Abstract
Background
Circular RNAs (circRNAs) have been found to have potential biological applications against tumors in humans. This study aimed to evaluate the diagnostic, prognostic, and clinicopathological value of circRNAs in head and neck squamous cell carcinoma (HNSCC).
Methods
The PubMed, Web of Science, EMBASE, and the Cochrane Library were comprehensively searched for the relevant studies before October 20, 2021. Statistical analysis was performed based on STATA 15.0, Meta‐DiSc 1.4, and RevMan 5.3 software.
Results
A total of 55 reports regarding 56 kinds of circRNA were studied in this meta‐analysis, including 23, 38, and 26 articles on diagnosis, prognosis, and clinicopathological features, respectively. The pooled sensitivity, specificity, and area under the curve (AUC) of the summary receiver‐operating characteristic curve (SROC) were 0.78, 0.84, and 0.87, respectively. Besides, the upregulation of oncogenic circRNAs was significantly associated with poorer overall survival (OS) (HR=2.25, p < 0.05) and disease‐free interval (DFS) (HR=1.92, p < 0.05). In contrast, the elevated expression of tumor suppressor circRNAs was associated with a favorable prognosis (HR=0.50, p < 0.05). In addition, the high expression of oncogenic circRNAs was associated with the tumor size (OR=3.59, p < 0.05), degree of differentiation (OR=1.89, p < 0.05), TNM stage (OR=2.35, p < 0.05), lymph node metastasis (OR=1.85, p < 0.05), and distant metastasis (OR=3.42, p < 0.05). Moreover, the expression of tumor suppressor circRNAs was associated with improved clinicopathological features (lymph node metastasis: OR=0.25, p < 0.05).
Conclusions
CircRNAs could serve as potential predictive indicators and be useful for the diagnosis, prognosis, and identification of clinicopathological features in HNSCC.
Keywords: biomarker, circular RNA, head and neck squamous cell carcinoma, diagnosis, prognosis
This study aimed to evaluate the diagnostic, prognostic, and clinicopathological value of circRNAs in patients with head and neck squamous cell carcinoma (HNSCC). The pooled sensitivity and specificity, diagnostic odds ratio (DOR), and overall area under the curve (AUC) of the summarizing receiver‐operating characteristics (SROC) were determined and used to assess the diagnostic value of circRNAs. In addition, subgroup analyses were performed to explore the main sources of heterogeneity. Pooled hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated to assess the association between circRNAs expression, overall survival (OS), and disease‐free interval (DFS). Pooled odds ratios (ORs) and 95% CIs were used to assess the relationship between circRNAs expression and clinicopathological parameters. (A) Forest plots of sensitivity and specificity for the diagnosis of circRNAs in HNSCC among 31 studies. (B) The AUC (SROC curve) for the diagnosis of circRNAs in HNSCC among 31 studies. (C) Forest plots of the association between the expression of circRNAs and OS in HNSCC. (D) Forest plots of the association between the expression of circRNAs and DFS in HNSCC.

1. INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the most common group of head and neck malignancies. Although this group of malignancies originates in different sites at the head and neck, including (1) nasal cavity and sinuses, (2) nasopharynx, (3) hypopharynx, larynx, and trachea, and (4) oral and oropharynx, their pathogenesis, staging system, treatment strategies, and prognosis are similar. Therefore, it is reasonable to group them together as HNSCC 1 . HNSCC is the fifth most common cancer occurring worldwide, with over 600,000 cases reported annually 2 , 3 . Despite advances in surgery, chemotherapy, immunotherapy, and radiotherapy, the 5 years survival rate of HNSCC patients still remains between 40%–50% 4 . Since the overall survival rate of patients with HNSCC has barely improved over the past few decades, it is critical to identify new molecular markers for the early detection and prognosis and identify new therapeutic targets for HNSCC addressing this dismal clinical situation 5 , 6 .
Circular RNAs (circRNAs), a new class of endogenous noncoding RNAs, are characterized by a closed‐loop structure formed by covalent bonds between the head and tail, and are usually generated by the exons of precursor mRNAs through reverse splicing 7 , 8 . CircRNAs may regulate carcinogenesis in different cancers by performing their complex biological functions, i.e., by acting as ceRNA or miRNA sponges, regulating regulatory gene transcription and expression, interacting with RNA‐binding proteins, and translating RNAs into proteins. Because circRNAs are also more stable and conserved than linear RNAs, numerous circRNAs can occur in exosomes, peripheral blood, or tissues 8 , 9 , 10 , 11 . CircRNAs may be suitable for use as novel biomarkers and therapeutic targets for human cancer.
Studies have shown that circRNAs are abnormally expressed in numerous human cancers including esophageal cancer 12 , osteosarcoma 13 , lung cancer 14 , and breast cancer 15 . Simultaneously, several studies have confirmed the role of circRNAs in the proliferation, migration and invasion, apoptosis, angiogenesis, deterioration, and recurrence of human cancer 16 , 17 , 18 , 19 . These results indicate that circRNAs have significant potential for use in human cancer prediction, and prognosis and clinical treatment. CircRNAs can act as both tumor suppressors and oncogenes in HNSCC 20 . Therefore, circRNAs may act as a new biomarker and therapeutic target for the prevention and treatment of HNSCC. However, inconsistent results from existing studies have become an obstacle to the application of circRNAs in clinical practice.
To our knowledge, no meta‐analysis has been performed till date to assess the diagnostic and prognostic value of circRNAs in HNSCC. Therefore, we conducted a systematic and comprehensive meta‐analysis of relevant studies, to explore the significance of circRNAs in the diagnosis and prognosis of HNSCC.
2. MATERIALS AND METHODS
2.1. Search strategy
This study was performed in accordance with the Preferred Reporting Items for Systematic Review and Meta‐analysis (PRISMA) Checklist 21 . As of October 20, 2021, we conducted a comprehensive search to identify studies that assessed the association of circRNAs with the diagnosis and prognosis or clinicopathological features of HNSCC using 4 electronic databases, i.e., PubMed, Web of Science, EMBASE, and the Cochrane Library database. The following terms were used in databases for report retrieval: (RNA, Circular OR circRNA OR Circular RNA OR ciRNA) AND (cancer OR tumor OR neoplasm OR tumor OR malignant OR metastasis OR carcinoma OR Squamous Cell Carcinoma OR SCC) AND (head and neck OR larynx OR oropharynx OR hypopharynx OR nasopharynx OR oral and cavity OR mouth OR laryngeal OR pharyngeal OR sinus OR sinonasal OR tongue OR NPC OR nasopharyngeal).
2.2. Study selection
Studies that met the following criteria are included: (1) cohort or case‐control studies; (2) studies in which HNSCC was histopathologically confirmed; (3) studies that evaluated the association between circRNAs expression, with the diagnosis, prognosis, and clinicopathological features of HNSCC.
The following reports were excluded: (1) studies not related to circRNAs or HNSCC; (2) reviews, case reports, or retracted studies; (3) studies involving animal experiments or cell line experiments; (4) studies lacking sufficient data; (5) studies that were not in English.
2.3. Data extraction and quality assessment
Two independent investigators (FHJ and WDT) evaluated the included studies and carefully extracted the data, and if disagreements occurred, a third investigator (LJP) was consulted to reach a consensus. The following data were extracted from the relevant studies: (a) basic characteristics: first author, publication date, country, sample size, sample type, circRNAs name, regulatory characteristic, cancer type, detection method, control type, and follow‐up time; (b) data acquired in diagnostic studies: TP, FP, FN, TN, sensitivity (SEN), specificity (SPE), area under the curve (AUC); (c) data for prognostic studies: hazard ratio (HR) values and 95% confidence interval (CI) of survival outcomes; and (4) clinicopathological features: age, sex, TNM stage, T stage, lymph node metastasis, distant metastasis, tumor size, and degree of differentiation.
The effect of the quality of included studies on diagnosis was assessed according to the Quality Assessment for Studies of Diagnostic Accuracy II (QUADAS II) checklist 22 . Studies on prognosis were rated by the Newcastle‐Ottawa Scale (NOS), as described previously 23 , 24 . Studies were considered to be of high quality if the QUADAS II score was ≥4 or the NOS score was ≥6.
