Abstract
Background
Colon cancer is highly prevalent, and cell proliferation and migration are major reasons for its progression to malignancy. The upregulation of INHBA, a glycoprotein hormone that regulates the secretion of pituitary hormones, is documented to be oncogenic in numerous cancers, consisting of breast, gastric, and ovarian cancer. Herein, we assessed the role of INHBA in the proliferation along with the migration of colon cancer cells.
Methods
TCGA datasets were used to assess INHBA expression and its correlation with prognosis in colon cancer patients. Analyses on JASPAR, PROMO, and ENCODE databases, uncovered high correlation between INHBA and BHLHE40. Western blot and RT‐qPCR analysis were used to determine protein and mRNA levels. Cell transfection inhibited the expression of INHBA and BHLHE40. Cell proliferation rates were determined using CCK8 analysis. Wound healing assays were adopted to explore cell migration.
Results
INHBA is markedly elevated in colon cancer tissues along with cells and is a predictive factor for patient's prognosis with colon cancer. INHBA silencing suppressed colon cancer cell proliferation and migration. Furthermore, we confirmed the association of INHBA with BHLHE40 in colon cancer cells. BHLHE40 could directly modulates INHBA expression. Here, we show that BHLHE40 modulates the expression of INHBA, which influences the proliferation, and migration of colon cancer cells.
Conclusion
INHBA acts as an oncogene in colon cancer and it can be regulated by the transcription factor BHLHE40.
Keywords: BHLHE40, colon cancer, INHBA, prognosis, tumor progression
TCGA database analysis showed that INHBA was highly expressed in colon cancer. Western blot and qPCR analysis confirmed that INHBA was overexpressed in colon cancer. Analysis on JASPAR, PROMO, and ENCODE databases revealed high correlation of INHBA with BHLHE40 in colon cancer. Then, we show that BHLHE40 modulates the expression of INHBA, which influences the proliferation, and migration of colon cancer cells.
![]()
1. INTRODUCTION
Colon cancer is among the most frequent malignancies of the digestive system. In China, improved living standards and dietary changes have led to increased colon cancer incidence and mortality. About 20% of colon cancer patients already have metastatic disease at the time of diagnosis, and at this stage, the disease is difficult to remove surgically, causing poor prognosis. 1 Although advances in surgical techniques, radiotherapy, chemotherapy, and targeted therapy, have greatly improved colon cancer prognosis, 2 disease recurrence and metastasis are major obstacles to successful colon cancer treatment. 3
INHBA (inhibin A), a glycoprotein hormone secreted by gonads, is made of an α subunit and a βA subunit. INHBA was previous thought to suppress the synthesis and secretion of pituitary follicle‐stimulating hormone (FSH). 4 INHBA has been associated with various cancers. In breast cancer, INHBA upregulation has been shown to promote metastasis and cancer recurrence by activating epithelial–mesenchymal transition. 5 INHBA is also upregulated in gastric cancer and its silencing affects TGF‐β signaling cascade, in turn dampening gastric cancer cell growth, migration along with invasion. 6 In lung cancer, INHBA upregulation suppresses Hippo signaling and promotes NSCLC invasion and metastasis. 7 INHBA upregulation in ovarian, 8 esophageal, 9 and colorectal cancer, 10 promotes cancer cell proliferation and migration. These findings indicate that INHBA has oncogenic functions. Nonetheless, the role of INHBA in colon cancer is unclear.
BHLHE40 is a basic helix–loop–helix (BHLH) transcription factor family member and is a key modulator of various cellular processes, consisting of cell proliferation, differentiation, and development. 11 Recent investigations have implicated BHLHE40 in tumor growth along with modulation of apoptosis. BHLHE40 expression in lung cancer is reported to be remarkably lower than in surrounding normal tissues and its elevation may inhibit tumor cell proliferation by downregulating Cyclin D1. 12 BHLHE40 overexpression is reported to antagonize the pro‐apoptotic effect of 8‐Methoxypsoralen (8‐MOP) and inhibit apoptotic in hepatocellular carcinoma. 13 BHLHE40 mRNA along with protein contents are markedly higher in gastric cancer tissues than in surrounding noncancerous tissue, and are associated with the degree of malignancy. 14 According to another study, BHLHE40 expression in colon cancer tissues is much higher than in surrounding normal tissues. 15 However, the mechanism by which BHLHE40 promotes colon cancer cell growth coupled with migration is unknown. Herein, we assessed the role along with the responsible mechanisms of INHBA in the regulation of proliferation, and migration in colon cancer cells. Findings from this study may uncover novel colon cancer therapeutic strategies.
We find that INHBA upregulation in colon cancer positively correlates with colon cancer malignant progression. We show that the effect of INHBA overexpression on the proliferation coupled with migration of colon cancer cells is influenced by the transcription factor, BHLHE40. These findings illustrate that INHBA plays an indispensable role in colon cancer and might be employed as a therapeutic target.
2. MATERIALS AND METHODS
2.1. Bioinformatic analysis
The colon cancer dataset was abstracted from TCGA (cancergenome.nih.gov) and used determine INHBA protein expression in colon cancer tissues vs. noncancerous intestinal tissues, and the prognosis of colon cancer subjects. The expression of INHBA in colon cancer was studied using GraphPad Prism software. The impact of INHBA expression on disease relapse along with survival of colon cancer subjects was determined using Kaplan–Meier analysis. JASPAR resource tool (https://jaspar.genereg.net/), PROMO (http://alggen.lsi.upc.es/cgi‐bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) and ENCODE (https://www.encodeproject.org/) were employed to assess INHBA‐associated transcription factors. Interaction between INHBA and BHLHE40 was confirmed using ChIP‐seq analysis.
