Abstract
Mutations in the plant homeodomain-like finger protein 6 (PHF6) gene are strongly associated with acute myeloid (AML) and T-cell acute lymphoblastic leukemia (T-ALL). In this study, we demonstrated that PHF6 can bind to H3K9me3 and H3K27me1 on the nucleolar chromatin and recruit histone methyltransferase SUV39H1 to the rDNA locus. The deletion of PHF6 caused a decrease in the recruitment of SUV39H1 to rDNA gene loci, resulting in a reduction in the level of H3K9me3 and the promotion of rDNA transcription. The knockdown of either SUV39H1 or PHF6 significantly attenuated the effects of increase in H3K9me3 and suppressed the transcription of rDNA induced by the overexpression of the other interacting partner, thereby establishing an interdependent relationship between PHF6 and SUV39H1 in their control of rRNA transcription. The PHF6 clinical mutants significantly impaired the ability to bind and recruit SUV39H1 to the rDNA loci, resulting in an increase in rDNA transcription activity, the proliferation of in vitro leukemia cells, and the growth of in vivo mouse xenografts. Importantly, significantly elevated levels of pre-rRNA were observed in clinical AML patients who possessed a mutated version of PHF6. The specific rDNA transcription inhibitor CX5461 significantly reduced the resistance of U937 AML cells deficient in PHF6 to cytarabine, the drug that is most commonly used to treat AML. Collectively, we revealed a novel molecular mechanism by which PHF6 recruits methyltransferase SUV39H1 to the nucleolar region in leukemia and provided a potential therapeutic target for PHF6-mutant leukemia.
Key words: PHF6, SUV39H1, AML, rDNA transcription, Epigenetic, CX5461, Methyltransferase, Leukemia
Abbreviations: AML, acute myeloid; BFLS, Borjeson–Forssman–Lehmann syndrome; ChIP, chromatin immunoprecipitation; CML, chronic myeloid leukemia; Co-IP, co-immunoprecipitation; DFC, dense fibrillar component; FC, fibrillar centers; FUrd, 5-fluorouridine; H3K27me1, histone H3 (mono-methyl K27); H3K27me2, histone H3 (di-methyl K27); H3K27me3, histone H3 (tri-methyl K27); H3K9me1, histone H3 (mono-methyl K9); H3K9me2, histone H3 (di-methyl K9); H3K9me3, histone H3 (tri-methyl K9); MPAL, mixed-phenotype acute leukemia; NLS, nuclear localization sequences; NoRC, nucleolar chromatin remodeling complex; NuRD, nucleosome remodeling and deacetylase; PHF6, plant homeodomain-like finger protein 6; pre-rRNA, pre-ribosme RNA; rDNA, ribosomal DNA; Re-ChIP, double chromatin immunoprecipitation; RNA Pol I, RNA polymerase I; rRNA, ribosome RNA; SUV39H1, suppressor of variegation 3-9 homolog 1; T-ALL, T-cell acute lymphoblastic leukemia; UBF1, upstream binding factor 1
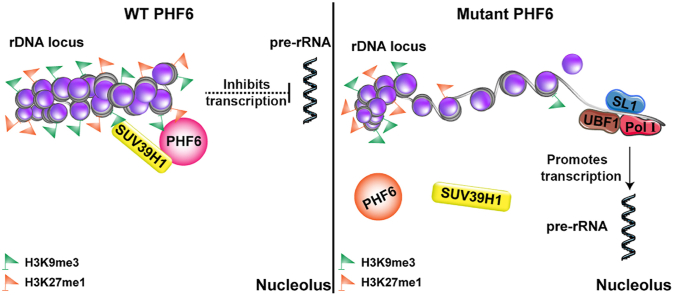
Graphical abstract
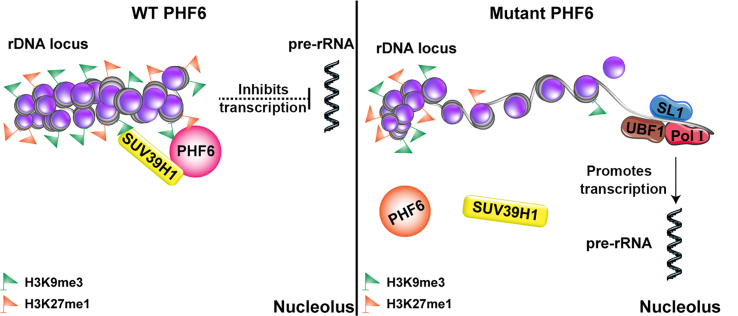
Wild-type PHF6 is recognized as H3K9me3 and H3K27me1, and it recruits SUV39H1 to catalyze the methylation of H3K9 and H3K27 to inhibit the transcription of rDNA. When PHF6 is mutated, SUV39H1 cannot be recruited for rDNA methylation, resulting in uncontrolled rDNA transcription.
1. Introduction
The plant homeodomain (PHD)-like finger protein 6 (PHF6) gene that is located on the X chromosome was originally identified as the gene responsible for Borjeson-Forssman-Lehmann syndrome (BFLS)1, 2, 3, a hereditary neurodevelopmental disorder that presents with mental retardation and physical deformities4. To date, loss-of-function PHF6 mutations are not only identified in genetic diseases but also in many types of cancer, including T-cell acute lymphoblastic leukemia (T-ALL)5, acute myeloid leukemia (AML)6, chronic myeloid leukemia (CML)7, mixed-phenotype acute leukemia (MPAL)8, and hepatocellular carcinoma9. In contrast to missense mutations that overwhelmingly exist in BFLS patients, neoplastic PHF6 mutations consist of deletions, insertions, frameshifts, nonsense, or missense mutations10,11.
Impressive advances have been made in understanding the functions and molecular mechanisms of PHF6 in recent years. Structurally, PHF6 comprises two nuclear localization sequences (NLS) and two nearly identical chromatin-binding zinc finger domains, including ZaP1 (aa 14–134) and ZaP2 (aa 209–332), each of which consists of a zinc knuckle and an atypical PHD finger (ZaP)6, indicating a potential function in transcriptional regulation. PHF6 has been shown to bind double stranded DNA (dsDNA) in vitro12 and can interact with several known transcriptional regulators, such as upstream binding factor 1 (UBF1) and the RNA polymerase II associated factor13. Furthermore, PHF6 was found to be a chromatin regulator protein that plays important roles in the regulation of developmental processes of neurogenesis, hematopoiesis, and lymphocyte lineage transition, as well as in leukemogenesis10. Significant differences in chromatin accessibility have been observed between hematopoietic stem cells (HSCs) from control and loss-of-function PHF6 mutant animals6. The deletion of PHF6 in B-cell leukemia results in a significant alteration in the chromatin landscapes at the transcriptional start sites of B- and T-cell-specific genes. In addition, it results in a lymphocyte lineage transition from B-cell leukemia to mixed-lineage lymphoma in vivo. These results highlight the potential capacity of PHF6 to bind modified histones and regulate chromatin epigenetics in a manner similar to the proteins found within canonical PHD domains3,12. PHF6 has recently been shown to physically interact with the nucleosome remodeling and deacetylase (NuRD) complex, a chromatin remodeling complex that acts as a transcriptional regulator that can epigenetically target several genes that influence embryogenesis, neurogenesis, hematopoiesis, and oncogenesis12. NuRD has been reported to inhibit the methyltransferase recruiter TIP5, a component of the nucleolar chromatin remodeling complex (NoRC) in the nucleolus, to maintain the demethylation state of rDNA promoters13. This is surprising, considering reports that the PHF6–NuRD complex is restricted to the nucleoplasm and is not present in the nucleolus13. However, preliminary research by both our group and other researchers has shown that PHF6 is predominantly localized in the fibrillar centers (FC) and dense fibrillar component (DFC) boundaries of the nucleoli, indicating that its key functions take place in the early-stage regulation of the ribosomal RNA (rRNA) transcription process14. The evidence accumulated to date has highlighted nucleolus dysfunction as a hallmark of several inherited and acquired human diseases, particularly intellectual development disorders and cancer15. The precise functions and molecular mechanisms by which PHF6 regulates chromatin epigenetics within the nucleoli remain unclear.
In this study, we found that PHF6 acts as a reader protein of histone lysine methylation to recognize and bind H3K9me3 and H3K27me1 within the nucleoli. PHF6 was also found to recruit the H3K9me2/3 methyltransferase SUV39H1 to methylate H3K9, maintain the heterochromatin status of rDNA gene loci, and suppress the transcription of rRNA. Furthermore, the interdependence of PHF6 and SUV39H1 in maintaining H3K9me3 levels, blocking rRNA transcription, and inhibiting leukemia cell proliferation was demonstrated. Importantly, PHF6 loss-of-function mutations from clinical patients significantly impaired the ability of PHF6 to bind to histone and recruit SUV39H1 to the rDNA loci. Excitingly, clinical sample screening showed that the pre-rRNA levels in AML patients who possessed a mutant PHF6 were significantly higher than those in AML patients that lacked this mutation. Suppressing rDNA transcription using the first-in-class selective inhibitor of rDNA transcription CX5461 significantly reduced the resistance of U937 AML deficient in PHF6 to cytarabine, the drug that is most commonly used in the treatment of AML. Thus, this mechanism constitutes a novel therapeutic strategy for diseases associated with PHF6 mutations.
2. Materials and methods
2.1. Clinical samples of patients with AML
Frozen bone marrow mononuclear cells from patients with AML were obtained from the biobank at the Zhujiang Hospital of the Southern Medical University, Guangzhou, China, in varying groups: PHF6 wild type (WT, n = 10) and PHF6 mutants (n = 5). All the patients gave informed consent for this study, and the study was approved by the Hematological Department at the Zhujiang Hospital of the Southern Medical University (ClinicalTrials.gov, No. 2019-KY-072-01).