2.4. Statistical analysis
Statistical analysis was performed using Stata 15.0, Revman 5.3, and Meta‐DiSc 1.4 software. The TP, FP, FN, and TN values were calculated to determine the pooled sensitivity, specificity, AUC, negative likelihood ratio (NLR), positive likelihood ratio (PLR), and diagnostic odds ratio (DOR) at the corresponding 95% CI, to evaluate the diagnostic value of circRNAs in HNSCC. The corresponding 95% CI value of the HRs was used to evaluate the relationship between circRNAs and the prognosis of HNSCC patients. The association between circRNAs expression and clinicopathological parameters was assessed using a combination of odds ratios (ORs) with a 95% CI. The threshold effect was evaluated using a Spearman’s correlation coefficient, and values were considered statistically significant if p < 0.05. The nonthreshold effect was tested using the Cochran's Q test and the I2 test, and the level of statistical significance was set as p < 0.01 or I2 >50%. When there is no heterogeneity between studies, fixed‐effect models can be used to merge data. Otherwise, the random‐effects model is used. The source of heterogeneity was traced using sensitivity analysis and meta‐regression tests. The Deek’s funnel plot asymmetry test for the diagnostic meta‐analysis, p < 0.01, was considered statistically significant. And publication bias between studies about prognosis was evaluated using the Begg’s test and Egger’s test, p < 0.05 was considered statistically significant.
3. RESULT
3.1. Search results
The process for the selection of research articles to be reviewed is shown in Figure 1. A total of 644 potential literatures were initially identified via database searches. After 159 duplicate publications were excluded the titles and abstracts of the remaining 485 articles were assessed. Among these, 294 articles were excluded after reviewing for various reasons, and only 191 articles were reviewed thoroughly. Finally, 55 articles that involved details regarding 56 unique circRNAs and 5,576 HNSCC cases (all cases were reliably diagnosed via histopathological analysis) were included in this meta‐analysis. To be specific, we included 31 diagnostic studies (from 23 articles 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 ), 38 prognostic studies (from 38 articles 25 , 26 , 27 , 28 , 29 , 30 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 ), and 27 clinical‐pathological feature‐related studies(from 26 articles 28 , 29 , 43 , 47 , 48 , 50 , 51 , 53 , 55 , 56 , 58 , 63 , 64 , 65 , 66 , 67 , 68 , 70 , 71 , 72 , 73 , 74 , 76 , 77 , 78 , 79 ).
FIGURE 1.

The flow chart of the research selection process
3.2. Study characteristics and quality assessment
Table 1 and Table 2 show the basic characteristics of the included studies. 55 articles were included, 54 articles were conducted in China and 1 article in Italy. The number of patients in each study had an individual range of 20–292. All studies were published between 2017 and 2021 and were conducted for 20–90 months. As shown in Table 1, the diagnostic meta‐analysis of 31 eligible studies was performed; of these, 23 articles involved reports regarding 26 types of circRNAs. The quality assessment of these 23 articles was performed as shown in Figure S1. The expression of circRNAs in all diagnostic studies was determined by quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis of tissues (n = 18), plasma (n = 9), serum (n = 3), and saliva (n = 1). There are a total of 16 upregulated circRNAs and 15 downregulated circRNAs. Tumor types included OSCC (n = 15), LSCC (n = 8), NPC (n = 4), HPSCC (n = 2), TSCC (n = 1), and HNSCC (n = 1). As shown in Table 2, we performed a prognostic meta‐analysis of 38 relevant studies that assessed the association between circRNAs and OS, and 7 studies that assessed the association between circRNAs and DFS. Our prognostic meta‐analysis showed a total of 30 circRNAs upregulated (tumor promoters) and 8 circRNAs downregulated (tumor suppressors) in HNSCC. The expression of circRNAs was calibrated in most studies using qRT‐PCR analysis, except for 3 studies in which the expression was calibrated using in situ hybridization (ISH). Species included serum (n = 3) and tumor (n = 35) samples. Tumor types included OSCC (n = 12), LSCC (n = 7), NPC (n = 10), HPSCC (n = 2), TSCC (n = 3), and HNSCC (n = 4).
TABLE 1.
Main characteristics of studies for diagnosis analysis in HNSCC
| Study | Year | Country | CircRNAs | Regulation | Sample size | Cancer | Specimen | Method | Diagnostic power | Source of the control group | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | SEN | SPE | AUC | |||||||||
| Fan C(a) et al | 2019 | China | CircMAN1A2 | Upregulated | 100 | 121 | NPC | Serum | qRT‐PCR | 0.81 | 0.86 | 0.91 | healthy controls |
| Wang(a) J et al | 2020 | China | Hsa_circ_0066755 | Upregulated | 16 | 19 | NPC | Tissue | qRT‐PCR | 0.88 | 0.84 | 0.90 | nasal polyps tissues |
| Wang(b) J et al | 2020 | China | Hsa_circ_0066755 | Upregulated | 86 | 86 | NPC | Plasma | qRT‐PCR | 0.86 | 0.79 | 0.85 | healthy controls |
| Shuai M et al | 2020 | China | Hsa_circ_001387 | Upregulated | 100 | 100 | NPC | Tissue | qRT‐PCR | 0.70 | 0.96 | 0.92 | adjacent normal tissues |
| Yao Y et al | 2020 | China | Hsa_circ_0001742 | Upregulated | 146 | 146 | TSCC | Tissue | qRT‐PCR | 0.78 | 0.81 | 0.87 | adjacent normal tissues |
| Wang X et al | 2020 | China | Hsa_circ_103862 | Upregulated | 62 | 62 | LSCC | Tissue | qRT‐PCR | 0.82 | 0.69 | 0.81 | adjacent normal tissues |
| Guo Y et al | 2020 | China | Hsa_circ_0036722 | Downregulated | 41 | 41 | LSCC | Tissue | qRT‐PCR | 0.61 | 0.95 | 0.84 | adjacent normal tissues |
| Han J(a) et al | 2021 | China | Hsa_circ_0019201 | Upregulated | 20 | 20 | LSCC | Plasma | qRT‐PCR | 0.95 | 0.85 | 0.93 | healthy controls |
| Han J(b) et al | 2021 | China | Hsa_circ_0019201 | Upregulated | 100 | 100 | LSCC | Plasma | qRT‐PCR | 0.64 | 0.95 | 0.77 | healthy controls |
| Han J(c) et al | 2021 | China | Hsa_circ_0011773 | Upregulated | 20 | 20 | LSCC | Plasma | qRT‐PCR | 1.00 | 0.75 | 0.91 | healthy controls |
| Han J(d) et al | 2021 | China | Hsa_circ_0011773 | Upregulated | 100 | 100 | LSCC | Plasma | qRT‐PCR | 0.78 | 0.98 | 0.86 | healthy controls |
| Han J(e) et al | 2021 | China | Hsa_circ_0122790 | Upregulated | 20 | 20 | LSCC | Plasma | qRT‐PCR | 0.85 | 0.95 | 0.97 | healthy controls |
| Han J(f) et al | 2021 | China | Hsa_circ_0122790 | Upregulated | 100 | 100 | LSCC | Plasma | qRT‐PCR | 0.83 | 0.95 | 0.91 | healthy controls |
| Guo Y(a) et al | 2020 | China | CircMORC3 | Downregulated | 33 | 33 | HPSCC | Tissue | qRT‐PCR | 0.81 | 0.69 | 0.83 | adjacent normal tissues |
| Guo Y(b) et al | 2020 | China | CircMORC3 | Downregulated | 22 | 22 | HPSCC | Plasma | qRT‐PCR | 0.72 | 0.68 | 0.77 | vocal cord polyps tissues |
| Shen Z et al | 2021 | China | Hsa_circ_0016148 | Downregulated | 137 | 137 | HNSCC | Tissue | qRT‐PCR | 0.92 | 0.87 | 0.91 | adjacent normal tissues |
