Abstract
The lowest concentration of an antimicrobial agent that can inhibit the visible growth of a microorganism after overnight incubation is called as minimum inhibitory concentration (MIC) and the drug prescriptions are made on the basis of MIC data to ensure successful treatment outcomes. Therefore, reliable antimicrobial susceptibility data is crucial, and it will help clinicians about which drug to prescribe. Although few prediction studies based on strategies have been conducted, however, no single machine learning (ML) modelling has been carried out to predict MICs in N. gonorrhoeae. In this study, we propose a ML based approach that can predict MICs of a specific antibiotic using unitigs sequences data. We retrieved N. gonorrhoeae genomes from European Nucleotide Archive and NCBI and analysed them combined with their respective MIC data for cefixime, ciprofloxacin, and azithromycin and then we constructed unitigs by using de Brujin graphs. We built and compared 35 different ML regression models to predict MICs. Our results demonstrate that RandomForest and CATBoost models showed best performance in predicting MICs of the three antibiotics. The coefficient of determination, R2, (a statistical measure of how well the regression predictions approximate the real data points) for cefixime, ciprofloxacin, and azithromycin was 0.75787, 0.77241, and 0.79009 respectively using RandomForest. For CATBoost model, the R2 value was 0.74570, 0.77393, and 0.79317 for cefixime, ciprofloxacin, and azithromycin respectively. Lastly, using feature importance, we explore the important genomic regions identified by the models for predicting MICs. The major mutations which are responsible for resistance against these three antibiotics were chosen by ML models as a top feature in case of each antibiotics. CATBoost, DecisionTree, GradientBoosting, and RandomForest regression models chose the same unitigs which are responsible for resistance. This unitigs-based strategy for developing models for MIC prediction, clinical diagnostics, and surveillance can be applicable for other critical bacterial pathogens.
Keywords: Machine learning, Antimicrobial resistance, Neisseria gonorrhoeae, Minimum inhibitory concentration
1. Introduction
Antimicrobial resistance (AMR) is a major threat to global health and development that affects millions of people each year. In October 2020, the World Health Organization (WHO) declared top-ten global public health threats faced by humankind and AMR was stated as one of them (Prestinaci et al., 2015). AMR spread is typically driven by the overuse and misuse of antimicrobials in clinical settings and agriculture sector. These two factors mainly drive the development of resistant pathogens in clinics as well as agriculture. Consequently, AMR causes significant morbidity and mortality for patients worldwide, with severe economic impacts. It is estimated that AMR could cause 10 million deaths each year by 2050 and it could force up to 24 million people into extreme poverty (WHO, 2019). Currently, 700,000 deaths occur worldwide each year due to drug-resistant diseases and in USA AMR costs about 55 billion dollars annually because of health care related expenses (WHO, 2019). With each passing day, the number of untreatable infectious diseases increases, such as sexually transmitted infections, urinary tract infections, and respiratory tract infections. Sexually transmitted diseases (STDs) are very common (WHO, 2019), and millions of new infections occur every year in USA and around the world.
After chlamydia, gonorrhea is the second most common STD in Europe, which is caused by N. gonorrhoeae and it can infect both women and men (ECDPC, 2019). Gonorrhea can cause infections in the throat, rectum, and genitals. Typically STDs affect individuals of all ages, however, STDs are very common among young people (ages 15–24 years) (ECDPC, 2019). Recently, the WHO estimated the worldwide prevalence of urogenital gonorrhea to a total of 30.6 million (0.7% in men and 0.9% in women) cases. By region, the occurrence among women was highest in the WHO African region (1.9%), followed by the Western Pacific (WP) region (0.9%), Americas (0.9%), and were lowest in the European region (0.3%). Likewise, gonorrhea occurrence was highest among men in the WHO African region (1.6%), followed by the Americas (0.8%), WP region (0.7%) and lowest in Europe (0.3%) (Kirkcaldy et al., 2019). Moreover, in the UK, an increase of 26% of gonorrhoeae infections was reported from 2017 to 2018 (Kirkcaldy et al., 2019). According to data (2005–2012) released by the ministry of health of Saudi Arabia, the annual incidence of STDs was high with an overall incidence of 92.1 infections per 100,000 of population (Kirkcaldy et al., 2019). Moreover, many infected people (especially women) experience no symptoms, which exacerbates the spread of the disease (WHO, 2019). However if the infection is left untreated, it can lead to infertility in women and can occasionally spread to other parts of the body such as joints, heart valves, the brain, or the spinal cord (Centers for Disease Control and Prevention, 2021).
