Abstract
A total of 32 Listeria monocytogenes strains (16 from a recent outbreak of invasive listeriosis and 16 from two outbreaks of noninvasive listeriosis, all three occurring in Italy) were characterized by PCR-ribotyping, arbitrarily primed PCR (AP-PCR), and the recently developed infrequent-restriction-site PCR (IRS-PCR). The discriminatory ability of the techniques, first evaluated on 29 unrelated L. monocytogenes food isolates using Simpson's index of diversity, was 0.714 for PCR-ribotyping, 0.690 for AP-PCR, and 0.919 for IRS-PCR. IRS-PCR was also more capable of distinguishing among strains from the invasive listeriosis outbreak: three different clusters were identified by IRS-PCR compared to two clusters identified by both PCR-ribotyping and AP-PCR. Within each of the two outbreaks of noninvasive listeriosis, the patterns were practically identical, as demonstrated by all three techniques. Only IRS-PCR succeeded in clearly discriminating the strains related to noninvasive listeriosis from all of the other strains included in this study, including those from the outbreak of invasive listeriosis. This finding may suggest the presence of unique differences in their DNA sequences.
Listeria monocytogenes has been recognized as a foodborne pathogen causing listeriosis in humans since the 1980s, when a number of listeriosis outbreaks were found to be associated with the consumption of contaminated foodstuffs. Until recently, food-borne listeriosis was commonly regarded as an invasive disease that affected only susceptible population groups (e.g., immunocompromised persons, newborn children, and pregnant women); it was considered to be associated with bacteremia only in certain target organs and was only rarely associated with gastrointestinal symptoms (24). However, recent reports of a new noninvasive form of listeriosis that causes febrile gastroenteritis clearly indicate that persons with no predisposing conditions may be affected. This finding increases the public health significance of L. monocytogenes (2, 3, 12, 19, 22), in that the emergence of this new form of food-borne listeriosis could affect the epidemiology of the disease both in terms of a higher attack rate and in terms of a wider range of potential vehicles. However, the factors that influence the infectious dose and the occurrence and course of infection still need to be clarified. It is possible that the clonal variants of the pathogen each interact with the host in a different way (6, 26). Of the high-resolution molecular typing methods for assessing genetic heterogeneity, fingerprinting techniques based on PCR have proven to be among the most effective (27). They are generally based on enzymatic amplification through PCR of DNA segments flanking either undetermined sequences (e.g., randomly amplified polymorphic DNA [RAPD] and arbitrarily primed PCR [AP-PCR] [15, 16]) or defined and conserved sequences (e.g., rRNA genes [PCR-ribotyping] [25] and the repetitive-element families [ERIC- and REP-PCR] [14, 23]).
A new PCR-based fingerprinting technique, known as infrequent-restriction-site PCR (IRS-PCR), based on the selective amplification of DNA restriction fragments, has recently been developed and used to type several bacterial species (17, 21, 30). We applied this technique to related and unrelated L. monocytogenes isolates and compared the results to those obtained by PCR-ribotyping and AP-PCR. The related strains were those isolated during the investigations of an outbreak of invasive listeriosis and two outbreaks of noninvasive listeriosis that recently occurred in Italy. The outbreak of invasive listeriosis occurred in 1998: six hospitalized patients, three of whom were heart transplanted, while the others, affected with Woldenstrom's syndrome, hepatic cirrhosis, and chronic hepatitis, respectively, had developed mild to severe (meningoencephalitis) symptoms of listeriosis while in the same hospital (P. Aureli and G. Franciosa, unpublished data). The two outbreaks of noninvasive listeriosis occurred in 1994 and 1997 (2, 22); both involved a large number of immunocompetent persons, and L. monocytogenes was detected from the clinical specimens and from the environmental samples collected where the suspected food sources had been prepared. However, whereas in the 1994 outbreak the implicated food remained uncertain (22), in the 1997 outbreak it was clearly identified as a cold salad of corn and tuna fish (2).
The main objective of the present study was to verify the causal relationships among the L. monocytogenes strains isolated from each outbreak and to compare the epidemic clones to isolates recovered from different types of foods.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
A total of 61 L. monocytogenes isolates were used in this study (Table 1). A total of 29 of them were unrelated strains and, of these, 25 were from different types of food recovered over a period of 2 years (i.e., 1997 and 1998) and were randomly selected from the culture collection at the Food Microbiology Laboratory of the Istituto Superiore di Sanità. L. monocytogenes ScottA (a clinical isolate of serotype 4b [7]), kindly provided by M. P. Doyle, University of Georgia, Griffin, was included in the experiments as a reference strain. Strains 26 through 28 were clinical isolates of serotype 4b obtained, respectively, from two elderly people (strains 26 and 27) and a newborn (strain 28). These strains were sent to our laboratory by some local health units of southern and northern Italy.
