Abstract
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide. This study was designed to evaluate biological patterns, explore molecular classification and correlate with survival outcome in treatment naïve CRC patients.
Methods
Over 11 years consecutive series of 435 CRC patients were operated on as primary surgical therapy. A total of 201 CRC patients were included, whose complete set of clinical information was available, and their good quality tumour blocks were retrieved. Immunohistochemistry was used for tumour analysis, and partitional clustering was performed using R software for cluster analysis.
Results
The median age was 43 (range 10–85) years; adenocarcinoma was the most commonly seen histological type. The great majority had positive CK20, CEA, E-Cadherin, Ki67, CDX2, and p53 expression. There were four distinct molecular classes found, whereas Ki67, CDX2, and p53 play the main role in partitioning. Younger age negatively impacted survival; overall and disease-specific survival was 26 months only with 50 months’ longest survival.
Conclusion
Colorectal cancer is a biologically heterogeneous disease with at least four distinct molecular patterns, where cell proliferation and gene repair mechanisms appear to play the key role.
1. Introduction
Colorectal cancer (CRC) is a heterogeneous disease, the second most common cancer in women and the third most common in men worldwide (Globocan [Internet], 2020). According to Globocan, 2020, it is the 4th top cancer reported around the world and 7th most common cancer in Pakistan with a rate of 5.3/100,000 population. Globocan stated to have ∼14,000 new cases of colorectal cancer in 2020 globally, and further rise is expected in the upcoming years where an absolute increase of 79% was predicted by 2040. However, with a geographical variation highest rise was predicted in Africa at 95%, followed by Latin America at 74%, Asia at 71%, Oceania at 57%, North America at 35%, Europe at 25% rise in the new cases by 2040. A similar rise in mortality is suspected, reported to be 915,880 in 2020 (Globocan [Internet], 2020).
Given the heterogeneous nature of CRC, at least three different pathogenesis pathways have been reported to be involved (Álvaro et al., 2019). Each pathway involves several markers, and these markers can be used as potential therapeutic targets to improve clinical outcome. Major prognostic and predictive factors routinely used include the clinical stage of the disease with the uncertain role of other potential factors. Molecular classification of colorectal cancer based on gene mutation (i.e., KRAS, BRAF) has reported at least five genetically distinct molecular classes (Alwers et al., 2019). KRAS mutant tumours have been reported not to respond to anti-EGFR therapy; thus, it can be suspected that CRC has low EGFR expression, while other classes were suggested to be related to DNA instability. Therefore studying gene repair mechanisms would be an essential aspect to be looked at, which can be represented by p53 protein in cancer cells (Alwers et al., 2019).
Similarly Carcinoembryonic antigen (CEA) has shown positive expression in a great majority of CRC patients, and there has been associated metastatic potential (Belov et al., 2011). Based on the heterogeneity of the disease, different patterns of molecular classes were reported; thus CRC subtyping consortium finally developed a consensus molecular classification with four consensus classes. However, their translational aspect is still far from being used in clinical practice (Belov et al., 2011).
The lesson learned from the molecular signature of breast cancer based on genetic signature and validated by immunohistochemistry showed a potential to bring predictive and prognostic factors in clinical practice with promising results in improving clinical outcome. Also, the combination of the biomarkers in the molecular class suggests more about pathogenesis and potentially predicting response to therapy than a single marker alone. Therefore, this study was conducted, including treatment naïve CRC patients who underwent primary surgery, and their tumours were analyzed using Immunohistochemistry (IHC). Given their role in colorectal cancer development, progression, or prognosis, the biological markers were chosen. A list of potential biomarkers was identified and finally included Oestrogen receptor (Salehi far et al., 2021, Qasim, 2011), Progesterone receptor (Salehi far et al., 2021, Qasim, 2011), Human Epidermal Growth Factor Receptor- 2 (HER2) (Hasan et al., 2018), Cell proliferation marker (Ki67) (Imaizumi et al., 2020), B-Cell Lymphoma-2 (Bcl2) (Tukaram Patil et al., 2019), E-Cadherin (Kim et al., 2021), p53 (Lakpa et al., 2021), CEA (Lakpa et al., 2021), Epidermal Growth Factor Receptor (EGFR) (Uhlyarik et al., 2020), Vascular Endothelial Growth Factor (VEGF) (Mohamed et al., 2019), Programmed Death Ligand-1 (PD-L1) (Pyo et al., 2020), Caudal Type Homeobox-2 (CDX2) (Aasebø et al., 2020) and Cytokeratin 20 (CK20) (Al-Maghrabi et al., 2018) were selected given their IHC protocol and their role in prognosis and progression of colorectal cancer. The aims were to evaluate:
-
1.