Widespread and higher levels of resistance in N. gonorrhoeae reported in recent times has compromised the management and control of gonorrhoeae. N. gonorrhoeae highly variable strains have rapidly developed resistance to penicillins, sulphonamides, macrolides, tetracyclines, early generation cephalosporins, and fluoroquinolones (Roberts, 2019). At present, in many countries of the world, injectable extended-spectrum cephalosporin and ceftriaxone is the only remaining empiric monotherapy for gonorrhoeae (Magnus and Shafer, 2014). Globally, most of the countries now recommend a dual therapy (azithromycin and ceftriaxone) to treat gonorrhoeae infections (Magnus and Shafer, 2014). However, azithromycin was removed from recommendations because of increasing levels of resistance to this antibiotic and later only ceftriaxone was left for prescriptions to the gonorrhoeae patients in the United Kingdom (Derbie et al., 2020, Fifer et al., 2018; Fifer et al., 2020). Recently, in February 2018, first case of ceftriaxone and azithromycin resistance has been reported (Whittles et al., 2018).
Simultaneously, with the growing prevalence of bacterial resistance against antimicrobials, there has been a consistent reduction in the discovery of novel antibiotics. It is more worrisome that the antibiotic pipeline has slowed to a trickle. Though scientists have paid more attention towards AMR, but the overall situation is increasingly deteriorating. Thus, we still need to design new combinational therapies, novel antimicrobial peptides, and effective resistance prediction strategies to combat and reduce AMR efficiently. Although few prediction studies based on strategies have been conducted, however, no single machine learning modelling has been carried out to predict MIC in N. gonorrhoeae (Whittles et al., 2018). In a study, conducted by Golparian et al, Oxford Nanopore MinION sequencer was used, and BLAST algorithms were used for the prediction of decreased susceptibility or resistance to recommended therapeutic antimicrobials in N. gonorrhoeae isolates (Golparian et al., 2018). In another study, authors had demonstrated a whole genome sequence-based MIC prediction approach that allowed prediction for different gonorrhoeae antimicrobials (Whittles et al., 2018).
Many bacterial infections are treated empirically, and doctors prescribe a standard antibiotic to treat the patients. Therefore, there’s a growing interest to know the antibiotic resistance profile before the treatment of the patient begins. In case of clinical perspective, fast diagnostics are crucial to improve patient care. However, some practical limitations are there. For a conventional microbiology laboratory testing, the total time for organism growth, isolation, taxonomic identification, and antimicrobial MIC determination may exceed 24 h (i.e., MRSA) (Giltner et al., 2014) to months (i.e., tuberculosis) (Forbes et al., 2018). Developments in DNA sequencing technology has offered scientists very effective tools to sequence whole genomes including resistant genes. After this breakthrough, sequencing is being widely used to help diagnostic, for health surveillance and patient care decisions and it can give us results in hours rather than days. The resistance mechanisms coded in the resistant bacterial DNAs could reveal us about resistance to different antibiotics and also give valuable information on rising novel resistance mechanisms. Although sequencing reveals resistance in these bacteria however, we don’t always know information on resistance. Since minimizing the time to optimal antimicrobial therapy significantly improves patient outcomes, rapid sequencing-based ML approaches for the prediction of MICs may have clinical utility. In this study, we have used ML algorithms to predict MICs using genome sequences (unitigs; are an efficient but flexible way of representing DNA variation in bacteria) which could enhance our understanding and ability to recognize and contain new resistant strains.
2. Materials and methods
2.1. Data pre-processing
We used genome sequences from N. gonorrhoeae bacterial species (Chisholm et al., 2016, Demczuk et al., 2015, Demczuk et al., 2016, Eyre et al., 2017, Fifer et al., 2018, Jacobsson et al., 2016, Lee et al., 2018, Sánchez-Busó et al., 2019, Simon et al., 2018, Unemo et al., 2016; Grad et al., 2014, Grad et al., 2016). The whole genome sequences and antibiotic resistance data of gonorrhoeae was gathered from the European Nucleotide Archive (ENA) and NCBI. To analyze the collected data, the related gathered MIC values of the azithromycin, cefixime, and ciprofloxacin antibiotics were used. We constructed unitigs according to Jaillard et al. (2018). Briefly, a De Brujin graph was built using the genomes as input in the GATB C++ library and contigs were used to compact the DBG graph using traversal algorithms and getting unitigs in the DBG (Jaillard et al., 2018, Wheeler, 2019). DBGs are directed graphs that efficiently represent all the information contained in a set of sequences. In these graphs, nodes represent all the unique k-mers (genome sequence substrings of length k) extracted from the input sequences while edges represent (k − 1)-exact-overlaps between k-mers. These graphs can be compacted into cDBGs by merging linear paths into a single node referred to as a unitig. Compaction yields a graph with locally optimal resolution: regions of the genome which are conserved across individuals are rep-resented by long unitigs, while regions which are highly variable are fractioned into shorter unitigs. The unitigs were further filtered to get the precise sequences associated with resistance. This process allowed us to represent the similarities and differences between these different bacteria in an efficient way (Jaillard et al., 2018). The filtering resulted in 8290 unitigs strongly associated with resistance.
2.2. Problem description
In this study we used observed MIC values as the target value. The machine learning models were trained to predict the MIC values. Therefore, we solved a regression problem where the target of the ML model is to predict exact value of MIC for the specific antibiotic under study.