TABLE 1.
L. monocytogenes isolates analyzed by PCR-fingerprinting techniques
| Category and strain | Serotype | Source | ||||
|---|---|---|---|---|---|---|
| Unrelated | ||||||
| 1 | 1/2a | Chicken livers | ||||
| 2 | 1/2a | Green salad | ||||
| 3 | 1/2a | Green salad | ||||
| 4 | 1/2a | Octopus | ||||
| 5 | 1/2a | Boiled ham | ||||
| 6 | 1/2a | Boiled ham | ||||
| 7 | 1/2a | Pesto sauce | ||||
| 8 | 1/2a | Cheese | ||||
| 9 | 1/2a | Octopus salad | ||||
| 10 | 1/2b | Mozzarella | ||||
| 11 | 1/2b | Ice cream | ||||
| 12 | 1/2b | Mixed salad | ||||
| 13 | 1/2b | Meat roll | ||||
| 14 | 1/2c | Meat | ||||
| 15 | 3a | Nut sauce | ||||
| 16 | 3a | Truffle sauce | ||||
| 17 | 3c | Surimi | ||||
| 18 | 4ab | Sandwich | ||||
| 19 | 4b | Sausage | ||||
| 20 | 4b | Green salad | ||||
| 21 | 4b | Cheese | ||||
| 22 | 4b | Pesto sauce | ||||
| 23 | 4b | Cured meat | ||||
| 24 | 4b | Frozen cod | ||||
| 25 | 4b | ScottA, clinical | ||||
| 26 | 4b | Clinical | ||||
| 27 | 4b | Clinical | ||||
| 28 | 4b | Clinical | ||||
| 29 | 4d | Smoked salmon | ||||
| Outbreak 1 (invasive listeriosis) | ||||||
| 30 | 1/2a | Body fluid | ||||
| 31 | 1/2b | Body fluid | ||||
| 32 | 1/2b | Body fluid | ||||
| 33 | 1/2b | Body fluid | ||||
| 34 | 1/2b | Body fluid | ||||
| 35 | 1/2b | Body fluid | ||||
| 36 | 1/2a | Frozen peas | ||||
| 37 | 1/2a | Frozen mushrooms | ||||
| 38 | 1/2a | Frozen potatoes | ||||
| 39 | 1/2b | Roasted chicken | ||||
| 40 | 1/2b | Cooked vegetables | ||||
| 41 | 1/2b | Cooked vegetables | ||||
| 42 | 1/2b | Boiled chicken | ||||
| 43 | 1/2b | Boiled trout | ||||
| 44 | 1/2b | Floor drain | ||||
| 45 | 1/2b | Trencher | ||||
| Outbreak 2a (noninvasive listeriosis) | ||||||
| 46 | 1/2b | Blood | ||||
| 47 | 1/2b | Blood | ||||
| 48 | 1/2b | Shrimp tart | ||||
| 49 | 1/2b | Cheese tart | ||||
| 50 | 1/2b | Fruit cake | ||||
| 51 | 1/2b | Freezer | ||||
| 52 | 1/2b | Mixer | ||||
| Outbreak 3b (noninvasive listeriosis) | ||||||
| 53 | 4b | Blood | ||||
| 54 | 4b | Stool | ||||
| 55 | 4b | Stool | ||||
| 56 | 4b | Stool | ||||
| 57 | 4b | Stool | ||||
| 58 | 4b | Stool | ||||
| 59 | 4b | Sweet corn salad | ||||
| 60 | 4b | Floor drain | ||||
| 61 | 4b | Sink drain |
Of the 61 outbreak isolates, 32 (strains 30 to 61) were taken from sources associated with three separate listeriosis outbreaks (Table 1). Strains 30 to 45 were from the outbreak of invasive listeriosis (here referred to as outbreak 1) (Aureli and Franciosa, unpublished). L. monocytogenes had been isolated from the body or tissue fluids of all six hospital patients involved, from some of the foods collected and stored for the hospital's routine quality control analyses (both cooked foods served to the patients and frozen foods), and from two environmental swabs from the hospital's kitchen. Strains 46 through 52 (outbreak 2) and 53 through 61 (outbreak 3) included some human, food, and environmental isolates recovered during the investigation of the 1994 and 1997 outbreaks of noninvasive listeriosis (2, 22). All of the strains had been identified by conventional procedures (5) at the time of isolation; they were further confirmed as L. monocytogenes by PCR detection of some virulence genes (i.e., hly, plcA, plcB, and their transcriptional activator prfA), as previously described (10).