2.2. Tissue samples
We obtained 20 colon cancer tissues along with para‐cancerous tissues from subjects who were treated with surgery for colon cancer at the Department of Colorectal Surgery, The Affiliated Lihuili Hospital, Ningbo University (Ningbo). Ethical approval for the research work was granted by the hospital's ethics committee. The subjects of this research premise gave written informed consent.
2.3. Cell culture and transfections
The human normal colon epithelial cell line CCD 841 CoN along with the colon cancer cell lines HCT116, SW480 and SW620 were commercially provided by the American Type Culture Collection (ATCC). HCT116 and SW480 were inoculated in RPMI 1640 (HyClone) for growth. SW620 and CCD 841 CoN were inoculated in DMEM (HyClone) enriched with 10% FBS (Gibco) at 37°C, 5% CO2.
2.4. Cell transfection
The siRNAs against INHBA, siINHBA‐1, siINHBA‐2 and negative control siRNA (NC) were supplied by Shanghai MEIXUAN Biological science and technology Ltd. Anti‐BHLHE40 siRNA, siBHLHE40 and negative control siRNA, siNC were provided by GenePharma. SiRNA transfection was done using Lipofectamine 2000 system (Invitrogen) as described by the manufacturer. The siRNA sequence shown in Table 4.
TABLE 4.
siRNA sequence
| siRNA | Sence | Antisence |
|---|---|---|
| siINHBA‐1 | 5’‐GCUCUUACCACAUGAUACATT‐3′ | 5’‐UGUAUCAUGUGGUAAGAGCTT‐3′ |
| siINHBA‐2 | 5’‐GCAGAAAUGAAUGAACUUATT‐3′ | 5’‐UAAGUUCAUUCAUUUCUGCTT‐3′ |
| siNC (for INHBA) | 5’‐UUCUCCGAACGUGUCACGUTT‐3′ | 5’‐ACGUGACACGUUCGGAGAATT‐3′ |
| siBHLHE40 | 5’‐CCCACAUGUACCAAGUGUATT‐3’ | 5’‐UACACUUGGUACAUGUGGGTT‐3’ |
| siNC (for BHLHE40) | 5’‐UUCUCCGAACGUGUCACGUTT‐3’ | 5’‐ACGUGACACGUUCGGAGAATT‐3’ |
2.5. Western blotting
Isolation of proteins from tumor tissues and cells was done with the RIPA buffer (Solarbio) supplemented with 1% protease/phosphatase inhibitor cocktail (Sangon Biotech, Shanghai, China). Fractionation of proteins was done on the 12.5% SDS‐PAGE and blotted onto PVDF membranes (Millipore). They were then blocked with 5% nonfat milk and inoculated overnight with specified primary antibodies at 4°C. We used the following primary antibodies: anti‐INHBA (Cell Signaling Technology), GAPDH (Cell Signaling Technology), and anti‐BHLHE40 (Abclonal). Membranes were then inoculated with HRP‐linked secondary antibody (Boster) and the signal developed using enhanced chemiluminescence.
2.6. RT‐qPCR analysis
Isolation of total RNA from cells was performed with the Trizol reagent (Invitrogen) and quantified using spectrophotometry. Generation of cDNA from the RNA was done with the transcription kit (Thermo). RT‐qPCR assessment was done in duplicate using a SYBR green RT‐qPCR mix (Roche) and the following primers (Table 5).
TABLE 5.
Primer sequences of INHBA for qRT‐PCR and CHIP assays
| Gene | Primer sequences |
|---|---|
| INHBA | F:5’‐TTTGCCGAGTCAGGAACAGC‐3′ |
| R:5’‐AAGAGCCAGACTTCTGCACG‐3’ | |
| GAPDH | F:5’‐AATGGGCAGCCGTTAGGAAA‐3′ |
| R:5’‐GCGCCCAATACGACCAAATC‐3’ | |
| INHBA (for CHIP) | F:5’‐GGTGGTTGCAGAGGGGTTG‐3′ |
| R:5’‐TGACTCTGAAACTCCGCTGG‐3′ |
2.7. Cell counting kit 8 (CCK8) assay
The CCK8 Kit (APExBIO Technology LLC) was adopted to measure cell proliferation as described by the manufacturer. In brief, 5 × 103 cells were cultured for 24 h in 96‐well plates, transfected, then inoculated in normal media. 10 μl of CCK8 were then introduced into every well and the cells incubated for 3 h. Cell proliferation rate was determined by measuring absorbance at 450 nm at 0, 24, 48, and 72 h on a microplate reader (Thermo Fisher).
2.8. Wound healing assays
Cell migration was assessed via the wound healing test by seeding 1 × 105 cells into each well of a 6‐well cell culture plate. A 95% confluence, we scratched the cell monolayer using micropipette tips, and the cells washed several times using PBS to remove cell debris. Serum‐free media was then introduced and the wound closure rate determined by imaging wound healing on an inverted microscope (Motic). The wound closure rate was assessed with the formula: rate of wound healing = [(width of wound at 0 hours – width of wound at 24 hours)/0‐hour wound width] × 100%. 16
2.9. Chromatin immunoprecipitation (ChIP)
For ChIP, about 1 × 107 cells were collected in media and fixed (in 1% formaldehyde) 10 min. Excess formaldehyde was then quenched using a glycine solution at a final level of 0.125 M. Fixed cells were then collected by spinning for 5 min at 500 g. ChIP was done with the ChIP assay kit (Cell Signaling Technology) as documented by the manufacturer. The IgG antibody was provided in the ChIP kit. Anti‐BHLHE40 was acquired from Abclonal, China. RT‐qPCR analysis using LightCycler 480 SYBR Green I Master was used to quantify immunoprecipitated DNA (Roche, US) with the following primers (Table 5).
