Abstract
Salinity is widespread environmental stress that poses great obstacles to rapeseed development and growth. Polyamines are key plant growth regulators that play a pivotal role in regulating salt tolerance. Rapeseed (Brassica napus L.) seedlings were treated by spermine (Spm) and spermidine (Spd) versus untreated control under salt stress conditions. It was detected that the Spd-treated plants had significantly elevated chlorophyll and proline content and maintained higher photosystem II (PSII) activity than those treated with Spm as well as untreated control under salt-stressed conditions. Similarly, Spd alleviated the devastating effects of NaCl stress on CO2 assimilation and significantly elevated Rubisco activity (ribulose 1,5-bisphosphate carboxylase/oxygenase). The application of Spd also enhanced the activities of different antioxidant enzymes under NaCl stress. It modulated their respective transcription levels, including ascorbate peroxidase (APX), catalase (CAT), superoxide dismutase (SOD), glutathione reductase (GR), and dehydroascorbate reductase (DHAR). In addition, exogenously sprayed Spd enhanced the polyamine pathway as observed by upregulated transcription of polyamine oxidase (PAO) and diamine oxidase (DAO). The Spd application enhanced expressions of Calvin cycle enzyme related genes such as Rubisco small subunit, Rubisco large subunit, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 3-phosphoglyceric acid kinase (PGK), triose-3-phosphate isomerase (TPI), fructose-1,6-bisphosphate aldolase (FBA), sedoheptulose-1,7-bisphosphatase (SBPase), and fructose-1,6-bisphosphate phosphatase (FBPase). Consequently, this study demonstrates that exogenous application of Spd has a valuable role in regulating antioxidant enzyme activity, polyamine pathway, and Calvin cycle enzyme-related genes to alleviate salt stress damage in the plants.
Keywords: Gene expression, Photosynthesis, Rapeseed, Salinity stress, Spermine, Spermidine
1. Introduction
Salinity is widespread environmental stress that destructively impacts geographical distribution and agriculture production (El-Mageed et al., 2021, FAOSTAT, 2021). Increased salt in soil or irrigation water decreases plant capacity of water take-up while increases absorption Na+ and Cl− (Deinlein et al., 2014, Mansour et al., 2020). Excessive salt accumulation in the root zone causes osmotic toxic stress and nutrient imbalance at plant cells (Baker and Rosenqvist, 2004, Desoky et al., 2021a). Subsequently, salt stress adversely affects cell elongation, metabolic process, and photosynthetic efficacy (Chen and Murata, 2011, Mansour et al., 2021a).
Rapeseed (Brassica napus L.) is an essential source for oil production worldwide (Raboanatahiry et al., 2021). Its oil has different health benefits as a result of containing oleic acid and linoleic acid (Piazza and Foglia, 2001). Rapeseed is moderately sensitive to salinity, and salt stress reduces tremendously its growth, especially in arid and semiarid regions (Musgrave, 2000). Thus, there is much focus on improving its growth under salinity stress (Munns, 2002, Moustafa et al., 2021a).
Enhancing seedling vigor is crucial for plant stand establishment and successful plant development particularly under stressful growing conditions (El-Sanatawy et al., 2021). Various efficient, cost-effective, and ecologically friendly strategies have been explored to mitigate the effects of environmental stresses, including salt, by plant growth regulators application, osmoprotectants, and different nutrition applications (Ahanger et al., 2017, Yakhin et al., 2017). The beneficial influences of growth regulators such as polyamines (PAs), amino acids, and phytohormones in mitigating against salinity stress on plants have been deduced in various studies (ElSayed et al., 2018).
Polyamines are polycationic low molecular weight compounds detected in all organisms and include spermine (Spm), putrescine (Put), and spermidine (Spd). Polyamines play an essential role at different development stages including cell division, nucleic acids stability, embryogenesis, dormancy termination, aging regulation, plant growth, and stress resistance (Todorova et al., 2007). Polyamines accumulation is essential in the plant reaction to salt stress and accordingly PAs has a decisive importance in salinity tolerance (Chen et al., 2019). Exogenous PAs of various types and concentrations displayed significant attenuation of the influences of salt stress in various crops and reduce resultant damage (Verma and Mishra, 2005). Spermine level in plant cells is an essential marker of salt tolerance (Liu et al., 2015). Exogenous PAs, particularly Spm and Spd, exhibited a considerable increase in scavenging the reactive oxygen species (ROS) and photosynthesis, improving plant growth, and decreasing the restrained impacts of salt stress (ElSayed et al., 2018). Overall, polyamines function as antioxidants, preventing oxidative damage of plant tissue and as a result of ROS damage and lipid peroxidation (Wu et al., 2018, Chen et al., 2019). The Calvin cycle, which is situated in the plastid stroma of photosynthetic eukaryotes, is a vital cycle of responses that capture the output of photosynthetic light reactions to fix CO2, which are then utilized for sucrose biosynthesis and starch (Quick and Neuhaus, 1997). There is currently little information in the literature about the potential mechanisms of how exogenous PAs alleviate salt stress inhibition of carbon assimilation in the plant.
This study aimed at investigating the effect of exogenous Spd and Spm on rapeseed plants grown under short-term NaCl stress (6 h) and long-term NaCl stress (15 days) by investigating the antioxidant defense, photosynthetic carbon assimilation, and the transcription of genes encoding enzymes in the Calvin cycle.
2. Materials and methods
2.1. Plant material and experimental treatments
Seeds of rapeseed (Brassica napus L. cv. Serw-4) were surface-sterilized for 10 min with a 3% (v/v) NaOCl solution, after that gently rinsed with deionized water several times. The seeds were then germinated on moist cotton bed at 24 °C in the culture room in the dark. The germinated seeds were placed in germination pots (7 × 8.5 × 10 cm) in perlite mixture and vermiculite and immersed in water for one week in darkness. After that, the seedlings were cultivated in a growth chamber with 16 and 8 hrs of light and dark photoperiod in the same order (450 μmol m−2 s−1 photosynthetic active radiation) at 25 °C and 20 °C of day and night and 60% relative humidity (RH) for another week. At the two true leaf stage (two weeks), seedlings were moved to a hydroponic box containing half-strength Hoagland's solution (Hoagland and Arnon, 1950). Seedlings with consistent growth were selected for the following treatments: control (half-strength Hoagland’s solution), 150 mM NaCl, 150 mM NaCl + 0.25 mM Spm and 150 mM NaCl + 0.25 mM Spd and began 14 days after rapeseed germination. NaCl was gradually added in increments of 50 mM per day to the 1/2 strength Hoagland's solution (to avoid osmotic shock) to the required level of 150 mM NaCl (this means all plants will receive the same concentration 150 mM NaCl). Each treatment was repeated thrice, and each replicate included eight plants. After 6 h and 15 days of treatment, (150 mM NaCl) samples of tissue were collected from the completely expanded third leaves and immediately placed in liquid nitrogen before being analyzed for each of the parameters measured.
2.2. Growth attributes
For determination of fresh mass (FM), rapeseed plants were weighed. The dray mass (DM) was recorded by drying samples in oven at 70 °C for 72 h and then were weighed.
2.3. Photosynthetic parameters
After 6 h and 15 days of NaCl treatment, leaves of rapeseed plants were collected, finely ground to powder with liquid nitrogen and extracted in acetone 80 (v/v). To make the chlorophyll extracts entirely clear, they were centrifuged for 3–5 min at 1000g. The absorption of chlorophyll a and b were assayed using spectrophotometric analysis of the resultant extracts Lichtenthaler (1987). The quantum yield (FV/FM) of PSII and CO2 fixation were assayed applying the photosynthetic analyzer (Li-6400, LI-COR BioSciences, Lincoln, NE, USA) (Lu et al., 2003). The CO2 assimilation rate was determined at a light intensity of 100 μmol/photons m2/s1, a RH of 50% and at 22 °C utilizing a portable gas exchange system (GFS-3000, Walz, Easton, MD, USA).
2.4. Proline content
Proline content in rapeseed leaves was measured after 6 h and 15 days of salt treatment (DAT) for stressed and control plants as outlined by Vicente et al. (2004). The proline content was determined utilizing L-proline as a standard. The amount of proline in the plant samples was measured in mol proline/g FW.
2.5. Estimation of lipid peroxidation and hydrogen peroxide (H2O2) concentrations
Lipid peroxidation was stated as thiobarbituric acid reactive substances (TBARS) content generated by the thiobarbituric acid (TBA) reaction as explained by Hodges et al. (1999). The TBARS (MDA) level was recorded as μmol mg−1 FW. Hydrogen peroxide concentrations were determined as outlined by Alexieva et al. (2001). The concentration of H2O2 was identified by comparing it to a standard calibration curve created earlier with varying H2O2 concentrations.
