Abstract
Objective
Oral squamous cell carcinoma (OSCC) is one of the most common oral malignant tumors. circ_0004872 can inhibit the progression of gastric cancer, but its effect on the growth and metastasis of OSCC is still unclear.
Methods
qRT‐PCR was used to detect the expression levels of circ_0004872 and miR‐424‐5p in cancer tissues of OSCC patients and adjacent normal tissues, OSCC cell lines, and human normal oral keratinocytes (HOK). CCK‐8, cell colony formation, flow cytometry, and transwell assay were used to detect cell proliferation rate, viability, apoptosis rate, and invasion ability. Use glucose/lactic acid kit to assay cell glycolysis ability. The dual‐luciferase reporter gene experiment and RIP experiment verified the relationship between circ_0004872 and miR‐424‐5p. The protein levels were examined by Western blot.
Results
The expression of circ_0004872 was significantly downregulated in OSCC tissues and cells, and the overexpression of circ_0004872 inhibited the proliferation, vitality, invasion, and glycolysis of OSCC cells, and promoted apoptosis. The expression of miR‐424‐5p was greatly upregulated in OSCC tissues and OSCC cells. circ_0004872 can adsorb miR‐424‐5p in OSCC cells, and circ_0004872 can reverse the promoting effect of miR‐424‐5p overexpression on the process of OSCC cells.
Conclusion
circ_0004872 suppresses the proliferation, invasion, and glycolysis of OSCC cells by sponged miR‐424‐5p, and promotes apoptosis, which can be used as a potential target for early diagnosis and targeted therapy of OSCC.
Keywords: circ_0004872, glycolysis, invasion, miR‐424‐5p, oral squamous cell carcinoma
Circular RNA circ_0004872 was significantly downregulated in oral squamous cell carcinoma (OSCC) tissues and cells. Circ_0004872 inhibited OSCC cell proliferation, invasion, and glycolysis, and promoted apoptosis through spongy adsorption of microRNA‐424‐5p, which in turn inhibited OSCC progression.
1. INTRODUCTION
Oral squamous cell carcinoma (OSCC) is one of the most common malignant tumors in oral and maxillofacial regions, accounting for about 3% of the total malignant tumors. 1 Low survival rates due to difficult early diagnosis and high incidence of lymph node metastasis. 2 So far, the incidence of oral squamous cell carcinoma has gradually increased globally. 3 At present, the treatment of OSCC is mainly surgery, radiotherapy, and chemotherapy, but the survival rate is still low, and the prognosis of most OSCC patients has not been significantly improved. In addition, the recurrence rate is high. 4 The pathogenic factors of OSCC are complex and diverse, such as smoking, drinking, and infection with high‐risk viruses, but the pathogenesis is not clear. Therefore, an in‐depth understanding of the molecular mechanism of the occurrence and development of OSCC is essential to explore new therapeutic strategies.
Circular RNA (circular RNA, circRNA is a kind of closed circular RNA, which belongs to a kind of noncoding RNA and exists stably in mammals. It has no 5′ end cap and 3′ end tail and can be spliced by exons or introns. 5 What's more, it is characterized by highly stable, conserved, and specific expressions. 6 , 7 In recent years, many studies have shown that circular RNAs regulate the function of target miRNAs by adsorbing microRNAs (miRNAs) and regulate gene expression at the post‐transcriptional level, 8 , 9 thus participating in the progress of various human diseases, including colon cancer, 10 ovarian cancer, 11 gastric cancer, 12 esophageal cancer, 13 glioma, 14 and oral squamous cell carcinoma. 15 Therefore, circular RNA is one of the potential targets for tumor diagnosis and treatment. Studies have found that circ_0004872 is involved in the development of gastric cancer and can be used as a target for the diagnosis of early gastric cancer. 16 However, the role and mechanism of circ_0004872 in OSCC have not been reported.
This study, thus, is the first to clarify the difference in the expression of circ_0004872 between OSCC and adjacent normal tissues, explore the relationship between circ_0004872 and miR‐424‐5p, and reveal the role of circ_0004872 and miR‐424‐5p in the malignant biological behavior of OSCC, so as to lay a molecular biological foundation for targeted therapy and the discovery of new markers.