| Sun S et al | 2018 | China | Hsa_Circ_001242 | Downregulated | 40 | 40 | OSCC | Tissue | qRT‐PCR | 0.73 | 0.78 | 0.78 | adjacent normal tissues |
| Li B et al | 2018 | China | Hsa_Circ_0008309 | Downregulated | 45 | 45 | OSCC | Tissue | qRT‐PCR | 0.51 | 0.91 | 0.76 | adjacent normal tissues |
| He T et al | 2018 | China | CircPVT1 | Upregulated | 50 | 50 | OSCC | Tissue | qRT‐PCR | 0.69 | 0.86 | 0.79 | adjacent normal tissues |
| Zhao S et al | 2018 | China | Hsa_circ_0001874+ Hsa_circ_0001971 | Upregulated | 93 | 85 | OSCC | Saliva | qRT‐PCR | 0.93 | 0.78 | 0.92 | healthy controls |
| Li X et al | 2019 | China | Hsa_circ_0004491 | Downregulated | 40 | 40 | OSCC | Tissue | qRT‐PCR | 0.73 | 0.68 | 0.75 | adjacent normal tissues |
| Xia B et al | 2019 | China | CircMMP9 | Upregulated | 25 | 16 | OSCC | Plasma | qRT‐PCR | 0.89 | 0.81 | 0.91 | healthy controls |
| Su W et al | 2019 | China | Hsa_circ_0005379 | Downregulated | 37 | 37 | OSCC | Tissue | qRT‐PCR | 0.70 | 0.61 | 0.68 | adjacent normal tissues |
| Dou Z et al | 2019 | China | Hsa_circ_0072387 | Downregulated | 63 | 63 | OSCC | Tissue | qRT‐PCR | 0.71 | 0.70 | 0.75 | adjacent normal tissues |
| Fan C(b) et al | 2019 | China | CircMAN1A2 | Upregulated | 55 | 121 | OSCC | Serum | qRT‐PCR | 0.67 | 0.92 | 0.78 | healthy controls |
| Wang Z et al | 2019 | China | Hsa_circ_009755 | Downregulated | 27 | 27 | OSCC | Tissue | qRT‐PCR | 0.70 | 0.78 | 0.78 | adjacent normal tissues |
| Zhang H et al | 2020 | China | Hsa_circ_0003829 | Downregulated | 60 | 60 | OSCC | Tissue | qRT‐PCR | 0.70 | 0.80 | 0.81 | adjacent normal tissues |
| Li L et al | 2020 | China | Hsa_circ_0086414 | Downregulated | 55 | 55 | OSCC | Tissue | qRT‐PCR | 0.66 | 0.87 | 0.75 | adjacent normal tissues |
| Chen G et al | 2020 | China | CircATRNL1 | Downregulated | 48 | 48 | OSCC | Tissue | qRT‐PCR | 0.85 | 0.51 | 0.71 | adjacent normal tissues |
| Zhang B et al | 2020 | China | Hsa_circ_009755 | Downregulated | 42 | 42 | OSCC | Tissue | qRT‐PCR | 0.69 | 0.89 | 0.83 | adjacent normal tissues |
| Fan X et al | 2021 | China | CircSPATA6 | Downregulated | 46 | 25 | OSCC | Serum | qRT‐PCR | 0.79 | 0.69 | 0.77 | healthy controls |
Abbreviations: AUC, area under the curve; HNSCC, head and neck squamous cell carcinoma; HPSCC, hypopharyngeal squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; OSCC, oral squamous cell carcinoma; SEN, sensitivity; SPE, specificity.
TABLE 2.
Main characteristics of studies for prognosis analysis in HNSCC
| Study | Year | CircRNAs | Country | High | Low | Test method | Type | Sample type | Regulation pattern | Follow‐up months | Survival indicators |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Shuai M et al | 2018 | Hsa_circ_0000285 | China | 105 | 45 | qRT‐PCR | NPC | Serum | Upregulated | 80 | OS |
| Chen L et al | 2019 | CircRNA_000543 | China | 75 | 48 | qRT‐PCR | NPC | Tissue | Upregulated | 100 | OS |
| Luo Y et al | 2020 | CircMYC | China | 148 | 62 | qRT‐PCR | NPC | Serum | Upregulated | 60 | OS, DFS |
| Shuai M et al | 2020 | Hsa_circ_001387 | China | 54 | 46 | qRT‐PCR | NPC | Tissue | Upregulated | 60 | OS |
| Hong X et al | 2021 | CircCRIM1 | China | 91 | 127 | qRT‐PCR | NPC | Tissue | Upregulated | 120 | OS, DFS |
| Ke Z et al | 2020 | CircHIPK3 | China | 32 | 31 | qRT‐PCR | NPC | Tissue | Upregulated | 150 | OS |
| Dong Q et al | 2020 | Hsa_circ_0028007 | China | 160 | 81 | qRT‐PCR | NPC | Tissue | Upregulated | 40 | OS |
| Fang X et al | 2021 | CircTRAF3 | China | 50 | 50 | qRT‐PCR | NPC | Tissue | Upregulated | 100 | OS, DFS |
| Li W et al | 2021 | CircTGFBR2 | China | 29 | 46 | ISH | NPC | Tissue | Downregulated | 100 | OS |
| Liu Z et al | 2021 | CircZNF609 | China | 35 | 25 | qRT‐PCR | NPC | Tissue | Upregulated | 60 | OS |
| Verduci L et al | 2017 | CircPVT1 | Italy | 71 | 35 | qRT‐PCR | HNSCC | Tissue | Upregulated | 70 | OS |
| Ju H et al | 2021 | CircGNG7 | China | 22 | 43 | ISH | HNSCC | Tissue | Downregulated | 60 | OS |
| Zhang S et al | 2021 | Hsa_circ_0032822 | China | 30 | 30 | qRT‐PCR | HNSCC | Tissue | Upregulated | 120 | OS, DFS |
| Shen Z et al | 2021 | Hsa_circ_0016148 | China | 65 | 72 | qRT‐PCR | HNSCC | Tissue | Downregulated | 60 | OS |
| Wang Z et al | 2020 | CircMATR3 | China | 24 | 26 | qRT‐PCR | HPSCC | Tissue | Upregulated | 60 | OS |
| Wu P et al | 2021 | CircCUX1 | China | 45 | 33 | qRT‐PCR | HPSCC | Tissue | Upregulated | 48 | OS, DFS |
| Yao Y et al | 2020 | Hsa_circ_0001742 | China | 73 | 73 | qRT‐PCR | TSCC | Tissue | Upregulated | 60 | OS |
| Qian C et al | 2021 | Hsa_circ_0043265 | China | 20 | 20 | qRT‐PCR | TSCC | Tissue | Downregulated | 60 | OS |
| Qian C et al | 2021 | Hsa_circ_0000003 | China | 20 | 20 | qRT‐PCR | TSCC | Tissue | Upregulated | 60 | OS |
| Wei Z et al | 2019 | Hsa_circ_0042666 | China | 18 | 17 | qRT‐PCR | LSCC | Tissue | Downregulated | 150 | OS |
| Wang J et al | 2019 | CircFLNA | China | 19 | 20 | qRT‐PCR | LSCC | Tissue | Upregulated | 200 | OS |
| Gao W et al | 2020 | CircPARD3 | China | 50 | 50 | qRT‐PCR | LSCC | Tissue | Upregulated | 70 | OS |
| Wang X et al | 2020 | Hsa_circ_103862 | China | 80 | 72 | ISH | LSCC | Tissue | Upregulated | 60 | OS |
| Zang Y et al | 2020 | CircCCND1 | China | 50 | 51 | qRT‐PCR | LSCC | Tissue | Upregulated | 80 | OS |
| Chu Y et al | 2020 | Hsa_circ_0067934 | China | 20 | 20 | qRT‐PCR | LSCC | Tissue | Upregulated | 90 | OS |
| Wu Y et al | 2021 | circCORO1C | China | 48 | 48 | qRT‐PCR | LSCC | Tissue | Upregulated | 70 | OS |
| Dou Z et al | 2019 | Hsa_circ_0072387 | China | 15 | 63 | qRT‐PCR | OSCC | Tissue | Downregulated | 60 | OS |
| Xia B et al | 2019 | CircMMP9 | China | 37 | 37 | qRT‐PCR | OSCC | Tissue | Upregulated | 80 | OS |
| Hao C et al | 2020 | CircITCH | China | 46 | 57 | qRT‐PCR | OSCC | Tissue | Downregulated | 60 | OS |
| Li K et al | 2020 | Hsa_circ_0000745 | China | 32 | 32 | qRT‐PCR | OSCC | Tissue | Upregulated | 60 | OS |
| Wang J et al | 2020 | CircEPSTI1 | China | 72 | 82 | qRT‐PCR | OSCC | Tissue | Upregulated | 60 | OS |
| Luo Y et al | 2020 | Hsa_circ_0000199 | China | 68 | 40 | qRT‐PCR | OSCC | Serum | Upregulated | 60 | OS, DFS |
| Yang Y et al | 2020 | Hsa_circ_002178 | China | 25 | 25 | qRT‐PCR | OSCC | Tissue | Upregulated | 60 | OS |
| Peng Q et al | 2020 | Hsa_circ_0000140 | China | 28 | 28 | qRT‐PCR | OSCC | Tissue | Downregulated | 50 | OS |
| Zhao W et al | 2020 | CircUHRF1 | China | 10 | 10 | qRT‐PCR | OSCC | Tissue | Upregulated | 50 | OS |
| Gao L et al | 2020 | Hsa_circ_0092125 | China | 50 | 36 | qRT‐PCR | OSCC | Tissue | Upregulated | 60 | OS |
| Chen H et al | 2021 | CircVAPA | China | 30 | 30 | qRT‐PCR | OSCC | Tissue | Upregulated | 50 | OS, DFS |
| Liu J et al | 2021 | CircIGHG | China | 64 | 105 | qRT‐PCR | OSCC | Tissue | Upregulated | 40 | OS |
Abbreviations: DFS, disease‐free interval; HNSCC, head and neck squamous cell carcinoma; HPSCC, hypopharyngeal squamous cell carcinoma; ISH, in situ hybridization; LSCC, laryngeal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; OS, overall survival; OSCC, oral squamous cell carcinoma; TSCC, tongue squamous cell carcinoma.