2.6. Antioxidant enzyme activities
Fresh leaves of rapeseed (0.5 g) were ground finely with liquid nitrogen and extracted with 5 ml reaction mixture containing 50 mM of a KP buffer, and 0.1 mM Na2-EDTA. The reaction mixture was centrifugated at 4 °C for 30 min at 20,000 g. The enzyme extract (supernatant) was applied to determine the activities of the antioxidant enzyme following Cakmak et al. (1993). Bradford method (Bradford, 1976) was applied to assay the soluble protein content. The variation in the absorbance of solution was reported at 560-nm for 1-min, and the activity of SOD was stated as unit mg−1 protein (the quantity of enzyme needed to restrain nitro blue tetrazolium (NBT) decrease by 50%). Catalase (CAT) was determined following Csiszár et al. (2007) by observing the decrease in the absorbance at 240-nm for 1-min, and due to degradation of H2O2.Ascorbate peroxidase (APX) activity was assessed following Hemeda and Klein (1990). The activity was measured spectrophotometrically at 470-nm using guaiaco1 and H2O2 as substrate and hydrogen donor. APX activity was stated as μmol AsA min−1 mg−1 protein. Glutathione reductase (GR) activity was assessed following Bela et al. (2018) by detecting the enzyme-dependent oxidation of nicotinamide-adenine dinucleotide phosphate (NADPH) by observing absorbance increment at 340 nm. The glutathione reductase (GR) activity was expressed as the quantity of enzyme that oxidized 1 μmol NADPH min−1.
2.7. Analysis of Rubisco (ribulose 1,5 biphosphate carboxylase/oxygenase)
The activity of Rubisco was determined following Wang et al. (2009). A spectrophotometer was utilized to determine the enzyme activity by recording a decrease in absorbance at 340-nm for 3-min.
2.8. Quantitative RT-PCR analysis
Genomic RNA was extracted from rapeseed plants at each stress time point (6 h and 15 DAT of NaCl treatment) using ZR-Plant RNA MiniPrep™ Kit (Zymo Research, Irvine, CA. USA). The complementary DNA (cDNA) was applied utilizing RevertAid H-Minus First Strand cDNA Synthesis Kit (Thermo-Fisher-Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Determining transcription levels of antioxidant-related genes were performed using qRT–PCR, which applied on the iCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA). The iQ SYBR Green Supermix (Bio-Rad) was used following the manufacturer’s instructions. The PCR conditions were applied according to ElSayed et al. (2021). The actin gene was chosen as an internal control for data normalization. The quantification of relative gene transcription was calculated by the 2−ΔΔCT method (Pfaffl, 2001).
2.9. Statistical analysis
The data were analyzed using two-way ANOVA, and the least significant difference (LSD) test was calculated at the 1% significance level to determine the difference between treatments. All experiments described were repeated three times. To study the differences and interrelations between stressed and untreated control concerning antioxidant enzyme activities and their corresponding gene transcript levels and transcript level for Calvin cycle-related genes, Principal Component Analysis (PCA) was applied using R statistical software version 4.1.1.
3. Results
3.1. Growth rate and photosynthesis parameters
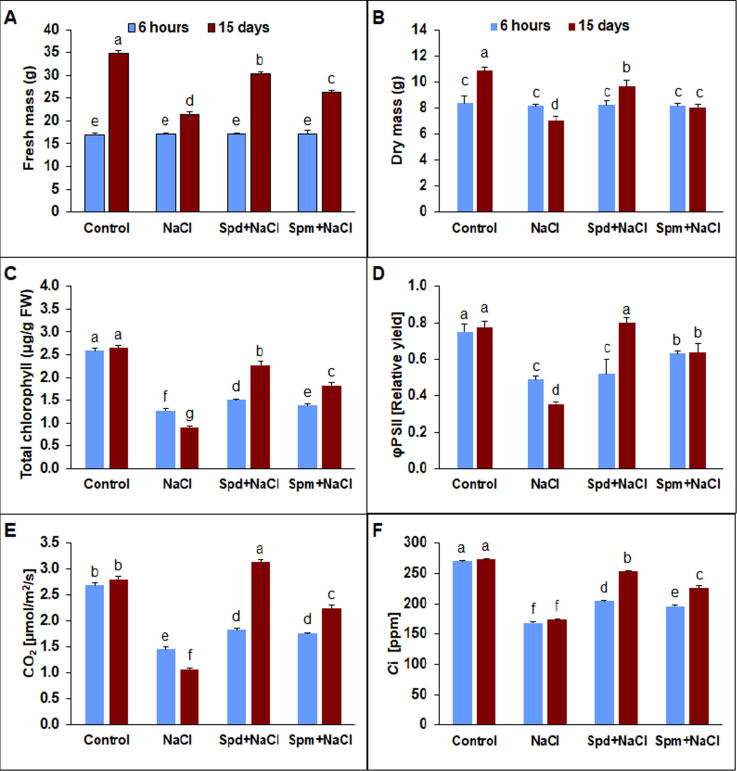
NaCl stress negatively impacted the development and growth of rapeseed plants. After 15 days of salt treatment (DAT) the fresh mass (FM) and dry mass (DM) were significantly declined compared with non-stressed control (Fig. 1A-B). However, the applications of Spm or Spd improved salt-stress inhibition of rapeseed growth at 15 DAT. The application of Spd and Spm enhanced FM by 41.8% and 23.2 as well as DM by 137.4 and 65.7%, in the same order compared to untreated plants under salt stress. Accordingly, alleviation of FM and DM was considerably more pronounced with exogenous Spd application compared to Spm treatment at 15 DAT (Fig. 1A-B).
Fig. 1.
Growth rate and photosynthesis parameters. A) Fresh mass (FM), B) Dry mass (DM), C) Total chlorophyll, D) Photosynthetic quantum yield (φPSII), E) Net photosynthetic rate, and F) intercellular CO2 concentration (Ci) in rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. The bars on the columns correspond to the SE, and distinct letters differ significantly by LSD (p < 0.01).
Total chlorophyll content, photosynthetic quantum yield, CO2 assimilation rate, and intercellular concentration (Ci) significantly reduced under NaCl stress compared with non-stressed control (Fig. 1C-F). In contrast, the applications of Spd and Spm improved total chlorophyll content in rapeseed plants grown under NaCl stress by 279.8 and 150.2%, respectively, compared to untreated control under salt stress at 15 DAT (Fig. 1C). Likewise, the applied exogenously Spd and Spm promoted the photosynthetic quantum yield of rapeseed plants under salt stress by 153.8 and 79.8%, respectively, compared to untreated stressed plants (Fig. 1D). Both CO2 assimilation rate and intercellular concentration (Ci) showed a significant (P < 0.01) decrease under NaCl stress compared to non-stressed conditions (Fig. 1E-F). The treated plants with Spd exhibited a substantial increase in the assimilation rate in particular at 15 DAT compared to untreated plants under salt stress (Fig. 1E). Moreover, exogenously sprayed Spd exhibited an increase in Ci in salt-stressed plants compared with untreated treatment under salt stress (Fig. 1F).
3.2. Rubisco activity and transcript level of RbcL and RbcS genes
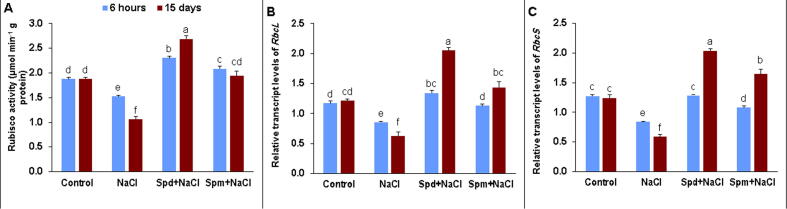
Rubisco activity substantially declined under salt stress at both time points compared to the non-stressed control (Fig. 2A). In contrast, the exogenous application of Spd and Spm substantially boosted Rubisco activity by 51.2 and 36.6% at 6 h and 153.4 and 82.6% at 15 DAT, in the same order, compared with untreated treatment under salt stress (Fig. 2A). Therefore, salt-stressed plants treated by exogenous Spd showed the highest Rubisco activity, particularly at 15 DAT. RbcL and RbcS transcript levels were significantly decreased in rapeseed plants exposed to NaCl at both time points compared with non-stressed ones (Fig. 2B-C). Notwithstanding, Spd and Spm applications increased significantly gene expression of RbcL and RbcS genes with superiority of the application of Spd at 15 DAT.
Fig. 2.
Rubisco activity and transcript level of RbcL and RbcS Genes. A) Rubisco activity, B) Transcript amounts of RbcL, and C) Transcript amounts of RbcS genes in rapeseed seed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. qPCR experiments were repeated thrice with two replications each to quantify transcript levels. Transcript levels were normalized to actin and GAPDH transcript levels. The bars on the columns correspond to the SE, and distinct letters differ significantly by LSD (p < 0.01).
