Abstract
Purpose
Fast and accurate auto-segmentation on daily images is essential for magnetic resonance imaging (MRI)–guided adaptive radiation therapy (ART). However, the state-of-the-art auto-segmentation based on deep learning still has limited success, particularly for complex structures in the abdomen. This study aimed to develop an automatic contour refinement (ACR) process to quickly correct for unacceptable auto-segmented contours.
Methods and Materials
An improved level set–based active contour model (ACM) was implemented for the ACR process and was tested on the deep learning–based auto-segmentation of 80 abdominal MRI sets along with their ground truth contours. The performance of the ACR process was evaluated using 4 contour accuracy metrics: the Dice similarity coefficient (DSC), mean distance to agreement (MDA), surface DSC, and added path length (APL) on the auto-segmented contours of the small bowel, large bowel, combined bowels, pancreas, duodenum, and stomach.
Results
A portion (3%-39%) of the corrected contours became practically acceptable per the American Association of Physicists in Medicine Task Group 132 (TG-132) recommendation (DSC >0.8 and MDA <3 mm). The best correction performance was seen in the combined bowels, where for the contours with major errors (initial DSC <0.5 or MDA >8 mm), the mean DSC increased from 0.34 to 0.59, the mean MDA decreased from 7.02 mm to 5.23 mm, and the APL reduced by almost 20 mm, whereas for the contours with minor errors, the mean DSC increased from 0.72 to 0.79, the mean MDA decreased from 3.35 mm to 3.29 mm, and more than one-third (39%) of the ACR contours became clinically acceptable. The execution time for the ACR process on one subregion was less than 2 seconds using an NVIDIA GTX 1060 GPU.
Conclusions
The ACR process implemented based on the ACM was able to quickly correct for some inaccurate contours produced from MRI-based deep learning auto-segmentation of complex abdominal anatomy. The ACR method may be integrated into the auto-segmentation step to accelerate the process of MRI-guided ART.
Introduction
Magnetic resonance imaging (MRI)–guided adaptive radiation therapy (ART) is currently being introduced into clinics by taking advantage of the emerging MR-Linac technology.1,2 The MR-Linac is a hybrid system that integrates an MRI scanner and a linear accelerator (Linac), enabling superior soft-tissue contrast, functional information, and real-time MRI-guided radiation therapy (RT) delivery. In particular, the MR-Linac enables online adaptive replanning to account for patient interfraction changes at each fraction, substantially improving RT delivery accuracy. However, one of the bottleneck problems in the clinical practice of MRI-guided online adaptive replanning with current technology is the impractically long time required to segment the patient's anatomy of the day, which can exceed 30 minutes by conventional manual segmentation in a tumor site with complex anatomy (eg, abdomen).3, 4, 5
In recent years, deep learning (DL) techniques, particularly the convolutional neural networks (CNNs), have been successfully applied to automatically segment organs from medical images including MRI.6, 7, 8, 9, 10, 11 Although it has been well documented that such auto-segmentations can be more efficient compared with the time-consuming and labor-intensive manual delineations, available MRI-based auto-segmentation methods still have limited success, particularly for complex structures such as those in the abdomen. Fu et al6 reported their DL auto-segmentation on MRI in the abdomen achieved average Dice similarity coefficients (DSCs) of 0.953, 0.931, 0.850, 0.866, and 0.655 for the liver, kidneys, stomach, bowels, and duodenum, respectively. Bobo et al7 described their CNN for the multiorgan segmentation on abdominal MRI, with DSCs of 0.556 and 0.691 in the stomach and pancreas. Chen et al8 and Amjad et al9 reported similar results from their DL auto-segmentation solutions. These previous studies indicate that the DL auto-segmentation on MRI for complex organs (eg, bowels) is generally unacceptable for clinical use. Although improvements for the auto-segmentation of complex structures can be anticipated with continually developing robust DL algorithms and/or larger training data sets, they cannot be guaranteed. The unacceptable auto-segmented contours must be examined and edited manually before their clinical use.
Manual contour editing is generally labor-intensive and time-consuming and can be subjective, inevitably introducing inter- and intra-observer errors. A desirable solution to reduce or even replace the manual process is to automate the contour editing. Several techniques have been proposed for automatic contour refinement (ACR). One of these techniques is the dense conditional random field (CRF) model,12 which has been used as a postprocessing strategy after the CNN auto-segmentation.13,14 By formulating the final segmentation results using the soft label probability maps computed from the CNN as a maximum a posteriori inference problem, this method is capable of incorporating the contextual information coming from both local and global relationships between the image voxels.13 However, the dense CRF model might not be suitable for correcting the very inaccurate contours that need substantial editing. Christ et al13 applied the dense CRF model after their CNN auto-segmentation for the liver on CT and found that the DSC improved from 0.931 to 0.943. Kamnitsas et al14 also implemented dense CRF for final postprocessing of their CNN models for brain lesion segmentation on MRI, with a minimal (less than 0.01) increase in DSC. Although these studies demonstrated that the dense CRF model was very useful to smooth contour boundaries between different structures and removed small isolated wrong contours from the CNN predictions, the slight improvements measured by the DSC failed to sufficiently correct for the inaccurate contours of the complex abdominal organs. Another ACR technique is active contour models (ACMs),15,16 which include edge-based ACMs and region-based ACMs. The edge-based ACMs use the image gradient to identify the boundaries, whereas the region-based ACMs use the image statistical information inside and outside the contour to guide the evolution.15 In general, the region-based ACMs show several advantages over the edge-based ACMs because they are less sensitive to image noise and are more able to identify weak boundaries.15 The region-based ACMs can be implemented by the level set method, which provides more flexibility and convenience, and have been used in a variety of image segmentation tasks in combination with DL.16 However, the segmentation accuracy of the ACM methods can strongly rely on the parameters used in the models, which are usually chosen empirically. For the complex organs in the abdomen, especially the bowels with large size and shape variations, it is impractical to determine the appropriate parameters by manual adjustment for each specific bowel loop using the conventional methods.
