Abstract
An X-prolyl-dipeptidyl peptidase has been purified from Lactobacillus sakei by ammonium sulfate fractionation and five chromatographic steps, which included hydrophobic interaction, anion-exchange chromatography, and gel filtration chromatography. This procedure resulted in a recovery yield of 7% and an increase in specificity of 737-fold. The enzyme appeared to be a dimer with a subunit molecular mass of approximately 88 kDa. Optimal activity was shown at pH 7.5 and 55°C. The enzyme was inhibited by serine proteinase inhibitors and several divalent cations (Cu2+, Hg2+, and Zn2+). The enzyme almost exclusively hydrolyzed X-Pro from the N terminus of each peptide as well as fluorescent and colorimetric substrates; it also hydrolyzed X-Ala at the N terminus, albeit at lower rates. Km s for Gly-Pro- and Lys-Ala-7-amido-4-methylcoumarin were 29 and 88 μM, respectively; those for Gly-Pro- and Ala-Pro-p-nitroanilide were 192 and 50 μM, respectively. Among peptides, β-casomorphin 1-3 was hydrolyzed at the highest rates, while the relative hydrolysis of the other tested peptides was only 1 to 12%. The potential role of the purified enzyme in the proteolytic pathway by catalyzing the hydrolysis of peptide bonds involving proline is discussed.
Lactic acid bacteria constitute one of the most important group of microorganisms used in food fermentation. In the last decades, the metabolic traits of these bacteria have been the object of exhaustive studies, which evidence promising applications (12). The proteolytic properties of dairy lactic acid bacteria are among the best characterized to date (5, 13). This is a consequence of the impact of proteolysis on the physiology of these organisms as well as on the development of texture and flavor of dairy products (6, 18). Proteolysis on milk proteins is initiated by a cell wall-associated proteinase, which hydrolyzes caseins into oligopeptides. The generated peptides are mainly translocated via the oligopeptide transport system and, further, hydrolyzed by a pool of intracellular peptidases, which include endopeptidases, aminopeptidases, dipeptidases, tripeptidases, and dipeptidyl peptidases (13).
Lactobacillus sakei is the most competitive species in meat fermentation and therefore constitutes a frequently used starter culture. The proteolytic events that occur during meat processing also lead to the generation of small peptides and free amino acids, which are considered to be flavor compounds (1, 31). In fermented meat products, muscle as well as microbial enzymes are responsible for the proteolytic changes, but their roles remain elusive (21, 31). Particularly, the studies on the proteolytic system of meat lactobacilli are rather limited. It has been shown that several Lactobacillus spp. exhibit proteolytic activity on porcine muscle myofibrillar and sarcoplasmic proteins (7, 8, 28). These studies highlighted the potential role of L. sakei in amino acid and peptide generation especially from sarcoplasmic proteins. Attention has also been focused on intracellular peptidases of L. sakei, but only a limited number of general peptidases (a broad-specificity aminopeptidase, dipeptidase, and tripeptidase) have been purified and characterized so far (23, 24, 25).
Despite the fact that proline is not an abundant amino acid in the major muscle myofibrillar and sarcoplasmic proteins, the unique structure of proline in the polypeptide chains may restrict their susceptibility to proteolysis (13). Most peptidases with general specificity are, in fact, unable to split peptide bonds involving proline, and specific enzymes are required to avoid proline-peptide accumulation. The X-prolyl-dipeptidyl aminopeptidase (X-PDP, PepX; EC 3.4.14.5) is the most-studied proline-specific peptidase in dairy lactic acid bacteria (5, 13), but it has not been described in meat lactobacilli. The enzyme, also know as dipeptidyl peptidase IV, has its counterpart in mammals; for instance, it was recently purified from porcine skeletal muscle (29). The X-PDP is able to release specifically X-Pro dipeptides from the N termini of peptide chains. Proline is one of the free amino acids required for optimal growth by L. sakei (22), but its concentration is limited in raw meat (27). Thus, specific proline-specific peptidases could be also critical in the proteolytic chain for the supply of amino acids, which can be essential to support the growth and fermentative capability of lactobacilli.
In this work, we described the purification and characterization of an X-PDP from L. sakei. The properties of the purified enzyme are discussed in relation to those of the dipeptidyl peptidases previously characterized and the conditions inherent to the meat environment. To our knowledge, this is the first report on the presence of a proline-specific peptidase in a meat Lactobacillus species.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
L. sakei CECE (Colección Española de Cultivos Tipo) 4808, originally isolated from sausages (26), was routinely grown in MRS broth or agar (Oxoid, Hampshire, United Kingdom) at 30°C. For purification purposes, the organism was grown in 1.5-liter batch cultures of MRS broth (Oxoid) at 30°C, without agitation. The medium was inoculated with an overnight culture (0.3%) and was incubated to a final optical density of 3.0 to 3.2 at 660 nm.
