Abstract
Because they are totally transferred to the future generations until mutations occur, Y chromosome genetic markers are commonly utilised in forensics for the classification of male lineages for criminal justice purposes. The mutation rate of Rapidly Mutating Y-STRs (RM Y-STRs) markers is high. That is not seen in other Y-STRs markers, and they appear to be effective in distinguishing paternally related men. This study aimed to estimate the population and mutational parameters of 13 RM Y-STRs in 13 unrelated males born in Gilgit, Pakistan. Repeat there was no population substructure and strong discriminating capacity in the counts. In this population, there were higher mutation rates with the unusual structure of repeats. More research is needed to better characterize these loci in diverse Pakistani groups.
Keywords: Mutation rates, Rapidly mutating, STR, Y chromosome
Abbreviations: Y-STR, Y-Chromosomal Short Tandem Repeat; STRs, Short Tandem Repeats; RM Y-STR, Rapidly mutating Y-Chromosomal Short Tandem Repeat; PCR, Polymerase chain reaction; ISFG, International Society of Forensic Genetics; Bio-Edit, Biological sequence alignment editor
1. Introduction
Y-STRs (Y-chromosomal short tandem repeats) are STRs (also known as microsatellites) from the non-recombining region of the human Y chromosome that are used to determine paternity when the putative father is not present, as well as in forensic genetics (Chen et al., 2015). In the forensic field, short tandem repeats are the most commonly employed polymorphism markers (STRs) (Zhang et al., 2015, Collins et al., 2004). In many human populations, for example, Y-STR is used to differentiate lines; however, It is less effective in distinguishing between related males who share the same paternal lineage and cannot be identified on an individual level (Kayser et al., 2004). Consanguineous marriages are frequent in Pakistan, with first and second cousins accounting for 61% of all marriages. First cousin marriages are more common on the paternal side than on the maternal side (32%), (21%). Second cousins account for 8% of all marriages, while other relatives account for 7% and non-relatives account for 33%. There are a variety of consanguinity tendencies in both urban and rural areas. In urban regions, 40% of first-cousin marriages occur, but in rural areas, 57% occur (http://www.pbscensus.gov.pk). Due to these developments, genetic distinction between related and unrelated males in the Pakistani population is difficult (Adnan et al., 2019a, Adnan et al., 2019b). Sexual offences against the human population, paternity tests, and missing person reports all use Y-chromosome STRs. Because Y-STRs have paternal ancestry, each community has a significant structure that aids in understanding population history (Karafet et al., 2008). In these cases, the most latest Y-STR panels (Yfiler and PPY23) have proven to be beneficial (Adnan et al., 2018: Adnan et al., 2019a, Adnan et al., 2019b: Ye et al., 2015: Gibson-Daw et al., 2017: Rapone et al., 2016: Redd et al., 2002). This is primarily due to male-specific Y-chromosome markers being uniparental inherited and lacking recombination. According to several studies, Y-STR haplotype diversity can be increased using current Y-STR sets, and male lineage differentiation can be improved by carefully selecting extra Y-STRs (Hanson and Ballantyne, 2004, Hanson and Ballantyne, 2007, Vermeulen et al., 2009). A panel of 13 rapidly mutating (RM) YSTRs (DYF399S1, DYF387S1, DYS570, DYS576, DYS518, DYS526a + b, DYS626, DYS627, DYF403S1a + b, DYF404S1, DYS449, DYS547, and DYS612) has been reported. Most Y-STRs, including all currently used in forensics, have mutation rates in the order of 1 * 10−3 or below, and mutation rates in the order of 1 * 10−3 or lower have been observed. This RM Y-STR panel differentiates between males who are closely related and males who are distantly related extremely effectively (Ballantyne et al., 2012). The study aims to develop a 13 R-M Y-STRs database for the Pakistani population and to increase the global database for 13 R-M Y-STRs. Polymorphism is detected in Y-STRs through data analysis Burki (2015), Hall (1999), Tamura et al. (2013) and Weightman (2002) discuss such type of results in the publsihed works.
2. Methods
2.1. Sampling, DNA extraction, and quality estimation
Blood samples were collected from 13 unrelated healthy male individuals from different areas of the Gilgit population. Informed consent was signed by all members who contributed to the study. DNA was isolated from preserved blood using commercially available kit i.e. Genomic DNA kits (Thermo Fisher Scientific Inc., Waltham Massachusetts, USA) according to the manufacturer’s instructions. A NanoDrop™ 1000 spectrophotometer was used to determine parameters such as concentrations and purity for extracted DNA products.