2.10. Statistical analysis
GraphPad Prism v9.0 was utilized for statistical analysis. Kaplan–Meier analysis was used for survival analysis. Correlation between INHBA expression and clinicopathological features was assessed using the Chi‐square test. The prognostic value of INHBA was assessed using Cox regression analysis. Experimental data are given as mean ± SD. p < 0.05 denotes statistical significance.
3. RESULTS
3.1. Prediction of INHBA expression and prognosis in colon cancer patients
Analysis of INHBA expression in healthy intestinal tissue (n = 26) vs. colon cancer tissue (n = 258) on TCGA revealed that INHBA expression was markedly elevated in colon cancer tissue in contrast with healthy intestinal tissue (p < 0.0001, Figure 1A). Analysis of INHBA levels in 26 colon cancer tissues along with matched paracancerous tissue found that its expression was markedly elevated in colon cancer tissues in contrast with paracancerous tissue (p < 0.0001, Figure 1B). Next, we evaluated the relationship of INHBA contents with colon cancer prognosis in the TCGA cohort using Kaplan–Meier analysis and found that the survival and recurrence rates of colon cancer were remarkably elevated in subjects with high INHBA levels in contrast with those with low INHBA expression (Figure 1C,D).
FIGURE 1.
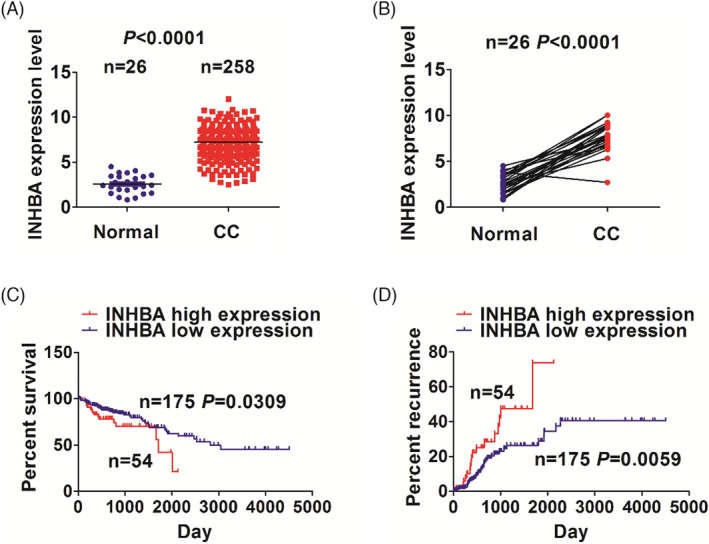
Prediction of INHBA expression and prognosis in colon cancer patients. (A) INHBA contents were markedly higher in colon cancer tissues (n = 258) when compared with normal tissues (n = 26). (B) INHBA contents in colon cancer tissues (n = 26) was markedly higher in contrast with paracancerous tissues. (C, D) Survival analysis based on the TCGA database exhibited that colon cancer patients with INHBA upregulation had lower survival and higher recurrence rates
To further investigate the significance of INHBA expression in colon cancer, we assessed the association between INHBA expression and clinicopathological characteristics in 229 colon cancer patients in the TCGA database and found that INHBA contents were remarkably linked with T stage (P < 0.05, Table 1), indicating that elevated INHBA expression promotes colon cancer progression.
TABLE 1.
Relationship of INHBA contents with clinicopathologic features in colon cancer subjects
| Variables | Cases (n) | INHBA | p value | |
|---|---|---|---|---|
| High | Low | |||
| Total | 229 | 54 | 175 | |
| Age (years) | ||||
| ≥60 | 151 | 33 | 118 | 0.414 |
| <60 | 78 | 21 | 57 | |
| Gender | ||||
| Male | 127 | 30 | 97 | 1.000 |
| Female | 102 | 24 | 78 | |
| Pathological stage | ||||
| I/II | 131 | 26 | 105 | 0.157 |
| III/IV | 98 | 28 | 70 | |
| T stage | ||||
| T1/T2 | 40 | 4 | 36 | 0.025 |
| T3/T4 | 189 | 50 | 139 | |
| N stage | ||||
| Negative | 135 | 26 | 109 | 0.082 |
| Positive | 94 | 28 | 66 | |
| M stage | ||||
| Negative | 160 | 39 | 121 | 0.736 |
| Positive | 69 | 15 | 54 | |
Next, we assessed clinicopathological features and prognostic value of INHBA using Cox regression assessment. Univariate assessment illustrated that survival (Table 2), pathological stage, N stage, M stage and INHBA expression were significant prognostic predictors of colon cancer (p < 0.05), while multivariate analysis identified M stage and INHBA as independent prognostic factors. Univariate analysis (Table 3) identified age, pathological stage, N stage, M stage, and INHBA expression as significant prognostic predictors of colon cancer (p < 0.05), while multivariate data illustrated INHBA as an independent prognostic factor.
TABLE 2.