Enzyme assay.
X-PDP activity was determined throughout the purification and characterization work using l-glycine-l-proline-7-amido-4-methylcoumarin (AMC; Sigma, St. Louis, Mo.) as substrate. The reaction mixture consisted of 250 μl of 50 mM Tris-HCl (pH 7.5) containing 0.2 mM substrate and 50 μl of enzyme. The release of fluorescence was determined after 10 min of incubation at 37°C in a multiscan fluorimeter (Fluoroscan II; Labsystems, Oy, Finland) at excitation and emission wavelengths of 360 and 440 nm, respectively. Three replicas were measured for each experimental point. One unit of enzyme activity was defined as the amount of enzyme that hydrolyzes 1 μmol of substrate per h at 37°C.
Purification.
The cell extract used for purification was obtained by lysozyme and ultrasonic treatments as described elsewhere (7, 28). The purification procedure consisted of the following steps.
(i) Ammonium sulfate fractionation.
The cell extract was fractionated with ammonium sulfate in two steps by addition of the reagent at 4°C and further incubation for 20 min. The precipitate obtained between 50 and 70% saturation was collected by centrifugation (15,000 × g for 20 min at 4°C) and dissolved in 50 mM Tris-HCl (pH 7.5).
(ii) Hydrophobic interaction chromatography.
The protein fraction obtained by ammonium sulfate precipitation was applied to a phenyl-Sepharose Fast Flow column (23 by 1.3 cm; Pharmacia, Uppsala, Sweden) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 0.5 M (NH4)2SO4. The retained proteins were eluted at 4 ml/min, using a linear ammonium sulfate gradient from 0.5 to 0 M (NH4)2SO4 (340 ml) and a final isocratic step at 0 M (NH4)2SO4, in 50 mM Tris-HCl (pH 7.5) (160 ml). The eluant was collected in 6-ml fractions.
(iii) First strong anion-exchange chromatography.
The active sample from the previous chromatographic step was applied to a Resource Q anion-exchange column (6 ml; Pharmacia) equilibrated with 20 mM sodium phosphate buffer (pH 6.0). Proteins were eluted at 6 ml/min, applying an initial isocratic step in the equilibration buffer (18 ml) followed by a linear gradient from 0 to 0.3 M NaCl in the same buffer (120 ml). The eluant was collected in 2-ml fractions.
(iv) Weak anion-exchange chromatography.
The active sample obtained from the first anion-exchange chromatography was applied to a Biosep-DEAE column (75 by 7.8 mm; Phenomenex, Torrance, Calif.) equilibrated with 20 mM sodium phosphate buffer (pH 6.0). Proteins were eluted at 1 ml/min using a linear gradient from 0 to 0.3 M NaCl in the equilibration buffer, and fractions of 1 ml were collected.
(v) Gel filtration chromatography.
The active sample concentrated to 1.5 ml was applied to a Sephacryl 200 HR column (100 by 1.5 cm, Pharmacia) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 0.1 M NaCl. Proteins were eluted at 10 ml/h, and fractions of 2 ml were collected.
(vi) Second strong anion-exchange chromatography.
The active sample was again applied to a Resource Q anion-exchange column (6 ml; Pharmacia) equilibrated with 20 mM sodium phosphate buffer (pH 6.0) containing 0.1 M NaCl. Proteins were eluted at 4 ml/min in a narrower salt gradient from 0.1 to 0.25 M NaCl (80 ml) in the same buffer. Fractions of 1 ml were collected.
Every chromatographic separation was carried out in a fast protein liquid chromatography system (Pharmacia) except for the gel filtration step, which was performed by classical chromatography. When required, active fractions from each purification step were concentrated by ultrafiltration through a 30-kDa-cutoff membrane (Millipore, Bedford, Mass.). Desalting and buffer exchange of active fractions were carried out by gel filtration on a PD10 column (Pharmacia).
Determination of protein concentration.
The protein concentration was determined by the BCA (bicinchoninic acid) method with the BCA protein assay reagent (Pierce, Rockford, Ill.).
Determination of molecular mass.
Purification was monitored by polyacrylamide gel electrophoresis (PAGE) under denaturing conditions, using sodium dodecyl sulfate (SDS), and under native conditions (14). In both cases, 10% polyacrylamide gels were used. The molecular mass of the denatured enzyme was estimated by using a broad-range molecular weight protein standard (Bio-Rad, Richmond, Calif.). Proteins were visualized after Coomassie brilliant blue R-250 staining. The relative molecular mass of the native enzyme was determined by gel filtration on a Sephacryl 200 HR column as described above. The column was calibrated with the following standard proteins (Sigma): β-amylase (200 kDa), aldolase (158 kDa), albumin (68 kDa), chymotrypsinogen A (25 kDa), and cytochrome c (12.4 kDa).
