Del Rosario and Gabrielle et al. show that TMEM120A/TACAN inhibits mechanically activated PIEZO2 channels both in heterologous systems and in dorsal root ganglion neurons. TMEM120A does not inhibit PIEZO1 and TREK1 channel activity and its expression alone does not lead to appearance of mechanically activated currents.
Abstract
PIEZO2 channels mediate rapidly adapting mechanically activated currents in peripheral sensory neurons of the dorsal root ganglia (DRG), and they are indispensable for light touch and proprioception. Relatively little is known about what other proteins regulate PIEZO2 activity in a cellular context. TMEM120A (TACAN) was proposed to act as a high threshold mechanically activated ion channel in nociceptive DRG neurons. Here, we find that Tmem120a coexpression decreased the amplitudes of mechanically activated PIEZO2 currents and increased their threshold of activation. TMEM120A did not inhibit mechanically activated PIEZO1 and TREK1 channels and TMEM120A alone did not result in the appearance of mechanically activated currents above background. Tmem120a and Piezo2 expression in mouse DRG neurons overlapped, and siRNA-mediated knockdown of Tmem120a increased the amplitudes of rapidly adapting mechanically activated currents and decreased their thresholds to mechanical activation. Our data identify TMEM120A as a negative modulator of PIEZO2 channel activity, and do not support TMEM120A being a mechanically activated ion channel.
Introduction
PIEZO2 is a nonselective cation channel that is responsible for the rapidly adapting mechanically activated currents in dorsal root ganglia (DRG) neurons (Coste et al., 2010). PIEZO2 plays crucial roles in light touch and proprioception both in mice and in humans (Kefauver et al., 2020). In humans, loss of function mutations in PIEZO2 lead to impaired proprioception, loss of discriminative touch perception, as well as ataxia and muscular dystrophy (Chesler et al., 2016). In mice, combined deletion of Piezo2 in DRG neurons and in Merkel cells resulted in a profound loss of light touch (Ranade et al., 2014). Selective deletion of Piezo2 in proprioceptive neurons in mice recapitulated not only ataxia, but also much of the skeletal abnormalities observed in human loss of function patients (Assaraf et al., 2020).
In contrast to its well-established roles in light touch and proprioception, the role of PIEZO2 channels in detecting painful stimuli is less clear. Genetic deletion of Piezo2 in DRG neurons and Merkel cells resulted in no change in sensitivity to noxious mechanical stimuli and no change in mechanical allodynia in inflammation (Ranade et al., 2014). Deletion of Piezo2 in a different subset of DRG neurons, on the other hand, reduced sensitivity to noxious mechanical stimuli, as well as reduced inflammatory and neuropathic mechanical allodynia (Murthy et al., 2018b). In another study, deletion of Piezo2 in DRG neurons increased threshold to light touch, but paradoxically reduced the threshold to painful mechanical stimuli, indicating that activation of PIEZO2 channels may reduce pain (Zhang et al., 2019), in accordance with the gate control theory of pain (Melzack and Wall, 1965). These data highlight the potentially complex role of PIEZO2 in detecting noxious mechanical stimuli.
PIEZO2 is not the only mechanically activated channel in DRG neurons, and it is not the sole sensor responsible for all aspects of somatosensory touch and mechanical pain (Kefauver et al., 2020). Several novel putative mechanically activated channels have been identified in recent years, including Tentonin3 (TMEM150C; Hong et al., 2016), Elkin1 (TMEM87A; Patkunarajah et al., 2020), OSCAs (TMEM63; Murthy et al., 2018a), and TACAN (TMEM120A; Beaulieu-Laroche et al., 2020). The physiological roles of these novel putative mechanically activated channels are not very well understood.
Tentonin3 (TMEM150C) was proposed to be a component of slowly adapting mechanically activated currents in proprioceptive DRG neurons, and its genetic deletion resulted in abnormal gait and loss of motor coordination (Hong et al., 2016). Later its function as a mechanically activated channel was debated (Dubin et al., 2017; Hong et al., 2017; Parpaite et al., 2021) and it was proposed that Tentonin3 acts as a modulator of PIEZO2, PIEZO1, and TREK1 channel activity (Anderson et al., 2018).
TMEM120A was also proposed to be responsible for the slowly adapting mechanically activated currents in small-to-medium nonpeptidergic nociceptive DRG neurons (Beaulieu-Laroche et al., 2020). Cells transfected with Tmem120a displayed currents in response to negative pressure in the cell-attached mode that were above those in control cells, but not when stimulated by indentation with a glass probe in whole-cell patch-clamp experiments (Beaulieu-Laroche et al., 2020). Inducible genetic deletion of Tmem120a in nonpeptidergic Mrgprd positive DRG neurons in mice resulted in reduced behavioral responses to painful mechanical stimuli. siRNA-mediated knockdown of Tmem120a reduced the proportion of ultra-slowly-adapting mechanically activated (MA) currents in Trpv1 expressing DRG neurons, but it was not tested whether their proportion also decreases in neurons with conditional deletion of Tmem120a (Beaulieu-Laroche et al., 2020). Tmem120a knockdown in DRG neurons by intrathecal administration of antisense oligodeoxynucleotides in rats reduced mechanical hyperalgesia induced by intradermal administration of various proinflammatory compounds, but had no effect on mechanical hyperalgesia in chemotherapy-induced neuropathic pain (Bonet et al., 2020).
Recently, the cryo-EM structure of TMEM120A was reported by several groups (Niu et al., 2021; Rong et al., 2021; Xue et al., 2021). All three groups found that the protein forms a dimer, and located a coenzyme-A molecule in the structure. The protein showed structural homology to the ELOVL family of lipid modifying enzymes (Nie et al., 2021), indicating that TMEM120A may function as a lipid modifying enzyme. Furthermore, all three publications found that expression of Tmem120a did not induce the appearance of mechanically activated currents, in contrast to the findings of Beaulieu-Laroche et al. (2020). A fourth group also determined the structure of TMEM120A, which did not contain coenzyme-A, and they found that expressing a mutant (M207A) of Tmem120a resulted in the appearance of small mechanically activated currents evoked by negative pressure in the cell-attached configuration (Chen et al., 2022).
Here, we examined whether TMEM120A can act as a modulator of PIEZO channels. We found that coexpression of Tmem120a with Piezo2 resulted in a robust decrease in the amplitudes of mechanically activated currents and an increase in their mechanical thresholds in whole-cell patch-clamp experiments compared to cells expressing Piezo2 alone. On the other hand, coexpressing Tmem120a with Piezo1 did not have a significant effect on mechanically induced currents either when the cells were stimulated with a blunt glass probe in the whole-cell configuration, or when negative pressure was used in cell-attached patches. Expressing Tmem120a alone did not induce mechanically activated currents above background levels observed in control cells. Tmem120a showed broad expression in most DRG neurons, including neurons expressing Piezo2. siRNA-mediated knockdown of Tmem120a did not decrease the proportion of slowly adapting currents, but it increased the amplitudes and decreased the thresholds of rapidly adapting mechanically activated currents in DRG neurons. Our work identifies TMEM120A as a negative modulator of PIEZO2 channels.
Materials and methods
Cell culture
Human Embryonic Kidney 293 (HEK293) cells were purchased from American Type Culture Collection (ATCC; catalogue number CRL-1573), RRID: CVCL_0045. Cell identity was verified by short tandem repeat analysis at ATCC. Additional cell authentication was not performed, but passage number of the cells was monitored, and cells were used up to passage number 25–30 from purchase, when a new batch of cells was thawed with low passage number. HEK293 cells were cultured in MEM (Gibco) with 10% FBS (Gemini) and 100 IU/ml penicillin plus 100 µg/ml streptomycin (Gemini) in 5% CO2 at 37°C.
Piezo1 deficient Neuro2A (N2A) cells, in which endogenous Piezo1 was deleted by CRISPR (Moroni et al., 2018; Romero et al., 2020), were cultured in Dulbecco’s Modified Eagle Medium (Gibco), 1% penicillin–streptomycin, and 10% FBS.
HEK293 cells were transfected with Effectene reagent (Qiagen) in 35-mm tissue culture dishes with 200 ng Piezo1 or Piezo2 and 200 ng Tmem120a or 50 ng tdTomato. N2A-Pz1-KO cells used for patch-clamp experiments in the whole-cell configuration were transfected using the same reagent with 500 ng Piezo2 and 500 ng Tmem120a-tdTomato or 125 ng tdTomato. N2A-Pz1-KO cells used for experiments in the cell-attached configuration were transfected with 1,500 ng Piezo1 or 1,500 ng TREK1 and 1,500 ng Tmem120a-Tom or 375 ng tdTomato (listed concentrations of Tmem120a-tdTomato and tdTomato cDNA were used for experiments when transfected by themselves). The day after transfection, the cells were split and plated on poly-Lysine–coated 12-mm glass cover slips for patch-clamp experiments, which were performed 48–72 h after transfection.
DRG neurons
DRG neurons were isolated as described previously (Borbiro et al., 2015; Del Rosario et al., 2020). All animal procedures were approved by the Institutional Animal Care and Use Committee at Rutgers New Jersey Medical School. Mice were kept in a barrier facility under a 12/12-h light/dark cycle. Wild-type C57BL6 mice (2–4 mo old) from either sex (The Jackson Laboratory) were anesthetized with i.p. injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) and perfused via the left ventricle with ice-cold Hank’s buffered salt solution (HBSS; Life Technologies). DRGs were harvested from all spinal segments after laminectomy and removal of the spinal column and maintained in ice-cold HBSS for the duration of the isolation. Isolated ganglia were cleaned from excess nerve tissue and incubated with type 1 collagenase (2 mg/ml; Worthington) and dispase (5 mg/ml; Sigma-Aldrich) in HBSS at 37°C for 30 min, followed by mechanical trituration. Digestive enzymes were then removed after centrifugation of the cells at 100 g for 5 min.
Isolated DRG neurons were resuspended in Nucleofector Solution (Mouse Neuron Nucleofactor Kit, VPG-1001; Lonza) and predesigned fluorescently labelled siRNA directly against mouse Tmem120a from Qiagen-FlexiTube (cat. S101311919) or AllStars Neg. siRNA (cat. 1027294) was added at a final concentration of 300 nM. The transfection reactions were transferred to aluminum cuvettes for Nucleofection via Nucleofector 2b device (Lonza). After Nucleofection, DRG neurons were seeded onto laminin-coated glass coverslips and cultured in serum containing Dulbecco’s Modified Eagle Medium, and fresh medium was added every 24 h. Whole-cell patch-clamp experiments were performed 48–72 h after transfection using neurons displaying siRNA-mediated fluorescence.
Whole-cell patch clamp
Whole-cell patch-clamp recordings were performed at room temperature (22–24°C) as described previously (Borbiro et al., 2015). Briefly, patch pipettes were prepared from borosilicate glass capillaries (Sutter Instrument) using a P-97 pipette puller (Sutter instrument) and had a resistance of 2–7 MΩ. After forming gigaohm-resistance seals, the whole-cell configuration was established, and the MA currents were measured at a holding voltage of −60 mV using an Axopatch 200B amplifier (Molecular Devices) and pClamp 10. Currents were filtered at 2 kHz using low-pass Bessel filter of the amplifier and digitized using a Digidata 1,440 unit (Molecular Devices). All measurements were performed with extracellular solution containing 137 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, and 10 mM glucose (pH adjusted to 7.4 with NaOH). The patch pipette solution contained 140 mM K+ gluconate, 1 mM MgCl2, 2 mM Na2ATP, 5 mM EGTA, and 10 mM HEPES (pH adjusted to 7.2 with KOH).