2.2. Cell cultures
HeLa cells, HEK 293 cells, and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) medium with 10% fetal bovine serum, 100 mg/mL streptomycin, and 100 U/mL penicillin at 37 °C in a humidified incubator that contained 5% CO2. Jurkat and U937 cells were maintained in RPMI 1640 medium that contained 10% fetal bovine serum, 100 mg/mL streptomycin, and 100 U/mL penicillin at 37 °C in a humidified incubator that contained 5% CO2. All the cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA).
2.3. Plasmids
The pHrD-IRES-Luc plasmid was provided by Ke Yang and Du Xiaojuan (Peking University, Beijing, China). The PHF6 shRNA, SUV39H1 shRNA, Over- PHF6, Over- SUV39H1 plasmids were provided by Huang Sidong (McGill University, Montreal, Canada). To obtain the plasmids for human PHF6, cDNA was extracted from HEK293T, amplified by PCR, and cloned into pFlag-CMV2 and pEGFP-N1 vectors. Mutants of PHF6, including R342X, R274X, R116X, C99F, C215F, ΔPHD1, ΔPHD2, and ΔPHD1/2 were constructed from the WT PHF6. The plasmids for human SUV39H1, cDNA was extracted from HEK293T, amplified by PCR, and cloned into a pcDNA3.1/V5-His vector. SUV39H1 (R235H) was constructed from the WT SUV39H1 plasmid.
2.4. Lentiviral production and infection
To produce the shRNA lentivirus against PHF6 and SUV39H1, recombinant packaging plasmids were co-transfected with the shPHF6 and shSUV39H1 plasmids into HEK 293T cells. Culture supernatants that contained the virus were collected 12 and 36 h after transfection. To infect the cells with lentivirus, HeLa, Jurkat, and U937 cells were cultured with the lentiviral solution for 24 h in the presence of 1 μg/mL polybrene (Sigma–Aldrich, St. Louis, MO, USA).
2.5. Synthetic sequence of the mimetic polypeptide
The mimetic peptides were synthesized by Shenzhen Hanyu Co., Ltd. (Shenzhen, China). These polypeptides were labeled with biotin. The specific sequences are listed in Supporting Information Table S1.
2.6. Real-time RT-PCR analysis
Total RNA was extracted from HEK293, HeLa, and Jurkat cells using RNAiso Plus (TaKaRa, Dalian, China) and reverse transcribed to cDNA using a Primescript RT Reagent Kit (TaKaRa) following the manufacturer's instructions and then subjected to quantitative PCR analysis using 2 × SYBR Green qPCR Master Mix (TransGen Biotech, China). The primers for quantitative PCR amplification were as follows: pre-rRNA (human), sense: 5′-GAACGGTGGTGTGTCGTTC-3′; anti-sense: 5′-GCGTCTCGTCTCGTCTCACT-3′; β-actin (human), sense: 5′-CGTCACCAACTGGGACGACA-3′; anti-sense: 5′-CTTCTCGCGGTTGGCCTTGG-3′.
2.7. Western blotting
The protein fractions were separated using a polyacrylamide gel (SDS-PAGE) and then transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk for 1 h at room temperature and incubated with the desired primary antibodies overnight at 4 °C. After 1 h incubation at room temperature with an HRP-coupled secondary antibody, and the specific proteins were detected using an ECL reagent (Protein Tech, China).
2.8. Nucleolus fractionation
The nucleoli were isolated from Jurkat cells as described above16. The cells were resuspended in 10 volumes of hypotonic buffer (1 mmol/L MgCl2, 10 mmol/L Tris-HCl, and 10 mmol/L NaCl, pH 7.4) and incubated on ice for 30 min. NP-40 (final concentration of 0.3%) was added, and the cells were homogenized and lysed with a Dounce homogenizer with a gap of 0.4 mm. The nucleus was separated from the cytoplasm by centrifugation at 1200 × g for 5 min, then passed through 1200 × g for purification. The sample was centrifuged for 10 min in solution I (880 mmol/L sucrose and 5 mmol/L MgCl2). The purified cell nuclei were suspended in solution II (340 mmol/L sucrose and 5 mmol/L MgCl2) and then sonicated several times. Each ultrasound lasted for 30 s with intervals of 5 min between them until the nuclear membrane of the cell had ruptured completely. The nucleoli were intact when examined under a microscope. The nucleolus and nucleoplasm (supernatant) were collected by centrifugation at 2000 × g. A volume of 880 mmol/L sucrose lasted for 20 min. In addition, the nuclear and nucleolar fractions were subjected to Western blotting using the antibodies described above.
2.9. Luciferase reporter assay
To evaluate the effect of PHF6 knockdown or overexpression on the activity of rDNA promoter, shPHF6 or over-PHF6 HEK293 cells were grown on 12-well plates, followed by the addition of 1 μg pHrD-IRES-Luc into the media. The cells were incubated for 24 h and then harvested in the lysis buffer. The relative rDNA promoter activity was measured in cell lysates using a dual luciferase assay kit (Promega, Madison, WI, USA). The cells were harvested, and the relative activities of rDNA promoter were measured as described above.
2.10. 5-Fluorouridine (FUrd) incorporation assay
To monitor nascent rRNA, a FUrd incorporation assay was performed. HeLa or Jurkat cells were grown on glass coverslips, and FUrd (Sigma–Aldrich) was incubated at 10 μmol/L for 15 min and fixed using pre-chilled methanol for 5 min. The cells were washed with phosphate-buffered saline (PBS) and blocked with 3% BSA in PBS for 1 h at room temperature. After incubation with primary antibodies at 4 °C overnight, the coverslips were incubated with fluorescein or rhodamine-conjugated secondary antibodies for 1 h at room temperature. The nuclei were counterstained with DAPI. Finally, the cells were examined and imaged using a confocal microscope.
2.11. Co-immunoprecipitation assay (Co-IP)
HeLa cells were transfected with FLAG-PHF6 or V5-SUV39H1, lysed in RIPA buffer, and then sonicated for 10 s. The cell lysates were clarified by centrifugation at 12,000 × g for 30 min. Protein extracts were then mixed with the indicated primary antibodies and protein A/G agarose (Sigma–Aldrich) or anti-FLAG M2 agarose overnight at 4 °C. The complexes were collected and washed with TBST. The resolved proteins were analyzed using Western blotting.
Flag-PHF6 bound to the histone methylation to mimic peptides. The PHF6 protein that had been tagged with Flag was produced in HeLa cells that had been transfected with the Flag-PHF6 plasmid and then extracted from ultrasonic cells. Next, Flag beads and excessive amounts of PHF6 protein and peptides were co-incubated overnight. The beads were separated, and the supernatant was obtained by centrifugation at 3000 × g at 4 °C. The supernatant was discarded, and the beads were washed three times with PBS. The beads were then boiled for 5 min and prepared for a Western blot assay.
2.12. Chromatin immunoprecipitation (ChIP) and double chromatin immunoprecipitation (Re-ChIP)
ChIP assays were performed as previously described17 with minor modifications. Briefly, the cells were fixed with 1% formaldehyde at room temperature for 15 min. Cross-linking reactions were quenched by the addition of 125 mmol/L glycine. The cells were then washed with 1 × PBS, and after collection, they were resuspended in a ChIP lysis buffer (0.1% deoxycholic acid, 5 mmol/L EDTA, 50 mmol/L HEPES, 150 mmol/L NaCl, 1% Triton X-100, pH 8.0, and a protease inhibitor cocktail). After obtaining the 200–1000 bp DNA fragments, the sheared chromatin was immunoprecipitated with the indicated antibodies or control IgG in combination with Dynabeads protein G magnetic beads (Invitrogen, Carlsbad, CA, USA). The beads were resuspended with 10% Chelex 100 after several washes and boiled for 10 min. Proteinase K was added after the tubes had been cooled on ice. The tubes were then incubated in a shaker (1 h, 55 °C at 1400 × g) followed by boiling for 10 min. After centrifugation, the suspension was collected to perform a qPCR analysis, and the fold enrichment relative to the IgG control antibody was calculated. Primers (H0, H1, H4, H8, H13, H18, H23, and H42.9) for the rDNA used in the ChIP assays were synthesized as previously described16,18.
The complexes from each main ChIP sample of beads were eluted in 10 mmol/L DTT at 37 °C for 30 min for the re-ChIP experiment. After centrifugation, the supernatant was diluted with re-CHIP buffer (20 mmol/L Tris-HCl, 150 mmol/L NaCl, 2 mmol/L EDTA, and 1% Triton X-100, pH 8.0). The diluted complexes were examined using a second primary antibody as described for the first ChIP.
2.13. Colony formation assay
Each group was removed from 1 × 104 cells in a six-well plate, and the cells were cultured with the corresponding drugs. After one week, the cells were fixed using paraformaldehyde (PFA) and stained with Crystal Violet Staining Solution (Beyotime Institute of Biotechnology, Hainan, China).
2.14. CCK-8 assay
The proliferation of cells was evaluated using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) following the manufacturer's instructions. Briefly, the cells were plated in 96-well plates in a medium that contained 10% FBS at ∼5 × 103 cells/well. A volume of 10 μL CCK-8 solution was then added to each well, and they were incubated for 2 h. The absorbance at 450 nm was measured using a microplate reader.
2.15. Xenograft tumor model
Female BALB/c nude mice were obtained from the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Science (Beijing, China). All the animal studies were conducted based on the protocols approved by the Administrative Committee on Animal Research of the Laboratory Animal Center, Sun Yat-sen University, Guangzhou, China (SYSU-LACUC-2019-B633).
The mice were randomly divided into five groups (n = 5). HeLa or U937 cell xenograft models were established by the subcutaneous injection of 5 × 106 of the control, overexpressed PHF6, or shPHF6 cells. After the tumors had formed, a tumor growth curve was evaluated in terms of tumor volume (V), which was estimated using the following Eq. (1):
| (1) |
where a and b are the major and minor axes of the tumor, respectively, as measured by a caliper. After 3 weeks of observation, the nude mice were sacrificed; the tumor tissues were removed, and the tumor weights were measured. The results were considered statistically significant if the two-tailed P-values were < 0.05.