To minimize or eliminate manual contour editing, and thus to accelerate the segmentation process for MRI-guided ART, this study aimed to develop an ACR process based on an ACM method that does not require manual parameter adjustment to quickly refine unacceptable DL auto-segmented contours of the complex abdominal organs, including the small and large bowels, pancreas, duodenum, and stomach. Clinical MRI data were used to test the ACM, and the performance of the ACR process was evaluated based on clinical criteria.
Methods and Materials
This study was approved by the Institutional Review Board of Medical College of Wisconsin.
MRI data set
A total of 80 abdominal MRI scans acquired during routine RT for 71 patients with abdominal cancers were used for this study, including 65 scans from RT simulation (MR-SIM) and 15 from MRI-guided ART with an MR-Linac (MRL). The MR-SIM images were acquired on a 3T scanner (Verio, Siemens, Germany) using an axial T2-weighted half-Fourier singleshot turbo spin-echo (HASTE) sequence17 with the following parameters: repetition time (TR) = 2000 msec; echo time (TE) = 95∼98 msec; flip angle = 150°∼160°; matrix size = 320 × 212 ∼ 320 × 288; pixel size = 1.0625 × 1.0625 ∼ 1.3125 × 1.3125 mm2; and slice thickness = 3∼5 mm. The MRL images were the motion-average MRIs derived from 4-dimensional (4D) MRIs acquired on a 1.5T MR-Linac (Unity, Elekta Inc, Sweden) using either turbo field echo or balanced turbo field echo sequences.3 The acquisition parameters were as follows: TR = 4.30∼5.32 msec; TE = 1.85∼2.21 msec; flip angle = 25°∼50°; matrix size = 256 × 256 ∼ 352 × 352; pixel size = 1.5909 × 1.5909 ∼ 1.6406 × 1.6406 mm2; and slice thickness = 2.38∼2.5 mm.
All MRIs were first standardized using a preprocessing workflow developed based on MIM (MIM Software Inc, Cleveland, Ohio),18 including the following steps: (1) bias correction for magnetic field inhomogeneity using a nonparametric nonuniform intensity normalization algorithm19; (2) image denoising using the anisotropic diffusion filter20 to smooth out noise while preserving edge features; and (3) image intensity normalization to the range 0 to 255. For the ACR application described below, a contrast-limited adaptive histogram equalization method21 was applied to the MRIs to further enhance image contrast.
Auto-segmentation and data preparations
The obtained MRIs were then input into a DL auto-segmentation research tool (Admire, Elekta Inc), previously developed based on a 3D deep CNN architecture (a modified 3D-ResUNet)22,23 to generate auto-segmented contours of the small bowel, large bowel, combined bowels, pancreas, duodenum, and stomach. To assess contour accuracy, the auto-segmented contours were compared with the ground truth contours delineated manually by an experienced researcher and independently verified by 2 radiation oncologists.
Each slice of the MRI was cropped into multiple 2D subregions based on the dilated initial auto-segmented contours, which were generated by convoluting with a 2D square kernel of size 25 × 25. For the bowels, each cropped subregion on a slice included at least a complete loop of the initial contour; for the pancreas, duodenum, and stomach, only 1 cropped subregion was obtained on each slice. Based on the American Association of Physicists in Medicine Task Group 132 (TG-132) report,24 contours with a DSC <0.8 or a mean distance to agreement (MDA) >3 mm are considered inaccurate. Only the subregions with inaccurate contours were used as the initial contours to test the ACR process described in the next section. In addition to the initial contour, the ACM method described in the next section used the probability map (ie, the probability of each pixel belonging to the segmented organ) generated from the DL algorithm. Meanwhile, using a trial-and-error method, we tested whether adding a fudge factor to enhance low probability regions in the probability maps would improve the ACR performance, because those low-probability regions would be often associated with contour inaccuracy. Fudge factors ranging from 0.1 to 0.5 were tested. Based on the test results, we observed that the best performance of the ACR process occurred by increasing by 0.4 on the probability map of each voxel, with the maximum probability set to 1.0. This fudge factor of 0.4 was included as a part of the input data for the ACR.