Mechanically activated currents were measured from isolated DRG neurons, transiently transfected HEK293 cells, and transiently transfected Piezo1 deficient N2A cells, as previously described (Borbiro et al., 2015). Briefly, mechanical stimulation was performed using a heat-polished glass pipette (tip diameter, about 3 µm), controlled by a piezoelectric crystal drive (Physik Instrumente) positioned at 60°C to the surface of the cover glass. The probe was positioned so that 10-µm movement did not visibly contact the cell but an 11.5-µm stimulus produced an observable membrane deflection. Measurements from cells that showed significant swelling after repetitive mechanical stimulation or had substantially increased leak current were discarded. The inactivation kinetics from MA currents were measured by fitting the MA current with an exponential decay function in the Clampfit software, which measured the inactivation time constant (tau). We used the currents evoked by the third stimulation after the threshold in most experiments, except in cells where only the two largest stimuli evoked a current, where we used the current evoked by the largest stimulus, provided it reached 40 pA.
For the DRG neuron experiments, to categorize mechanically activated currents as rapidly, intermediate, and slowly adapting, we used the criteria from our previous publications (Borbiro et al., 2015; Del Rosario et al., 2020). We considered currents to be rapidly adapting if they fully inactivated before end of the 200-ms mechanical stimulus. The inactivation time constants (tau) of these currents were 14.80 ± 3.17 ms (mean ± SD, range 8–22 ms), similar to that in our earlier work (Borbiro et al., 2015; Del Rosario et al., 2020). This inactivation time constant was very similar to those in HEK293 or Piezo1 deficient N2A cells transfected with Piezo2 (Fig. S1, F, L, and R), indicating that these currents were predominantly mediated by PIEZO2. Note that these inactivation time constants are longer than the originally reported <10 ms for PIEZO2 (Coste et al., 2010), but the inactivation time constants of both PIEZO1 and PIEZO2 show considerable variability between different research laboratories (Szczot et al., 2017; Wu et al., 2017; Romero et al., 2019).
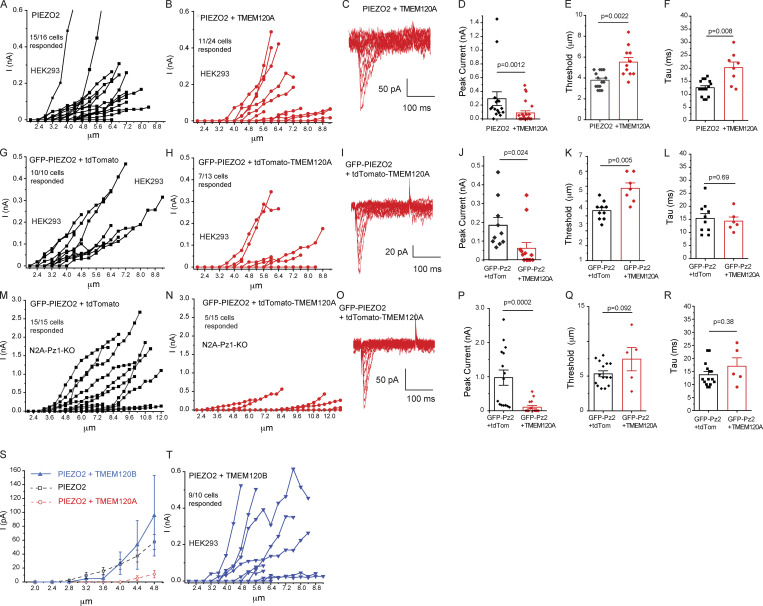
Figure S1.
TMEM120A but not TMEM120B inhibits PIEZO2 currents. Data from Fig. 1 showing the full range of measurements with indentation depth increased until seals were lost. (A and B) Data from HEK293 cells transfected with Piezo2 and Tmem120A. (C) Representative traces for increasing indentation depth till 7.2 μm. (D) Peak current amplitudes regardless of indentation depth. (E) Mechanical threshold of responding cells. (F) Inactivation time constant (tau). (G and H) Data from HEK293 cells transfected with GFP-Piezo2 and tdTomato-Tmem120A. (I) Representative traces for indentation depths till 8.4 μm. (J) Peak current amplitudes regardless of indentation depth. (K) Mechanical threshold of responding cells. (L) Inactivation time constant (tau). (M and N) Data from Piezo1 deficient N2A cells transfected with GFP-Piezo2 and tdTomato-Tmem120A. (O) Representative traces for indentation depths till 11.6 μm. (P) Peak current amplitudes regardless of indentation depth. (Q) Mechanical threshold of responding cells. (R) Inactivation time constant (tau). (S) Whole-cell patch-clamp data from HEK293 cells transfected with Piezo2 and Tmem120B, mean ± SEM of current amplitudes as a function of indentation depth. For PIEZO2 alone and PIEZO2 + TMEM120A data were replotted from Fig. 1 (dashed lines). (T) Data for full the range of indentations. Statistical significance was calculated with two-sample t test (F, K, L, and Q) or Mann-Whitney test (D, E, J, P, and R). Data are shown as mean ± SEM and scatter plots.
Intermediate adapting currents did not fully inactivate, but the leftover current at the end of the mechanical simulation was <50% of the peak current. The inactivation time constant for these currents was 36.35 ± 12.4 ms (range 22–55 ms). Slow adapting currents also did not fully inactivate, and the leftover current at the end of the mechanical simulation was >50% of the peak current, and/or the time constant was over 55 ms. The inactivation time constant for these currents was 88.14 ± 34.14 (range 58–151 ms).
Cell-attached patch clamp
Patch-clamp recordings in the cell-attached configuration were performed at room temperature (22–24°C) similar to that described previously (Lewis and Grandl, 2015). After forming gigaohm-resistance seals, MA currents were measured at a holding voltage of −80 mV for PIEZO1 currents and at 0 mV for TREK1 currents, using an Axopatch 200B amplifier (Molecular Devices) and pClamp 10. Currents were filtered at 2 kHz using low-pass Bessel filter of the amplifier and digitized using a Digidata 1,440 unit (Molecular Devices). Mechanical stimulation in cell-attached patches was performed using a high-speed pressure clamp (Besch et al., 2002; HSPC-1, ALA Scientific) controlled by pClamp 11.1 software (Molecular Devices), as described earlier (Borbiro et al., 2015).
All measurements were performed with a bath solution containing, 140 mM KCl, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose (pH adjusted to 7.4 with KOH) to bring the membrane potential of the cells close to zero. The patch pipette solution contained 130 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM HEPES, and 10 mM TEA-Cl (pH adjusted to 7.4 with NaOH).
TIRF microscopy
HEK293 cells were transiently transfected with cDNA encoding Tmem120a-tdTomato, or tdTomato, and GFP-Piezo1 or GFP-Piezo2. The next day, the transfected cells were plated on poly-L-lysine–coated 35-mm round coverslip (#1.5 thickness; Thermo Fisher Scientific). The cells were used for TIRF imaging 2 d after transfection. Cells plated on the coverslip were placed into a recording chamber filled with extracellular solution containing (in mM) 137 NaCl, 5 KCl, 1 MgCl2, 10 HEPES, and 10 glucose (pH 7.4). TIRF images were obtained at room temperature using a Nikon Eclipse Ti2 microscope. Fluorescence excitation was performed using a 15-mW solid state 488- and 561-nm laser at 90% of the maximal power through a CFI Apochromat TIRF 60X oil objective (NA of 1.49). Images were captured using an ORCA-Fusion Digital CMOS camera (Hamamatsu) through emission filters 525/50 and 600/50 nm for the green and red channel, respectively.
To visualize actin and tubulin cytoskeleton, the cells were labeled with either Sir-Actin (CY-SC001) or Spy650-tubulin (CY-SC503; Cytoskeleton, Inc.) according to the manufacturer’s instructions. Live cells were incubated for 1 h at 37°C with either Sir-Actin (2 μM) or Spy650-tubulin (500× dilution of the stock solution recommended by the manufacturer). The cells were washed with the extracellular solution before imaging to remove the excess dyes. For cytoskeletal disruption, cells were incubated in either 10 µM colchicine (C-9754; Millipore Sigma) for 1 h or 1 µM cytochalasin D (11330; Cayman Chemicals) for 24 h. To visualize the probes, the cells were illuminated by the 640-nm 15 mW solid state laser, and emission was collected using a 700/75-nm emission filter.
The images were analyzed using Nikon NIS-Elements AR Analysis software. Regions of interest (ROIs) were generated by hand drawing the outline of individual cells allowing for analysis over the total area of each cell in the TIRF angle. Mean fluorescence intensities within these single-cell ROIs were determined via Nikon NIS-Elements AR Analysis software. To correct for background fluorescent signal, a single rectangular ROI was generated on each image over an area absent of any cell or cell debris, and mean fluorescent intensity was calculated. Background mean intensity was averaged from each image per coverslip and subtracted from the mean intensity of the single-cell ROIs. The mean fluorescent intensities from each cell on a single coverslip were averaged (5–22 total cells/coverslip) and plotted. To determine colocalization, the same single-cell ROIs were used. NIS-Elements AR Analysis software generated Pearson’s correlation coefficients between designated fluorescent channels as a measure of colocalization. The Pearson’s correlation coefficients from each cell on a single coverslip was averaged and plotted. GFP-PIEZO2 and GFP-PIEZO1 fluorescent puncta were identified using the “Spot Detector” feature of NIS-Elements AR Analysis software for each cell. The number of puncta detected was normalized by their respective cell’s area. Values from each cell on individual coverslips were averaged and plotted.
Confocal microscopy
Measurements were conducted with an Olympus FluoView-1000 confocal microscope in the frame scan mode using a 60X water immersion objective at room temperature. Green fluorescence was measured using excitation wavelength of 473 nm; emission was detected through a 515/50-nm band-pass filter. Red fluorescence was measured using excitation wavelength of 559 nm; emission was detected through a 585/50-nm band-pass filter. Image analysis was performed using Olympus FluoView-1000 and ImageJ.
Western blotting
Piezo1 deficient N2A cells (106) were transfected with siRNA against mouse Tmem120a from Qiagen-FlexiTube (cat. S101311919) or AllStars Neg. siRNA (cat. 1027294) at a final concentration of 300 nM. Nucleofection was used as previously described. The cells were plated on 35-mm tissue culture dishes and harvested 48 h after transfection via scraping in Cell-Lytic-M lysis buffer (Cat. No.: C2978; MilliporeSigma) containing Halt Protease Inhibitor Cocktail (Cat. No.: 78438; Thermo Fisher Scientific). Protein samples were denatured in Laemmli sample buffer (Cat. No.: 161-0747; BioRad) and 2-mercaptoethanol for 1 h in a 37°C water bath. Samples were run on BioRad 4–15% mini-protean gels (Cat. No.: 4561084). Semidry transfers were performed using BioRad Trans-blot Turbo Transfer System onto nitrocellulose membranes (Cat. No.: 1704158; BioRad). The membranes were blocked for 1 h at room temperature in Tris-buffered saline with 0.1% Tween-20 (TBS-T) with 5% milk and incubated with 1:500 rabbit anti-TMEM120A antibodies (Cat. No.: MBS3223965; MyBioSource) in TBS-T with 5% milk overnight at 4°C. The membranes were probed with 1:10,000 goat anti-rabbit HRP antibodies (Cat. No.: G21234; Invitrogen) in TBS-T with 5% milk for 1 h at room temperature. Super Signal West Femto Max Sensitivity substrate (Cat. No.: 34095; Thermo Fisher Scientific) was used to develop the membranes and they were imaged with a FlourChem-8800 imager. The membranes were stripped using Restore Stripping buffer (Cat. No.: 46430; Thermo Fisher Scientific) then reblocked and probed with 1:5,000 rabbit anti-βTubulin antibodies (Cat. No.: NB600-936; Novus) in TBS-T with 5% milk overnight at 4°C. The membranes were developed and imaged as previously described. Images were analyzed using ImageJ.
RNAScope in situ hybridization
DRG isolation was performed as described before with transcardial perfusion under deep anesthesia first with 10 ml of HBSS and after with 10 ml of 4% formaldehyde. L3–L5 DRGs were collected from mice; the tissues were postfixed for 1 h in 4% formaldehyde and dehydrated in a series of gradient sucrose concentration (10, 20, and 30%). After freezing in O.C.T. compound block (Sakura Finetech), DRGs were sectioned into 12-µm-thick slices, and RNAScope assay was then carried out as previously described (Su et al., 2020).