The mice were randomly divided into three groups (n = 5), which were subjected to peri-tumoral injection of saline as a control and either cytarabine (2 mg/kg) or CX5461 (2 mg/kg). The mice were closely observed for clinical signs and behavior. The volumes of tumors were measured by two experienced observers every second day using a Vernier caliper. The mice were subjected to peritumoral injections on Days 0, 3, 6, and 9. Subcutaneous xenograft tumor tissues were isolated and fixed in 4% PFA, and then stained with hematoxylin–eosin (H&E). An immunohistochemical analysis for ki67 was then performed.
2.16. Statistics
All the experiments were conducted in triplicate or higher unless otherwise stated. The results are expressed as the mean ± standard deviation (SD), and the statistical significance of all results was determined using a one-way analysis of variance (ANOVA) followed by a Tukey test and a Student's t-test. P < 0.05 is considered to be statistically significant (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
3. Results
3.1. PHF6 localizes to the nucleolus and regulates transcription of rDNA
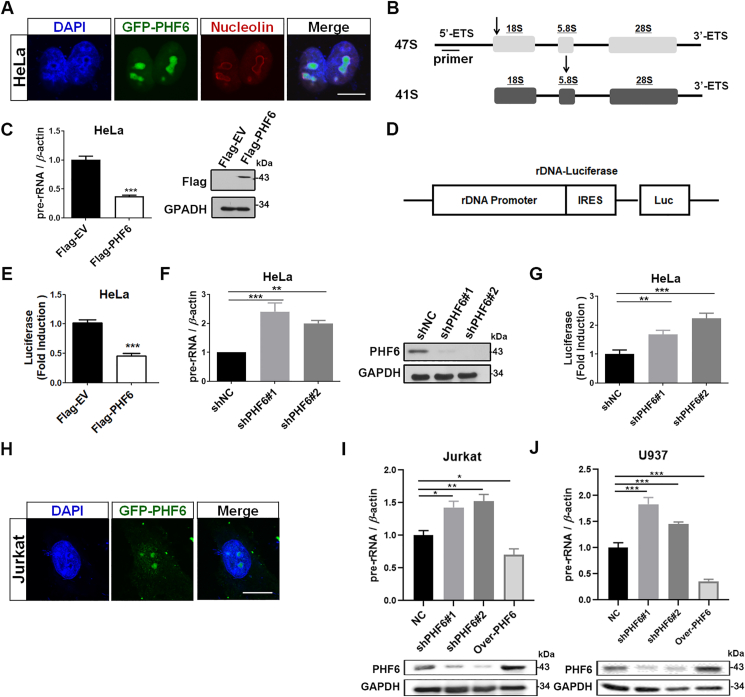
As previously described, we used confocal microscopy to reveal that PHF6 preferentially localizes to the nucleolus of HeLa cells (Fig. 1A). 47S ribosomal RNA precursors (pre-rRNA) were synthesized, processed, and assembled to ribosomal subunits in the nucleolus. The overexpression of PHF6 significantly decreased the activity of the 47S pre-rRNA level and rDNA promoter in HeLa cells (Fig. 1B–E), whereas the knockdown of PHF6 promoted the activity of rDNA promoter and restored 47S to its pre-rRNA levels of transcription (Fig. 1F and G). PHF6 is thought to play an important role in the occurrence and development of T-ALL and AML. Thus, the subcellular localization of PHF6 and its effects on rRNA transcription in Jurkat and U937 cells were investigated. As expected, the accumulation of PHF6 was prevalent in the nucleolus, and it was able to regulate the transcription of rDNA genes in Jurkat and U937 cells (Fig. 1H–J).
Figure 1.
PHF6 localizes to the nucleolus and regulates rDNA transcription. (A) Confocal images of HeLa cells transfected with GFP-PHF6 plasmids using nucleolin as a nucleolar marker. Scale bar, 10 μm. (B) Arrows indicate the position of qPCR primers on 47S pre-rRNA. (C) qPCR analysis of 47S pre-rRNA from HeLa cells transfected with Flag-empty vector (EV) or Flag-PHF6 plasmid. Western blots show the protein levels of Flag-PHF6. (D) Schematic representation of pHrD-IRES-Luc. (E) Flag-EV, Flag-PHF6, and PHrD-IRES-Luc were co-transfected into HeLa cells for 24 h, and the activity of luciferase was measured. (F) qPCR analysis of 47S pre-rRNA from HeLa cells silenced with negative control shRNA (shNC) and PHF6 shRNAs (shPHF6#1 and shPHF6#2). Immunoblot of PHF6 in the indicated HeLa cells. (G) Jurkat cells were silenced with negative shRNA (shNC), PHF6 shRNAs (shPHF6#1 and shPHF6#2), and PHrD-IRES-Luc was then transfected into the cells. Luciferase activity was measured after an additional 24 h. (H) Confocal images of Jurkat cells transfected with the GFP-PHF6 plasmid. Scale bar, 10 μm. (I) and (J) qPCR analysis of 47S pre-rRNA from Jurkat (I) and U937 cells (J) infected with negative shRNA (shNC), PHF6 shRNAs (shPHF6#1 and shPHF6#2), and over-PHF6. Immunoblot of PHF6 in the indicated Jurkat and U937 cells. GAPDH was used as loading control. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. All the values are expressed as the mean ± SD, n = 3.
3.2. PHF6 selectively recognizes and binds to the heterochromatin-associated histone marks of H3K9me3 and H3K27me1
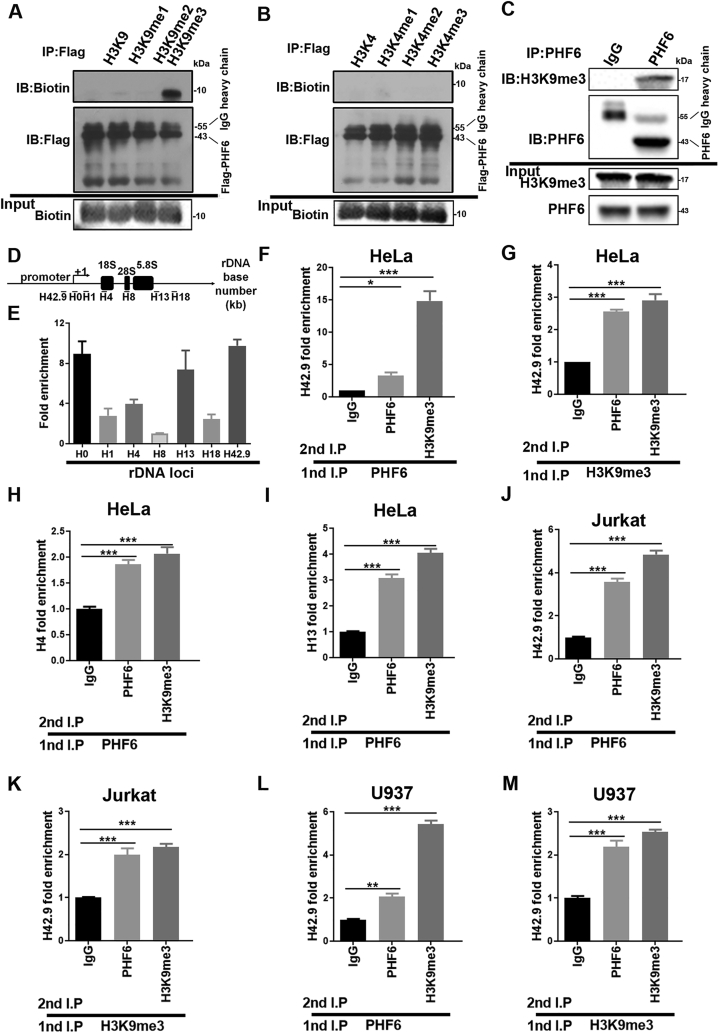
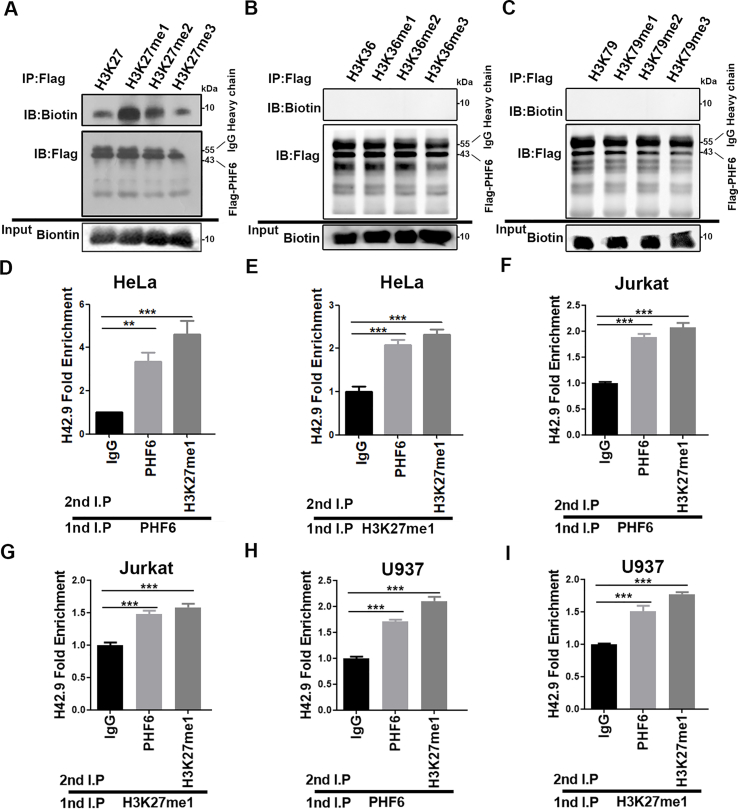
The transcription of rDNA gene, a crucial step in ribosome biogenesis, is strictly regulated by the epigenetic state of chromatin within the nucleolus19. Structurally, PHF6 contains two atypical PHD domains, a motif common to many chromatin-regulatory proteins3, indicating that PHF6 might function as an epigenetic reader or effector of histone H3 tails. Interestingly, a previous study showed that PHF6 interacts with the NuRD complex and that this PHF6–NuRD complex is restricted to the nucleoplasm and is not present in the nucleolus2. In this study, the relationship between PHF6 and the epigenetic modification of histone methylation was explored by incubating PHF6 with synthetic histone H3 mimetic peptides that were non-, mono-, di-, or trimethylated at specific lysine residues and then analyzing their abilities to bind PHF6. We identified the specificity of PHF6 toward the heterochromatin-associated histone marks H3K9me3 (Fig. 2A) and H3K27me1 (Supporting Information Fig. S1A), but not active chromatin marks H3K4, H3K36, and H3K79 (Fig. 2B, Fig. S1B and S1C). Co-IP assays also revealed an obvious interaction between PHF6 and histone H3K9me3 (Fig. 2C). To examine the protein occupancies of PHF6 at the rDNA gene loci, ChIP assays were performed. As expected, PHF6 was associated with the rDNA gene throughout the rDNA repeats, and it occupied the rRNA coding region, the intergenic spacer (IGS) region, and the rDNA promoter region (H42.9) (Fig. 2D and E). A ChIP-re-ChIP assay was then performed to determine the in vivo colocalization of PHF6 interaction, or whether it was in close contact with H3K9me3. The results show the colocalization of PHF6 interaction with H3K9me3 in the rDNA loci (Fig. 2F–M). Similarly, PHF6 and H3K27me1 were also demonstrated to strongly co-localize in the rDNA loci (Fig. S1D–S1I), suggesting that PHF6 selectively recognizes and binds to both H3K9me3 and H3K27me1 at the rDNA gene loci in HeLa, Jurkat and U937 cells.