Active contour model
An ACM method based on an improved level set25 was implemented in this work for ACR. This method was originated from the Chan-Vese model,26 a region-based level set algorithm in which the contour evolves by minimizing an energy functional. Because it is generally not practical to manually adjust various ACM parameters for a variety of complex inaccurate contours, we followed the approach proposed by Hatamizadeh et al25 to generalize the scalar parameters in the ACM to 2D parameter maps using the probability maps obtained from the DL models. In addition, we converted probability maps to signed distance maps to initialize the level set in the ACM. The contours to be corrected were then evolved iteratively to minimize the energy functional and better match to the desired boundary. For the ACR application, the number of iterations was set to be 600 because large changes were expected for the initial inaccurate contours. More details on the ACM method are provided in the Supplementary materials and the relevant publications.25, 26, 27
Automatic contour refinement
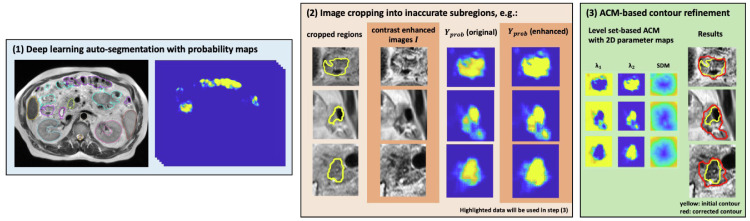
Figure 1 shows the workflow of the proposed ACR process based on the ACM algorithm. The process includes the following 3 steps: (1) inputting the test image, the DL auto-segmented contours, and the DL probability map; (2) preprocessing the input data by cropping each image slice into multiple 2D subregions and enhancing the probability map with the determined fudge factor; and (3) executing the ACM to correct or refine the contours and postprocessing the obtained contours if any obvious geometric inaccuracy is detected, eg, contour smoothing of imperfect contours, removal of isolated regions with low probability, and filling of empty holes.
Figure 1.
The 3-step workflow of the proposed automatic contour refinement process. Abbreviations: ACR = automatic contour refinement; SDM = signed distance map.
The performance of the ACR process was evaluated based on the DSC, MDA, and 2 recently introduced metrics, surface DSC (sDSC)28 and added path length (APL).29 The sDSC measures the surface overlap of 2 contours instead of the volumetric overlap as measured by DSC. The APL calculates the surface length of the ground truth contour that is not captured by the initial (auto-segmented) contour, which is the distance that the cursor needs to travel when correcting the inaccurate contour. Compared with traditional metrics (eg, DSC, MDA), the sDSC and APL have been shown to be more clinically relevant because they are better correlated with the contour editing time.29 The values of the DSC, MDA, sDSC, and APL were calculated for the initial and ACR (corrected) contours with respect to the ground truth contours. Because a distance difference of 2 mm between the ground truth contour and the initial or ACR contour is generally considered to be practically acceptable, a 2-mm tolerance was assumed in the calculation of the sDSC and APL. To measure the performance of the ACR at the different levels of contour inaccuracy, the inaccurate subregions were further divided into 2 groups, a major error group with subregions of initial DSC <0.5 or MDA >8 mm and a minor error group with remaining subregions of 0.5 ≤ DSC < 0.8 or 3 mm < MDA ≤ 8 mm. To assess whether the improvements from the ACR were statistically significant, changes of the 4 accuracy metrics for the contours obtained before and after the ACR were analyzed using the paired t test.
Results
The execution time to correct each inaccurate subregion with the developed ACR was less than 2 seconds on an NVIDIA GTX 1060 GPU. Table 1 compares the DSC, MDA, sDSC, and APL values obtained before and after the ACR for both the major and the minor error groups of the small bowel, large bowel, combined bowels, pancreas, duodenum, and stomach, along with the P values from the paired t test. The accuracies of the refined contours in the major error group were significantly improved for all organs (P < .001), with the best improvement for the combined bowels and pancreas. For the minor error group, the improvements as measured by the APL were significant for all the organs (P < .001 for decreases of the APL). However, these improvements as measured by other metrics (DSC, MDA, and sDSC) were mixed. A portion (3%-39%) of the corrected contours become practically acceptable after the ACR process per the TG-132 recommendation (DSC ≥ 0.8 and MDA ≤ 3 mm). The best correction performance was seen for the combined bowels: (1) for the major error group, the mean DSC increased from 0.34 to 0.59, the mean MDA decreased from 7.02 mm to 5.23 mm, and more notably, the APL was reduced by almost 20 mm and the improvement was observed in 84% of the subregions; (2) for the minor error group, the mean DSC increased from 0.72 to 0.79, the mean MDA decreased from 3.35 mm to 3.29 mm, and more than one-third (39%) of the corrected contours became clinically acceptable after the ACR. Among all the structures, the largest improvement was seen in the major error group of the pancreas, where the APL was reduced by approximately one-third after the ACR. For the duodenum and stomach, although the mean DSC, MDA, and sDSC were slightly changed after the ACR, particularly for contours with minor errors, the APLs decreased. The decreases in APLs for all the contours in both major and minor error groups indicate that the use of the ACR reduces the manual editing times required to correct for all the unacceptable contours of all the organs.
Table 1.