Simultaneous detection of mouse RNA transcripts for Piezo2, Tmem120a, and various neuronal markers was performed on fixed, frozen DRG sections using Advanced Cell Diagnostics (ACD) RNAScope Multiplex Fluorescent Reagent Kit v2 (cat number: 323110) according to the manufacturer’s instructions, and commercially available probes for Mm-Tmem120a (cat number: 513211), Mm-Piezo2 (cat number: 400191-C2), Mm-Trpv1 (cat number: 313331-C3), Mm-Th (cat number: 317621-C3), Mm-Nefh (cat number: 443671-C3), and Calcb (CGRP2; cat number: 425511-C3) were purchased from ACD.
In short, the sections were postfixed in prechilled 4% paraformaldehyde in PBS for 15 min at 4°C, washed three times with PBS for 5 min each before dehydration through 50, 70, and 100 and 100% ethanol for 5 min each. We then treated slides with a protease 4 for 20 min and washed in distilled water. Probe hybridization and signal amplification were performed according to manufacturer’s instructions. The following fluorophores were used to detect corresponding RNAScope probes: Opal 520, 570, and 690 reagent kits (Akoya Biosciences). Cells were stained with DAPI (ACD) and mounted on the slide with Gold Antifade Mountant. Slides were imaged on a Nikon A1R confocal microscope with a 20× Plan Apo air objective, NA 0.75; and images were quantified in Nikon NIS-Elements.
Totally, three mice were used in the experiment, from each animal were harvested 6 L3–5 DRG ganglions; after sectioning of the tissue, six slices from each animal were treated, and two to three slices from each animal were randomly selected for analysis. A total number of 2,075 neurons for the Trpv1 probe, 1,863 neurons for the Th probe, 1,821 neurons for Nefh, and 1,839 neurons for the Calcb probe were analyzed.
Neuronal cell borders were determined and segmented manually dependently on the level of the fluorescent probes signal and DIC images. Cells were considered as signal positive if the mean fluorescence signal in the ROI exceeded 0.5 times the SD of the fluorescence signal in this channel.
cDNA constructs
The Piezo1-IRES-GFP and the Piezo2 pcDNA clones were from Dr. Ardem Patapoutian, Scripps Research, San Diego, CA (Coste et al., 2010). The following cDNA clones were purchased from Origene: myc-tagged mouse Tmem120A (MR205146, NM_172541), myc-tagged mouse Tmem120b (MR205067, NM_001039723), and myc-tagged mouse Trek1 (KCNK2; MR206535, BC062094). The GFP-tagged Piezo1 construct (GFP-Piezo1) was generated by subcloning the mouse Piezo1 to the pcDNA3.1(-) vector from the original IRES-GFP vector then PCR cloning GFP and ligating it to the N-terminus of Piezo1 (Jiang et al., 2021). The GFP-tagged Piezo2 construct (GFP-Piezo2) was generated by PCR cloning GFP and ligating it to the N-terminus of Piezo2 in the pCMV SPORT6 vector. The tdTomato-tagged Tmem120a construct was generated by PCR cloning Tmem120a using the Origene MR205146 clone as a template and subcloning it to the ptdTomato-N1 vector (Clontech), placing the tdTomato tag to the C-terminus of Tmem120a. For PCR cloning, the Pfu-Ultra proofreading enzyme (Agilent) was used and the constructs were verified with sequencing.
Statistics
Data are represented as mean ± SEM plus scatter plots. For normally distributed data, statistical significance was calculated either with two sample t test (two tailed), or ANOVA, with Bonferroni post hoc test. Normality was assessed by the Shapiro Wilk test or the Lilliefors test. For non-normally distributed data, Wilcoxon or Mann-Whitney tests were used as appropriate. Variance was assessed using Levene’s test for homogeneity of variance (centered on median). For normally distributed data with unequal variance, Welch’s t tests were used as appropriate. The specific tests for each experiment are described in the figure legends. No statistical method was used to predetermine sample sizes, but our sample sizes are similar to those generally employed by the field. Experiments were performed in a random order. Most statistical calculations and data plotting were performed using the Origin 2021 software.
Online supplemental material
Fig. S1 shows additional data analysis and plots from the data shown in Fig. 1, and data indicating that TMEM120B does not inhibit Piezo2 currents. Fig. S2 shows additional data analysis and plots from the data shown in Fig. 2. Fig. S3 shows surface expression data with TIRF, Fig. S4 shows representative confocal images for the cellular localization of TMEM120A, Figs. S5 and S6 show TIRF images with cytoskeletal probes for actin and tubulin, and Fig. S7 shows additional analysis and supporting data for Fig. 4.
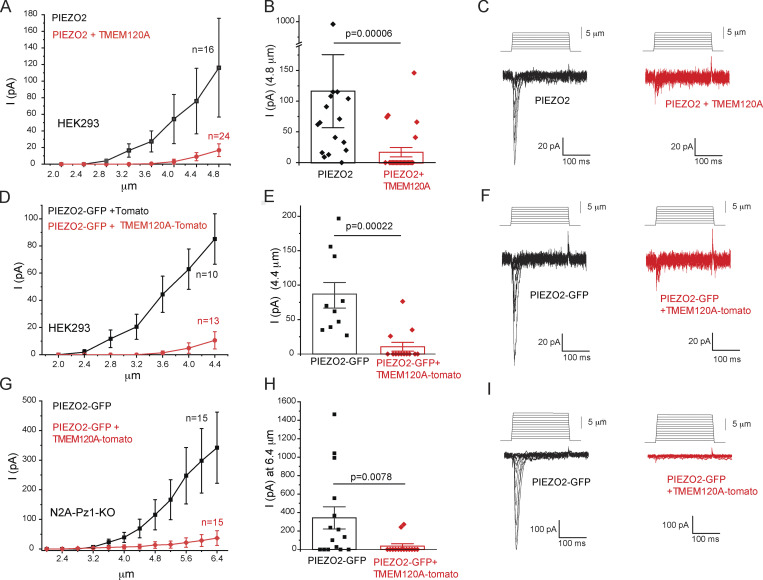
Figure 1.
TMEM120A inhibits PIEZO2 currents. Whole-cell patch-clamp experiments at −60 mV in cells transiently transfected with Piezo2 with or without Tmem120a were performed as described in the Materials and methods section. (A) HEK293 cells were transfected with Piezo2 and GFP with or without Tmem120a. In some cells, GFP-tagged Piezo2 was used instead of Piezo2 plus GFP. Current amplitudes are plotted (mean ± SEM) for Piezo2 expressing cells (black) and for cells expressing Piezo2 and Tmem120a (red). (B) Scatter plots and mean ± SEM for current amplitudes at 4.8 μm indentation. Statistical significance was calculated with the Mann-Whitney test. (C) Representative current traces. (D) HEK293 cells were transfected with GFP-tagged Piezo2 and with tdTomato-tagged Tmem120a or tdTomato. Current amplitudes are plotted (mean ± SEM) for Piezo2 expressing cells (black) and for cells expressing Piezo2 and Tmem120a (red). (E) Scatter plots and mean ± SEM for current amplitudes at 4.4 μm indentation. Statistical significance was calculated with the Mann-Whitney test. (F) Representative current traces. (G) Piezo1 deficient N2A cells were transfected with GFP-tagged Piezo2 and with tdTomato-tagged Tmem120a or tdTomato. Current amplitudes (mean ± SEM) are plotted for Piezo2 expressing cells (black) and for cells expressing Piezo2 and Tmem120a (red). (H) Scatter plots and mean ± SEM for current amplitudes at 6.4 μm indentation. Statistical significance was calculated with the Mann-Whitney test. (I) Representative current traces.
Figure S2.
TMEM120A does not inhibit PIEZO1 and TREK1 currents. Data from Fig. 2 showing the full range of measurements with indentation depth or negative pressure increased until the seal was lost. (A and B) Data from whole-cell patch-clamp experiments in HEK293 cells transfected with Piezo1 alone or Tmem120a + Piezo1. (C and D) Data from cell-attached patch clamp experiments in Piezo1 deficient N2A cells transfected with Piezo1 alone or Tmem120a + Piezo1. (E) Representative traces for cell-attached patch clamp experiments in N2A cell transfected with tdTomato-Tmem120a. (F) Full range of data from cell-attached patch-clamp experiments in N2A cell transfected with tdTomato-Tmem120a. (G) Representative traces for cell-attached patch-clamp experiments in N2A cell transfected with tdTomato. (H) Full range of data from cell-attached patch-clamp experiments in N2A cells transfected with tdTomato. (I–L) Full range of data from cell-attached patch-clamp experiments at 0 mV in N2A cells transfected with TREK1 + tdTomato (I), TREK1 + tdTomato-Tmem120a (J), tdTomato-Tmem120a alone (K), and tdTomato alone (L). The slight discrepancy in the number of cells compared to Fig. 2 in C, D, F, and I–L is because these panels include cells where the seal was lost before the negative pressure reached 55 mmHg, and those cells were excluded from Fig. 2.
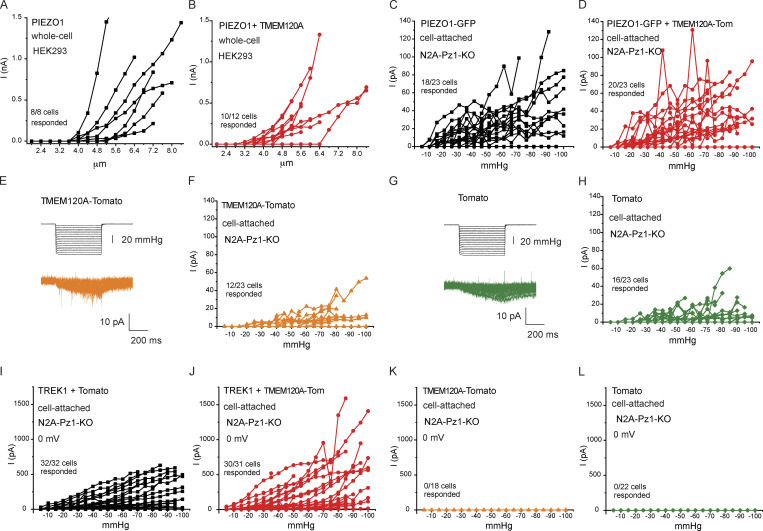
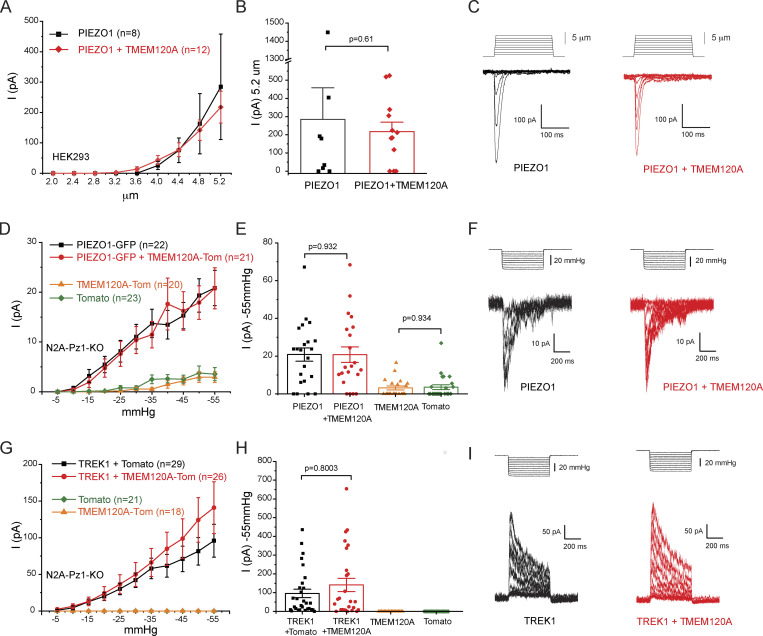
Figure 2.