Figure 2.
PHF6 selectively recognizes and binds to the heterochromatin-associated histone marks. (A)–(B) Co-IP analysis of Flag-PHF6 binding to histone methylation mimic peptides. The upper graph shows the amount of various mimic polypeptides that interact with Flag-PHF6. The middle graph shows the amount of Flag-PHF6 in the Flag antibody conjugate. The lower graph shows the amounts of mimic polypeptides added. (C) Co-IP analysis of histone H3 containing the trimethylated lysine 9 (H3K9) with PHF6 in HeLa cells. (D) Schematic diagram of human rDNA gene repeat sequence and positions of ChIP-qPCR primers. (E) ChIP assays were performed using control IgG and PHF6 antibodies, and the precipitated DNA was then analyzed using qPCR with the aforementioned primers. The relative rDNA fold-enrichment was normalized to control IgG treatment. (F)–(M) Re-ChIP experiments detected the binding of PHF6 to H3K9me3 at the rDNA loci using H4, H13, and H42.9 primers. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. All values are expressed as the mean ± SD, n = 3.
3.3. PHF6 interacts with SUV39H1 in the nucleolus, and they accumulate together at the rDNA loci
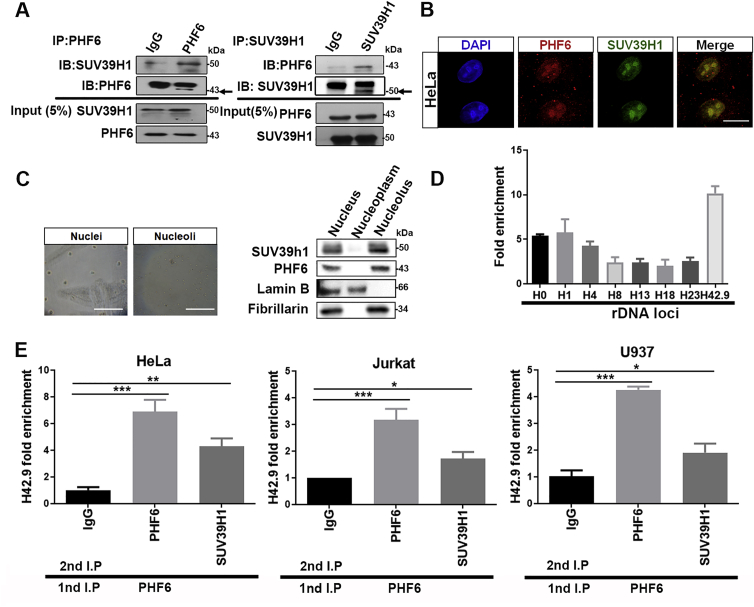
Surprisingly, most PHD proteins contain histone modifying catalytic domains, whereas PHF6 lacks such a catalytic domain20. Thus, we hypothesize that other histone modifying enzymes coordinate with PHF6 at the rDNA gene loci. An analysis of PHF6 interactors using immunoprecipitation and mass spectrometry (Supporting Information Table S2) led to the identification of SUV39H1, a histone methylase involved in the trimethylation of histone H3 on lysine 9. Co-IP further confirmed the interaction of PHF6 with SUV39H1 in HeLa (Fig. 3A). Confocal microscopy (Fig. 3B) and cell-fractionation experiments (Fig. 3C) supported the co-presence of PHF6 and SUV39H1 in nucleoli, suggesting that PHF6 and SUV39H1 play important roles in the epigenetic modification of nucleolar chromatin. To monitor the binding of SUV39H1 to rDNA, ChIP assays were performed using SUV39H1 antibodies. SUV39H1 was also bound to whole rDNA gene repeats in a manner similar to that of PHF6 (Fig. 3D). Furthermore, ChIP-re-ChIP assays in HeLa, Jurkat and U937 cells suggested that PHF6 and SUV39H1 bound to the rDNA gene as a protein complex (Fig. 3E).
Figure 3.
PHF6 interacts with SUV39H1 in the nucleolus, and they accumulate together in the rDNA loci. (A) Co-IP analysis of SUV39H1 with PHF6 in HeLa cells. (B) Confocal images of HeLa cells to show PHF6 (red fluorescence) colocalized with SUV39H1 (green fluorescence) in the nucleoli. Scale bar, 10 μm. (C) HeLa cells were fractioned into nuclear (left image), nucleoplasm and nucleolar (right image) fractions, and the proteins were detected by Western blotting using the antibodies described. Lamin B and fabrillarin are the nuclear and nucleolar markers, respectively. Scale bar, 50 μm. (D) ChIP assays were performed with control IgG and SUV39H1 antibodies, and the precipitated DNA was then analyzed using qPCR with the forementioned primers. The relative rDNA fold-enrichment was normalized to control the IgG treatment. (E) Re-ChIP experiments detected the binding of PHF6 to SUV39H1 in HeLa, Jurkat and U937 cells. The primer H42.9 was used for detection using real-time quantitative reverse transcription PCR. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. All the values are expressed as the mean ± SD, n = 3.
3.4. PHF6 and SUV39H1 cooperatively regulate H3K9 methylation and rDNA transcription in an interdependent manner
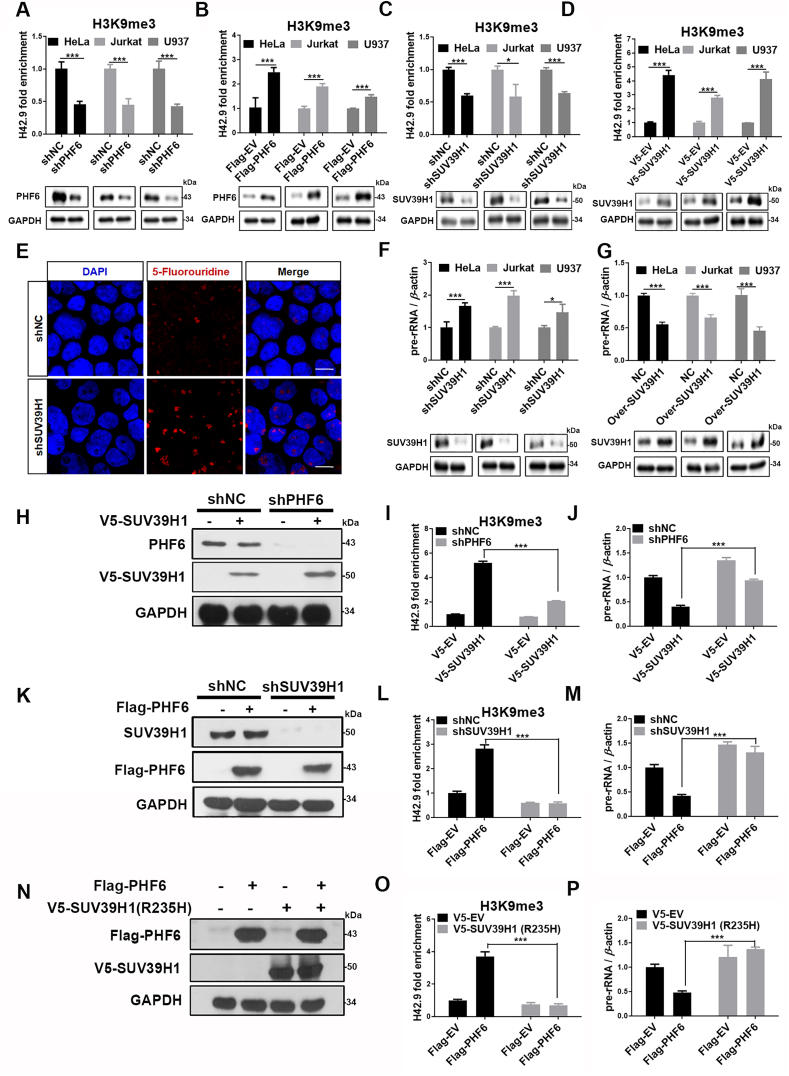
SUV39H1 is a well-known histone methyltransferase that specifically trimethylates the ‘Lys-9’ of histone H3 using monomethylated H3 ‘Lys-9’ as a substrate21. The interaction of PHF6 with SUV39H1 in the nucleolus raised the possibility that PHF6 may cooperate with SUV39H1 to regulate the heterochromatic state of rDNA genes. Thus, the effects of PHF6 and SUV39H1 on the levels of histone H3K9 trimethylation of nucleolar chromatin were investigated. As expected, the knockdown of PHF6 by shRNA led to a reduction in the levels of H3K9me3 at the rDNA gene loci in HeLa, Jurkat and U937 cells (Fig. 4A), while the overexpression of PHF6 significantly increased the levels of H3K9me3 (Fig. 4B). Similarly, the knockdown or overexpression of SUV39H1 resulted in the reduction or increase in the levels of H3K9me3 in the rDNA gene loci, respectively in HeLa, Jurkat and U937 cells (Fig. 4C and D). Subsequently, the depletion of SUV39H1 significantly promoted pre-rRNA synthesis as detected by BrdU incorporation assays and qPCR (Fig. 4E and F), whereas the overexpression of SUV39H1 led to an inhibition of the expression of pre-rRNA (Fig. 4G), indicating that SUV39H1 and PHF6 serve important roles in maintaining the heterochromatic state of rDNA.