Cox regression assessment of INHBA expression as a survival predictor
| Variables | Univariate Cox regression analysis | Multivariate Cox regression analysis | ||
|---|---|---|---|---|
| RR (95% CI) | p value | RR (95% CI) | p value | |
| Age (years) | ||||
| <60 vs. ≥60 | 1.376 (0.761 to 2.488) | 0.291 | 2.463 (1.275 to 4.757) | 0.007 |
| Gender | ||||
| Male vs. Female | 1.651 (0.952 to 2.862) | 0.074 | 1.520 (0.862 to 2.680) | 0.148 |
| Pathological stage | ||||
| III/IV vs. I/II | 2.847 (1.654 to 4.903) | <0.0001 | 2.129 (0.514 to 8.816) | 0.297 |
| T stage | ||||
| T3 + T4 vs. T1 + T2 | 2.388 (0.844 to 6.481) | 0.102 | 1.351 (0.456 to 4.009) | 0.857 |
| N staging | ||||
| Positive vs. Negative | 2.716 (1.586 to 4.650) | <0.0001 | 1.192 (0.302 to 4.704) | 0.802 |
| M stage | ||||
| Positive vs. Negative | 2.994 (1.754 to 5.111) | <0.0001 | 3.078 (1.736 to 5.459) | <0.0001 |
| INHBA expression | ||||
| High vs. Low | 1.910 (1.051 to 3.473) | 0.034 | 1.912 (1.031 to 3.545) | 0.040 |
TABLE 3.
Cox regression assessment of INHBA expression as a recurrence predictor
| Variables | Univariate Cox regression analysis | Multivariate Cox regression analysis | ||
|---|---|---|---|---|
| RR (95% CI) | p value | RR (95% CI) | p value | |
| Age (years) | ||||
| <60 vs. ≥60 | 0.567 (0.326 to 0.986) | 0.044 | 0.533 (0.283 to 1.003) | 0.051 |
| Gender | ||||
| Male vs. Female | 1.663 (0.934 to 2.961) | 0.084 | 2.311 (1.240 to 4.309) | 0.008 |
| Pathological stage | ||||
| III/IV vs.I/II | 2.078 (1.194 to 3.619) | 0.010 | 0.528 (0.101 to 2.749) | 0.448 |
| T stage | ||||
| T3 + T4 vs. T1 + T2 | 2.487 (0.896 to 6.906) | 0.080 | 1.767 (0.600 to 5.204) | 0.302 |
| N staging | ||||
| Positive vs. Negative | 2.187 (1.257 to 3.803) | 0.006 | 2.766 (0.541 to 14.140) | 0.222 |
| M stage | ||||
| Positive vs. Negative | 1.980 (1.113 to 3.522) | 0.020 | 1.723 (0.937 to 3.169) | 0.080 |
| INHBA expression | ||||
| High vs. Low | 2.261 (1.245 to 4.107) | 0.007 | 1.982 (1.079 to 3.639) | 0.027 |
3.2. INHBA is elevated in colon cancer tissues and cells
Next, we examined INHBA expression in normal intestinal epithelial cells, and the colon cancer cell lines. SW620, SW480, and HCT116 using Western blot along with RT‐qPCR analyses. These analyses exhibited that INHBA contents were much elevated in colon cancer cell lines in contrast with healthy intestinal epithelial cells (p < 0.05), with HCT116 cells having the highest levels (Figure 2A,B). Western blotting assessment of INHBA contents in 20 patient colon cancer samples and adjacent paracancerous tissues revealed higher INHBA levels in the tumor tissues (Figure 2C). Together, these data exhibit that INHBA is elevated in colon cancer tissues and cell lines.
FIGURE 2.
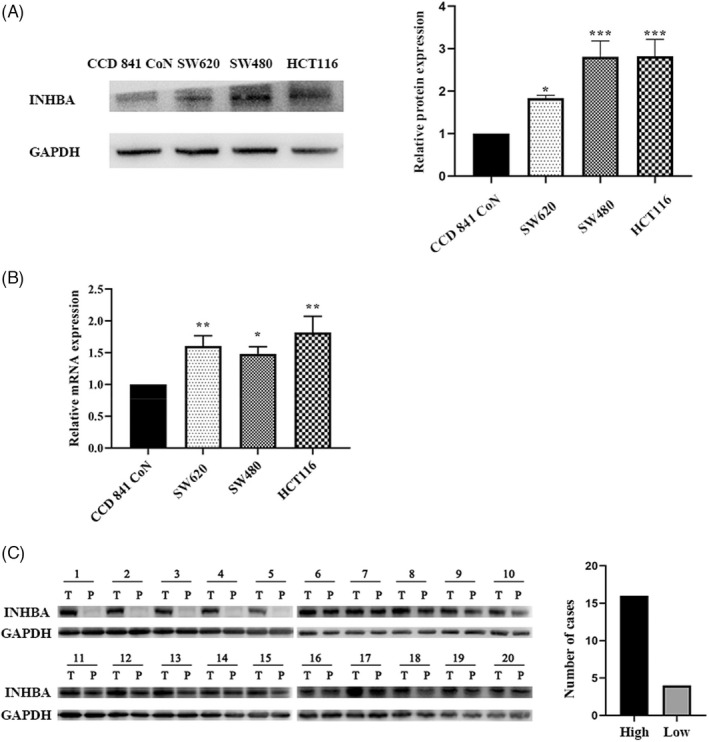
INHBA is elevated in human colon cancer. Western blotting along with RT‐qPCR showed that INHBA protein (A), and mRNA (B) levels are remarkably higher in colon cancer cells (HCT116, SW620 and SW480) than in the healthy intestinal epithelial cell line, CCD 841 CoN. Western blot (C) analysis showed that INHBA is frequently upregulated in the colon cancer tissues (T) than in paracancerous tissues (P). Data are given as mean ± SD of three independent experiments. *, **, and *** indicate p = < 0.05, <0.01, and <0.001 vs. CCD 841 CoN cells
3.3. INHBA silencing dampens the proliferation along with migration of colon cancer cells
To assess if INHBA affects colon cancer cell proliferation, and migration, we first silenced it using siRNA (siINHBA‐1 and siINHBA‐2) in SW620 and HCT116 colon cancer cell lines. Because Western blotting data revealed that siINHBA‐1 suppressed INHBA expression most effectively (Figure 3A), it was used in all further experiments. CCK8 analysis showed that INHBA knockdown with siINHBA‐1 remarkably suppressed cell proliferation in contrast with the NC group (cell transfects of siRNA; p < 0.01, Figure 3B). Next, cell migration analysis using wound healing assays revealed that INHBA silencing markedly suppressed the migration of colon cancer cells (p < 0.01, Figure 3C). These findings indicate that INHBA modulates colon cancer cell proliferation along with migration.
