Abstract
The pathway of propionate conversion in a syntrophic coculture of Smithella propionica and Methanospirillum hungatei JF1 was investigated by 13C-NMR spectroscopy. Cocultures produced acetate and butyrate from propionate. [3-13C]propionate was converted to [2-13C]acetate, with no [1-13C]acetate formed. Butyrate from [3-13C]propionate was labeled at the C2 and C4 positions in a ratio of about 1:1.5. Double-labeled propionate (2,3-13C) yielded not only double-labeled acetate but also single-labeled acetate at the C1 or C2 position. Most butyrate formed from [2,3-13C]propionate was also double labeled in either the C1 and C2 atoms or the C3 and C4 atoms in a ratio of about 1:1.5. Smaller amounts of single-labeled butyrate and other combinations were also produced. 1-13C-labeled propionate yielded both [1-13C]acetate and [2-13C]acetate. When 13C-labeled bicarbonate was present, label was not incorporated into acetate, propionate, or butyrate. In each of the incubations described above, 13C was never recovered in bicarbonate or methane. These results indicate that S. propionica does not degrade propionate via the methyl-malonyl-coenzyme A (CoA) pathway or any other of the known pathways, such as the acryloyl-CoA pathway or the reductive carboxylation pathway. Our results strongly suggest that propionate is dismutated to acetate and butyrate via a six-carbon intermediate.
In methanogenic environments propionate is oxidized by acetogenic bacteria to acetate and carbon dioxide (16, 18). Methanogenic archaea make this reaction energetically favorable by removing reducing equivalents either as hydrogen or as formate (1, 3, 19). Syntrophic propionate oxidation mainly occurs via the randomizing methyl-malonyl-coenzyme A (CoA) pathway, as was demonstrated for several Syntrophobacter species (6, 7, 11), as well as for mixed methanogenic cultures (2, 5, 8, 13, 14, 15, 22). However, other pathways of propionate degradation are possible as well, such as a nonrandomizing pathway via butyrate (9, 22, 23). In these studies, evidence was provided that part of the propionate is carboxylated to butyrate which is then degraded to acetate. Alternative possible pathways of propionate conversion were recently documented by Textor et al. (21).
Recently, a novel syntrophic propionate-oxidizing bacterium was isolated which may possess a propionate-degradation pathway via butyrate (10). Cocultures of Smithella propionica and a hydrogen- and formate-utilizing methanogen produce less methane and more acetate than cocultures with Syntrophobacter strains. In addition, the cocultures with S. propionica produce small amounts of butyrate. It was suggested that this organism dismutates propionate to acetate and butyrate followed by syntrophic β-oxidation of butyrate to acetate. We report here the results of 13C-nuclear magnetic resonance (NMR) studies to elucidate the pathway of propionate oxidation in S. propionica.
MATERIALS AND METHODS
Organisms and cultivation.
Methanospirillum hungatei JF1T was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen in Braunschweig, Germany. The MS medium (3) with 0.5 g of casein tryptic peptone and 0.5 g of yeast extract per liter, but with 1 mM l-cysteine instead of 1 mM mercaptoethane sulfonate, was used to grow syntrophic cultures of S. propionica and M. hungatei. The methanogens were pregrown on H2 and CO2 in 120-ml serum vials with 50 ml of medium. After growth the gas atmosphere was replaced by N2 and CO2 (80:20), and S. propionica (in coculture with Methanospirillum hungatei) was inoculated into these M. hungatei cultures. The cocultures were incubated at 37°C with 10 mM propionate.
NMR spectroscopy.