Dependence of pH and temperature.
The dependence of pH was determined in the range from 4.0 to 8.5 using the following buffers: 50 mM sodium acetate, pH 4.5 to 5.5; 50 mM sodium phosphate, pH 6.0 to 7.0; and Tris-HCl, pH 7.5 to 8.5. The dependence of temperature was determined at optimum pH in the range from 5 to 60°C as previously described (24). In every case, activity was expressed as a percentage of the activity obtained at either optimum pH or temperature.
Effects of chemical agents and metal cations on activity.
Effects of potential inhibitors on X-PDP activity were assayed by the addition of several chemical agents and metal salts, at 0.1 or 1 mM, to the reaction buffer. Activity was assayed as described above and expressed as a percentage of the activity obtained in the absence of the added compound.
Substrate specificity.
The relative activity of the X-PDP against several fluorescent substrates was determined according to the standard activity assay. The relative hydrolysis of several dipeptidyl–p-nitroanilide (pNA) substrates was also determined. The reaction mixture consisted of 250 μl of 50 mM Tris-HCl (pH 7.5) containing 0.5 mM substrate and 50 μl of enzyme. Absorbance was determined at 405 nm in a multiplate reader (ELX800; BioTek Instruments, Madrid, Spain) after 15 min of incubation. The relative hydrolysis of several peptides was estimated by measuring the disappearance of the substrate by capillary electrophoresis (24, 25).
Determination of kinetic parameters.
Kinetic parameters of the purified enzyme were estimated for Gly-Pro-AMC and Lys-Ala-AMC, using concentrations ranging from 0.005 to 0.06 and 0.01 to 0.15 mM, respectively; those for Ala-Pro-pNA and Gly-Pro-pNA were also determined, using concentrations ranging from 0.01 to 1 mM. Activity was measured continuously at 37°C as described above, and kinetic parameters were calculated from Lineweaver-Burk plots.
RESULTS
Purification of the enzyme.
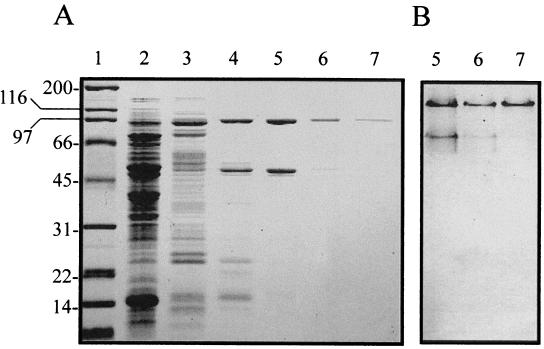
The X-PDP of L. sakei was purified by selective fractionation with ammonium sulfate and five chromatographic steps. Results of the purification procedure are summarized in Table 1. The highest X-PDP activity precipitated at 50 to 70% ammonium sulfate saturation, with a recovery of almost 90%. From the phenyl-Sepharose column, the unique peak of X-PDP activity partially coeluted with aminopeptidase activity at 0 M (NH4)2SO4 (data not shown). Chromatographic separation on the strong anion-exchange column (Resource Q) allowed further isolation of X-PDP activity, which eluted as a unique peak at 0.195 M NaCl (data not shown). In this purification step, an important enrichment in specific activity was obtained (Table 1). From the Biosep-DEAE column, the enzyme eluted at 0.22 M NaCl. This chromatographic step resulted in 148-fold purification, but still two major protein bands coeluted in the active fractions, as shown by electrophoretic analysis under native (Fig. 1B) and denaturing (Fig. 1A) conditions. Electrophoretic analysis of the active fractions eluting from the Sephacryl 200 HR gel filtration chromatography showed almost complete disappearance of the protein band of lower molecular mass, suggesting that the protein band of approximately 88 kDa corresponded to the enzyme (Fig. 1). The final chromatographic step on the strong anion-exchange column (Resource Q), using a narrower salt gradient, resulted in a single protein band on both an SDS-containing and a native polyacrylamide gel (Fig. 1). The complete purification procedure yielded 6.9% of the total activity, with an increase in specificity of 716.6-fold.
TABLE 1.
Purification of X-PDP from L. sakei
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 1,137.50 | 22,563.8 | 19.8 | 100 | 1.0 |
| 50–70% ammonium sulfate cut | 392.50 | 20,216.6 | 51.5 | 89.6 | 2.6 |
| Phenyl-Sepharose | 54.89 | 16,262.3 | 296.3 | 72.1 | 15.0 |
| Resource Q (0–0.3 M NaCl) | 4.74 | 9,636.4 | 2,033.1 | 42.7 | 102.7 |
| Biosep-DEAE | 1.74 | 5,128.3 | 2,947.3 | 22.7 | 148.8 |
| Sephacryl 200 HR | 0.52 | 2,827.8 | 5,438.1 | 12.5 | 274.6 |
| Resource Q (0.1–0.25 M NaCl) | 0.11 | 1,560.7 | 14,188.2 | 6.9 | 716.6 |
FIG. 1.