2.2. PCR amplification
Thirteen single copy or multi-copy primers were selected from the rapidly mutating RM-YSTRs reviewed from the literature and STRBase database (Balassa et al., 2021, Ruitberg et al., 2001). UCSC genome browser provides In-Silico PCR online tool (Kuhn et al., 2008, Sabit et al., 2021) which is used for retrieving PCR amplicon lengths for related primers. Selected primers were used for PCR amplification to gain amplified products for sequencing. Primer sequences and library length used for PCR amplification are given in Table 1. PCR amplification was done using a 25 μl reaction mixture containing: 6 μl of master mix (Go Taq Green, Thermo Fisher Scientific), 2 μl of genomic sample DNA, 1.5 μl of each primer (Forward and Reverse), and double distilled water to level the value of reaction mixture. The amplification was run on SimpliAmp Thermal Cycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: Initial denaturation of 15 min at 95 °C followed by 40 cycles of 1 min at 95 °C and 1 min at 60 °C. Then the final extension of 10 min was set at 72 °C. PCR products were separated and visualized by Gel electrophoresis. 4 μl of PCR product and 3ul of DNA loading dye (TFS) was subjected to 1.5% of agarose gel. A voltage gradient of 100 was applied for 30–35 min. DNA bands were visualized by using UV light.
Table 1.
In PCRs, RM Y-STRs primer sequences and library length were used.
| RM Y-STR | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Library length (bp) |
|---|---|---|---|
| DYS576 | TTGGGCTGAGGAGTTCAATC | GGCAGTCTCATTTCCTGGAG | 191 |
| DYS570 | GAACTGTCTACAATGGCTCACG | TCAGCATAGTCAAGAAACCAGACA | 256 |
| DYF387S1 | GCCTGGGTGACAGAGCTAGA | GCCACAGTGTGAGAAGTGTGA | 257, 261 |
| DYF399S1 | GGGTTTTCACCAGTTTGCAT | CCATGTTTTGGGACATTCCT | 289, 302, 293 |
| DYS626 | GCAAGACCCCATAGCAAAAG | AGAAGAATTTTGGGACATGTTT | 253 |
| DYS627 | CTAGGTGACAGCGCAGGATT | GGATAATGAGCAAATGGCAAG | 337 |
| DYS526a + b | TCTGGTGAACTGATCCAAACC | GGG TTACTTCGCCAGAAGGT | 370 |
| DYS518 | GGCAACACAAGTGAAACTGC | TCAGCTCTTACCATGGGTGAT | 279 |
| DYS612 | CCCCATGCCAGTAAGAATA | GTGAGGGAAGGCAAAAGAAAA | 204 |
| DYF404S1 | GGCTTAAGAAATTTCAACGCATA | CCATGATGGAACAATTGCAG | 197, 189 |
| DYS449 | TGGAGTCTCTCAAGCCTGTTCTA | CCTGGAAGTGGAGTTTGCTGT | 355 |
| DYS547 | TCCATGTTACTGCAAAATACAC | TGACAGAGCATAAACGTGTC | 438 |
| DYF403S1 | CAAAAT TCATGTGGATAATGA | ACAGAGCAGGATCCATCTA | 312, 316, 437 |
2.3. Sanger sequencing
Following the manufacturer's instructions, the PCR product was purified using the kit method (Gene JET Genomic DNA Purification Kit). Sanger sequencing of PCR amplicons was done through MACROGEN Korea.
2.4. RM Y-STR nomenclature
The RM Y-STR nomenclature used in this study is in agreement with the DNA commission's recommendations from the International Society of Forensic Genetics (ISFG) (Gusmao et al., 2006). The allele designation and repeat structure of each locus are shown in Table 2.
Table 2.