Stable isotopes (minimum, 99% 13C) were obtained from Campro Scientific B.V. (Veenendaal, The Netherlands). Serum vials were prepared with 10 mM concentrations of either [1-13C]propionate, [2-13C]propionate, [3-13C]propionate, or [2,3-13C]propionate as substrates in 50 ml of medium. To test the incorporation of H13CO31−, the coculture was grown on 10 mM unlabeled propionate in the presence of 50 mM H13CO31−. The combination of 10 mM unlabeled propionate and 4 mM [1-13C]acetate or [2-13C]acetate was also tested. After 10, 20, and 30 days 3-ml samples were withdrawn for analysis. Cells were removed by centrifugation at 10,000 × g, and D2O and dioxane were added to 2 ml of supernatant to give a final volume of 2.5 ml in 10-mm (outer-diameter) NMR tubes containing 10% D2O and 100 mM dioxane. The proton-decoupled 13C-NMR spectra of the samples were recorded at 75.47 MHz on a Bruker AMX-300 NMR spectrometer. For each spectrum 7,200 transients (2 h) were accumulated and stored on disk using 32,000 datum points, a 45° pulse angle (pulse duration, 9 μs), and a delay time of 1 s between the pulses. The measuring temperature was maintained at 25°C, and the chemical shift belonging to the dioxane carbon nuclei (67.4 ppm) was used as an internal standard. The deuterium in the samples (10% [vol/vol]) was used for the field lock. A balance of 13C-labeled compounds was calculated by relating the areas of the observed resonances to the areas in the spectrum of a sample containing propionate, butyrate, and acetate (100 mM concentrations of each; 1.11% natural abundance) measured under identical conditions with dioxane as an internal standard.
Other analytical techniques.
The remainder of the 3-ml samples withdrawn for NMR measurements was analyzed for organic acids. Also, 0.4-ml gas samples were withdrawn to determine the amount of CH4 produced. Organic acids acids were measured with a Spectrasystem HPLC system equipped with an autosampler and Refractomonitor. The acids were separated on a Polyspher OAHY column (30 cm by 6.5 mm; Merck, Darmstadt, Germany) in 0.01 N H2SO4 at a flow rate of 0.6 ml/min and a column temperature of 60°C. The acids eluting from the column were quantified by differential refractometry (17).
Methane levels were measured chromatographically with a Packard-Becker 417 gas chromatograph equipped with a thermal conductivity detector and molecular sieve 13X (60/80 mesh). The column temperature was 50°C, and the carrier gas was argon at a flow rate of 30 ml/min.
RESULTS
Growth experiments.
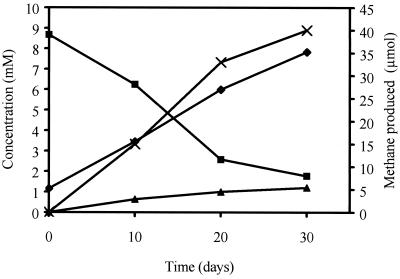
Growth of the syntrophic coculture of S. propionica and M. hungatei is shown in Fig. 1. After 30 days of incubation, the culture produced 0.1 mol of methane, 1 mol of acetate, and 0.1 mol of butyrate per mol of propionate degraded (Fig. 1). In control bottles without propionate and in bottles to which 5 mM bromoethane sulfonate was added, no measurable changes in the organic acid concentration were observed and no methane was produced.
FIG. 1.
Growth of S. propionica in coculture with M. hungatei JF1 in 50-ml batches. ■, Propionate; ⧫, acetate; ▴, butyrate; ×, methane produced.
NMR measurements.