2. MATERIALS AND METHODS
2.1. Tissue specimens
The tumor tissues and adjacent normal tissues of 60 patients with oral squamous cell carcinoma treated in our hospital from June 2017 to December 2019 were collected. All tissue samples were confirmed by the pathology department of the hospital. All patients did not receive chemotherapy or radiotherapy and signed the informed consent. This study was approved by the Medical Ethics Committee of The Affiliated Stomatological Hospital of Nanchang University.
2.2. Cell culture and transfection
Human normal oral epithelial keratinocytes (HOK) and five OSCC cell lines SCC‐6, HN4, SCC‐9, CAL‐27, and SCC‐4 were purchased from the Shanghai Institute of Cells. All cells were cultured in RPMI‐1640 medium supplemented with 10% FBS and 1% penicillin‐streptomycin at 37℃ in a 5% CO2 incubator. A549 cells in the logarithmic growth phase were collected, diluted to 2 × 106 cells/ml, and inoculated in 6‐well plates. When the fusion degree was 80%–90%, the cells were transfected according to Lipofectamine transfection reagent (Invitrogen). circ_0004872 overexpression plasmid (circ_0004872) and control vector (circ_NC), miR‐424‐5p mimics (miR), and mimics NC (miNC) were designed and synthesized by Guangzhou Ruibo Biology.
2.3. Real‐time fluorescent quantitative PCR (RT‐qPCR)
Total RNA was extracted by a total RNA extraction kit. cDNA was synthesized on the basis of reverse transcription PCR kit instructions. The cDNA was subjected to reaction according to the instructions of real‐time PCR, and the reaction system was as follows: 95°C for 1 min; 95°C 40 s, 58°C 40 s, 72°C 45 s, 40 cycles. With U6 and GAPDH as an internal reference, the 2−ΔΔCt method was used for data analysis. 17 The primer sequences were shown in Table 1.
TABLE 1.
Primer sequence
| Primer | Sequence |
|---|---|
| circ_0004872 | F: 5′‐CACACAGGGTTCCTGACAGA‐3′ |
| R: 5′‐TCTCCCTCAGGGTTCTCTGG‐3′ | |
| MiR‐424‐5p | F: 5′‐ACACTCCAGCTGGGCAGCAGCAATTCATGT‐3′ |
| R: 5′‐TGGTGTCGTGGAGTCG‐3′ | |
| U6 | F: 5′‐CAGCCACAAAAGAGCACAAT‐3′ |
| R: 5′‐CAGCCACAAAAGAGCACAAT‐3′ | |
| GAPDH | F: 5′‐GTCAAGGCTGAGAACGGGAA‐3′ |
| R: 5′‐AAATGAGCCCCAGCCTTCTC‐3′ |
2.4. CCK‐8 detection
The treated SCC‐9 and CAL‐27 cells were inoculated in 96‐well plates at a density of 2000 cells/well, and 10‐μl CCK‐8 solution was added to each well. The 96‐well plate was cultured in the incubator for 1 h, and the optical density at 450 nm was detected by an enzyme‐labeled instrument.
2.5. Flow cytometry
Annexin V‐allophycocyanin (APC) apoptosis detection kit (BD Pharmingen) was used to detect apoptosis. Cells in each group were digested with trypsin and centrifuged. The cells were washed twice with precooled PBS buffer, and 1 × 106 cells/ml suspension was prepared with 1 × binding buffer. 5‐μl Annexin V‐FITC and 5‐μl PI were added to 100‐μl cell suspension, mixed, and incubated for 15 min at room temperature (20–25℃) in the dark. The results were measured by FACScan flow cytometry (Becton Dickinson).
2.6. Transwell assay
SCC‐9 and CAL‐27 cells in each group were added to the Transwell chamber coated with Matrigel, and a 500‐μl 10% FBS medium was added to the lower chamber. Cultured for 48 h, wiped Matrigel and uninvaded cells, fixed with methanol, stained with crystal violet, observed, and counted under a microscope.