TMEM120A does not inhibit PIEZO1 and TREK1 currents. (A) HEK293 cells were transfected with Piezo1 in IRES-GFP vector, with or without Tmem120a. Mechanically activated currents were evoked by increasing indentations with a blunt glass probe in whole-cell patch-clamp experiments. Current amplitudes are plotted (mean ± SEM) for Piezo1 expressing cells (black) and for cells expressing Piezo1 and Tmem120a (red). (B) Scatter plots and mean ± SEM for current amplitudes at 5.2 μm indentation. Statistical significance was calculated with the Mann-Whitney test. (C) Representative current traces. (D) Piezo1 deficient N2A cells were transfected with GFP-Piezo1 with or without Tmem120a-tdTomato, and with tdTomato-Tmem120a alone or tdTomato alone. Mechanically activated currents were evoked by applying increasing negative pressures through the patch pipette in cell-attached patch-clamp experiments. Measurements were performed at −80 mV holding potential. Current amplitudes are plotted (mean ± SEM) for cells expressing Piezo1 (black), Piezo1 and Tmem120a (red), Tmem120a alone (orange), and tdTomato alone (green). (E) Scatter plots and mean ± SEM for current amplitudes at −55 mmHg. Statistical significance was calculated with the Mann-Whitney test. (F) Representative current traces. (G) Piezo1 deficient Neuro2A cells were transfected with TREK1 with, or without Tmem120a-tdTomato, and with Tmem120a-tdTomato alone or tdTomato alone. Mechanically activated currents were evoked by applying increasing negative pressures through the patch pipette in cell-attached patch-clamp experiments. Measurements were performed at 0 mV holding potential. Current amplitudes are plotted (mean ± SEM) for cells expressing TREK1 (black), TREK1 and Tmem120a (red), Tmem120a alone (orange), and tdTomato alone (green). (H) Scatter plots and mean ± SEM for current amplitudes at −55 mmHg. Statistical significance was calculated with the Mann-Whitney test. (I) Representative current traces.
Figure S3.
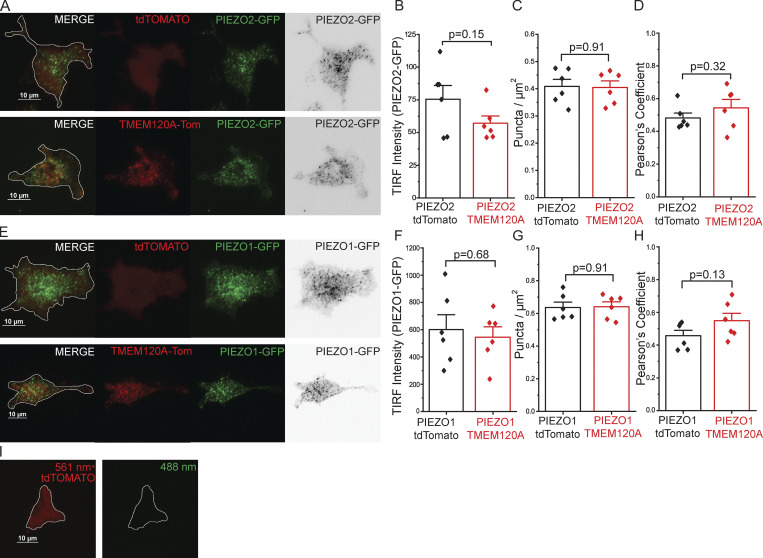
TMEM120A shows only weak colocalization with PIEZO1 or PIEZO2, and does not change their cell-surface expression. TIRF microscopy was performed as described in the Materials and methods section. (A) Representative TIRF images for HEK293 cell transfected with tdTomato-Tmem120a and GFP-Piezo2 (bottom) and tdTomato and GFP-Piezo2 (top). (B) Summary data for the fluorescence intensity for GFP-PIEZO2 in the TIRF mode for cells cotransfected with tdTomato or tdTomato-Tmem120a. (C) Number of GFP-PIEZO2 puncta per μm. (D) Pearson’s coefficient for colocalization of tdTomato-TMEM120A with GFP-PIEZO2 and tdTomato with GFP-PIEZO2. (E) Representative TIRF images for HEK293 cell transfected with tdTomato-Tmem120a and GFP-Piezo1 (bottom) and tdTomato and GFP-Piezo1 (top). (F) Summary data for the fluorescence intensity for GFP-PIEZO1 in the TIRF mode for cell cotransfected with tdTomato or tdTomato-Tmem120a. (G) Number of GFP-PIEZO1 puncta per μm. (H) Pearson’s coefficient for colocalization of tdTomato-TMEM120A with GFP-PIEZO1 and tdTomato with GFP-PIEZO1. (I) Representative image of cell expressing only tdTomato with 561-nm laser (left) and 488-nm laser (right). Bar graphs show mean ± SEM and scatter plots. Individual symbols show the average value of cells for one coverslip (5–22 cells/coverslip) from two independent transfection. Statistical significance was calculated with two-sample t test.
Figure S4.
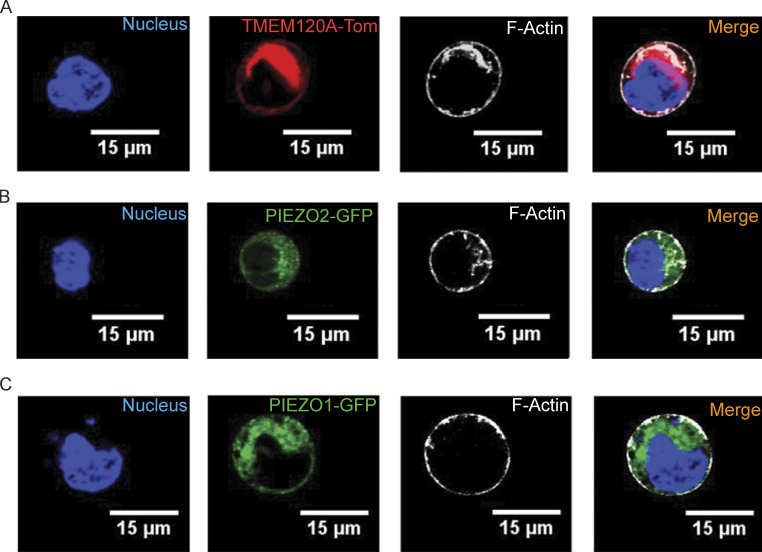
TMEM120A is broadly distributed throughout cells. HEK293 cells were transfected with tdTomato-Tmem120a, GFP-Piezo1 or GFP-Piezo2, labeled with Sir-Actin, and confocal images were obtained as described in the Materials and methods section. (A) Representative confocal images of tdTomato-TMEM120A and Sir-Actin. (B) Representative confocal images of GFP-PIEZO2 and Sir-Actin, which labels F-Actin. (C) Representative confocal images of GFP-PIEZO1 and Sir-Actin. Two independent transfections were performed and 26–30 cells per group were imaged.
Figure S5.
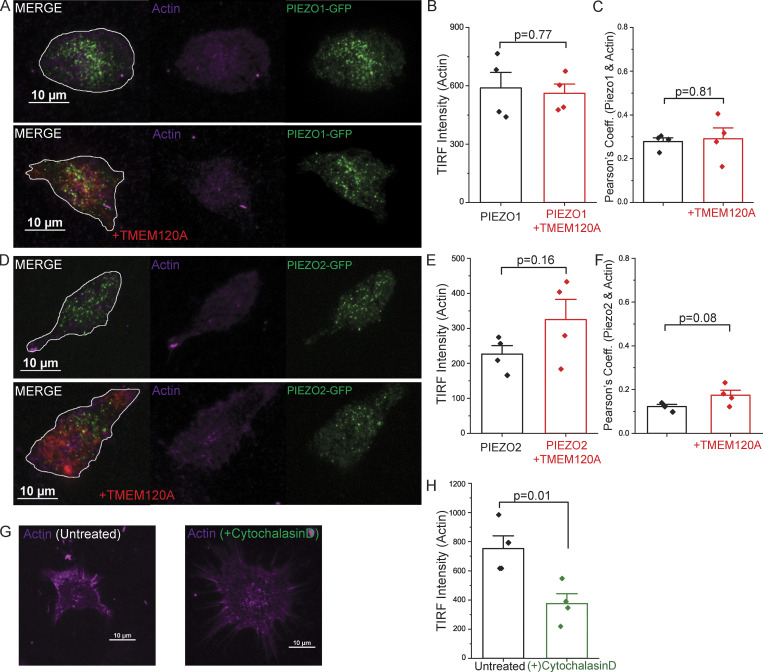
TMEM120A does not affect the actin cytoskeleton. HEK293 cells were transfected with tdTomato-Tmem120A and GFP-Piezo1 or GFP-Piezo2, labeled with Sir-Actin, and TIRF images were obtained as described in the Materials and methods section. (A) Representative TIRF images for GFP-PIEZO1 and Sir-Actin. Cell outlines displayed in white on merged images. (B) TIRF intensity of Sir-Actin with and without TMEM120A. (C) Pearson’s coefficient for colocalization of GFP-PIEZO1 and Sir-Actin with and without TMEM120A. (D) Representative TIRF images for GFP-PIEZO2 and Sir-Actin. Cell outlines displayed in white on merged images. (E) TIRF intensity of Sir-Actin with and without TMEM120A. (F) Pearson’s coefficient for colocalization of GFP-PIEZO2 and Sir-Actin with and without TMEM120A. (G) Representative TIRF images for Sir-Actin in untreated and cytochalasin D treated cells. (H) TIRF intensity of Sir-Actin with and without cytochalasin D treatment. Bar graphs show mean ± SEM and scatter plots. Individual symbols show the average value of cells for one coverslip (5–22 cells/coverslip) from two independent transfections. Statistical significance was calculated with the two-sample t test.
Figure S6.
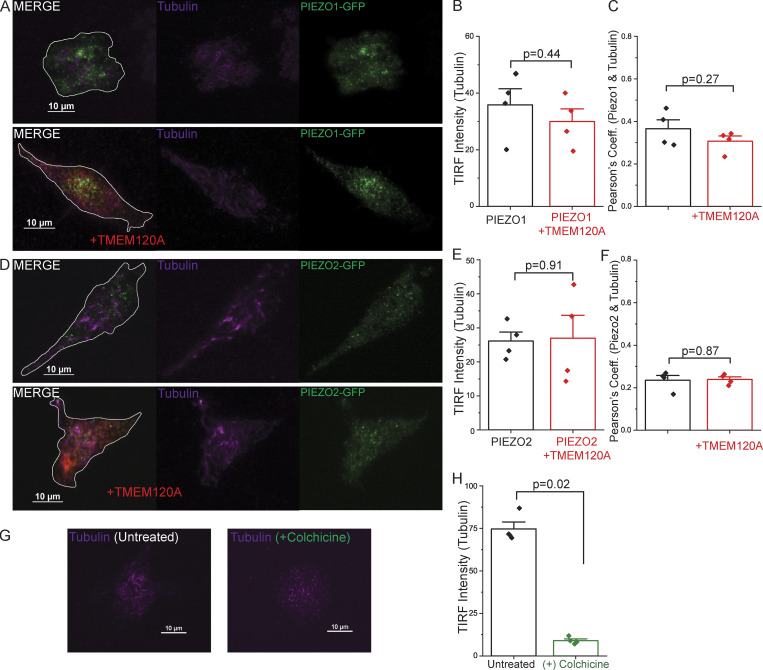
TMEM120A does not affect the tubulin cytoskeleton. HEK293 cells were transfected with tdTomato-Tmem120A and GFP-Piezo1 or GFP-Piezo2, labeled with Spy650-tubulin, and TIRF images were obtained as described in the Materials and methods section. (A) Representative TIRF images for GFP-PIEZO1 and Spy650-tubulin. Cell outlines displayed in white on merged images. (B) TIRF intensity of Spy650-tubulin with and without TMEM120A. Statistical significance was calculated using the two-sample t test. (C) Pearson’s coefficient for colocalization of GFP-PIEZO1 and Spy650-tubulin with and without TMEM120A. Statistical significance was calculated using the two-sample t test. (D) Representative TIRF images for GFP-PIEZO2 and Spy650-tubulin. Cell outlines displayed in white on merged images. (E) TIRF intensity of Spy650-tubulin. Statistical significance was calculated using the Welch’s t test. (F) Pearson’s coefficient for colocalization of GFP-PIEZO2 and Spy650-tubulin with and without TMEM120A. Statistical significance was calculated using the two-sample t test. (G) Representative TIRF images for Spy650-tubulin in untreated and colchicine treated cells. (H) TIRF intensity of Spy650-tubulin with and without colchicine treatment. Statistical significance was calculated with the Mann-Whitney test. Bar graphs show mean ± SEM and scatter plots. Individual symbols show the average value of cells for one coverslip (5–22 cells/coverslip) from two independent transfections.