RM Y-STRs chromosomal band locations and Repeat structure.
| RM Y-STR | Chromosomal location | Repeat structure |
|---|---|---|
| DYS576 | Yp11.2 | (AAAG)2-20 |
| DYS570 | Yp11.2 | (TTTC)3-16 |
| DYF387S1 | Yq11.2, Yq11.23 | *(AAAG)n(GTAG)n(GAAG)nN(GAAG)n(AAAG)o |
| DYF399S1 | q11.223, Yq11.23, Yq11.2 | (GAAA)3N11(GAAA)16 |
| DYS626 | Yq11.223 | (GAAA)14–16 N24(GAAA)3N6(GAAA)5(AAA)1(GAAA)2(GAAG)1(GAAA)3 |
| DYS627 | Yp11.2 | (AGAA)3N16(AGAG)3(AAAG)11–13 |
| DYS526a + b | Yp11.2 | (CTTT)5N12-24(CCTT)7–9 N113(CCTT)11–14 |
| DYS518 | Yq11.221 | (AAAG)11–12(GGAG)1(AAAG)4 (GAAG)1N2(AAAG)16 N27(AAGG)4 |
| DYS612 | Yq11.221 | *(CCT)(CTT)(TCT)(CCT)(TCT) |
| DYF404S1 | Yq11.23, Yq12 | (TTTC)11–13N18–42(TTTC)3–9 |
| DYS449 | Yp11.2 | (TTTC)7-16N50(TTTC)17 |
| DYS547 | Yq11.221 | A: (CCTT)9 T(CAAT)4N1(AAAG)5N96(GGAA)6N19(CCTT)3(TCTC)1 N12(TTTC)5 N21(TTTC)B: (AAAG)6 N8 (AAAG)4 N180(AAAG)20 |
| DYF403S1 | Yp11.2 | (TTCT)5 N8 (TTCT)3 N39 (TTCT)3 |
*Indicate loci have no repeat units in our samples. “N” represents the number of bp present upstream and downstream regions of repeat motifs. A and B represent sample numbers 1 and 2 respectively. The column under the Repeat structure heading represents the repeat range of two different samples for each locus as every marker was applied on two different samples.
2.5. Data analysis
Sequencing outputs were subjected to various analysis steps to study STR repeat variation. Biological sequence alignment editor (Bio-Edit 7.2.6.1) is an open-source alignment tool that was used for refining and trimming unnecessary parts in sequences. Repeat motifs were estimated by direct counting.
3. Results
We studied 13 RM Y-STR loci in 13 unrelated males from Pakistan's Gilgit province in this study. RM Y-STR markers were discovered to be DYF387S1, DYF399S1, DYF403S1, DYF404S1, DYS449, DYS518, DYS526, DYS547, DYS570, DYS576, DYS612, DYS626, and DYS627 (all with mutation rates > 1 × 10−2). Nine of the 13 RM Y-STR markers were single-copy (however, six of them included multiple Y-STR loci in one amplicon). Five markers are included in multicopy systems (DYS526 with two copies, DYF387S1 with two copies, DYF404S1 with two copies, DYF399S1 with three copies, and DYF403S1 with four copies). Table 2 provides the observed allele ranges of the 13 RM Y-STRs, as well as other variables, as well as detailed information on the repeat structure and chromosomal position. Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6 depicts the graphical representation of the obtained results.
Fig. 1.
Visualization of Extracted DNA from Gilgit Samples on Electrograph of Ethidium Bromide Stained 1% of Agarose Gel.
Fig. 2.
Amplified PCR products for RM-YSTR Markers visualized on Electrograph of Ethidium Bromide Stained 1.5% Agarose Gel.
Fig. 3.
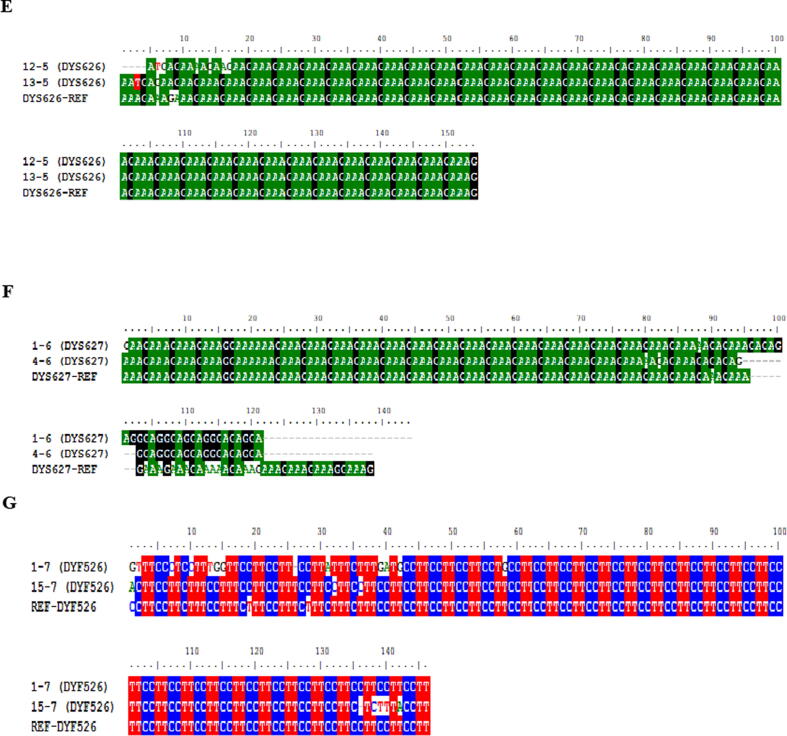
Multiple sequence alignment for RM-YSTR Primers using MEGA7 showing repeats among various sequences against reference. (A) DYS576, (B) DYS570, (C) DYF387S1, (D) DYF399S1. Gap in alignment highlights variation in STR Repeat. Visualization in Black color view via BioEdit.