2.7. Detection of glucose consumption and lactic acid production
According to the glucose detection kit and lactic acid detection kit instructions for testing, the supernatant of the cell‐culture medium was collected, diluted, and added to the 96‐well plate. Meanwhile, the standard pore was set, and the working solution was added. After reaction at 37℃ for 20 min, the optical density of each well at 570 nm was measured, and the standard curve was plotted and the contents of glucose and lactic acid were calculated. 18
2.8. Dual‐luciferase reporter assay
When the cell fusion degree reached 80%–90%, transfection was performed. The constructed wild‐type (circ_0004872‐WT) and mutant‐type (circ_0004872‐MUT) double luciferase reporter vectors of circ_0004872 were co‐transfected with NC or miR‐424‐5p mimics into cells, respectively, and cultured for 48 h. The cells were collected and added to the luciferase substrate, and the luciferase activity was detected by a luminescence instrument. The relative luciferase activity of the firefly was calculated with the luciferase activity of the sea kidney as the internal reference. 19
2.9. RNA immune co‐precipitation (RIP) assay
The binding of circ_0004872 and miR‐424‐5p to AGO2 protein 20 was detected on the ground of the Magna RIP RNA‐Binding Protein Immunoprecipitation kit (Millipore). The cells were washed with precooling PBS, and the supernatant was discarded. Cells were lysed with RIPA and centrifuged. The supernatant was taken as a cell extract. Some cell extracts were used as the input group, and the other parts were incubated with magnetic beads and antibody AGO2 (ab32381, 1:50, Abcam) or negative control IgG (ab109489, 1:100, Abcam) at 4°C overnight. The samples were placed on the magnetic pedestal to collect the magnetic beads‐protein complexes, and the samples and input were digested by proteinase K to extract RNA. Ultimately, the expression of circ_0004872 and miR‐424‐5p was detected by RT‐qPCR with GAPDH as a negative control.
2.10. Western blot
Cells in each group were lysed with RIPA lysate, and protein concentrations were detected by a BCA kit. The protein was separated by SDS‐PAGE and transferred to a PVDF membrane. The membrane was blocked at room temperature for 1 h and incubated overnight at 4°C with anti‐GLUT1 (ab14683, 1:100, Abcam), LDHA (ab47010, 1:100, Abcam), Ki67 (ab15580, 1:100, Abcam), MMP‐2 (ab92536, 1:100, Abcam), MMP‐9 (ab283575, 1:100, Abcam), and GAPDH (ab8245, 1:100, Abcam). Wash the membrane twice the next day, add diluted enzyme‐labeled secondary antibody, and incubate at room temperature for 1 h. The chemiluminescence reagent was added to develop the protein, and the image was collected in the gel imaging system. The protein level was analyzed by Image J software. GAPDH was used as the internal reference to calculate the relative expression of the protein.
2.11. Statistical analysis
SPSS 22.0 software was used for statistical analysis of the data. t test was used for comparison between the two groups, and one‐way analysis of variance was used for comparison among multiple groups. Results were expressed as mean ± standard deviation (SD). p < 0.05 indicated that the difference was statistically significant.
3. RESULTS
3.1. Low expression of circ_0004872 in OSCC
The expression of circ_0004872 in OSCC tissues and cell lines was detected. The results showed that the expression of circ_0004872 in the Tumor group was conspicuously lower than that in the Normal group (Figure 1A). Compared with the HOK group, circ_0004872 was lowly expressed in SCC‐6, HN4, SCC‐9, CAL‐27, and SCC‐4 cells of OSCC cells, and the lowest expression was found in CAL‐27, while the highest expression was found in SCC‐9 (Figure 1B).
FIGURE 1.