3.3. Malondialdehyde, proline, and H2O2 contents
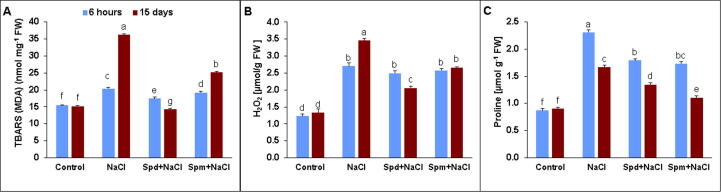
Thiobarbituric acid reactive substances (TBARS) amount was used to determine membrane damage resulted from lipid peroxidation in the leaves of rapeseed plants (Fig. 3A). Salt stress significantly increased the amount of TBARS (MDA) compared with non-stressed control (Fig. 3A). However, the applications of Spd and Spm considerably diminished TBARS (MDA) level in salt-stressed plants by 60.7 and 30.6%, respectively, compared with untreated ones under salt stress at 15 DAT. The H2O2 content of rapeseed leaves was considerably elevated at both time points in salt-stressed plants compared to non-stressed ones (Fig. 3B). However, the exogenous applications of Spd and Spm reduced H2O2 content in salt-stressed plants by 40.6% and 23.1%, respectively, compared with untreated ones under salt stress (Fig. 3B). The proline content increased significantly in rapeseed leaves of salt-stressed rapeseed plants compared with non-stressed plants (Fig. 3C). Proline content was relatively decreased at 15 DAT compared to 6 h in untreated stressed plants. Applications of exogenous Spd and Spm for salt-stressed plants declined accumulation in proline content by 22.5 and 25.0% at 6 h and 19.1 and 33.5% at 15 DAT, in the same order, compared with untreated ones under salt stress (Fig. 3C). Hence, Spm application exhibited the highest decline in the amount of proline content at 15 DAT compared to Spd as well as untreated plants (Fig. 3C).
Fig. 3.
Malondialdehyde, H2O2, and Proline Contents. A) Thiobarbituric acid reactive substances (TBARS) (MDA), B) H2O2 and C) Proline contents in leaves of rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. The bars on the columns correspond to the SE, and distinct letters differ substantially by LSD (p < 0.01).
3.4. Antioxidant enzymes activities
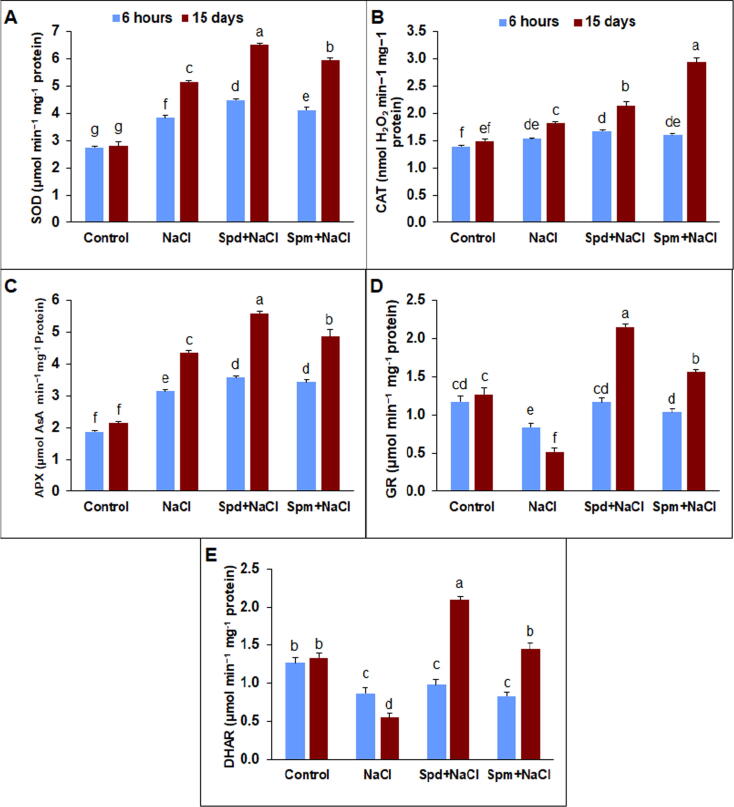
Salinity caused oxidative stress and thereby increased significantly the activity of antioxidant enzymes comprising of ascorbate peroxidase (APX), catalase (CAT), and superoxide dismutase (SOD), in salt-stressed rapeseed plants compared to non-stressed control (Fig. 4). The applications of Spd and Spm enhanced SOD activity by 16.8 and 7.3% at 6 h and 26.4 and 15.1% at 15 DAT, in the same order, compared to untreated control under salt stress (Fig. 4A). Likewise, the exogenous applications of Psd and Spm exhibited a significant enhancement in CAT activity by 17.4 and 61.5%, respectively, compared with untreated stressed rapeseed plants (Fig. 4B). There was a substantial boost in APX activity in NaCl-stressed plants compared with non-stressed control (Fig. 4C). Otherwise, the foliar-supplied Spd and Spm boosted the APX activity in salt-stressed plants by 28.4 and 11.7%, respectively, compared with untreated stressed plants. Glutathione reductase (GR) activity was inhibited substantially under salt stress compared to non-stressed control (Fig. 4D). The applications of Spd and Spm considerably increased GR activity by 39.5 and 23.6% at 6 h and 316.1 and 202.0% at 15 DAT, in the same order, compared with untreated stressed plants. Dehydroascorbate reductase (DHAR) activity was considerably reduced under salt stress compared with non-tressed plants (Fig. 4E). Both Spd and Spm applications enhanced DHAR activity in salt-stressed plants by 280.6 and 162.7%, respectively compared to untreated salt-stressed ones at 15 DAT.
Fig. 4.
The activities of antioxidant enzymes in rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. A) superoxide dismutase (SOD); B) catalase (CAT); C) ascorbate peroxidase (APX); D) glutathione reductase (GR); and E) dehydroascorbate reductase (DHAR). The bars on the columns correspond to the SE, and distinct letters differ substantially by LSD (p < 0.01).
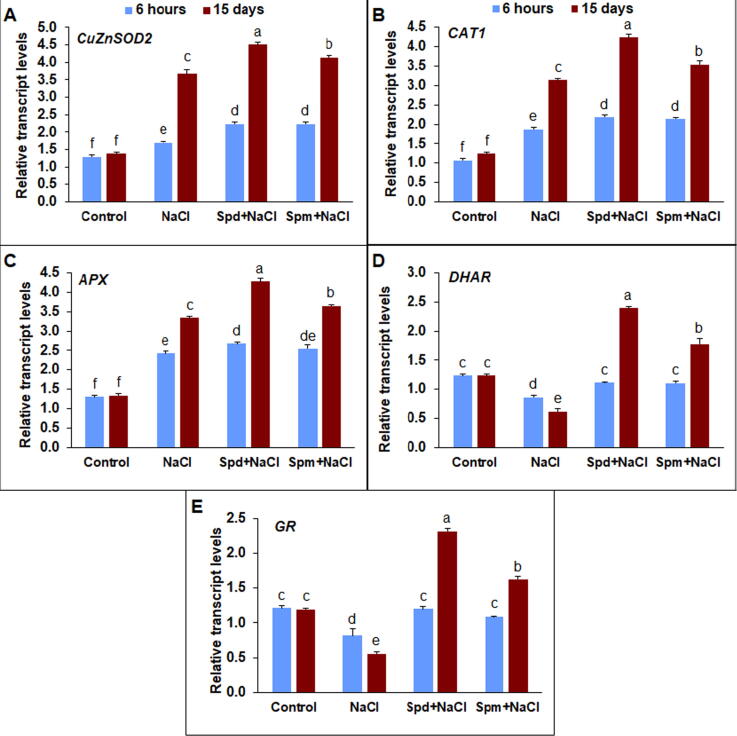
3.5. Transcript levels of antioxidant enzyme related genes
The detoxification reaction in rapeseed plants under salt stress either with or without application of Spd or Spm was evaluated by assessing the expression of CuZnSOD2, CAT1, APX, DHAR, and GR genes (Fig. 5). There was a substantial increase in CuZnSOD2 transcript level in salt-stressed rapeseed plants, in particular at 15 DAT compared with non-stressed control (Fig. 5A). Otherwise, the applications of Spm and Spd stimulated the transcript level of CuZnSOD2 by 23.1 and 13.0%, respectively, compared to untreated plants under salt stress at 15 DAT (Fig. 5A). CAT1 expression was also significantly upregulated in salt-stressed compared with non-stressed control (Fig. 5B). Exogenous applications of Spd and Spm induced considerable enhancement in the expression of CAT1 gene by 34.9 and 12.9%, respectively, compared with untreated plants at 15 DAT (Fig. 5B). Regarding APX transcript level, similar up-regulation trends were detected in salt-stressed rapeseed plants compared to non-stressed control (Fig. 5C). The applications of Spd and Spm enhanced APX transcript level by 28.2 and 9.4%, respectively, compared to untreated plants at 15 DAT. Under salt stress transcript level of DHAR gene transcript significantly declined compared to non-stressed control (Fig. 5D). The applications of Spd and Spm reinforced the transcript level of DHAR by 288.6 and 188.0%, respectively, in comparison with untreated stressed plants. Salt stress declined GR transcript levels compared to non-stressed control (Fig. 5E). The applications of Spd and Spm boosted the transcript level of GR by 317.2 and 192.5%, respectively, compared to untreated plants under salt stress at 15 DAT.