All cultures of the listeria strains were checked for purity on Oxford agar plates (Oxoid, Basingstoke, United Kingdom) prior to the experiments. Single colonies were inoculated into tryptone soy broth (TSB; Oxoid) and incubated at 37°C for 24 h. Then, 1 ml of these broth cultures was used for DNA extraction.
Serotyping.
All of the strains were serotyped on the basis of somatic (O) and flagellar (H) antigens by a commercial kit (Listeria O Antisera; Denka Seiken, Tokyo, Japan) according to the manufacturer's instructions.
Preparation of template DNA.
Either purified DNA or DNA from listeria cell lysates was used directly as the template in all PCR amplification assays except for the IRS-PCR assay, which requires the pretreatment of DNA, performed as described below.
Genomic DNA was extracted and purified as reported elsewhere (8). The DNA from each strain was dissolved in 150 μl of 1× TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0). Yields were estimated by using a UV spectrophotometer (GeneQuant II; Pharmacia Biotech, Cambridge, England), and DNA samples were stored at −20°C until use.
Listeria cell lysates were obtained by centrifuging 1 ml of 24-h TSB cultures; the pellets were washed twice with 1× TE buffer, suspended in 1 ml of sterile distilled water, and subjected to ultrasound treatment (model T460; Trans-Sonic, Singen, Germany) for 10 min to facilitate lysis. Lysates were freshly prepared for each experiment.
To obtain the appropriate template for the IRS-PCR experiments, we adopted the procedure of Riffard et al. (21), with some modifications. Bacterial DNA (either pure [2 μg] or from the crude lysates [18 μl], without further purification) was first cleaved with 40 U of XbaI and 40 U of PstI (Boehringer, Mannheim, Germany) in a total reaction volume of 25 μl overnight at 37°C. The resulting restriction fragments were stored at −20°C until subsequent ligation to double-stranded restriction halfsite-specific adapters (AX and PS) (17, 21). Specifically, the adapters were the partially complementary oligonucleotides AX1 (5′-CTA GTA CTG GCA GAC TCT-3′) and AX2 (5′-GCCAGT A-3′) (for XbaI) and PS1 (5′-GAC TCG ACT CGC ATG CA-3′) and PS2 (5′-TGC GAG T-3′) (for PstI) (Biogen, Rome, Italy). Equal molar amounts of AX1 (previously phosphorylated at the 5′ end by T4 Polynucleotide Kinase 3′-Phosphatase Free [Boehringer], as recommended by the manufacturer) and AX2 and of PS1 and PS2 were mixed in 1× PCR buffer (Perkin-Elmer Cetus, Branchburg, N.J.): annealing was performed with a thermal cycler (Model PTC-150 Minicycler; M. J. Research, Inc., Watertown, Mass.) programmed to decrease the temperature from 80 to 4°C over 1 h. The restriction fragments were ligated to the adapters in accordance with the procedures of Riffard et al. (21). Finally, the ligation products were redigested with 10 U of XbaI and 10 U of PstI at 37°C for 30 min.
XbaI-PstI restriction fragments tagged with specific adapters were kept at −20°C until their use as templates in the IRS-PCR experiments.
PCR conditions.
All reaction mixtures contained, in a total volume of 50 μl, 1× PCR buffer II (10 mM Tris-HCl, 50 mM KCl [pH 8.3]; Perkin-Elmer Cetus), 200 μM concentrations of each deoxynucleoside triphosphate (Pharmacia), and 1.25 U of Taq polymerase (Perkin-Elmer Cetus). The final concentrations of MgCl2 (Perkin-Elmer Cetus) were adjusted to 1.5 mM in the IRS-PCR experiments and to 3 mM in the PCR-ribotyping and AP-PCR experiments.
The PCR-ribotyping and AP-PCR experiments were performed with both purified DNA (2 ng) and crude lysates (2.5 μl); when crude lysates were used, PCR mixtures were heated in a thermal cycler at 99°C for 10 min prior to the addition of the polymerase for a better release of the DNA. The IRS-PCR experiments were performed using as templates for amplification 5 μl of the ligation products obtained after restriction either of purified DNA or of the genetic material released from the lysed bacteria.
We used different oligonucleotide primers (Biogen) and amplification parameters for each technique, as follows.
(i) PCR-ribotyping was carried out according to the method of Sontakke and Farber (25). The primers for the amplification of the DNA spacer regions between the 16S and 5S rRNA genes were P (5′-TTG TAC ACA CCG CCC GTC A-3′) and M (5′-GCTTAA CTT CCG TGT TCG GTA TGG G-3′). Amplification was achieved after initial denaturation of the template at 94°C for 2 min, followed by 35 cycles at 94°C for 1 min, 35°C for 1 min, and 72°C for 2.5 min, with a ramp time of 2 min between 35 and 72°C; a final extension phase was performed at 72°C for 5 min.
