA damage. Cancer Cell 21:25–35. [DOI] [PubMed] [Google Scholar]
- Ganguly D, Chen J. 2015. Modulation of the disordered conformational ensembles of the P53 transactivation domain by cancer-associated mutations. PLoS Comput Biol. 11:e1004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. 2002. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21:6225–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughran SJ, Pless E, Stearns SC. 2016. How elephants beat cancer. Elife 5:e21864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haronikova L, Olivares-Illana V, Wang L, Karakostis K, Chen S, Fahraeus R. 2019. The p53 mRNA: an integral part of the cellular stress response. Nucleic Acids Res. 47:3257–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Haupt Y. 2017. P53 at the start of the 21st century: lessons from elephants. F1000Res 6:2041. [DOI] [PMC free article] [PubMed]
- Hendler A, Akiva E, Sandhu M, Goldberg D, Arbely E, Jackson CJ, Aharoni A. 2021. Human SIRT1 multispecificity is modulated by active-site vicinity substitutions during natural evolution. Mol Biol Evol. 38:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hientz K, Mohr A, Bhakta-Guha D, Efferth T. 2017. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 8:8921–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiue EH, Wright KM, Douglass J, Hwang MS, Mog BJ, Pearlman AH, Paul S, DiNapoli SR, Konig MF, Wang Q, et al. 2021. Targeting a neoantigen derived from a common TP53 mutation. Science 371:eabc8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Wilcken R, Andreeva A. 2014. Tracing the evolution of the p53 tetramerization domain. Structure 22:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannakos A, Adamaki M, Tsintarakis A, Vojtesek B, Fåhraeus R, Zoumpourlis V, Karakostis K. 2022. Targeting oncogenic pathways in the era of personalized oncology: a systemic analysis reveals highly mutated signaling pathways in cancer patients and potential therapeutic targets. Cancers 14:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakostis K, Fahraeus R. 2019. Shaping the regulation of the p53 mRNA tumour suppressor: the co-evolution of genetic signatures. BMC Cancer 19:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakostis K, Ponnuswamy A, Fusee LT, Bailly X, Laguerre L, Worall E, Vojtesek B, Nylander K, Fahraeus R. 2016. P53 mRNA and p53 protein structures have evolved independently to interact with MDM2. Mol Biol Evol. 33:1280–1292. [DOI] [PubMed] [Google Scholar]
- Karakostis K, Vadivel Gnanasundram S, Lopez I, Thermou A, Wang L, Nylander K, Olivares-Illana V, Fahraeus R. 2019. A single synonymous mutation determines the phosphorylation and stability of the nascent protein. J Mol Cell Biol. 11:187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. 2011. P53 isoforms: an intracellular microprocessor? Genes Cancer 2:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Vassilev LT. 2004. Targeting the p53–MDM2 interaction to treat cancer. Br J Cancer. 91:1415–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of P53 stability By Mdm2. Nature 387:299–303. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Ludwig RL, Ashcroft M, Vousden KH. 1998. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 18:5690–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. 1996. Structure of the MDM2 oncoprotein bound to the P53 tumor suppressor transactivation domain. Science 274:948–953. [DOI] [PubMed] [Google Scholar]
- Labute P. 2008. The generalized born/volume integral implicit solvent model: estimation of the free energy of hydration using london dispersion instead of atomic surface area. J Comput Chem. 29:1693–1698. [DOI] [PubMed] [Google Scholar]
- Lai CW, Xie C, Raufman JP, Xie G. 2022. Targeting post-translational regulation of p53 in colorectal cancer by exploiting vulnerabilities in the p53–MDM2 axis. Cancers (Basel) 14:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Levine A. 2010. P53 research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2:a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levandowski CB, Jones T, Gruca M, Ramamoorthy S, Dowell RD, Taatjes DJ. 2021. The Delta40p53 isoform inhibits p53-dependent eRNA transcription and enables regulation by signal-specific transcription factors during p53 activation. PLoS Biol. 19:e3001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. 2020. P53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 20:471–480. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tavana O, Gu W. 2019. P53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 11:564–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logotheti S, Michalopoulos I, Sideridou M, Daskalos A, Kossida S, Spandidos DA, Field JK, Vojtesek B, Liloglou T, Gorgoulis V, et al. 2010. Sp1 binds to the external promoter of the p73 gene and induces the expression of TAp73gamma in lung cancer. FEBS J. 277:3014–3027. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. 2009. P53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 9:758–762. [DOI] [PubMed] [Google Scholar]