3.3. Expression of circRNAs with diagnosis in HNSCC
3.3.1. Data analysis
Thirty‐one relevant studies from 23 articles were included in the meta‐analysis. As shown in Figure 2, there was significant heterogeneity in the pooled sensitivity (I2=71.19%, p < 0.001) and specificity (I2=81.29%, p < 0.001) values. Therefore, the random‐effects model was used to analyze diagnostic parameters. The forest diagram shows the value of circRNAs in the diagnosis of HNSCC; the pooled sensitivity was 0.78 (95% CI=0.74–0.82), specificity was 0.84 (95% CI=0.79–0.88), PLR was 4.86 (95% CI=3.77–6.27), NLR was 0.26 (95% CI=0.22–0.31), and the combined DOR was 19 (95% CI=13–26) (Figure 2A,B,C). In addition, Figure 2D shows a summary receiver operator characteristic (SROC) curve with an AUC of 0.87 (95% CI=0.84–0.90).
FIGURE 2.

Forest plots of sensitivity, specificity, PLR, NLR, DOR, and AUC for diagnosis of circRNAs in HNSCC among 31 studies. (A) Sensitivity and specificity; (B) PLR and NLR; (C) DOR; (D) AUC (SROC curve)
3.3.2. Threshold effect, heterogeneity, and subgroup analysis
The Spearman’s correlation coefficient value was 0.313, and the p value was 0.086, indicating that the threshold effect was not observed. Figure 2D shows that there was no typical shoulder and arm, indicating that there was no threshold effect. This can also be equated with the fact that the threshold effect is not a source of heterogeneity.
We have also shown the construction of a bivariate boxplot, which is a useful tool for detecting heterogeneity in each study (Figure 3C). Three studies did not occur in the boxplot, including studies 2, 20, and 26. Studies 26 involved the use of plasma, and studies 2 and 20 involved the use of tissue. This implies that the sample source could be the main cause of heterogeneity. Meta‐regression analysis showed that the sample size, specimen, circRNAs expression, and tumor type might decide the source of heterogeneity (Figure 3B).
FIGURE 3.
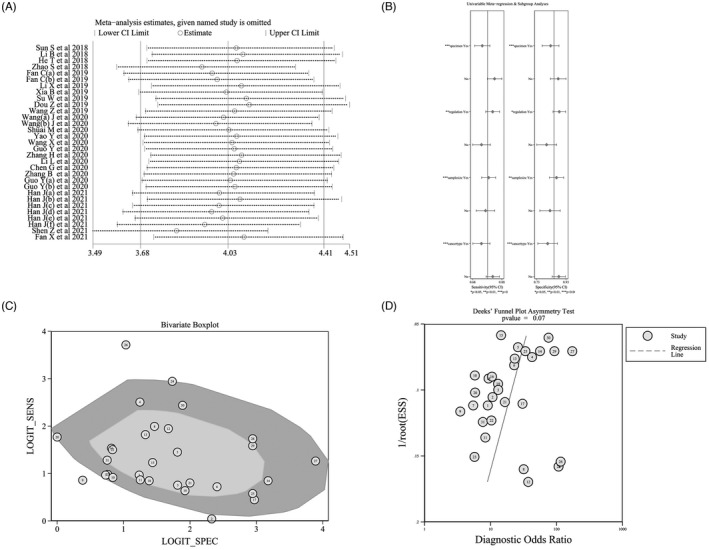
Assessment of the diagnostic accuracy of circRNAs in HNSCC. (A) Sensitivity analysis; (B) meta‐regression analysis; (C) bivariate boxplot; (D) Deek’s funnel plot
Then, subgroup analysis was performed based on the circRNAs expression level, sample size, specimen, control source, and tumor type; results are shown in Table 3. The diagnostic performance of carcinogenic circRNAs was higher than that of tumor‐inhibiting circRNAs (AUC: 0.91 vs 0.82). The diagnostic performance of circRNAs in studies involving large samples was higher than that in studies involving small samples (AUC: 0.89 vs 0.84). With regard to the source of circRNAs extraction, plasma sample‐based studies exhibited the highest sensitivity (0.84), specificity (0.89), DOR (43), and AUC (0.92) values, compared with values in studies based on tissue or serum/saliva samples. In addition, circRNAs analysis was diagnostically effective for distinguishing patients with HNSCC from healthy individuals than for distinguishing HNSCC tissues from adjacent noncancerous tissues (AUC: 0.91 vs 0.83). Finally, the subgroup analysis of HNSCC based on tumor types from multiple parts indicated that circRNAs showed good diagnostic value for the detection of LSCC (AUC: 0.93), NPC (AUC: 0.90), and OSCC (AUC: 0.83). These results suggest that circRNAs may be an ideal diagnostic biomarker for HNSCC.
TABLE 3.
Subgroup analysis of diagnostic accuracy of circRNAs for HNSCC
| Variable | No | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) | Heterogeneity | |
| I2 | p | ||||||||
| Overall | 31 | 0.78 (0.74–0.82) | 0.84 (0.79–0.88) | 4.9 (3.8–6.3) | 0.26 (0.22–0.31) | 19 (13–26) | 0.87 (0.84–0.90) | 98.0% | <0.001 |
| Regulation | |||||||||
| Upregulated | 16 | 0.81 (0.76–0.86) | 0.88 (0.83–0.92) | 6.7 (4.8–9.5) | 0.21 (0.17–0.27) | 32 (22–46) | 0.91 (0.88–0.93) | 96.0% | <0.001 |
| Downregulated | 15 | 0.74 (0.68–0.79) | 0.78 (0.71–0.84) | 3.4 (2.5–4.5) | 0.33 (0.27–0.41) | 10 ( 7–15) | 0.82 (0.79–0.85) | 93.0% | <0.001 |
| Sample size | |||||||||
| >100 | 14 | 0.79 (0.73–0.83) | 0.88 (0.82–0.92) | 6.4 (4.4–9.3) | 0.24 (0.19–0.30) | 26 (17–42) | 0.89 (0.86–0.92) | 96.0% | <0.001 |
| <100 | 17 | 0.76 (0.70–0.81) | 0.79 (0.72–0.84) | 3.6 (2.7–4.7) | 0.30 (0.24–0.38) | 12 ( 8–17) | 0.84 (0.80–0.87) | 94.0% | <0.001 |
| Specimen | |||||||||
| Tissue | 18 | 0.74 (0.69–0.79) | 0.81 (0.75–0.86) | 4.0 (3.0–5.3) | 0.31 (0.26–0.38) | 13 ( 9–18) | 0.84 (0.80–0.87) | 96.0% | <0.001 |
| Plasma | 9 | 0.84 (0.76–0.89) | 0.89 (0.81–0.94) | 7.8 (4.4–13.8) | 0.18 (0.12–0.27) | 43 (23–81) | 0.92 (0.89–0.94) | 93.0% | <0.001 |
| Serum/Saliva | 4 | 0.82 (0.71–0.89) | 0.84 (0.77–0.90) | 5.3 (3.7–7.5) | 0.21 (0.14–0.34) | 25 (16–39) | 0.90 (0.87–0.92) | 84.0% | =0.001 |
| Source of control | |||||||||
| Adjacent | 17 | 0.74 (0.69–0.79) | 0.81 (0.74–0.86) | 3.9 (2.9 –5.3) | 0.32 (0.27–0.39) | 12 (8–18) | 0.83 (0.80–0.86) | 96.0% | <0.001 |
| Healthy/other | 14 | 0.83 (0.78–0.88) | 0.87 (0.81–0.91) | 6.4 (4.4 –9.4) | 0.19 (0.15–0.25) | 33 (22–52) | 0.91 (0.89–0.93) | 95.0% | <0.001 |
| Cancer type | |||||||||
| OSCC | 15 | 0.74 (0.68–0.79) | 0.79 (0.68–0.79) | 3.5 (2.7 –4.5) | 0.33 (0.27–0.40) | 11 (7–15) | 0.83 (0.79–0.86) | 95.0% | <0.001 |
| LSCC | 8 | 0.82 (0.71–0.89) | 0.92 (0.83–0.96) | 9.8 (5.0 –19.3) | 0.20 (0.13–0.32) | 49 (24 –101) | 0.93 (0.90–0.95) | 94.0% | <0.001 |
| NPC | 4 | 0.81 (0.72–0.87) | 0.88 (0.78–0.94) | 6.6 (3.7 –11.6) | 0.22 (0.16–0.31) | 30 (18–51) | 0.90 (0.87–0.92) | 83.0% | =0.002 |
| Other | 4 | 0.82 (0.71–0.89) | 0.80 (0.73–0.86) | 4.1 (2.7 –6.3) | 0.22 (0.13–0.39) | 18 (7–47) | 0.87 (0.84–0.90) | 0 | =0.336 |
Abbreviations: AUC, area under the curve; CI, confidence interval; DOR, diagnostic odds ratio; HNSCC, head and neck squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; NLR, negative likelihood ratio; NPC, nasopharyngeal carcinoma; OSCC, oral squamous cell carcinoma; PLR, positive likelihood ratio; SEN, sensitivity; SPE, specificity.