Fig. 5.
Transcript levels of antioxidant enzymes encoding genes in leaves of rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. A) CuZnSOD2, B) CAT1, C) APX, D) DHAR, and E) GR. Transcript levels were determined by qPCR and standardized against actin and GAPDH transcript levels. The experiments of qPCR were replicated three times with two technical replications. The bars on the columns correspond to the SE, and distinct letters differ substantially by LSD (p < 0.01).
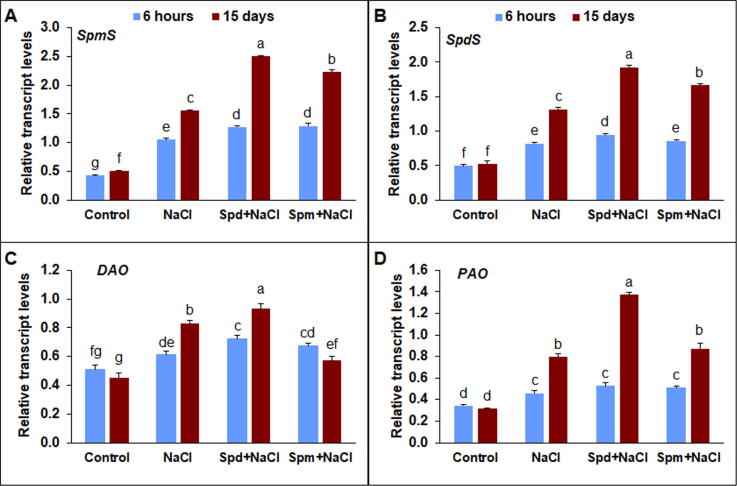
3.6. Transcript levels of polyamine synthase and polyamine pathway catabolic related genes
In rapeseed plants subjected to NaCl stress, the expression of spermidine synthase (SpdS) and spermine synthase (SpmS) significantly improved (Fig. 6A-B). Nevertheless, exogenous Spd application significantly enhanced the transcript level of SpdS and SpmS by 46.5 and 60.9% compared to NaCl-stressed untreated treatment (Fig. 6A-B). Similarly, Spm application significantly boosted the transcript level of SpdS and SpmS by 26.8 and 43.3% compared to stressed untreated plants. NaCl-stressed plants treated with Spd at 15 DAT showed a significantly higher transcript level of diamine oxidase (DAO) by 12.0% compared to stressed untreated plants (Fig. 6C). The transcript level of polyamine oxidase (PAO) improved under salt stress in comparison with the non-stressed control (Fig. 6D). Exogenous Spd application had considerable amelioration in PAO expression by 72.5 at 15 DAT compared to stressed untreated plants. On the other hand, exogenous Spm application displayed a similar performance to salt-stressed untreated treatment (Fig. 6D).
Fig. 6.
Expression of polyamine biosynthesis-related genes and polyamine pathway catabolic-related genes. A) spermine synthase (SpmS), B) spermidine synthase (SpdS), C) diamine oxidase (DAO), and D) polyamine oxidase (PAO) in rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. Transcript levels were determined by qPCR and normalized against actin and GAPDH transcript levels. The qPCR experiments were repeated three times with two technical replications. The bars on the columns correspond to the SE, and distinct letters differ substantially by LSD (p < 0.01).
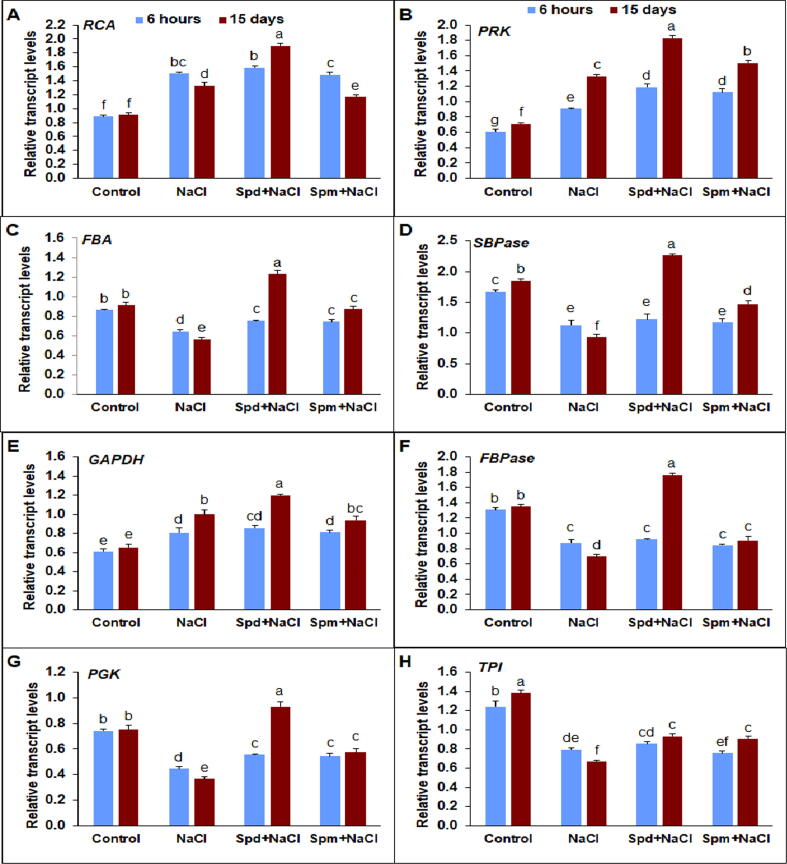
3.7. Transcript levels of Calvin cycle related genes
Transcriptions of eight key genes involved in carbohydrate metabolism associated with the Calvin cycle considerably differed by treatments of NaCl and Spd or Spm (Fig. 7A-H). The transcript levels of ribulose-bisphosphate carboxylase/oxygenase activase (RCA) increased significantly in salt-stressed rapeseed plants compared with non-stressed ones (Fig. 7A). Applied exogenously Spd and Spm increased transcription of RCA with superiority of Spd at 15 DAT with an increase by 42.8% compared to untreated stressed plants. The level of transcription of ribulose-5-phosphate kinase (PRK) also improved in stressed plants compared with non-stressed control (Fig. 7B). The Spd and Spm applications displayed substantial enhancement in the transcription level of PRK by 37.5 and 13.0 compared to untreated stressed plants. The transcription levels of fructose-1,6-bisphosphate aldolase (FBA) were significantly decreased in untreated plants under salt stress at both time points compared to the non-stressed control (Fig. 7C). Exogenous Spd application displayed the highest level of FBA transcription at 15 DAT. The transcription level of sedoheptulose-1,7-bisphosphatase (SBPase) gene significantly decreased in untreated stressed plants compared to non-stressed control (Fig. 7D). The Spd and Spm applications improved SBPase by 143.0 and 57.9%, respectively, compared to untreated stressed plants at 15 DAT. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was up-regulated under salt stress treatment compared to the non-stressed control (Fig. 7E). Spd application boosted the relative transcript level of GAPDH by 20.0% compared to untreated stressed plants. The transcription level of fructose-l,6-bisphosphate phosphatase (FBPase) considerably diminished in salt-stressed compared with non-stressed control (Fig. 7F). The Spd and Spm applications stimulated relative transcription of FBPase by 152.6 and 29.6%, respectively, compared to untreated stressed plants at 15 DAT. The relative transcript level of 3-phosphoglyceric acid kinase (PGK) transcript level decreased significantly in comparison with the non-stressed control (Fig. 7G). Exogenous applications of Spd and Spm enhanced relative PGK transcript level by 155.0 and 56.6% in the same order, compared to untreated stressed plants at 15 DAT. Under salt stress, the rapeseed plants had considerably decreased relative transcription levels for triose-3-phosphate isomerase (TPI) at both time points compared with non-stressed control (Fig. 7H). The applications of Spd and Spm enhanced transcription of TPI gene by 40.0 and 36.1%, respectively, compared to untreated plants under salt stress at 15 DAT.
Fig. 7.
Transcriptions of key genes involved in carbohydrate metabolism related to the Calvin cycle in rapeseed plants treated with 150 mM NaCl, 0.25 mM Spd + 150 mM NaCl or 0.25 mM Spm + 150 mM NaCl compared to untreated plants. A) ribulose-bisphosphate carboxylase/oxygenase activase (RCA), B) ribulose-5-phosphate kinase (PRK), C) fructose-1,6-bisphosphate aldolase (FBA), D) sedoheptulose-1,7-bisphosphatase (SBPase), E) glyceraldehyde-3-phosphate dehydrogenase (GAPDH), F) fructose-l,6-bisphosphate phosphatase (FBPase), G) 3-phosphoglyceric acid kinase (PGK), and H) triose-3-phosphate isomerase (TPI). Transcript levels were determined by qPCR and standardized against actin and GAPDH transcript levels. The experiments of qPCR were replicated thrice with two replications each. The bars on the columns correspond to the SE, and distinct letters differ substantially by LSD (p < 0.01).