Electrophoretic analysis of X-PDP active fractions obtained at different purification steps. (A) SDS-PAGE; (B) native PAGE. Lane 1, molecular weight markers (positions indicated in kilodaltons); lane 2, cell extract; lane 3, Phenyl-Sepharose chromatography; lane 4, Resource Q chromatography (0 to 0.3 M NaCl); lane 5, Biosep-DEAE chromatography; lane 6, Sephacryl 200 HR chromatography; lane 7, Resource Q chromatography (0.1 to 0.25 M NaCl).
Molecular mass.
The molecular mass of the purified enzyme estimated by SDS-PAGE was approximately 88 kDa (Fig. 1). The relative molecular mass of the native enzyme estimated by gel filtration was around 170 kDa, suggesting that the X-PDP is composed of two subunits of equal molecular mass.
Dependence of pH and temperature.
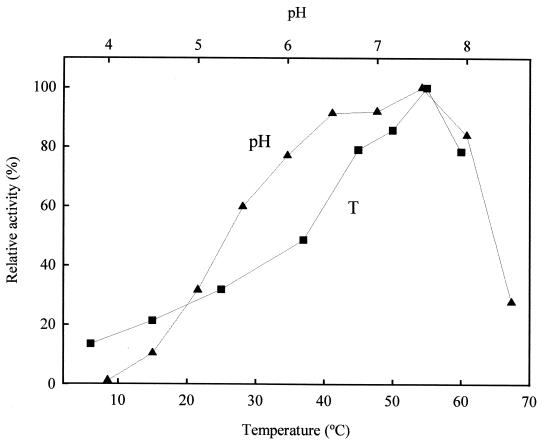
The purified enzyme is active in a broad pH range from 4.0 to 8.5, with an optimum at pH 7.5 (Fig. 2). Considerable activity (10 to 60%) was retained at pHs between 4.5 and 5.5. The optimum temperature was 55°C. Enzyme activity decreased significantly below 45°C, although substantial activity (around 20 to 30%) was observed at 15 to 25°C (Fig. 2).
FIG. 2.
Effects of pH (▴) and temperature (■) on X-PDP activity from L. sakei using Gly-Pro-AMC as substrate. The activities at optimal pH and temperature were given a value of 100%, which corresponded to 0.011 and 0.012 U (μmol h−1), respectively. The degree of purification of the enzyme preparation used was of 716.6-fold.
Effects of chemical agents and metal cations.
The presence of chelating agents such as EDTA and o-phenanthroline had no effect on activity, indicating that the purified X-PDP is not a metalloenzyme (Table 2). The reducing agents dithiothreitol and 2-mercaptoethanol as well as the sulfhydryl group reagents iodoacetate and trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E-64) did not have significant effect on activity (Table 2). These results indicate that sulfhydryl groups are not involved in the catalytic activity. Activity was reduced to almost 50% in the presence of phenylmethylsulfonyl fluoride (1.0 mM) and completely abolished in the presence of 3,4-dichloroisocoumarin, which are both specific inhibitors of serine proteinases (Table 2). The effects of amastatin, bestatin, and puromycin, which are typical inhibitors of exopeptidases, were negligible (Table 2). Pepstatin A, which is an aspartic proteinase inhibitor, had no effect on activity (Table 2). The presence of Cu2+, Hg2+, and Zn2+ caused 53 to 78% inhibition, while the other divalent cations tested had no significant effect on enzyme activity (Table 3).
TABLE 2.
Effects of chemical agents on the activity of purified (615.1-fold) X-PDP
| Chemical agent | Relative activitya
|
|
|---|---|---|
| 0.1 mM | 1.0 mM | |
| EDTA | 104 | 101 |
| o-Phenanthroline | 104 | 103 |
| Dithiothreitol | 106 | 105 |
| 2-Mercaptoethanol | 103 | 102 |
| Iodoacetate | 107 | 89 |
| E-64 | 91 | 112 |
| Pepstatin A | 100 | 113 |
| Amastatin | 96 | 100 |
| Bestatin | 93 | 98 |
| Puromycin | 89 | 94 |
| Phenylmethylsulfonyl fluoride | 90 | 57 |
| 3,4-Dichloroisocoumarin | 0 | 0 |
Expressed as a percentage of that obtained in the absence of any added chemical agent, which was given a value of 100%, corresponding to 0.014 U (μmol · h−1).
TABLE 3.