- Lynsdale CL, Mumby HS, Hayward AD, Mar KU, Lummaa V. 2017. Parasite-associated mortality in a long-lived mammal: variation with host age, sex, and reproduction. Ecol Evol. 7:10904–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Collavin L, Del Sal G. 2019. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 26:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, Olivier M, Hall J, Mollereau B, Hainaut P, et al. 2011. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 18:1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Perrier S, Aoubala M, Ageorges S, Groves MJ, Diot A, Fernandes K, Tauro S, Bourdon JC. 2010. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS Lett. 584:4463–4468. [DOI] [PubMed] [Google Scholar]
- Marcel V, Van Long FN, Diaz JJ. 2018. 40 years of research put P53 in translation. Cancers (Basel) 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. 2009. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 1:a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Campbell H, Drummond CJ, Li K, Murray K, Slatter T, Bourdon JC, Braithwaite AW. 2021. Adaptive homeostasis and the P53 isoform network. EMBO Rep. 22:e53085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moding EJ, Min HD, Castle KD, Ali M, Woodlief L, Williams N, Ma Y, Kim Y, Lee CL, Kirsch DG. 2016. An extra copy of P53 suppresses development of spontaneous Kras-driven but not radiation-induced cancer. JCI Insight. 1:e86698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Petrenko O. 2003. The MDM2–p53 interaction. Mol Cancer Res. 1:1001–1008. [PubMed] [Google Scholar]
- Naski N, Gajjar M, Bourougaa K, Malbert-Colas L, Fahraeus R, Candeias MM. 2009. The p53 mRNA–Mdm2 interaction. Cell Cycle 8:31–34. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Grimm SA, Bushel PR, Li J, Li Y, Bennett BD, Lavender CA, Ward JM, Fargo DC, Anderson CW, et al. 2018. Revealing a human p53 universe. Nucleic Acids Res. 46:8153–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota A, Nakao H, Sawada Y, Karnan S, Wahiduzzaman M, Inoue T, Kobayashi Y, Yamamoto T, Ishii N, Ohashi T, et al. 2017. Delta40p53alpha suppresses tumor cell proliferation and induces cellular senescence in hepatocellular carcinoma cells. J Cell Sci. 130:614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padariya M, Kote S, Mayordomo M, Dapic I, Alfaro J, Hupp T, Fahraeus R, Kalathiya U. 2021. Structural determinants of peptide-dependent TAP1–TAP2 transit passage targeted by viral proteins and altered by cancer-associated mutations. Comput Struct Biotechnol J. 19:5072–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Jackson JG, Post SM, Abbas HA, Quintas-Cardama A, Hamir AN, Lozano G. 2013. The p53–Mdm2 feedback loop protects against DNA damage by inhibiting P53 activity but is dispensable for P53 stability, development, and longevity. Genes Dev. 27:1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 28:622–629. [DOI] [PubMed] [Google Scholar]
- Picksley SM, Vojtesek B, Sparks A, Lane DP. 1994. Immunochemical analysis of the interaction of P53 with MDM2; –fine mapping of the MDM2 binding site on P53 using synthetic peptides. Oncogene 9:2523–2529. [PubMed] [Google Scholar]
- Reddy PC, Sinha I, Kelkar A, Habib F, Pradhan SJ, Sukumar R, Galande S. 2015. Comparative sequence analyses of genome and transcriptome reveal novel transcripts and variants in the Asian elephant elephas maximus. J Biosci. 40:891–907. [DOI] [PubMed] [Google Scholar]
- Rozan LM, El-Deiry WS. 2007. P53 downstream target genes and tumor suppression: a classical view in evolution. Cell Death Differ. 14:3–9. [DOI] [PubMed] [Google Scholar]
- Rutkowski R, Hofmann K, Gartner A. 2010. Phylogeny and function of the invertebrate P53 superfamily. Cold Spring Harb Perspect Biol. 2:a001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomao N, Karakostis K, Hupp T, Vollrath F, Vojtesek B, Fahraeus R. 2021. What do we need to know and understand about p53 to improve its clinical value? J Pathol. 254:443–453. [DOI] [PubMed] [Google Scholar]
- Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Gross M, Backofen R, Diederichs S. 2019. A pan-cancer analysis of synonymous mutations. Nat Commun. 10:2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siau JW, Coffill CR, Zhang WV, Tan YS, Hundt J, Lane D, Verma C, Ghadessy F. 2016. Functional characterization of p53 pathway components in the ancient metazoan trichoplax adhaerens. Sci Rep. 6:33972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli JA, Boddy AM, Gardner HL, DeWitt SB, Tuohy J, Megquier K, Sheth MU, Hsu SD, Thorne JL, London CA, et al. 2020. Improving cancer drug discovery by studying cancer across the tree of life. Mol Biol Evol. 37:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli JA, Gardner H, Cannataro VL, Gunady EF, Boddy AM, Johnson NA, Fisk JN, Gaffney SG, Chuang JH, Li S, et al. 2020. Molecular biology and evolution of cancer: from discovery to action. Mol Biol Evol. 37:320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkin D. 2015. Expression signature based on TP53 target genes doesn't predict response To TP53–MDM2 inhibitor in wild type TP53 tumors. Elife 4:e10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakyte K, Rotheneder M, Lammens K, Bartho JD, Gradler U, Fuchss T, Pehl U, Alt A, van de Logt E, Hopfner KP. 2021. Molecular basis of human ATM kinase inhibition. Nat Struct Mol Biol. 28:789–798. [DOI] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5:e11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada-Martinez D, Avelar RA, Lopes I, Zhang B, Novoa G, de Magalhaes JP, Trizzino M. 2022. Positive selection and enhancer evolution shaped lifespan and body mass in great apes. Mol Biol Evol. 39:msab369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion A, Rouanet P, Thezenas S, Detournay D, Grenier J, Lopez-Crapez E. 2002. Interest of investigating p53 status in breast cancer by four different methods. Oncol Rep. 9:1167–1172. [PubMed] [Google Scholar]
- Tollis M, Boddy AM, Maley CC. 2017. Peto's paradox: how has evolution solved the problem of cancer prevention? BMC Biol. 15:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Ferris E, Campbell MS, Harris VK, Rupp SM, Harrison TM, Kiso WK, Schmitt DL, Garner MM, Aktipis CA, et al. 2021. Elephant genomes reveal accelerated evolution in mechanisms underlying disease defenses. Mol Biol Evol. 38:3606–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Robbins J, Webb AE, Kuderna LFK, Caulin AF, Garcia JD, Berube M, Pourmand N, Marques-Bonet T, O'Connell MJ, et al. 2019. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol Biol Evol. 36:1746–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schneider-Utaka AK, Maley CC. 2020. The evolution of human cancer gene duplications across mammals. Mol Biol Evol. 37:2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. 2002. P53 mutant mice that display early ageing-associated phenotypes. Nature 415:45–53. [DOI] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. 2018. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 24:1765–1776. [DOI] [PubMed] [Google Scholar]
- Vilar S, Cozza G, Moro S. 2008. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 8:1555–1572. [DOI] [PubMed] [Google Scholar]
- Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. 2006. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 23:251–263. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 104:18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Salvi R, Argentini M, Dureuil C, Delumeau I, Abecassis J, Debussche L, Wasylyk B. 1999. P53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with P53. Oncogene 18:1921–1934. [DOI] [PubMed] [Google Scholar]
- Wheeler LC, Harms MJ. 2021. Were ancestral proteins less specific? Mol Biol Evol. 38:2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Prives C. 2018. Relevance of the p53–MDM2 axis to aging. Cell Death Differ. 25:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lou G, Jin WL. 2021. The arsenal of TP53 mutants therapies: neoantigens and bispecific antibodies. Signal Transduct Target Ther. 6:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim HS, Cho YS, Guang X, Kang SG, Jeong JY, Cha SS, Oh HM, Lee JH, Yang EC, Kwon KK, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 46:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan YA, Wu H, Powell AT, Daughdrill GW, Ytreberg FM. 2013. Impact of The K24N mutation on the transactivation domain of P53 and its binding to murine double-minute clone 2. Proteins 81:1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Pan WY, Chen J. 2019. P53 and its isoforms in DNA double-stranded break repair. J Zhejiang Univ Sci B. 20:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wu L, Yue X, Zhang C, Wang J, Li J, Sun X, Zhu Y, Feng Z, Hu W. 2018. A polymorphism in the tumor suppressor P53 affects aging and longevity in mouse models. Elife 7 e34701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The article complies with the MBE's policy on the availability of data and materials.