Figure S7.
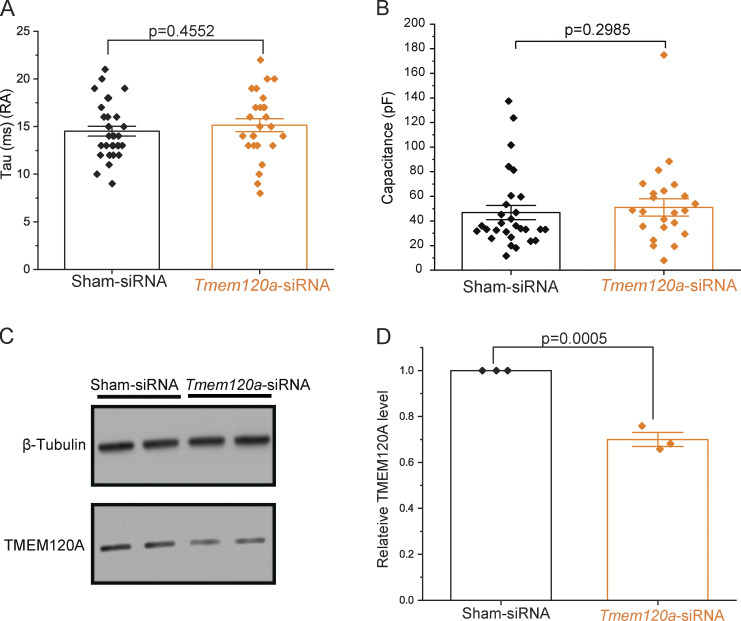
TMEM120A negatively regulates rapidly adapting mechanically activated currents in mouse DRG neurons. Data from Fig. 4 showing further analysis of Tmem120a-siRNA in mouse DRG neurons and knockdown confirmation in N2A cells. (A) Scatter plots and mean ± SEM for the inactivation time constant (tau) for rapidly adapting (RA) currents. Statistical significance was calculated with two-sample t test. (B) Scatter plots and mean ± SEM of capacitance for neurons displaying RA currents. Statistical significance calculated with the Mann-Whitney test. (C) Piezo1 deficient Neuro2A cells were transfected with Sham-siRNA or Tmem120a-siRNA for Western blot analysis as described in the methods section. Representative Western blot image with β-tubulin antibody application (top panel) and TMEM120A antibody application (bottom panel). (D) The ratio of TMEM120A band intensity to β-tubulin for each Western blot (three independent transfections) was normalized to Sham-siRNA, scatter plots, and mean ± SEM. Statistical significance calculated with two-sample t test. Source data are available for this figure: SourceData FS7.
Figure 4.
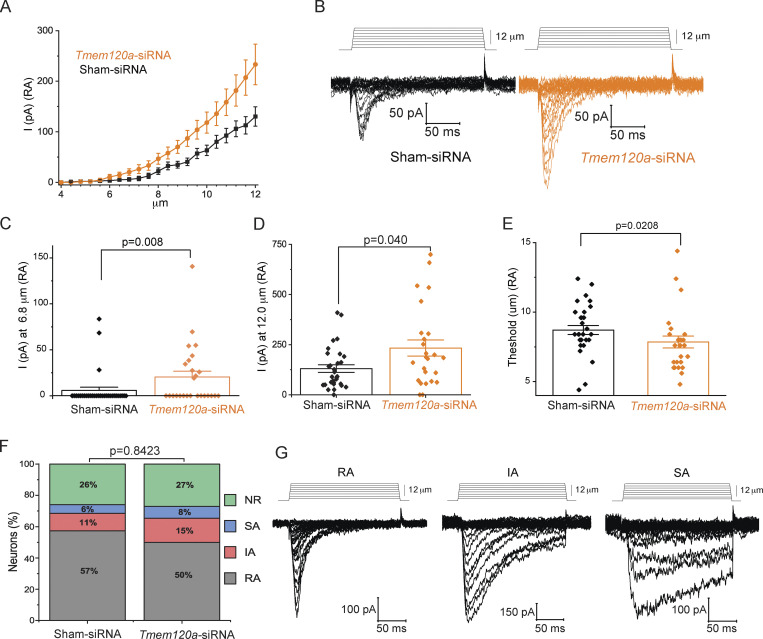
TMEM120A negatively regulates rapidly adapting mechanically activated currents in mouse DRG neurons. Mouse DRG neurons were transfected with Tmem120a-siRNA or nontargeting negative control siRNA (Sham-siRNA) as described in the Materials and methods section. Whole-cell patch-clamp experiments were performed at −60 mV with mechanically activated currents evoked by increasing indentation with a blunt glass probe. (A) DRG neurons with rapidly adapting (RA) inactivation kinetics indicative of native Piezo2 were measured. Current amplitudes are plotted (mean ± SEM) for Sham-siRNA neurons with RA kinetics (black) and for Tmem120a -siRNA neurons with RA kinetics (orange). (B) Representative RA-type current traces. (C and D) Scatter plots and mean ± SEM for current amplitudes at 6.8 and 12.0 μm indentations. (E) Scatter plots and mean ± SEM for mechanical threshold (blunt glass probe indentation depth required to elicit RA type currents). (F) Percentage of cells displaying RA, intermediate adapting (IA), and slow adapting (SA) currents and nonresponding neurons (NR) for those transfected with Sham-siRNA and Tmem120a -siRNA. The electrophysiology data are from four independent DRG neuron preparations and transfections, n = 54 for Sham-siRNA, and n = 52 for Tmem120a -siRNA. Statistical significance was assessed using the chi-squared test. (G) Representative RA, IA, and SA current traces. Statistical significance for C–E was calculated with the Mann-Whitney test.
Results
We first transiently transfected HEK293 cells with Piezo2 and Tmem120a, and measured mechanically activated currents in response to indentation of the cell membrane with a blunt glass probe in whole-cell patch-clamp experiments. Cells expressing Piezo2 alone displayed rapidly adapting mechanically activated currents that showed increased amplitudes in response to deeper indentations (Fig. 1, A–C). When Tmem120a was cotransfected with Piezo2, the majority of the cells did not show responses to mechanical indentations, with a small number of cells responding to stronger stimuli, resulting in significantly decreased average current amplitudes (Fig. 1, A–C).
To ensure that all cells we patched indeed expressed Tmem120a and Piezo2, we transfected HEK293 cells with Tmem120a tagged with tdTomato, and Piezo2 tagged with GFP, and patched cells displaying both red and green fluorescence. Coexpression of Tmem120a-tdTomato robustly decreased GFP-PIEZO2 current amplitudes (Fig. 1, D–F) indicating that the responses in the Tmem120a transfected group were unlikely to be due to the lack of TMEM120A, and the lack of responses were not due to the lack of PIEZO2. The thresholds for mechanical activation among the responding cells were significantly higher in HEK cells transfected with Tmem120a compared to control cells (Fig. S1, E and K).
To ensure that the reduction in PIEZO2 channel activity is not specific to HEK293 cells, we also coexpressed Tmem120a with Piezo2 in N2A cells in which Piezo1 was deleted with CRISPR (Moroni et al., 2018; Romero et al., 2020). In these cells, we could perform deeper indentations of the cell without losing the seal, thus we detected larger currents. Similar to HEK293 cells, coexpression of Tmem120a-tdTomato with GFP-Piezo2 strongly reduced mechanically activated currents (Fig. 1, G–I).
Both in HEK293 cells and in N2A cells, we stimulated all cells with increasing indentations until the seal was lost, but we only plotted data in Fig. 1 up to the indentation depth where all cells still had intact seals. Fig. S1 displays the full data that includes cells that responded at higher indentation depths, and representative traces for mechanically activated currents in Tmem120a expressing cells at higher indentation levels. The overall responsiveness of control cells was 94–100%, while the Tmem120a expressing cells was 33–54% (Fig. S1, A, B, G, H, M, and N). The peak current amplitudes regardless of the indentation depth, i.e., just before the seal was lost, were also significantly lower in Tmem120a expressing cells in all three groups (Fig. S1, D, J, and P).
PIEZO2 currents in cells expressing either GFP-Piezo2 or with tomato-tagged Tmem120a showed similar inactivation kinetics in both HEK293 cells and in N2A cells (Fig. S1, L and R). In HEK293 cells, cotransfected with Piezo2 and nontagged Tmem120a, the time constant of inactivation increased slightly but significantly (Fig. S1 F).
We also tested if TMEM120A’s close homologue TMEM120B had any effect on PIEZO2 current amplitudes. TMEM120B shares 69% sequence identity with TMEM120A, and the two proteins have a very similar homodimeric structure with six transmembrane helices in each monomer (Ke et al., 2021). Fig. S1, S and T, shows that PIEZO2 current amplitudes were similar in HEK293 cells transfected with, or without Tmem120b.
We also patched HEK293 cells expressing Tmem120a-tdTomato alone, and we did not observe any mechanically activated currents in response to indentation with a blunt glass probe up to 9.2 μm (data not shown, n = 11), in accordance with the original report describing TMEM120A/TACAN (Beaulieu-Laroche et al., 2020).
Next, we tested if TMEM120A modulates the closely related PIEZO1 channels. Coexpression of Tmem120a in HEK293 cells did not inhibit PIEZO1 channel activity evoked by indentation with a blunt glass probe in whole-cell patch-clamp experiments (Fig. 2, A–C; and Fig. S2, A and B). Coexpressing Tmem120b also did not affect PIEZO1 activity in the whole-cell patch-clamp mode (data not shown).
Unlike PIEZO2 (Shin et al., 2019), PIEZO1 currents can be reliably evoked by negative pressure in the cell-attached mode (Coste et al., 2010). To test the effect of TMEM120A in this modality, we expressed GFP-Piezo1 with Tmem120a-tdTomato or tdTomato in Piezo1 deficient Neuro2A cells. Negative pressures applied through the patch pipette reproducibly evoked mechanically activated currents. Current amplitudes in the Tmem120a expressing cells were similar to those without Tmem120a (Fig. 2, D–F). We also performed experiments in cells only expressing Tmem120a, or tdTomato. In contrast to Piezo1 expressing cells, both groups displayed no or negligible currents at low pressures. Increasing negative pressures evoked small currents in both Tmem120a-tdTomato and in tdTomato transfected cells, but the amplitudes of those were very similar in the two groups (Fig. 2, D and E; and Fig. S2, E–H).
We also tested if TMEM120A had any effect on the activity of TREK1, a mechanically activated K+ selective ion channel. We evoked outward TREK1 currents in cell-attached patches in Piezo1 deficient N2A cells by negative pressures at 0 mV (Fig. 2, G–I). Currents in cells cotransfected with tdTomato-Tmem120a and TREK1 showed similar current amplitudes to those transfected with TREK1 and tdTomato (Fig. 2, G–I; and Fig. S2, I and J). In cells expressing Tmem120a-tdTomato, or tdTomato alone, negative pressures did not induce any currents in these conditions (Fig. 2, G and H; and Fig. S2, K and L).