2.3. Comparison between machine learning models
We first used 35 models to check and compare the prediction accuracies of different models using unitigs (Table S1). The random seed parameter was set to 313 in order to maintain consistency in the splitting of the dataset for comparative analysis of the results. Several machine learning models exist which can be used for solving a regression task. We tested the performance of 35 machine learning models to predict the exact value of MIC of three antibiotics. The names of these models are listed in Table S1.
2.4. Train test split
We split the data into a 80% training and 20% test set. The splitting was done randomly to avoid any bias. The training data was further split into training and vali-dation sets during K-Fold cross validation. The process of K-Fold cross validation was used for hyperparameter optimization. During cross validation, the data is split into training and validation subsets. Splitting data into training and validation subsets can be done using various methods, such as leave-one-out, leave-p-out, k-fold, and Monte-Carlo sampling. The model is trained on the training set, while its accuracy is measured on the validation set. The aim of this process is to assess the ability of the machine learning algorithm to generalize to new data and select hyperparameters. The value of K was 10 during K-Fold cross validation. After selecting the best hyperparameters for the model, we evaluated it’s performance on test set. The test data was not seen by the model during training. In this way, the generalization ability of the model on unseen data was assessed. The RMSE and R2 values reported here for different models are of test dataset.
2.5. Hyperparameter optimization
The performance of machine learning algorithm is strongly affected by the choice of hyperparameters used to build it. Several algorithms exist for optimization of hyperparameters of machine learning algorithms. These include, grid search, random search and Bayesian optimization algorithm. We optimized the performance of models using Bayesian optimization algorithm. The most important set of parameters for each ML model were chosen and optimized.
2.6. Performance metrics
We used mean squared error (MSE) for training of machine learning models. On the other hand, we used coefficient of determination (R2) during optimization of hyperparameters. Since the optimization problem was solved as a minima problem, we used (1-R2) as objective function to be minimized. We also recorded the bias in the prediction of the models. The positive value of bias indicate the predicted MIC value is higher than the true MIC value while the negative bias shows that the predicted MIC value is lower than true MIC value. The formula to calculate MSE, BIAS and R2 is given below;
(Aslam et al., 2021)where oi and pi are the observed and predicted MIC, respectively, and n indicates the number of datasets.
2.7. Python libraries
We used XGBoost, LightGBM, CatBoost and Scikit-learn libraries to build the machine learning models. The hyperparameters were optimized using scikit-optimize library which implemented Bayesian optimization algorithm. The complete machine learning pipeline from data-preprocessing, to building and training of models, prediction of MICs and analysis of results was performed using AI4Water which is a python based framework for performing aforementioned tasks (Abbas et al., 2021).
2.8. Code availability
The code to reproduce the results presented in this article is available at GitHub repository (link: https://github.com/Asadmalic/mic_prediction_ml).
3. Results
3.1. Performance of models
We used 35 different ML models to predict the MICs for three antibiotics (cefixime, ciprofloxacin, azithromycin). Table 1 shows a brief summary of accuracies of different models and show how a model relate to observations in terms of their RMSE (root mean square error) and coefficient of determination. Among all the used ML models, five models (5/35) showed high accuracies as compared to the other models (Table S1). The value of accuracies were high for all the antibiotics under study for these five models. RandomForest, XGBoost, CATBoost, HitsGradientBoosting, GradientBoosting and ADARegressor models showed high accuracies for three antibiotics as compared to other models (Table 1). Among three antibiotics, the accuracies for azithromycin were highest for all the models (Table 1). RandomForest and XGBoost showed higher accuracies for azithromycin (R2 = 0.79009, 0.76031) and cefixime (R2 = 0.75787, 0.79708) respectively. Moreover, MIC prediction is complex process, so we observed some models with very poor accuracies. Among these models, ElasticNet (R2 = 1.8281e−19) and Lasso (R2 = 1.883e−15) showed very poor accuracies.
Table 1.
Metrics comparison of 35 different ML regression models. Models were compared on the basis of their performance to predict MIC values of three antibiotics on test datasets.