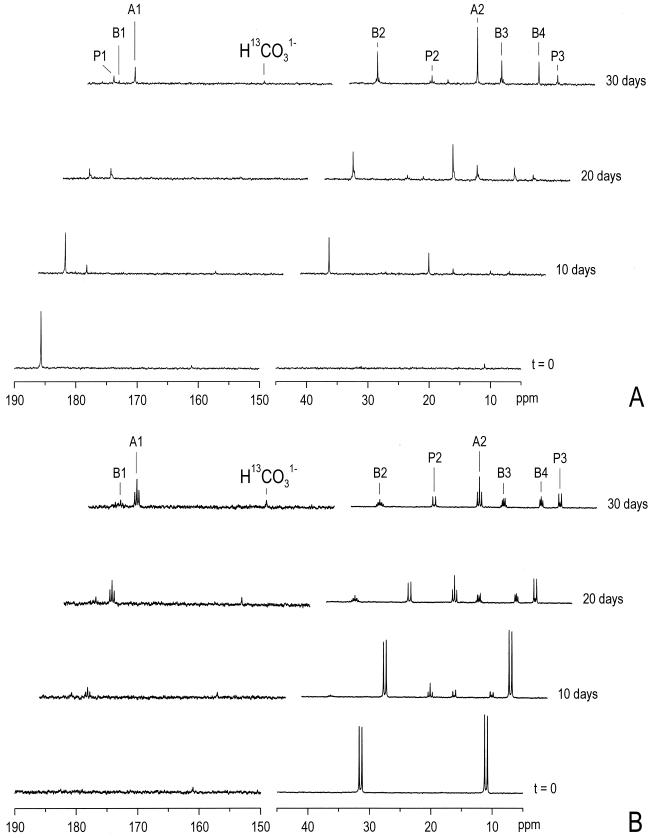
When S. propionica was grown with [3-13C]propionate, both [2-13C]acetate and unlabeled acetate were produced, while [1-13C]acetate was not formed (Tables 1 and 2). Label initially appeared mainly at the C4 position of butyrate, but after 30 days of incubation, label was recovered at the C2 and C4 positions of butyrate, in a ratio of about 1:1.5 (Table 1). [2-13C]propionate yielded [1-13C]acetate as well as unlabeled acetate, though small amounts of [2-13C]acetate and [1,2-13C]acetate were also detected. Throughout this experiment, nearly equal amounts of label were detected at the C1 and C3 positions of butyrate (Tables 1 and 2). The batches fed with [1-13C]propionate yielded nearly equal amounts of [1-13C]acetate, [2-13C]acetate, and unlabeled acetate (Fig 2A; Tables 1 and 2). In this experiment the label initially appeared at the C2 position of butyrate, while after 30 days of incubation label was distributed more evenly over all carbon atoms (Fig. 2A). Double-labeled propionate at the C2 and C3 positions yielded [1-13C]acetate, [2-13C]acetate, and [1,2-13C]acetate in a ratio near 1:1:1. Butyrate was initially mainly double labeled at the C3 and C4 positions, but after 30 days of incubation substantial amounts of single labeled butyrate and other combinations were also detected (Fig. 2B, Tables 1 and 2).
TABLE 1.
13C recoveries from propionate conversion by S. propionica and M. hungatei after 30 days of incubationa
| Isotope recovered |
13C recovery (%) with a substrate:
|
||||
|---|---|---|---|---|---|
| [1-13C]P | [2-13C]P | [3-13C]P | [2,3-13C]P | [2-13C]A | |
| [1-13C]propionate | 11 | ||||
| [2-13C]propionate | 2 | 14 | 1 | ||
| [3-13C]propionate | 2 | 17 | 1 | 6 | |
| [1,2-13C]propionate | 1 | 2 | |||
| [2,3-13C]propionate | 1 | 15 | |||
| [1-13C]acetate | 29 | 53 | 16 | ||
| [2-13C]acetate | 27 | 1 | 62 | 18 | 57 |
| [1,2-13C]acetate | 2 | 2 | 35 | ||
| [1-13C]butyrate | 4 | 9 | 3 | ||
| [1,2-13C]butyrate | 3 | ||||
| [2-13C]butyrate | 5 | 9 | 1 | 9 | |
| [2,3-13C]butyrate | 2 | 3 | |||
| [3-13C]butyrate | 3 | 10 | 1 | ||
| [3,4-13C]butyrate | 5 | ||||
| [4-13C]butyrate | 4 | 13 | 2 | 10 | |
| Total | 93 | 91 | 101 | 104 | 82 |
Propionate (P) was tested with label at the C1, C2, or C3 atom or double-labeled at the C2 and C3 atoms. Acetate (A) was introduced with label at the C2 atom. The inaccuracy of the quantified signals is usually >5% for methyl and methylene carbon atoms present at a concentration of at least 0.5 mM (values of >10% in the table). Quantification of signals present at lower concentrations and especially those of carboxylic acids is less accurate. However, the inaccuracy of those signals was usually only between 5 and 20%.
TABLE 2.