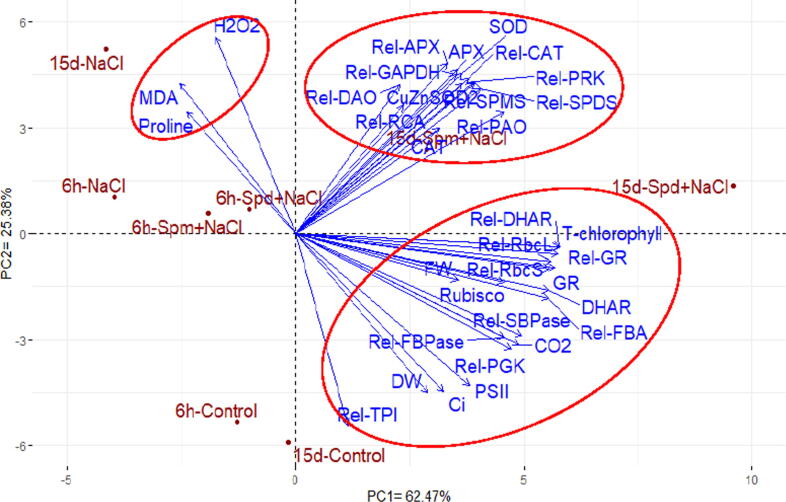
3.8. Interrelationship among evaluated characters
Principal components estimated the interrelationships among evaluated parameters under different treatments. The first two principal components presented about 87.85 of variability (62.47% by PCA1 and 25.38% by PCA2). Consequently, the first two principal components were applied to perform the biplot (Fig. 8). The parameters that have parallel vectors or are close to each other display a robust positive association, while those are situated near opposite demonstrate a negative relationship. The evaluated parameters could be separated into three groups. The first group included fresh mass, dry mass, total chlorophyll, φPSII, CO2, Ci, GR, DHAR, relative transcript level of DHAR, the relative transcript level of GR, Rubisco activity, the relative transcript level of RbcL, RbcS, TPI, PGK, SBPase, FBPase, and FBA genes. The second group contained CAT1, SOD, APX, the relative transcript level of CuZnSOD2, CAT1, APX, SPMS SPDS DAO PAO PRK RCA, and GAPDH genes. The third group comprised proline content, TBARS (MDA), and H2O2. A strong positive correlation was observed among parameters included in each group. Additionally, a positive relationship was noticed between the first and second groups, while a negative relationship was realized between the first and third groups.
Fig. 8.
PC-biplot based on physiological and molecular parameters of salt-stressed rapeseed plants treated with NaCl versus non-stressed conditions, compared with exogenously sprayed Spd and Spm.
The salt-stressed plants treated with exogenous Spd application associated with the first group, including plant biomass (FM and DM), photosynthesis parameters (chlorophyll content, ɸPSII, CO2 assimilation rate, Ci, Rubisco activity) and DHAR, GR activity, and their gene expression. Likewise, stressed plants treated with exogenous Spm associated with the second group containing antioxidant enzyme (CAT, APX, and SOD) activity and their gene expression, polyamine synthase, and catabolic related genes. Conversely, the third group includes proline content, H2O2, and TBARS (MDA) associated with salt-stressed plants without Spd and Spm treatments.
The PC1 seems to correspond with applications of Spd and Spm, untreated treatment is situated on the negative side of PC1 followed by application of Spm, and Spd is located on the end positive side of PC1. Thereby, PC biplot reinforced the obtained aforementioned results that Spd exhibited the highest enhancement for rapeseed plants under salt stress conditions.
4. Discussion
Salinity is estimated to affect approximately half of the world's irrigated lands and 20% of its cultivated lands (Kamboj et al., 2015). There is a critical need to develop salt-tolerant genotypes as well as find sustainable strategies to alleviate the diverse impact of salt stress to ensure global food security (Moustafa et al., 2021b). Furthermore, better understanding tolerance mechanisms and identifying important and reliable biochemical and physiological parameters associated with salt tolerance is required (ElSayed et al., 2018, Desoky et al., 2021b). The photosynthesis, CO2 assimilation and plant growth are frequently hampered by salt stress (ElSayed et al., 2021, Selem et al., 2022). In the current study, salinity decreased biomass, total chlorophyll, CO2 assimilation rate, and PSII (ΦPSII) in rapeseed plants (Fig. 1). Reduction of chlorophyll content and damage of photosynthesis activity are crucial factors leading to a lower photosynthetic capacity. Consequently, the current study hypothesized that one of the potential mechanisms responsible for low Chl content might be a stomatal limitation associated with a reduction in the CO2 assimilation in the cellular leaf space (Ci) (Praxedes et al., 2006). The applied-foliar of Spd significantly boosted Chl content and PSII in salt-stressed rapeseed plants, thus enhancing plant growth under stressed conditions compared with untreated plants under salt stressed conditions (Fig. 1). The increased photosynthesis could be ascribed to Spd's capability to increase stomatal conductance and thus CO2 concentration in plant cells (Shu et al., 2014). Moreover, Spd prevented degradation of Chl in salt-stressed plants and protected the PSII's integrity (Tang et al., 2018). These findings were confirmed by the Rubisco activity which increased in treated plants by application of Spd or Spm under salt stress compared with untreated ones (Fig. 2A). Rubisco activity reached its peak by using Spd application, implying its ability in avoiding inhibition of Rubisco activity in rapeseed plants in stressed environments.
Salinity can influence the photosynthesis process by stomatal constraints and lead to a reduction in carbon assimilation (Hernández and Almansa, 2002). Salt exposure can cause rapid termination of plant growth, even after a short period (Parida and Das, 2005). The accumulation of salts in the young leaves also affects the photosynthetic process in the long run (Munns and Tester, 2008) and even halophyte plants exhibited a reduction in chlorophyll and carotenoid content (Stepien and Johnson, 2009). The photosynthesis rate (Pn) could be declined by stomatal closure, or further non-stomatal restrictions, as suppression of the Calvin Cycle enzymes like phosphoenolpyruvate carboxylase (PECP), Rubisco, ribulose-5-phosphate glyceraldehyde-3-phosphate dehydrogenase, kinase, or fructose-1,6-bisphosphatase (Chaves et al., 2009).
Spermidine has a decisive role in attenuating the deleterious impacts of environmental stress in the plants (Kasukabe et al., 2006) by protecting the plasma membrane and membrane structure (Roy et al., 2005, Stassinos et al., 2021), suppressing RNase and protease activity (Chattopadhayay et al., 2002). Moreover, Kasukabe et al., 2004, Lu et al., 2009, deduced that restriction in Rubisco activity is one of the essential biochemical constraints engaged in salt-related photosynthesis downregulation. In the present study, salt stress treatment declined RbcL and RbcS gene expression. Still, the decrease was mitigated by the applications of Spd or Spm, with the highest expression of both genes was assigned for Spd application (Fig. 2B-C). As a result, we suggest that applied exogenously Spd adjusts the expression of RbcL and RbcS genes which could additionally influence the function and structure of Rubisco (Spreitzer, 2003).
Our findings showed that TBARS (MDA) and H2O2 levels increased in rapeseed plants subjected to salt stress. While, TBARS (MDA) and H2O2 were significantly lower in treated plants with Spd and Spm under salt stress conditions (Fig. 3A-B). This suggests that Spm and Spd are likely important in ROS scavenging and inhibiting lipid peroxidation under salt stress. Furthermore, Tang and Newton, 2005, Hsu and Kao, 2007 proposed that using Spm could facilitate plasmalemma stabilization in the plant and diminish oxidative stress. In this context, Kasukabe et al. (2004) suggested that Spd has an important role in stress signaling pathways. Proline accumulation is a common induced adaptive reaction in stressed plants which can help in sustaining photosynthetic efficiency (Duan et al., 2008). Furthermore, besides its osmoprotective role, proline has antioxidant properties and it protects macromolecules against oxidation during dehydration which ameliorates plant tolerance to environmental stresses (Tang et al., 2015). In accordance with the current study, rapeseed plants under salt stress accumulated significant amounts of proline compared to non-stressed plants (Fig. 3). Conversely, there was significantly less increase in proline level in treated rapeseed plants with both applications of Spd and Spm compared with untreated plants under salt-stressed conditions (Fig. 3C). The proline content was considerably higher in treated plants with exogenous Spd and Spm than non-stressed plants under salt stress. Within the plant cell, the proline performs many physiological and biochemical functions. Among these functions, it reduces osmotic potential and toxic ion uptake, which it does as an osmolyte and a ROS scavenger (Woodward and Bennett, 2005, El-Badri et al., 2021a).