Performance of the automatic contour refinement process
| Organs | Error groups of initial DL auto-segmentation based on DSC and MDA | Percentage of subregions with improved DSC and MDA after ACR | Percentage of subregions with DSC ≥0.8 and MDA ≤3 mm after ACR | Mean DSC change after ACR | Mean MDA change (mm) after ACR | Mean sDSC change after ACR | Mean APL change (mm) after ACR |
|---|---|---|---|---|---|---|---|
| Small bowel | Major errors | 956/1218 (78%) | 104/1218 (9%) | 0.33 → 0.55* | 8.10 → 6.42* | 0.28 → 0.42* | 101.75 → 83.89* |
| Minor errors | 1116/2150 (52%) | 534/2150 (25%) | 0.70 → 0.75* | 3.92 → 3.98 (P = .049)† |
0.54 → 0.57* | 98.10 → 90.72* | |
| Large bowel | Major errors | 654/898 (73%) | 76/898 (8%) | 0.34 → 0.54* | 9.13 → 7.85* | 0.29 → 0.43* | 81.68 → 65.26* |
| Minor errors | 955/1689 (57%) | 500/1689 (30%) | 0.72 → 0.76* | 3.94 → 4.03 (P = .056)† |
0.56 → 0.60* | 70.25 → 60.04* | |
| Combined bowels | Major errors | 753/900 (84%)‡ | 105/900 (12%)‡ | 0.34 → 0.59* | 7.02 → 5.23* | 0.31 → 0.49* | 96.64 → 76.86* |
| Minor errors | 1706/2925 (58%)‡ | 1144/2925 (39%)‡ | 0.72 → 0.79* | 3.35 → 3.29 (P = .047) |
0.58 → 0.64* | 100.72 → 94.83* | |
| Pancreas | Major errors | 160/204 (78%) | 23/204 (11%) | 0.32 → 0.55* | 6.87 → 4.70* | 0.24 → 0.44* | 76.51 → 51.27* |
| Minor errors | 416/767 (54%) | 216/767 (28%) | 0.70 → 0.73* | 3.55 → 3.51 (P = .550) |
0.47 → 0.52* | 65.40 → 50.82* | |
| Duodenum | Major errors | 184/302 (61%) | 9/302 (3%) | 0.33 → 0.49* | 6.71 → 6.08* | 0.27 → 0.37* | 66.58 → 52.37* |
| Minor errors | 295/808 (37%) | 143/808 (18%) | 0.69 → 0.69 (P = .397)† |
3.43 → 4.13*,† | 0.53 → 0.51*,† | 51.31 → 44.77* | |
| Stomach | Major errors | 54/111 (49%) | 6/111 (5%) | 0.44 → 0.56* | 9.97 → 9.78 (P = .546) |
0.27 → 0.33 (P = .001) |
84.97 → 72.89* |
| Minor errors | 234/601 (39%) | 99/601 (16%) | 0.78 → 0.77 (P = .558)† |