Figure 4.
PHF6 and SUV39H1 cooperatively regulate the methylation of H3K9 and rDNA transcription in an interdependent manner. (A) and (B) ChIP analysis of H3K9me3 binding to the rDNA loci in PHF6-silenced (A) or PHF6-overexpressed (B) HeLa, Jurkat and U937 cells. Immunoblot of PHF6 or Flag-PHF6 in the indicated HeLa, Jurkat or U937 cells. (C) and (D) ChIP analysis of H3K9me3 binding to the rDNA loci in SUV39H1-silenced (C) or PHF6-overexpressed (D) HeLa, Jurkat and U937 cells. Immunoblot of SUV39H1 or V5-SUV39H1 in the indicated HeLa, Jurkat and U937 cells. (E) Confocal images of HeLa cells silenced using shNC, shSUV39H1, and then pulsed with FUrd for 15 min to stain with anti-BrdU (red). Scale bar, 10 μm. (F) and (G) qPCR analysis of 47S pre-rRNA from SUV39H1-silenced (F) or overexpressed (G) HeLa, Jurkat and U937. Immunoblot of SUV39H1 in the indicated HeLa, Jurkat and U937 cells. GAPDH was used as loading control. (H) PHF6-silenced (shPHF6) or control (shNC) HeLa cells were transfected with the V5-SUV39H1 plasmid (+) or a control without the plasmid (−), and then (I) ChIP analysis of H3K9me3 binding to the rDNA loci, and (J) qPCR analysis of 47S pre-rRNA were performed. (K) SUV39H1-silenced (shSUV39H1) or control (shNC) HeLa cells was transfected with the Flag-PHF6 plasmid (+) or a control without the plasmid (−), and then (L) ChIP analysis of H3K9me3 binding to the rDNA loci, and (M) qPCR analysis of 47S pre-rRNA was performed. (N) HeLa cells were co-transfected with Flag-PHF6 and V5-SUV39H1 (R235H). Cell lysates were prepared to analyze Flag/V5-SUV39H1 (R235H) levels using Western blotting. (O) ChIP analysis of the binding of H3K9me3 to the rDNA loci in (N) cells. (P) qPCR analysis of 47S pre-rRNA from (N) cells. All values are expressed as the mean ± SD, n = 3; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
To investigate the relationship between SUV39H1 and PHF6 in the regulation of levels of methylation of H3K9 and rDNA transcription, double knockdown and overexpression experiments were performed. The knockdown of PHF6 (Fig. 4H) significantly attenuated the upregulated levels of expression of H3K9me3 at the rDNA loci caused by overexpression of SUV39H1 (Fig. 4I), thereby diminishing the inhibition of pre-rRNA transcription that was induced by the overexpression of SUV39H1 (Fig. 4J). Intriguingly, the knockdown of SUV39H1 (Fig. 4K) almost completely inhibited the increase in levels of expression of H3K9me3 caused by the overexpression of PHF6 (Fig. 4L) and diminished the inhibition of pre-rRNA transcription induced by the overexpression of PHF6 (Fig. 4M), indicating that PHF6 regulates the modification of H3K9 by trimethylation and the rDNA transcription in an SUV39H1-dependent manner. To further confirm this hypothesis, the impact of a dominant negative mutant SUV39H1 (R235H) on the level of H3K9me3 at rDNA repeats and rDNA transcription was examined. The overexpression of SUV39H1 (R235H) significantly inhibited the levels of expression of H3K9me3 and promoted the transcription of rDNA, indicating that SUV39H1 (R235H) interferes with the function of WT SUV39H1. More importantly, SUV39H1 (R235H) totally suppressed the upregulated expression of H3K9me3 induced by the overexpression of PHF6 and diminished the inhibition of pre-rRNA transcription (Fig. 4N–P). Overall, this led to the conclusion that PHF6 and SUV39H1 cooperatively regulate the methylation of H3K9 and rDNA transcription in an interdependent manner as a nucleolar protein complex.
3.5. PHF6 mutants have a reduced ability to recruit SUV39H1, modify the methylation of H3K9, and regulate rDNA transcription
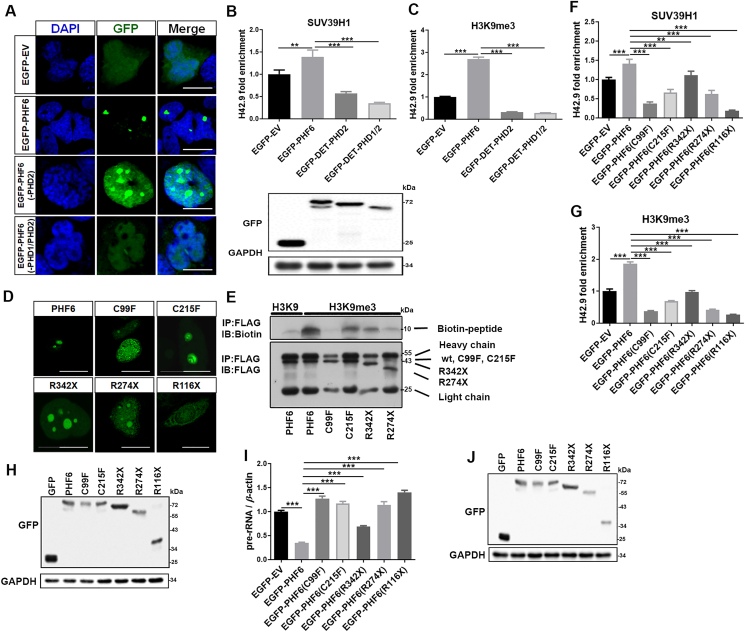
Most of the PHD proteins contain PHD domains (Cys4–His–Cys3) that recognize and bind methylated histones and modulate chromatin structures22. PHF6 proteins contain two atypical PHD domains (PHD1, Cys4–His–Cys–His in residues 82–131 and PHD2, Cys4–His–Cys–His in residues 280–329). A truncation experiment showed that PHF6 proteins that lacked PHD2 or both PHD1 and PHD2 diffused into the nucleoplasm and affected the recruitment of SUV39H1 to rDNA gene repeats (Fig. 5A–C). This, in turn, reduced the levels of expression of H3K9me3 in the nucleolar histone region, indicating that the PHD domains play important roles in regulating the structure of nucleolar chromatin. Furthermore, several clinical mutants that corresponded to BFLS (C99F and C215F) and leukemia (C99F, R342X, R274X, and R116X) were constructed to evaluate their effects on histone modification along the nucleolar region. As shown by confocal microscopy, WT PHF6 was predominantly distributed in the nucleolus in small, localized clusters. In comparison, the subcellular localization and arrangement of PHF6 mutants differed significantly. For example, C215F fused into larger clusters in the nucleolus; C99F and R342X were diffusely distributed throughout the nucleolus (i.e., discrete clusters were less prevalent); they were also increasingly distributed in the nucleoplasm; R274X completely diffused into the nucleoplasm; and R116X even dispersed into the cytoplasm (Fig. 5D). The ability of these mutants to bind H3K9me3 mimetic peptides decreased significantly compared with the WT PHF6 (Fig. 5E). As expected, these mutants also affected the recruitment of SUV39H1 to rDNA locus (Fig. 5F–H), thus, reducing the levels of H3K9me3 in the nucleolar histone region. Correspondingly, the overexpression of these mutants resulted in a loss of ability to inhibit the transcription of pre-rRNA as was observed with the WT PHF6 (Fig. 5I and J).
Figure 5.