In stressed plants, oxidative damage caused by ROS is mitigated by the antioxidant system that comprises antioxidant enzymes which scavenge ROS (Foyer and Noctor, 2011, Mansour et al., 2021b). The obtained findings revealed salt stress enhanced SOD, CATAPX, DHAR, and GR activities in rapeseed plants compared with non-stressed control (Fig. 4A, 4B, 4C). Similarly, previous studies disclosed a significant improvement in the antioxidant activities in Brassica napus (El-Badri et al., 2021b, Stassinos et al., 2021), Oryza sativa (Khan and Panda, 2008), Triticum aestivum (ElSayed et al., 2018), and Phaseolus vulgaris (ElSayed et al., 2021). The antioxidant enzymes are considered as the main scavenging components in regulating oxidative stress by adjusting the concentrations of H2O2 and O2•− during stressful growing conditions (Mishra et al., 2013, Desoky et al., 2021c). Besides scavenging O2•− radicals, they maintain the cell membrane from damage (Alscher et al., 2002, Sattar et al., 2021). Accordingly, their increased activities are adaptive traits and possibly have a considerable role in the repair of tissue by lowering toxic levels of H2O2 (Vital et al., 2008). Consequently, high antioxidant enzyme activity has an integral role in cell defense against ROS and plant salt tolerance. The application of Spm and Spd enhanced significantly CAT, APX, SOD, DHAR, and GR activities compared to untreated stressed plants (Fig. 4A, 4B, 4C). The treated plants with Spd experienced less ROS-caused oxidative stress, which could be caused by upregulated activities of the antioxidant enzymes for scavenging H2O2. The endo/exogenous increment of Spd, regulates salt-induced oxidative stress by increasing plants resistance, which includes a reduction in H2O2 production, activation of antioxidative defense system that led to an amelioration of free radical content and improvement in cell viability (Ebeed et al., 2017). Likewise, cucumber plants treated with Spd substantially boosted the content of Spd and Spm, which resulted in enhanced antioxidant enzyme activity, improved ROS scavenging capability, and decreased membrane lipid peroxidation that eventually led to improved stress tolerance (Wu et al., 2018).
The obtained findings of qRT-PCR revealed that the application of Spm and Spd increased gene expression of CuZnSOD2, CAT1, APX, DAHR, and GR in salt-stressed rapeseed plants, especially at 15 DAT. There was a consistent pattern in the observed changes for the transcript levels of genes in rapeseed plants and antioxidant activities. Moreover, salt-stressed rapeseed plants treated with Spd and Spm showed increased relative transcription levels for SpmS and SpdS (Fig. 6) which are part of the polyamine biosynthesis pathway. Furthermore, the application of Spd increased DAO and PAO transcription levels, particularly at 15 DAT (Fig. 6). According to Eller et al. (2006), catabolic enzymes such as PAO and DAO are localized in the plant cell walls, and H2O2 produced by polyamines catabolism may has substantial importance in cross-linking responses in stressed and non-stressed plants. Furthermore, Δ1-pyrroline is produced by polyamine degradation through PAO and DAO enzymes. Δ1-pyrroline could be catabolised to γ-aminobutyric acid, which induced by salt stress, then transaminated to succinate before being introduced into the Krebs cycle (Wang et al., 2017). Polyamine exogenous application increases endogenous polyamines level, positive effects related to maintaining membrane integrity; gene expression regulation for the synthesis of osmotic solutes; while decreases ROS production; and limit accumulation of Na+ and Cl− ion in different plant organs (Yiu et al., 2009).
Calvin cycle enzymes play a crucial role in enhancing photosynthetic efficiency by producing mediates for glycolysis and/or developing blocks for cellular elements (Furbank and Taylor, 1995, Raines, 2003, Uematsu et al., 2012). The DNA of all Calvin cycle enzymes has been isolated and sequenced. The Calvin cycle is introduced by Rubisco which catalyzes the carboxylation of the CO2 acceptor molecule. In the present study, the applied-foliar Spd considerably improved the activity of Rubisco in NaCl-stressed rapeseed plants. Based on these results, it appears that Spd hastened carbon assimilation through the Calvin cycle, boosted photosynthetic efficiency, which enhanced plant withstand against salt stress (Shu et al., 2014). Calvin cycle consists of 11 different enzymes that catalyze 13 different reactions (Raines, 2003). Rubisco, SBPase, FBPase, and PRKase are key enzymes in the Calvin cycle. These enzymes were probable to be the most important in regulating the rate of CO2 fixation (Raines, 2003). Eight genes related to enzymes engaged in the Calvin cycle were assessed in rapeseed plants after the application of Spm and Spd under salt-stressed conditions. The expressions of PGK, FBA, FBPase, SBPase, and TPI genes were significantly decreased under NaCl stress, although the application of Spd boosted the relative gene transcription level, particularly at 15 DAT (Fig. 7, Fig. 8). In contrast, NaCl stress increased transcription of the GAPDH, RCA, and PRK genes. Burzyński and Żurek (2007) proposed that GAPDH, RCA, and PRK enzymes are required to reduce the gene expression of PGK, FBA, FBPase, SBPase, and TPI indicating that salinity stress affected carbon assimilation (Shu et al., 2014). Nevertheless, the application of Spd attenuated the downregulation of these genes. According to Lv et al. (2017) improving the activity of FBA could increase CO2 assimilation in plant leaf tissues. Furthermore, the FBA gene family is important in plant reactions to environmental stresses such as salt, heat, drought, and low temperature (Lv et al., 2017). Koßmann et al. (1994) manifested that FBPase has a decisive function in the Calvin cycle and transport of photosynthetic products. Furthermore, Miyagawa et al. (2001) elucidated that FBP/ SBPase-overexpressing accumulates more sucrose, hexose, and starch. Also, Tamoi et al. (2006) depicted that increasing the activity of SBPase in tobacco overexpressing improved photosynthesis and increased plant growth. In the current study, the up-regulation of PRK, GAPDH, and RCA in rapeseed plants might be described by acclimatization to salt stress. The exogenously sprayed Spd under salt stress could regulate the salt stress-induced accumulation of transcripts of genes related to the Calvin cycle.
5. Conclusions
The use of exogenous Spm or Spd improved the growth rate and photosynthesis of salt-stressed rapeseed plants. Application of Spm or Spd upregulated the gene expression of antioxidant enzymes related genes (CuZnSOD2, CAT1, APX, GR, and DHAR) under salt stress. Furthermore, the exogenous Spd improved the polyamine pathway by up-regulated DAO and PAO transcriptions. The Spd application improved expressions of Calvin cycle enzyme-related genes. Spd is important to regulate the activities of antioxidant enzymes, the pathway to polyamine, and genes linked to the Calvin Cycle in order to alleviate the damage of plants from salt stress. The stimulatory effect processes of Spm or Spd can be separately functioned or incorporated in a single strategy to promote cell growth or maintain cell survival in response to stress. More research could be applied to understand the impact of different times of application using Spm and/or Spd to take full advantage of their agricultural benefits.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The author thanks Zagazig University for supporting the research. Also, the authors wish to express their appreciation to Taif University for funding this work through Taif University Researchers Supporting Project number (TURSP-2020/111), Taif University, Taif, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ahanger M.A., Akram N.A., Ashraf M., Alyemeni M.N., Wijaya L., Ahmad P. Plant responses to environmental stresses—from gene to biotechnology. AoB Plants. 2017;9:1–17. doi: 10.1093/aobpla/plx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexieva V., Sergiev I., Mapelli S., Karanov E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant, Cell Environ. 2001;24:1337–1344. [Google Scholar]
- Alscher R.G., Erturk N., Heath L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002;53:1331–1341. [PubMed] [Google Scholar]
- Baker N.R., Rosenqvist E. Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J. Exp. Bot. 2004;55:1607–1621. doi: 10.1093/jxb/erh196. [DOI] [PubMed] [Google Scholar]
- Bela K., Riyazuddin R., Horváth E., Hurton Á., Gallé Á., Takács Z., Zsigmond L., Szabados L., Tari I., Csiszár J. Comprehensive analysis of antioxidant mechanisms in Arabidopsis glutathione peroxidase-like mutants under salt-and osmotic stress reveals organ-specific significance of the AtGPXL’s activities. Environ. Exp. Bot. 2018;150:127–140. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burzyński M., Żurek A. Effects of copper and cadmium on photosynthesis in cucumber cotyledons. Photosynthetica. 2007;45:239–244. [Google Scholar]