4.21 → 4.90*,† | 0.46 → 0.45 (P = .032)† |
96.71 → 90.40* |
Abbreviations: ACR = automatic contour refinement; APL = added path length; DL, deep learning; DSC = Dice similarity coefficient; MDA = mean distance to agreement; sDSC = surface Dice similarity coefficient.
Indicates a paired t test P value <.001.
Metrics changes had minimal or no improvement.
The best percentage numbers achieved in combined bowels.
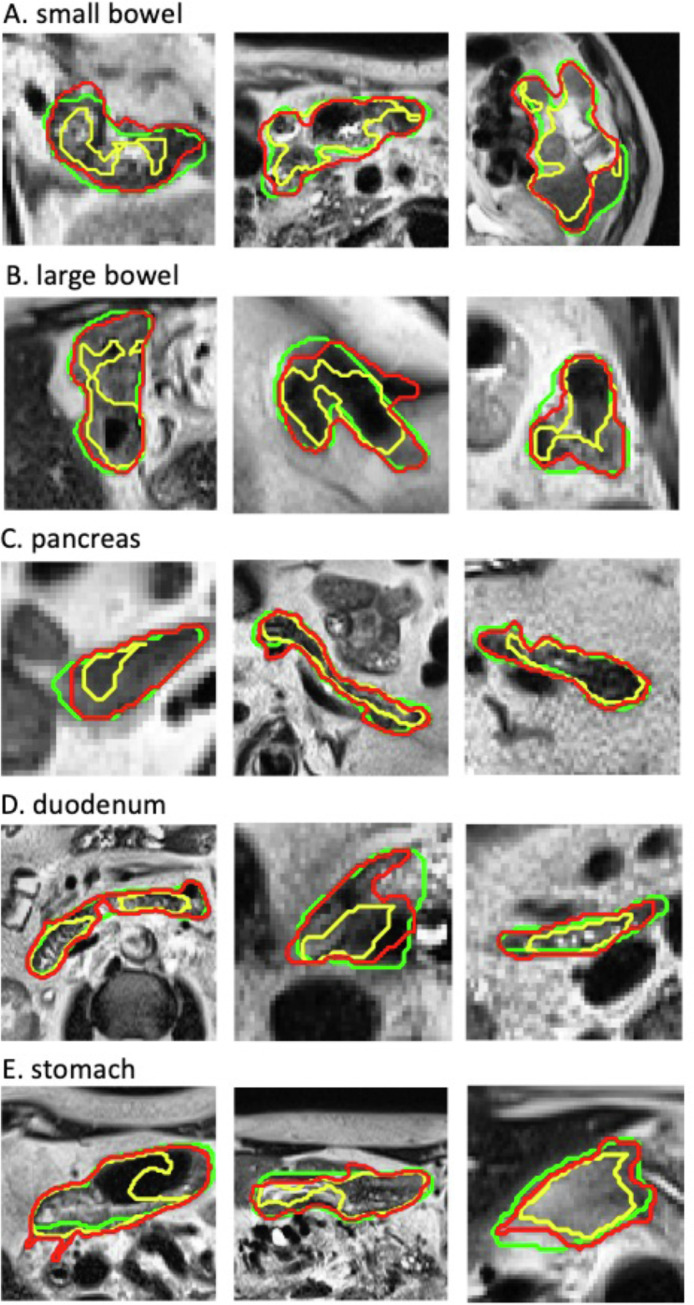
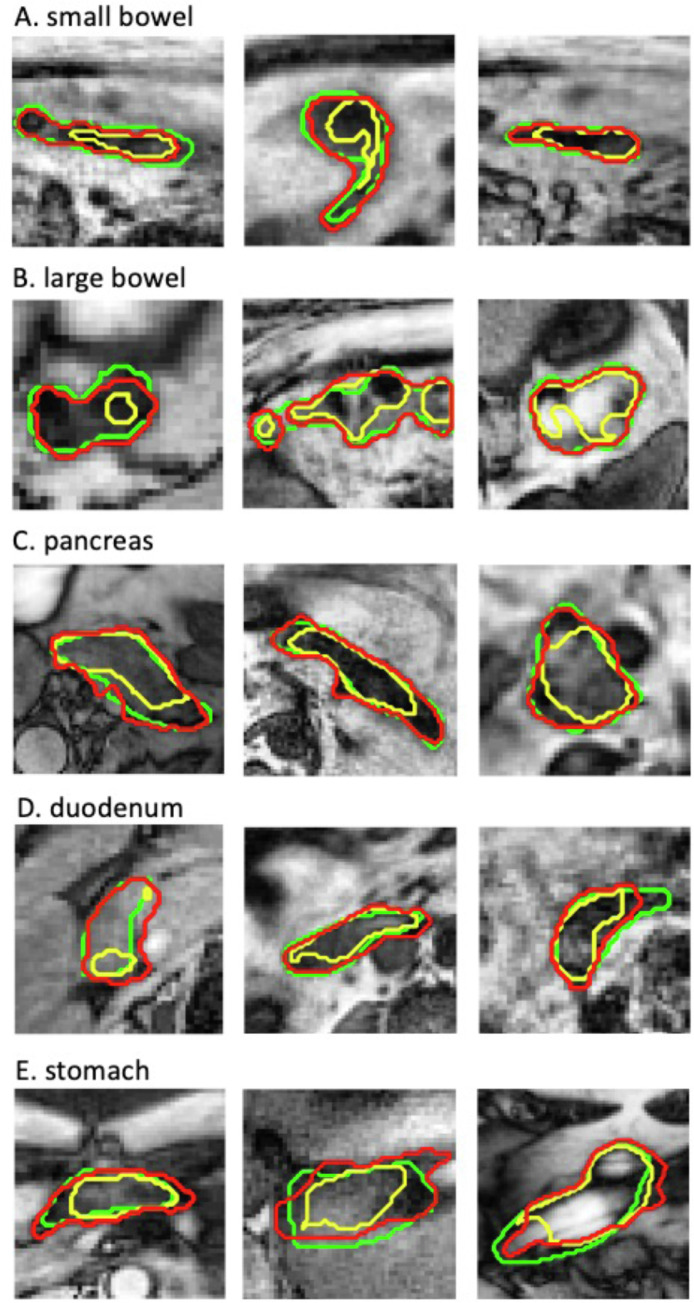
To demonstrate the contour quality improvements with the ACR, the scatter plots of the 4 contour accuracy metrics for each organ and each error group obtained before and after the ACR process are shown in Figure 2. For most contours, except for the duodenum and stomach contours with minor errors, the contour accuracy was improved after the ACR; as shown in the figure, after the ACR, more data points shifted toward the origin of each plot (eg, a higher DSC or sDSC and a lower MDA or APL). Figures 3 and 4 compare the contours of the initial DL auto-segmentation, ACR, and ground truth for representative MR-SIM and MRL cases, respectively. As is shown, the ACR process improved contour accuracy even for the very irregularly shaped contours of the complex organs.
Figure 2.
Comparisons of various contour accuracy metrics (DSC vs MDA, DSC vs sDSC, and DSC vs APL) for the A, small bowel, B, large bowel, C, combined bowels, D, pancreas, E, duodenum, and F, stomach before and after the ACR process. Note that the axes of the DSC and sDSC were reversed so contours with higher quality would shift toward the origin for all plots. Abbreviations: ACR = automatic contour refinement; APL = added path length; DSC = Dice similarity coefficient; MDA = mean distance to agreement; sDSC = surface Dice similarity coefficient.
Figure 3.