The pattern of biomarkers, i.e.,ER, PR, HER2, Ki67, Bcl2, E-Cadherin, p53, CEA, EGFR, VEGF, PD-L1, CDX2 and CK20 in CRC by using IHC.
-
2.
Partitional Clustering of CRC by using K-means and Partitioning Around Medoids (PAM) methods.
-
3.
Correlating biological characteristics with survival outcome.
2. Patients and methods
2.1. Patients
Over 11 years (i.e., 2008 to 2018), a total of 435 consecutive patients with colorectal cancer were diagnosed and treated at a single center at Liaquat University of Medical & Health Sciences (LUMHS), Jamshoro, Pakistan, and their clinical information available from diagnosis until death/ last follow-up at NIMRA cancer hospital, LUMHS, Jamshoro. These patients received primary surgical therapy without neo-adjuvant systemic or radiotherapy (n = 201). Those who received neoadjuvant chemotherapy or radiotherapy or incomplete clinical information and missing tumour blocks were excluded (n = 234). After their diagnosis, they received treatment as per hospital policy following International guidelines. The patients with good quality tumor samples available and a complete set of clinical information and follow-up were included in this study. All the patients were treatment naïve, including those who underwent emergency surgery due to obstruction without any prior diagnosis and those who were operated on after diagnosis and staging but without receiving any neo-adjuvant (both systemic or radiotherapy) therapy.
2.2. Tumour analysis
Formalin-Fixed Paraffin-embedded (FFPE) tumor blocks were retrieved from the cancer tissue archive. Hematoxylin and Eosin (H& E) staining was done to identify the most representative tumor block. Whole tumour block were used to analyze biomarkers, including ER, PR, HER 2-neu, Ki-67, Bcl-2, E-Cadherin, P53, CEA, EGFR, and VEGF. PDL1, CDX-2, and CK 20 by using indirect IHC. For IHC one, cut section of 3–5 μm thickness was used for each block. The preparation process was done by using PT-Link while primary antibodies were ready to use (RTU) by DAKO. Incubation time, dilution, and temperature are summarized in Supplementary Table 1. Envision Flex High pH (Link) secondary antibody was used. Finally treated with chromogen DAB and counterstained with Haematoxylin.
Table 1.
Demographic and biological characteristics of colorectal cancer.
| Factor | N(%) |
|---|---|
| Age | |
| ≤25 | 37(18.4) |
| 26–50 | 95(47.3) |
| 51–75 | 63(31.3) |
| ≥75 | 6(3.0) |
| Tumour stage | |
| T1 | 2(1.1) |
| T2 | 50(27.5) |
| T3 | 89(48.9) |
| T4 | 15(8.2) |
| Lymph node stage | |
| N0 | 41(22.5) |
| N1 | 49(26.9) |
| N2 | 9(4.9) |
| Metastatic status | |
| M0 | 169 (92.4) |
| M1 | 14 (7.7) |
| Grade | |
| Well differentiated | 59(29.6) |
| Moderately differentiated | 97(48.7) |
| Poorly differentiated | 43(21.6) |
| Histological types | |
| Adenocarcinoma | 123(61.5) |
| Mucinous adenocarcinoma | 50(25.0) |
| Micro-papillary carcinoma | 14(7.0) |
| Signet ring cell carcinoma | 7(1.5) |
| Other types | 6(3.0) |
| Biological markers | |
| Ki67 Positive | 165(82.1) |
| CDX2 Positive | 171(85.1) |
| ER positive | 6(3.0) |
| PR positive | 5(2.5) |
| HER2 positive | 9(4.5) |
| EGFR positive | 17(8.5) |
| Bcl2 positive | 5(2.5) |
| P53 positive | 87(43.3) |
| CK20 positive | 199(99.0) |
| CEA positive | 198(98.5) |
| E-Cadherin positive | 199(99.0) |
| PDL-1 positive | 29(14.4) |
| VEGF positive | 11(5.5) |
2.3. Scoring
Immunohistochemistry staining of biomarkers assessed by the percentage of cells stained, as well as McCarty’s immunohistochemical scoring (H-score) was done (range 0–300) (Howell et al., 1984). The cutoff of the percentage of cells was used to define positivity/negativity. The scoring was done by FM, each section was scored three times, and an average of the scores was taken as the biomarker's final score. For inter-observer concordance, 25% of slides were randomly scored by BMS. Kappa statistics was performed for all markers for intra and inter- observer concordance. The Kappa score was 0.9–1.0, 0.8–1.0, respectively. The scoring was done using a Euromax simple microscope at 40x magnification size.