| Names of models used | Ciprofloxacin |
Cefixime |
Azithromycin |
||||
|---|---|---|---|---|---|---|---|
| RMSE | R2 | RMSE | R2 | RMSE | R2 | ||
| 1 | ADABoostRegressor | 13.61701 | 0.12067 | 0.04143 | 0.66328 | 6.28431 | 0.14474 |
| 2 | ADARegressor | 9.58793 | 0.62305 | 0.04086 | 0.76301 | 1.61560 | 0.78313 |
| 3 | BaggingRegressor | 6.10417 | 0.69089 | 0.04038 | 0.67140 | 1.37854 | 0.7369 |
| 4 | BayesianRidge | 6.06775 | 0.69462 | 0.03931 | 0.68590 | 1.54538 | 0.70676 |
| 5 | CATBoostRegressor | 3.87531 | 0.77393 | 0.03807 | 0.74570 | 1.40781 | 0.79317 |
| 6 | DecisionTreeRegressor | 7.82284 | 0.54359 | 0.04396 | 0.61863 | 1.91145 | 0.66770 |
| 7 | DummyRegressor | 9.12458 | 0.31455 | 0.07010 | 1.2696e-32 | 2.84645 | −0.00093 |
| 8 | ElasticNet | 7.39072 | 0.58821 | 0.07011 | 1.8281e-19 | 2.72904 | 0.65383 |
| 9 | ElasticNetCV | 0.03914 | 0.68890 | 0.68433 | 0.68433 | 1.51770 | 0.71596 |
| 10 | ExtraTreeRegressor | 8.01472 | 0.53616 | 0.04531 | 0.59814 | 1.81783 | 0.68063 |
| 11 | ExtraTreesRegressor | 0.04073 | 0.66861 | 0.04373 | 0.62071 | 1.75033 | 0.70090 |
| 12 | GaussianProcessRegressor | 0.04942 | 0.68436 | 0.04259 | 0.65792 | 1.99184 | 0.54625 |
| 13 | GradientBoostingRegressor | 6.02614 | 0.70108 | 0.03914 | 0.68890 | 1.44105 | 0.74478 |
| 14 | HistGradientBoostingRegressor | 5.71240 | 0.73384 | 0.03816 | 0.70486 | 1.35686 | 0.77633 |
| 15 | HuberRegressor | 6.68917 | 0.65186 | 0.04073 | 0.66861 | 1.59841 | 0.68702 |
| 16 | KNeighborsRegressor | 0.08358 | 0.63807 | 3.49214 | 0.63424 | 1.21033 | 0.71268 |
| 17 | KernelRidge | 9.60091 | 0.46778 | 0.03990 | 0.67779 | 1.62867 | 0.68134 |
| 18 | LarsCV | 9.51242 | 0.45764 | 0.04045 | 0.66715 | 1.55067 | 0.70463 |
| 19 | Lasso | 7.58347 | 0.57166 | 0.07010 | 1.883e-15 | 2.84645 | 0.73652 |
| 20 | LassoCV | 8.64752 | 0.54124 | 0.03942 | 0.68436 | 1.50917 | 0.71937 |
| 21 | LassoLarsCV | 0.03989 | 0.67792 | 0.04104 | 0.65787 | 1.52546 | 0.71292 |
| 22 | LassoLarsIC | 6.72328 | 0.63309 | 0.04039 | 0.66812 | 1.53725 | 0.70890 |
| 23 | LinearRegression | 8.41567 | 0.54621 | 0.34524 | 0.01717 | 1.76178 | 0.64022 |
| 24 | MLPRegressor | 6.61081 | 0.68461 | 0.05356 | 0.53807 | 1.57986 | 0.73443 |
| 25 | NuSVR | 7.65251 | 0.58619 | 0.03863 | 0.69966 | 2.20857 | 0.66841 |
| 26 | OrthogonalMatchingPursuit | 11.75760 | 0.31796 | 0.03991 | 0.67679 | 1.55638 | 0.70262 |
| 27 | OrthogonalMatchingPursuitCV | 6.73327 | 0.62534 | 0.04989 | 0.66592 | 1.54569 | 0.70547 |
| 28 | PoissonRegressor | 6.10017 | 0.69149 | 0.06490 | 0.58745 | 2.30000 | 0.46240 |
| 29 | RandomForestRegressor | 2.69214 | 0.77241 | 0.04104 | 0.75787 | 1.33418 | 0.79009 |
| 30 | Ridge | 9.60075 | 0.46806 | 0.03989 | 0.67792 | 1.62883 | 0.68132 |
| 31 | RidgeCV | 6.79794 | 0.64527 | 0.03931 | 0.68599 | 1.53935 | 0.70838 |
| 32 | SGDRegressor | 4.29214 | 0.68241 | 0.04483 | 0.67187 | 1.82504 | 0.70766 |
| 33 | SVR | 7.71597 | 0.58373 | 0.07486 | 0.57187 | 2.21276 | 0.66886 |
| 34 | XGBoostRegressor | 6.01090 | 0.70446 | 0.03859 | 0.79708 | 1.44787 | 0.76031 |
| 35 | XGBoostRFRegressor | 5.81138 | 0.72186 | 0.44421 | 0.68538 | 1.47658 | 0.73711 |
RMSE: root mean square error, R2: Coefficient of determination, NuSVR: Nu Support Vector Regression, SVR: Support Vector Regression.
3.2. Comparison of R2 and RMSE of models
Among 35 regression models used for azithromycin, we chose 3 models with best accuracies. CATBoost, RandomForest, and BaggingRegressor performed best among all the tested models as shown in Table 1 and Fig. S5. The accuracy of the different models was compared by the comparison of the RMSE, R2, and relative error between the predicted values and true values for each of the drug resistance prediction models. The accuracy of the different models for ciprofloxacin and cefixime is given in the Figs. S6 and S7 respectively. Moreover, among 35 used ML models, five top performing models predicted the MIC values, which were similar to true MIC values as shown in Figs. 1 and S1 and S2. These Figures show ML based predicted and true MIC values of azithromycin, ciprofloxacin, and cefixime for N. gonorrhoeae and the results presented in this figure are for CATBoost model only (Figs. 1, S1 and S2).