Comparisons of representative initial DL auto-segmentation (yellow), ACR (red), and ground truth (green) contours for the small bowel, large bowel, pancreas, duodenum, and stomach on MR-SIM data. Images shown are rescaled for better display (they do not reflect their original sizes). Abbreviations: ACR = automatic contour refinement; DL = deep learning; MR-SIM, magnetic resonance scans from radiation therapy simulation.
Figure 4.
Comparisons of representative initial DL auto-segmentation (yellow), ACR (red), and ground truth (green) contours for the small bowel, large bowel, pancreas, duodenum, and stomach on MRL data. Images shown are rescaled for better display (they do not reflect their original sizes). Abbreviations: ACR = automatic contour refinement; DL = deep learning; MRL = magnetic resonance imaging and linear accelerator.
Additional details on the performance of the ACR for MR-SIM images and MRL images, respectively, are provided in Supplementary Tables S1 and S2. In general, the performance of the ACR for the MR-SIM scans was better than that for the MRL scans. Comparisons of the accuracy metrics for these 2 data sets before and after the ACR are shown in Supplementary Figures S1 and S2.
Discussion
In this study, an automatic contour refinement process was developed based on an improved ACM algorithm to quickly correct for inaccurate contours generated by DL auto-segmentation. The effectiveness of the ACR process was demonstrated for complex abdominal structures including the bowels, pancreas, duodenum, and stomach. The ACR process can be implemented as a step after DL auto-segmentation to minimize subsequent manual editing effort, substantially accelerating the recontouring during MRI-guided ART, particularly for tumor sites with complex anatomy (eg, the abdomen).
Although DL-based segmentation methods have enabled the organ auto-contouring and achieved great success in many clinical applications, the current auto-segmented contours of challenging organs, such as the bowels, can still be clinically unacceptable.6, 7, 8, 9 Inevitably, manual editing needs to be performed subsequently to make the contours acceptable. The manual editing is generally time-consuming and labor-intensive, especially for inaccurate contours with irregular shapes (eg, complex abdominal organs). The presented ACR process can efficiently improve contour accuracy by reducing contour errors (eg, converting contours with major errors to those with minor errors) or even making a portion of the inaccurate auto-segmented contours practically acceptable. Thus, the adoption of the ACR will reduce the workload for manual editing, which is clearly indicated by the reduction of the APL. For the contours with major errors, average improvements of more than 0.2 increases in the DSC and 20-mm reductions in the APL after the ACR were seen for bowels and pancreas. For the contours with minor errors, more than 25% of the unacceptable bowel and pancreas contours became acceptable after the ACR. Although there were no obvious improvements for the duodenum and stomach contours if measured by other metrics, their APLs were reduced after the ACR, as seen in Table 1.
The ACR workflow was fast, with the execution time less than 2 seconds for each subregion and less than 4 minutes for each MRI set. By decreasing the number of iterations from 600 to 400 during the ACR execution, the processing time for each MRI set can be reduced to less than 2 minutes, with minimal effect on the ACR performance (eg, the average DSC and sDSC decreased <0.01, the average MDA increased <0.1 mm, and the average APL increased <1 mm). The time may be further shortened by applying smaller iteration numbers.
The ACM method that uses probability maps to define the per-pixel parameters and to initialize the contour evolution eliminates the need of the manual adjustment for the parameters, which is one of the major limitations with the traditional ACM methods.16 For our purpose of correcting a large variety of contour inaccuracies for the complex abdominal organs, it would be impractical to determine and adjust ACM parameters for each specific case during the correction. Instead of fixing scalar parameters, the ACM method implemented in the ACR process establishes 2D parameter maps using the probability maps produced from the CNN models, which provide clues for the contours to be adjusted based on the actual organ boundaries. Therefore, the performance of the ACR depends on the quality of the probability maps from the DL auto-segmentation. This may explain why a better ACR performance was achieved for the combined bowels compared with the small bowel and large bowel separately, because the adopted CNN models had some difficulty in differentiating the small and large bowels.30,31
The ACR performed better for the MR-SIM scans compared with the MRL scans. This is primarily owed to the motion artifacts in the MRL sets, which were motion average images derived from 4D MRIs.3 In contrast, the MR-SIM sets were acquired with a respiration trigger. In addition, the quality of the probability maps for the MRL sets was poorer compared with those for the MR-SIM sets, because a smaller number of MRL sets was used in the training of the DL auto-segmentation model. The poor probability maps affected the ACR performance for the MRL sets. Nonetheless, the ACR still achieved very promising results, particularly for the contours with major errors (as shown in Table S2).