2.4. Cluster analysis
The biological patterns were characterized by partitional clustering methods as described in (Syed et al., 2013), using R, a data analysis software. The H-score of the biomarkers was used for cluster analysis. K-means and PAM clustering algorithms were run over the data, varying the number of clusters between 2 and 20. Validity indices (external validation criteria) were used to suggest the best number of clusters to consider. When running K-means between 2 and 20 clusters, the algorithm stopped after the split in 4 groups, and clusters with 0 elements were returned. This suggests that a split in more than four groups may not be ideal.
2.5. Statistical analysis
The X-tile Bio-informatics software was used to define cutoffs (Camp et al., 2004). The Statistical Package for Social Sciences (SPSS, version 21.0, Chicago) was used for data collection and analysis. Chi-square test used for comparisons of biomarker expression between groups. Survival analysis was performed using Kaplan–Meier methods with the application of log-rank and generalized Wilcoxon tests as appropriate. A p-value of <0.05 was considered significant. Overall survival was calculated as the time from date of diagnosis till death from any cause. In contrast, disease-specific survival was calculated as the time from the date of diagnosis till death from metastases due to colorectal cancer.
3. Results
A total of 201 patients with histopathologically confirmed colorectal cancer were included in this study. The median age of the patients was 43 (range = 10–85) years. Out of which 110 (54.7%) were males, and 91 (45.3%) were females. All patients underwent primary surgery without any prior intervention. 33 (16%) had radiotherapy postoperatively, while 95(47%) had adjuvant chemotherapy. Adenocarcinoma was the most commonly seen cancer (61.5%), followed by mucinous variety and a small proportion of other subtypes. Most of the cancers were well differentiated. A summary is presented in Table 2. Colon was involved in 119 (59.5%) including cecum (n = 6), ascending colon (n = 16), hepatic flexure (n = 15), transverse colon (n = 8), splenic flexure (n = 6), descending colon (n = 11), sigmoid colon (n = 31), recto-sigmoid junction (n = 26). Rectum was involved in 81 (40.5%) patients.
Table 2.
Clinical and biological characteristics of novel molecular classes of Colorectal cancer.