Fig. 4.
Multiple sequence alignment for RM-YSTR Primers using MEGA7 showing repeats among various sequences against reference. (A) DYS626, (B) DYS627, (C) DYS526. Gap in alignment highlights variation in STR Repeat. Visualization in Black color view via BioEdit.
Fig. 5.
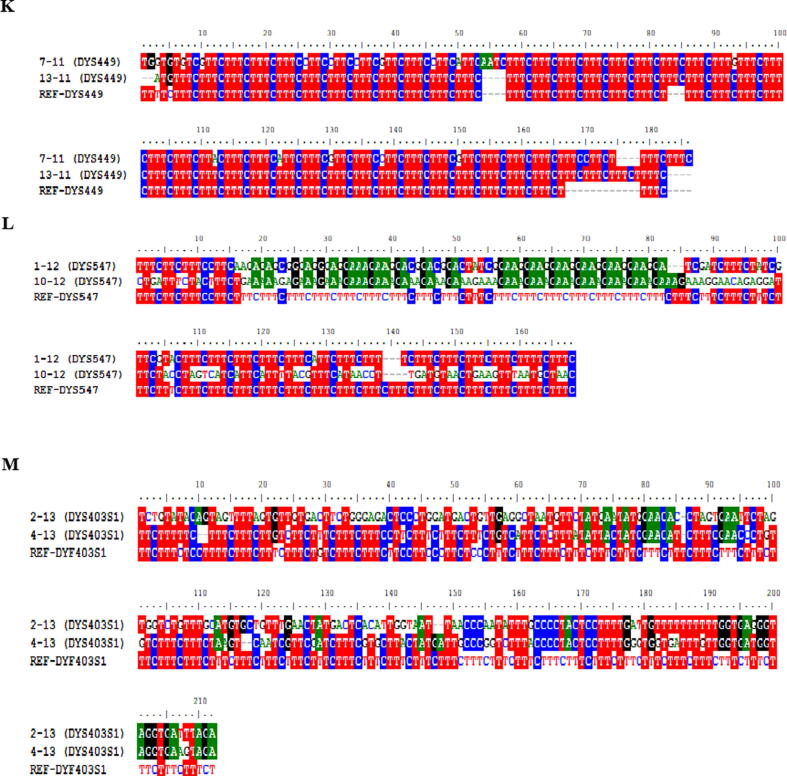
Multiple sequence alignment for RM-YSTR Primers using MEGA7 showing repeats among various sequences against reference. (A) DYS518, (B) DYS612, (C) DYF404S1, (D). Gap in alignment highlights variation in STR Repeat. Visualization in Black color view via BioEdit.
Fig. 6.
Multiple sequence alignment for RM-YSTR Primers using MEGA7 showing repeats among various sequences against reference. (A) DYS449, (B) DYS547, (C) DYS403S1. Gap in alignment highlights variation in STR Repeat. Visualization in Black color view via BioEdit.