FIGURE 3.
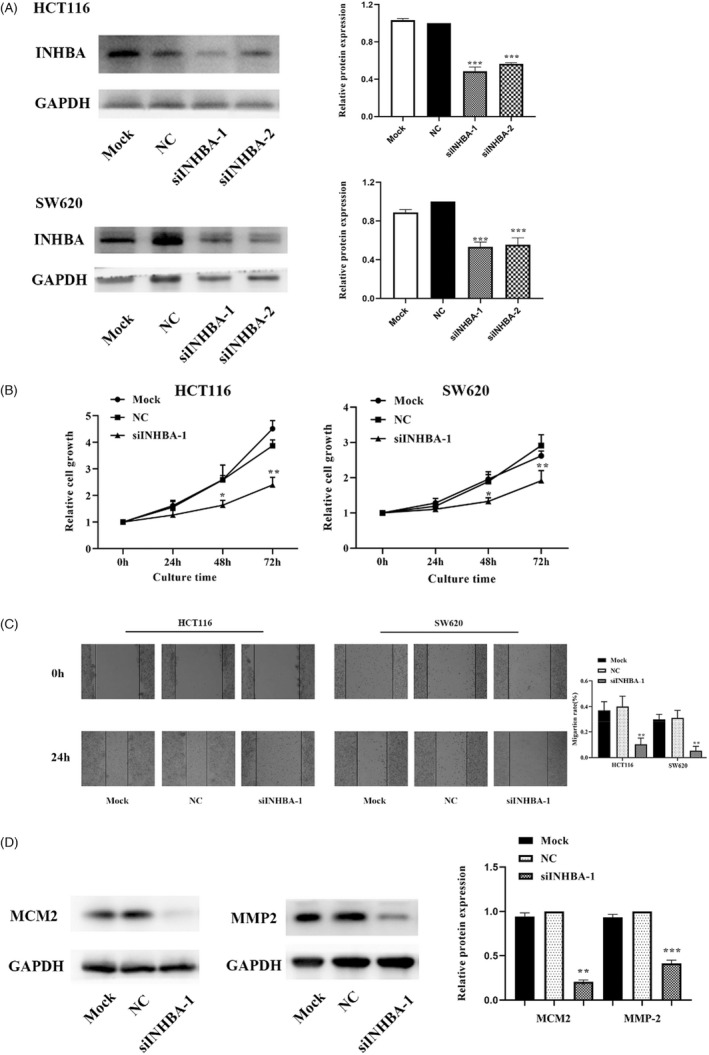
INHBA silencing suppressed the proliferation and migration of colon cancer cells. Western blot analysis revealed that transfection with siINHBA effectively reduced INHBA expression in HCT116 along with SW620 cells (A). CCK8 assessment exhibited that INHBA silencing dampened the proliferation of HCT116 along with SW620 cells (B). Wound healing assessment illustrated that INHBA silencing dampened the migration of HCT116 along with SW620 (C). Western blotting assessment illustrated that INHBA silencing reduced the contents of the cell proliferation‐associated protein MCM2 and the cell migration‐associated protein MMP‐2 (D). Data are given as mean ± SD of three independent experiments. *, **, and *** indicate p = < 0.05, <0.01 and <0.001 compared with NC
MCM2, a member of the minichromosome maintenance protein (MCM) family, is a sensitive and specific indicator of cells entering the proliferative phase, and is upregulated in proliferating cells. 17 Next, we examined MCM2 levels in HCT116 cells using Western blot assessment and observed in contrast with the NC group, INHBA‐silenced cells had remarkably lower MCM2 levels (Figure 3D).
Matrix metalloproteinases (MMPs) may promote cancer metastasis and infiltration by disrupting extracellular matrix and vascular basement membrane. MMP‐2 and MMP‐9 are highly expressed in malignant tumors and are key mediators of tumor invasion and metastasis. 18 Western blot analysis of HCT116 cells showed MMP‐2 expression was markedly reduced in INHBA‐silenced cells when compared with NC cells (Figure 3D).
3.4. INHBA is a BHLHE40 target
To examine the mechanism by which INHBA functions in colon cancer carcinogenesis, we used the bioinformatics softwares, JASPAR, PROMO, and ENCODE, to predict transcription factors that act upstream of the INHBA gene. After superimposing the results from the three softwares, we identified the transcription factor, BHLHE40, as a potential modulator of INHBA gene expression (Figure 4A). This observation was validated by ChIP‐seq analysis that revealed a BHHE40 binding site near the INHBA promoter region (Figure 4B). Together, these results show that the transcription factor, BHLHE40, may regulate the INHBA expression.
FIGURE 4.