3.3.3. Publication bias and sensitivity analysis
Sensitivity analysis showed that the results of the meta‐analysis did not change when studies were omitted item by item (Figure 3A). The Deek’s funnel plot asymmetry test is a useful tool for assessing the potential publication bias in studies. The results of the use of this test showed that there was no significant publication bias, and the p value was 0.07 (Figure 3D).
3.4. Expression of circRNAs with prognosis in HNSCC
3.4.1. Data analysis
Survival analysis showed that oncogenic circRNAs overexpression was significantly associated with a worsened OS (HR=2.25, 95% CI: 1.99–2.55) and DFS (HR=1.92, 95%CI: 1.53–2.40), as shown in Figure 4A and 4C, respectively. In addition, the increased expression of tumor‐inhibiting circRNAs caused a prediction of improved OS (HR=0.50, 95%CI: 0.38–0.66), as shown in Figure 4A. These studies were all fixed‐effect models without significant heterogeneity.
FIGURE 4.

Forest plots and sensitivity analysis of the association between the expression of circRNAs and the prognosis of patients with HNSCC. (A) Forest plots for OS; (B) sensitivity analysis for OS; (C) forest plots for DFS; (D) sensitivity analysis for DFS
3.4.2. Heterogeneity and subgroup analysis
Subgroup analysis was further conducted according to the sample size, sample source, circRNAs detection method, and tumor type, to explore the source of heterogeneity. The results are shown in Table 4. The prognostic significance of upregulated circRNAs in OS was evaluated in 30 studies with 3058 HNSCC patients and its pooled HR was 2.25 (95%CI=1.99–2.55, I2=0.0, Phet=0.870, fixed‐effects model), it suggested that the HNSCC patients with higher expression of circRNAs had shorter overall survival time than those with lower expression of circRNAs among tumor‐oncogene circRNAs, and among the downregulated circRNAs, the pooled HR for OS was 0.50 (95%CI=0.38‐0.66, I2=0.0, Phet=0.999, fixed‐effects model), and it suggested that the higher expression of tumor suppressor circRNAs in HNSCC was associated with longer overall survival time. In terms of tumor type, a high level of circRNAs expression was associated with OSCC (OS: HR=1.70; 95%Cl=1.06–2.73), LSCC (OS: HR=2.12; 95%Cl=1.52–2.96), NPC (OS: HR=2.04; 95%Cl=1.67–2.48), and HPSCC (OS: HR=2.56; 95%Cl=1.63–4.01) and was associated with a poor prognosis. Though a high level of expression of circRNAs was associated with TSCC (OS: HR=1.22; 95%Cl=0.58–2.55), no defined correlation was observed. Only seven studies were related to DFS and tumor‐oncogene circRNAs, so we were unable to conduct further analysis.
TABLE 4.
Subgroup analysis of prognostic outcomes of circRNAs for HNSCC
| Variable | No | Patients | HR (95%CI) | p‐value | Heterogeneity | ||
| I2 (%) | P HET | Model | |||||
| Overall Survival | |||||||
| Overall | 38 | 3647 | 1.74 (1.40–2.15) | <0.001 | 67.8 | <0.001 | Random |
| Regulation | |||||||
| Upregulated | 30 | 3058 | 2.25 (1.99–2.55) | <0.001 | 0 | 0.870 | Fixed |
| Downregulated | 8 | 589 | 0.50 (0.38–0.66) | <0.001 | 0 | 0.999 | Fixed |
| Sample size | |||||||
| >100 | 14 | 2118 | 1.89 (1.37– 2.60) | <0.001 | 78.1 | 0.2908 | Random |
| <100 | 24 | 1529 | 1.62 (1.21–2.16) | 0.005 | 56.8 | 0.2736 | Random |
| Specimen | |||||||
| Tissue | 35 | 3179 | 1.65 (1.32–2.06) | <0.001 | 66.4 | <0.001 | Random |
| Serum | 3 | 468 | 2.73 (1.71–4.35) | <0.001 | 58.7 | 0.089 | Random |
| Test method | |||||||
| qRT‐PCR | 35 | 3355 | 1.84 (1.49–2.28) | <0.001 | 65.9 | <0.001 | Random |
| ISH | 3 | 292 | 0.77 (0.28–2.16) | 0.625 | 73.0 | 0.025 | Random |
| Cancer type | |||||||
| OSCC | 12 | 1022 | 1.70 (1.06–2.73) | 0.028 | 76.1 | <0.001 | Random |
| LSCC | 7 | 563 | 2.12 (1.52–2.96) | <0.001 | 42.0 | 0.111 | Fixed |
| NPC | 10 | 1340 | 2.04 (1.67–2.48) | <0.001 | 30.4 | 0.166 | Fixed |
| HNSCC | 4 | 368 | 1.03 (0.42–2.52) | 0.947 | 82.5 | 0.001 | Random |
| TSCC | 3 | 226 | 1.22 (0.58–2.55) | 0.599 | 53.7 | 0.115 | Random |
| HPSCC | 2 | 128 | 2.56 (1.63–4.01) | <0.001 | 0 | 0.475 | Fixed |
| Disease‐free survival | |||||||
| Overall (upregulated) | 7 | 834 | 1.92 (1.53–2.4) | <0.001 | 0 | 0.986 | Fixed |
Abbreviations: CI, confidence interval; Fixed, fixed‐effects model; HNSCC, head and neck squamous cell carcinoma; HPSCC, hypopharyngeal squamous cell carcinoma; HR, hazard ratio; LSCC, laryngeal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; OSCC, oral squamous cell carcinoma; Phet, p value of heterogeneity; Random, random‐effects model; TSCC, tongue squamous cell carcinoma.
3.4.3. Publication bias and sensitivity analysis
The results of sensitivity analysis showed that no single study could affect the combined HRs of the OS and DFS (Figure 4B, D). To track the potential publication bias during the meta‐analysis, we conducted certain tests (Figure 5A,B,C,D). The p values of Begg's and Egger's tests for the OS and DFS were all greater than 0.05, indicating that there was no publication offset.
FIGURE 5.