| Characteristics | CRC Novel Cluster 1 (n = 67) | CRC Novel Cluster 2 (n = 38) | CRC Novel Cluster 3 (n = 28) | CRC Novel Cluster 4 (n = 50) |
|---|---|---|---|---|
| Mean age in years (range) | 42.8 (10-78 years) | 48.6 (10-85 years) | 42.8 (12-65 years) | 37.6 (10-77 years) |
| Gender n(%) | ||||
| Male Females |
39(58.2) 28(41.8) |
22(57.9) 16(42.1) |
14(50.0) 14(50.0) |
27(54.0) 23(46.0) |
| Tumour location n(%) | ||||
| Right Colon Transverse Left colon Sigmoid Rectum Recto-sigmoid |
8(11.9) 4(6.0) 10(14.9) 10(14.9) 33(49.3) 8(11.9) |
9(24.3) 2(5.4) 4(10.8) 7(18.9) 11(29.7) 4(10.8) |
3(10.7) 2(7.1) 2(7.1) 4(14.3) 10(35.7) 7(25.0) |
11(22.0) 0 0 6(12.0) 26(52.0) 7(14.0) |
| Grade n(%) | ||||
| 1 | 19(28.4) | 12(32.4) | 11(39.3) | 12(24.5) |
| 2 | 31(46.3) | 20(54.1) | 13(46.4) | 25(51.0) |
| 3 | 17(25.4) | 5(13.5) | 4(14.3) | 12(24.5) |
| Lymphocytic infiltration n(%) | ||||
| Present | 52(77.6) | 32(84.2) | 26(92.9) | 36(72.0) |
| Not present | 15(22.4) | 6(15.8) | 2(7.1) | 14(28.0) |
| Median disease specific survival (months) | 30 | 25 | 26 | 23 |
3.1. Biological characteristics
CK20, CEA, and E-Cadherin were found positive in ∼99% of cases, while Ki67 was positive in 82%, CDX2 in 85% of patients, and p53 was positive in 43%. However, ER (3%), PR (2.5%), HER2 (4.5%), EGFR (8.5%), Bcl2 (2.5%), PDL1 (14.4%) and VEGF (5.5%) were found positive in a small proportion of patients (Table 2). Age between 51 and 75 showed a differing pattern in males and females where a higher rate of male patients of CRC was seen (Fig. 1a). With advancing age, poorly differentiated cancers appear to decline (Fig. 1b). Grade showed significant association with p53 positivity (Fig. 1c).
Fig. 1a.
Gender specific pattern of age groups of colorectal cancer.
Fig. 1b.
Age standardized pattern of histological grade in colorectal cancer.
Fig. 1c.
Association of histological grade and p53 in colorectal cancer.
3.2. Molecular classification
Four distinct molecular classes were identified (Fig. 2). The key differentiating markers were Ki67, CDX2 and p53. Cluster 4 was characterized by younger patients (mean age 37 years), while relatively older patients fell into cluster 2 (mean age 48 years). Clusters 3 and 4 were predominantly observed in the rectum and sigmoid region. Table 3 describes the characteristics of molecular classes.
Fig. 2a.
Biplots K-means of molecular classification of colorectal cancer. b. Novel molecular classification of colorectal cancer in Pakistani population c. Immunohistochemical pattern of novel molecular classification of colorectal cancer in Pakistani population (Magnification size −10× using Euromax Microscope with camera).
3.3. Survival
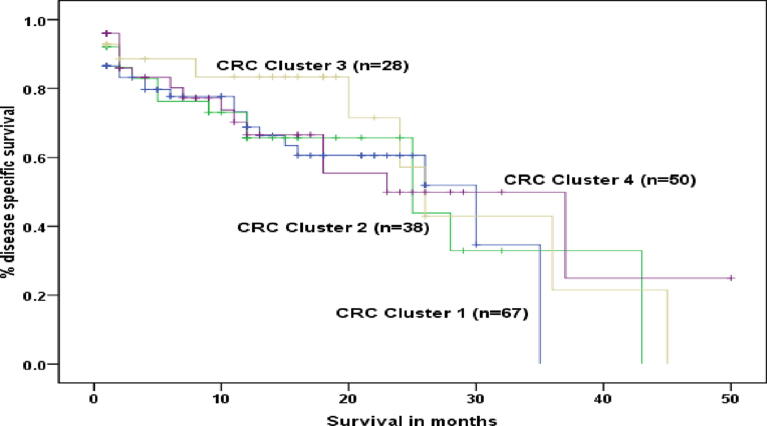
The median overall survival was 26 months, and disease-specific survival was 26 months. The longest survival was seen at 50 months in one patient. Younger age (<50) was significantly associated with poor disease-specific survival, and Ki67 positive status showed borderline significance (p = 0.06) with inferior disease-specific survival. Histological grade, nodal status, ER, PR, HER 2-neu, Bcl-2, E-Cadherin, P53, CEA, EGFR, VEGF, PDL1, CDX-2, and CK 20 did not show any association with survival (p-value > 0.05). Molecular classes did not show any significant association with clinical outcome (Fig. 3).
Fig. 3a.