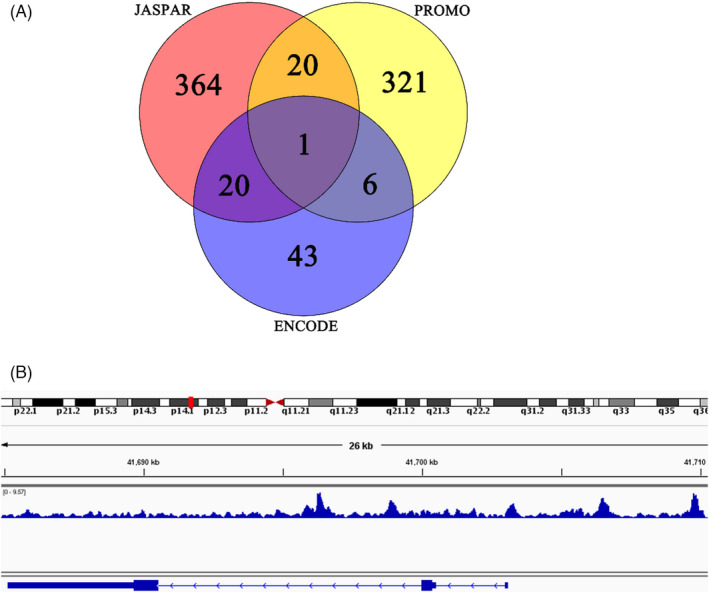
The transcription factor, BHLHE40 targets INHBA gene. (A) Bioinformatics analyses showed that BHLHE40 acts upstream of INHBA. (B) Potential BHLHE40 binding sites in INHBA were identified using ChIP‐seq
3.5. BHLHE40 directly regulates INHBA expression
To investigate if BHLHE40 influences colon cancer cell proliferation by modulating INHBA expression, we first examined BHLHE40 protein contents in colon cancer cells. This analysis showed that BHLHE40 levels were markedly elevated in colon cancer cells in contrast with healthy intestinal epithelial cells (p < 0.05, Figure 5A). These data indicate that BHLHE40 may directly influence colon cancer cell proliferation in vitro. To test if BHLHE40 regulates INHBA expression, we first silenced BHLHE40 in HCT116 cells. Western blotting data of the expression of BHLHE40 and INHBA in BHLHE40‐silenced cells showed that BHLHE40 knockdown correlated with reduced INHBA protein levels (Figure 5B). ChIP revealed that BHLHE40 promotes INHBA expression by docking to its promoter (Figure 5C). Altogether, these data illustrate that BHLHE40 directly modulates INHBA expression.
FIGURE 5.
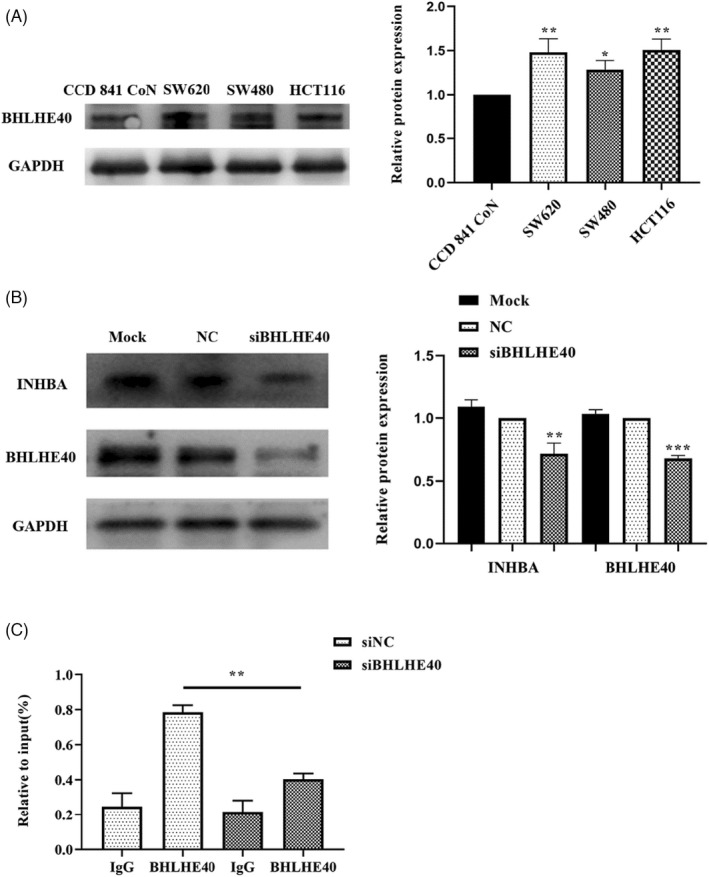
BHLHE40 directly regulates INHBA expression. (A) Western blot showed that BHLHE40 protein levels in the colon cancer cells (HCT116, SW480, SW620) were remarkably elevated in contrast with normal intestinal epithelial cells (CCD 841 CoN). (B) INHBA expression was suppressed by silencing BHLHE40 expression in colon cancer cells. (C) ChIP showed that BHLHE40 might bind to the promoter region of INHBA. Data are shown as mean ± SD of three independent experiments. *, **, and *** indicate p = < 0.05, <0.01, and <0.001, respectively
3.6. BHLHE40 regulates colon cancer cell proliferation and migration via INHBA
To explore the influence of BHLHE40‐mediated, INHBA expression on colon cancer cell proliferation along with migration, we silenced BHLHE40 in HCT116, as well as SW620 cells and examined their effect on cell proliferation using CCK8 analysis. This analysis exhibited that in contrast with the NC group, BHLHE40 knockdown for 2 days remarkably suppressed cell proliferation (Figure 6A). Thereafter, we adopted wound healing assessment to assess the impact of BHLHE40 on colon cancer cells. The data indicated that in contrast with the NC group, 24‐h cell migration was remarkably suppressed by BHLHE40 silencing (Figure 6B). Moreover, Western blotting assessment revealed that in contrast with the NC group, the contents of the proliferation‐associated factor, MCM2, and the migration‐associated factor, MMP‐2, were remarkably suppressed upon BHLHE40 silencing (Figure 6C).