PHF6 mutants are less able to bind H3K9me3 peptides and recruit SUV39H1. (A) Confocal images of HeLa cells that were transfected with EGFP-EV, EGFP-PHF6, EGFP-PHF6 (-PHD2), and EGFP-PHF6 (-PHD1/PHD2) plasmids. Scale bar, 10 μm. (B) and (C) ChIP analysis of SUV39H1 (B) and H3K9me3 (C) at the rDNA loci in (A) cells. Western blot results show the protein levels of EGFP-PHF6. (D) Confocal images of HeLa cells that were transfected with the clinical PHF6 mutants indicated. Scale bar, 10 μm. (E) Co-IP analysis of Flag-PHF6 and its mutants binding to peptides that mimic H3K9me3. (F) and (G) ChIP analysis of SUV39H1 (F) and H3K9me3 (G) binding to rDNA loci in HeLa cells that were transfected with the plasmids indicated. (H) Immunoblot of GFP in indicated cell lysates (F and G). (I) qPCR analysis of 47S pre-rRNA from HeLa cells that were transfected with the plasmids indicated. (J) Immunoblot of GFP in HeLa cells transfected with the indicated plasmids. All values are expressed as the mean ± SD, n = 3; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6. PHF6 and SUV39H1 coordinately regulate cell proliferation in an interdependent manner
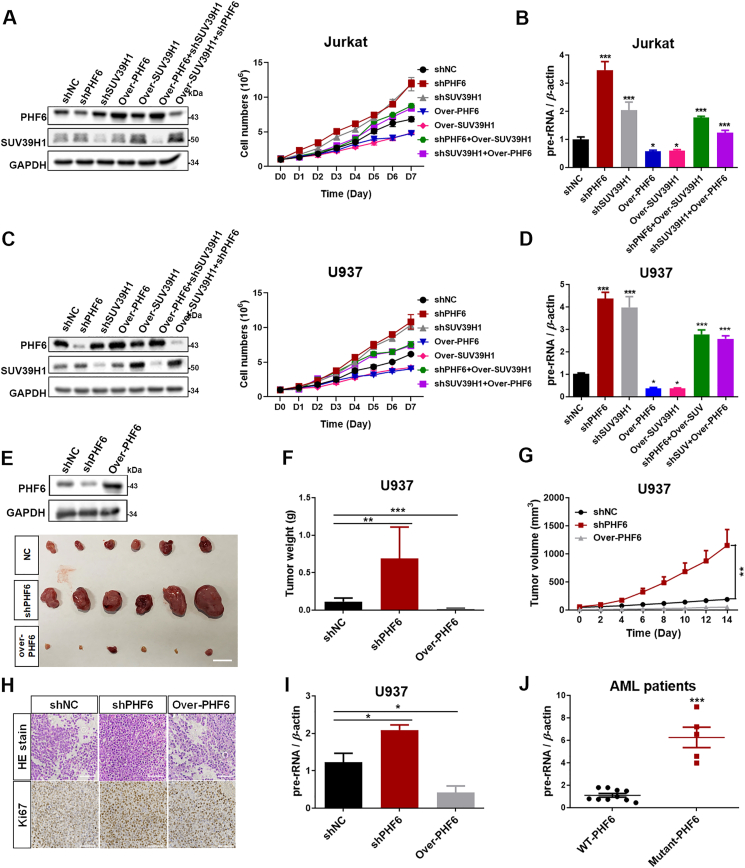
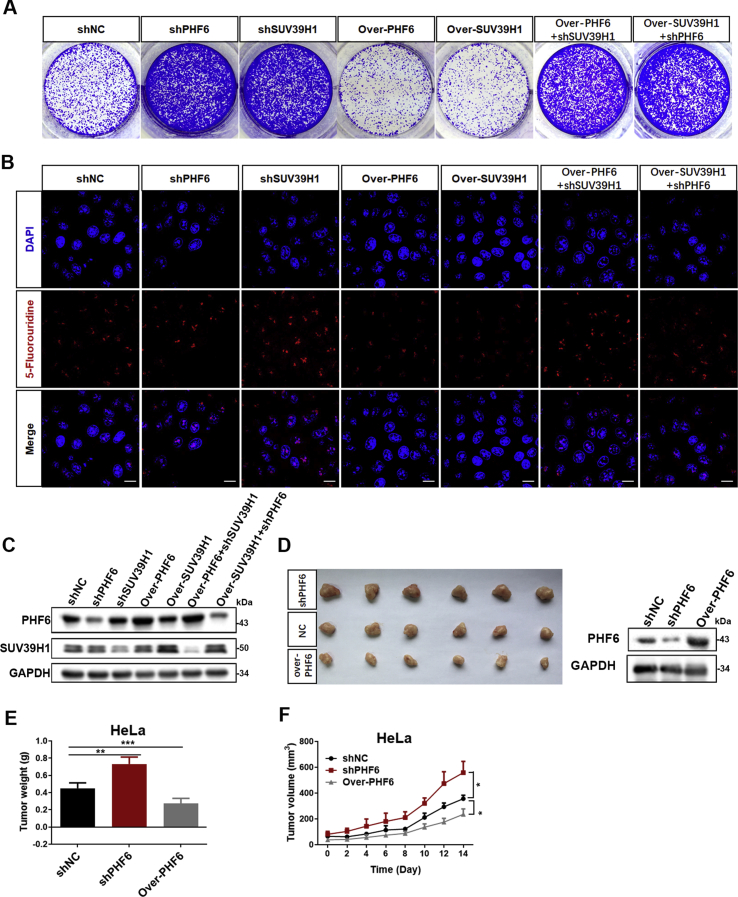
The relationship between PHF6 and SUV39H1 in regulating rDNA transcription and cell proliferation was further investigated. As expected, knockdown of either PHF6 or SUV39H1 significantly promoted rDNA transcription and proliferation of Jurkat and U937 cells, whereas rDNA transcription and proliferation were dramatically decreased following the overexpression of PHF6 or SUV39H1 (Fig. 6A–D). Impressively, the individual overexpression of either PHF6 or SUV39H1 did not effectively suppress SUV39H1 or PHF6 knockdown-induced the increase of rDNA transcription and proliferation in both Jurkat and U937 cells, indicating an interdependent relationship between PHF6 and SUV39H1 in the regulation of the rDNA transcription and cell proliferation. These results were confirmed in more detail in HeLa cell line (Supporting Information Fig. S2A–S2C), whereas the transcription of rDNA and the formation of colonies decreased dramatically following the overexpression of PHF6 or SUV39H1 (Fig. S2A and S2B), thus indicating an interdependent relationship between PHF6 and SUV39H1 in the regulation of transcription of rDNA and proliferation of cells.
Figure 6.
PHF6 and SUV39H1 coordinatively regulate cell growth and proliferation in an interdependent manner. (A) The proliferation of Jurkat cells with the treatments indicated was monitored using a CCK-8 assay (n = 3). Immunoblots of PHF6 and SUV39H1 in the indicated Jurkat cells. (B) qPCR analysis of 47S pre-rRNA from (A) cells (n = 3). (C) The proliferation of U937 cells with the treatments indicated was monitored using a CCK-8 assay (n = 3). Immunoblots of PHF6 and SUV39H1 in the indicated U937 cells. (D) qPCR analysis of 47S pre-rRNA from (C) cells (n = 3). (E)–(G) Xenograft tumor growth of U937 AML cells infected with shNC, shPHF6, and over-PHF6 lentivirus (n = 5). Immunoblot of PHF6 in the indicated tumors. GAPDH was used as loading control. Scale bar, 10 mm. (H) H&E and ki67 staining for U937 xenograft tissues. Scale bar, 100 μm. (I) qPCR analysis of 47S pre-rRNA from U937 xenograft tissues (n = 5). (J) qPCR analysis of 47S pre-rRNA of PBMCs from AML patients (WT-PHF6 = 10 and Mutant-PHF6 = 5). All values are expressed as the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
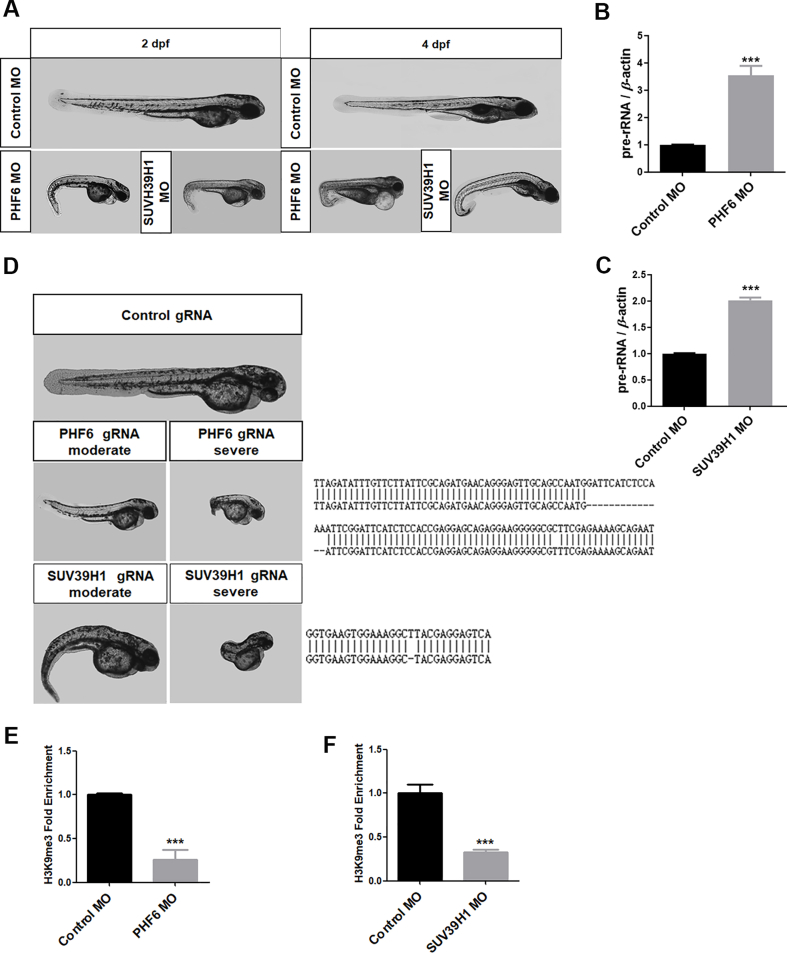
Zebrafish are a popular model vertebrate to investigate gene functions in development. Morpholino or CRSPR technology was used to explore the roles of PHF6 and SUV39H1 during the development of zebrafish. Both deletion of PHF6 and SUV39H1 resulted decreased of H3K9me3 levels in nucleolar chromatin and in the elevation of pre-rRNA transcription (Supporting Information Fig. S3A–S3C), which was accompanied by typical morphological defects, particularly a severe tail bend (Fig. S3D–S3F). This zebrafish model provides additional evidence for the importance of interdependence of PHF6 and SUV39H1 in the regulation of epigenetic status of nucleolar chromatin and subsequent cell proliferation.