Effects of metal cations on the activity of purified (615.1-fold) X-PDP
| Metal salt | Relative activitya
|
|
|---|---|---|
| 0.1 mM | 1.0 mM | |
| CaCl2 | 94 | 88 |
| CoCl2 | 94 | 81 |
| CuCl2 | 83 | 31 |
| HgCl2 | 43 | 22 |
| MgCl2 | 88 | 85 |
| NaCl | 93 | 90 |
| ZnCl2 | 50 | 47 |
Expressed as a percentage of that obtained in the absence of any added metal salt, which was given a value of 100%, corresponding to 0.016 U (μmol h−1).
Substrate specificity.
The specificity of the purified enzyme was essentially confined to X-Pro- and X-Ala-AMC or -pNA substrates (Table 4). Maximal hydrolysis rates were obtained when proline was in the N-penultimate position. The hydrolysis of substrates containing alanine in the N-penultimate position was at best 10% of the activity on X-Pro N-terminal substrates (Table 4). The enzyme did not show aminopeptidase activity since it did not hydrolyze amino acyl-AMC substrates (Arg-, Gly-, Leu-, and Pro-AMC [Table 4]). The lack of hydrolysis of the substrate Ala-Ala-Phe-AMC also indicated that the purified enzyme did not have tripeptidyl peptidase activity (Table 4). Among colorimetric substrates, Ala-Pro-pNA was hydrolyzed at the highest rates. The relative activities toward these substrates showed that the N-terminal residue exerted an effect on the specificity with the following order of preference: alanine, arginine, and glycine. The activity of the X-PDP against several peptides was analyzed by capillary electrophoresis (Table 5). Dipeptidase activity was not detected against Gly-Pro. Several peptides with sequences X-Pro or X-Ala at the N terminus were hydrolyzed. Among those, β-casomorphin fragment 1-3 was hydrolyzed at the highest rates, while only 1 to 12% relative activity was observed on the other hydrolyzed peptides (Table 5). In general, the enzyme showed preference for X-Pro N-terminal peptides (Table 5). Moreover, diprotin A, which also has been considered as an inhibitor of these enzymes, was hydrolyzed at a similar rate as tetraalanine. The enzyme did not display specificity for X-Pro-Pro N-terminal peptides such as bradykinin. Further studies would be required to distinguish the influence of the amino acid residue located at every position of a peptide on hydrolytic rates.
TABLE 4.
Relative activity of purified (716.6-fold) X-PDP on various fluorescent (AMC derivatives) and colorimetric (pNA derivatives) substrates
| Substrate | Relative activitya |
|---|---|
| Gly-Pro-AMC | 100.0 |
| Gly-Arg-AMC | 0.0 |
| Ala-Arg-AMC | 0.0 |
| Arg-Arg-AMC | 0.0 |
| Pro-Arg-AMC | 0.0 |
| Gly-Gly-AMC | 0.0 |
| Gly-Ala-AMC | 0.4 |
| Lys-Ala-AMC | 9.9 |
| Ala-Ala-Phe-AMC | 0.0 |
| Arg-AMC | 0.0 |
| Gly-AMC | 0.0 |
| Leu-AMC | 0.0 |
| Pro-AMC | 0.0 |
| Ala-Pro-pNA | 100.0 |
| Arg-Pro-pNA | 60.5 |
| Gly-Pro-pNA | 25.4 |
| Ala-Ala-pNA | 1.7 |
Expressed as a percentage of the rates of hydrolysis of Gly-Pro-AMC and Ala-Pro-pNA, which were given a value of 100%, corresponding to 0.011 and 0.089 U (μmol h−1), respectively.
TABLE 5.
Relative activity of purified (615.1-fold) X-PDP on various peptides
| Peptide | Relative activitya |
|---|---|
| Gly-Pro | 0.0 |
| Tyr-Pro-Phe (β-casomorphin fragment 1–3) | 100.0 |
| Gly-Pro-Ala | 11.7 |
| Ile-Pro-Ile (diprotin A) | 2.0 |
| Ala-Ala-Ala | 1.1 |
| Gly-Pro-Gly-Gly | 5.0 |
| Gly-Pro-Arg-Pro | 4.1 |
| Ala-Ala-Ala-Ala | 2.2 |
| Arg-Pro-Pro-Gly-Phe (bradykinin fragment 1–5) | 0.0 |
Expressed as a percentage of the rate of hydrolysis of Tyr-Pro-Phe, which was given a value of 100%.
Kinetic parameters.
The Km and Vmax values determined for several fluorescent and colorimetric substrates are shown in Table 6.
TABLE 6.
Kinetic parameters of purified (716.6-fold) X-PDP for fluorescent (AMC derivatives) and colorimetric (pNA derivatives) substrates
| Substrate | Km (μM) | Vmax (μmol h−1 mg−1) |
|---|---|---|
| Gly-Pro-AMC | 29 | 47,438 |
| Lys-Ala-AMC | 88 | 4,239 |
| Gly-Pro-pNA | 192 | 170 |
| Ala-Pro-pNA | 50 | 658 |
DISCUSSION
In this study, we describe the purification and characterization of an X-PDP from L. sakei, a species commonly involved in meat fermentation. The biochemical properties of this enzyme and its possible role in nitrogen metabolism are discussed.