FIGURE 6.
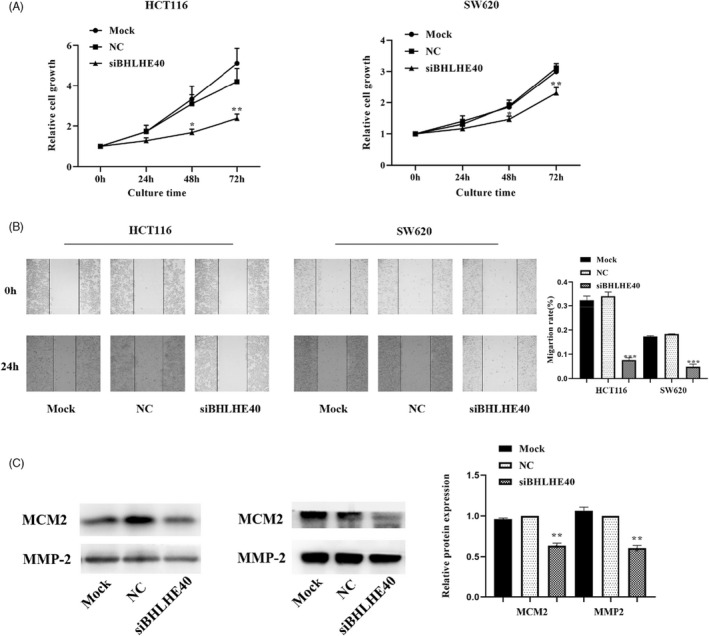
BHLHE40 regulates colon cancer cell proliferation along with migration via INHBA. CCK8 coupled with wound healing assays illustrated that BHLHE40 knockdown suppressed growth, as well as migration in HCT116 and SW620 cells, respectively (A, B). Western blotting assessmnet illustrated that BHLHE40 silencing dampened the expression of the proliferation‐associated protein, MCM2, and the migration‐associated protein, MMP‐2 (C). Data are given as mean ± SD of three independent experiments. *, **, *** indicates P = < 0.05, <0.01, and <0.001, relative to NC
4. DISCUSSION
Cancer is a remarkable public health challenge, worldwide. According to 2022 statistics from the American Cancer Society, colorectal cancer is the second leading cause of cancer deaths, and the incidence of colorectal cancer in adults aged <50 years is rising. 19 Therefore, effective therapies against colon cancer are urgently needed for improved survival outcomes.
INHBA has been implicated in the carcinogenesis along with the progression of numerous cancers, 5 , 6 , 7 , 8 , 9 , 10 including colon cancer. However, its role in colon cancer is unknown. Using Western blot and RT‐qPCR analyses, we show that INHBA is aberrantly expressed in the colon cancer cell lines, HCT116, SW480, and SW620, which is consistent with our bioinformatics analysis results. Through database analysis, Li et al. 20 identified INHBA as an independent risk factor for DFS in colon adenocarcinoma and showed that a prediction model including INHBA expression levels can improve prognosis prediction in colon adenocarcinoma patients. Here, using COX analysis, we show that high INHBA expression is an independent risk factor for survival and recurrence of colon cancer. Future studies will seek to confirm the impact of INHBA on colon cancer prognosis on a larger sample size.
Cancer cell proliferation and migration are major drivers of cancer progression. Here, we show that INHBA expression promotes the proliferation along with the migration of colon cancer cells. CCK8 and wound healing assays exhibited that INHBA knockdown strongly suppressed the proliferation coupled with the migration of colon cancer cells, indicating that high INHBA expression promotes colon cancer progression. These findings highlight INHBA as a possible diagnostic along with therapeutic factor against colon cancer.
Studies have implicated BHLHE40 in the onset and progression of numerous cancers. 12 , 13 , 14 , 15 Analysis on JASPAR, PROMO, and ENCODE databases revealed high correlation of INHBA with BHLHE40 in colon cancer. INHBA is also a target of the transcription factor BHLHE40, according to ChIP data. Analysis of BHLHE40 expression showed that it was markedly upregulated in colon cancer cells in contrast with the normal epithelial intestinal cells. BHLHE40 is reported to play a key role in the onset and progress of gastric cancer. 14 We therefore reasoned that BHLHE40 may promote colon cancer invasion by modulating INHBA expression. This possibility was supported by the observation that INHBA expression was suppressed by BHLHE40 silencing in the colon cancer cells, which also remarkably suppressed their proliferation along with migration. Thus, BHLHE40 may affect the proliferation, and migration of colon cancer cells by regulating INHBA expression. These findings indicate that INHBA may have therapeutic value against colon cancer.
In conclusion, our study finds that high INHBA expression promotes the proliferation along with migration of colon cancer cells through its modulation by BHLHE40. However, the study has certain limitations. Moreover, we only validated our conclusions in vitro. In future investigations, we will validate our findings in vivo using an animal model of colon cancer. A previous investigation showed that circulating levels of INHBA could predict ovarian cancer and used it for postoperative monitoring, indicating that INHBA is a sensitive and specific indicator of ovarian epithelial cancer. 21 In future clinical studies, we will evaluate the diagnostic and prognostic value of circulating INHBA in blood samples from colon cancer patients before surgery, after surgery, and during follow‐up. However, because INHBA is extensively expressed in various human tissues where it performs a variety of physiological roles, further research is required before it can be clinically applied. Nonetheless, it has potential diagnostic and therapeutic value against colon cancer.