Overall survival in patients with colorectal cancer. b. Disease specific survival in patients with colorectal cancer c. Disease specific survival of patients with colorectal cancer – a comparison of <50 and older then 50 years. d. Disease specific survival of patients with colorectal cancer – Comparison of novel molecular classes.
4. Discussion
The study presented a novel molecular classification of colorectal cancer where Ki67, CDX2, and p53 IHC expression plays a crucial role in partitioning. Overall survival was observed to be poor, and the same was observed with disease-specific survival. Our study showed that younger patients tend to have more age preponderance in clusters such that the mean age for cluster 4 was 37 years while for cluster 2 was 48 years. That was an interesting finding linked with younger age as a poor prognostic factor. This was previously reported in a study that included two age groups according to the age of onset of CRC. The classification was done based on MSI and BRAF mutation. More younger patients tend to have more left colon cancers and there was a comparison made based on a mutation in CpG island Methylator Phenotype (CIMP) (Perea et al., 2014). More younger patients tend to have CIMP- high type tumours with a mean age of 36 years, while combined classes showed MSI/CIMP- high type with a mean of 29 years (Perea et al., 2014). However, the clinical correlation was not reported in this study.
Another reported classification included the tumour microenvironment, and 167 gene signature identified four distinct molecular classes (Perez Villamil et al., 2012). This model of CRC classification microsatellite instability, histological types, high stromal content, β-Catenin, and BRAF significantly influenced hierarchical clustering. MSS subtypes, BRAF, and KRAS mutations were associated with the worst survival (Alwers et al., 2019).
A previously published IHC based molecular classification of CRC including four independent cohorts (including AMC-AJCC-II, LUMC, CAIRO, and CAIRO2) including CDX2, FRMD6, HTR2B, and ZEB1 by using IHC (Trinh et al., 2017) and validated the presence of four distinct molecular classes where there were two basic classes were seen as epithelial-like and mesenchymal-like. Another model analyzed gene patterns mainly looking at RAS where four molecular classes were reported with distinct response to FOLFIRI (Stintzing et al., 2019). However, a previously reported study included paraffin-embedded tissue sections and analyzed genes, and classification of CRC was made on the expression pattern of KRAS, BRAF, and CIMP, which reported five molecular classes. There was a significant association of change of pattern of CRC molecular classes with advancing age, gender, family history of CRC, and the tumour site (Phipps et al., 2015). Similar findings were reported when only stage 2 CRC was evaluated using consensus molecular classification (Purcell et al., 2019). The molecular classes showed a significant influence of age, gender, site of the tumour, and stage of the disease.
P53 is a tumour suppressor gene located on chromosome 17 (Nasierowska-Guttmejer et al., 2000). Its mutations have been linked with many cancers, including CRC. In our study, p53 appears to be a key governing factor in classifying molecular patterns. P53 mutation was previously associated with depressed neoplasms (DNs) more than other phenotypes (Konda et al., 2014). Similarly, p53 was also significantly associated with the worst prognosis (Xu et al., 2007). A study that used Polymerase Chain Reaction (PCR) for molecular classification of CRC showed that p53 mutation was associated with subclass 2 out of five subclasses (Sugai et al., 2017). P53 mutation was present in 44% of patients out of 753 tested CRC (Domingo et al., 2013). Thus our findings are consistent with already existing literature. Previously reported data on HER2 showed membranous expression in 3% (3 out of 100 cases), which is relatively consistent with our data where around 4% showed membranous expression (Osako et al., 1998).
Programmed Cell Death Protein-1 (PD-L1) has been associated with BRAF mutations and liked with poor differentiation (Azcue et al., 2021). High expression of PD-L1 was reported to be 20%, while low expression was 35%. It was significantly associated with consensus molecular classification over-expressed in CMS1 and under-expressed in CMS4. It was also reported to be concomitantly found with BRAF mutation (Shiovitz and Grady, 2015). Thus protein expression differentiating molecular classification can correspond to the BRAF mutant class. In our study, the novel molecular classification pattern was not directly dependent on PD-L1, but in novel CRC class 2, it was not positive in any patient.