(ii) For AP-PCR, the primer M13 (5′-GTT GTA AAA CGA CGG CCA GT-3′), derived from the core sequence of the bacteriophage M13 genome, was used at a final concentration of 0.5 μM. This primer was selected from among 10 different 20-mer oligonucleotides, previously tested with different serotypes of L. monocytogenes, for its higher discriminatory power. The use of primers selected from the bacteriophage M13 sequence has been recommended for detecting DNA polymorphisms in virtually all species (4). The temperature profile consisted of two cycles at 94°C for 5 min, 40°C for 5 min, and 72°C for 5 min, followed by 40 high-stringency cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. Both the primer sequence and the temperature profile were adapted from the method of Vila et al. (28).
(iii) For IRS-PCR, primers PS1 and PX-G (5′-AGA GTC TGC CAG TAC TAG AG-3′) (17) for the selective amplification of the XbaI-PstI restriction fragments were used at a final concentration of 1 μM. The cycling conditions were the same as those described by Mazurek et al. (17). After initial denaturation at 94°C for 5 min, the PCR mixtures were subjected to 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 90 s; a final extension was performed at 72°C for 5 min.
The amplification experiments were performed in duplicate with a thermal cycler (model PT100; M. J. Research) to assess the reproducibility of the patterns.
Analysis of the amplification products.
The PCR products (35 μl) were separated by electrophoresis on a 1.5 or 2% agarose gel containing ethidium bromide (0.5 μg/ml) at 60 V for 4 h in 1× TAE (Tris-acetate-EDTA) buffer. Gel images were digitized through a UV-gel image acquisition camera (Gel Doc 1000; Bio-Rad Laboratories, Hercules, Calif.). Intergel comparison was performed with the GelCompar 3.1 system (Applied Maths, Kortrijk, Belgium) (9). Following normalization and background subtraction with mathematical algorithms, three separate lists were created for the reconstructed patterns of PCR-ribotyping, AP-PCR, and IRS-PCR. Finally, the similarities between profiles within each list were calculated by applying the Pearson product-moment correlation coefficient (r), and clustering was performed by the unweighted pair-group method using arithmetic averages (UPGMA). An intralinkage homology level of ≥80% between patterns was assumed as the cutoff for defining a close genetic relationship between strains and was used to define the clusters.
RESULTS AND DISCUSSION
Unrelated isolates.
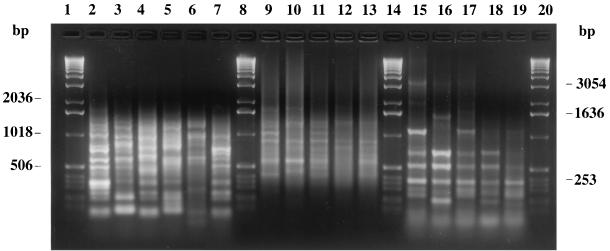
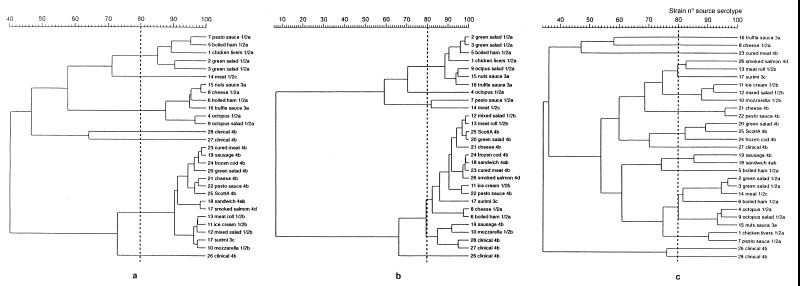
Based on the results of serotyping, the 25 unrelated L. monocytogenes food strains were divided into eight different serovars, distributed as follows: 1/2a (n = 9), 4b (n = 6), 1/2b (n = 4), 3a (n = 2), 1/2c (n = 1), 3c (n = 1), 4ab (n = 1), and 4d (n = 1) (Table 1). Hence, serovars 1/2a, 1/2b, and 4b, which commonly cause the clinical disease, were the most frequently detected. In no case was the prevalence of a serovar associated with a specific food. The clinical isolates were confirmed as serotype 4b. The number of bands varied according to the specific PCR-based fingerprinting technique: PCR-ribotyping showed from 4 to 11 bands, AP-PCR showed from 5 to 13 bands, and the amplification of the XbaI-PstI restriction fragments by IRS-PCR showed from 6 to 11 bands. All bands were evenly distributed along the tracks and shorter than 2,036 bp (Fig. 1). PCR-ribotyping, AP-PCR, and IRS-PCR revealed percentages of similarity greater than 83, 87, and 90%, respectively, when comparing the analysis using purified DNA to that using crude cell lysates. Since these differences did not affect the final clustering of the genetic profiles, we used the patterns obtained with cell lysates to construct the UPGMA dendrograms. Computer analysis showed that PCR-ribotyping and AP-PCR produced, respectively, 7 and 6 clusters of strains and that neither technique was capable of distinguishing among serotypes (Fig. 2a and b), whereas IRS-PCR produced 13 clusters and was generally able to distinguish among serotypes. Specifically, the degree of differentiation was greatest among the most common serovars (i.e., 1/2a, 1/2b, and 4b), whereas the single