X-PDP activity is widely distributed in dairy lactic acid bacteria but has not been described in meat lactobacilli. The enzyme is the most-studied proline-specific peptidase, and it has been purified from lactococci and several dairy Lactobacillus species (5). In L. sakei, a separate enzyme appeared to be responsible for the majority of Gly-Pro-AMC-hydrolyzing activity, as the first purification step on the phenyl-Sepharose column resulted in a unique peak of activity. This substrate could also be sequentially hydrolyzed by the major aminopeptidase of L. sakei described to date (24). This enzyme eluted as a separated peak at 0.33 M ammonium sulfate (data not shown). However, the low hydrolysis rates of this general aminopeptidase on N-terminal proline and glycine residues likely prevented the detection of a second peak of activity. To our knowledge, the complete purification procedure applied in this study resulted in a specific activity enrichment superior to the one obtained for previously purified X-PDP from dairy lactic acid bacteria (5).
The molecular mass of L. sakei X-PDP was in good agreement with data previously reported for other lactic acid bacteria. Most of the X-PDP enzymes are considered to be dimers with a subunit molecular mass of 80 to 95 kDa (3, 11, 15, 16, 32, 33). However, the enzymes of Lactobacillus casei and of some strains of Lactobacillus helveticus and Lactobacillus delbrueckii have been found to be monomers or trimers (2, 9, 17, 19, 20).
The optimum pH of L. sakei X-PDP was in the pH range (6.5 to 7.5) reported for the corresponding enzymes of other lactic acid bacteria (2, 3, 9, 10, 11, 16, 19, 20, 30, 32). The activity of L. sakei X-PDP at acidic pH may favor its implication in peptide hydrolysis during meat fermentation. Remarkably, this activity is even higher than the activity displayed by the muscle dipeptidyl peptidase IV at acidic pH (29). The purified peptidase showed a relatively high optimal temperature (55°C), as it was detected for the corresponding peptidase from Lactobacillus lactis (16). However, most of these enzymes have an optimum between 45 and 50°C (2, 3, 9, 11, 16, 19, 20, 30, 32, 33). The purified X-PDP retained substantial activity at temperatures around those used in meat fermentation processes (15 to 25°C). This fact makes also feasible the participation of L. sakei X-PDP in degradation of muscle-derived peptides during meat fermentation.
The use of inhibitors of the different classes of proteases indicated that the purified X-PDP belongs to the serine protease group. The corresponding enzymes purified from dairy lactic acid have been unequivocally assigned to the same protease class. Analysis of the active site of the enzyme from L. lactis revealed that the consensus sequences differ from those of other serine protease families. The lactococcal enzyme and the mammalian dipeptidyl peptidase IV were proposed to be classified in a new group of serine proteases related to prolyl endopeptidases (4). Several divalent cations behaved as inhibitors of the purified enzyme from L. sakei. The same cations (Cu2+, Hg2+, and Zn2+) have also been reported to be strong inhibitors of the X-PDP activity of L. helveticus CNRZ 32, L. delbrueckii subsp. bulgaricus, and L. acidophilus (3, 10).
The enzyme hydrolyzed almost exclusively substrates with an X-Pro N-terminal sequence. The slight hydrolysis of X-Ala N-terminal substrates seems to be also a general characteristic of this enzyme (5, 29). In fact, the affinity of L. sakei X-PDP for Gly-Pro-AMC was three times higher than the affinity for Lys-Ala-AMC, as deduced from Kms. The X-PDP of other lactic acid bacteria showed Kms for Gly-Pro-AMC and Lys-Ala-AMC of the same order of magnitude (15, 16). The highest activity of the X-PDP from dairy organisms has been reported to be against substrates with N-terminal uncharged (Ala or Gly) or basic (Arg) residues. Specificity studies of L. sakei X-PDP on pNA derivatives indicated that the nature of the N-terminal residue exerts an important effect on activity. Thus, the rate of hydrolysis of Gly-Pro-pNA was fourfold lower than that of Ala-Pro-pNA, likewise, the Kms reflected about fourfold-higher affinity for the last substrate. This is not in agreement with what has been stated by other authors (15). The differences between the Kms for Gly-Pro-AMC and Gly-Pro-pNA were even higher (more than sixfold), indicating that the affinity greatly depends on the C-terminal group as well. The influence of this C-terminal group on Kms was also pointed out by Lloyd et al. (15). The substrate bradykinin, which contains a X-Pro-Pro sequence, was not hydrolyzed, confirming the specificity of the enzyme (15). Comparison of the specificities of L. sakei X-PDP and the porcine muscle dipeptidyl peptidase IV (29) reveals important differences. While Kms for fluorescent and colorimetric substrates are similar, Vmaxs, and therefore maximum catalytic activities, are higher with L. sakei X-PDP. Nevertheless, the significance of these differences in peptide degradation should be determined by systematic specificity studies.