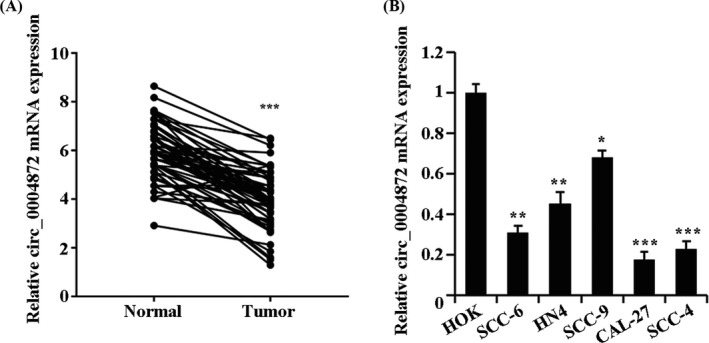
Downregulation of circ_0004872 expression in OSCC tissues and cells. (A) qRT‐PCR detects the expression of circ_0004872 in OSCC tissues and adjacent normal tissues, ***p < 0.001. (B) The expression of circ_0004872 in OSCC cell lines (SCC‐6, HN4, SCC‐9, CAL‐27, and SCC‐4) and human normal oral epithelial keratinocytes (HOK) was detected by qRT‐PCR, *p < 0.05, **p < 0.01, and ***p < 0.001 vs. HOK group
3.2. circ_0004872 inhibits the proliferation and invasion, and promotes apoptosis of OSCC cells
To investigate the effect of circ_0004872 expression on the biological function of OSCC cells, we transfected circ_0004872 overexpression vectors into CAL‐27 and SCC‐9 cells. The transfection efficiency was verified by qRT‐PCR (Figure 2A). Compared with the circ‐NC group, the overexpression of circ_0004872 significantly inhibited the proliferation, viability, and invasion of CAL‐27 and SCC‐9 but promoted cell apoptosis (Figure 2B–E). Western blot results showed that Ki67, MMP‐2, and MMP‐9 proteins closely related to cell invasion and metastasis in the circ_0004872 group were significantly decreased (Figure 2F). These results suggest that the overexpression of circ_0004872 can inhibit the proliferation and invasion of OSCC cells, and promote apoptosis.
FIGURE 2.
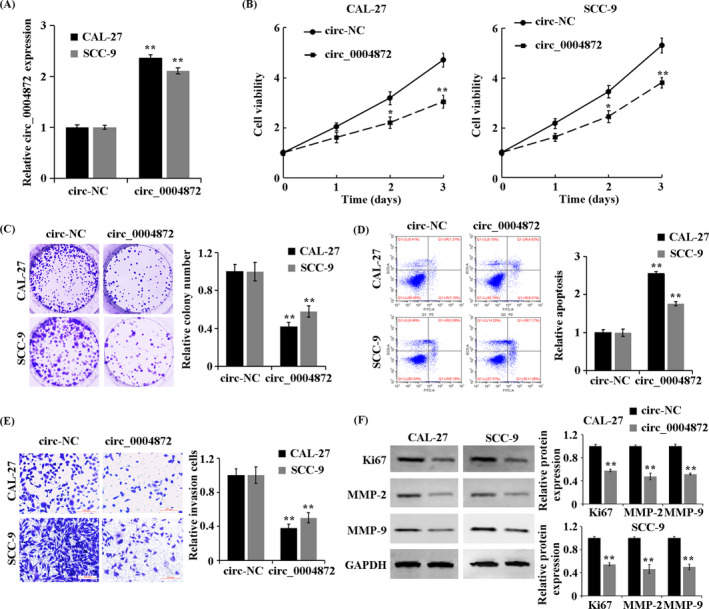
circ_0004872 inhibits the proliferation and invasion, and promotes apoptosis of OSCC cells. (A) qRT‐PCR detects the expression of circ_0004872 in the treatment group. (B) CCK‐8 method to detect the cell proliferation rate in the treatment group. (C) cell colony formation experiment to observe the cell viability of the treatment group. (D) detection of apoptosis rate by flow cytometry. (E) Transwell assay to detect the invasion ability of cells in the treatment group. (F) Western blot was used to detect the protein expressions of Ki67, MMP‐2, and MMP‐9 in the treated cells. **p < 0.01 vs. circ‐NC group
3.3. circ_0004872 inhibits glycolysis of OSCC cells
In CAL‐27 and SCC‐9 cells, compared with the circ‐NC group, the glucose consumption and lactate production levels in the circ_0004872 group were significantly decreased (Figure 3A,B). The protein levels of GLUT1 and LDHA in the circ_0004872 group were also significantly decreased (Figure 3C, p < 0.05). The above results showed that circ_0004872 could inhibit the glycolysis of OSCC cells.