3.1. Repeat motif counting
Every two unrelated male individual samples had each of the 13 RM Y-STR markers applied. The tetranucleotide markers DYS570 and DYS576 are also simple tetranucleotide markers. DYS570 and DYS576 have the structures (TTTC)n and (AAAG)n, respectively. For DYS570, the number of repeats was (TTTC)3–16, while for DYS576 it was (AAAG)2–20. The structures reported for DYS449 and DYF404S1 were given the same designation. DYF399S1 is a repeat-structured tetranucleotide complex locus (GAAA). The DYF399S1 marker has the same number of repetitions (GAAA)3N11(GAAA)16 in both samples. The DYS626 locus is a tetra nucleotide amplicon with a complicated structure. A length polymorphism was discovered downstream from the major (GAAA) polymorphism in the repeat unit (GAAA). The structure of this locus results (GAAA)nN(GAAA)nN(GAAA)(AAA)(GAAA)(GAAG)(GAAA). The total number of repeats in our sample was (GAAA)14-16N24(GAAA)3N6(GAAA)5(AAA)1(GAAA)2(GAAG)1(GAAA)3. A previous research of 51 populations from around the world found a similar genetic structure (HGDP-CEPH) (Ballantyne et al., 2012). (AGAA)nN(AGAG)(AAAG)N(AAGG) is the reported structure for DYS627, and the allele identification for this locus in our samples is (AGAA)3N16(AGAG)3(AAAG)11–13. The hypothesised structure for locus DYF526 (a + b) is (CCTT)(CTTT)(CCTT)N(CCTT)n, with (CTTT)5N12-24(CCTT)7–9 N113(CCTT)11–14 as the observed number of repetitions.
3.2. Novel variations in standard repeat structures
Sequencing analysis revealed internal sequence variations in regular repeat units for loci DYS518 and DYS547 in our population dataset. For instance, for locus DYS518 repeat units (AAAG)11–12(GGAG)1(AAAG)4 (GAAG)1N2(AAAG)16 N27(AAGG)4 were found which are different than the ISFG recommended repeat structure (AAAG)n(GAAG)(AAAG)n(GGAG)(AAAG)N(AAAG). DYS547 is the same way. The following tetranucleotides are used instead of ordinary repeat units (CCTT)nT(CTTC)N(TTTC)N(CCTT)n(TCTC)n(TTTC) the following tetranucleotides (CCTT)9T(CAAT)4N1(AAAG)5N96(GGAA)6N19(CCTT)3(TCTC)1N12(TTTC)5N21(TTTC)3 were found. At this locus, a novel repeat unit, GGAA, was discovered. In our population, one sample revealed an entirely different repeat structure for the DYS547 locus (AAAG)6N8(AAAG)4N180(AAAG)20. In our dataset, no repeat units were discovered for loci DYF387S1 and DYS612.
4. Discussion
The unique RM Y-STR panel has demonstrated the capacity to significantly increase the difference between related and unrelated males for use in forensic proceedings due to the high mutation rates of the markers used. The better efficacy of the RM Y-STR array in discriminating male people, in particular, compared to others like Yfiler, is attributable to the manner the markers were determined. The majority of the previous panels were made up of Y-STRs that showed high levels of variability within a community. The diversity represented by a particular marker or haplotype, on the other hand, is strongly dependent on a community's demographic history. As a result, Y-STRs identified in one group may perform very well within that population, but have limited utility when applied to other population samples that lack the source population's unique demographic characteristics. In this study, instead of using diversity measurements as ascertainment criteria, we used an alternate strategy of examining mutation rates, which are driven by the molecular properties of the repeat unit in question (Ballantyne et al., 2012). The repetition motif of each locus is one of the potential drawbacks in using RM Y-STRs in forensic investigations. We have described repeat motif variation in the 13 RM Y-STRs within 13 male samples of the Gilgit population. For DYS518 the reference assembly contains an array of AAAG followed by GAAG. However, we observe GGAG in place of GAAG and an extra repeat motif AAGG also found in our samples. DYS547 in reference assembly is represented as (CCTT)nT(CTTC)N(TTTC)N(CCTT)n(TCTC)n(TTTC) but we observe three different repeat motifs (CAAT)(AAAG)(GGAA) in our sample. But other samples showed a completely different structure with one repeat unit AAAG other than TTTC. The small sample size of our study is one of its drawbacks. While some haplogroups are represented many times and so give evidence for coherent relationships with specific RM Y-STR sequence variants, others are singletons, leaving the status of detected variants unclear. We have reported repeat motif variations in sequence-based markers and further study is required for the confirmation of these variants in our population.
5. Conclusion
This study offered loci polymorphism for 13 RM Y-STR makers in the Gilgit population group of Pakistan. These RM loci have proven to be ideal in the identification and discrimination of unrelated as well as paternally related male individuals in forensic investigations. The variations in the repeat structure of Loci DYS518 and DYS547 in this population will be confirmed further by sequencing. It can be proposed soon that currently used Y-STR kits will be replaced by RM Y-STR because of their higher diversity and discriminating power. Further study is needed to identify the haplotype frequencies and diversities for these RM Y-STRS to assess their full potential for identification in the Gilgit population.