Distribution of 13C in acetate recovered from propionate conversion by S. propionica and M. hungatei after 30 days of incubation
| Substrate | Mean concn (mM) of acetate isotope recovered
|
|||
|---|---|---|---|---|
| [1-13C] | [2-13C] | [1,2-13C] | Unlabeled | |
| [1-13C]propionate | 2.5 ± 0.2 | 2.3 ± 0.1 | 0.1 ± 0.03 | 2.1 ± 0.5 |
| [2-13C]propionate | 4.7 ± 0.3 | 0.1 ± 0.03 | 0.2 ± 0.05 | 2.1 ± 0.5 |
| [3-13C]propionate | 0 | 5.2 ± 0.2 | 0 | 1.7 ± 0.3 |
| [2,3-13C]propionate | 2.3 ± 0.2 | 2.7 ± 0.1 | 2.6 ± 0.3 | 0 |
FIG. 2.
Time courses of propionate conversion by S. propionica as measured by 1H-decoupled 13C-NMR. P, propionate; A, acetate; B, butyrate. The numbers refer to the position of the 13C atoms. (A) Incubation with [1-13C]propionate. (B) Incubation with [2,3-13C]propionate. The resonances within the area of the carboxyl-groups (150 to 190 ppm) in this spectrum are enlarged by a factor 4.
Label was also recovered in butyrate and propionate when the culture was grown on propionate in the presence of labeled acetate. Label from [1-13C]acetate was detected at the C1 and C3 positions of butyrate and at the C2 of propionate (data not shown), whereas [2-13C]acetate yielded label at the C2 and C4 positions of butyrate and at the methyl group of propionate (Table 1).
Although H13CO31− was visible in all of the NMR spectra due to natural abundance (approximately 0.5 mM; Fig. 2), there were no substantial increases of the bicarbonate area observed. Therefore, we did not make further attempts to quantify H13CO31− or 13CH4. In addition, when the coculture was grown in the presence of 50 mM H13CO31− we could not detect incorporation of label, since all the observed areas were due to the natural abundance of the compounds present, as was calculated from the high-pressure liquid chromatography data.
DISCUSSION
The stoichiometry of propionate conversion by the coculture of S. propionica and M. hungatei was similar, as reported previously (10). Our results obtained with 13C-NMR support the theory that propionate is dismutated to acetate and butyrate, followed by syntrophic β-oxidation of butyrate to acetate. In addition, the results enabled us to propose a pathway of propionate conversion by S. propionica. A randomizing pathway, which was found for several Syntrophobacter species, could be excluded since there was no exchange in label due to symmetry in any of the intermediates (6, 7, 11). Initially, we expected to find an acryloyl-CoA-like pathway in combination with reductive carboxylation, as reported in previous studies (9, 22, 23). However, [1-13C]propionate did not yield H13CO31−, and experiments with [2,3-13C]propionate showed that at least half of the methyl-methylene bonds were broken. Furthermore, H13CO31− was not incorporated into propionate, indicating that the C1 of butyrate is introduced either via transcarboxylation or via Claisen condensation. Condensations involving propionyl-CoA were reported by Reeves and Ajl (12) and by Tabuchi et al. (20). However, these pathways both lead to the formation of acetyl-CoA via decarboxylation (of pyruvate) and do not explain the breakage of the methyl-methylene bonds either. Incubations with labeled acetate showed that an acetyl-CoA condensation pathway is present in S. propionica, most likely one similar to the pathway found for Syntrophomonas wolfei (24). However, the majority of the butyrate is produced in a different fashion, since single-labeled propionate initially yielded mainly single-labeled butyrate and [2,3-13C]propionate initially yielded mainly [3,4-13C]butyrate.