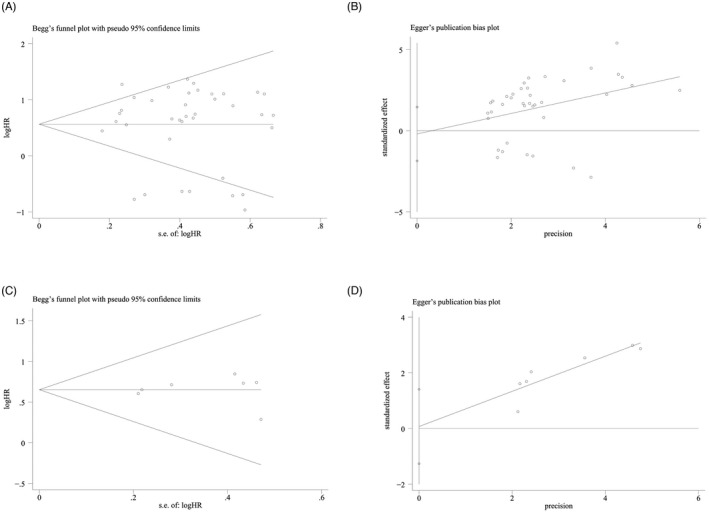
Publication bias of the association between the expression of circRNAs and the prognosis of patients with HNSCC. (A) Begg’s funnel plot for OS; (B) Egger's test plot for OS; (C) Begg’s funnel plot for DFS; (D) Egger’s test plot for DFS
3.5. Expression of circRNA with clinicopathological parameters in HNSCC
In 27 included studies on clinicopathological parameters from 26 articles, a total of 27 circRNAs were described. The correlation between circRNAs and clinicopathological parameters of HNSCC patients is shown in Table 5. Our results showed that among these the clinicopathological features, oncogenic circRNA upregulation was associated with tumor size (OR=3.59, 95%CI=2.48–5.19, p < 0.001), degree of differentiation (OR=1.89, 95%CI=1.36–2.61, p < 0.001), TNM staging (OR=2.35, 95%CI=1.94–2.85, p < 0.001), lymph node metastasis (OR=1.85, 95%CI=1.23–2.78, p = 0.003), and distant metastasis (OR=3.42, 95%CI=2.42–4.84, p < 0.001). The upregulation of circRNA was associated with an improvement in clinicopathological features and lymph node metastasis (OR=0.25, 95%CI=0.14–0.47, p < 0.001). There was no statistical correlation between the expression of tumor suppressor gene circRNAs and age, sex, tumor size, tumor stage, and differentiation.
TABLE 5.
Association between expression of circRNAs and clinicopathological features
| Categories | Tumor promoter | Tumor suppressor | ||||||||||||
| Studies | Patients | OR (95% CI) | p | I2 | P HET | Model | Studies | Patients | OR (95% CI) | p | I2 | p het | Model | |
| Age (old/young) | 20 | 1864 | 1.15 (0.95–1.40) | 0.141 | 0.0% | 0.636 | Fixed | 4 | 234 | 0.80 (0.48–1.35) | 0.408 | 0.0% | 0.487 | Fixed |
| Gender (M/W) | 22 | 2182 | 1.03 (0.85–1.24) | 0.772 | 0.0% | 0.949 | Fixed | 4 | 234 | 1.01 (0.60–1.70) | 0.983 | 0.0% | 0.905 | Fixed |
| Tumor size (large/small) | 8 | 550 | 3.59 (2.48–5.19) | <0.000 | 0.0% | 0.939 | Fixed | 1 | 40 | 0.44 (0.12–1.57) | 0.207 | – | – | – |
| Differentiation grade (poor/well) | 9 | 1052 | 1.89 (1.36–2.61) | <0.000 | 0.0% | 0.872 | Fixed | 1 | 56 | 0.38 (0.12–1.19) | 0.098 | – | – | – |
| TNM stage (III+IV/I+II) | 20 | 1952 | 2.35 (1.94–2.85) | <0.000 | 55.2% | 0.002 | Random | 3 | 131 | 0.87 (0.12–6.57) | 0.895 | 86.1% | 0.001 | Random |
|
T classification (T3 + T4/T1 + T2) |
13 | 1453 | 1.53 (0.94–2.47) | 0.085 | 76.4% | <0.000 | Random | – | – | – | – | – | – | – |
| Lymph node metastasis (Y/N) | 15 | 1500 | 1.85 (1.23–2.78) | 0.003 | 67.7% | <0.000 | Random | 3 | 194 | 0.25 (0.14–0.47) | <0.000 | 49.4% | 0.139 | Fixed |
| Distant metastasis (Y/N) | 7 | 659 | 3.42 (2.42–4.84) | <0.000 | 10.6% | = 0.348 | Fixed | 1 | 103 | 0.39 (0.17–0.91) | 0.029 | – | – | – |
Abbreviations: CI, confidence interval; Fixed, fixed‐effects model; Random, random‐effects model; OR, odds ratio; Phet, p value of heterogeneity.
4. DISCUSSION
CircRNAs seem to have good prospects as an ideal biomarker for human cancer diagnosis or prognosis over the last decade due to their special advantages as a biomarker, which include their stable and continuous covalent closed loops, high stability in cells and body fluids, and close association between its complex biological functions and carcinogenesis 20 .
Four previous meta‐analyses by Wang 80 , Ding 81 , Li 82 , and Tan 83 examined the association between circRNAs and cancer, and confirmed that circRNAs might play an important role in the diagnosis and prognosis of human cancer. The predictive role of circRNAs in different malignancies, including esophageal cancer 84 , lung cancer 85 , and colorectal cancer 86 , has also been confirmed recently. However, there are few articles on HNSCC tumors in these meta‐analyses. A growing number of studies have shown that some circRNAs are abnormally expressed in HNSCC 32 , 37 , 39 , 65 , 66 , 67 , 68 . However, the predictive value of circRNAs in HNSCC is still unclear. To our knowledge, this is the first meta‐analysis to address the relationship between the expression of circRNAs and the diagnosis, prognosis, and clinicopathological features of HNSCC.
In our analysis, overall, the pooled sensitivity and specificity of circRNAs for the diagnosis of HNSCC were 0.78 and 0.84, respectively, and the AUC was 0.87. Furthermore, the overall DOR was 19, while the combined PLR and NLR were 4.86 and 0.26, respectively. In addition, through subgroup analysis, we found that circRNAs were effective for the diagnosis of different HNSCC tumor types, especially LSCC (AUC: 0.93), NPC (AUC: 0.90), and OSCC (AUC: 0.83). The SROC curve and Spearman correlation coefficient indicated that there were no threshold effects. This indicates that the threshold effect is not the source of heterogeneity. Considering the significant heterogeneity, we chose the random‐effects model. However, the results of the bivariate boxplot and meta‐regression analysis indicate that the sources of heterogeneity between the included studies may be the sample size, tumor type, circRNAs expression, control source, and sample. These results suggest that circRNAs may be suitable for use as potential biomarkers for the diagnosis of HNSCC.
To determine the relationship between circRNA, OS, and DFS in HNSCC patients, a total of 38 eligible prognostic studies were included. Overall, the high expression of oncogenic RNA resulted in a significant deterioration in the OS, whereas the high expression of inhibited circRNAs resulted in a significantly better OS in HNSCC patients. In addition, when grouped by tumor types, the high expression of circRNAs was indicative of a worsened prognosis for patients with OSCC, LSCC, NPC, and HPSCC, but not those with TSCC. This may be attributable to the limited number of studies on individuals with TSCC (n = 3). During our search, seven studies examined the association of circRNAs with DFS, and we found that the overexpression of oncogenic circRNAs was associated with a shorter DFS.
The upregulation of circRNAs was significantly correlated with the tumor size, degree of differentiation, TNM stage, lymph node metastasis, and distant metastasis. The downregulation of circRNAs, a tumor suppressor gene, led to poor lymph node metastasis.
However, certain limitations are associated with our meta‐analysis. First, most of the demographic data included in the meta‐analysis were from China; hence, our conclusions were more applicable to the Chinese or Asian population, which may affect the applicability of our findings across different regions. Second, the number of included studies on circRNA, a tumor suppressor gene, is relatively small, and more studies need to be conducted in the future, to further confirm the results. In addition, some studies do not clearly state the sensitivity, specificity, or HR values. We extracted essential data from the ROC and KM curves provided, which could lead to potential deviations. Finally, although we performed a hierarchical analysis, heterogeneity was still observed in some subgroups.