Fig. 1.
Graph shows machine-learning based predicted and true MIC values of azithromycin in the dataset for CATBoost model. Blue dots show the true MIC values of azithromycin antibiotic for N. gonorrhoeae, while orange dots show the predicted MIC values by the CATBoost machine learning-based regression model.
3.3. Feature importance
The feature importance of ML models is shown in Figs. 2 and S3, S4 for MICs prediction of azithromycin, ciprofloxacin and cefixime. Fig. 2 shows that which unitig sequence was how much important for the model for predicting MICs. We found that the unitig sequence (GGGTTTAAAACGTCGTGAGACAGTTT-GGTCCCTATCTGCAGTGGGCGTTGGAAGTTTGACG) was the most important feature for prediction of MICs for azithromycin (Fig. 2). This unitig contained a mutation in the 23S ribosomal RNA which is responsible for most high-level azithromycin resistance. The following models chose this sequence of unitigs as the most important feature; ADABoost, CATBoost, DecisionTree, ExtraTree, ExtraTrees, GradientBoosting, RandomForest, XGBoost Regressors. Moreover, in case of ciprofloxacin, the major mutation (GTGCGACAGCAAAGTCCAAACCAGCGTCCCCGCC) which is responsible for resistance was chosen by following models as a top feature; ADABoost, CATBoost, DecisionTree, ExtraTree, GradientBoosting, RandomForest, and XGBoost RF Regressor (Fig. S3). Similarly, in Fig. S4 in case of cefixime, the major mutation (CGAACAGGCGACGATGTCTTTCGGTTACGGCCTGCA) that drives resistance was chosen as a top feature by these models; CATBoost Regressor, DecisionTreeRegressor, ExtraTreeRegressor, GradientBoostingRegressor, and RandomForestRegressor.
Fig. 2.
Most important features (unitigs) selected by different machine learning-based models for MIC predictions of Azithromycin. Figure shows the feature importance results selected by ADABoostRegressor model.
3.4. Cross validation
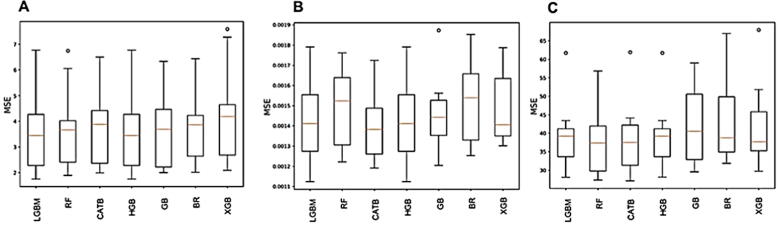
We monitored the performance of ML models during each of the 10 folds of cross validation. The results are plotted as box and whisker plots for the best performing models for each of the three antibiotics are shown in Fig. 3A–C. The minimum variation is shown by BaggingRegressor model while the largest variation is shown by GradientBoost model for azithromycin (Fig. 3A). In case of ciprofloxacin and cefixime, the minimum variation was shown by GradientBoost and HitsGradientBoost respectively (Fig. 3 B and C).
Fig. 3.
Performance of ML models during each of the 10 folds of cross validation. The results are plotted as box and whisker plots for the best performing models for each of the three antibiotics. The minimum variation is shown by BR model while the largest variation is shown by GB model for azithromycin (A). In case of ciprofloxacin and cefixime, the minimum variation was shown by GB (B) and HGB (C) respectively. Orange lines within the box show median MSE values. LGBM: Light Gradient Boosted Machine, RF: Random Forest, CATB: CATBoost, HGB: Hits Gradient Boosting, GB: Gradient Boosting, BR: Bagging Regressor, XGB: XGBoost.
4. Discussion
We built, using 35 models, machine learning-based MIC prediction models for N. gonorrhoeae. Importantly, our proposed models offer an approach for performing prediction of MIC directly from sequence (genome) data using unitigs and actual MIC values that could be applied to more veterinary or human bacterial pathogens.