Overall, this study provides substantial information on the potential role of the X-PDP from L. sakei in peptide degradation on the basis of its biochemical properties. It is also anticipated that this activity may have two major consequences during meat fermentation: (i) physiological, through the release of essential or stimulating amino acids required for optimal growth, and (ii) technological, by accelerating the whole process and modifying the composition of flavor compounds.
ACKNOWLEDGMENTS
This work was supported by grant ALI97-0353 from CICYT (Spain). The postdoctoral contract to Y. Sanz from MEC (Spain) is acknowledged.
REFERENCES
- 1.Aristoy M-C, Toldrá F. Isolation of flavour peptides from raw pork meat and dry-cured ham. In: Charalambous G, editor. Food flavours: generation, analysis and process influence. Amsterdam, The Netherlands: Elsevier Science B.V.; 1995. pp. 1323–1344. [Google Scholar]
- 2.Atlan D, Laloi P, Portalier R. X-prolyl dipeptidyl aminopeptidase of Lactobacillus delbrueckii subsp. bulgaricus: characterization of the enzyme and isolation of deficient mutants. Appl Environ Microbiol. 1990;56:2174–2179. doi: 10.1128/aem.56.7.2174-2179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockelmann W, Fobker M, Teuber M. Purification and characterization of the X-prolyl-dipeptidyl-aminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus acidophilus. Int Dairy J. 1991;1:51–66. [Google Scholar]
- 4.Chich J-F, Chapot-Chartier M-P, Ribadeau-Dumas B, Gripon J-C. Identification of the active site serine of the X-prolyl dipeptidyl aminopeptidase from Lactococcus lactis. FEBS Lett. 1992;314:139–142. doi: 10.1016/0014-5793(92)80960-o. [DOI] [PubMed] [Google Scholar]
- 5.Christensen J E, Dudley E G, Pederson J A, Steel J L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- 6.El-Soda M. The role of lactic acid bacteria in accelerated cheese ripening. FEMS Microbiol Rev. 1993;12:239–252. [Google Scholar]
- 7.Fadda S, Sanz Y, Vignolo G, Aristoy M-C, Oliver G, Toldrá F. Hydrolysis of pork muscle sarcoplasmic proteins by Lactobacillus curvatus and Lactobacillus sakei. Appl Environ Microbiol. 1999;65:578–584. doi: 10.1128/aem.65.2.578-584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fadda S, Sanz Y, Vignolo G, Aristoy M-C, Oliver G, Toldrá F. Characterization of pork muscle protein hydrolysis caused by Lactobacillus plantarum. Appl Environ Microbiol. 1999;65:3540–3546. doi: 10.1128/aem.65.8.3540-3546.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habibi-Najafi M B, Lee B H. Purification and characterization of X-prolyl-dipeptidyl peptidase from Lactobacillus casei subsp. casei LLG. Appl Microbiol Biotechnol. 1994;42:280–286. doi: 10.1007/BF00902729. [DOI] [PubMed] [Google Scholar]
- 10.Khalid N M, Marth E H. Purification and partial characterization of a prolyl-dipeptidyl aminopeptidase from Lactobacillus helveticus CNRZ 32. Appl Environ Microbiol. 1990;56:381–388. doi: 10.1128/aem.56.2.381-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiefer-Partsch B, Bockelmann W, Geis A, Teuber M. Purification of an X-prolyl-dipeptidyl aminopeptidase from the cell wall proteolytic system of Lactococcus lactis subsp. cremoris. Appl Microbiol Biotechnol. 1989;31:75–78. [Google Scholar]
- 12.Konings W N, Kok J, Kuipers O P, Poolman B. Lactic acid bacteria: the bugs of the new millennium. Curr Opin Microbiol. 2000;3:276–282. doi: 10.1016/s1369-5274(00)00089-8. [DOI] [PubMed] [Google Scholar]
- 13.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd R J, Graham G, Pritchard G. Characterization of X-prolyl dipeptidyl aminopeptidase from Lactococcus lactis subsp. lactis. J Gen Microbiol. 1991;137:49–55. [Google Scholar]
- 16.Meyer J, Jordi R. Purification and characterization of prolyl-dipeptidyl-aminopeptidase from Lactobacillus lactis and from Streptococcus thermophilus. J Dairy Sci. 1987;70:738–745. doi: 10.3168/jds.S0022-0302(87)80068-1. [DOI] [PubMed] [Google Scholar]