FIGURE 3.
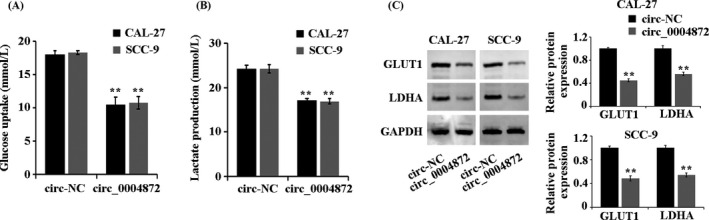
circ_0004872 inhibits glycolysis of OSCC cells. (A) Detection of glucose consumption in treated cells by kits. (B) detection of lactic acid production in treated cells by kits. (C) Western blot was used to detect the protein expression of GLUT1 and LDHA in the treated cells. **p < 0.01 indicates vs. circ‐NC group
3.4. circ_0004872 can adsorb miR‐424‐5p in OSCC cells
The prediction results of the Starbase database (https://starbase.sysu.edu.cn/) showed that circ_0004872 had an interaction with miR‐424‐5p (Figure 4A). The dual‐luciferase reporter gene experiment confirmed that miR‐424‐5p mimics significantly inhibited the luciferase activity of the circ_0004872‐WT vector, confirming the interaction between circ_0004872 and miR‐424‐5p (Figure 4B). The results of the RIP test showed that circ_0004872 and miR‐424‐5p were significantly enriched in the RNA pulled by the AGO2 antibody, which further verified the relationship between them (Figure 4C).
FIGURE 4.
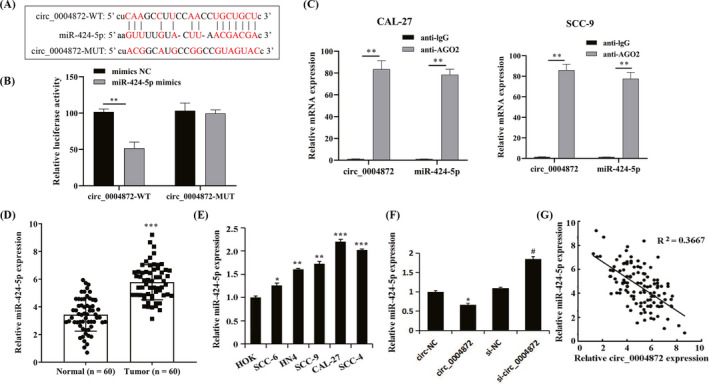
circ_0004872 adsorbs miR‐424‐5p in OSCC cells. (A) Interaction diagram of circ_0004872 and miR‐424‐5p sequences. (B) The dual‐luciferase reporter gene experiment verified the interaction between circ_0004872 and miR‐424‐5p, **p < 0.01. (C) RIP experiment verified the interaction between circ_0004872 and miR‐424‐5p, **p < 0.01. (D) the expression of miR‐424‐5p in OSCC tissues and adjacent normal tissues was detected by qRT‐PCR, and ***p < 0.001. (E) qRT‐PCR was used to detect the expression of miR‐424‐5p in OSCC cell lines, *p < 0.05, **p < 0.01, and *** p < 0.001 vs. HOK. (F) qRT‐PCR was used to detect the expression of miR‐424‐5p in each treatment group, *p < 0.05 vs. circ‐NC group, and #p < 0.05 vs. si‐NC group. (G) circ_0004872 was negatively correlated with the expression of miR‐424‐5p in OSCC
Compared with the Normal group, the expression level of miR‐424‐5p in the Tumor group was significantly increased (Figure 4D, p < 0.05). Compared with the HOK group, the expression levels of miR‐424‐5p in SCC‐6, HN4, SCC‐9, CAL‐27, and SCC‐4 cells were greatly increased (Figure 4E). In CAL‐27 and SCC‐9 cells, compared with the circ‐NC group, the expression level of miR‐424‐5p in the circ_0004872 group was significantly decreased. Compared with the si‐NC group, the expression level of miR‐424‐5p in the si‐circ_0004872 group was evidently increased (Figure 4F). The expression of circ_0004872 was negatively correlated with miR‐424‐5p in oral squamous cell carcinoma (Figure 4G). The above results showed that miR‐424‐5p was upregulated in OSCC tissues and cells, while circ_0004872 significantly reduced the expression level of miR‐424‐5p by adsorbing miR‐424‐5p in OSCC cells.