5. CONCLUSION
Taken together, our meta‐analysis showed that circRNAs can be used as promising biomarkers for the diagnosis of patients with HNSCC, and especially for those with LSCC and NPC. Furthermore, our study also found that there is a significant association between circRNAs overexpression, prognostic outcomes, and clinicopathological values in patients with HNSCC. This implies that circRNAs might play an important role in the occurrence and development of HNSCC. However, more comprehensive, high‐quality, and large‐scale studies involving populations from more regions need to be performed, to elucidate the roles of circRNAs in HNSCC.
7. CONFLICT OF INTEREST
None declared.
8.
Supporting information
Figure S1
6. ACKNOWLEDGMENTS
The authors thank all patients for providing the clinical data and samples.
Feng H, Wang D, Liu J, et al. Diagnostic and prognostic value of circRNAs expression in head and neck squamous cell carcinoma: A meta‐analysis. J Clin Lab Anal. 2022;36:e24496. doi: 10.1002/jcla.24496
Huajun Feng, Dingting Wang and Jinping Liu, contributed equally to this work.
8.1. DATA AVAILABILITY STATEMENT
The original contributions presented in the study are included in the article/supplementary material; further reasonable inquiries can be directed to the corresponding author/s.
REFERENCES
- 1. Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Audet N, Beasley NJ, MacMillan C, et al. Lymphatic vessel density, nodal metastases, and prognosis in patients with head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131(12):1065‐1070. [DOI] [PubMed] [Google Scholar]
- 3. Hussey DH, Latourette HB, Panje WR. Head and neck cancer: an analysis of the incidence, patterns of treatment, and survival at the University of Iowa. Ann Otol Rhinol Laryngol Suppl. 1991;152:2‐16. [DOI] [PubMed] [Google Scholar]
- 4. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9‐22. [DOI] [PubMed] [Google Scholar]
- 5. Conley BA. Treatment of advanced head and neck cancer: what lessons have we learned? J Clin Oncol. 2006;24(7):1023‐1025. [DOI] [PubMed] [Google Scholar]
- 6. Noorlag R, van der Groep P, Leusink FK, et al. Nodal metastasis and survival in oral cancer: Association with protein expression of SLPI, not with LCN2, TACSTD2, or THBS2. Head Neck. 2015;37(8):1130‐1136. [DOI] [PubMed] [Google Scholar]
- 7. Hsu MT, Coca‐Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339‐340. [DOI] [PubMed] [Google Scholar]
- 8. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rybak‐Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870‐885. [DOI] [PubMed] [Google Scholar]
- 12. Huang E, Fu J, Yu Q, et al. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR‐339‐5p/CDC25A axis. Epigenomics. 2020;12(7):587‐603. [DOI] [PubMed] [Google Scholar]
- 13. Xiao‐Long M, Kun‐Peng Z, Chun‐Lin Z. Circular RNA circ_HIPK3 is down‐regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856‐1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang L, Ma H, Kong W, et al. Up‐regulated circular RNA VANGL1 contributes to progression of non‐small cell lung cancer through inhibition of miR‐195 and activation of Bcl‐2. Biosci Rep. 2019;39(6):BSR20182433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple‐negative breast cancer progression. Theranostics. 2018;8(14):4003‐4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou LH, Yang YC, Zhang RY, et al. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297‐2303. [DOI] [PubMed] [Google Scholar]
- 17. Wang Q, Wang T, Hu Y, et al. Circ‐EIF4G3 promotes the development of gastric cancer by sponging miR‐335. Pathol Res Pract. 2019;215(9):152507. [DOI] [PubMed] [Google Scholar]
- 18. Liu X, Abraham JM, Cheng Y, et al. Synthetic circular RNA functions as a miR‐21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. 2018;13:312‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y, Zhang C, Xiong J, et al. Emerging important roles of circRNAs in human cancer and other diseases. Genes Dis. 2021;8(4):412‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou DN, Ye CS, Deng YF. CircRNAs: potency of protein translation and feasibility of novel biomarkers and therapeutic targets for head and neck cancers. Am J Transl Res. 2020;12(5):1535‐1552. [PMC free article] [PubMed] [Google Scholar]
- 21. Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta‐analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529‐536. [DOI] [PubMed] [Google Scholar]
- 23. Tu C, Ren X, He J, et al. The value of lncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers. J Cancer. 2019;10(24):5992‐6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang C, Ren X, He J, et al. The prognostic value of long noncoding RNA SNHG16 on clinical outcomes in human cancers: a systematic review and meta‐analysis. Cancer Cell Int. 2019;19:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dou Z, Li S, Ren W, et al. Decreased expression of hsa_circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Dis. 2019;25(5):1302‐1308. [DOI] [PubMed] [Google Scholar]
- 26. Shen ZS, Wang LQ, Ye D. The expression profile and clinical application value of hsa_circ_0016148 in head and neck squamous cell carcinoma. J Clin Lab Anal. 2021;35(11):e23997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shuai MX, Huang L. High expression of hsa_circRNA_001387 in nasopharyngeal carcinoma and the effect on efficacy of radiotherapy. Oncotargets and Therapy. 2020;13:3965‐3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X, Wu TY, Wang P, et al. Circular RNA 103862 promotes proliferation and invasion of laryngeal squamous cell carcinoma cells through the miR‐493‐5p/GOLM1 Axis. Front Oncol. 2020;10:1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia B, Hong T, He X, et al. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplantation. 2019;28(12):1614‐1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao Y, Bi L, Zhang CG. Circular RNA_0001742 has potential to predict advanced tumor stage and poor survival profiles in tongue squamous cell carcinoma management. J Clin Lab Anal. 2020;34(8):e23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen G, Li Y, He Y, et al. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Nucleic Acids. 2020;19:961‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan CM, Wang JP, Tang YY, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110(7):2180‐2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan X, Wang Y. Circular RNA circSPATA6 inhibits the progression of oral squamous cell carcinoma cells by regulating TRAF6 via miR‐182. Cancer Manag Res. 2021;13:1817‐1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo Y, Huang Q, Zheng J, et al. Diagnostic significance of downregulated circMORC3 as a molecular biomarker of hypopharyngeal squamous cell carcinoma: a pilot study. Cancer Manag Res. 2020;12:43‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo Y, Huang Q, Zheng J, et al. Diagnostic role of dysregulated circular RNA hsa_circ_0036722 in laryngeal squamous cell carcinoma. Onco Targets Ther. 2020;13:5709‐5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han J, Lin Q, Dong C. Plasma cell‐free circRNAs panel act as fingerprint predicts the occurrence of laryngeal squamous cell carcinoma. Aging (Albany NY). 2021;13(13):17328‐17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He T, Li X, Xie D, et al. Overexpressed circPVT1 in oral squamous cell carcinoma promotes proliferation by serving as a miRNA sponge. Mol Med Rep. 2019;20(4):3509‐3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li BW, Wang F, Li X, Sun S, Shen Y, Yang H. Hsa_circ_0008309 may be a potential biomarker for oral squamous cell carcinoma. Disease Markers. 2018;2018:7496890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li L, Zhang ZT. Hsa_circ_0086414 might be a diagnostic biomarker of oral squamous cell carcinoma. Med Sci Monit. 2020;26:e919383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X, Zhang HY, Wang YF, Sun S, Shen Y, Yang H. Silencing circular RNA hsa_circ_0004491 promotes metastasis of oral squamous cell carcinoma. Life Sci. 2019;239:116883. [DOI] [PubMed] [Google Scholar]
- 41. Su W, Wang Y, Wang F, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19(1):400. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Sun S, Li B, Wang Y, et al. Clinical significance of the decreased expression of hsa_circ_001242 in oral squamous cell carcinoma. Dis Markers. 2018;2018:6514795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J, Kong J, Nie Z, et al. Circular RNA Hsa_circ_0066755 as an oncogene via sponging miR‐651 and as a promising diagnostic biomarker for nasopharyngeal carcinoma. Int J Med Sci. 2020;17(11):1499‐1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Tang J, Wang Y, et al. Circular RNA hsa_circ_009755 downregulation correlates with clinicopathology in oral squamous cell carcinoma. OncoTargets and Therapy. 2019;12:4025‐4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang BR, Wang ZH, Shen YH, et al. Silencing circular RNA hsa_circ_009755 promotes growth and metastasis of oral squamous cell carcinoma. Genomics. 2020;112(6):5275‐5281. [DOI] [PubMed] [Google Scholar]