Next, we performed dual color TIRF imaging to assess cell-surface expression and colocalization of PIEZO2, PIEZO1, and TMEM120A. In TIRF imaging, the intensity of the excitation light decreases exponentially with the distance from the cover glass, only illuminating a narrow layer at the bottom of the cell, representing the plasma membrane and a narrow subplasma membrane region, up to 200 nm (Fish, 2009; Martin-Fernandez et al., 2013; Yamamura et al., 2015). We cotransfected HEK293 cells with GFP-Piezo1 or GFP-Piezo2, and Tmem120a-tdTomato or tdTomato. Fig. S3, A and B, shows that TIRF intensity of GFP-PIEZO2 was similar in cells cotransfected with Tmem120a-tdTomato or tdTomato. Fig. S3, E and F, shows that coexpression of Tmem120a-tdTomato did not change fluorescence intensity of GFP-PIEZO1 either. These data suggest that coexpression of Tmem120a did not change the cell-surface expression of PIEZO2 or PIEZO1.
Both GFP-PIEZO2 and GFP-PIEZO1 showed punctate localization in the TIRF images (Fig. S3, A and E), in accordance with earlier reports (Gottlieb and Sachs, 2012; Ellefsen et al., 2019; Ridone et al., 2020; Jiang et al., 2021). The number of GFP-PIEZO2 and GFP-PIEZO1 puncta per area was not different in Tmem120a transfected and control cells (Fig. S3, C and G). TMEM120A-tdTomato also showed inhomogeneous distribution and displayed very weak colocalization with PIEZO2 and PIEZO1 with Person’s coefficients of ∼0.55. tdTomato alone showed a more homogenous distribution in TIRF images, and its Pearson’s coefficient with PIEZO2 and PIEZO1 was only slightly, and not statistically significantly lower than that for TMEM120A-tdTomato (Fig. S3, D and H).
TMEM120A was shown earlier to be present in the plasma membrane using cell-surface biotinylation (Beaulieu-Laroche et al., 2020; Chen et al., 2022), but it was also shown to be localized to the endoplasmic reticulum (Li et al., 2021 Preprint) and the nuclear envelope (Batrakou et al., 2015). Consistent with these earlier data, TMEM120A-tdTomato showed not only plasma membrane, but also substantial intracellular localization in confocal microscopy experiments (Fig. S4 A). GFP-PIEZO2 as well as GFP-PIEZO1 also showed substantial localization to intracellular membranes (Fig. S4, B and C), most likely reflecting trafficking to the plasma membrane.
PIEZO2 has been shown to be modulated by both the actin cytoskeleton and microtubules (Jia et al., 2016; Chang and Gu, 2020). To test if TMEM120A acts via altering the organization of the plasma membrane associated cytoskeleton, we labeled HEK293 cells expressing Tmem120a-tdTomato and GFP-Piezo1 or GFP-Piezo2 with the cell permeable live cell dyes Sir-Actin and Spy650-tubulin and performed TIRF microscopy. Both actin (Fig. S5) and tubulin (Fig. S6) showed a very weak colocalization with PIEZO2 and PIEZO1 (Pearson’s coefficients between 0.1–0.4), and TMEM120A did not change this colocalization. TMEM120A also did not change the intensity of actin or tubulin labeling. As positive controls, we treated cells with the actin disrupting agent cytochalasin D, and with the microtubule disrupting colchicine. Cytochalasin D reduced the intensity of actin labeling (Fig. S5, G and H) and colchicine dramatically reduced the intensity of tubulin labeling (Fig. S6, G and H). These data indicate that TMEM120A does not act via altering the cytoskeletal elements adjacent to the plasma membrane.
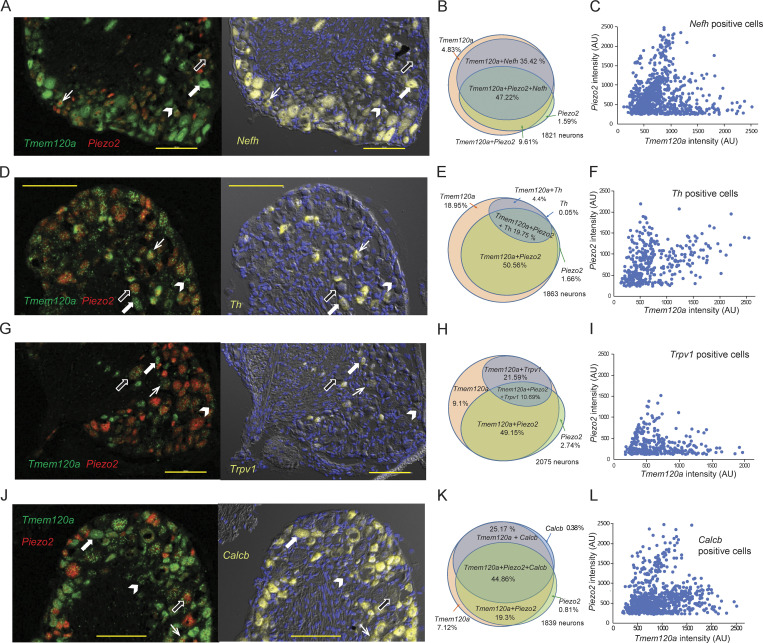
Next, we tested if Tmem120a is expressed in the same neurons as Piezo2 using RNAScope fluorescence in situ hybridization. We performed triple labeling experiments with probes for Piezo2, Tmem120a and one of the following neuronal markers: Neurofilament heavy chain (Nefh), Tyrosine hydroxylase (Th), Transient receptor potential vanilloid 1 (Trpv1), and calcitonin gene-related peptide 2 (Calcb; Fig. 3). Nefh labels neurons corresponding to myelinated Aβ and Aδ fibers and proprioceptors, Th labels a subset of nonpeptidergic C fiber low-threshold mechanoreceptors, Trpv1 noxious heat-sensitive C-fiber nociceptors, while Calcb small peptidergic C-fibers, and Aδ fiber nociceptors, with some expression in a subset of nonpeptidergic high-threshold mechanoreceptors (Le Pichon and Chesler, 2014; Usoskin et al., 2015). Tmem120a was expressed in >90% of neurons. Piezo2 was found in 60–70% of neurons and >95% of the Piezo2 positive neurons also expressed Tmem120a. Piezo2 showed a variable level of coexpression with the different neuronal markers (Fig. 3, B, E, H, and K), the highest being Th (Fig. 3 E), consistent with earlier work showing high Piezo2 levels in these neurons (Usoskin et al., 2015) and these neurons being low-threshold mechanoreceptors (Le Pichon and Chesler, 2014). Piezo2 showed the lowest coexpression with Trpv1, which is consistent with low responsiveness of TRPV1 positive cells to mechanical stimuli (Borbiro et al., 2015).
Figure 3.
Tmem120a and Piezo2 are coexpressed in various DRG populations. RNAScope fluorescence in situ hybridization on mouse DRGs was performed as described in the methods section, with probes for Tmem120a, Piezo2, and four different neuronal markers. The data represent three independent DRG preparations, and two to three slices per condition for each preparation. Arrows in representative images show cells that express: Tmem120a and neuronal marker (white wide arrow), Tmem120a and Piezo2 and neuronal marker (white arrow), Tmem120a (arrowhead), and Tmem120a + Piezo2 (black wide arrow). Horizontal yellow lines indicate 100 μm on each image. (A) Left panel: Representative image for DRGs labeled with Tmem120a (green) and Piezo2 (red). Right panel: The same section labeled with Nefh (yellow). The section was also stained with DAPI to label nuclei. (B) Venn diagram showing coexpression of Tmem120a, Piezo2, and Nefh. (C) Intensity of Piezo2 labeling as a function of Tmem120a signal for individual cells that was positive for Nefh. For both Tmem120a and Piezo2, cells that were below the threshold for counting as positive for Tmem120a or Piezo2 are not shown. (D) Representative images for DRGs labeled with Tmem120a (green) and Piezo2 (red) and Th (yellow). (E) Venn diagram showing coexpression of Tmem120a, Piezo2, and Th. (F) Intensity of Piezo2 labeling as a function of Tmem120a signal for individual cells that were positive for Th. (G) Representative images for DRGs labeled with Tmem120a (green) and Piezo2 (red) and Trpv1 (yellow). (H) Venn diagram showing coexpression of Tmem120a, Piezo2, and Trpv1. (I) Intensity of Piezo2 labeling as a function of Tmem120a signal for individual cells that were positive for Trpv1. (J) Representative images for DRGs labeled with Tmem120a (green) and Piezo2 (red) and Calcb (CGRP2; yellow). (K) Venn diagram showing coexpression of Tmem120a, Piezo2, and Calcb. (L) Intensity of Piezo2 labeling as a function of Tmem120a signal for individual cells that was positive for Calcb. AU, arbitrary units.
We also plotted the intensity of the Piezo2 RNAScope signal intensity as a function of the Tmem120a signal intensity of each individual neuron that also showed staining with the neuronal markers (Fig. 3, C, F, I, and L). Each neuronal subpopulation displayed cells with different Tmem120a/Piezo2 ratios, with a tendency of having cell populations with higher Piezo2 and lower Tmem120a, as well as higher Tmem120a and lower Piezo2 expression levels. Th positive cells had relatively large proportions of high Piezo2 low Tmem120a expressing cells, which are consistent with their roles as low-threshold mechanoreceptors (Le Pichon and Chesler, 2014). Nefh positive neurons showed a similar, but less pronounced trend, which is consistent with many of these neurons serving as low-threshold mechanoreceptors or proprioceptors (Le Pichon and Chesler, 2014). Trpv1 and Calcb positive neurons, on the other hand, had more neurons with low Piezo2 and high Tmem120a, consistent with these neurons being nociceptors and not playing major roles in mechanosensation, or having high mechanical thresholds (Le Pichon and Chesler, 2014). These data suggest that Tmem120a/Piezo2 ratios may be a determinant of Piezo2 current levels.
To assess the role of endogenous TMEM120A, we transfected isolated DRG neurons with fluorescently labeled siRNA against Tmem120a, or noncoding control siRNA. We patched cells that displayed red fluorescence, and mechanically stimulated them with a blunt glass probe. We found that the amplitudes of the rapidly adapting currents were higher in the Tmem120a siRNA group compared to the control group (Fig. 4, A–D), which is compatible with TMEM120A acting as a negative regulator of PIEZO2 in DRG neurons. The difference was more pronounced at low-indentation depths, where much fewer cells responded in the control group compared to the Tmem120a siRNA group (Fig. 4 C). Consistent with this, the thresholds for mechanical activation of rapidly adapting MA in neurons transfected with Tmem120a siRNA were also significantly lower than in the sham siRNA group (Fig. 4 E). There was no difference in the inactivation time constants (Fig. S7 A) or the cell capacitance (Fig. S7 B) between the Tmem120a siRNA group and control. Tmem120a siRNA did not change the proportion of the rapid, intermediate, and slowly adapting mechanically activated currents in DRG neurons (Fig. 4, F and G). To assess the efficiency of the siRNA against Tmem120a, we transfected Piezo1 deficient N2A cells with siRNA against Tmem120a and found a statistically significant ∼30% decrease at the protein level by Western blot compared to sham siRNA (Fig. S7, C and D). Given that the transfection efficiency in N2A cells was ∼50% based on the number of fluorescent cells, the knockdown of Tmem120a in individual cells that took up the siRNA is likely to be around 60% at the protein level.
Overall our data show that TMEM120A inhibits the activity of PIEZO2 channels when the two proteins are heterologously expressed together, and endogenous TMEM120A inhibits the activity of endogenous PIEZO2 currents in DRG neurons.