3.7. Inhibition of rRNA transcription may be a potential drug target for PHF6 mutations in cancer
Next, the effects of PHF6 on xenograft growth derived from HeLa and AML cell line U937 were evaluated in nude mice. The knockdown of PHF6 significantly promoted the growth of xenografts, whereas its overexpression suppressed the growth of both U937 and HeLa xenografts (Fig. 6E–G and Fig. S2D–S2F). H&E staining and the expression of ki67 also showed that cell proliferation in the PHF6-knockdown U937 tumors was more active than those in the control and U937 tumors in which PHF6 was overexpressed (Fig. 6H). Importantly, the knockdown of PHF6 in U937 tumors resulted in a more significant increase in rRNA levels compared with the control or U937 tumors in which PHF6 was overexpressed (Fig. 6I). Furthermore, we identified five PHF6-mutant AMLs out of 6242 AML patients from mononuclear cell samples derived from bone marrow obtained from the biobank of Zhujiang Hospital of Southern Medical University (Guangzhou, China). Excitingly, the pre-rRNA expression of AML cells from patients who had a mutated form of PHF6 was significantly higher than that from the wild type patients (Fig. 6J). These results indicate that PHF6 and SUV39H1 coordinately regulate the transcription of rDNA and the proliferation of cells and that rDNA transcription might be a potential therapeutic target for leukemia owing to mutant PHF6.
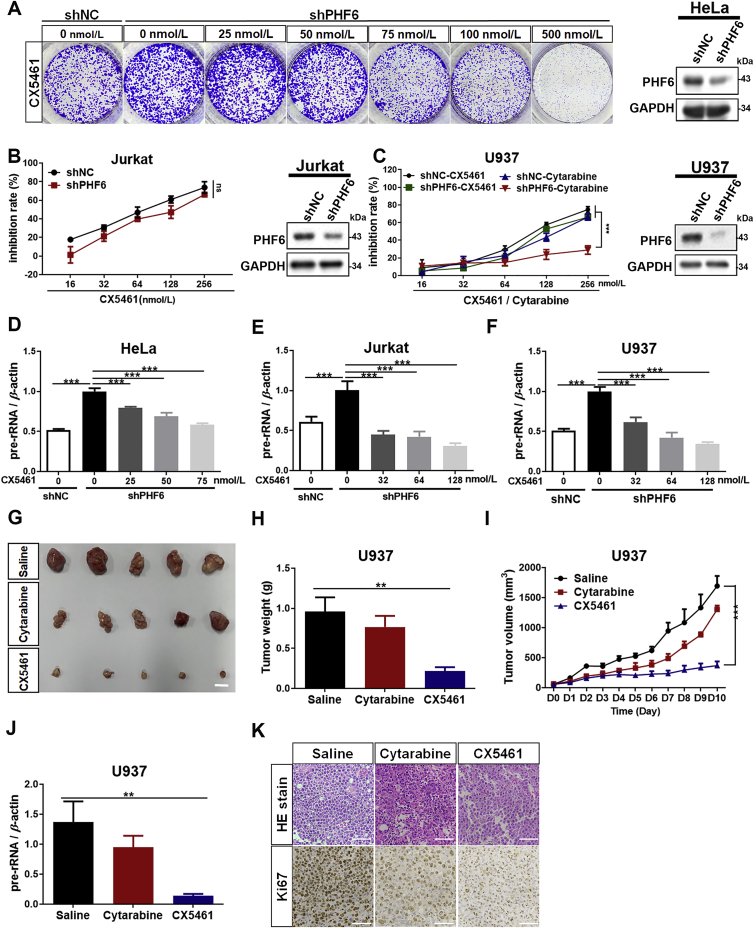
CX5461 is a small molecule drug that targets RNA polymerase I and specifically inhibits the transcription of rDNA. It is currently under evaluation in an FDA approved phase I clinical trial for breast cancer23. The effects of CX5461 on the proliferation in PHF6-knockdown HeLa, Jurkat and U937 cells were evaluated (Fig. 7A–C). CX5461 effectively inhibited the proliferation of HeLa, Jurkat and U937 cells in which PHF6 had been knocked down and was accompanied by a dose-dependent decrease in the levels of transcription of rRNA (Fig. 7D–F). Importantly, compared with the WT cells, Jurkat and U937 cells in which PHF6 had been knocked down did not display significant resistance to treatment with CX5461 (Fig. 7B and C). Cytarabine is the most commonly used to treat different forms of leukemia, including AML, CML, T-ALL, APL and Hodgkin's lymphoma. Although cytarabine is the drug most commonly used to treat AML, a significant proportion of patients do not respond to treatment with cytarabine, as a result of underlying mechanisms of action that have yet to be fully clarified24. In this study, we found that U937 AML cells that were deficient in PHF6 displayed significant resistance to cytarabine compared with the WT U937 cells (Fig. 7C). This resistance was successfully overcome using CX5461 (Fig. 7C and G–K). Accordingly, the expression of pre-rRNA in tumors was also significantly lower in the CX5461 group than those in the cytarabine and saline groups (Fig. 7J). These results were further confirmed by H&E staining and the levels of expression of ki67, indicating that CX5461 was a potent inhibitor of tumors that were deficient in PHF6 (Fig. 7K). These data suggest that the increase in the activity of rRNA transcription induced by mutated PHF6 plays a key role in cytarabine resistance, and ribogenesis might be a potential therapeutic target for PHF6 mutant leukemia.
Figure 7.
CX5461 may be a potential drug for PHF6 mutant leukemia. (A) Cell colonies of PHF6-silenced (shPHF6) or control (shNC) HeLa and (B) Cell viability of PHF6-silenced (shPHF6) or control (shNC) Jurkat cells were treated with different concentrations of CX5461 (0, 16, 32, 64, 128, and 256 nmol/L). Inhibition rate of cell proliferation was monitored using a CCK-8 assay (n = 3). (C) Inhibition rate of cell proliferation in PHF6-silenced (shPHF6) or control (shNC) U937 cells were treated with CX5461 or cytarabine in different concentrations of (0, 16, 32, 64, 128, and 256 nmol/L). The inhibition rate of CX5461 and cytarabine was monitored using a CCK-8 assay (n = 3). Western blots show the protein levels of PHF6 in HeLa, Jurkat and U937 cells. (D)–(F) qPCR analysis of pre-rRNA in PHF6-silenced (shPHF6) or control (shNC) HeLa cells (D), Jurkat (E) and U937 (F) cells treated with different concentration of CX5461, respectively (n = 3). (G)–(I) Xenograft tumor growth of PHF6-silenced U937 AML cells that were treated with saline, CX5461 (1 mg/kg), or cytarabine (1 mg/kg). (J) qPCR analysis of 47S pre-rRNA from (H) tumors. Scale bar, 10 mm. (K) H&E staining and ki67 expression for (H) tumor tissues (n = 5). Scale bar, 100 μm. All the values are expressed as the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
4. Discussion
The transcription of rRNA by RNA polymerase I comprises more than 50% of nuclear transcription (depending on the organism and growth conditions) and is a process central to the growth and proliferation of cells25. Recent evidence clearly suggests that epigenetic mechanisms influence the transcription of rRNA26. For example, NoRC can mediate the silencing of rRNA transcription by recruiting DNA methyltransferase and histone deacetylase to rRNA promoters, which in turn, drive histone H4 deacetylation, H3K9 demethylation, and de novo DNA methylation, and thereby establishes the heterochromatic status at the rDNA promoter region27. Yang et al.28 also identified an energy-dependent nucleolar silencing complex (eNoRC) that contains nucleomethylin, SIRT1, and SUV39H1 that can regulate the repression of energy-dependent rRNA transcription by establishing silent chromatin in the rDNA loci29.
Recently, the nucleolus-localized proteins that contain PHD have been implicated in epigenetic modification. Canonical PHD-containing proteins are common structural components of the chromatin remodeling complex and act as epigenetic decoders to bind post-translationally modified histones, particularly at specific methylation sites30. Most proteins that contain PHD also have a lysine demethylase domain, such as the known nucleolus-localized proteins PHF2 and PHF8 that contain PHD. PHF8 has been shown to specifically recognize modifications in H3K4me2/3 and demethylate the adjacent H3K9me1/2 to activate rDNA transcription17. In contrast, PHF6 is a distinct member of the PHD family that has two atypical PHD-like zinc fingers and does not contain a defined catalytic domain, thus, indicating that there is a different epigenetic modification mechanism of PHF6 in the nucleolus region. In this study, PHF6 was found to function as a reader of histone epigenetic modifications to specifically bind H3K9me3 and H3K27me1 and then recruit methyltransferase SUV39H1 to catalyze the methylation of H3K9 to both establish and maintain the heterochromatin status of rDNA gene loci. The knockdown of SUV39H1 or PHF6 significantly attenuated the increase in the H3K9me3 levels of rDNA gene regions induced by the overexpression of another protein, suggesting an interdependent relationship between SUV39H1 and PHF6 in maintaining the heterochromatin organization of rDNA loci and controlling the transcription of rRNA.
Mutations in the PHF6 gene have recently been described in BFLS, AML, and T-ALL31, 32, 33. In this study, we found that the subcellular localization and pattern of PHF6 mutations identified in BFLS and leukemia patients were significantly altered, indicating that the occurrence or development of BFLS and PHF6-mutant leukemia is closely related to the structural change of chromatin in the rDNA loci and the subsequent activation of rDNA transcription induced by mutant PHF6. In addition, our findings facilitate the development of therapeutic interventions for leukemia caused by PHF6 mutations.
Mutations in the PHF6 gene have recently been described in AML, T-ALL, prostate, bladder, uterine, ovarian, liver cancer, lung cancer, and skin cancer31, 32, 33, 34. Therefore, we speculated that the regulation of rDNA transcription by PHF6 and SUV39H1 might be pan-cancer, but further investigation is needed. In this study, we found that the subcellular localization and pattern of PHF6 mutations identified in BFLS and leukemia patients were significantly altered, indicating that the occurrence or development of BFLS and PHF6-mutant leukemia is closely related to the structural change of chromatin in the rDNA loci and the subsequent activation of rDNA transcription induced by mutant PHF6. In addition, our findings facilitate the development of therapeutic interventions for leukemia caused by PHF6 mutations.