- 17.Meyer-Barton E C, Klein J R, Imam M, Plapp R. Cloning and sequence analysis of the X-prolyl-dipeptidyl-aminopeptidase gene (pepX) from Lactobacillus delbrueckii subsp. lactis DSM7290. Appl Microbiol Biotechnol. 1993;40:82–89. doi: 10.1007/BF00170433. [DOI] [PubMed] [Google Scholar]
- 18.Mierau I, Kunji E R S, Venema G, Kok J. Casein and peptide degradation in lactic acid bacteria. Biotechnol Gen Eng Rev. 1997;14:279–301. doi: 10.1080/02648725.1997.10647945. [DOI] [PubMed] [Google Scholar]
- 19.Miyakawa H, Kobayashi S, Shimamura S, Tomita M. Purification and characterization of X-prolyl-dipeptidyl-aminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus LBU-147. J Dairy Sci. 1991;74:2375–2381. [Google Scholar]
- 20.Miyakawa H, Hashimoto I, Nakamura T, Ishibashi N, Shimamura S, Igoshi K. Purification and characterization of X-prolyl-dipeptidyl-aminopeptidase from Lactobacillus helveticus LHE-511. Milchwissenschaft. 1994;9:670–673. [Google Scholar]
- 21.Molly K, Demeyer D I, Johansson G, Raemaekers M, Guistelinck M, Greenen I. The importance of meat enzymes in ripening and flavor generation in dry fermented sausages. First results of an European project. Food Chem. 1997;59:539–545. [Google Scholar]
- 22.Montel M-C, Labadie J. Specific nutritional requirements of lactobacilli spp. from meat. Zentbl Bakteriol Hyg B. 1986;183:23–27. [PubMed] [Google Scholar]
- 23.Montel M-C, Seronine M-P, Talon R, Hébraud M. Purification and characterization of a dipeptidase from Lactobacillus sakei. Appl Environ Microbiol. 1995;61:837–839. doi: 10.1128/aem.61.2.837-839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz Y, Toldrá F. Purification and characterization of an aminopeptidase from Lactobacillus sakei. J Agric Food Chem. 1997;45:1552–1558. [Google Scholar]
- 25.Sanz Y, Mulholland F, Toldrá F. Purification and characterization of a tripeptidase from Lactobacillus sakei. J Agric Food Sci. 1998;46:349–353. doi: 10.1021/jf970629u. [DOI] [PubMed] [Google Scholar]
- 26.Sanz Y, Hernandez M, Ferrus M A, Hernández J. Identification and differentiation of Lactobacillus sakei strains from meat origin by restriction fragment length polymorphism of the 16S rRNA genes. J Appl Microbiol. 1998;84:600–606. doi: 10.1046/j.1365-2672.1998.00387.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanz Y, Toldrá F. The role of exopeptidases from Lactobacillus sakei in dry fermented sausages. In: Pandali S G, editor. Recent research and developments in agricultural and food chemistry. Vol. 3. Trivandrum, India: Research Signpost; 1999. pp. 11–21. [Google Scholar]
- 28.Sanz Y, Fadda S, Vignolo G, Aristoy M-C, Oliver G, Toldrá F. Hydrolysis of muscle myofibrillar proteins by Lactobacillus curvatus and Lactobacillus sakei. Int J Food Microbiol. 1999;53:115–125. doi: 10.1016/s0168-1605(99)00134-8. [DOI] [PubMed] [Google Scholar]
- 29.Sentandreu, M. A., and F. Toldrá. Dipeptidyl peptidase IV from porcine skeletal muscle: purification and biochemical properties. Food Chem., in press. [DOI] [PubMed]
- 30.Tsakalidou E, Anastasiou R, Papadimitriou K, Manolopoulou E, Kalantzopoulos G. Purification and characterization of an intracellular X-prolyl dipeptidyl aminopeptidase from Streptococcus thermophilus ACA-DC 4. J Biotechnol. 1998;59:203–211. doi: 10.1016/s0168-1656(97)00157-0. [DOI] [PubMed] [Google Scholar]
- 31.Verplaetse A. Proceeding of the 40th International Congress on Meat Science and Technolnology, The Hague, The Netherlands. 1994. Influence of raw meat properties and processing technology on aroma quality of raw fermented meat products; pp. 45–65. [Google Scholar]
- 32.Vesanto E, Savijoki K, Rantanen T, Steel J L, Palva A. An X-prolyl dipeptidyl aminopeptidase (pepX) gene from Lactobacillus helveticus. Microbiology. 1995;141:3067–3075. doi: 10.1099/13500872-141-12-3067. [DOI] [PubMed] [Google Scholar]
- 33.Zevaco C, Monnet V, Gripon J-C. Intracellular X-prolyl dipeptidyl peptidase from Lactococcus lactis subsp. lactis: purification and properties. J Appl Bacteriol. 1990;68:357–366. [Google Scholar]