Discussion
Here, we report that TMEM120A inhibits mechanically activated PIEZO2 channels. TMEM120A was proposed to function as an ion channel responsible for slowly adapting mechanically activated currents in DRG neurons. Overexpression of Tmem120a increased the amplitudes of currents evoked by negative pressure applied through the patch pipette in the cell-attached configuration in several cell lines (Beaulieu-Laroche et al., 2020). These currents, however, were very small, only 1–2 pA on average, approximately doubling the amplitudes of background currents observed in mock transfected cells (Beaulieu-Laroche et al., 2020). In our hands, overexpression of Tmem120a in Piezo1 deficient N2A cells did not increase the amplitudes of currents evoked by negative pressures over background levels in the cell-attached configuration, but overexpression of Piezo1 with or without Tmem120a induced a 5.5-fold increase over background current levels. The lack of appearance of mechanically activated currents after Tmem120a expression is consistent with several recent publications (Niu et al., 2021; Rong et al., 2021; Xue et al., 2021). Overexpression of Tmem120a, on the other hand, evoked a robust decrease in Piezo2 currents evoked by indentation of the cell membrane with a blunt glass probe in the whole-cell configuration. Interestingly, a recent paper showed that expressing the M207A mutant of Tmem120a resulted in a sixfold increase in mechanically activated currents compared to mock transfected cells, while wild-type Tmem120a expressing cells were not markedly different from mock-transfected controls (Chen et al., 2022).
In the whole-cell configuration, neither the original publication describing TMEM120A (TACAN; Beaulieu-Laroche et al., 2020), nor we, observed increased mechanically activated currents after overexpression of Tmem120a. Treatment of DRG neurons in the Trpv1 lineage with siRNA against Tmem120a, on the other hand, decreased the proportion of mechanically activated currents with ultra-slow inactivation kinetics in the whole-cell configuration (Beaulieu-Laroche et al., 2020). In our hands, Tmem120a siRNA had no significant effect on the proportion of slowly adapting MA currents, which is consistent with a recent report, which found no effect of Tmem120a siRNA on the proportion and amplitudes of slowly and ultra-slowly adapting currents (Parpaite et al., 2021). Tmem120a siRNA, on the other hand, increased the amplitudes of rapidly adapting MA currents in DRG neurons, which is consistent with TMEM120A suppressing PIEZO2 activity. In recent single-cell RNA sequencing experiments, Tmem120a was found to have the highest expression in a subpopulation of DRG neurons that did not respond to mechanical indentation, also raising doubts about TMEM120A serving as a mechanically activated ion channel in DRG neurons (Michel et al., 2020).
TMEM120A was shown by immunohistochemistry to be present predominantly in small nonpeptidergic DRG neurons with a significant expression also in tyrosine hydroxylase positive neurons, but no expression in myelinated NF200 neurons (Beaulieu-Laroche et al., 2020). A recent report, however, showed a broader expression of Tmem120a, with substantial expression also in myelinated DRG neurons (Parpaite et al., 2021). In our hands, Tmem120a expression with RNAScope in situ hybridization was detected in more than 90% of neurons, both in cells expressing a marker of myelinated neurons, and in neurons expressing Calcb (CGRP2), Trpv1, or Th. Tmem120a and Piezo2 expression also showed substantial overlap, with varying Tmem120a/Piezo2 ratios, indicating that TMEM120A may, in principle, be an important determinant of PIEZO2 activity in DRG neurons. Consistent with this, we found that siRNA-mediated knockdown of Tmem120a increased the amplitudes of rapidly adapting mechanically activated currents in DRG neurons. The effect was more pronounced at lower indentation depths, and the mechanical threshold was shifted to the left in neurons transfected with Tmem120a siRNA. When we plotted the maximal current amplitudes before the seals were lost, the difference between the Tmem120a siRNA group and the control was not statistically significant (P = 0.15, data not shown), consistent with a recent report (Parpaite et al., 2021).
The effect of Tmem120a siRNA on current amplitudes (∼50%) was substantially smaller than the almost complete elimination of the currents by overexpression of Tmem120a, indicating that endogenous TMEM120A exerts a smaller effect than overexpressed TMEM120A. The relatively small increase in current can also be partly due to incomplete knockdown of TMEM120A by the siRNA at the protein level. Tmem120a overexpression reduced PIEZO2 current amplitudes, shifted the thresholds towards stronger mechanical stimuli, and reduced the number of cells responding to mechanical stimuli. A large fraction of the reduction in current amplitudes comes from the lack of responsiveness of many Tmem120a expressing cells. It is hard to tell if the nonresponding cells would have displayed MA currents if the seals would have stayed intact for deeper indentations, and if they responded what the maximum currents would have been. This hinders drawing firm conclusions on the mechanism of inhibition when Tmem120a is overexpressed. The common observation between siRNA knockdown and overexpression of Tmem120a was the shifted threshold of mechanical activation, suggesting that this may be the primary effect of TMEM120A. It is possible therefore that the lack of responsiveness was due to a large shift in threshold to mechanical forces that cannot be attained without disrupting seal integrity in a large fraction of cells.
When Tmem120a was conditionally knocked out from nonpeptidergic DRG neurons that express Mrgprd in a tamoxifen-inducible fashion, mice showed reduced nocifensive responses to von-Frey filaments (greater or equal to 1 g), but they retained reflexive withdrawal (Beaulieu-Laroche et al., 2020). Paw withdrawal from painful pinprick stimuli, on the other hand, was not affected by Tmem120a deletion (Beaulieu-Laroche et al., 2020). Tmem120a knockdown by spinal intrathecal injection of antisense oligodeoxynucleotides into rats, reduced inflammatory, but not chemotherapy-induced mechanical hyperalgesia (Bonet et al., 2020). These data support the idea that TMEM120A regulates some, but not all forms of mechanonociception.
Our data, however, indicate that TMEM120A inhibits PIEZO2 currents. Can this be reconciled with the behavioral findings in Tmem120a deficient mice? As mentioned earlier, the role of PIEZO2 in detecting noxious mechanical stimuli is complex. One study found that paradoxically, conditional deletion of Piezo2 in DRG neurons using an Advillin-cre mouse line reduced the threshold to painful mechanical stimuli in the Randall-Selitto test, even though the mice had defective gentle touch (Zhang et al., 2019). Ectopic expression of Piezo1 in DRG neurons, on the other hand, decreased sensitivity in the Randall-Selitto test (Zhang et al., 2019). These data indicate that activation of PIEZO2 may inhibit mechanical pain. This is compatible with classical gate control theory of pain (Melzack and Wall, 1965), where stimulation of light touch receptors reduces pain. In this framework, increasing PIEZO2-mediated currents by Tmem120a knockdown can potentially reduce pain.
What is the mechanism of PIEZO2 inhibition by TMEM120A? Tmem120a coexpression did not significantly change TIRF signal for GFP-PIEZO2, suggesting that TMEM120A does not decrease cell-surface expression of PIEZO2. While TIRF illuminates not only the plasma membrane but also a narrow subplasmalemmal cytoplasmic region, given the exponential decay of the illumination intensity by distance, a displacement of a large fraction of PIEZO2 from the plasma membrane to the cytoplasm should be detectable with TIRF even if the channel remains close to the plasma membrane. The increase in the mechanical threshold of PIEZO2 when TMEM120A is present also argues against decreased surface expression being the major mechanism of inhibition, which would be expected to evoke a similar decrease of currents at every stimulation level. While TIRF imaging does not have sufficient resolution to detect protein–protein interaction, the weak colocalization of TMEM120A with PIEZO2 makes it unlikely that the mechanism is direct interaction with the channel. TMEM120A also did not induce a major reorganization of the actin and tubulin cytoskeleton, and it did not inhibit the activity of two other mechanosensitive channels PIEZO1 and TREK1. These data indicate that the inhibition of PIEZO2 is not due to a general change in mechanical properties of the cell, or a general decrease of the ability of the cell to transduce mechanical forces to ion channels.
TMEM120A was also characterized earlier as a fat-specific nuclear envelope transmembrane protein 29 (Malik et al., 2010; Batrakou et al., 2015), and it was shown that its overexpression can alter gene expression (de Las Heras et al., 2017). Thus, it is possible that TMEM120A increases or decreases the expression of other proteins that regulate PIEZO2 properties. Consistent with its reported nuclear envelope localization, we also find that substantial amount of TMEM120A is intracellular.
Recent publications showed that the structure of TMEM120A determined by cryo-EM showed similarity to the fatty acid elongase ELOVL7, and the structure of TMEM120A also contained a coenzyme-A molecule (Niu et al., 2021; Rong et al., 2021; Xue et al., 2021). PIEZO2 as well as PIEZO1 were shown to be regulated by a variety of lipids (Borbiro et al., 2015; Narayanan et al., 2018; Romero et al., 2019; Romero et al., 2020), therefore it is possible that TMEM120A modulates PIEZO2 activity through modifying the lipid content of the cell. Exploring this possibility will require future research efforts characterizing lipid changes induced by TMEM120A and identifying potential lipid species that modify the function of PIEZO2.
Supplementary Material
contains original blots for Fig. S7.
Acknowledgments
Jeanne M. Nerbonne served as editor.
The Piezo1 IRES GFP and the Piezo2 pcDNA clones were kind gifts from Dr. Ardem Patapoutian, Scripps Research. The Piezo1 deficient Neuro2A cell line was a kind gift from Dr. Valeria Vasquez (University of Tennessee Health Science Center, Memphis, TN) with permission from Dr. Gary Lewin (Max Delbruck Center for Molecular Medicine, Berlin, Germany). The authors are grateful to Dr. Sabine Hilfiker for giving access to her Olympus confocal microscope, and to Rachel Fasiczka for help with this microscope.
This study was supported by National Institutes of Health grants R01-NS055159 to T. Rohacs and F31-NS100484 and F99-NS113422 to J.S. Del Rosario.
The authors declare no competing financial interests.
Author contributions: J.S. Del Rosario, M. Gabrielle, and Y. Yudin conceived, designed and performed the experiments, analyzed and visualized the data, and reviewed and edited the manuscript. T. Rohacs supervised the project, conceived and designed experiments, analyzed and visualized the data, and wrote and edited the manuscript. All authors approved the final version of the manuscript.