representatives of the rarer serovars was not distinguished (i.e., 1/2c and 3a clustered with a 1/2a strain, 4d clustered with a 1/2b strain, and 4ab clustered with a 4b strain) (Fig. 2c). These results are consistent with the divisions of L. monocytogenes strains obtained with other techniques, specifically, restriction fragment length polymorphism analysis (29), multilocus enzyme electrophoresis (MEE) (11), ribotyping (11), pulsed-field gel electrophoresis (PFGE) (20), and the recently developed amplified fragment length polymorphism (AFLP) technique (1). AFLP is conceptually similar to IRS-PCR, in that both methods are based on the selective amplification by PCR of double-digested genomic DNA. However, AFLP has a number of drawbacks, all of which can be overcome with IRS-PCR; specifically, in AFLP, the two adapters are of equal length, neither is phosphorylated (hence, they are not prevented from acting as primers), one of the primers is radioactively or fluorescently labeled, denaturing polyacrylamide gel electrophoresis is required to resolve the large number of products generated, and sophisticated detection systems must be employed to detect the fingerprints. All replications with IRS-PCR showed identical profiles, demonstrating this technique's high level of reproducibility.
FIG. 1.
Representative fingerprints of outbreak-related L. monocytogenes strains produced by IRS-PCR (lanes 2 to 7), AP-PCR (lanes 9 to 13), and PCR-ribotyping (lanes 15 to 19). Lanes 2, 9, and 15, strain 27; lanes 3, 10, and 16, strain 33; lanes 4, 11, and 17, strain 40; lanes 5, 12, and 18, strain 49; lane 6, strain 43; lanes 7, 13, and 19, strain 54; lanes 1, 8, 14, and 20, 1-kb DNA ladder (Gibco-BRL).
FIG. 2.
Dendrograms representing genetic relationships between unrelated L. monocytogenes isolates based on profiles obtained by PCR-ribotyping (a), AP-PCR (b), and IRS-PCR (c).
Interestingly, both PCR-ribotyping and IRS-PCR separated all the unrelated clinical isolates of serotype 4b but L. monocytogenes ScottA from the food isolates (Fig. 2a and c). At an 80% clone cutoff value for the Pearson coefficient, the highest discriminatory ability, as determined by Simpson's index of diversity (13), was found for IRS-PCR (0.919 compared to 0.714 for PCR-ribotyping and 0.690 for AP-PCR). We used the core sequence of bacteriophage M13 as the random primer for the AP-PCR experiments in order to assess this primer's suitability in discriminating among L. monocytogenes isolates. However, low-resolution bands with a high background were mostly obtained, with a consequent low degree of discrimination.
Isolates from the outbreak of invasive listeriosis (outbreak 1).
One clinical L. monocytogenes isolate and all of the isolates recovered from the frozen foods (Table 1) were identified as serotype 1/2a, whereas the remaining isolates, including those from the environment, were serotype 1/2b. The dendrograms of the profiles obtained with PCR-ribotyping and AP-PCR also consisted of two clusters. However, whereas the grouping obtained with PCR-ribotyping was identical to that of serotyping, when using AP-PCR, the clinical isolate of serotype 1/2a clustered with the 1/2b isolates. In contrast to the other techniques, IRS-PCR revealed three different clusters. Specifically, five clinical isolates and all of the isolates from the cooked foods and the environment, all serotype 1/2b, were in the same cluster (89% intralinkage homology level), suggesting that this outbreak was likely due to cross-contamination. A second cluster consisted of all of the isolates from frozen foods, all serotype 1/2a, which were strictly interrelated (87% homology level). The third cluster consisted of the one clinical isolate of serotype 1/2a, which showed a homology level of <70% with the second cluster and could therefore be discriminated. These results can be considered as plausible, since the frozen foods were boiled before being served and thus probably did not account for any cases of listeriosis. The clinical isolate of serotype 1/2a may have originated from a different unidentified source of infection. Figure 1 shows representative IRS-PCR, AP-PCR, and PCR-ribotyping profiles of the outbreak-related L. monocytogenes strains.