Ethics approval and consent to participate
The study was commenced after getting the approval from Institutional Review Board of Quaid-I-Azam University, Islamabad, Pakistan. Written consent forms were filled from all the members participating in this study, your study has been assigned protocol #BEC-FBS-QAU2020-277.
Consent for publication
Yes, all patients included in this research gave written informed consent to publish the data contained within this study. If the patient was less than 16 years old, deceased, or unconscious when consent for publication was requested, written informed consent for the publication of this data was given by their parent or legal guardian.
6. Availability of data and material
The Availability of data and materials section I were used the raw data in this study and all data is contained within the manuscript and no additional files.
Competing interests: The authors declare no conflict of interest.
Funding: Muhammad Ramzan Khan Designed the study, provided funds, and reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University, Abha, Saudi Arabia for funding this work through research groups program under grant number R.G.P-2/135/42.
Footnotes
Peer review under responsibility of King Saud University.
References
- Adnan A., Rakha A., Kasim K., Noor A., Nazir S., Hadi S., Pang H. Genetic characterization of Y-chromosomal STRs in Hazara ethnic group of Pakistan and confirmation of DYS448 null allele. Int. J. Legal Med. 2019;133(3):789–793. doi: 10.1007/s00414-018-1962-x. doi.org/10.1007/s00414-018-1962-x. [DOI] [PubMed] [Google Scholar]
- Adnan A., Rakha A., Nazir S., Khan M.F., Hadi S., Xuan J. Evaluation of 13 rapidly mutating Y-STRs in endogamous Punjabi and Sindhi ethnic groups from Pakistan. Int. J. Legal Med. 2019;133:799–802. doi: 10.1007/s00414-018-01997-9. doi.org/10.1007/s00414-018-01997-9. [DOI] [PubMed] [Google Scholar]
- Adnan A., Rakha A., Noor A., van Oven M., Ralf A., Kayser M. Population data of 17 Y-STRs (Yfiler) from Punjabis and Kashmiris of Pakistan. Int. J. Legal Med. 2018;132(1):137–138. doi: 10.1007/s00414-017-1611-9. doi.org/10.1007/s00414-017-1611-9. [DOI] [PubMed] [Google Scholar]
- Ballantyne K.N., Keerl V., Wollstein A., Choi Y., Zuniga S.B., Ralf A., Kayser M. A new future of forensic Y-chromosome analysis: rapidly mutating Y-STRs for differentiating male relatives and paternal lineages. Forens. Sci. Int.: Gen. 2012;6(2):208–218. doi: 10.1016/j.fsigen.2011.04.017. doi.org/10.1016/j.fsigen.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Burki S.J. Rowman & Littlefield; 2015. Historical Dictionary of Pakistan. [Google Scholar]
- Chen, M.Y., Pu, C.E., Wu, F.C., Lai, H.Y., Ho, C.W., 2015, September. Rapidly mutating Y-STRs population data in Taiwan and haplotype probability estimation for forensic purp:oses. In: 2015 International Carnahan Conference on Security Technology (ICCST). IEEE, pp. 395–402. doi.10.1109/CCST.2015.7389717.
- Collins P.J., Hennessy L.K., Leibelt C.S., Roby R.K., Reeder D.J., Foxall P.A. Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: the AmpFlSTR Identifiler PCR Amplification Kit. J. Forensic Sci. 2004;49(6):1265–1277. doi: 10.1520/JFS2002195. [DOI] [PubMed] [Google Scholar]
- Gibson-Daw G., Albani P., Gassmann M., McCord B. Rapid microfluidic analysis of a Y-STR multiplex for screening of forensic samples. Anal. Bioanal. Chem. 2017;409(4):939–994. doi: 10.1007/s00216-016-9950-9. 10.1007/s00216-016-9950-9. [DOI] [PubMed] [Google Scholar]
- Gusmao L., Butler J.M., Carracedo A., Gill P., Kayser M., Mayr W.R., Morling N., Prinz M., Roewer L., Tyler-Smith C., Schneider P.M. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int. J. Legal Med. 2006;120(4):191–200. doi: 10.1007/s00414-005-0026-1. [DOI] [PubMed] [Google Scholar]
- Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic acids symposium series.