3.5. circ_0004872 can reverse the promoting effect of miR‐424‐5p overexpression on the OSCC cell process
To further explore the biological relationship between circ_0004872 and miR‐424‐5p and their roles in the development of OSCC, the experimental results showed that in CAL‐27 and SCC‐9 cells, compared with the ciNC+miNC group, the proliferation rate, viability, invasion ability, glucose consumption, and lactic acid production of the cells in the ciNC+miR group were significantly increased, and the apoptosis rate was obviously decreased. The protein expression levels of Ki67, MMP‐2, MMP‐9, GLUT1, and LDHA were significantly increased. Compared with the circ‐NC+miR group, the overexpression of circ_0004872 significantly inhibited the promotion of miR‐424‐5p on the functions of CAL‐27 and SCC‐9 cells (Figure 5A–H, p < 0.05). According to the above results, the overexpression of miR‐424‐5p can promote the proliferation, invasion, and glycolysis of OSCC cells and inhibit apoptosis. The overexpression of circ_0004872 can reverse the role of miR‐424‐5p in promoting OSCC.
FIGURE 5.
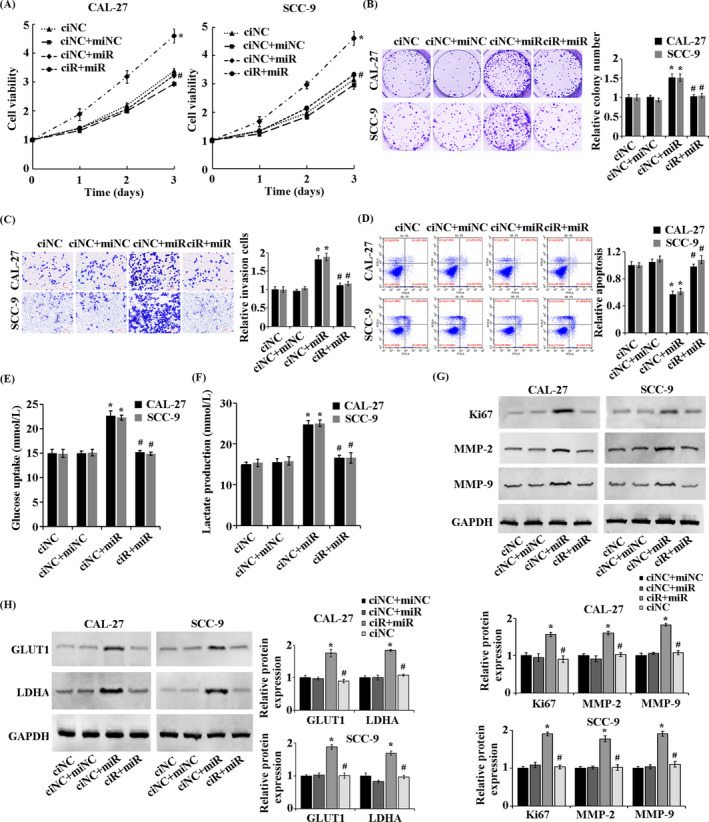
circ_0004872 reverses the role of miR‐424‐5p overexpression in OSCC cell progression. (A) CCK‐8 assay for cell proliferation. (B) cell colony formation experiment to detect cell viability. (C) Transwell assay to detect the invasion ability of cells in the treatment group. (D) detection of apoptosis rate by flow cytometry. (E) detection of glucose consumption in treated cells by kits. (F) the production of lactic acid in the treated cells was detected by kit. (G) Western blot was used to detect the protein expressions of Ki67, MMP‐2, and MMP‐9 in the treated cells. (H) Western blot was used to detect the protein expressions of GLUT1 and LDHA in the treated cells. *p < 0.05 vs. ciNC+miNC group; #p < 0.05 vs. ciNC+miR group
4. DISCUSSION
OSCC is one of the most common tumor diseases in the world. The incidence rate of OSCC varies from region to region. 21 Due to the special anatomical structure of the oral and maxillofacial region, cervical lymph node metastasis and worse biological behavior occur. Although medical technology continues to progress, the diagnosis and treatment of tumors have also made substantial progress but still cannot solve the problem of low survival rate. 22 Therefore, OSCC remains a fatal disease in the head and neck and has gradually become a major public health problem.