Isolates from the outbreaks of noninvasive listeriosis (outbreaks 2 and 3).
The clinical, food, and environmental isolates from outbreaks 2 and 3 were confirmed as belonging to serotypes 1/2b and 4b, respectively (Table 1). Highly homologous PCR-based fingerprints were obtained for all of the isolates associated with outbreak 2, except for isolate 52 (isolated from a mixer), which had a homology level of <20% according to IRS-PCR and of <50% according to both PCR-ribotyping and AP-PCR. These findings are consistent with the previous subtyping of the same strains by MEE, PFGE, and RAPD, and they substantiate the lack of causal relationship for the isolate from the mixer (8, 22).
All of the strains from outbreak 3 showed nearly identical profiles with all PCR-based typing techniques, resulting in homology levels among isolates of >90, 97, and 93% when IRS-PCR, AP-PCR, and PCR-ribotyping, respectively, were used. Previous analysis of the same strains, using PFGE and RAPD, also showed indistinguishable patterns (2).
These results strongly support the hypothesis of extensive cross-contamination as the primary cause of infection in both outbreaks (2, 22); indeed, some environmental isolates showed the same profiles as those of the food and clinical isolates.
Comparison between related and unrelated strains.
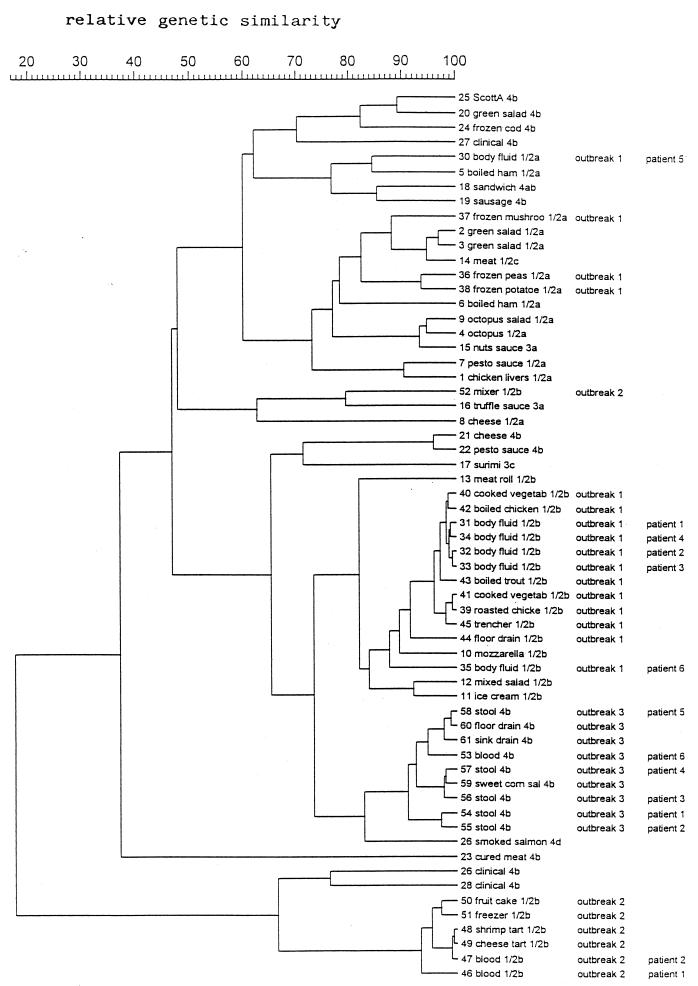
The comparison between the IRS-PCR patterns of the related and unrelated isolates revealed the following: (i) the strains from outbreak 1 (invasive listeriosis) were divided into three clusters, all of which included some unrelated food isolates of similar serotypes (i.e., 1/2a and 1/2b); (ii) all but one of the related strains of outbreak 2 (noninvasive listeriosis) clustered apart from all of the other isolates tested in this study, whereas the isolate from the mixer, which was not apparently implicated in the outbreak, clustered with a strain of serotype 3a; and (iii) the strains from outbreak 3 (noninvasive listeriosis) clustered apart from all other strains tested, although the homology level was approximately 75% with respect to some strains of serotype 1/2b, including those involved in outbreak 1 (Fig. 3). The strains from outbreak 3 were also distinguished from L. monocytogenes ScottA and the other clinical isolates of serotype 4b, the relative genetic similarity level between them being <50% (Fig. 3).
FIG. 3.