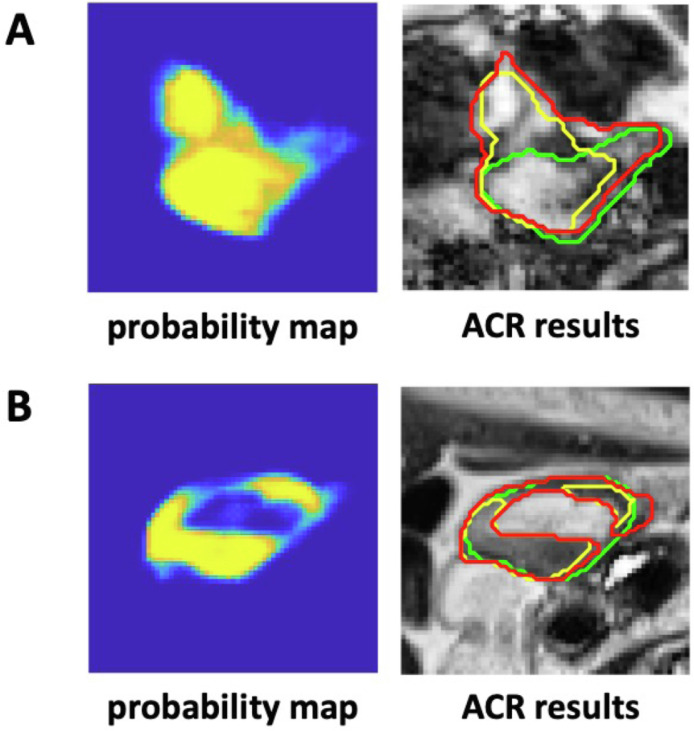
There are 2 major limitations in this study. First, the achievable accuracy by the ACR process is limited by the accuracy of the probability maps from the DL auto-segmentation. For example, if a probability map mislabels a background region as a part of an organ or wrongly includes a region of another organ, the ACR process would unlikely be able to correct for the contour. As an example, Figure 5A shows that the probability map inaccurately labeled some background pixels as small bowel (the values of these pixels on the probability map was equal or close to 1); the contour evolution would still be initialized and performed based on these incorrect regions, making it hard to correct these mistaken pixels. This could be a problem in a region with complicated anatomy (eg, the bowels and duodenum) where it is difficult to distinguish the organ from its surrounding background. Second, the ACR process relies on a region-based ACM, which has an intrinsic limitation in handling contours with inhomogeneous intensities. Figure 5B illustrates an example of a stomach contour where the ACR method failed to encompass the bright region, even though the probability map gave some hints. These 2 limitations explain why the ACR method was ineffective in correcting some inaccurate contours of complex organs.
Figure 5.
Comparison of the sample probability maps and the contours of the initial DL auto-segmentation (yellow), ACR (red), and ground truth (green) of the small bowel (A) and stomach (B), showing the limitations of the ACR process. Images shown are rescaled for better display (they do not reflect their original sizes). Abbreviations: ACR = automatic contour refinement; DL = deep learning.
Clearly, more robust methods for automatic contour refinement are needed. It is anticipated that as more advanced DL auto-segmentation algorithms and/or larger training data sets become available, the accuracy of the auto-segmentation will be continually increased. Demand for correcting the auto-segmented contours may be primarily for complex structures such as abdominal organs. The presently developed ACR process may still be applicable.
This study on ACR is a part of our effort to develop a 4-step segmentation pipeline for MRI-guided ART, including (1) auto-segmentation of MRI based on DL22; (2) auto-check of the obtained auto-segmented contours to detect their inaccuracies32; (3) auto-refinement of the detected inaccurate contours; and (4) manual editing using robust tools for the uncorrectable contours. Such a segmentation pipeline would effectively address the current slowness in the recontouring process, making MRI-guided daily online adaptive replanning more practical.
Conclusion
This work demonstrates the feasibility of using an improved ACM method to automatically refine inaccurate contours from the DL auto-segmentation of the complex abdominal organs, including the bowels, pancreas, duodenum, and stomach. This automatic contour refinement process is fast and efficient without the need for manual parameter adjustment. The developed ACR method can be integrated into the recontouring process to improve segmentation accuracy, minimize the subsequent tedious manual editing, and accelerate the execution of MRI-guided ART.
Acknowledgments
Input from Ergun Ahunbay, PhD, Haidy Nasief, PhD, William Hall, MD, Beth Erickson, MD, and Virgil Willcut, MS, is appreciated.
Footnotes
Sources of support: This research was partially supported by the Medical College of Wisconsin (MCW) Cancer Center and Froedtert Hospital Foundation, the MCW Meinerz and Fotsch Foundations, and the National Cancer Institute of the National Institutes of Health under award number R01CA247960. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures: The Medical College of Wisconsin received institutional research support from Elekta AB. Dr Xu and Mr Thill are employees of Elekta AB. All other authors have no disclosures to declare.
Research data are stored in an institutional repository and will be shared upon request to the corresponding author.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.adro.2022.100968.