Based on the genomic data, sequence analysis was used to obtain unitigs information and ML methods were applied to establish prediction models for the MIC values of N. gonorrhea. Based on MIC values and unitigs, in this study, 35 machine learning model were used. By feature importance, we proposed a top feature-based CATBoostregression model, which had the best predictive performance for all three antibiotics. According to recent studies, we found that gene mutations might affect drug resistance of bacteria; therefore, we make an effort to find the relevant unitigs causing resistance (Naha et al., 2021). We used 8290 unitigs for prediction, and the prediction results above show that the mean accuracy of the unitigs was above 85% which shows that the performance of the regression models is best (Pataki et al., 2020, Pesesky et al., 2020, ValizadehAslani et al., 2020). To evaluate our models, we compared MIC prediction models used by other related studies. In these published studies, different machine learning models were used to predict MICs or antibiotic resistance in bacteria (Pesesky et al., 2020, ValizadehAslani et al., 2020, Yang et al., 2018, Nguyen et al., 2019, Li et al., 2021). In a study, the authors used the XGBoost model with k-mers features, and their results reveals a precision of around 91% for MIC predictions, and our results are also close to the previously published results (Nguyen et al., 2019). In our study, among 35 used models, the best accuracies were obtained for 5 models including RandomForest, and CATBoostRegressor models. Similarly, according to a study, XGBoost model to predict MICs for non-typhoidal Salmonella, resulted in a good accuracy without a large number of samples (Nguyen et al., 2019, Li et al., 2021).
We also used classification models for MIC prediction on the same samples; how-ever, the mean accuracy of classification models was less than the mean accuracy of the regression models, which shows that the performance of the classification models was not good as compared to regression models. This result demonstrate that the regression model performed very well. Similar results were reported by Tan et al. for the prediction of MIC of meropenem against Klebsiella pneumoniae using metagenomic data (Tan et al., 2021).
Our study shows that the unitig sequences which are very important features for high-level of resistance in N. gonorrhoeae against different antibiotics were truly selected by five models for each antibiotic. For MIC prediction of azithromycin, 9 regression models predicted same unitig which is responsible for resistance. Similarly, 5 regression models predicted the specific unitig that is responsible for the cefixime resistance. Moreover, in case of ciprofloxacin, 7 models predicted the important resistance determinant unitig that is causing high level of resistance in gonorrhoeae and similar results were obtained by other studies for different bacterial species using MIC predictive models (Tan et al., 2021). This establishes that the important features obtained from our models may help to understand the reasons for the development of resistance in gonorrhoeae. Moreover, these three antibiotics belongs to different classes of antibiotics. Azithromycin belongs to macrolides, ciprofloxacin belongs to fluoroquinolones and cefixime belongs to cephalosporin class of antibiotics. Thus, the unitigs selected by our predictive models should be considered when determining their MICs.
Among 35 used ML models, some of them had lower accuracy while predicting MICs. The machine learning modelling requires adequate input data to train the ML models to form a training dataset and a “testing dataset” to assess the performance of the model (Macesic et al., 2017, Li et al., 2020). Among the three antibiotics, the resistant background of N. gonorrhoeae was different for each drug, therefore, after randomly split the limited data into training dataset or testing dataset, different ML models could not have enough to learn from the training dataset and therefore lead to a relative lower accuracy while predicting the testing set of the model.
5. Conclusions
In summary, our used models predicted the MICs which are close to real MIC values of these antibiotics. Moreover, there can be a lot of resistant genes in gonorrhoeae that can be missed in the predictive models but this study can help to annotate and study novel resistant hypothetical genes. The 35 used models, being trained exclusively on the presence-absence of the unitigs and MIC values, demonstrate promising performance in our study. Moreover, currently we have used publicly available collections of gonorrhoeae genomes with MIC data from different countries. Since the resistant gene content may vary across pathogen populations, validation of the gonorrhoeae models using strains from different countries is important to its application in global health. Additionally, in clinical practice while prescriptions, machine learning based prediction models can perform well by selecting important feature values and can meaningfully improve detection efficiency compared to experimental methods of measuring MIC values, providing doctors with a faster access to information on drug resistance profile of the patient for drug prescription and administration. This will im-prove the usefulness of antibiotic, avoiding future resistance and allowing patients to take medication promptly and finally reducing the time and cost of the laboratory experiment.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number IFPRC-103-141-2020 and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2022.02.047.
Contributor Information
Muhammad Yasir, Email: yasirkhattak.mrl@gmail.com.
Asad Mustafa Karim, Email: asadkarim@suwon.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abbas, A., Boithias, L., Pachepsky, Y., Kim, K., Chun, J.A., Cho, K.H., 2021. AI4Water v1.0: an open source python package for modeling hydrological time series using data-driven methods, Geosci. Model. Dev. Discuss. doi:10.5194/gmd-2021-139 [preprint].
- Aslam M., Lee S.-J., Khang S.-H., Hong S. Two-stage attention over LSTM with bayesian optimization for day-ahead solar power forecasting. IEEE Access. 2021;9:107387–107398. [Google Scholar]
- Centers for Disease Control and Prevention, 2021. Gonorrhea – CDC Fact Sheet. <https://www.cdc.gov/std/gonorrhea/stdfact-gonorrhea-detailed.htm> (Accessed on 13 February 2022).