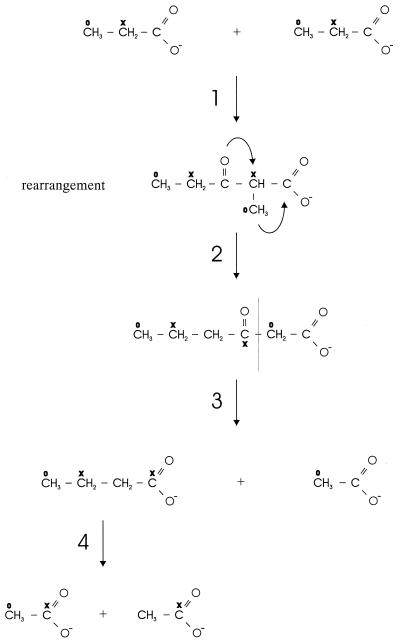
A pathway which could explain the observed labelling pattern is depicted in Fig. 3. The high levels of [1-13C]butyrate from [2-13C]propionate and [2,3-13C]propionate suggest that the C2 of propionate is coupled to the carboxyl group of a second propionate molecule. A rearrangement of the six-carbon intermediate to give an unbranched molecule followed by cleavage of acetate would explain the ratios of labeled acetate, as well as the ratios of labeled to unlabeled acetate (Fig. 3). The residual four-carbon molecule (butyrate) is then further oxidized syntrophically to acetate, a result which agrees with the amounts of methane produced. The presence of such pathway is strongly favored by the fact that we could not demonstrate incorporation or excretion of H13CO31−. The incubations in the presence of labeled acetate revealed that the pathway is reversible. This explains the observed shift of label in time toward a more equal distribution in butyrate. It also explains why small amounts of label are recovered in [2-13C]acetate from [2-13C]propionate and double-labeled acetate from either [1-13C]- or [2-13C]propionate, while [3-13C]propionate yielded exclusively [2-13C]acetate. In addition, it explains the distribution of label in butyrate, as well as the formation of [2,3-13C]butyrate from [2-13C]propionate.
FIG. 3.
Proposed pathway for propionate conversion by S. propionica. Step 1, condensation of the C2 of propionate to the carboxyl of another propionate molecule or derivative; step 2, rearrangement of the methyl group and transfer of the oxygen to the C3 of the intermediate; step 3, cleavage of 3-ketohexanoate yielding butyrate and acetate; step 4, syntrophic β-oxidation of butyrate to acetate.
Most likely all steps in the proposed pathway require CoA derivatives, as occurs during butyrate oxidation. The initial activation of propionate may be accomplished by CoA transfer from acetyl-CoA or another CoA-containing intermediate. Like other rearrangement reactions, the isomerization of the two-methyl group to an unbranched molecule is likely a coenzyme B12-dependent reaction. The mechanism of this rearrangement may be identical to the reaction catalyzed by methyl-malonyl-CoA mutase (4). Transfer of the keto group would require a reduction and a dehydration, yielding a double bond between C3 and C4, followed by the addition of H2O and oxidation to 3-ketohexanoate. Possibly the last two steps are catalyzed by crotonase and butyryl-CoA dehydrogenase, enzymes also required for the cleavage of butyrate.
S. propionica is the first syntrophic propionate-oxidizer that may account for the nonrandomizing pathway observed in methanogenic habitats. The results seem to fit in previous studies with methanogenic biomass and enrichment cultures in which the presence of an alternative route for propionate oxidation was clearly demonstrated by the use of 13C-NMR (9, 22, 23). The isolation of S. propionica enabled us to study this pathway into detail without interference of the randomizing methyl-malonyl-CoA pathway which also occurs in complex microbial communities. It might be interesting to study the occurrence of S. propionica or microorganisms with a similar pathway in anaerobic digesters and other methanogenic environments.