References
- Anderson, E.O., Schneider E.R., Matson J.D., Gracheva E.O., and Bagriantsev S.N.. 2018. TMEM150C/Tentonin3 is a regulator of mechano-gated ion channels. Cell Rep. 23:701–708. 10.1016/j.celrep.2018.03.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaraf, E., Blecher R., Heinemann-Yerushalmi L., Krief S., Carmel Vinestock R., Biton I.E., Brumfeld V., Rotkopf R., Avisar E., Agar G., and Zelzer E.. 2020. Piezo2 expressed in proprioceptive neurons is essential for skeletal integrity. Nat. Commun. 11:3168. 10.1038/s41467-020-16971-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batrakou, D.G., de Las Heras J.I., Czapiewski R., Mouras R., and Schirmer E.C.. 2015. TMEM120A and B: Nuclear envelope transmembrane proteins important for adipocyte differentiation. PLoS One. 10:e0127712. 10.1371/journal.pone.0127712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu-Laroche, L., Christin M., Donoghue A., Agosti F., Yousefpour N., Petitjean H., Davidova A., Stanton C., Khan U., Dietz C., et al. 2020. TACAN is an ion channel involved in sensing mechanical pain. Cell. 180:956–967.e17. 10.1016/j.cell.2020.01.033 [DOI] [PubMed] [Google Scholar]
- Besch, S.R., Suchyna T., and Sachs F.. 2002. High-speed pressure clamp. Pflugers Arch. 445:161–166. 10.1007/s00424-002-0903-0 [DOI] [PubMed] [Google Scholar]
- Bonet, I.J.M., Araldi D., Bogen O., and Levine J.D.. 2021. Involvement of TACAN, a mechanotransducing ion channel, in inflammatory but not neuropathic hyperalgesia in the rat. The J. Pain. 22:498–508. 10.1016/j.jpain.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbiro, I., Badheka D., and Rohacs T.. 2015. Activation of TRPV1 channels inhibits mechanosensitive Piezo channel activity by depleting membrane phosphoinositides. Sci. Signal. 8:ra15. 10.1126/scisignal.2005667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W., and Gu J.G.. 2020. Role of microtubules in Piezo2 mechanotransduction of mouse Merkel cells. J. Neurophysiol. 124:1824–1831. 10.1152/jn.00502.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Wang Y., Li Y., Lu X., Chen J., Li M., Wen T., Liu N., Chang S., Zhang X., et al. 2022. Cryo-EM structure of the human TACAN in a closed state. Cell Rep. 38:110445. 10.1016/j.celrep.2022.110445 [DOI] [PubMed] [Google Scholar]
- Chesler, A.T., Szczot M., Bharucha-Goebel D., Ceko M., Donkervoort S., Laubacher C., Hayes L.H., Alter K., Zampieri C., Stanley C., et al. 2016. The role of PIEZO2 in human mechanosensation. N. Engl. J. Med. 375:1355–1364. 10.1056/NEJMoa1602812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste, B., Mathur J., Schmidt M., Earley T.J., Ranade S., Petrus M.J., Dubin A.E., and Patapoutian A.. 2010. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 330:55–60. 10.1126/science.1193270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Heras, J.I., Zuleger N., Batrakou D.G., Czapiewski R., Kerr A.R.W., and Schirmer E.C.. 2017. Tissue-specific NETs alter genome organization and regulation even in a heterologous system. Nucleus. 8:81–97. 10.1080/19491034.2016.1261230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosario, J.S., Yudin Y., Su S., Hartle C.M., Mirshahi T., Rohacs T., and Rohacs T.. 2020. Gi-coupled receptor activation potentiates Piezo2 currents via Gβγ. EMBO Rep. 21:e49124. 10.15252/embr.201949124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, A.E., Murthy S., Lewis A.H., Brosse L., Cahalan S.M., Grandl J., Coste B., and Patapoutian A.. 2017. Endogenous Piezo1 can confound mechanically activated channel identification and characterization. Neuron. 94:266–270.e3. 10.1016/j.neuron.2017.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefsen, K.L., Holt J.R., Chang A.C., Nourse J.L., Arulmoli J., Mekhdjian A.H., Abuwarda H., Tombola F., Flanagan L.A., Dunn A.R., et al. 2019. Myosin-II mediated traction forces evoke localized Piezo1-dependent Ca(2+) flickers. Commun. Biol. 2:298. 10.1038/s42003-019-0514-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, K.N. 2009. Total internal reflection fluorescence (TIRF) microscopy. Curr. Protoc. Cytom. Chapter 12:Unit12.18. 10.1002/0471142956.cy1218s50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb, P.A., and Sachs F.. 2012. Piezo1: Properties of a cation selective mechanical channel. Channels. 6:214–219. 10.4161/chan.21050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, G.S., Lee B., and Oh U.. 2017. Evidence for mechanosensitive channel activity of Tentonin 3/TMEM150C. Neuron. 94:271–273.e2. 10.1016/j.neuron.2017.03.038 [DOI] [PubMed] [Google Scholar]
- Hong, G.S., Lee B., Wee J., Chun H., Kim H., Jung J., Cha J.Y., Riew T.R., Kim G.H., Kim I.B., and Oh U.. 2016. Tentonin 3/TMEM150c confers distinct mechanosensitive currents in dorsal-root ganglion neurons with proprioceptive function. Neuron. 91:708–710. 10.1016/j.neuron.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Jia, Z., Ikeda R., Ling J., Viatchenko-Karpinski V., and Gu J.G.. 2016. Regulation of Piezo2 mechanotransduction by static plasma membrane tension in primary afferent neurons. J. Biol. Chem. 291:9087–9104. 10.1074/jbc.M115.692384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Del Rosario J.S., Botello-Smith W., Zhao S., Lin Y.C., Zhang H., Lacroix J., Rohacs T., and Luo Y.L.. 2021. Crowding-induced opening of the mechanosensitive Piezo1 channel in silico. Commun. Biol. 4:84. 10.1038/s42003-020-01600-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, M., Yu Y., Zhao C., Lai S., Su Q., Yuan W., Yang L., Deng D., Wu K., Zeng W., et al. 2021. Cryo-EM structures of human TMEM120A and TMEM120B. Cell Discov. 7:77. 10.1038/s41421-021-00319-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefauver, J.M., Ward A.B., and Patapoutian A.. 2020. Discoveries in structure and physiology of mechanically activated ion channels. Nature. 587:567–576. 10.1038/s41586-020-2933-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pichon, C.E., and Chesler A.T.. 2014. The functional and anatomical dissection of somatosensory subpopulations using mouse genetics. Front. Neuroanat. 8:21. 10.3389/fnana.2014.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, A.H., and Grandl J.. 2015. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife. 4:e12088. 10.7554/eLife.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Huang S., Li X., Yang X., Xu N., Qu J., and Mak H.Y.. 2021. The endoplasmic reticulum-resident protein TMEM-120/TMEM120A promotes fat storage in C. elegans and mammalian cells. bioRxiv. (Preprint posted June 29, 2021). 10.1101/2021.06.29.450322 [DOI] [Google Scholar]
- Malik, P., Korfali N., Srsen V., Lazou V., Batrakou D.G., Zuleger N., Kavanagh D.M., Wilkie G.S., Goldberg M.W., and Schirmer E.C.. 2010. Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol. Life Sci. 67:1353–1369. 10.1007/s00018-010-0257-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fernandez, M.L., Tynan C.J., and Webb S.E.D.. 2013. A “pocket guide” to total internal reflection fluorescence. J. Microsc. 252:16–22. 10.1111/jmi.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzack, R., and Wall P.D.. 1965. Pain mechanisms: A new theory. Science. 150:971–979. 10.1126/science.150.3699.971 [DOI] [PubMed] [Google Scholar]
- Michel, N., Narayanan P., Shomroni O., and Schmidt M.. 2020. Maturational changes in mouse cutaneous touch and Piezo2-mediated mechanotransduction. Cell Rep. 32:107912. 10.1016/j.celrep.2020.107912 [DOI] [PubMed] [Google Scholar]
- Moroni, M., Servin-Vences M.R., Fleischer R., Sanchez-Carranza O., and Lewin G.R.. 2018. Voltage gating of mechanosensitive PIEZO channels. Nat. Commun. 9:1096. 10.1038/s41467-018-03502-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, S.E., Dubin A.E., Whitwam T., Jojoa-Cruz S., Cahalan S.M., Mousavi S.A.R., Ward A.B., and Patapoutian A.. 2018a. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. Elife. 7:e41844. 10.7554/eLife.41844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, S.E., Loud M.C., Daou I., Marshall K.L., Schwaller F., Kuhnemund J., Francisco A.G., Keenan W.T., Dubin A.E., Lewin G.R., and Patapoutian A.. 2018b. The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci. Translat. Med. 10:eaat9897. 10.1126/scitranslmed.aat9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, P., Hutte M., Kudryasheva G., Taberner F.J., Lechner S.G., Rehfeldt F., Gomez-Varela D., and Schmidt M.. 2018. Myotubularin related protein-2 and its phospholipid substrate PIP2 control Piezo2-mediated mechanotransduction in peripheral sensory neurons. Elife. 7:e32346. 10.7554/eLife.32346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, L., Pascoa T.C., Pike A.C.W., Bushell S.R., Quigley A., Ruda G.F., Chu A., Cole V., Speedman D., Moreira T., et al. 2021. The structural basis of fatty acid elongation by the ELOVL elongases. Nat. Struct. Mol. Biol. 28:512–520. 10.1038/s41594-021-00605-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y., Tao X., Vaisey G., Olinares P.D.B., Alwaseem H., Chait B.T., and MacKinnon R.. 2021. Analysis of the mechanosensor channel functionality of TACAN. Elife. 10:e71188. 10.7554/eLife.71188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parpaite, T., Brosse L., Sejourne N., Laur A., Mechioukhi Y., Delmas P., and Coste B.. 2021. Patch-seq of mouse DRG neurons reveals candidate genes for specific mechanosensory functions. Cell Rep. 37:109914. 10.1016/j.celrep.2021.109914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkunarajah, A., Stear J.H., Moroni M., Schroeter L., Blaszkiewicz J., Tearle J.L., Cox C.D., Furst C., Sanchez-Carranza O., Ocana Fernandez M.D.A., et al. 2020. TMEM87a/Elkin1, a component of a novel mechanoelectrical transduction pathway, modulates melanoma adhesion and migration. Elife. 9:e53308. 10.7554/eLife.53308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade, S.S., Woo S.H., Dubin A.E., Moshourab R.A., Wetzel C., Petrus M., Mathur J., Begay V., Coste B., Mainquist J., et al. 2014. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature. 516:121–125. 10.1038/nature13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridone, P., Pandzic E., Vassalli M., Cox C.D., Macmillan A., Gottlieb P.A., and Martinac B.. 2020. Disruption of membrane cholesterol organization impairs the activity of PIEZO1 channel clusters. J. Gen. Physiol. 152:e201912515. 10.1085/jgp.201912515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, L.O., Caires R., Nickolls A.R., Chesler A.T., Cordero-Morales J.F., and Vasquez V.. 2020. A dietary fatty acid counteracts neuronal mechanical sensitization. Nat. Commun. 11:2997. 10.1038/s41467-020-16816-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, L.O., Massey A.E., Mata-Daboin A.D., Sierra-Valdez F.J., Chauhan S.C., Cordero-Morales J.F., and Vasquez V.. 2019. Dietary fatty acids fine-tune Piezo1 mechanical response. Nat. Commun. 10:1200. 10.1038/s41467-019-09055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y., Jiang J., Gao Y., Song D., Song D., Liu W., Xiao B., Zhao Y., Xiao B., and Liu Z.. 2021. TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells. Elife. 10:e71474. 10.7554/eLife.71474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, K.C., Park H.J., Kim J.G., Lee I.H., Cho H., Park C., Sung T.S., Koh S.D., Park S.W., and Bae Y.M.. 2019. The Piezo2 ion channel is mechanically activated by low-threshold positive pressure. Sci. Rep. 9:6446. 10.1038/s41598-019-42492-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S., Yudin Y., Kim N., Tao Y.X., and Rohacs T.. 2021. TRPM3 channels play roles in heat hypersensitivity and spontaneous pain after nerve injury. J. Neurosci. 41:2457–2474. 10.1523/JNEUROSCI.1551-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczot, M., Pogorzala L.A., Solinski H.J., Young L., Yee P., Le Pichon C.E., Chesler A.T., and Hoon M.A.. 2017. Cell-type-specific splicing of Piezo2 regulates mechanotransduction. Cell Rep. 21:2760–2771. 10.1016/j.celrep.2017.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin, D., Furlan A., Islam S., Abdo H., Lonnerberg P., Lou D., Hjerling-Leffler J., Haeggstrom J., Kharchenko O., Kharchenko P.V., et al. 2015. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18:145–153. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- Wu, J., Young M., Lewis A.H., Martfeld A.N., Kalmeta B., and Grandl J.. 2017. Inactivation of mechanically activated Piezo1 ion channels is determined by the C-terminal extracellular domain and the inner pore helix. Cell Rep. 21:2357–2366. 10.1016/j.celrep.2017.10.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J., Han Y., Baniasadi H., Zeng W., Pei J., Grishin N.V., Wang J., Tu B.P., and Jiang Y.. 2021. TMEM120A is a coenzyme A-binding membrane protein with structural similarities to ELOVL fatty acid elongase. Elife. 10:e71220. 10.7554/eLife.71220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura, H., Suzuki Y., and Imaizumi Y.. 2015. New light on ion channel imaging by total internal reflection fluorescence (TIRF) microscopy. J. Pharmacol. Sci. 128:1–7. 10.1016/j.jphs.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Zhang, M., Wang Y., Geng J., Zhou S., and Xiao B.. 2019. Mechanically activated piezo channels mediate touch and suppress acute mechanical pain response in mice. Cell Rep. 26:1419–1431.e4. 10.1016/j.celrep.2019.01.056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. S7.