- Cakmak I., Strbac D., Marschner H. Activities of Hydrogen Peroxide-Scavenging Enzymes in Germinating Wheat Seeds. J. Exp. Bot. 1993;44:127–132. [Google Scholar]
- Chattopadhayay M.K., Tiwari B.S., Chattopadhyay G., Bose A., Sengupta D.N., Ghosh B. Protective role of exogenous polyamines on salinity-stressed rice (Oryza sativa) plants. Physiol. Plant. 2002;116:192–199. doi: 10.1034/j.1399-3054.2002.1160208.x. [DOI] [PubMed] [Google Scholar]
- Chaves M.M., Flexas J., Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Shao Q., Yin L., Younis A., Zheng B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019;9:1945. doi: 10.3389/fpls.2018.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.H., Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant, Cell Environ. 2011;34:1–20. doi: 10.1111/j.1365-3040.2010.02232.x. [DOI] [PubMed] [Google Scholar]
- Csiszár J., Lantos E., Tari I., Madosa E., Wodala B., Vashegyi A., Horváth F., Pécsváradi A., Szabó M., Bartha B. Antioxidant enzyme activities in Allium species and their cultivars under water stress. Plant Soil Environ. 2007;53:517. [Google Scholar]
- Deinlein U., Stephan A.B., Horie T., Luo W., Xu G., Schroeder J.I. Plant salt-tolerance mechanisms. Trends Plant. Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoky E.-S.-M., Elrys A.S., Mansour E., Eid R.S., Selem E., Rady M.M., Ali E.F., Mersal G.A., Semida W.M. Application of biostimulants promotes growth and productivity by fortifying the antioxidant machinery and suppressing oxidative stress in faba bean under various abiotic stresses. Sci. Hortic. 2021;288 [Google Scholar]
- Desoky E.-S.-M., Mansour E., El-Sobky E.-S.-E., Abdul-Hamid M.I., Taha T.F., Elakkad H.A., Arnaout S.M., Eid R.S., El-Tarabily K.A., Yasin M.A. Physio-biochemical and agronomic responses of faba beans to exogenously applied nano-silicon under drought stress conditions. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.637783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoky E.-S.-M., Merwad A.-R., Abo El-Maati M.F., Mansour E., Arnaout S.M., Awad M.F., Ramadan M.F., Ibrahim S.A. Physiological and biochemical mechanisms of exogenously applied selenium for alleviating destructive impacts induced by salinity stress in bread wheat. Agronomy. 2021;11:926. [Google Scholar]
- Duan J., Li J., Guo S., Kang Y. Exogenous spermidine affects polyamine metabolism in salinity-stressed Cucumis sativus roots and enhances short-term salinity tolerance. J. Plant Physiol. 2008;165:1620–1635. doi: 10.1016/j.jplph.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Ebeed H.T., Hassan N.M., Aljarani A.M. Exogenous applications of Polyamines modulate drought responses in wheat through osmolytes accumulation, increasing free polyamine levels and regulation of polyamine biosynthetic genes. Plant Physiol. Biochem. 2017;118:438–448. doi: 10.1016/j.plaphy.2017.07.014. [DOI] [PubMed] [Google Scholar]
- El-Badri, A.M., Batool, M., AA Mohamed, I., Wang, Z., Khatab, A., Sherif, A., Ahmad, H., Khan, M.N., Hassan, H.M., Elrewainy, I.M., 2021b. Antioxidative and Metabolic Contribution to Salinity Stress Responses in Two Rapeseed Cultivars during the Early Seedling Stage. Antioxidants 10, 1227. [DOI] [PMC free article] [PubMed]
- El-Badri A.M., Batool M., Mohamed I.A., Khatab A., Sherif A., Wang Z.K., Salah A., Nishawy E., Ayaad M., Kuai J. Modulation of salinity impact on early seedling stage via nano-priming application of Zinc oxide on rapeseed (Brassica napus, L.) Plant Physiol. Biochem. 2021;166:376–392. doi: 10.1016/j.plaphy.2021.05.040. [DOI] [PubMed] [Google Scholar]
- El-Mageed A., Taia A., Belal E.E., Rady M.O., El-Mageed A., Shimaa A., Mansour E., Awad M.F., Semida W.M. Acidified biochar as a soil amendment to drought stressed (Vicia faba L.) plants: influences on growth and productivity, nutrient status, and water use efficiency. Agronomy. 2021;11:1290. [Google Scholar]
- El-Sanatawy A.M., El-Kholy A.S., Ali M., Awad M.F., Mansour E. Maize seedling establishment, grain yield and crop water productivity response to seed priming and irrigation management in a Mediterranean arid environment. Agronomy. 2021;11:756. [Google Scholar]
- Eller M.H., Warner A.L., Knap H.T. Genomic organization and expression analyses of putrescine pathway genes in soybean. Plant Physiol. Biochem. 2006;44:49–57. doi: 10.1016/j.plaphy.2006.01.006. [DOI] [PubMed] [Google Scholar]
- ElSayed A., Rafudeen M., Gomaa A., Hasanuzzaman M. Exogenous melatonin enhances the ROS metabolism, antioxidant defense-related gene expression and photosynthetic capacity of Phaseolus vulgaris L. to confer salt stress tolerance. Physiol. Plant. 2021;1–13 doi: 10.1111/ppl.13372. [DOI] [PubMed] [Google Scholar]
- ElSayed A.I., Rafudeen M.S., El-Hamahmy M.A., Odero D.C., Hossain M.S. Enhancing antioxidant systems by exogenous spermine and spermidine in wheat (Triticum aestivum) seedlings exposed to salt stress. Funct. Plant Biol. 2018;45:745–759. doi: 10.1071/FP17127. [DOI] [PubMed] [Google Scholar]
- FAOSTAT, 2021. Food and Agriculture Organization of the United Nations. Statistical Database. Available online: http://www.fao.org/faostat/en/#data (accessed on 22 January 2022).
- Foyer C.H., Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank R.T., Taylor W.C. Regulation of photosynthesis in C3 and C4 plants: a molecular approach. Plant Cell. 1995;7:797. doi: 10.1105/tpc.7.7.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemeda H., Klein B. Effects of naturally occurring antioxidants on peroxidase activity of vegetable extracts. J. Food Sci. 1990;55:184–185. [Google Scholar]
- Hernández J.A., Almansa M.S. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol. Plant. 2002;115:251–257. doi: 10.1034/j.1399-3054.2002.1150211.x. [DOI] [PubMed] [Google Scholar]
- Hoagland, D.R., Arnon, D.I., 1950. The water-culture method for growing plants without soil. Circular. California Agricultural Experiment Station 347.
- Hodges D.M., DeLong J.M., Forney C.F., Prange R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta. 1999;207:604–611. doi: 10.1007/s00425-017-2699-3. [DOI] [PubMed] [Google Scholar]
- Hsu Y.T., Kao C.H. Cadmium-induced oxidative damage in rice leaves is reduced by polyamines. Plant Soil. 2007;291:27–37. [Google Scholar]
- Kamboj A., Ziemann M., Bhave M. Identification of salt-tolerant barley varieties by a consolidated physiological and molecular approach. Acta Physiol. Plant. 2015;37:1716. [Google Scholar]
- Kasukabe Y., He L., Nada K., Misawa S., Ihara I., Tachibana S. Overexpression of Spermidine Synthase Enhances Tolerance to Multiple Environmental Stresses and Up-Regulates the Expression of Various Stress-Regulated Genes in Transgenic Arabidopsis thaliana. Plant Cell Physiol. 2004;45:712–722. doi: 10.1093/pcp/pch083. [DOI] [PubMed] [Google Scholar]
- Kasukabe Y., He L., Watakabe Y., Otani M., Shimada T., Tachibana S. Improvement of environmental stress tolerance of sweet potato by introduction of genes for spermidine synthase. Plant Biotechnol. 2006;23:75–83. [Google Scholar]
- Khan M., Panda S. Alterations in root lipid peroxidation and antioxidative responses in two rice cultivars under NaCl-salinity stress. Acta Physiol. Plant. 2008;30:81–89. [Google Scholar]
- Koßmann J., Sonnewald U., Willmitzer L. Reduction of the chloroplastic fructose-1, 6-bisphosphatase in transgenic potato plants impairs photosynthesis and plant growth. Plant J. 1994;6:637–650. [Google Scholar]
- Lichtenthaler H.K. [34] Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Liu J.-H., Wang W., Wu H., Gong X., Moriguchi T. Polyamines function in stress tolerance: from synthesis to regulation. Front. Plant Sci. 2015;6:827. doi: 10.3389/fpls.2015.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Jiang G., Wang B., Kuang T. Photosystem II photochemistry and photosynthetic pigment composition in salt-adapted halophyte Artimisia anethifolia grown under outdoor conditions. J. Plant Physiol. 2003;160:403–408. doi: 10.1078/0176-1617-00839. [DOI] [PubMed] [Google Scholar]
- Lu K., Cao B., Feng X., He Y., Jiang D. Photosynthetic response of salt-tolerant and sensitive soybean varieties. Photosynthetica. 2009;47:381–387. [Google Scholar]