5. CONCLUSIONS
In summary, our findings indicate that INHBA is oncogenic in colon cancer and that its upregulation, mediated by the transcription factor BHLHE40, enhances cancer cell proliferation along with migration.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Wang J, Li B, Yang S, et al.. Upregulation of INHBA mediated by the transcription factor BHLHE40 promotes colon cancer cell proliferation and migration. J Clin Lab Anal. 2022;36:e24539. doi: 10.1002/jcla.24539
Funding information
This study was funded by Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (Grant No. 2020KY275, 2022KY1081), Ningbo Natural Science Foundation (Grant No. 2019A610334, 2021 J291), and Ningbo medical key discipline (Grant No. 2022‐F01)
REFERENCES
- 1. Kampoli K, Foukas PG, Ntavatzikos A, Arkadopoulos N, Koumarianou A. Interrogating the interplay of angiogenesis and immunity in metastatic colorectal cancer. World J Methodol. 2022;12(1):43‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Łukaszewicz‐Zając M, Mroczko B. Circulating Biomarkers of Colorectal Cancer (CRC)‐Their Utility in Diagnosis and Prognosis. J Clin Med. 2021;10(11):2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chandra R, Karalis JD, Liu C, et al. The colorectal cancer tumor microenvironment and its impact on liver and lung metastasis. Cancers (Basel). 2021;13:6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lorenzen JR, Channing CP, Schwartz NB. Partial characterization of FSH suppressing activity (folliculostatin) in porcine follicular fluid using the metestrous rat as an in vivo bioassay model. Biol Reprod. 1978;19(3):635‐640. [DOI] [PubMed] [Google Scholar]
- 5. Yu Y, Wang W, Lu W, Chen W, Shang A. Inhibin β‐A (INHBA) induces epithelial‐mesenchymal transition and accelerates the motility of breast cancer cells by activating the TGF‐β signaling pathway. Bioengineered. 2021;12(1):4681‐4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen ZL, Qin L, Peng XB, Hu Y, Liu B. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF‐β signaling pathway. J Cell Physiol. 2019;234(10):18065‐18074. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y, Yan S, Li Y, et al. Inhibin βA is an independent prognostic factor that promotes invasion via Hippo signaling in non‐small cell lung cancer. Mol Med Rep. 2021;24(5):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X, Yang Z, Xu S, et al. Targeting INHBA in ovarian cancer cells suppresses cancer xenograft growth by attenuating stromal fibroblast activation. Dis Markers. 2019;2019:7275289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lyu S, Jiang C, Xu R, Huang Y, Yan S. INHBA upregulation correlates with poorer prognosis in patients with esophageal squamous cell carcinoma. Cancer Manag Res. 2018;10:1585‐1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He Z, Liang J, Wang B. Inhibin, beta A regulates the transforming growth factor‐beta pathway to promote malignant biological behaviour in colorectal cancer. Cell Biochem Funct. 2021;39(2):258‐266. [DOI] [PubMed] [Google Scholar]
- 11. Shen M, Kawamoto T, Yan W, et al. Molecular characterization of the novel basic helix‐loop‐helix protein DEC1 expressed in differentiated human embryo chondrocytes. Biochem Biophys Res Commun. 1997;236(2):294‐298. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y, Wang L, Lin XY, et al. The transcription factor DEC1 (BHLHE40/STRA13/SHARP‐2) is negatively associated with TNM stage in non‐small‐cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol. 2013;34(3):1641‐1650. [DOI] [PubMed] [Google Scholar]
- 13. Peng Y, Liu W, Xiong J, et al. Down regulation of differentiated embryonic chondrocytes 1 (DEC1) is involved in 8‐methoxypsoralen‐induced apoptosis in HepG2 cells. Toxicology. 2012;301(1–3):58‐65. [DOI] [PubMed] [Google Scholar]
- 14. Zheng Y, Jia Y, Wang Y, et al. The hypoxia‐regulated transcription factor DEC1 (Stra13, SHARP‐2) and its expression in gastric cancer. Omics. 2009;13(4):301‐306. [DOI] [PubMed] [Google Scholar]
- 15. Li Y, Zhang H, Xie M, et al. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation‐induced apoptosis and selective inhibition of procaspase activation. Biochem J. 2002;367(Pt. 2):413‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo J, Liu Y. INHBA promotes the proliferation, migration and invasion of colon cancer cells through the upregulation of VCAN. J Int Med Res. 2021;49(6):3000605211014998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alison MR, Hunt T, Forbes SJ. Minichromosome maintenance (MCM) proteins may be pre‐cancer markers. Gut. 2002;50(3):290‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon SO, Park SJ, Yun CH, Chung AS. Roles of matrix metalloproteinases in tumor metastasis and angiogenesis. J Biochem Mol Biol. 2003;36(1):128‐137. [DOI] [PubMed] [Google Scholar]
- 19. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 20. Li X, Yu W, Liang C, et al. INHBA is a prognostic predictor for patients with colon adenocarcinoma. BMC Cancer. 2020;20(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsigkou A, Marrelli D, Reis FM, et al. Total inhibin is a potential serum marker for epithelial ovarian cancer. J Clin Endocrinol Metab. 2007;92(7):2526‐2531. [DOI] [PubMed] [Google Scholar]