Given the experience of breast cancer research, IHC based molecular classification can potentially play a significant role in identifying therapeutic targets and providing proper precision medicine to improve clinical outcome. As the incidence of CRC is feared to rise in upcoming years, it is of utmost importance to control disease survival.
This study was a single-center study with a consecutive series of CRC patients, including treatment naïve tumours; thus, a natural pattern of biomarkers without the interference of chemo and radiotherapy has been presented. The method of molecular classification using R software is also a well-established method in breast cancer, as reported previously. However, there are limited biomarkers, and smaller sample size is appreciated as a limitation of the study. Correlation with gene signature is also recommended in the future, given that the immunohistochemical classification did not show any significant association with survival outcome. However, it has pointed out distinct pathways of colorectal cancer that might have a relationship with the development of colorectal cancer and genetic relationships. Therefore, further studies on risk factors and genetic exploration of these pathways are recommended.
5. Conclusion
In conclusion, CRC is a heterogeneous disease with at least four distinct molecular types. Gene repair mechanism and cell proliferation markers (i.e., p53 and Ki67). Intestine-specific transcription factor (CDX2) has also shown an association with the molecular classification of CRC. Thus, this is now observed that multiple intracellular mechanisms are working together, taking part in tumorigenesis and disease progression. Other associated markers can be traced by following these key pathways.
Ethical consideration
The study was approved by the local research ethics committee of Liaquat University of Health & Sciences Jamshoro Sindh Pakistan under Ethical approval Number LUMHS/REC/641 dated 26-12-2017 The clinical data were retrospectively collected from the Institutional database and tumour blocks were retrieved from the Institutional tissue archive, where tissues are preserved for research purpose. The dataset includes cancer patients from 2008 and a considerable number is not alive; thus, informed consent from individual patients was waived.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Funding
The study was funded by the Higher Education Commission of Pakistan under NRPU 3883 to PI Binafsha Manzoor Syed and Co-PI Jawaid Naeem Qureshi. Fayaz Hussain Mangi worked as a Ph.D. scholar under the supervision of PI.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Globocan [Internet], 2020. 2020 is an online, for all cancer sites combined. <https://www.uicc.org/news/globocan-2020-new-global-cancer-data#:∼:text=GLOBOCAN>.
- Álvaro E., Cano J.M., García J.L., Brandáriz L., Olmedillas-López S., Arriba M., et al. Clinical and molecular comparative study of colorectal cancer based on age-of-onset and tumor location: two main criteria for subclassifying colorectal cancer. Int. J. Mol. Sci. 2019;20(4) doi: 10.3390/ijms20040968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwers E., Bläker H., Walter V., Jansen L., Kloor M., Arnold A., et al. External validation of molecular subtype classifications of colorectal cancer based on microsatellite instability, CIMP BRAF and KRAS. BMC Cancer. 2019;19(1):1–10. doi: 10.1186/s12885-019-5842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov L., Zhou J., Christopherson R.I. Cell surface markers in colorectal cancer prognosis. Int. J. Mol. Sci. 2011;12:78–113. doi: 10.3390/ijms12010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi far, S., Soltani, M., Zardast, M., Ghasemian Moghaddam, M.R., 2021. Investigating the factors associated with the level of expression of estrogen and progesterone receptors in patients suffering from colorectal cancer. In: Antwi, S. (Ed.), J Cancer Epidemiol [Internet]. 15, 1–6. <https://www.hindawi.com/journals/jce/2021/4478155/>. [DOI] [PMC free article] [PubMed]