- Lv G.-Y., Guo X.-G., Xie L.-P., Xie C.-G., Zhang X.-H., Yang Y., Xiao L., Tang Y.-Y., Pan X.-L., Guo A.-G., Xu H. Molecular Characterization, Gene Evolution, and Expression Analysis of the Fructose-1, 6-bisphosphate Aldolase (FBA) Gene Family in Wheat (Triticum aestivum L.) Front. Plant Sci. 2017;8:1030. doi: 10.3389/fpls.2017.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour E., Mahgoub H.A., Mahgoub S.A., El-Sobky E.-S.-E., Abdul-Hamid M.I., Kamara M.M., AbuQamar S.F., El-Tarabily K.A., Desoky E.-S.-M. Enhancement of drought tolerance in diverse Vicia faba cultivars by inoculation with plant growth-promoting rhizobacteria under newly reclaimed soil conditions. Sci. Rep. 2021;11:1–20. doi: 10.1038/s41598-021-02847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour E., Moustafa E.S., Abdul-Hamid M.I., Ash-shormillesy S.M., Merwad A.-R.-M., Wafa H.A., Igartua E. Field responses of barley genotypes across a salinity gradient in an arid Mediterranean environment. Agric. Water Manag. 2021;258 [Google Scholar]
- Mansour E., Moustafa E.S., Desoky E.-S.-M., Ali M., Yasin M.A., Attia A., Alsuhaibani N., Tahir M.U., El-Hendawy S. Multidimensional evaluation for detecting salt tolerance of bread wheat genotypes under actual saline field growing conditions. Plants. 2020;9:1324. doi: 10.3390/plants9101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Bhoomika K., Dubey R. Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma. 2013;250:3–19. doi: 10.1007/s00709-011-0365-3. [DOI] [PubMed] [Google Scholar]
- Miyagawa Y., Tamoi M., Shigeoka S. Overexpression of a cyanobacterial fructose-1, 6-/sedoheptulose-1, 7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat. Biotechnol. 2001;19:965–969. doi: 10.1038/nbt1001-965. [DOI] [PubMed] [Google Scholar]
- Moustafa E.S., Ali M., Kamara M.M., Awad M.F., Hassanin A.A., Mansour E. Field screening of wheat advanced lines for salinity tolerance. Agronomy. 2021;11:281. [Google Scholar]
- Moustafa E.S., El-Sobky E.-S.-E., Farag H.I., Yasin M.A., Attia A., Rady M.O., Awad M.F., Mansour E. Sowing date and genotype influence on yield and quality of dual-purpose barley in a salt-affected arid region. Agronomy. 2021;11:717. [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant, Cell Environ. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Musgrave M.E. Realizing the Potential of Rapid-Cycling Brassica as a Model System for Use in Plant Biology Research. J. Plant Growth Regul. 2000;19:314–325. doi: 10.1007/s003440000036. [DOI] [PubMed] [Google Scholar]
- Parida A.K., Das A.B. Salt tolerance and salinity effects on plants: a review. Ecotoxicol. Environ. Saf. 2005;60:324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza G.J., Foglia T.A. Rapeseed oil for oleochemical usage. Eur. J. Lipid Sci. Technol. 2001;103:450–454. [Google Scholar]
- Praxedes S.C., DaMatta F.M., Loureiro M.E., Ferrao M.A., Cordeiro A.T. Effects of long-term soil drought on photosynthesis and carbohydrate metabolism in mature robusta coffee (Coffea canephora Pierre var. kouillou) leaves. Environ. Exp. Bot. 2006;56:263–273. [Google Scholar]
- Quick W.P., Neuhaus H.E. In: A Molecular Approach to Primary Metabolism in Higher Plants. Foyer C.H., Quick W.P., editors. Taylor and Francis London; 1997. The regulation and control of photosynthetic carbon assimilation; pp. 41–62. [Google Scholar]
- Raboanatahiry N., Li H., Yu L., Li M. Rapeseed (Brassica napus): processing, utilization, and genetic improvement. Agronomy. 2021;11:1776. [Google Scholar]
- Raines C.A. The Calvin cycle revisited. Photosynth. Res. 2003;75:1–10. doi: 10.1023/A:1022421515027. [DOI] [PubMed] [Google Scholar]
- Roy P., Niyogi K., Sengupta D.N., Ghosh B. Spermidine treatment to rice seedlings recovers salinity stress-induced damage of plasma membrane and PM-bound H+-ATPase in salt-tolerant and salt-sensitive rice cultivars. Plant Sci. 2005;168:583–591. [Google Scholar]
- Sattar A., Wang X., Abbas T., Sher A., Ijaz M., Ul-Allah S., Irfan M., Butt M., Wahid M.A., Cheema M. Combined application of zinc and silicon alleviates terminal drought stress in wheat by triggering morpho-physiological and antioxidants defense mechanisms. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0256984. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Selem E., Hassan A.A., Awad M.F., Mansour E., Desoky E.-S.-M. Impact of exogenously sprayed antioxidants on physio-biochemical, agronomic, and quality parameters of potato in salt-affected soil. Plants. 2022;11:210. doi: 10.3390/plants11020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S., Chen L., Lu W., Sun J., Guo S., Yuan Y., Li J. Effects of exogenous spermidine on photosynthetic capacity and expression of Calvin cycle genes in salt-stressed cucumber seedlings. J. Plant. Res. 2014;127:763–773. doi: 10.1007/s10265-014-0653-z. [DOI] [PubMed] [Google Scholar]
- Spreitzer R.J. Role of the small subunit in ribulose-1, 5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 2003;414:141–149. doi: 10.1016/s0003-9861(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Stassinos P.M., Rossi M., Borromeo I., Capo C., Beninati S., Forni C. Enhancement of Brassica napus Tolerance to High Saline Conditions by Seed Priming. Plants. 2021;10:403. doi: 10.3390/plants10020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P., Johnson G.N. Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol. 2009;149:1154–1165. doi: 10.1104/pp.108.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi M., Nagaoka M., Miyagawa Y., Shigeoka S. Contribution of fructose-1, 6-bisphosphatase and sedoheptulose-1, 7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol. 2006;47:380–390. doi: 10.1093/pcp/pcj004. [DOI] [PubMed] [Google Scholar]
- Tang S., Zhang H., Li L., Liu X., Chen L., Chen W., Ding Y. Exogenous spermidine enhances the photosynthetic and antioxidant capacity of rice under heat stress during early grain-filling period. Funct. Plant Biol. 2018;45:911. doi: 10.1071/FP17149. [DOI] [PubMed] [Google Scholar]
- Tang W., Newton R.J. Polyamines reduce salt-induced oxidative damage by increasing the activities of antioxidant enzymes and decreasing lipid peroxidation in Virginia pine. Plant Growth Regul. 2005;46:31–43. [Google Scholar]
- Tang X., Mu X., Shao H., Wang H., Brestic M. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2015;35:425–437. doi: 10.3109/07388551.2014.889080. [DOI] [PubMed] [Google Scholar]
- Todorova D., Sergiev I., Alexieva V., Karanov E., Smith A., Hall M. Polyamine content in Arabidopsis thaliana (L.) Heynh during recovery after low and high temperature treatments. Plant Growth Regul. 2007;51:185–191. [Google Scholar]
- Uematsu K., Suzuki N., Iwamae T., Inui M., Yukawa H. Increased fructose 1, 6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 2012;63:3001–3009. doi: 10.1093/jxb/ers004. [DOI] [PubMed] [Google Scholar]
- Verma S., Mishra S.N. Putrescine alleviation of growth in salt stressed Brassica juncea by inducing antioxidative defense system. J. Plant Physiol. 2005;162:669–677. doi: 10.1016/j.jplph.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Vicente, O., Boscaiu, M., Naranjo, M.Á., Estrelles, E., Bellés, J.M.a., Soriano, P., 2004. Responses to salt stress in the halophyte Plantago crassifolia (Plantaginaceae). J. Arid Environ. 58, 463-481.
- Vital S.A., Fowler R.W., Virgen A., Gossett D.R., Banks S.W., Rodriguez J. Opposing roles for superoxide and nitric oxide in the NaCl stress-induced upregulation of antioxidant enzyme activity in cotton callus tissue. Environ. Exp. Bot. 2008;62:60–68. [Google Scholar]
- Wang H., Gu M., Cui J., Shi K., Zhou Y., Yu J. Effects of light quality on CO2 assimilation, chlorophyll-fluorescence quenching, expression of Calvin cycle genes and carbohydrate accumulation in Cucumis sativus. J. Photochem. Photobiol. B: Biol. 2009;96:30–37. doi: 10.1016/j.jphotobiol.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gu W., Meng Y., Xie T., Li L., Li J., Wei S. γ-Aminobutyric acid imparts partial protection from salt stress injury to maize seedlings by improving photosynthesis and upregulating osmoprotectants and antioxidants. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep43609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.J., Bennett I.J. The effect of salt stress and abscisic acid on proline production, chlorophyll content and growth of in vitro propagated shoots of Eucalyptus camaldulensis. Plant Cell Tiss. Org. Cult. 2005;82:189–200. [Google Scholar]
- Wu J., Shu S., Li C., Sun J., Guo S. Spermidine-mediated hydrogen peroxide signaling enhances the antioxidant capacity of salt-stressed cucumber roots. Plant Physiol. Biochem. 2018;128:152–162. doi: 10.1016/j.plaphy.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Yakhin O.I., Lubyanov A.A., Yakhin I.A., Brown P.H. Biostimulants in plant science: A global perspective. Front. Plant Sci. 2017;7:2049. doi: 10.3389/fpls.2016.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu J.-C., Juang L.-D., Fang D.-Y.-T., Liu C.-W., Wu S.-J. Exogenous putrescine reduces flooding-induced oxidative damage by increasing the antioxidant properties of Welsh onion. Sci. Hortic. 2009;120:306–314. [Google Scholar]