Cytarabine is most commonly used to treat different forms of leukemia32,34. Among the patients with AML mutation in Fig. 6M, we found that three patients carrying PHF6 mutations were resistant to cytarabine treatment. Interestingly, we also found that shPHF6 were resistant to high concentration of cytarabine treatment when compared to shNC control cells (Fig. 7C), which is consistent with the clinical observation. In contrast, shPHF6 were less resistant to CX5461 treatment when compared to shNC control cells (Fig. 7C). These data together indicate that AML patients with PHF6 loss of function mutations may be resistant to cytarabine treatment, and CX5461 alone or in combination with other therapeutic agents has the potential to overcome this drug resistance.
5. Conclusions
Our study demonstrates that PHF6 functions as a tumor suppressor by recognizing H3K9me3 and H3K27me1, recruiting SUV39H1 to catalyze H3K9 methylation, and establishing/maintaining the heterochromatin status of rDNA loci (Supporting Information Fig. S4). The dysregulation of rDNA transcription caused by PHF6 mutations may be a driver of BFLS and PHF6-related leukemia, and the specific rDNA transcription inhibitor CX5461 is a potential drug for overcoming cytarabine resistance in PHF6 mutant leukemia.
Acknowledgments
We are grateful to Sidong Huang for the gift of PHF6 and SUV39H1 shRNA plasmids (McGill University, Canada). And pHrD-IRES-Luc plasmid was provided by Yang Ke and Xiaojuan Du (Peking University, China). This research was supported by the National Natural Science Foundation of China (81702750, 81670141, 81970145and 82001698); Natural Science Foundation of Guangdong Province (2020A1515011465and 2020A151501467, China); Science, Technology & Innovation Commission of Shenzhen Municipality ( JCYJ20180307154700308, JCYJ20170818163844015, JCYJ20180307151420045, JCYJ20190807151609464, JCYJ20200109142605909 and JCYJ20210324120007020, China); Sun Yat-sen University (20ykzd17 and 20ykpy122, China); International Collaboration of Science and Technology of Guangdong Province (2020A0505100031, China); Guangdong Provincial Key Laboratory of Digestive Cancer Research (No. 2021B1212040006, China); The Social Development Foundation of Jiangsu Province (BE2018691, China) and Sigrid Jusélius foundation in Finland for funding the project (Finland).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.10.025.
Contributor Information
Fang Cheng, Email: chengf9@mail.sysu.edu.cn.
Laiqiang Huang, Email: huanglq@mail.tsinghua.edu.cn.
Hongbo Chen, Email: chenhb7@mail.sysu.edu.cn.
Author contributions
Hsiang-i Tsai, Yanping Wu, Dandan Su, Yingyi Wu, Xiaoyan Liu and Linglu Wang, Chao He and Fan Shu performed the experiments and analyzed the data. Rui Huang and Zhanxue Xu prepared the clinical and pathological data of AML patients. Yuxin Pang, Chong Sun, Haitao Zhu and Dongqing Wang provided experimental materials. Hsiang-i Tsai, Yanping Wu, Fang Cheng and Hongbo Chen wrote the manuscript. Fang Cheng, Laiqiang Huang and Hongbo Chen designed and supervised this project. The author(s) read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig S1.
Fig S2.
Fig S3.
Fig S4.
References
- 1.Lower K.M., Turner G., Kerr B.A., Mathews K.D., Shaw M.A., Gedeon A.K., et al. Mutations in PHF6 are associated with Borjeson–Forssman–Lehmann syndrome. Nat Genet. 2002;32:661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 2.Bellad A., Bandari A.K., Pandey A., Girimaji S.C., Muthusamy B. A novel missense variant in PHF6 gene causing Borjeson–Forssman–Lehman syndrome. J Mol Neurosci. 2020;70:1403–1409. doi: 10.1007/s12031-020-01560-5. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Li F., Ruan K., Zhang J., Mei Y., Wu J., et al. Structural and functional insights into the human Borjeson–Forssman–Lehmann syndrome-associated protein PHF6. J Biol Chem. 2014;289:10069–10083. doi: 10.1074/jbc.M113.535351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahani-Asl A., Cheng C., Zhang C., Bonni A. Pathogenesis of borjeson–forssman–lehmann syndrome: insights from PHF6 function. Neurobiol Dis. 2016;96:227–235. doi: 10.1016/j.nbd.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendorff A.A., Quinn S.A., Rashkovan M., Madubata C.J., Ambesi-Impiombato A., Litzow M.R., et al. Phf6 loss enhances HSC self-renewal driving tumor initiation and leukemia stem cell activity in T-ALL. Cancer Discov. 2019;9:436–451. doi: 10.1158/2159-8290.CD-18-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ntziachristos P. PHF6: it is written in the stem cells. Blood. 2019;133:2461–2462. doi: 10.1182/blood.2019000810. [DOI] [PubMed] [Google Scholar]
- 7.Mori T., Nagata Y., Makishima H., Sanada M., Shiozawa Y., Kon A., et al. Somatic PHF6 mutations in 1760 cases with various myeloid neoplasms. Leukemia. 2016;30:2270–2273. doi: 10.1038/leu.2016.212. [DOI] [PubMed] [Google Scholar]
- 8.Xiao W., Bharadwaj M., Levine M., Farnhoud N., Pastore F., Getta B.M., et al. PHF6 and DNMT3A mutations are enriched in distinct subgroups of mixed phenotype acute leukemia with T-lineage differentiation. Blood Adv. 2018;2:3526–3539. doi: 10.1182/bloodadvances.2018023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo N.J., Kim Y.R., Lee S.H. Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncol. 2012;51:107–111. doi: 10.3109/0284186X.2011.592148. [DOI] [PubMed] [Google Scholar]
- 10.Todd M.A., Ivanochko D., Picketts D.J. PHF6 degrees of separation: the multifaceted roles of a chromatin adaptor protein. Genes. 2015;6:325–352. doi: 10.3390/genes6020325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng C., Deng P.Y., Ikeuchi Y., Yuede C., Li D., Rensing N., et al. Characterization of a mouse model of Borjeson–Forssman–Lehmann syndrome. Cell Rep. 2018;25:1404–14014.e6. doi: 10.1016/j.celrep.2018.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 13.Todd M.A., Picketts D.J. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res. 2012;11:4326–4337. doi: 10.1021/pr3004369. [DOI] [PubMed] [Google Scholar]
- 14.Todd M.A., Huh M.S., Picketts D.J. The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events. Eur J Hum Genet. 2016;24:1453–1459. doi: 10.1038/ejhg.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Leung J.W., Gong Z., Feng L., Shi X., Chen J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J Biol Chem. 2013;288:3174–3183. doi: 10.1074/jbc.M112.414839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Sullivan A.C., Sullivan G.J., McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 18.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 19.Catez F., Dalla Venezia N., Marcel V., Zorbas C., Lafontaine D.L.J., Diaz J.J. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem Pharmacol. 2019;159:74–81. doi: 10.1016/j.bcp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 20.McCann T.S., Sobral L.M., Self C., Hsieh J., Sechler M., Jedlicka P. Biology and targeting of the Jumonji-domain histone demethylase family in childhood neoplasia: a preclinical overview. Expert Opin Ther Targets. 2019;23:267–280. doi: 10.1080/14728222.2019.1580692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritsch L., Robin P., Mathieu J.R., Souidi M., Hinaux H., Rougeulle C., et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Baker L.A., Allis C.D., Wang G.G. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilton J., Cescon D., Bedard P., Ritter H., Tu D., Soong J., et al. CCTG IND. 231: a phase 1 trial evaluating CX-5461 in patients with advanced solid tumors. Ann Oncol. 2018;29:iii8. [Google Scholar]
- 24.Galmarini C.M., Thomas X., Calvo F., Rousselot P., El Jafaari A., Cros E., et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk Res. 2002;26:621–629. doi: 10.1016/s0145-2126(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 25.Sharifi S., Bierhoff H. Regulation of RNA polymerase I transcription in development, disease, and aging. Annu Rev Biochem. 2018;87:51–73. doi: 10.1146/annurev-biochem-062917-012612. [DOI] [PubMed] [Google Scholar]
- 26.Rossetti S., Wierzbicki A.J., Sacchi N. Mammary epithelial morphogenesis and early breast cancer. Evidence of involvement of basal components of the RNA polymerase I transcription machinery. Cell Cycle. 2016;15:2515–2526. doi: 10.1080/15384101.2016.1215385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forestan C., Farinati S., Rouster J., Lassagne H., Lauria M., Dal Ferro N., et al. Control of maize vegetative and reproductive development, fertility, and rRNAs silencing by histone deacetylase 108. Genetics. 2018;208:1443–1466. doi: 10.1534/genetics.117.300625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L., Song T., Chen L., Kabra N., Zheng H., Koomen J., et al. Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol Cell Biol. 2013;33:3835–3848. doi: 10.1128/MCB.00476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musselman C.A., Lalonde M.E., Cote J., Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 31.Chao M.M., Todd M.A., Kontny U., Neas K., Sullivan M.J., Hunter A.G., et al. T-cell acute lymphoblastic leukemia in association with Borjeson–Forssman–Lehmann syndrome due to a mutation in PHF6. Pediatr Blood Cancer. 2010;55:722–724. doi: 10.1002/pbc.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Vlierberghe P., Patel J., Abdel-Wahab O., Lobry C., Hedvat C.V., Balbin M., et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130–134. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Vlierberghe P., Palomero T., Khiabanian H., Van der Meulen J., Castillo M., Van Roy N., et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowenberg B., Pabst T., Vellenga E., van Putten W., Schouten H.C., Graux C., et al. Cytarabine dose for acute myeloid leukemia. N Engl J Med. 2011;364:1027–1036. doi: 10.1056/NEJMoa1010222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.