- Qasim B.J. Immunohistochemical expression of estrogen and progesterone receptors in human colorectal adenoma and carcinoma using specified automated cellular image analysis system: a clinicopathological study. Oman Med. J. [Internet] 2011;26(5):307–314. doi: 10.5001/omj.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan R, Bhatt D, Khan S, Khan V, Verma AK, Anees A, et al. Association of Her-2 Expression and Clinicopathological Parameters in Colorectal Carcinoma in Indian Population. Open Access Maced J Med Sci [Internet]. 2018 Dec 22;7(1):6–11. Available from: https://spiroski.migration.publicknowledgeproject.org/index.php/mjms/article/view/oamjms.2019.008. [DOI] [PMC free article] [PubMed]
- Imaizumi K, Suzuki T, Kojima M, Shimomura M, Sakuyama N, Tsukada Y, et al. Ki67 expression and localization of T cells after neoadjuvant therapies as reliable predictive markers in rectal cancer. Cancer Sci [Internet]. 2020 Jan 18;111(1):23–35. Available from: https://onlinelibrary.wiley.com/doi/10.1111/cas.14223. [DOI] [PMC free article] [PubMed]
- Tukaram Patil S., Wilfred Devadass C., Shetty Badila P. Histopathological evaluation and analysis of immunohistochemical expression of bcl-2 oncoprotein in colorectal carcinoma. Iran J Pathol [Internet]. 2019;14(4):317–321. doi: 10.30699/ijp.2019.102982.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Yang Y., Byeon S., Jeong Y., Kwon J., Lee K.H., et al. E-cadherin and angiopoietin-2 as potential biomarkers for colorectal cancer with peritoneal carcinomatosis. Anticancer Res [Internet]. 2021;41(9):4497–4504. doi: 10.21873/anticanres.15260. [DOI] [PubMed] [Google Scholar]
- Lakpa S.C., Kumar R.V., Lilly M. Role of immunohistochemistry markers (p53 and CEA), in study of colorectal tumours. J. Pharm. Res. Int. [Internet] 2021:94–106. [Google Scholar]
- Uhlyarik A., Piurko V., Vizkeleti L., Pápai Z., Rásó E., Lahm E., et al. EGFR protein expression of KRAS wild-type colorectal cancer: predictive value of the sidedness for efficacy of anti-EGFR therapy. Pathol Oncol Res [Internet]. 2020;26(3):1429–1434. doi: 10.1007/s12253-018-00572-2. [DOI] [PubMed] [Google Scholar]
- Mohamed, S.Y., Mohammed, H.L., Ibrahim, H.M., Mohamed, E.M., Salah, M., 2019. Role of VEGF, CD105, and CD31 in the prognosis of colorectal cancer cases. J. Gastrointest Cancer [Internet] 50(1), 23–34. <https://link.springer.com/10.1007/s12029-017-0014-y>. [DOI] [PubMed]
- Pyo J-S, Ko SH, Ko YS, Kim NY. Clinicopathological significance of PD-L1 expression in colorectal cancer: Impact of PD-L1 expression on pFOXO1 expression. Pathol - Res Pract [Internet]. 2020 Feb;216(2):152764. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0344033819320047. [DOI] [PubMed]
- Aasebø, K., Dragomir, A., Sundström, M., Mezheyeuski, A., Edqvist, P.-H., Eide, G.E., et al., 2020. CDX2: A Prognostic Marker in Metastatic Colorectal Cancer Defining a Better BRAF Mutated and a Worse KRAS Mutated Subgroup. Front Oncol [Internet], 10. <https://www.frontiersin.org/article/10.3389/fonc.2020.00008/full>. [DOI] [PMC free article] [PubMed]
- Al-Maghrabi, J., Emam, E., Gomaa, W., 2018. Immunohistochemical staining of cytokeratin 20 and cytokeratin 7 in colorectal carcinomas: four different immunostaining profiles. Saudi J. Gastroenterol. [Internet] 24(2), 129. <http://www.saudijgastro.com/text.asp?2018/24/2/129/229487>. [DOI] [PMC free article] [PubMed]
- Howell A., Harland R.N.L., Bramwell V.H.C., Swindell R., Barnes D.M., Redford J., et al. Steroid-hormone receptors and survival after first relapse in breast cancer. Lancet. 1984;323(8377):588–591. doi: 10.1016/s0140-6736(84)90995-4. [DOI] [PubMed] [Google Scholar]
- Syed B.M., Green A.R., Paish E.C., Soria D., Garibaldi J., Morgan L., et al. Biology of primary breast cancer in older women treated by surgery: with correlation with long-term clinical outcome and comparison with their younger counterparts. Brit. J. Cancer. 2013;108(5) doi: 10.1038/bjc.2012.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- Perea, J., Rueda, D., Canal, A., Rodríguez, Y., Álvaro, E., Osorio, I., et al., 2014. Age at onset should be a major criterion for subclassification of colorectal cancer. J. Mol. Diagnos. [Internet] 16(1), 116–26. <http://dx.doi.org/10.1016/j.jmoldx.2013.07.010>. [DOI] [PubMed]