Studies have shown that a variety of noncoding RNAs are abnormally expressed in OSCC tumor tissues, which may be related to the occurrence and development of OSCC, indicating that the development of OSCC is regulated by both coding genes and noncoding genes. 23 Circ RNA can regulate gene expression by targeting sponge to adsorb miRNAs 8 , 10 and then participate in the development of tumors. Zhu et al 24 have confirmed that circ‐BANP is overexpressed in colon cancer tissues, and the disorder of circ‐BANP plays an important role in colon cancer cells, which can be used as a marker for the prognosis and treatment of colon cancer. Du et al 25 found that the expression level of circ‐Foxo3 in breast cancer tissues and cell lines was significantly decreased, and its overexpression could promote apoptosis and inhibit tumor growth. Chen et al 26 found that the expression of hsa_circ_100395 in lung cancer tissues was decreased, which could regulate the proliferation, migration, and invasion of lung cancer cells by regulating the miR‐1228/TCF21 pathway. Therefore, circ RNA is a potential therapeutic target and tumor biomarker. In the aspect of OSCC, Chen et al 27 reported that circ_100290 could regulate the proliferation of OSCC cells by combining endogenous competition with miR‐29. But until now, the research on circ_0004872 has only been carried out in gastric cancer, and the mechanism of OSCC has not been reported. In this study, we found that the expression of circ0004872 was significantly downregulated in OSCC tissues and OSCC cells, suggesting that the overexpression of circ_0004872 could affect the occurrence and development of OSCC. To further explore the effect of circ_0004872 on the biological activities of OSCC cells, it was found that the upregulation of circ_0004872 could inhibit the proliferation, vitality, invasion, and glycolysis of OSCC cells, and promote cell apoptosis. Therefore, circ_0004872 is an important molecule in the occurrence and development of OSCC, which can inhibit the malignant biological behavior of OSCC cells and has the potential to become a therapeutic target and biomarker for OSCC.
According to the results of Shengxin prediction, it was suggested that circ_0004872 had a targeting effect on miR‐424‐5p and further confirmed that circ_0004872 could sponge adsorb miR‐424‐5p in OSCC cells through the dual‐luciferase reporter gene and RIP experiment. In order to further clarify the role of circ_0004872 and miR‐424‐5p in OSCC and the relationship between them, it was found that miR‐424‐5p was highly expressed in OSCC tissues and OSCC cells, and was negatively correlated with the expression level of circ_0004872. Similar to Li et al, 28 they confirmed that miR‐424‐5p was highly expressed in laryngeal squamous cell carcinoma and promoted its proliferation, migration, and invasion. Zhang et al 29 found circGDI2 could inhibit OSCC progression by targeting miR‐424‐5p/SCAI. We also found that the overexpression of miR‐424‐5p could promote the proliferation, invasion, and glycolysis of OSCC cells and inhibit apoptosis, which was contrary to the trend of circ_0004872. Overexpression of circ_0004872 could reverse the promoting effect of miR‐424‐5p on the function of OSCC cells. Based on the above results, it is speculated that circ_0004872 regulates the biological behavior of OSCC cells through sponge adsorption of miR‐424‐5p, including the proliferation, vitality, invasion, glycolysis, and apoptosis of OSCC cells, which provides a new clue for the study of the mechanism of occurrence and development of OSCC cells.
In summary, we confirm that circ_0004872 can regulate the biological behavior of OSCC cells by sponge miR‐424‐5p, which can be used as a potential target for early diagnosis and targeted therapy of OSCC. However, this study only involves one of the complex functions of circ_RNA and is not explored through in vivo experiments. Further studies are needed to clarify the relationship between circ_0004872 and OSCC, so as to better play its role in the clinic.