REFERENCES
- 1.Boone D R, Bryant M P. Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from methanogenic ecosystems. Appl Environ Microbiol. 1980;40:626–632. doi: 10.1128/aem.40.3.626-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boone D R. Propionate exchange reactions in methanogenic ecosystems. Appl Environ Microbiol. 1984;48:863–864. doi: 10.1128/aem.48.4.863-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boone D R, Johnson R L, Liu Y. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and implications in the measurement of Km for H2 and formate uptake. Appl Environ Microbiol. 1989;55:1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halpern J. Mechanisms of coenzyme B12-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- 5.Houwen F P, Dijkema C, Schoenmakers C H H, Stams A J M, Zehnder A J B. 13C-NMR study of propionate degradation by a methanogenic coculture. FEMS Microbiol Lett. 1987;41:269–274. [Google Scholar]
- 6.Houwen F P, Plokker J, Stams A J M, Zehnder A J B. Enzymatic evidence for involvement of the methyl-malonyl-CoA pathway in propionate oxidation by Syntrophobacter wolinii. Arch Microbiol. 1990;155:52–55. [Google Scholar]
- 7.Houwen F P, Dijkema C, Stams A J M, Zehnder A J B. Propionate metabolism in anaerobic bacteria: determination of carboxylation reactions with 13C-NMR spectroscopy. Biochim Biophys Acta. 1991;1056:126–132. [Google Scholar]
- 8.Koch M, Dolfing J, Wuhrmann K, Zehnder A J B. Pathway of propionate degradation by enriched methanogenic cocultures. Appl Environ Microbiol. 1983;45:1411–1414. doi: 10.1128/aem.45.4.1411-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lens P N L, O'Flaherty V, Dijkema C, Colleran E, Stams A J M. Propionate degradation by mesophilic anaerobic sludge: degradation pathways and effects of other volatile fatty acids. J Ferm Bioeng. 1996;82:387–391. [Google Scholar]
- 10.Liu Y, Balkwill D L, Aldrich H C, Drake G R, Boone D R. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int J Syst Bacteriol. 1999;49:545–556. doi: 10.1099/00207713-49-2-545. [DOI] [PubMed] [Google Scholar]
- 11.Plugge C M, Dijkema C, Stams A J M. Acetyl-CoA cleavage pathway in a syntrophic propionate oxidizing bacterium growing on fumarate in the absence of methanogens. FEMS Microbiol Lett. 1993;110:71–76. [Google Scholar]
- 12.Reeves H C, Ajl S J. Alpha-hydroxyglutaric acid synthethase. J Bacteriol. 1962;84:186–187. doi: 10.1128/jb.84.1.186-187.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins J E. 13C nuclear magnetic resonance studies of propionate catabolism in methanogenic cultures. Appl Environ Microbiol. 1987;53:2260–2261. doi: 10.1128/aem.53.9.2260-2261.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbins J E. A proposed pathway for catabolism of propionate in methanogenic cocultures. Appl Environ Microbiol. 1988;54:1300–1301. doi: 10.1128/aem.54.5.1300-1301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schink B. Mechanisms and kinetics of succinate and propionate degradation in anoxic freshwater sediments and sewage sludge. J Gen Microbiol. 1985;131:643–650. [Google Scholar]
- 16.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stams A J M, van Dijk J B, Dijkema C, Plugge C M. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol. 1993;59:1114–1119. doi: 10.1128/aem.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stams A J M. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Leeuwenhoek. 1994;66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 19.Stams A J M, Dong X. Role of formate and hydrogen in the degradation of propionate and butyrate by defined suspended cocultures of acetogenic and methanogenic bacteria. Antonie Leeuwenhoek. 1995;68:281–284. doi: 10.1007/BF00874137. [DOI] [PubMed] [Google Scholar]
- 20.Tabuchi T, Serizawa N, Uchiyama H. A novel pathway for the partial oxidation of propionyl-CoA to pyruvate via seven-carbon tricarboxylic acids in yeasts. Agric Biol Chem. 1974;38:2571–2572. [Google Scholar]
- 21.Textor S, Wendisch V F, de Graaf A A, Müller U, Linder M I, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 22.Tholozan J L, Samain E, Grivet J P, Moletta R, Dubourguier H C, Albagnac G. Reductive carboxylation of propionate to butyrate in methanogenic ecosystems. Appl Environ Microbiol. 1988;54:441–445. doi: 10.1128/aem.54.2.441-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tholozan J L, Samain E, Grivet J P, Albagnac G. Propionate metabolism in a methanogenic enrichment culture. Direct reductive carboxylation and acetogenesis pathways. FEMS Microbiol Ecol. 1990;73:291–298. [Google Scholar]
- 24.Wofford N Q, Beaty P S, McInerney M J. Preparation of cell-free extracts and the enzymes involved in the fatty acid metabolism in Syntrophomonas wolfei. J Bacteriol. 1986;167:179–185. doi: 10.1128/jb.167.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]