- Perez Villamil, B., Romera Lopez, A., Hernandez Prieto, S., Lopez Campos, G., Calles, A., Lopez Asenjo, J.A., et al., 2012. Colon cancer molecular subtypes identified by expression profiling and associated to stroma, mucinous type and different clinical behaviour, BMC Cancer [Internet] 12(1), 1. Available from: BMC Cancer. [DOI] [PMC free article] [PubMed]
- Alwers, E., Jia, M., Kloor, M., Bläker, H., Brenner, H., Hoffmeister, M., 2019. Associations between molecular classifications of colorectal cancer and patient survival: a systematic review, Clin. Gastroenterol. Hepatol. [Internet] 17(3), 402–410.e2. doi:10.1016/j.cgh.2017.12.038. [DOI] [PubMed]
- Trinh A., Trumpi K., De Sousa E., Melo F., Wang X., De Jong J.H., Fessler E., et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin. Cancer Res. 2017;23(2):387–398. doi: 10.1158/1078-0432.CCR-16-0680. [DOI] [PubMed] [Google Scholar]
- Stintzing S., Wirapati P., Lenz H.J., Neureiter D., Fischer von Weikersthal L., Decker T., et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019;30(11):1796–1803. doi: 10.1093/annonc/mdz387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps A.I., Limburg P.J., Baron J.A., Burnett-Hartman A.N., Weisenberger D.J., Laird P.W., et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148(1):77–87.e2. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell R.V., Schmeier S., Lau Y.C., Pearson J.F., Frizelle F.A. Molecular subtyping improves prognostication of Stage 2 colorectal cancer. BMC Cancer. 2019;19(1):1–9. doi: 10.1186/s12885-019-6327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasierowska-Guttmejer A., Trzeciak L., Nowacki M.P., Ostrowski J. P53 protein accumulation and P53 gene mutation in colorectal cancer. Pathol. Oncol. Res. 2000;6(4):275–279. doi: 10.1007/BF03187331. [DOI] [PubMed] [Google Scholar]
- Konda K., Konishi K., Yamochi T., Ito Y.M., Nozawa H., Tojo M., et al. Distinct molecular features of different macroscopic subtypes of colorectal neoplasms. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0103822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Wang F., Di M., Huang Q., Wang M., Hu H., et al. Classification based on the combination of molecular and pathologic predictors is superior to molecular classification on prognosis in colorectal carcinoma. Clin. Cancer Res. 2007;13(17):5082–5088. doi: 10.1158/1078-0432.CCR-07-0597. [DOI] [PubMed] [Google Scholar]
- Sugai T., Eizuka M., Takahashi Y., Fukagawa T., Habano W., Yamamoto E., et al. Molecular subtypes of colorectal cancers determined by PCR-based analysis. Cancer Sci. 2017;108(3):427–434. doi: 10.1111/cas.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Ramamoorthy R., Oukrif D., Rosmarin D., Presz M., Wang H., et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J. Pathol. 2013;229(3):441–448. doi: 10.1002/path.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osako T., Miyahara M., Uchino S., Inomata M., Kitano S., Kobayashi M. Immunohistochemical study of c-erbB-2 protein in colorectal cancer and the correlation with patient survival. Oncology. 1998;55(6):548–555. doi: 10.1159/000011911. [DOI] [PubMed] [Google Scholar]
- Azcue P., Encío I., Setas D.G., Alecha J.S., Galbete A., Mercado M., et al. Pd-l1 as a prognostic factor in early-stage colon carcinoma within the immunohistochemical molecular subtype classification. Cancers (Basel). 2021;13(8):1–18. doi: 10.3390/cancers13081943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiovitz S., Grady W.M. Molecular markers predictive of chemotherapy response in colorectal cancer. Curr. Gastroenterol. Rep. 2015;17(2):1–19. doi: 10.1007/s11894-015-0431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.