Dendrogram representing the genetic relationships between total L. monocytogenes isolates based on IRS-PCR fingerprinting.
The results of IRS-PCR indicate that the strains implicated in the two outbreaks of noninvasive listeriosis can be separated from all of the other isolates considered in this study. For outbreak 2, this could be partly due to the fact that the strains had been isolated at least 3 years earlier than all of the other strains. Alternatively, it has been hypothesized that the different pathways of the disease (invasive and noninvasive) might be related to differences in the pathogenicity of the causative L. monocytogenes strains, which is determined by specific virulence-associated genes (19); consequently, differences in these genes' sequences and/or in the sequences of the genes that control their phenotypic expression might account for the unique patterns. On the other hand, strains involved in the outbreak of invasive listeriosis generated IRS-PCR profiles comparable to those from some food isolates randomly selected from our culture collection. This may suggest that strains potentially capable of causing invasive listeriosis are more widespread than those responsible for noninvasive listeriosis, thus explaining why noninvasive disease has been reported only rarely. Another currently accepted hypothesis is that the occurrence of noninvasive listeriosis may be underestimated because L. monocytogenes is not among the pathogens routinely investigated in outbreaks of gastrointestinal illness (2, 3, 22). These conclusions cannot be drawn from the results obtained with PCR-ribotyping and AP-PCR, probably because of the lower discriminative power of these techniques.
To verify the findings of this study, further analyses are currently being conducted with additional L. monocytogenes strains from different sources and using other sensitive molecular techniques.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Program of the Ministry of Health.
We thank Peter Ben Embarek of the Fisheries Department, Food and Agriculture Organization of the United Nations (FAO), Rome, Italy, for revision of the manuscript and Mark Kanjeff, Laboratorio Epidemiologia e Biostatistica, ISS, for English editing.
REFERENCES
- 1.Aarts H J M, Hakemulder L E, Van Hoef A M A. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int J Food Microbiol. 1999;49:95–102. doi: 10.1016/s0168-1605(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 2.Aureli P, Fiorucci G C, Caroli D, Marchiaro G, Novara O, Leone L, Salmaso S. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N Engl J Med. 2000;342:1236–1241. doi: 10.1056/NEJM200004273421702. [DOI] [PubMed] [Google Scholar]
- 3.Dalton C, Austin C, Sobel J, Hayes P S, Bibb W F, Graves L M, Swaminathan B, Proctor M E, Griffin P M. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N Engl J Med. 1997;336:100–105. doi: 10.1056/NEJM199701093360204. [DOI] [PubMed] [Google Scholar]
- 4.Desmarais E, Lanneluc I, Lagnel J. Direct amplification of length polymorphisms (DALP), or how to get and characterize new genetic markers in many species. Nucleic Acids Res. 1998;26:1458–1465. doi: 10.1093/nar/26.6.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly C W. Conventional methods to detect and isolate Listeria monocytogenes. In: Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1999. pp. 225–260. [Google Scholar]
- 6.Farber J M. An introduction to the hows and whys of molecular typing. J Food Prot. 1996;59:1091–1101. doi: 10.4315/0362-028X-59.10.1091. [DOI] [PubMed] [Google Scholar]
- 7.Fleming D W, Cochi S L, MacDonald K L, Brondum J, Hayes P S, Plikaytis B D, Holmes M B, Audurier A, Broome C V, Reingold A L. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N Engl J Med. 1985;312:404–407. doi: 10.1056/NEJM198502143120704. [DOI] [PubMed] [Google Scholar]
- 8.Franciosa G, Pourshaban M, Gianfranceschi M, Aureli P. Genetic typing of human and food isolates of Listeria monocytogenes from episodes of listeriosis. Eur J Epidemiol. 1998;14:205–210. doi: 10.1023/a:1007448210169. [DOI] [PubMed] [Google Scholar]
- 9.Gerner-Smidt P, Graves L M, Hunter S, Swaminathan B. Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages. J Clin Microbiol. 1998;36:1318–1323. doi: 10.1128/jcm.36.5.1318-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gianfranceschi M, Franciosa G, Gattuso A, Aureli P. Detection of two phospholipases C by means of plate tests for the rapid identification of pathogenic Listeria monocytogenes. Arch Lebensmittelhyg. 1998;49:54–57. [Google Scholar]