- Hanson E.K., Ballantyne J. A highly discriminating 21 locus Y-STR “megaplex” system designed to augment the minimal haplotype loci for forensic casework. J. Forens. Sci. 2004;49(1):40–51. Paper id JFS2003209–491. [PubMed] [Google Scholar]
- Hanson, E.K., Ballantyne, J., 2007. An ultra-high discrimination Y chromosome short tandem repeat multiplex DNA typing system. PloS One 2(8), e688. doi.org/10.1371/journal.pone.0000688. [DOI] [PMC free article] [PubMed]
- Sabit H., Abdel-Ghany S., Al-Dhafar Z., Said O.A., Ali Al-Saeed J., Ahmed Alfehaid Y., Aly Osman M. Molecular characterization and phylogenetic analysis of Rhynchophorus ferrugineus (Olivier) in Eastern Province, Saudi Arabia. Saudi J. Biol. Sci. 2021;28(10):5621–5630. doi: 10.1016/j.sjbs.2021.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T.M., Mendez F.L., Meilerman M.B., Underhill P.A., Zegura S.L., Hammer M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18(5):830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balassa K., Balassa G., Gondor O.K., Janda T., Almási A., Rudnóy S. Changes in physiology, gene expression and ethylene biosynthesis in MDMV-infected sweet corn primed by small RNA pre-treatment. Saudi J. Biol. Sci. 2021;28(10):5568–5578. doi: 10.1016/j.sjbs.2021.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M., Kittler R., Erler A., Hedman M., Lee A.C., Mohyuddin A., Sajantila A. A comprehensive survey of human Y-chromosomal microsatellites. Am. J. Human Gene. 2004;74(6):1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Wang T., Smith K.E., Rosenbloom K.R., Rhead B., Raney B.J., Pohl A., Pheasant M., Meyer L., Hsu F., Hinrichs A.S., Harte R.A., Giardine B., Fujita P., Diekhans M., Dreszer T., Clawson H., Barber G.P., Haussler D., Kent W.J. The UCSC genome browser database: update 2009. Nucl. Acids Res. 2009;37(suppl_1):D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapone C., D’Atanasio E., Agostino A., Mariano M., Papaluca M.T., Cruciani F., Berti A. Forensic genetic value of a 27 Y-STR loci multiplex (Yfiler® Plus kit) in an Italian population sample. Forens. Sci. Int.: Gene. 2016;21:e1–e5. doi: 10.1016/j.fsigen.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Redd A.J., Agellon A.B., Kearney V.A., Contreras V.A., Karafet T., Park H., Hammer M.F. Forensic value of 14 novel STRs on the human Y chromosome. Forens. Sci. Int. 2002;130(2–3):97–111. doi: 10.1016/s0379-0738(02)00347-x. doi.org/10.1016/S0379-0738(02)00347-X. [DOI] [PubMed] [Google Scholar]
- Ruitberg C.M., Reeder D.J., Butler J.M. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucl. Acids Res. 2001;29(1):320–322. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., Wollstein A., van der Gaag K., Lao O., Xue Y., Wang Q., Roewer L., Knoblauch H., Tyler-Smith C., de Knijff P., Kayser M. Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forens. Sci. Int.: Gene. 2009;3(4):205–213. doi: 10.1016/j.fsigen.2009.01.009. doi.org/10.1016/j.fsigen.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weightman B.A. John Wiley; New York: 2002. Dragons and Tigers: A Geography of South, East, and Southeast Asia. [Google Scholar]
- Ye Y., Gao J., Fan G., Liao L., Hou Y. Population genetics for 23 Y-STR loci in Tibetan in China and confirmation of DYS448 null allele. Forens. Sci. Int.: Gene. 2015;16:e7–e10. doi: 10.1016/j.fsigen.2014.11.018. [DOI] [PubMed] [Google Scholar]
- Zhang S., Bian Y., Tian H., Wang Z., Hu Z., Li C. Development and validation of a new STR 25-plex typing system. Forens. Sci. Int.: Gene. 2015;17:61–69. doi: 10.1016/j.fsigen.2015.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Availability of data and materials section I were used the raw data in this study and all data is contained within the manuscript and no additional files.
Competing interests: The authors declare no conflict of interest.
Funding: Muhammad Ramzan Khan Designed the study, provided funds, and reviewed the manuscript.