AUTHOR CONTRIBUTIONS
YHD and HYX involved in the study concept and design; YLZ involved in the acquisition of data; YHD and YLZ involved in the analysis and interpretation of data; YHD and HYX involved in the drafting of the manuscript; All authors have read and approved the manuscript.
Dai Y, Zhu Y, Xu H. circ_0004872 inhibits proliferation, invasion, and glycolysis of oral squamous cell carcinoma by sponged miR‐424‐5p. J Clin Lab Anal. 2022;36:e24486. doi: 10.1002/jcla.24486
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1. Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26:123‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keshavarzi M, Darijani M, Momeni F, et al. Molecular imaging and oral cancer diagnosis and therapy. J Cell Biochem. 2017;118:3055‐3060. [DOI] [PubMed] [Google Scholar]
- 3. Sasahira T, Kirita T, Kuniyasu H. Update of molecular pathobiology in oral cancer: a review. Int J Clin Oncol. 2014;19:431‐436. [DOI] [PubMed] [Google Scholar]
- 4. Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301‐308. [DOI] [PubMed] [Google Scholar]
- 5. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829‐1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Zhang X‐O, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792‐806. [DOI] [PubMed] [Google Scholar]
- 10. Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down‐regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahmed I, Karedath T, Andrews SS, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366‐36381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208‐219. [DOI] [PubMed] [Google Scholar]
- 13. Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta‐catenin signaling pathway. Oncotarget. 2016;7:63449‐63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao L, Zhao C, Li S, et al. circ‐PKD2 inhibits carcinogenesis via the miR‐204‐3p/APC2 axis in oral squamous cell carcinoma. Mol Carcinog. 2019;58:1783‐1794. [DOI] [PubMed] [Google Scholar]
- 16. Cunying M. The molecular mechanism of hsa_circ_0004872 in regulating the occurrence and development of gastric cancer. 2019. [Google Scholar]
- 17. He Z, Wang X, Huang C, et al. The FENDRR/miR‐214‐3P/TET2 axis affects cell malignant activity via RASSF1A methylation in gastric cancer. Am J Transl Res. 2018;10:3211‐3223. [PMC free article] [PubMed] [Google Scholar]
- 18. Yuanyuan L, Bin C, Yun W. Effects of long non‐coding RNA lnc‐p26090 on the glycolysis and proliferation in oral squamous cell carcinoma. Int J Stomatol. 2018;45:628‐634. [Google Scholar]
- 19. Zou P, Zhu M, Lian C, et al. miR‐192‐5p suppresses the progression of lung cancer bone metastasis by targeting TRIM44. Sci Rep. 2019;9:19619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L, He KE, Chen S, et al. Circ_0001175 promotes hepatocellular carcinoma cell proliferation and metastasis by regulating miR‐130a‐5p. Onco Targets Ther. 2020;13:13315‐13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scully C, Bagan JV. Recent advances in oral oncology 2008; squamous cell carcinoma imaging, treatment, prognostication and treatment outcomes. Oral Oncol. 2009;45:e25‐e30. [DOI] [PubMed] [Google Scholar]
- 22. Bolha L, Ravnik‐Glavac M, Glavac D. Circular RNAs: biogenesis, function, and a role as possible cancer biomarkers. Int J Genomics. 2017;2017:6218353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ouyang S, Zhang P, Wang J, Huang Z, Liao L. Expression of long non‐coding RNA colon cancer associated transcript 2 and its clinicopathologic significance in oral squamous cell carcinoma. Zhonghua Kou Qiang Yi Xue Za Zhi. 2016;51:286‐291. [DOI] [PubMed] [Google Scholar]
- 24. Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138‐144. [DOI] [PubMed] [Google Scholar]
- 25. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR‐1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen L, Zhang S, Wu J, et al. Retraction note: circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR‐29 family. Oncogene. 2019;38:5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Liu J, Hu W, et al. miR‐424‐5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. 2019;12:10441‐10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Tang K, Chen L, Du M, Qu Z. Exosomal CircGDI2 suppresses oral squamous cell carcinoma progression through the regulation of MiR‐424‐5p/SCAI axis. Cancer Manag Res. 2020;12:7501‐7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