- 11.Graves L M, Swaminathan B, Reeves M W, Hunter S B, Weaver R E, Plikaytis B D, Schuchat A. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J Clin Microbiol. 1994;32:2936–2943. doi: 10.1128/jcm.32.12.2936-2943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitmann M, Gerner-Smidt P, Heltberg O. Gastroenteritis caused by Listeria monocytogenes in a private day-care facility. Pediatr Infect Dis J. 1997;16:827–828. doi: 10.1097/00006454-199708000-00025. [DOI] [PubMed] [Google Scholar]
- 13.Hunter P A, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jersěk B, Gilot P, Gubina M, Klun N, Mehle J, Tcherneva E, Rijpens N, Herman L. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J Clin Microbiol. 1999;37:103–109. doi: 10.1128/jcm.37.1.103-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence L M, Harvey J, Gilmour A. Development of a random amplification of polymorphic DNA typing method for Listeria monocytogenes. Appl Environ Microbiol. 1993;59:3117–3119. doi: 10.1128/aem.59.9.3117-3119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louie M, Jayaratne P, Luchsinger I, Devenish J, Yao J, Schlech W, Simor A. Comparison of ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for molecular typing of Listeria monocytogenes. J Clin Microbiol. 1996;34:15–19. doi: 10.1128/jcm.34.1.15-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazurek G H, Reddy V, Marston B J, Haas W H, Crawford J T. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996;34:2386–2390. doi: 10.1128/jcm.34.10.2386-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng J, Doyle M P. Emerging issues in microbiological food safety. Annu Rev Nutr. 1997;17:255–275. doi: 10.1146/annurev.nutr.17.1.255. [DOI] [PubMed] [Google Scholar]
- 19.Miettinen M K, Siitonen A, Heiskanen P, Haajanen H, Björkroth K J, Korkeala H J. Molecular epidemiology of an outbreak of febrile gastroenteritis caused by Listeria monocytogenes in cold-smoked rainbow trout. J Clin Microbiol. 1999;37:2358–2360. doi: 10.1128/jcm.37.7.2358-2360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore M A, Datta A R. DNA fingerprinting of Listeria monocytogenes strains by pulsed-field gel electrophoresis. Food Microbiol. 1994;11:31–38. [Google Scholar]
- 21.Riffard S, Lo Presti F, Vandenesch F, Forey F, Reyrolle M, Etienne J. Comparative analysis of infrequent-restriction-site PCR and pulsed-field gel electrophoresis for epidemiological typing of Legionella pneumophila serogroup 1 strains. J Clin Microbiol. 1998;36:161–167. doi: 10.1128/jcm.36.1.161-167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salamina G, Dalle Donne E, Niccolini A, Poda G, Cesaroni D, Bucci M, Fini R, Maldini M, Schuchat A, Swaminathan B, Bibb W, Rocourt J, Binkin N, Salmaso S. A foodborne outbreak of gastroenteritis involving Listeria monocytogenes. Epidemiol Infect. 1996;117:429–436. doi: 10.1017/s0950268800059082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciacchitano C J. DNA fingerprinting of Listeria monocytogenes using enterobacterial repetitive intergenic consensus (ERIC) motifs—polymerase chain reaction/capillary electrophoresis. Electrophoresis. 1998;19:66–70. doi: 10.1002/elps.1150190112. [DOI] [PubMed] [Google Scholar]
- 24.Slutsker L, Schuchat A. Listeriosis in humans. In: Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1999. pp. 75–95. [Google Scholar]
- 25.Sontakke S, Farber J M. The use of PCR ribotyping for typing strains of Listeria spp. Eur J Epidemiol. 1995;11:665–673. doi: 10.1007/BF01720301. [DOI] [PubMed] [Google Scholar]
- 26.Struelens M J, De Gheldre Y, Deplano A. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect Control Hosp Epidemiol. 1998;19:565–569. doi: 10.1086/647874. [DOI] [PubMed] [Google Scholar]
- 27.van Belkum A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin Microbiol Rev. 1994;7:174–184. doi: 10.1128/cmr.7.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vila J, Marcos A, Llovet T, Coll P, Jimenez De Anta T. A comparative study of ribotyping and arbitrarily primed polymerase chain reaction or investigation of hospital outbreaks of Acinetobacter baumannii infection. J Med Microbiol. 1994;41:244–249. doi: 10.1099/00222615-41-4-244. [DOI] [PubMed] [Google Scholar]
- 29.Vines A, Reeves M W, Hunter S, Swaminathan B. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res Microbiol. 1992;143:281–294. doi: 10.1016/0923-2508(92)90020-o. [DOI] [PubMed] [Google Scholar]
- 30.Yoo J H, Choi J H, Shin W S, Huh D H, Cho Y K, Kim K M, Kim M Y, Kang M W. Application of infrequent-restriction-site PCR to clinical isolates of Acinetobacter baumannii and Serratia marcescens. J Clin Microbiol. 1999;37:3108–3112. doi: 10.1128/jcm.37.10.3108-3112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]