- Chisholm S.A., Wilson J., Alexander S., Tripodo F., Al-Shahib A., Schaefer U., Lythgow K., Fifer H. An out-break of high-level azithromycin resistant Neisseria gonorrhoeae in England. Sex Transm Infect. 2016;92(5):365–367. doi: 10.1136/sextrans-2015-052312. [DOI] [PubMed] [Google Scholar]
- Eyre D.W., De Silva D., Cole K., Peters J., Cole M.J., Grad Y.H., Demczuk W., Martin I., Mulvey M.R., Crook D.W., Walker A.S., Peto T.E.A., Paul J. WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2017;72(7):1937–1947. doi: 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk W., Lynch T., Martin I., Van Domselaar G., Graham M., Bharat A., Allen V., Hoang L., Lefebvre B., Tyrrell G., Horsman G., Haldane D., Garceau R., Wylie J., Wong T., Mulvey M.R., Munson E. Whole-genome phylo-genomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J. Clin. Microbiol. 2015;53(1):191–200. doi: 10.1128/JCM.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk W., Martin I., Peterson S., Bharat A., Van Domselaar G., Graham M., Lefebvre B., Allen V., Hoang L., Tyrrell G., Horsman G., Wylie J., Haldane D., Archibald C., Wong T., Unemo M., Mulvey M.R., Munson E. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant neisseria gonorrhoeae in Canada from 1997 to 2014. J. Clin. Microbiol. 2016;54(5):1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbie A., Mekonnen D., Woldeamanuel Y., Abebe T. Azithromycin resistant gonococci: a literature review. Antimicrob. Resist. Infect. Contr. 2020;9:138. doi: 10.1186/s13756-020-00805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control, 2019. <https://www.ecdc.europa.eu/en/news-events/gonorrhoea-cases-rise-across-europe> (Accessed on 16 January 2022).
- Fifer H., Cole M., Hughes G., Padfield S., Smolarchuk C., Woodford N., Wensley A., Mustafa N., Schaefer U., Myers R., Templeton K., Shepherd J., Underwood A. Sustained transmission of high-level azithromy-cin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet. Infect. Dis. 2018;18(5):573–581. doi: 10.1016/S1473-3099(18)30122-1. [DOI] [PubMed] [Google Scholar]
- Fifer H., Saunders J., Soni S., Sadiq S.T., FitzGerald M. UK national guideline for the management of in-fection with Neisseria gonorrhoeae. Int. J. STD. AIDS. 2020;31(1):4–15. doi: 10.1177/0956462419886775. [DOI] [PubMed] [Google Scholar]
- Forbes B.A., Hall G.S., Miller M.B., Novak S.M., Rowlinson M.C., Salfinger M., Somoskövi A., Warshauer D.M., Wilson M.L. Practical guidance for clinical microbiology laboratories: mycobacteria. Clin. Microbiol. Rev. 2018;31:e00038–e117. doi: 10.1128/CMR.00038-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner C.L., Kelesidis T., Hindler J.A., Bobenchik A.M., Humphries R.M. Frequency of susceptibility testing for patients with persistent methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 2014;52(1):357–361. doi: 10.1128/JCM.02081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golparian D., Donà V., Sánchez-Busó L., Foerster S., Harris S., Endimiani A., Low N., Unemo M. Antimicrobial resistance prediction and phylogenetic analysis of Neisseria gonorrhoeae isolates using the Oxford Nanopore MinION sequencer. Sci. Rep. 2018;8:17596. doi: 10.1038/s41598-018-35750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y.H., Kirkcaldy R.D., Trees D., Dordel J., Harris S.R., Goldstein E., Weinstock H., Parkhill J., Hanage W.P., Bentley S., Lipsitch M. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lan. Infect. Dis. 2014;14(3):220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y.H., Harris S.R., Kirkcaldy R.D., Green A.G., Marks D.S., Bentley S.D., Trees D., Lipsitch M. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquin-olones in the United States, 2000–2013. J. Infect. Dis. 2016;214(10):1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S., Golparian D., Cole M., Spiteri G., Martin I., Bergheim T., Borrego M.J., Crowley B., Crucitti T., Van Dam A.P. WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC >2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J. Antimicrob. Chemother. 2016;11:3109–3116. doi: 10.1093/jac/dkw279. [DOI] [PubMed] [Google Scholar]
- Jaillard, M., Lima, L., Tournoud, M., Mahé, P., Van, B.A., Lacroix, V., Jacob, L., 2018. A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events. PLoS. Genet. 14, e1007758. [DOI] [PMC free article] [PubMed]
- Kirkcaldy R.D., Weston E., Segurado A.C., Hughes G. Epidemiology of gonorrhoea: a global perspective. Sex Health. 2019;16:401–411. doi: 10.1071/SH19061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.S., Seemann T., Heffernan H., Kwong J.C., da Gonçalves S.A., Carter G.P., Woodhouse R., Dyet K.H., Bulach D.M., Stinear T.P., Howden B.P., Williamson D.A. Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J. Antimicrob. Chemother. 2018;73:353–364. doi: 10.1093/jac/dkx405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lin J., Hu Y., Zhou J. PARMAP: a pan-genome-based computational framework for predicting anti-microbial resistance. Front. Microbiol. 2020;11:578795. doi: 10.3389/fmicb.2020.578795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Z., Liang B. A review: antimicrobial resistance data mining models and prediction methods study for pathogenic bacteria. J. Antibiot. 2021;74:838–849. doi: 10.1038/s41429-021-00471-w. [DOI] [PubMed] [Google Scholar]