- 46. Zhang H, Shen Y, Zhang B, et al. Hsa_circ_0003829 serves as a potential diagnostic predictor for oral squamous cell carcinoma. J Int Med Res. 2020;48(9):300060520936880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao SY, Wang J, Ouyang SB, et al. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell Physiol Biochem. 2018;47(6):2511‐2521. [DOI] [PubMed] [Google Scholar]
- 48. Chen H, Zhang Y, Wu K, et al. circVAPA promotes the proliferation, migration and invasion of oral cancer cells through the miR‐132/HOXA7 axis. J Int Med Res. 2021;49(6):3000605211013207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen L, Zhou H, Guan Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR‐9/platelet‐derived growth factor receptor B axis. Biochem Biophys Res Commun. 2019;512(4):786‐792. [DOI] [PubMed] [Google Scholar]
- 50. Chu YL. Circ‐0067934 correlates with poor prognosis and promotes laryngeal squamous cell cancer progression by sponging miR‐1324. Eur Rev Medi Pharmacol Sci. 2020;24(8):4320‐4327. [DOI] [PubMed] [Google Scholar]
- 51. Dong QN, Zhang JF, Hao YL, et al. Implication of hsa_circ_0028007 in reinforcing migration, invasion, and chemo‐tolerance of nasopharyngeal carcinoma cells. J Clin Lab Anal. 2020;34(9):e23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang X, Huang W, Wu P, et al. CircRNA circTRAF3 promotes nasopharyngeal carcinoma metastasis through targeting miR‐203a‐3p/AKT3 axis. Pathol Res Pract. 2021;221:153438. [DOI] [PubMed] [Google Scholar]
- 53. Gao L, Wang QB, Zhi Y, et al. Down‐regulation of hsa_circ_0092125 is related to the occurrence and development of oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2020;49(3):292‐297. [DOI] [PubMed] [Google Scholar]
- 54. Gao W, Guo HN, Niu M, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI‐Akt‐mTOR pathway. Mol Cancer. 2020;19(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hao C, Wangzhou K, Liang Z, et al. Circular RNA ITCH suppresses cell proliferation but induces apoptosis in oral squamous cell carcinoma by regulating miR‐421/PDCD4 axis. Cancer Manag Res. 2020;12:5651‐5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hong XH, Liu N, Liang YL, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ju HY, Hu ZR, Wei DL, et al. A novel intronic circular RNA, circGNG7, inhibits head and neck squamous cell carcinoma progression by blocking the phosphorylation of heat shock protein 27 at Ser78 and Ser82. Cancer Commun. 2021;41(11):1152‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ke Z, Xie F, Zheng C, et al. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR‐4288‐induced ELF3 inhibition. J Cell Physiol. 2019;234(2):1699‐1706. [DOI] [PubMed] [Google Scholar]
- 59. Li K, Fan X, Yan Z, et al. Circ_0000745 strengthens the expression of CCND1 by functioning as miR‐488 sponge and interacting with HuR binding protein to facilitate the development of oral squamous cell carcinoma. Cancer Cell Int. 2021;21(1):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li WP, Lu HY, Wang H, et al. Circular RNA TGFBR2 acts as a ceRNA to suppress nasopharyngeal carcinoma progression by sponging miR‐107. Cancer Letters. 2021;499:301‐313. [DOI] [PubMed] [Google Scholar]
- 61. Liu JP, Jiang X, Zou AL, et al. circIGHG‐induced epithelial‐to‐mesenchymal transition promotes oral squamous cell carcinoma progression via miR‐142‐5p/IGF2BP3 signaling. Cancer Res. 2021;81(2):344‐355. [DOI] [PubMed] [Google Scholar]
- 62. Liu ZL, Liu FF, Wang F, et al. CircZNF609 promotes cell proliferation, migration, invasion, and glycolysis in nasopharyngeal carcinoma through regulating HRAS via miR‐338‐3p. Mol Cell Biochem. 2021;476(1):175‐186. [DOI] [PubMed] [Google Scholar]
- 63. Luo Y, Liu F, Guo J, et al. Upregulation of circ_0000199 in circulating exosomes is associated with survival outcome in OSCC. Sci Rep. 2020;10(1):13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo YW, Ma JQ, Liu FX, et al. Diagnostic value of exosomalcircMYCin radioresistant nasopharyngeal carcinoma. Head Neck. 2020;42(12):3702‐3711. [DOI] [PubMed] [Google Scholar]
- 65. Peng QS, Cheng YN, Zhang WB, et al. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR‐31 to inhibit Hippo signaling pathway. Cell Death Dis. 2020;11(2):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qian CJ, Chen SH, Li S, et al. Circ_0000003 regulates glutamine metabolism and tumor progression of tongue squamous cell carcinoma via the miR‐330‐3p/GLS axis. Oncol Rep. 2021;45(4):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qian CJ, Yang YS, Lan TC, et al. Hsa_circ_0043265 restrains cell proliferation, migration and invasion of tongue squamous cell carcinoma via targeting the miR‐1243/SALL1 axis. Pathol Oncol Res. 2021;27:587130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shuai MX, Hong JW, Huang DH, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16(5):6495‐6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Verduci L, Ferraiuolo M, Sacconi A, et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription‐competent complex. Genome Biol. 2017;18(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J, Jiang C, Li N, et al. The circEPSTI1/mir‐942‐5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 2020;11(8):682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang JX, Liu Y, Jia XJ, et al. Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA‐miR‐486‐3p‐FLNA axis. Cancer Cell Int. 2019;19:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang ZW, Wei P, Wei DM, et al. Effect of up‐regulation of circMATR3 on the proliferation, metastasis, progression and survival of hypopharyngeal carcinoma. J Cell Mol Med. 2020;24(8):4687‐4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei ZX, Chang KP, Fan CS. Hsa_circ_0042666 inhibits proliferation and invasion via regulating miR‐223/TGFBR3 axis in laryngeal squamous cell carcinoma. Biomed Pharmacother. 2019;119:109365. [DOI] [PubMed] [Google Scholar]
- 74. Wu P, Fang X, Liu YL, et al. N6‐methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12(4):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu YY, Zhang YL, Zheng XW, et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let‐7c‐5p/PBX3 axis. Mol Cancer. 2020;19(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang Y, Ci HS, Mao YL, et al. CircRNA‐002178 promotes the proliferation and migration of oral squamous cell carcinoma cells by activating the Akt/mTOR pathway. Eur Rev Med Pharmacol Sci. 2020;24(11):6122‐6130. [DOI] [PubMed] [Google Scholar]
- 77. Zang YZ, Li J, Wan BL, et al. circRNA circ‐CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR‐646. J Cell Mol Med. 2020;24(4):2423‐2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang SJ, Han JH, Fu J. The circ_0032822 promotes the proliferation of head and neck squamous cell carcinoma cells through miR‐141/EF3 signaling axis. Front Oncol. 2021;11:662496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhao W, Cui Y, Liu L, et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27(3):919‐933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang M, Yang Y, Xu J, et al. CircRNAs as biomarkers of cancer: a meta‐analysis. BMC Cancer. 2018;18(1):303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ding HX, Lv Z, Yuan Y, et al. The expression of circRNAs as a promising biomarker in the diagnosis and prognosis of human cancers: a systematic review and meta‐analysis. Oncotarget. 2018;9(14):11824‐11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li Y, Zeng X, He J, et al. Circular RNA as a biomarker for cancer: A systematic meta‐analysis. Oncol Lett. 2018;16(3):4078‐4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tan H, Gan L, Fan X, et al. Diagnostic value of circular RNAs as effective biomarkers for cancer: a systematic review and meta‐analysis. Onco Targets Ther. 2019;12:2623‐2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lin H, Yuan J, Liang G, et al. Prognostic and diagnostic significance of circRNA expression in esophageal cancer: a meta‐analysis. Gastroenterol Res Pract. 2020;2020:8437250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang X, Tian W, Wang S, et al. CircRNAs as promising biomarker in diagnostic and prognostic of lung cancer: An updated meta‐analysis. Genomics. 2021;113(1 Pt 1):387‐397. [DOI] [PubMed] [Google Scholar]
- 86. Yuan J, Guo D, Li X, et al. Prognostic and diagnostic value of circRNA expression in colorectal carcinoma: a meta‐analysis. BMC Cancer. 2020;20(1):448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further reasonable inquiries can be directed to the corresponding author/s.