Appendix. Supplementary materials
References
- 1.Mutic S, Dempsey JF. The ViewRay system: Magnetic resonance–guided and controlled radiotherapy. Semin Radiat Oncol. 2014:196–199. doi: 10.1016/j.semradonc.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Lagendijk JJ, Raaymakers BW, Van Vulpen M. The magnetic resonance imaging–linac system. Semin Radiat Oncol. 2014:207–209. doi: 10.1016/j.semradonc.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Paulson ES, Ahunbay E, Chen X, et al. 4D-MRI driven MR-guided online adaptive radiotherapy for abdominal stereotactic body radiation therapy on a high field MR-Linac: Implementation and initial clinical experience. Clinical and translational radiation oncology. 2020;23:72–79. doi: 10.1016/j.ctro.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamb J, Cao M, Kishan A, et al. Online adaptive radiation therapy: Implementation of a new process of care. Cureus. 2017;9:e1618. doi: 10.7759/cureus.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Güngör G, Serbez İ, Temur B, et al. Time analysis of online adaptive magnetic resonance–guided radiation therapy workflow according to anatomical sites. Pract Radiat Oncol. 2021;11:e11–e21. doi: 10.1016/j.prro.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Mazur TR, Wu X, et al. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiation therapy. Med Phys. 2018;45:5129–5137. doi: 10.1002/mp.13221. [DOI] [PubMed] [Google Scholar]
- 7.Bobo MF, Bao S, Huo Y, et al. Fully convolutional neural networks improve abdominal organ segmentation. Proc SPIE Int Soc Opt Eng. 2018;10574 doi: 10.1117/12.2293751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Ruan D, Xiao J, et al. Fully automated multi-organ segmentation in abdominal magnetic resonance imaging with deep neural networks. Med Phys. 2020;47:4971. doi: 10.1002/mp.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amjad A, Xu J, Thill D, et al. Deep learning-based auto-segmentation on CT and MRI for abdominal structures. Int J Radiat Oncol Biol Phys. 2020;108:S100–S101. [Google Scholar]
- 10.Savenije MH, Maspero M, Sikkes GG, et al. Clinical implementation of MRI-based organs-at-risk auto-segmentation with convolutional networks for prostate radiation therapy. Radiat Oncol. 2020;15:1–12. doi: 10.1186/s13014-020-01528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elguindi S, Zelefsky MJ, Jiang J, et al. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiation therapy. Phys Imaging Radiat Oncol. 2019;12:80–86. doi: 10.1016/j.phro.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krähenbühl P, Koltun V. Efficient inference in fully connected crfs with gaussian edge potentials. Adv Neural Inf Process Syst. 2011;24:109–117. [Google Scholar]
- 13.Christ PF, Elshaer MEA, Ettlinger F, et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. Med Image Comput Comput Assist Interv. 2016:415–423. [Google Scholar]
- 14.Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhang K, Zhang L, Song H, Zhou W. Active contours with selective local or global segmentation: A new formulation and level set method. Image and Vis Comput. 2010;28:668–676. [Google Scholar]
- 16.Hoang Ngan Le T, Luu K, Duong CN, et al. Active contour model in deep learning era: A revise and review. Applications of Hybrid Metaheuristic Algorithms for Image Processing. 2020:231–260. [Google Scholar]
- 17.Semelka RC, Kelekis NL, Thomasson D, Brown MA, Laub GA. HASTE MR imaging: Description of technique and preliminary results in the abdomen. J Magn Res Imaging. 1996;6:698–699. doi: 10.1002/jmri.1880060420. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Paulson E, Lim S, et al. A Patient-Specific Autosegmentation Strategy Using Multi-Input Deformable Image Registration for Magnetic Resonance Imaging–Guided Online Adaptive Radiation Therapy: A Feasibility Study. Advances in radiation oncology. 2020;5:1350–1358. doi: 10.1016/j.adro.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Machine Intelligence. 1990;12:629–639. [Google Scholar]
- 21.Zuiderveld K. Contrast limited adaptive histogram equalization. Graphics Gems. 1994:474–485. [Google Scholar]
- 22.Amjad A, Xu J, Thill D, et al. General and custom deep learning autosegmentation models for organs in head and neck, abdomen, and male pelvis. Medical Physics. 2022;49:1686–1700. doi: 10.1002/mp.15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Yang X, Chen H, Qin J, Heng PA. Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17) 2017:66–72. [Google Scholar]
- 24.Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiation therapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys. 2017;44:e43–e76. doi: 10.1002/mp.12256. [DOI] [PubMed] [Google Scholar]
- 25.Hatamizadeh A, Hoogi A, Sengupta D, et al. Deep active lesion segmentation. International Workshop on Machine Learning in Medical Imaging. 2019:98–105. [Google Scholar]
- 26.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Birdwell JD, Djouadi SM. Efficient implementation of the Chan-Vese models without solving PDEs. 2006 IEEE Workshop on Multimedia Signal Processing. 2006:350–354. [Google Scholar]
- 28.Nikolov S, Blackwell S, Zverovitch A, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiation therapy. arXiv preprint arXiv:1809.04430.2018.
- 29.Vaassen F, Hazelaar C, Vaniqui A, et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiation therapy. Phys Imaging Radiat Oncol. 2020;13:1–6. doi: 10.1016/j.phro.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amjad A, Xu J, Thill D, et al. Improving Deep Learning Auto-Segmentation Using an Adaptive Spatial Resolution Approach. MEDICAL PHYSICS. 482021.
- 31.Amjad A, Xu J, Zhu X, et al. Deep Learning Auto-Segmentation on Multi-Sequence MRI for MR-Guided Adaptive Radiation Therapy. American Society for Radiation Oncology (ASTRO) Annual Meeting. 2021 [Google Scholar]
- 32.Zhang Y, Plautz TE, Hao Y, Kinchen C, Li XA. Texture‐based, automatic contour validation for online adaptive replanning: a feasibility study on abdominal organs. Medical physics. 2019;46:4010–4020. doi: 10.1002/mp.13697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.