- Macesic N., Polubriaginof F., Tatonetti N.P. Machine learning: novel bioinformatics approaches for combating antimicrobial resistance. Curr. Opin. Infect. Dis. 2017;30:511–517. doi: 10.1097/QCO.0000000000000406. [DOI] [PubMed] [Google Scholar]
- Magnus U., Shafer W.M. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbio. Rev. 2014;27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naha, S., Sands, K., Mukherjee, S., Saha, B., Dutta, S., Basu, S., 2021. OXA181-like carbapenemases in Klebsiella pneumoniae ST14, ST15, ST23, ST48, and ST231 from septicemic neonates: coexistence with NDM-5, resistome, transmissibility, and genome diversity. mSphere. 6, e01156-20. [DOI] [PMC free article] [PubMed]
- Nguyen M., Long S.W., McDermott P.F., Olsen R.J., Olson R., Stevens R.L., Tyson G.H., Zhao S., Davis J.J. Using machine learning to predict antimicrobial mics and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol. 2019;57:e01260–e1318. doi: 10.1128/JCM.01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataki, B.A., Matamoros, S., van der Putten, B.C.L., Remondini, D., Giampieri, E., Aytan-Aktug, D., Hen-driksen, R.S., Lund, O., Csabai, I., Constance, S., SPS, C.M.A., group., 2020. Understanding and predicting ciprofloxa-cin minimum inhibitory concentration in Escherichia coli with machine learning. Sci. Rep. 10, 15026. [DOI] [PMC free article] [PubMed]
- Pavan, A.M., Lughi, V., 2012. Photovoltaics in Italy: toward grid parity in the residential electricity market. In: Proc. 24th Int. Conf. Microelectron. pp. 1–4.
- Pesesky M.W., Hussain T., Wallace M., Patel S., Andleeb S., Burnham C.D., Dantas G. Evaluation of ma-chine learning and rules-based approaches for predicting antimicrobial resistance profiles in gram-negative Bacilli from whole genome sequence data. Front. Microbiol. 2020;7:1887. doi: 10.3389/fmicb.2016.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestinaci F., Pezzotti P., Pantosi A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M., 2019. Sex diseases on the rise in England. <https://www.bbc.com/news/health-48509969> (Accessed on 16 January 2022).
- Sánchez-Busó L., Golparian D., Corander J., Gard Y.H., Ohnishi M., Flemming R. The impact of antimicrobials on gonococcal evolution. Nat. Microbiol. 2019;4:1941–1950. doi: 10.1038/s41564-019-0501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H.R., Michelle J.C., Gianfranco S., Leonor S.B., Daniel G., Susanne J., Richard G., Khalil A., Corin A.Y. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic sur-vey. Lan. Infect. Dis. 2018;18:758–768. doi: 10.1016/S1473-3099(18)30225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R., Yu A., Liu Z., Liu Z., Jiang R., Wang X., Liu J., Gao J., Wang X. Prediction of minimal inhibitory concentration of meropenem against klebsiella pneumoniae using metagenomic data. Front. Microbiol. 2021;12:712886. doi: 10.3389/fmicb.2021.712886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Golparian D., Sánchez-Busó L., Grad Y., Jacobsson S., Ohnishi M., Lahra M.M., Limnios A., Si-kora A.E., Wi T., Harris S.R. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J. Antimi-crob. Chemother. 2016;71:3096–3108. doi: 10.1093/jac/dkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ValizadehAslani T., Zhao Z., Sokhansanj B.A., Rosen G.L. Amino acid k-mer feature extraction for Quantitative Antimicrobial Resistance (AMR) prediction by machine learning and model interpretation for biological insights. Biology. 2020;9:365. doi: 10.3390/biology9110365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, N., 2019. Building Machine Learning Models for Predicting Antibiotic Resistance. <https://towardsdatascience.com/building-machine-learning-models-for-predicting-antibiotic-resistance-7640046a91b6> (Accessed on 16 January 2022).
- Whittles L.K., White P.J., Paul J., Didelot X. Epidemiological trends of antibiotic resistant gonorrhoea in the United Kingdom. Antibiot. (Basel) 2018;7:60. doi: 10.3390/antibiotics7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2019. https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (Accessed on 16 January 2022).
- Yang Y., Niehaus K.E., Walker T.M., Iqbal Z., Walker A.S., Wilson D.J., Peto T.E.A., Crook S.W., Smith E.G., Zhu T., Clifton D.A. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics. 2018;34:1666–1671. doi: 10.1093/bioinformatics/btx801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.